Abstract
In contrast to other retroviruses, Mason-Pfizer monkey virus (M-PMV) assembles immature capsids in the cytoplasm. We have compared the ability of minimal assembly-competent domains from M-PMV and human immunodeficiency virus type 1 (HIV-1) to assemble in vitro into virus-like particles in the presence and absence of nucleic acids. A fusion protein comprised of the capsid and nucleocapsid domains of Gag (CANC) and its N-terminally modified mutant (ΔProCANC) were used to mimic the assembly of the viral core and immature particles, respectively. In contrast to HIV-1, where CANC assembled efficiently into cylindrical structures, the same domains of M-PMV were assembly incompetent. The addition of RNA or oligonucleotides did not complement this defect. In contrast, the M-PMV ΔProCANC molecule was able to assemble into spherical particles, while that of HIV-1 formed both spheres and cylinders. For M-PMV, the addition of purified RNA increased the efficiency with which ΔProCANC formed spherical particles both in terms of the overall amount and the numbers of completed spheres. The amount of RNA incorporated was determined, and for both rRNA and MS2-RNA, quantities similar to that of genomic RNA were encapsidated. Oligonucleotides also stimulated assembly; however, they were incorporated into ΔProCANC spherical particles in trace amounts that could not serve as a stoichiometric structural component for assembly. Thus, oligonucleotides may, through a transient interaction, induce conformational changes that facilitate assembly, while longer RNAs appear to facilitate the complete assembly of spherical particles.
All retroviruses encode a single structural polyprotein precursor, Gag, that directs the assembly of the immature retroviral capsid and can direct the formation of retrovirus-like particles in mammalian cells and in bacteria. In addition to domains common to all retroviruses (matrix [MA], capsid [CA] and nucleocapsid [NC]), the Gag polyprotein of Mason-Pfizer monkey virus (M-PMV) also contains a phosphoprotein, pp16-18, an internal scaffold protein, p12, and a C-terminal domain, p4.
The CA domain plays a key role in the assembly of both immature retroviral capsids and the mature cores of infectious virions. The CA protein of human immunodeficiency virus type 1 (HIV-1) dimerizes in solution, while other CA proteins such as those of Rous sarcoma virus (RSV), equine infectious anemia virus, and human T-cell leukemia virus type 1 are monomeric in solution (5, 13, 27, 43, 44). Structural studies of retroviral CA proteins have shown that they consist of two structural domains, the N-terminal assembly domain and the C-terminal dimerization domain (1, 4, 6, 13, 15, 20, 27, 31, 43, 44, 57, 65, 76, 77). Several studies have focused on mutual interactions between CA proteins; both C-terminal-C-terminal and N-terminal-C-terminal interaction seem to be involved (47, 48). In addition, a recent model suggests that interactions may occur between CA molecules by domain swapping, similar to those of mammalian SCAN domains (39, 45).
Several studies have shown that the RSV and HIV-1 CA proteins can assemble in vitro into tubular structures (23, 36) or conical particles resembling a mature core (23, 28, 36, 44). The latter protein also assembles into hollow cylinders at high salt concentrations (22, 28, 36, 50), and structures assembled on lipid from a His-tagged HIV-1 CA protein show a cage-like lattice consisting of hexamers (2). CA proteins of several retroviruses form a β-hairpin stabilized by a salt bridge between the N-terminal proline and conservative aspartates in positions 50 to 57 of the protein, consistent with a molecular rearrangement of CA upon proteolytic maturation (26, 31, 37, 69). Extension of the N terminus would therefore prevent the hairpin formation and direct assembly of spherical rather than tubular structures (44, 61, 76). In concert with this, HIV-1 CA forms variously sized spherical particles in the presence of four or more flanking residues of MA, while HIV-1 CA particles lacking any MA residues are tubular (37, 76). Similarly, the C-terminal residues of the RSV p10 protein, which is located just N terminal of CA, differentiate between the formation of tubes or spheres in the assembly process of this virus (11, 41).
In agreement with these observations, we have shown previously, using a bacterial expression/assembly system, that M-PMV CA with an intact N-terminal proline directs the assembly of sheets from a CANC fusion protein (69). Deletion of the proline or replacement with alanine converts assembly to spheres; this is consistent with a transition between the precursor form of CA when the N-terminal proline is not available for interaction, allowing spherical immature capsids to form and the subsequent formation of a tubular core after its proteolytic release (69).
The role of RNA in the assembly of the retroviral immature particle remains ambiguous. It is clear that the NC protein is crucial for specific incorporation of genomic RNA and can facilitate Gag-Gag interactions. The majority of retroviral NC proteins contain two conserved zinc finger domains that are responsible for genome encapsidation but dispensable for assembly (3, 7, 16, 19, 21, 33, 35, 55). Deletions of zinc finger domains as well as changes of zinc concentrations had no effect on assembly of RSV particles in vitro, but deletion of the basic residues between them abrogated assembly (81, 83). These basic residues contribute to RNA binding (17, 66) and are critical for NC-RNA interactions (16, 18, 42, 64, 72). In vitro studies indicate that the electrostatic interactions of nucleic acids with NC are required for efficient assembly (10, 11, 36, 81), and mutation of basic residues results in assembly defects and decreased processing of Gag (14). Moreover, RNA facilitates the in vitro assembly of HIV-1 and RSV cores (11, 36), and RNase treatment of viral core preparations disrupts the cores (10, 12, 60). The fact that particles form when NC is replaced by a protein interaction motif such as a leucine zipper suggests that RNA might act in part by facilitating Gag dimerization (1, 40, 82). While viral genomic RNA is not essential for particle assembly (32, 33, 35, 49, 52, 67), these particles do contain small cellular RNAs (34, 49, 51, 56). Although RSV, HIV-1, and M-PMV Gag have been reported to assemble without the addition of RNA, the presence of contaminating nucleic acids was not fully excluded (11, 12, 46).
In this report, we reanalyze the role of RNA and oligonucleotides in the assembly of the M-PMV CANC fusion protein and compare these properties to those of the HIV-1 CANC protein. The results of these studies show that the M-PMV CANC protein, in contrast to that of HIV-1, is unable to assemble in vitro in the presence or absence of nucleic acid. Deletion of the N-terminal proline from M-PMV CANC, however, results in the assembly of spherical particles in the absence of any detectable nucleic acid. Addition of rRNA or MS2-RNA increased both the quantity and quality of the particles and resulted in packaging of genome amounts of nucleic acid. In contrast, while oligonucleotides could also facilitate assembly, nonstoichiometric amounts were packaged, arguing for a catalytic rather than structural role.
MATERIALS AND METHODS
Preparation of DNA constructs.
Preparation of expression plasmids for ΔProCANC and CANC of M-PMV was described previously (69). ΔProCANC and CANC HIV-1 expression plasmids are based on the pET22b vector. All cloning steps were carried out by established techniques that are described elsewhere (71). The cloning strategies and details of the PCR primers can be obtained upon request from the authors. No mutations were introduced by the cloning strategy. The 5′- and 3′-terminal regions of the newly created plasmids were verified by DNA sequencing. The correct sizes of all expressed proteins were confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the N termini were verified by N-terminal sequencing.
Expression of M-PMV and HIV-1 genes.
Luria-Bertani medium containing ampicillin (final concentration of 100 μg/ml) was inoculated with Escherichia coli BL21(DE3) cells carrying the appropriate construct to achieve an optical density at 590 nm of ∼0.1 and grown at 37°C. Expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 0.4 mM, when the cells reached an optical density at 590 nm of ∼0.6 to 0.8. The cells were harvested 4 h postinduction by low-speed centrifugation and were stored at −20°C.
Purification of M-PMV proteins CANC and ΔProCANC.
