Abstract
Human T-cell leukemia virus type 1 (HTLV-1) Tax protein activates viral transcription from the long terminal repeats (LTR). Mechanisms through which Tax activates LTR have been established, but coactivators of this process remain to be identified and characterized. Here we show that all three members of the TORC family of transcriptional regulators are coactivators of Tax for LTR-driven expression. TORC coactivation requires CREB, but not ATF4 or other bZIP factors. Tax physically interacts with TORC1, TORC2, and TORC3 (TORC1/2/3), and the depletion of TORC1/2/3 inhibited Tax activity. TORC coactivation can be further enhanced by transcriptional coactivator p300. In addition, coactivators in the p300 family are required for full activity of Tax independently of TORC1/2/3. Thus, both TORC and p300 families of coactivators are essential for optimal activation of HTLV-1 transcription by Tax.
Human T-cell leukemia virus type 1 (HTLV-1) is etiologically associated with adult T-cell leukemia, an aggressive and fatal malignancy of CD4+ T lymphocytes (32). HTLV-1 encodes a 40-kDa oncoprotein, Tax, which initiates the process of leukemogenesis (10, 16). Tax is a potent transcriptional activator of the viral long terminal repeats (LTR) as well as a subset of cellular genes, including various cytokine genes and proto-oncogenes (10, 23). The mechanisms through which Tax activates the viral LTR have been well studied. Thus, we understand that Tax acts as a homodimer (18, 39) that interacts with CREB and contacts a stretch of DNA to activate the three 21-bp repeats, also known as Tax-responsive elements (TRE), on HTLV-1 LTR (24, 29, 41). Optimal activation of LTR by Tax requires the HTLV-1 core promoter CREB and the 21-bp repeats (6). However, the molecular details before and after the formation of the ternary complex remain largely unknown.
Coordinated activation of transcription requires both DNA-binding activators, such as Tax and CREB, and coactivators which act through chromatin modification and/or the stimulation of preinitiation complex formation (36). Several transcriptional coactivators that are histone or factor acetyltransferases, including CREB-binding protein (CBP), p300, and P/CAF, have been shown to play roles in Tax-mediated transcription (11, 12, 17, 22, 27). Because Tax activation of the LTR is potent and Tax can influence multiple steps of transcription both prior and subsequently to TATA-binding factor recruitment (6), it likely interacts with additional coactivators.
A new family of CREB coactivators, termed transducers of regulated CREB activity (TORCs), has recently been identified and characterized (2, 8, 14, 37). Currently there are three members, TORC1, TORC2, and TORC3 (TORC1/2/3), in this family. All three activate CREB-dependent transcription (8, 14) but are differentially expressed and regulated by upstream signals such as AMP-activated protein kinase and LKB1 (26, 38). A recent study has suggested that TORC3 also serves to enhance Tax activation of HTLV-1 LTR (25). However, whether TORC3 is specifically required and whether Tax might generally interact with other TORC factors remain unclear. In particular, it will be of interest to understand whether TORC1 and TORC2 are also involved in mediating the action of Tax.
In this study, we investigated the contributory roles of TORC1, TORC2, and TORC3 in Tax activation of HTLV-1 LTR. We also explored the requirement for CREB in TORCs' coactivator function and in Tax-TORC interaction. We found that all three TORCs provide essential coactivator function for Tax activation of the HTLV-1 LTR and that Tax directly binds TORCs. In addition, we demonstrated that the p300 coactivator cooperates with TORCs and is required for their full activity in the activation of HTLV-1 LTR. Our work presents a new mechanistic facet to Tax-dependent regulation of gene transcription.
MATERIALS AND METHODS
Plasmids.
Human cDNAs containing complete coding regions for TORC1, TORC2, and TORC3 were derived from expressed sequence tag clones (IMAGE clone identification no. 4938995, 6188068, and 6470060) obtained from RZPD Deutsches Ressourcenzentrum für Genomforschung GmbH (Berlin, Germany). Eukaryotic expression plasmids for TORC1, TORC2, and TORC3 were based on pcDNA3.1/V5 (Invitrogen) or pEGFP (Clontech). The expression plasmid for myc-tagged CREB was constructed by inserting both a myc tag (via HindIII and BamHI) and the CREB gene (via EcoRI and EcoRV) into pcDNA3.1/V5-His6B (Invitrogen). Expression vectors for Gal-CREB, Gal-ATF4, Gal-LZIP, and Gal-CREB-H were derived from pM (Clontech). Reporter plasmids pLTR-Luc and pGal-Luc as well as expression plasmids for Tax, Gal-Tax, A-CREB, A-ATF4, A-LZIP, and A-CREB-H have been previously described in detail (5, 6, 18-21). Tax expression plasmid pIEX is driven by a cytomegalovirus promoter (19).
A-CREB, A-ATF4, A-LZIP, and A-CREB-H were constructed by fusing a designed acidic amphipathic extension onto the N terminus of the leucine zipper region (1). A-CREB was a gift from Charles Vinson (National Cancer Institute, Maryland) and contains 274 to 341 amino acids of mouse CREB (1). A-ATF4 contains 304 to 352 amino acids of human ATF4 (6). A-LZIP contains 175 to 223 amino acids of human LZIP (21). A-CREB-H contains 267 to 312 amino acids of human CREB-H (5).
The short hairpin RNA (shRNA) expression vector pSHAG-1 (34) was kindly provided by Greg Hannon (Cold Spring Harbor Laboratory, New York). The expression plasmid for E1A-12S (31) was from James Lundblad (Oregon Health and Science University, Oregon). Expression plasmid for p300 (30, 40) was originally obtained from Xiang-Jiao Yang (McGill University Health Center, Montréal, Canada) and provided to us by Zengguo Wu. Expression vectors for p300ΔHAT (33) and p300DY (15) mutants were provided by Y. Nakatani (Dana-Farber Cancer Institute, Massachusetts) and Tso-Pang Yao (Duke University Medical Center, North Carolina), respectively.
Reporter assay.
HeLa or 293T cells were cultured in Dulbecco's modified Eagle's medium and transfected using GeneJuice transfection reagent (Novagen). Jurkat cells were propagated in RPMI 1640 medium and transfected by electroporation (20). Cells were harvested 48 h after transfection. Luciferase activity was determined as described previously (5, 13) using dual-luciferase reagents (Promega). Transfection efficiencies were normalized to a control plasmid (pRLSV40 from Promega) expressing Renilla luciferase.
Coimmunoprecipitation.
HeLa cells grown in a 100-mm petri dish were harvested in 0.5 ml of immunoprecipitation buffer (25 mM HEPES, pH 7.5, 0.3 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol, 1% Triton X-100, 0.1% sodium dodecyl sulfate, 20 mM β-glycerophosphate, 1 mM sodium vanadate, and 1 mM phenylmethylsulfonyl fluoride). V5-tagged TORC protein was immunoprecipitated from the cleared lysate by overnight incubation at 4°C with rabbit anti-V5 (Sigma). Immunocomplex was precipitated by using protein A agarose (Invitrogen) and washed three times with immunoprecipitation buffer before being resuspended in sample buffer (60 mM Tris-Cl, 2% sodium dodecyl sulfate, 6% glycerol, 1% β-mercaptoethanol, and 0.002% bromophenol blue).
RNA interference (RNAi) knockdown of TORCs.
The shRNA expression cassettes comprising U6 promoter, shRNA (sense-loop-antisense), and termination signal T6 were PCR amplified and inserted into the vector by TA cloning. The three shRNAs targeting TORC1/2/3, namely shTORC1, shTORC2, and shTORC3, target nucleotides 1709 to 1729 of TORC1 (GACTCGCAGCAACTGGGATAC), nucleotides 452 to 472 of TORC2 (CAGCGAGATCCTCGAAGAATG), and nucleotides 1779 to 1807 of TORC3 (GTCACTTAACATGTTGAGTCCATCCAGGC), respectively. For the knockdown experiment, 5 million Jurkat cells were electroporated at 960 μF and 300 V with plasmids in serum-free RPMI 1640 medium.
RESULTS AND DISCUSSION
TORC proteins are coactivators of Tax.
TORC3 was recently shown to stimulate Tax activation of HTLV-1 LTR (25). TORC1 and TORC2 (TORC1/2) share only ∼30% amino acid identity with TORC3 (14). In addition, TORC1/2 transcripts are found in thymus, CD4+ lymphocytes, and Molt4 leukemic cells (8). Actually, TORC2 is most abundantly expressed in thymus (8). Thus, we asked whether TORC1/2 might be physiological coactivators in the context of Tax activation of HTLV-1 LTR.
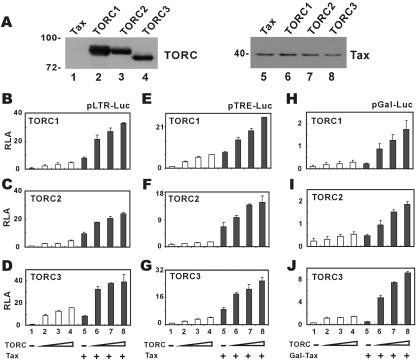
We expressed V5-tagged TORC1, TORC2, and TORC3 (TORC1/2/3) in HeLa cells (Fig. 1A) and noted that, in the absence of Tax, all three TORC proteins activated basal transcription from HTLV-1 LTR in a dose-dependent manner (Fig. 1B to D, compare groups 2 through 4 to group 1). In addition, TORC1/2/3 also cooperated with Tax in activating the LTR (Fig. 1B to D, compare groups 6 to 8 to group 5).
FIG. 1.
TORC proteins are transcriptional coactivators of HTLV-1 Tax. (A) Expression of TORCs in HeLa cells. Cells were transfected with Tax expression plasmid pIEX alone (lanes 1 and 5) or with pIEX plus pcDNA3.1-V5-TORC1/2/3 (lanes 2 to 4 and 6 to 8). Cell lysates were analyzed by Western blotting with rabbit anti-V5 (Invitrogen) or mouse anti-Tax (11). The expression of TORCs did not affect the activity of the cytomegalovirus promoter. (B to D) Cooperation of TORCs and Tax in the activation of HTLV-1 LTR. HeLa cells (2 × 105 per well) were transfected with reporter plasmid pLTR-Luc (1 μg) plus the indicated combinations of expression plasmids. A fixed amount of Tax expression plasmid pIEX (500 ng) and escalating amounts of expression plasmids (60 to 250 ng) for TORCs were used. (E to G) Cooperation of TORCs and Tax in the activation of TRE. HeLa cells (2 × 105 per well) were transfected with reporter plasmid pTRE-Luc (1 μg) plus the indicated combinations of expression plasmids. A fixed amount of Tax expression plasmid pIEX (500 ng) and escalating amounts of expression plasmids (60 to 250 ng) for TORCs were used. (H to J) Stimulation of Gal-Tax-mediated transcriptional activation by TORCs. HeLa cells (2 × 105 per well) were cotransfected with reporter plasmid pGal-Luc (1 μg) and expression plasmids for Gal-Tax (500 ng) and TORCs (60 to 250 ng). Cells were harvested 48 h after transfection, and relative luciferase activities (RLA) in arbitrary units were calculated from the readouts of firefly luciferase normalized to the readouts of Renilla luciferase. The results represent three independent experiments, and the error bars indicate standard deviations.