The bacterial pellet (from 1 liter of cell culture) was resuspended in 30 ml of buffer A (50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH 8.0) containing lysozyme (1 mg/ml), 0.05% 2-mercaptoethanol, phenylmethylsulfonyl fluoride (PMSF) (100 μg/ml final concentration), and 1.2 ml of Complete (Roche) protease inhibitor mix (1 tablet per 2 ml of water), and the mixture was stirred at room temperature for 30 min. The cells were sonicated on ice and then treated with sodium deoxycholate (0.1% final concentration) at 4°C for 30 min. The cell lysate was centrifuged at 10,000 × g for 10 min at 4°C. The pellet was resuspended in 10 ml of buffer A containing 0.5% Triton X-100 and 1 M NaCl and then was centrifuged at 10,000 × g for 10 min at 4°C. The pellet was resuspended in the same volume of buffer A containing 0.5% Triton X-100, 1.5 M NaCl, 0.05% 2-mercaptoethanol and centrifuged again as mentioned above. The supernatants containing CANC or ΔProCANC were dialyzed against buffer Z (50 mM phosphate buffer, pH 7.5, containing 500 mM NaCl) overnight at 4°C. Dialyzed material was loaded on top of a Zn2+-chelating fast flow Sepharose chromatography column (volume of the resin was 5 ml; Amersham Pharmacia Biotech) equilibrated in buffer Z. After three washing steps with 50 ml of buffer Z, the bound proteins were eluted with 35 ml of 2 M NH4Cl in buffer Z. The fractions containing desired protein were combined and dialyzed against buffer B (50 mM Tris-HCl, 100 mM NaCl, 0.01% 2-mercaptoethanol, 1 μM ZnCl2, pH 7.5) overnight at 4°C. Dialyzed material was then loaded on the phosphocellulose column (Whatman). The purified proteins were eluted using a NaCl gradient (100 mM to 2 M NaCl in buffer B), and the fractions were analyzed by SDS-PAGE. The fractions containing the required protein were combined, dialyzed overnight against 2 liters of buffer C (50 mM phosphate, 500 mM NaCl, 0.01% 2-mercaptoethanol, 1 μM ZnCl2, pH 7.5), concentrated to 1 to 2 mg/ml by Millipore Centriplus membranes, aliquoted, and stored at −20°C. The protein was determined by SDS-PAGE (Fig. 1), and the presence of nucleic acids was determined by spectrophotometry (A260 and A280) and by the Ribogreen Assay (Molecular Probes). To achieve the total elimination of nucleic acids, the protocol was modified by NaCl addition (1 M) to the Zn Sepharose loading buffer. Thus, the nucleic acids were successfully separated from the protein during a few washing steps. Proteins were then eluted by pH gradient (pH 7.5 to 4.0) in 50 mM phosphate buffer containing 1 M NaCl. Another approach for elimination of nucleic acids involved the presence of 8 M urea in the Zn Sepharose loading buffer. Proteins were then eluted by pH gradient (pH 7.5 to 4.0) of phosphate buffer, dialyzed against decreased concentrations of urea, and finally stored in the same buffer as that in the first-mentioned method. Both methods provided samples of the same purity.
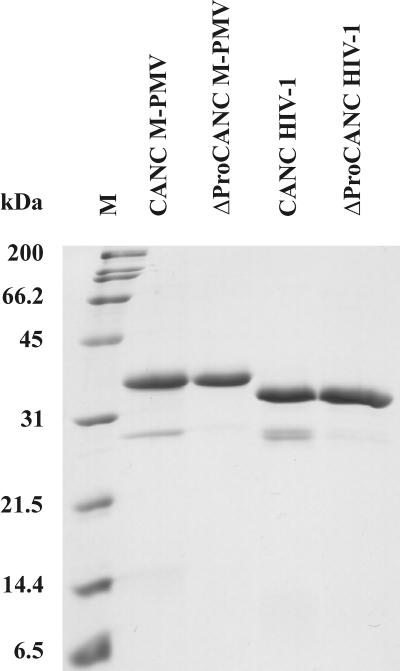
FIG. 1.
SDS-PAGE analysis of purified M-PMV and HIV-1 CANC and ΔProCANC proteins. All proteins were occasionally partially cleaved by bacterial proteases at positions that were identified close to the viral protease cleavage site between CA and NC proteins. If protease inhibitors were added, the fractions representing cleaved protein were minimal, thus not affecting the assembly process. M, molecular weight marker.
Purification of HIV-1 CANC and ΔProCANC proteins.
We have used modifications of the methods published by Campbell and Vogt as well as Ma and Vogt (10, 54) for purification of HIV-1 proteins. Frozen bacterial pellets (obtained from 1 liter of cell culture) were resuspended in 25 ml of buffer D (20 mM Tris-HCl, pH 8, 0.5 M NaCl, 10% glycerol, 1 mM EDTA, 1 mM PMSF, 10 mM dithiothreitol [DTT], 1 mM Triton X-100) on ice. The cells were disrupted by sonication (four disruptions for 15 s each on ice). Insoluble debris and nucleic acids were removed by ultracentrifugation (Beckman TLA 100.3; 65,000 rpm, 3 h, 4°C) after addition of 0.3% (wt/vol) polyethyleneimine. The protein was precipitated with 25% saturated ammonium sulfate. After 30 min at 4°C, the precipitate was collected by centrifugation for 10 min at 12,000 × g and then resuspended in buffer E (20 mM Tris-HCl, pH 8, 10 mM DTT, 1 mM PMSF, 0.1 M NaCl, 50 μM ZnCl2) at 5 ml/liter of cell culture. Insoluble material was removed by centrifugation for 4 min at 6,000 × g, and the supernatant was applied on a DEAE-cellulose column. After washing with buffer E, the flowthrough and washing fractions were pooled and loaded onto a phosphocellulose (Whatman) column. The resin with bound protein was washed with buffer E and then with buffer E containing 0.3 M NaCl. The protein was eluted with buffer E containing 0.5 M NaCl and then with the same buffer containing 1 M NaCl. Proteins were concentrated by ultrafiltration to 1 to 2 mg/ml, aliquoted, and stored at −20°C. The purity of proteins was determined by SDS-PAGE (Fig. 1), and the contamination with nucleic acids was assessed by spectrophotometry (A260 and A280) and by Ribogreen Assay (Molecular Probes).
In vitro assembly of M-PMV and HIV-1 particles.
An aliquot of 60 μg of purified CANC or ΔProCANC was mixed with oligodeoxyribonucleotides (8-mer GT8, 18-mer GT18, 22-mer GT22, and 22-mer GTAC22) or RNA (16S plus 32S E. coli rRNA or RNA of bacteriophage MS2). For RNA, a 10:1 (wt/wt) ratio of protein:RNA was prepared in a final volume of 100 μl; ratios of 100:1 and 60:1 (wt/wt) were used for oligonucleotides, and the final reaction volume was also 100 μl. The mixture was dialyzed against buffer containing 50 mM Tris-HCl, pH 8, 100 mM NaCl, 1 μM ZnCl2 for 2 h at room temperature. The dialysate was used for electron microscopy (EM) observation and for gradient ultracentrifugation. After ultracentrifugation on a linear sucrose gradient (10 to 60% [wt/wt], 50,000 rpm, 40 min, 4°C; Beckman TLS55 rotor), a total of 11 fractions of 200 μl each were collected. Any pelleted material was also resuspended in 200 μl of the aforementioned buffer and analyzed (fraction no. 12). Fractions were analyzed by SDS-PAGE, incorporated nucleic acids were quantified, and fractions containing particles or aggregates were used for transmission electron microscopy. For assembly of CANC or ΔProCANC in the absence of nucleic acids, the same dialysis conditions and analyses were employed.
Electron microscopy of negatively stained material.
Particles formed during assembly dialysis were negatively stained with 4% sodium silicotungstate (pH 7.2) on carbon-coated grids and studied by transmission electron microscopy using a JEOL JEM-1010 at magnifications ranging from ×10,000 to ×400,000 at 80 kV.
Protein and nucleic acid quantification.
Synthetic oligonucleotides labeled with fluorescein were used for in vitro assembly experiments. Fractions of the sucrose gradient (200 μl) used for analyzing assembly were placed in 96-well plates, and fluorescence was measured at 520 nm. The intensity of fluorescence was quantified using an AIDA LAS1000 system and corrected for sucrose presence.
A Ribogreen Quantitation kit (Molecular Probes) was used to quantify RNA according to the procedure recommended by the manufacturer. Sucrose gradient fractions were mixed with equal volumes of properly diluted Ribogreen reagent to a final volume of 200 μl in 96-well plates. The emission was measured at 520 nm.
The amounts of CANC proteins were assessed by the Bradford and Biuret method, and their distribution in sucrose gradient fractions was determined by SDS-PAGE using the LAS1000 (Fujifilm, Japan, and Raytest, Germany) analysis system and the AIDA computer program (Raytest, Germany).
RESULTS
Expression and purification of M-PMV proteins.
We have shown previously that, following expression in Escherichia coli, the CANC fusion protein of Mason-Pfizer monkey virus could assemble into sheets and that the mutant lacking the N-terminal proline, i.e., ΔProCANC, could assemble mostly into spherical structures (62, 69).
To study assembly in an in vitro system, which makes it possible to investigate the role of nucleic acids in this process, we have expressed both proteins in bacteria and optimized methods for their purification. Special attention was paid to the elimination of contaminating bacterial nucleic acids in the protein samples. In contrast to M-PMV Gag, which forms insoluble inclusion bodies (46), both CANC and ΔProCANC products were found in the supernatant fraction of the bacterial lysate. Affinity purification using the interaction of immobilized Zn2+ on the chelating Sepharose with two zinc fingers within NC yielded ∼90% pure protein that was essentially free of nucleic acids. Following further purification steps described in Materials and Methods, both proteins were purified to an estimated purity of >95% and concentrated to about 1 to 2 mg/ml (Fig. 1). The identity of purified proteins was confirmed by SDS-PAGE and by N-terminal sequencing, which also provided evidence that the N-terminal methionine was efficiently removed from both products (more than 95%). This was especially important for the CANC construct, since a residual initiating methionine would prevent the formation of the Pro1-Asp50 (or Pro1-Asp57) salt bridge, resulting in a phenotype similar to that of the ΔProCANC or Ala-ProCANC construct described previously (69). In spite of using protease inhibitors during the purification process, both proteins were occasionally partially cleaved by bacterial proteases. According to the sizes of these minor products and N-terminal protein sequencing, it was determined that multiple cleavage events occurred near the cleavage site between CA and NC proteins.
Nucleic acids significantly promote the assembly of M-PMV ΔProCANC particles but are dispensable as building components. (i) Assembly in the absence of nucleic acids.
In order to investigate the effect of various oligodeoxyribonucleotides and RNA, the ΔProCANC protein was purified as described in Materials and Methods. To ensure that the nucleic acids were efficiently eliminated from the protein, the purification step on phosphocellulose or use of high concentrations of NaCl or urea during purification were included. Two methods were used to quantify potential contamination with nucleic acids, a spectrophotometric method measuring the A260/A280 ratio and a fluorescence method using the Ribogreen reagent (Molecular Probes). The A260/A280 ratio of the purified ΔProCANC protein was below 0.69 for all preparations, which was comparable with preparations published for RSV Gag mutants used for similar purposes, where the spectrophotometrically determined value of A260/A280 corresponding to 0.7 was reported as negligible nucleic acid contamination (53, 81). By using the Ribogreen solution, we confirmed that the presence of nucleic acid was below 0.1 ng of nucleic acid per 1 μg of purified protein.
The purified nucleic acid-free ΔProCANC protein assembled particles in vitro without the addition of nucleic acids to the assembly mix, as documented both by the electrophoretic analysis of fractions from gradient ultracentrifugation (Fig. 2, fractions 8 to 12) and by electron microscopy of negatively stained material (Fig. 3A), respectively. However, the efficiency was quite low, and a majority of the particles pelleted to the bottom of the gradient in interconnected clusters (Fig. 3A). Most of the particles showed aberrant morphology, and fragments of spherical shells or partially closed shells prevailed. The ΔProCANC protein in the fractions corresponding to the peak of particles did not contain any nucleic acids detectable by the methods used (detection limits of 250 ng per ml for A260/A280 and 1 ng per ml for fluorescent determination of nucleic acids [data not shown]).
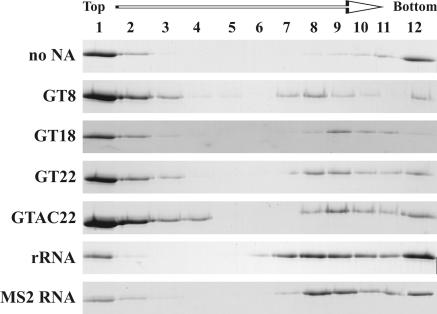
FIG. 2.
SDS-PAGE analysis of assembled M-PMV proteins in sucrose gradient fractions. ΔProCANC M-PMV protein (concentration of 0.6 mg/ml, i.e., 17 μM) was assembled without addition of nucleic acids or with oligodeoxyribonucleotides GT8, GT18, GT22, and GTAC22 at a ratio of 60:1 (wt/wt) or with RNA at a ratio of 10:1 (wt/wt). The reaction mixtures were centrifuged to equilibrium through a 10 to 60% (wt/wt) sucrose gradient, and fractions were analyzed by SDS-PAGE. Fraction 12 represents aggregated material that was pelleted at the bottom of the tube. The sucrose densities of the fractions were as follows: 1, 0.058 g/ml; 2, 0.069 g/ml; 3, 0.084 g/ml; 4, 1.10 g/ml; 5, 1.12 g/ml; 6, 1.15 g/ml; 7, 1.18 g/ml; 8, 1.21 g/ml; 9, 1.23 g/ml; 10, 1.26 g/ml; and 11, 1.28 g/ml. NA, nucleic acid.
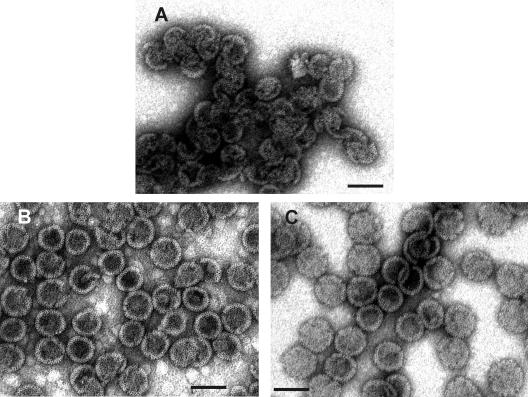
FIG. 3.