Because the three 21-bp repeats (or TREs) in HTLV-1 LTR are recognized by Tax and CREB (39, 41), we asked whether these TRE enhancers would also be targeted by TORCs. Indeed, TORC1/2/3 were capable of activating transcription from TRE alone and they cooperated with Tax in the activation of TRE (Fig. 1E to F, compare groups 2 through 4 to group 1 and compare groups 6 through 8 to group 5). Thus, the transcriptional activity of TORCs is mediated through TRE.
To verify the coactivator function of TORCs on Tax, we repeated the experiments in the context of a fusion protein comprising the Gal4 DNA binding domain and Tax (Gal-Tax) and a luciferase reporter construct under the control of Gal4-binding enhancer elements (pGal-Luc). In this setting, the escalation of the amount of TORC1/2/3 also led to substantial potentiation of Tax-dependent transcription (Fig. 1H to J, compare groups 6 through 8 to group 5). We also appreciated that TORCs had a weak general coactivator activities on pGal-Luc in the absence of Gal-Tax (Fig. 1H to J, compare groups 2 through 4 to group 1). Hence, TORC proteins preferentially coactivate Tax and HTLV-1 LTR.
Association of TORCs and Tax.
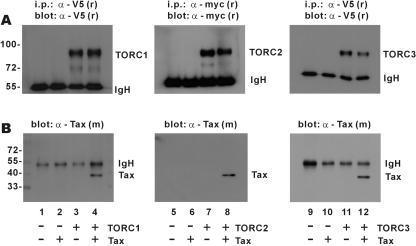
TORC3 has previously been shown to interact through its N-terminal coiled-coil domain with Tax (25). This finding agrees with our prediction that Tax preferentially interacts with a subset of cellular coiled-coil proteins, including mitotic checkpoint protein MAD1 and IκB kinase regulatory subunit IKK-γ/NEMO (7, 13, 19, 20). To investigate whether TORC1/2 also form a complex with Tax in cells, we precipitated them from cultured HeLa cells (Fig. 2A, lanes 3, 4, 7, 8, 11, and 12). In cells expressing both Tax and TORC1/2/3, the α-V5/myc precipitates also contained Tax (Fig. 2B, lanes 4, 8, and 12). By contrast, the Tax-TORC complex was not found in mock-, Tax-, or TORC-transfected cells (Fig. 2B, lanes 1 to 3, 5 and 6, and 9 to 11). Our results suggest that all three TORCs can complex with Tax in cultured mammalian cells.
FIG. 2.
Association of Tax with TORCs. HeLa cells were transfected with the indicated combinations of expression plasmids. Lysates of cells expressing V5-tagged TORC1 (lanes 3 and 4), myc-tagged TORC2 (lanes 7 and 8), V5-tagged TORC3 (lanes 11 and 12), and Tax (lanes 2, 4, 6, 8, 10, and 12) were immunoprecipitated (i.p.) with anti-V5 (α-V5) or anti-myc (α-myc) (Santa-Cruz), and the precipitates were analyzed by Western blotting with rabbit anti-V5 (panel A, lanes 1 to 4 and 9 to 12), rabbit anti-myc (panel A, lanes 5 to 8), or mouse anti-Tax (α-Tax) (B). Expression vector for myc-tagged TORC2 was derived from pcDNA4 (Invitrogen). Immunoglobulin H (IgH), heavy chain of immunoglobulin G. −, absence of indicated protein; +, presence of indicated protein.
CREB is required for Tax/TORC-induced activation of LTR.
While CREB is needed for optimal activation of HTLV-1 LTR by Tax (6), other bZIP proteins, including ATF4 and LZIP can also contribute to Tax activation of LTR (6, 9, 35). Two important questions are currently unanswered. First, is CREB uniquely essential for the transcriptional activation by Tax and TORCs? Second, can TORC proteins also serve as coactivators for ATF4, LZIP, and other bZIP factors?
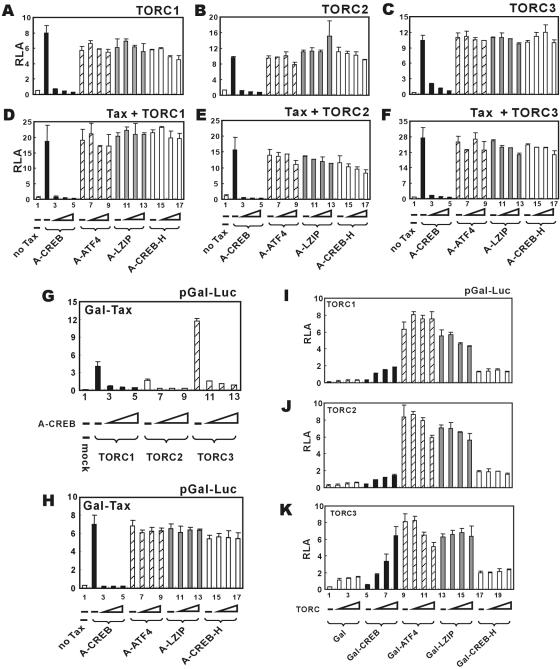
To address these questions, we employed a group of four dominant inactive bZIP proteins (A-CREB, A-ATF4, A-LZIP, and A-CREB-H) that were constructed using the same strategy of fusing an acidic peptide with the leucine zipper region (1). The specific dominant inhibitory activities of these mutants have been previously verified (1, 6). Particularly, the expression of A-CREB leads to a dramatic suppression of Tax activation of LTR, with more modest reduction observed with A-ATF4 and A-LZIP (6). When we carried out similar sets of experiments using TORC1/2/3, we found that only A-CREB completely inhibited the transcriptional activity of LTR (Fig. 3A to C, compare groups 3 to 5 to group 2), while A-ATF4, A-LZIP, and A-CREB-H did not significantly affect TORC-dependent activation (Fig. 3A to C, groups 6 through 17), with only small inhibitions observed at the highest doses (Fig. 3A, groups 16 and 17, and B, groups 9 and 17). Consistent with this, A-CREB could also erase the transcriptional activity from Tax plus TORC1/2/3, whereas the other three dominant inactive bZIP proteins had minimal effects (Fig. 3D to F, compare groups 6 through 17 to groups 2 through 5). Moreover, the coactivator function of TORC1/2/3 in the context of Gal-Tax and pGal-Luc was also blunted by A-CREB (Fig. 3G). To address whether CREB is required after the recruitment of Tax to the promoter, we tested the activity of Gal-Tax in the presence of A-CREB. We observed that Gal-Tax activity was abrogated by only A-CREB but not other dominant inactive bZIP proteins (Fig. 3H, compare groups 3 through 5 to groups 7 through 9, 11 through 13, and 15 through 17). Thus, we interpret CREB, but not ATF4 or other bZIP factors, to be responsible for Tax/TORC-mediated activation of HTLV-1 LTR.
FIG. 3.
TORC coactivation with Tax is specific for CREB. (A to F) CREB is required for transcriptional activities of TORC and Tax plus TORC. HeLa cells (2 × 105 per well) were transfected with reporter plasmid pLTR-Luc (1 μg) plus the indicated combinations of expression plasmids. Escalating amounts (60 to 250 ng) of expression vectors for dominant inactive forms of CREB, ATF4, LZIP, and CREB-H were used. (G and H) CREB is essential for TORC coactivation of Gal-Tax and for Gal-Tax activity. HeLa cells (2 × 105 per well) were transfected with reporter plasmid pGal-Luc (1 μg) plus the indicated combinations of expression plasmids. Escalating amounts (60 to 250 ng) of expression vector for dominant inactive form of CREB (A-CREB) were used. (I to K) TORC proteins are CREB-specific transcriptional coactivators. HeLa cells were cotransfected with reporter plasmid pGal-Luc and expression plasmids for TORC1 (I), TORC2 (J), and TORC3 (K) as well as expression plasmids for Gal (groups 1 to 4), Gal-CREB (groups 5 to 8), Gal-ATF4 (groups 9 to 12), Gal-LZIP (groups 13 to 16), and Gal-CREB-H (groups 17 to 20). The same amount (500 ng) of Gal plasmids and escalating amounts (60 to 250 ng) of TORC plasmids were used. The results represent three independent experiments, and the error bars indicate standard deviations. RLA, relative luciferase activity.
We also addressed whether TORC proteins act as coactivators for bZIP factors, such as ATF4 (9, 35), LZIP (21, 42), and CREB-H (5). We observed that fusion proteins Gal-CREB, Gal-ATF4, Gal-LZIP and Gal-CREB-H could activate pGal-Luc to different degrees (Fig. 3I to K, compare groups 5, 9, 13, and 17 to group 1). However, the expression of TORC1/2/3 enhanced the transcriptional activity of only Gal-CREB, not Gal-ATF4, Gal-LZIP, or Gal-CREB-H (Fig. 3I to K, compare groups 5 through 8 to groups 9 through 20). Taken together, TORC coactivators are active with only CREB, not other CREB-related bZIP proteins.
TORC coactivators were newly identified from genome-wide screening of transcriptional coregulators specific for CREB (8, 14). Our present work, together with a recent report on TORC3 (25), established that TORCs are also needed for the transcriptional activity mediated by Tax. We noted that TORC3 overexpressed in HeLa cells exhibited a relatively strong transcriptional activity on pGal-Luc compared to those of TORC1 and TORC2 (Fig. 1J compared to H and I and Fig. 3G and K compared to I and J). In contrast, TORC1/2/3 showed comparable coactivator activity on pLTR-Luc and pTRE-Luc (Fig. 1B to G). Further experiments conducted with recombinant TORC proteins are required to clarify whether any of the three TORC proteins is particularly active in interacting with Tax and/or stimulating transcription.
Our findings support the notion that the TORC-mediated coactivation of Tax is specific for CREB (Fig. 3). Thus, Tax plus CREB requires TORCs, while Tax plus other cellular transcription activator, may utilize a different set of coactivators. We suggest that Tax plus activator plus coactivators ultimately determines the specificity of the ternary complex for the viral LTR or for cellular promoters.
Consistent with our previous finding that CREB is preferred over ATF4, c-Jun, and other bZIP proteins for Tax activation of HTLV-1 LTR (6), here we showed that only CREB, not ATF4 or other bZIP factors, is required for the recruitment and activity of TORCs (Fig. 3). We further demonstrated that TORCs exert coactivator function on only CREB, not other CREB-related bZIP factors (Fig. 3). The recruitment and activation of TORCs by CREB could explain the preference for CREB by Tax in the activation of HTLV-1 LTR.
TORC coactivators are required for Tax activation of HTLV-1 LTR in T lymphocytes.
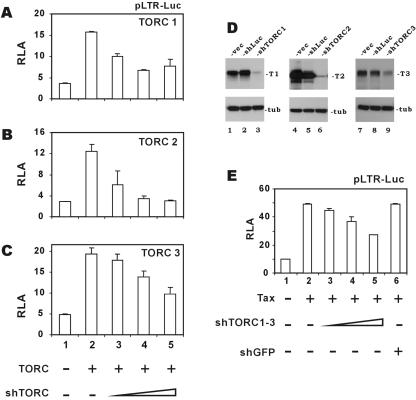
Above we characterized the role of TORC coactivators in Tax-mediated LTR transcription in cultured HeLa cells. To ask whether similar requirements can be shown in suspension T lymphocytes that are susceptible to physiological infection of HTLV-1, we used RNAi knockdown to investigate the roles of TORC coactivators. We designed three shRNAs targeting TORC1/2/3 (shTORC1/2/3). When we expressed these shRNAs in Jurkat cells, a 35 to 70% inhibition of exogenously expressed TORC1/2/3 was achieved (Fig. 4A to C). These shRNAs could also effectively knockdown TORC expression in HeLa cells as shown by Western blotting (Fig. 4D). Since shTORCs (e.g., shTORC1) inhibited only the desired isoform (e.g., TORC1), not the other two paralogs (e.g., TORC2/3), our RNAi depletion of TORCs was isotype specific (data not shown).