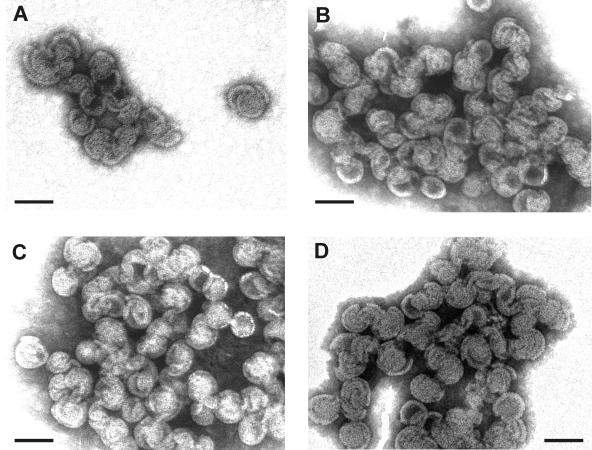
Transmission EM images of negatively stained M-PMV particles assembled from ΔProCANC protein. (A) Particles assembled from ΔProCANC protein without addition of nucleic acids. In this case, about 80% represented fragments not fully closed and interconnected particles. (B) Particles assembled from ΔProCANC protein and bacteriophage MS2 RNA at a ratio of 10:1 (wt/wt). (C) Particles assembled from ΔProCANC protein and 16S plus 32S rRNA of E. coli in a ratio of 10:1 (wt/wt). More than 90% of particles assembled in the presence of both MS2 RNA and rRNA were fully closed spheres. The average diameter of all assembled spheres was 75 nm ± 4 nm. Approximately 15%, 65%, and 75% of protein was efficiently assembled into particles in the absence of nucleic acid and in the presence of MS2 RNA and rRNA, respectively. The gross estimate (based on the number of molecules in the particle and the protein amount) is that 100% of assembled protein corresponds to 3 × 1011 particles. All assembly reactions were performed as mentioned in Materials and Methods. Bar, 100 nm.
(ii) Assembly of M-PMV ΔProCANC in the presence of nucleic acids.
To compare the effect of nucleic acids on the assembly of M-PMV oligodeoxyribonucleotides GT8-mer, GT18-mer, and GT22-mer, a self-complementary 22-mer, GTAC22, RNA of bacteriophage MS2 (3,569 nucleotides), and a mixture of 16S and 23S rRNA of E. coli (∼1,500 and 2,900 nucleotides, respectively) were included in the assembly mixtures. The oligonucleotides were selected based on previously published data that defined the requirements for in vitro assembly of the RSV-truncated Gag (ΔMBDΔPR) (53).
To investigate the homogeneity of the assembled particles, the material was centrifuged through a 10 to 60% sucrose gradient and the protein distribution in collected fractions was analyzed by SDS-PAGE. As expected, the assembled ΔProCANC nonenveloped particles were found in fractions corresponding to a density of approximately 1.2 to 1.3 g/ml (Fig. 2, fractions 7 to 11 from top of the gradient). In addition, all the samples contained a portion of free nonassembled protein that remained on top of the gradient. In contrast to assembly in the absence of nucleic acid, much less of the protein was found as aggregates or particles connected with aggregated material that pelleted at the bottom of the tube.
The stimulatory effect of RNA on the ΔProCANC assembly was significant both in the presence of bacteriophage MS2 RNA and the mixture of E. coli 16S rRNA and 23S rRNA, since under these assembly conditions a majority of the protein was found in fractions of a density corresponding to properly assembled particles (1.18 to 1.23 g/ml; Fig. 2 and 4A and B). Moreover, an electron microscopic analysis of these assembled structures revealed a majority of uniform spherical particles that appeared for the most part to have completed assembly (Fig. 3B and C).
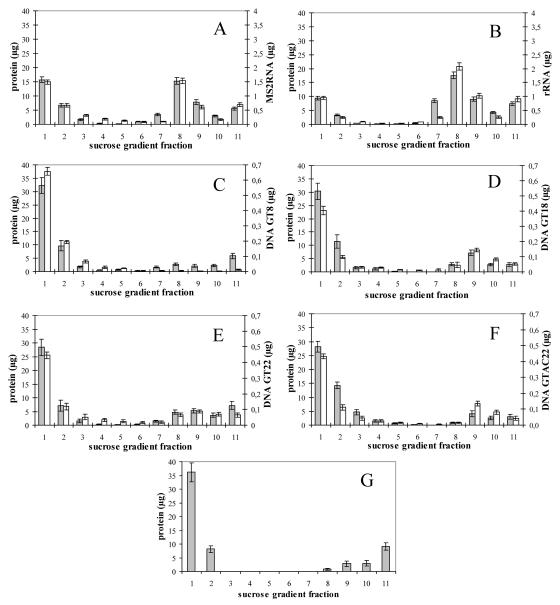
FIG. 4.
Protein and nucleic acid content of assembled M-PMV ΔProCANC in sucrose gradient fractions. Average values from five independent measurements are shown. Fraction 11 represents aggregated (pelleted) material. Error bars indicate standard deviations. ΔProCANC M-PMV protein (60 μg) was assembled in the presence of DNA oligonucleotides or RNA in the ratios 60:1 and 10:1 (wt/wt), respectively. The reaction mixtures were centrifuged to equilibrium in a 10 to 60% (wt/wt) sucrose gradient, and amounts of protein and DNA were quantified (see Materials and Methods). The sucrose densities of the fractions were as follows: 1, 0.058 g/ml; 2, 0.069 g/ml; 3, 0.084 g/ml; 4, 1.10 g/ml; 5, 1.12 g/ml; 6, 1.15 g/ml; 7, 1.18 g/ml; 8, 1.21 g/ml; 9, 1.23 g/ml; 10, 1.26 g/ml; and 11, 1.28 g/ml. Shown are (A) RNA of bacteriophage MS2; (B) 16S plus 32S rRNA of E. coli; (C) GT8; (D) GT18; (E) GT22; (F) GTAC22; and (G) ΔProCANC M-PMV protein assembled in the absence of nucleic acids. Grey bars, protein; white bars, DNA oligonucleotides or RNA.
In order to quantify the amount of incorporated nucleic acids into the assembled particles, the ratio of protein to nucleic acid was determined for all fractions collected from the sucrose gradient (Fig. 4). The amounts of incorporated deoxyribonucleic acids were assessed by fluorescence for fluorescein-labeled oligonucleotides and by Ribogreen for RNA (Table 1). Quantification analyses were performed in five independent experiments for each type of nucleic acid.
TABLE 1.
Quantification of oligonucleotides and RNA incorporated into M-PMV particles assembled in vitroa
| NA | Molar ratio of ΔProCANC/NA | ntd/particle (kb) | ntd/protein |
|---|---|---|---|
| GT22 | 13 | 2.5 | 1.7 |
| GT18 | 11 | 2.5 | 1.7 |
| GT8 | 48 | 0.2 | 0.2 |
| GTAC22 | 6 | 5.3 | 3.5 |
| rRNA | 180 | 18.3 | 12.2 |
| MS2 RNA | 343 | 15.8 | 10.5 |
NA, nucleic acid; ntd/particle and ntd/protein, approximate number of nucleotides per particle or protein molecule, respectively.
The amount of nucleic acids in one particle was determined based on our calculations (see Discussion) that an immature particle consists on average of ∼1,500 molecules of Gag (ΔProCANC in our case). Based on this, the ΔProCANC particles formed in the presence of MS2 RNA and rRNAs incorporated approximately 16 and 18 kb of RNA, respectively (Table 1). This corresponds well with the size of diploid genomic M-PMV RNA, which is approximately 17 kb. The uniformity of total length of incorporated RNA suggests a spatial control of incorporated genetic material by the virus-like particle.