FIG. 4.
TORC coactivators are required for optimal activation of the HTLV-1 LTR by Tax. (A to C) Inhibition of TORC-dependent transcriptional activity by shRNAs. Jurkat cells were transfected with reporter plasmid pLTR-Luc (8 μg), a fixed amount (2 μg) of expression plasmids for TORC1 (A), TORC2 (B), and TORC3 (C), and escalating amounts (2 to 10 μg) of plasmids expressing shRNA against TORC1, TORC2, or TORC3 (shTORC). (D) Depletion of TORCs by shRNAs. HeLa cells were transfected with expression plasmids for GFP-tagged TORC1 (lanes 1 to 3), TORC2 (lanes 4 to 6) and TORC3 (lanes 7 to 9) as well as empty shRNA expression vector (vec; lanes 1, 4 and 7), expression vector for shRNA targeting luciferase (shLuc; lanes 2, 5, and 8), and expression plasmid for shRNA against TORC1, TORC2, and TORC3 (shTORC1/2/3; lanes 3, 6, and 9), respectively. Expression of GFP-TORC proteins (upper panels) and antitubulin (tub; lower panels) was analyzed by Western blotting. (E) TORCs are required for Tax activation in T lymphocytes. Jurkat lymphocytes were electroporated with reporter plasmid pLTR-Luc (8 μg), Tax expression plasmid pIEX (1 μg), and increasing amounts (2 to 10 μg) of a mixture of plasmids expressing shRNAs targeting TORC1, TORC2, and TORC3 (shTORC1 to -3). The shRNA against GFP (shGFP) was also included as a control. Cells were harvested 48 h after electroporation, and relative luciferase activities (RLA) were measured. The results represent three independent experiments and the error bars indicate standard deviations. −, absence of indicated protein; +, presence of indicated protein.
When each of the three shTORCs was expressed singularly, we did not observe appreciable inhibition of Tax activation of LTR (data not shown). This might be explained by incomplete knockdown and/or redundant function of the three TORC proteins. We thus expressed all three shTORCs simultaneously in Jurkat cells and observed a moderate dose-dependent suppressive effect on Tax activity (Fig. 4E, compare groups 3 through 5 to group 2). The specificity of this effect was supported by a control experiment in which the shRNA targeting green fluorescent protein (GFP) failed to affect Tax activation of the LTR (Fig. 4E, compare group 6 to group 2). These results indicated that TORC coactivators may redundantly cooperate with Tax and that simultaneous knockdown of all three endogenous TORCs in T cells is needed for significant inhibition of Tax.
The incompleteness of the RNAi-induced silencing effect prevents us from establishing the physiological roles of TORCs and their exact function in the context of Tax and HTLV-1 LTR. In these regards, the creation of TORC-null mice could provide new opportunities for functional studies of TORCs. Nevertheless, our RNAi data did implicate functional redundancies among the three TORC coactivators.
Both TORCs and p300 family coactivators are required for Tax activation of LTR.
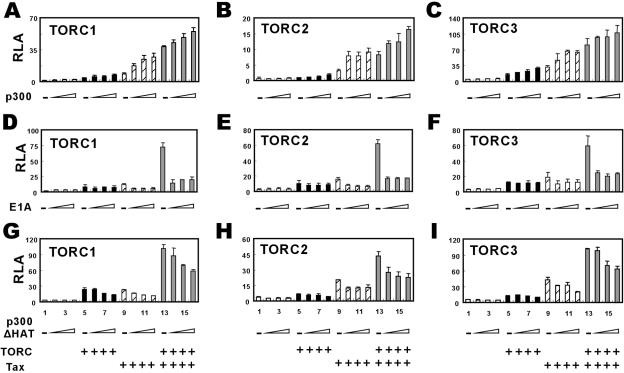
p300 and related coactivators in the same family have also been shown to play an important role in Tax-induced transcriptional activation (11, 12, 17, 22, 27). On the other hand, suppression of all three TORCs by RNAi resulted in only a partial inhibition of Tax-mediated activation of LTR (Fig. 4), suggesting that other coactivators might also contribute to this process. In light of this, we set out to investigate the relationship between the p300 and TORC families of coactivators in the context of Tax and HTLV-1 LTR. When we coexpressed p300 and TORC1/2/3 in human 293T cells in the absence of Tax, p300 was able to enhance TORC activity on HTLV-1 LTR modestly but in a dose-dependent manner (Fig. 5A to C, compare groups 6 through 8 to group 5). Consistent with previous reports (17, 23), p300 could also coactivate Tax in this experimental setting (Fig. 5A to C, compare groups 10 through 12 to group 9). In addition, in cells expressing p300, Tax and TORC1/2/3 simultaneously, p300 further increased the transcriptional activity mediated by Tax and TORC1/2/3 (Fig. 5A to C, compare groups 14 through 16 to group 13). Thus, p300 and TORC1/2/3 act additively in mediating Tax activation of LTR.
FIG. 5.
The role of p300 family coactivators in Tax/TORC-mediated activation of HTLV-1 LTR. (A to C) Additive action of TORC1/2/3 and p300. 293T cells (1.5 × 105 per well) were transfected with reporter plasmid pLTR-Luc (250 ng) plus the indicated combinations of expression plasmids. Escalating amounts of p300 expression plasmid (60 to 250 ng) were used with fixed amounts of plasmids for TORCs (15 ng) and Tax (250 ng). (D to F) Influence of the transcriptional activity of Tax and TORCs by E1A-12S. HeLa cells (4 × 104 per well) were transfected with reporter plasmid pLTR-Luc (250 ng) plus the indicated combinations of expression plasmids. Escalating amounts of E1A-12S plasmid (15 to 60 ng) were used with fixed amounts of TORC (15 ng) and Tax (250 ng) plasmids. (G to I) Influence of a dominant inactive p300 mutant on the activity of Tax and TORCs. HeLa cells (4 × 104 per well) were transfected with reporter plasmid pLTR-Luc (250 ng) plus the indicated combinations of expression plasmids. Escalating amounts of p300ΔHAT deletion mutant (60 to 250 ng) were used with fixed amounts of TORC (15 ng) and Tax (250 ng) plasmids. The results represent three independent experiments, and the error bars indicate standard deviations. RLA, relative luciferase activity.
Next, we examined the effect of adenovirus E1A-12S, a well-characterized inhibitor of p300 and related coactivators, including CBP and P/CAF (12, 29), on the activity of TORCs in the activation of LTR. Generally consistent with previous findings (12), E1A-12S could inhibit Tax activation of LTR (Fig. 5D to F, compare groups 10 through 12 to group 9). In contrast, it did not significantly influence TORC1/2/3-induced activation of LTR in the absence of Tax (Fig. 5D to F, compare groups 6 through 8 to group 5). Notably, E1A-12S substantially repressed transcriptional activity when both Tax and TORC1/2/3 were expressed (Fig. 5D to F, compare groups 14 through 16 to group 13). These results suggest that coactivators in the p300 family are required for optimal activation of HTLV-1 LTR by Tax independently of TORCs.
To determine the role of p300 in Tax/TORC-mediated transcriptional activation, we assessed the impact of p300ΔHAT, a dominant inactive mutant of p300 lacking the acetyltransferase domain (33, 40), on the activity of TORCs and Tax. The expression of p300ΔHAT did not significantly affect the activity of TORC1/2/3 in the activation of LTR (Fig. 5G to I, compare groups 6 through 8 to group 5), but it modestly inhibited the activity of Tax both in the presence and in the absence of TORC1/2/3 (compare groups 10 through 12 to group 9, and compare groups 14 through 16 to group 13). Similar results were also obtained with p300DY (15), another dominant negative mutant of p300 with an inactivating point mutation in the acetyltransferase domain (data not shown). The repressive activity of p300 mutants was not as dramatic as that with E1A-12S, suggesting that, in addition to p300, other coactivators in the same family that are inhibited by E1A-12S might also contribute to the full activity of Tax. Taken together, coactivators in both TORC and p300 families are required for Tax activation of HTLV-1 LTR.
Emerging evidence suggests that TORCs represent another regulatory point at which different stimuli converge (2, 26, 37, 38). Several lines of data support the notion that Tax serves to trigger activation of TORCs, bypassing cellular regulatory mechanisms. First, Tax physically interacts with TORCs (Fig. 2). Second, Tax activation of HTLV-1 LTR can bypass the need for stimulation by cAMP/protein kinase A (27), which also induces activation of TORCs (2, 8). Finally, Tax activation of HTLV-1 LTR is potent (6), which is explained by constitutive activation of a major group of transcriptional coactivators.
Exactly how TORCs activate transcription from HTLV-1 LTR remains to be understood. One mechanism by which TORCs activate CREB-dependent transcription is through TAFII130 (23). Tax has been shown to interact with TATA-binding protein and TAFII28 (3, 4). Other TATA-binding protein-associated factors, such as TAFII250, have also been shown to be important in Tax activation of HTLV-1 LTR (28). Further experiments are required to clarify whether and how TAFII130 might be particularly influential on Tax.
In summary, in this study we demonstrated the coactivator function of TORC1/2 in the context of Tax and HTLV-1 LTR (Fig. 1 and 4), the association of Tax with TORC1/2 in cultured cells (Fig. 2), and the requirement of CREB and p300 family coactivators for Tax/TORC-dependent transcription (Fig. 3 and 5). Our findings refine the model for Tax action, in which Tax physically interacts with CREB, TORCs, and coactivators of the p300 family to stimulate transcription from HTLV-1 LTR.
Acknowledgments
We thank G. J. Hannon, J. R. Lundblad, Y. Nakatani, C. Vinson, Z. Wu, X. J. Yang, and T. P. Yao for gifts of plasmid, Y.-P. Ching for helpful comments and suggestions, and A. C. S. Chun for critical reading of the manuscript.
D.-Y. Jin is a Leukemia and Lymphoma Society Scholar. This work was supported by grants to D.-Y. Jin from the Hong Kong Research Grants Council (projects HKU 7249/01 M, HKU 7294/02 M, and HKU 7683/05 M), Concern Foundation, and the National Natural Science Foundation of China (Young Investigator Award 30029001).