To compare the effect of short fragments of DNA on the assembly of M-PMV ΔProCANC oligodeoxyribonucleotides, GT8, GT18, GT22, and GTAC22 were mixed in 1:100 or 1:60 ratios with M-PMV ΔProCANC, and the mixtures were dialyzed against the assembly buffer. Although the longer oligonucleotides clearly stimulated particle assembly (Fig. 2 and 4C to F), the particles formed in the presence of any oligonucleotides used in this study were often irregular (Fig. 5). Most of them were not fully closed, and large aggregates of particle fragments were frequently found. We also observed a shift in density of the protein within the sucrose gradient fractions toward the lower densities in the case of GT8 oligonucleotide compared to that of the other oligonucleotides used (Fig. 4). This is in agreement with higher proportions of fragments and incompletely closed particles that were observed by electron microscopy compared to the particles assembled in the presence of GT18 and GT22 (Fig. 5). The amounts of incorporated DNA, calculated as the length in kilobases per particle, ranged from 2.5 kb to 0.2 kb for GT22 and GT8, respectively (Table 1), under conditions where excess DNA remained unincorporated on top of the gradients. Approximately 5.3 kb of the GTAC22 oligonucleotide, which is expected to be double stranded under the conditions of the experiment, was incorporated in one particle. The fact that this number is double that of the single-stranded GT22 (∼2.5 kb) suggests that the DNA length defines the efficiency of incorporation regardless of its single- or double-stranded form.
FIG. 5.
Transmission EM images of negatively stained M-PMV particles assembled from ΔProCANC protein in the presence of GT8, GT18, GT22, and GTAC22 oligonucleotides in the ratio 60:1 (wt/wt). Particles assembled from ΔProCANC protein and GT8 (A), GT18 (B), GT22 (C), and GTAC22 (D). All assembly reactions were performed as described in Materials and Methods. Most of the particles were not fully closed, sometimes having more layers. The proportion of regularly shaped particles sequentially increased in the order of GT8, GTAC22, GT18, GT22 (A, D, B, and C, respectively). The average diameter of assembled particles (in all panels) was 75 nm ± 5 nm. Approximately 20%, 30%, 35%, and 20% of protein was assembled into particles in the presence of GT8, GT18, GT22, and GTAC22 oligonucleotides, respectively. All assembly reactions were performed as mentioned in Materials and Methods. Bar, 100 nm.
In order to investigate whether the low efficiency of DNA incorporation into the particles is caused by the limitation of nucleic acid availability, the ΔProCANC protein was titrated with GT22 oligonucleotides. The protein was mixed with oligonucleotides in ratios of 200:1, 60:1, and 10:1 (wt/wt) and dialyzed in the assembly buffer for 2 to 3 h. The resulting material was analyzed as in the previous experiments by SDS-PAGE and electron microscopy. It was found that varying amounts of GT22 affected the yield of particles; however, it did not affect the ratio of proteins to oligonucleotides in assembled particles (data not shown).
(iii) Assembly of M-PMV CANC in vitro.
In contrast to ΔProCANC, the CANC protein formed no detectable structures in vitro under these conditions. It should be noted here that three independent purification methods were used (including two avoiding denaturation; see Materials and Methods), and circular dichroism spectroscopy proved the significant proportion of α-helical motifs comparable to M-PMV ΔProCANC protein. The effect of pH (5 to 10), ionic strength (0 to 2 M NaCl), Zn2+ concentration (1 μM to 1 mM), protein concentration (0.1 to 10 mg/ml), and assembly temperature (4 to 42°C) was tested in order to optimize assembly conditions, but no structures were observed for any combination of these conditions tested. Based on our previous data showing that the same construct forms sheets in the cytoplasm of E. coli (69), we investigated whether some cellular factor could promote the in vitro assembly of M-PMV CANC. For this purpose we included E. coli cell extract in the CANC assembly mixture. Even under these conditions, we observed only protein aggregates without any evidence of organized structures (data not shown).
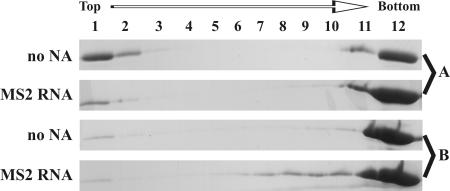
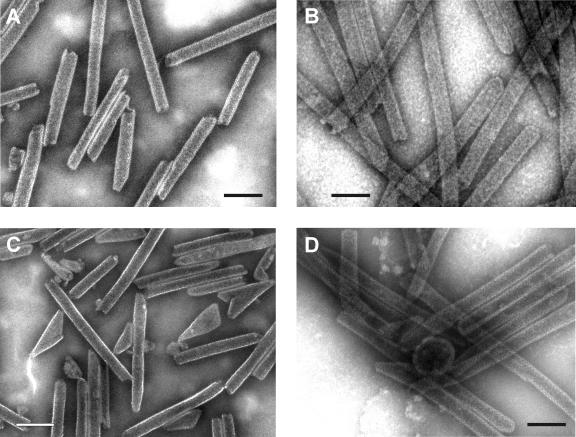
Since the results obtained with M-PMV CANC constructs vary from those described for a similar construct from RSV or HIV-1, we have used HIV-1 CANC and ΔProCANC as control proteins in order to determine if the assembly conditions used in our study would allow assembly of other retroviral Gag molecules. Both sedimentation analysis and electron microscopy were used to assess the efficiency of assembly. Figure 6 indicates that for both HIV-1 CANC and ΔProCANC, a majority of the protein that had assembled into tubular particles in the presence or in the absence of RNA sedimented at the bottom of the gradient. It is interesting that the HIV-1 ΔProCANC protein assembles into tubular particles, since it lacks the ability to form the N-terminal proline-aspartate salt bridge that has been reported to be the driving force for tube formation (76). Thus, additional interactions appear to be involved in this process, as is the case for RSV, where CANC proteins with short N-terminal extensions still assembled tubes and a stretch of 25 amino acids was required for efficient assembly of spheres (41). Short tubular particles (of a length comparable to sphere diameter) banded together with the spherical particles (Fig. 6, MS2 RNA, fractions 7 to 10). Under these conditions, the HIV-1 CANC formed predominantly (approximately 99%) tubes and only occasional core-like structures in the presence and in the absence of RNA. The diameter of the tubes was greater in the presence of RNA than in the tubes assembled from pure protein itself (Fig. 7A and B). Similarly, HIV-1 ΔProCANC in the absence of nucleic acid formed tubes (50 to 80%) and conical structures resembling cores. The presence of RNA during the assembly of HIV-1 ΔProCANC resulted in the rare formation of spherical particles (approximately 1%), with an average diameter of 80 to 85 nm, in addition to tubular structures and cores (10 to 40% of the total; Fig. 7C and D). The diameters of HIV-1 ΔProCANC tubular structures were also greater in the presence of RNA than in the absence of RNA. In the absence of nucleic acid, the diameter of CANC HIV-1 tubes was 35 nm with a standard deviation of ±4 nm, and that for ΔProCANC HIV-1 was 34 nm ± 3 nm. In the presence of RNA, the diameter of CANC HIV-1 tubes was 46 nm ± 5 nm, which is in good correlation with the data obtained for CANC HIV-1 tubular structures previously presented by several authors reporting 44 to 55 nm (8, 10, 36). ΔProCANC HIV-1 formed tubes of a diameter 44 nm ± 4 nm. These measurements were performed more than 100 times for each sample. All HIV-1 tubes discussed in this study were of variable lengths, ranging from 80 nm to 1,500 nm, which is in agreement with the published data (10).
FIG. 6.
SDS-PAGE analysis of assembled HIV-1 proteins in sucrose gradient fractions. ΔProCANC and CANC HIV proteins (concentration of 0.6 mg/ml, i.e., 19 μM) were assembled without addition of nucleic acids or with MS2 phage RNA at a ratio of 10:1 (wt/wt). The reaction mixtures were centrifuged to equilibrium through a 10 to 60% (wt/wt) sucrose gradient, and the fractions were analyzed by SDS-PAGE. Fraction 12 represents aggregated material that was pelleted at the bottom of the tube. The sucrose densities of the fractions were as follows: 1, 0.058 g/ml; 2, 0.069 g/ml; 3, 0.084 g/ml; 4, 1.10 g/ml; 5, 1.12 g/ml; 6, 1.15 g/ml; 7, 1.18 g/ml; 8, 1.21 g/ml; 9, 1.23 g/ml; 10, 1.26 g/ml; and 11, 1.28 g/ml. NA, nucleic acid. (A) CANC HIV-1 protein; (B) ΔProCANC HIV-1 protein.