REFERENCES
- 1.Ahn, S., M. Olive, S. Aggarwal, D. Krylov, D. Ginty, and C. Vinson. 1998. A dominate-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol. Cell. Biol. 18:967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bittinger, M. A., E. McWhinnie, J. Meltzer, V. Iourgenko, B. Latario, X. Liu, C. H. Chen, C. Song, D. Garza, and M. Labow. 2004. Activation of cAMP response element-mediated gene expression by regulated nuclear transport of TORC proteins. Curr. Biol. 14:2156-2161. [DOI] [PubMed] [Google Scholar]
- 3.Caron, C., R. Rousset, C. Béraud, V. Moncollin, J. M. Egly, and P. Jalinot. 1993. Functional and biochemical interaction of the HTLV-1 Tax1 transactivator with TBP. EMBO J. 12:4269-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caron, C., G. Mengus, V. Dubrowskaya, A. Roisin, I. Davidson, and P. Jalinot. 1997. Human TAFII28 interacts with the human T cell leukemia virus type I Tax transactivator and promotes its transcriptional activity. Proc. Natl. Acad. Sci. USA 94:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chin, K. T., H. J. Zhou, C. M. Wong, J. M. F. Lee, C. P. Chan, B. Q. Qiang, J. G. Yuan, I. O. L. Ng, and D. Y. Jin. 2005. The liver-enriched transcription factor CREB-H is a growth suppressor protein underexpressed in hepatocellular carcinoma. Nucleic Acids Res. 33:1859-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ching, Y. P., A. C. S. Chun, K. T. Chin, Z. Q. Zhang, K. T. Jeang, and D. Y. Jin. 2004. Specific TATAA and bZIP requirements suggest that HTLV-1 Tax has transcriptional activity subsequent to the assembly of an initiation complex. Retrovirology 1:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chun, A. C. S., Y. Zhou, C. M. Wong, H. Kung, K. T. Jeang, and D. Y. Jin. 2000. Coiled-coil motif as a structural basis for the interaction of HTLV type 1 Tax with cellular cofactors. AIDS Res. Hum. Retrovir. 16:1689-1694. [DOI] [PubMed] [Google Scholar]
- 8.Conkright, M. D., G. Canettieri, R. Screaton, E. Guzman, L. Miraglia, J. B. Hogenesch, and M. Montminy. 2003. TORCs: transducers of regulated CREB activity. Mol. Cell 12:413-423. [DOI] [PubMed] [Google Scholar]
- 9.Gachon, F., S. Thebault, A. Peleraux, C. Devaux, and J. M. Mesnard. 2000. Molecular interactions involved in the transactivation of the human T-Cell leukemia virus type 1 promoter mediated by Tax and CREB-2 (ATF-4). Mol. Cell. Biol. 20:3470-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gatza, M. L., J. C. Watt, and S. J. Marriott. 2003. Cellular transformation by the HTLV-1 Tax protein, a jack-of-all-trades. Oncogene 22:5141-5149. [DOI] [PubMed] [Google Scholar]
- 11.Harrod, R., Y. Tang, C. Nicot, H. S. Lu, A. Vassilev, Y. Nakatani, and C. Z. Giam. 1998. An exposed KID-like domain in human T-cell lymphotropic virus type 1 Tax is responsible for the recruitment of coactivators CBP/p300. Mol. Cell. Biol. 18:5052-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrod, R., Y. L. Kuo, Y. Tang, Y. Yao, A. Vassilev, Y. Nakatani, and C. Z. Giam. 2000. p300 and p300/cAMP-responsive element-binding protein associated factor interact with human T-cell lymphotropic virus type-1 Tax in a multi-histone acetyltransferase/activator-enhancer complex. J. Biol. Chem. 275:11852-11857. [DOI] [PubMed] [Google Scholar]
- 13.Huang, G. J., Z. Q. Zhang, and D. Y. Jin. 2002. Stimulation of IKK-γ oligomerization by the T-cell leukemia virus oncoprotein Tax. FEBS Lett. 531:494-498. [DOI] [PubMed] [Google Scholar]
- 14.Iourgenko, V., W. Zhang, C. Mickanin, I. Daly, C. Jiang, J. M. Hexham, A. P. Orth, L. Miraglia, J. Meltzer, D. Garza, G. W. Chirn, E. McWhinnie, D. Cohen, J. Skelton, R. Terry, Y. Yu, D. Bodian, F. P. Buxton, J. Zhu, C. Song, and M. A. Labow. 2003. Identification of a family of cAMP response element-binding protein coactivators by genome-scale functional analysis in mammalian cells. Proc. Natl. Acad. Sci. USA 100:12147-12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito, A., C. H. Lai, X. Zhao, S. Saito, M. H. Hamilton, E. Appella, and T. P. Yao. 2001. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 20:1331-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeang, K. T., C. Z. Giam, F. Majone, and M. Aboud. 2004. Life, death, and Tax: role of HTLV-1 oncoprotein in genetic instability and cellular transformation. J. Biol. Chem. 279:31991-31994. [DOI] [PubMed] [Google Scholar]
- 17.Jiang, H., H. Lu, R. L. Schiltz, C. A. Pise-Masison, V. V. Ogryzko, Y. Nakatani, and J. N. Brady. 1999. PCAF interacts with tax and stimulates tax transactivation in a histone acetyltransferase-independent manner. Mol. Cell. Biol. 19:8136-8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin, D. Y., and K. T. Jeang. 1997. HTLV-1 Tax self-association in optimal trans-activation function. Nucleic Acids Res. 25:379-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin, D. Y., F. Spencer, and K. T. Jeang. 1998. Human T-cell leukemia virus type I oncoprotein Tax targets the human mitotic checkpoint protein MAD1. Cell 93:81-91. [DOI] [PubMed] [Google Scholar]
- 20.Jin, D. Y., V. Giordano, K. V. Kibler, H. Nakano, and K. T. Jeang. 1999. Role of adaptor function in oncoprotein-mediated activation of NF-κB: HTLV-1 Tax interacts directly with IκB kinase γ. J. Biol. Chem. 274:17402-17405. [DOI] [PubMed] [Google Scholar]
- 21.Jin, D. Y., H. L. Wang, Y. Zhou, A. C. S. Chun, K. V. Kibler, Y. D. Hou, H. Kung, and K. T. Jeang. 2000. Hepatitis C virus core protein-induced loss of LZIP function correlates with cellular transformation. EMBO J. 19:729-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kashanchi, F., J. F. Duvall, R. P. S. Kwok, J. R. Lundblad, R. H. Goodman, and J. N. Brady. 1998. The coactivator CBP stimulates human T-cell lymphotropic virus type 1 Tax transactivation in vitro. J. Biol. Chem. 273:34646-34652. [DOI] [PubMed] [Google Scholar]
- 23.Kashanchi, F., and J. N. Brady. 2005. Transcriptional and post-transcriptional gene regulation of HTLV-1. Oncogene 24:5938-5951. [DOI] [PubMed] [Google Scholar]
- 24.Kimzey, A. L., and W. S. Dynan. 1998. Specific regions of contact between human T-cell leukemia virus type I Tax protein and DNA identified by photocross-linking. J. Biol. Chem. 273:13768-13775. [DOI] [PubMed] [Google Scholar]
- 25.Koga, H., T. Ohshima, and K. Shimotohno. 2004. Enhanced activation of Tax-dependent transcription of human T-cell leukemia virus type I (HTLV-1) long terminal repeat by TORC3. J. Biol. Chem. 279:52978-52983. [DOI] [PubMed] [Google Scholar]
- 26.Koo, S. H., L. Flechner, L. Qi, X. Zhang, R. A. Screaton, S. Jeffries, S. Hedrick, W. Xu, F. Boussouar, P. Brindle, H. Takemori, and M. Montminy. 2005. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature 437:1109-1111. [DOI] [PubMed] [Google Scholar]
- 27.Kwok, R. P., M. E. Laurance, J. R. Lundblad, P. S. Goldman, H. M. Shih, L. M. Connor, S. J. Marriott, and R. H. Goodman. 1996. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the co-activator CBP. Nature 380:642-646. [DOI] [PubMed] [Google Scholar]
- 28.Lemasson, I., N. J. Polakowski, P. J. Laybourn, and J. K. Nyborg. 2004. Transcription regulatory complexes bind the human T-cell leukemia virus 5′ and 3′ long terminal repeats to control gene expression. Mol. Cell. Biol. 24:6117-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lundblad, J. R., R. P. Kwok, M. E. Laurance, M. S. Huang, J. P. Richards, R. G. Brennan, and R. H. Goodman. 1998. The human T-cell leukemia virus-1 transcriptional activator Tax enhances cAMP-responsive element-binding protein (CREB) binding activity through interactions with the DNA minor groove. J. Biol. Chem. 273:19251-19259. [DOI] [PubMed] [Google Scholar]
- 30.Ma, K., J. K. Chan, G. Zhu, and Z. Wu. 2005. Myocyte enhancer factor 2 acetylation by p300 enhances its DNA binding activity, transcriptional activity, and myogenic differentiation. Mol. Cell. Biol. 25:3575-3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madison, D. L., P. Yaciuk, R. P. Kwok, and J. R. Lundblad. 2002. Acetylation of the adenovirus-transforming protein E1A determines nuclear localization by disrupting association with importin-α. J. Biol. Chem. 277:38755-38763. [DOI] [PubMed] [Google Scholar]
- 32.Matsuoka, M., and K. T. Jeang. 2005. Human T-cell leukemia virus type I at age 25: a progress report. Cancer Res. 65:4467-4470. [DOI] [PubMed] [Google Scholar]
- 33.Miyazato, A., S. Sheleg, H Iha, Y. Li, and K. T. Jeang. 2005. Evidence for NF-κB- and CBP-independent repression of p53's transcriptional activity by human T-cell leukemia virus type 1 Tax in mouse embryo and primary human fibroblasts. J. Virol. 79:9346-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paddison, P. J., A. A. Caudy, and G. J. Hannon. 2002. Stable suppression of gene expression by RNAi in mammalian cells. Proc. Natl. Acad. Sci. USA 99:1443-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reddy, T. R., H. Tang, X. Li, and F. Wong-Staal. 1997. Functional interaction of the HTLV-1 transactivator Tax with activating transcription factor-4 (ATF4). Oncogene 14:2785-2792. [DOI] [PubMed] [Google Scholar]
- 36.Roeder, R. G. 2005. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 579:909-915. [DOI] [PubMed] [Google Scholar]
- 37.Screaton, R. A., M. D. Conkright, Y. Katoh, J. L. Best, G. Canettieri, S. Jeffries, E. Guzman, S. Niessen, J. R. Yates III, H. Takemori, M. Okamoto, and M. Montminy. 2004. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell 119:61-74. [DOI] [PubMed] [Google Scholar]
- 38.Shaw, R. J., K. A. Lamia, D. Vasquez, S. H. Koo, N. Bardeesy, R. A. DePinho, M. Montminy, and L. C. Cantley. 2005. The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science 310:1642-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tie, F., N. Adya, W. C. Greene, and C. Z. Giam. 1996. Interaction of the human T-lymphotropic virus type 1 Tax dimer with CREB and the viral 21-base-pair repeat. J. Virol. 70:8368-8374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang, X. J., V. V. Ogryzko, J. Nishikawa, B. H. Howard, and Y. Nakatani. 1996. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382:319-324. [DOI] [PubMed] [Google Scholar]
- 41.Zhao, L. J., and C. Z. Giam. 1991. Interaction of the human T-cell lymphotrophic virus (HTLV) type I transcriptional activator Tax with cellular factors that bind specifically to the 21-base-pair repeats in the HTLV-1 enhancer. Proc. Natl. Acad. Sci. USA 88:11445-11449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou, H. J., C. M. Wong, J. H. Chen, B. Q. Qiang, J. G. Yuan, and D. Y. Jin. 2001. Inhibition of LZIP-mediated transcription through direct interaction with a novel host cell factor-like protein. J. Biol. Chem. 276:28933-28938. [DOI] [PubMed] [Google Scholar]