FIG. 7.
Transmission EM images of negatively stained tubes assembled from HIV-1 CANC and ΔProCANC proteins. (A) Tubes formed from nucleic acid-free CANC. (B) Tubes assembled from CANC protein and bacteriophage MS2 RNA at a ratio of 10:1 (wt/wt). (C) Nucleic acid-free ΔProCANC particles. (D) Particles assembled from ΔProCANC and bacteriophage MS2 RNA at a ratio of 10:1 (wt/wt). All assembly reactions were performed as mentioned in Materials and Methods. Predominantly tubes were observed for CANC both in the presence and the absence of nucleic acids (no spheres were observed, and occasional cores did not exceed 1%). Nucleic acid-free ΔProCANC particles assembled variable numbers of core-like structures (∼20 to 50%) versus tubes. Particles assembled from ΔProCANC and bacteriophage MS2 RNA assembled a majority of tubes as well as about 1% spheres and ∼10 to 40% core-like structures. Approximately 70% and 80% of CANC protein was efficiently assembled into particles without or in the presence of MS2 RNA, respectively. For ΔProCANC, approximately 80% and 90% of protein was efficiently assembled into particles without or with addition of MS2 RNA, respectively. Bar, 100 nm.
DISCUSSION
We have reported previously that a large portion of the M-PMV Gag polyprotein is dispensable for the bacterial assembly of immature capsid-like structures (69). As for other retroviruses, such as HIV-1 and RSV, the capsid (CA) and nucleocapsid (NC) domains were critical for particle formation in vitro. The presence of the N-terminal proline in the CA domain was shown to play a critically important role for M-PMV CANC assembly in E. coli, where CANC assembled in sheets and ΔProCANC mostly assembled into spheres (69). This observation was consistent with our prediction that CANC with a matured N terminus might represent the assembly-competent material for mature core formation. On the other hand, because of its inability to form the Pro1-Asp50 (or Asp57) salt bridge, the M-PMV ΔProCANC N terminus should resemble that of CA in the Gag precursor, which would then direct assembly into immature spherical particles.
In the present study, we have used an in vitro system to investigate the requirements for efficient assembly of particles from purified CANC and ΔProCANC proteins and to compare the differences in assembly of M-PMV and HIV-1. We have demonstrated that the absence of N-terminal proline was essential for M-PMV CANC to assemble in vitro, in contrast to the bacterial expression/assembly system, where this protein assembled into planar structures. Purified CANC protein did not assemble in vitro even under the most favorable conditions for ΔProCANC assembly (100 mM NaCl, 50 mM Tris-HCl, 1 μM ZnCl2, pH 8.0, in the presence of RNA). This suggests that the intracellular environment of E. coli facilitates the assembly of CANC structures, perhaps by providing a cellular factor(s) necessary for correct protein folding and CANC-CANC interactions. In contrast, several research groups have shown that HIV-1 CANC assembles in vitro into tubular structures (10, 22, 36, 37). Similarly, we were able to show that the HIV-1 CANC protein readily assembled into tubes under the same conditions where M-PMV CANC did not (Fig. 7). HIV-1 CANC assembled in the absence of RNA; however, RNA did increase the efficiency of its assembly, and an increase in tube diameter was also observed in the presence of nucleic acid. The inability of M-PMV CANC to form tubes may reflect some basic structural difference in the CA proteins of HIV-1 and M-PMV, since purified M-PMV CA aggregated without any defined structures in the presence of high salt (data not shown). This contrasts with the results with purified HIV-1 CA, which has previously been shown to form tubes under high protein and salt concentrations (22, 28, 29, 36, 50).
Unlike in RSV and HIV-1, the assembly of M-PMV ΔProCANC was independent of salt concentrations between 50 mM and 200 mM, temperatures between 5°C and 37°C, and pH within the range of 5.5 to 9 (data not shown). Purified M-PMV ΔProCANC formed spheres in a manner similar to that of CANC from other retroviruses, where short extensions as an N terminus of MA were necessary for the assembly of spherical particles (44, 76).
The exact role of the nucleic acid interactions with Gag proteins during the assembly process is still not completely understood. This study shows that assembly of ΔProCANC occurs even in the absence of nucleic acids, which is in agreement with previously published data for HIV-1 Gag (78, 79). Calculations based on experimental data show that the RSV NC binding site for nucleic acids spans eight nucleotides and therefore a 16-mer should be sufficient for bridging together two NC molecules (53, 54). HIV-1 and simian immunodeficiency virus NC were shown to have binding site sizes similar to those of RSV NC (73, 74, 80). For HIV-1 it has been proposed that binding of nucleic acid to NC concentrates the Gag polyproteins and induces CA-CA interactions that lead subsequently to the assembly of immature capsids. In this model, nucleic acids initially promote the interaction between carboxy-terminal domains of the CA protein and a spacer peptide between CA and NC (38, 68). In the case of M-PMV, an analogous spacer separating CA and NC has not been identified, and data from the bacterial expression system show that the minimal requirement for assembly of a C-terminally truncated ΔProCANC is the N-terminal 37 amino acids of NC (70).
The presence of residual nucleic acids in purified protein samples remains an issue in the experiments evaluating the role for nucleic acids in Gag assembly. In order to address this question, we have used a Ribogreen nucleic acid quantitation kit in addition to the standard spectrophotometric measurement of the A260/A280 ratio. Both methods showed that contamination with nucleic acids of bacterial origin was less than 10 ng of nucleic acid per assembly reaction (60 μg of protein in a total volume of 100 μl), which corresponds to the A260/A280 ratio of <0.7 that is considered negligible (53, 81). ΔProCANC deprived of nucleic acids assembled structures comparable to those formed from the same protein in the presence of oligonucleotides (Fig. 3 and 5). However, the yield of particles increased with the length of oligonucleotides used (Fig. 4). Our data presented here suggest that oligonucleotides promote M-PMV ΔProCANC assembly but are not incorporated in the stoichiometric amounts that would be necessary to serve as scaffolding units as proposed by Ma and Vogt (54). The lack of a requirement for nucleic acid as the structural component of the capsid and the enhancement of assembly efficiency by oligonucleotides as short as GT8 could be described by a model that is consistent with two previously published observations. First, the mechanism proposed by Ma and Vogt (54) originates from the observation that GT16 promotes assembly of CANC dimers; however, the oligonucleotide per se is dispensable for the dimer stability after its formation (54). Second, the oligonucleotides bound to the assembled particles are in dynamic equilibrium, as suggested by the exchange of bound nucleic acids for unbound ones (24). It therefore seems feasible that nucleic acid binding promotes a cooperative structural change recruiting an additional interaction domain in M-PMV ΔProCANC. Such a structural change could then induce the release of nucleic acid, which would become available for another CANC interaction. This would explain how a low nucleic acid/protein molar ratio promotes assembly. In contrast to the model proposed by Ma and Vogt (54), the role of oligonucleotides would not be in linking two molecules of ΔProCANC together but rather in promoting the conformational change of ΔProCANC monomers. The fact that the M-PMV NC structured core domain contains two independently folded, rotationally uncorrelated globular domains connected by a seven-residue flexible linker (30) makes it likely that significant changes in their orientation will be induced by nucleic acid binding. The loose binding of nucleic acids after such change would also allow its release from the core for reverse transcription.
The sufficient length of nucleic acid seems to play an important role in the assembly, as M13 phage DNA (our unpublished data) and the longer RNA of MS2 phage were better catalysts for particle assembly than DNA oligonucleotides, and its presence promoted formation of fully closed spherical particles of uniform size (Fig. 3). This is in agreement with the findings of Morikawa et al., who showed that RNA could support the assembly of virus-like particles from isolated HIV-1 Gag (58, 59). The M-PMV ΔProCANC particles contained significantly larger amounts of RNA compared to any of the oligonucleotides in this study (Table 1). The calculations were done for particles containing ∼1,500 ΔProCANC molecules. Such an estimate, which is lower than recently published data for HIV-1 (9), was based on our calculation of protein subunits in EM pictures (approximately 500 particles were evaluated) and previously published data (25, 63, 75).
In summary, we have shown here that in vitro assembly of M-PMV ΔProCANC can be stimulated by both DNA oligonucleotides and nonviral RNA. These studies point to a model in which nucleic acid, through a transient interaction with the NC domain, induces a conformational change that promotes assembly of Gag. Such a model may be broadly applicable to retroviruses that employ different morphogenetic pathways.
Acknowledgments
This work was supported by Grant Agency of the Czech Republic 203/03/P094, Grant Agency of the Academy of Sciences Grant A4055304, and research projects supported by the Czech Ministry of Education: 1M6837805002, Z 40550506, and MSM 6046137305.
REFERENCES
- 1.Accola, M. A., B. Strack, and G. Gottlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barklis, E., J. McDermott, S. Wilkens, S. Fuller, and D. Thompson. 1998. Organization of HIV-1 capsid proteins on a lipid monolayer. J. Biol. Chem. 273:7177-7180. [DOI] [PubMed] [Google Scholar]
- 3.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177-218. [DOI] [PubMed] [Google Scholar]
- 4.Berthtet-Colominas, C., S. Monaco, A. Novelli, G. Sibai, F. Mallet, and S. Cusack. 1999. Head-to-tail dimers and interdomain flexibility revealed by the crystal structure of HIV-1 capsid protein (p24) complexed with monoclonal antibody Fab. EMBO J. 18:1124-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birkett, A. J., B. Yelamos, I. Rodriguez-Crespo, F. Gavilanes, and D. L. Peterson. 1997. Cloning, expression, purification and characterization of the major core protein (p26) from equine infectious anemia virus. Biochim. Biophys. Acta 1339:62-72. [DOI] [PubMed] [Google Scholar]
- 6.Borsetti, A., A. Ohagen, and H. G. Gottlinger. 1998. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J. Virol. 72:9313-9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowles, N. E., P. Damay, and P.-F. Spahr. 1993. Effect of rearrangements and duplications of the Cys-His motifs of Rous sarcoma virus nucleocapsid protein. J. Virol. 67:623-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briggs, J. A. G., T. Wilk, R. Welker, H. G. Kräusslich, and S. D. Fuller. 2003. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 22:1707-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Briggs, J. A. G., M. N. Simon, I. Gross, H. G. Kräusslich, S. D. Fuller, V. M. Vogt, and M. C. Johnson. 2004. The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 11:672-675. [DOI] [PubMed] [Google Scholar]
- 10.Campbell, S., and V. M. Vogt. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69:6487-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell, S., and V. M. Vogt. 1997. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J. Virol. 71:4425-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell, S., and A. Rein. 1999. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 73:2270-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campos-Olivas, R., J. L. Newman, and M. F. Summers. 2000. Solution structure and dynamics of the Rous sarcoma virus capsid protein and comparison with capsid proteins of other retroviruses. J. Mol. Biol. 296:633-649. [DOI] [PubMed] [Google Scholar]
- 14.Cimarelli, A., and J. L. Darlix. 2002. Assembling the human immunodeficiency virus type 1. Cell. Mol. Life Sci. 59:1166-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornilescu, C. C., F. Bouamr, X. Yao, C. Carter, and N. Tjandra. 2001. Structural analysis of the N-terminal domain of the human T-cell leukemia virus capsid protein. J. Mol. Biol. 306:783-797. [DOI] [PubMed] [Google Scholar]
- 16.Dannull, J., A. Surovoy, G. Jung, and K. Moelling. 1994. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 13:1525-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darlix, J. L., M. Lapadat-Tapolsky, H. de Rocquigny, and B. P. Roques. 1995. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J. Mol. Biol. 254:523-537. [DOI] [PubMed] [Google Scholar]
- 18.De Rocquigny, H., C. Gabus, A. Vincent, M. C. Fournie-Zaluski, B. Roques, and J. L. Darlix. 1992. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc. Natl. Acad. Sci. USA 89:6472-6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dorfman, T., J. Luban, S. P. Goff, W. A. Haseltine, and H. G. Gottlinger. 1993. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 67:6159-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dorfman, T., A. Bukovsky, A. Ohagen, S. Hoglund, and H. G. Gottlinger. 1994. Functional domains of the capsid protein of human immunodeficiency virus type 1. J. Virol. 68:8180-8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dupraz, P., S. Oertle, C. Méric, P. Damay, and P.-F. Spahr. 1990. Point mutations in the proximal Cys-His box of Rous sarcoma virus nucleocapsid protein. J. Virol. 64:4978-4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ehrlich, L. S., B. E. Agresta, and C. A. Carter. 1992. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J. Virol. 66:4874-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ehrlich, L. S., T. Liu, S. Scarlata, B. Chu, and C. A. Carter. 2001. HIV-1 capsid protein forms spherical (immature-like) and tubular (mature-like) particles in vitro: structure switching by pH-induced conformational changes. Biophys. J. 81:586-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng, Y. X., T. Li, S. Campbell, and A. Rein. 2002. Reversible binding of recombinant human immunodeficiency virus type 1 Gag protein to nucleic acids in virus-like particle assembly in vitro. J. Virol. 76:11757-11762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuller, S. D., T. Wilk, B. D. Gowen, H. G. Kräusslich, and V. M. Vogt. 1997. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr. Biol. 7:729-738. [DOI] [PubMed] [Google Scholar]
- 26.Gamble, T. L., F. F. Vajdos, S. Yoo, D. K. Worthylake, M. Houseweart, W. I. Sundquist, and C. P. Hill. 1996. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87:1285-1294. [DOI] [PubMed] [Google Scholar]
- 27.Gamble, T. L., S. Yoo, F. F. Vajdos, U. K. von Schwedler, D. K. Worthylake, H. Wang, J. P. McCutcheon, W. I. Sundquist, and C. P. Hill. 1997. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849-853. [DOI] [PubMed] [Google Scholar]
- 28.Ganser, B. K., S. Li, V. Y. Klishko, J. T. Finch, and W. I. Sundquist. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80-83. [DOI] [PubMed] [Google Scholar]
- 29.Ganser-Pornillos, B. K., U. K. von Schwedler, K. M. Stray, C. Aiken, and W. I. Sundquist. 2004. Assembly properties of the human immunodeficiency virus type 1 CA protein. J. Virol. 78:2545-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao, Y., K. Kaluarachchi, and D. P. Giedroc. 1998. Solution structure and backbone dynamics of Mason-Pfizer monkey virus, MPMV, nucleocapsid protein. Prot. Sci. 7:2265-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gitti, R. K., B. M. Lee, J. Walker, M. F. Summers, S. Yoo, and W. I. Sundquist. 1996. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science 273:231-235. [DOI] [PubMed] [Google Scholar]
- 32.Gorelick, R. J., S. M. Nigida, L. O. Arthur, L. E. Henderson, and A. Rein. 1988. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc. Natl. Acad. Sci. USA 85:8420-8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorelick, R. J., S. M. J. Nigida, J. W. J. Bess, L. O. Arthur, L. E. Henderson, and A. Rein. 1990. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J. Virol. 64:3207-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorelick, R. J., S. M. Nigida, L. O. Arthur, L. E. Henderson, and A. Rein. 1991. Role of nucleocapsid cysteine arrays in retroviral assembly and replication: possible mechanisms in RNA encapsidation. In A. Kumar (ed.), Advances in molecular biology and targeted treatment for AIDS. Plenum Press, New York, N.Y.
- 35.Gorelick, R. J., D. J. Chabot, A. Rein, L. E. Henderson, and L. O. Arthur. 1993. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J. Virol. 67:4027-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gross, I., H. Hohenberg, and H. G. Kräusslich. 1997. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur. J. Biochem. 249:592-600. [DOI] [PubMed] [Google Scholar]
- 37.Gross, I., H. Hohenberg, C. Huckhagel, and H. G. Kräusslich. 1998. N-terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J. Virol. 72:4798-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gross, I., H. Hohenberg, T. Wilk, K. Wiegers, M. Grättinger, B. Mūller, S. Fuller, and H. G. Kräusslich. 2000. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 19:103-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ivanov, D., J. R. Stone, J. L. Maki, T. Collins, and G. Wagner. 2005. Mammalian SCAN domain dimer is a domain-swapped homolog of the HIV capsid C-terminal domain. Mol. Cell 17:137-143. [DOI] [PubMed] [Google Scholar]
- 40.Johnson, M. C., H. M. Scobie, Y. M. Ma, and V. M. Vogt. 2002. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J. Virol. 76:11177-11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joshi, S. M., and V. M. Vogt. 2000. Role of the Rous sarcoma virus p10 domain in shape determination of Gag virus-like particles assembled in vitro and within Escherichia coli. J. Virol. 74:10260-10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karpel, R. L., L. E. Henderson, and S. Oroszlan. 1987. Interactions of retroviral structural proteins with single-stranded nucleic acids. J. Biol. Chem. 262:4961-4967. [PubMed] [Google Scholar]
- 43.Khorasanizadeh, S., R. Campos-Olivas, and M. F. Summers. 1999. Solution structure of the capsid protein from the human T-cell leukemia virus type-1. J. Mol. Biol. 291:491-505. [DOI] [PubMed] [Google Scholar]
- 44.Kingston, R., E. Z. Eisenmesser, T. Fitzon-Ostendorp, G. W. Schatz, V. M. Vogt, C. B. Post, and M. G. Rossmann. 2000. Structure and self-association of the Rous sarcoma virus capsid protein. Structure 8:617-628. [DOI] [PubMed] [Google Scholar]
- 45.Kingston, R. L., and V. M. Vogt. 2005. Domain swapping and retroviral assembly. Mol. Cell 17:166-167. [DOI] [PubMed] [Google Scholar]
- 46.Klikova, M., S. S. Rhee, E. Hunter, and T. Ruml. 1995. Efficient in vivo and in vitro assembly of retroviral capsids from Gag precursor proteins expressed in bacteria. J. Virol. 69:1093-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lanman, J., J. Sexton, M. Sakalian, and P. E. Prevelige, Jr. 2002. Kinetic analysis of the role of intersubunit interactions in human immunodeficiency virus type 1 capsid protein assembly in vitro. J. Virol. 76:6900-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lanman, J., T. T. Lam, M. R. Emmett, A. G. Marshall, M. Sakalian, and P. E. Prevelige, Jr. 2004. Key interactions in HIV-1 maturation identified by hydrogen-deuterium exchange. Nat. Struct. Mol. Biol. 11:676-677. [DOI] [PubMed] [Google Scholar]
- 49.Levin, J. G., P. M. Grimley, J. M. Ramseur, and I. K. Berezesky. 1974. Deficiency of 60 to 70S RNA in murine leukemia virus particles assembled in cells treated with actinomycin D. J. Virol. 14:152-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li, S., C. P. Hill, W. I. Sundquist, and J. T. Finch. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407:409-413. [DOI] [PubMed] [Google Scholar]
- 51.Linial, M., E. Medeiros, and W. S. Hayward. 1978. An avian oncovirus mutant (SE21Q1b) deficient in genomic RNA: biological and biochemical characterization. Cell 15:1371-1381. [DOI] [PubMed] [Google Scholar]
- 52.Linial, M. L., and A. D. Miller. 1990. Retroviral RNA packaging: sequence requirements and implications. Curr. Top. Microbiol. 157:125-152. [DOI] [PubMed] [Google Scholar]
- 53.Ma, Y. M., and V. M. Vogt. 2002. Rous sarcoma virus Gag protein-oligonucleotide interaction suggests a critical role for protein dimer formation in assembly. J. Virol. 76:5452-5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma, Y. M., and V. M. Vogt. 2004. Nucleic acid binding-induced Gag dimerization in the assembly of Rous sarcoma virus particles in vitro. J. Virol. 78:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Méric, C., and P.-F. Spahr. 1986. Rous sarcoma virus nucleic acid-binding protein p12 is necessary for viral 70S RNA dimer formation and packaging. J. Virol. 60:450-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Méric, C., and S. P. Goff. 1989. Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitutions in the Cys-His box of the nucleocapsid protein. J. Virol. 63:1558-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Momany, C., L. C. Kovari, A. J. Prongay, W. Keller, R. K. Gitti, B. M. Lee, A. E. Gorbalenya, L. Tong, J. McClure, L. S. Ehrlich, M. F. Summers, C. Carter, and M. G. Rossmann. 1996. Crystal structure of dimeric HIV-1 capsid protein. Nat. Struct. Biol. 3:763-770. [DOI] [PubMed] [Google Scholar]
- 58.Morikawa, Y., T. Goto, and K. Sano. 1999. In vitro assembly of human immunodeficiency virus type 1 Gag protein. J. Biol. Chem. 274:27997-28002. [DOI] [PubMed] [Google Scholar]
- 59.Morikawa, Y., T. Goto, and F. Momose. 2004. Human immunodeficiency virus type 1 gag assembly through assembly intermediates. J. Biol. Chem. 279:31964-31972. [DOI] [PubMed] [Google Scholar]
- 60.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 98:5246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nandhagopal, N., A. A. Simpson, M. C. Johnson, A. B. Francisco, G. W. Schatz, M. G. Rossman, and V. M. Vogt. 2004. Dimeric Rous sarcoma virus capsid protein structure relevant to immature Gag assembly. J. Mol. Biol. 335:275-282. [DOI] [PubMed] [Google Scholar]
- 62.Nermut, V. N., P. Bron, D. Thomas, M. Rumlova, T. Ruml, and E. Hunter. 2002. Molecular organization of Mason-Pfizer monkey virus capsids assembled from Gag polyprotein in Escherichia coli. J. Virol. 76:4321-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parker, S. D., J. S. Wall, and E. Hunter. 2001. Analysis of Mason-Pfizer monkey virus Gag particles by scanning transmission electron microscopy. J. Virol. 75:9543-9548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Poon, D. T., J. Wu, and A. Aldovini. 1996. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J. Virol. 70:6607-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reicin, A. S., S. Paik, R. D. Berkowitz, J. Luban, I. Lowy, and S. P. Goff. 1995. Linker insertion mutations in the human immunodeficiency virus type 1 gag gene: effects on virion particle assembly, release, and infectivity. J. Virol. 69:642-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rein, A. 1994. Retroviral RNA packaging: a review. Arch. Virol. Suppl. 9:513-522. [DOI] [PubMed] [Google Scholar]
- 67.Rein, A., D. P. Harvin, J. Mirro, S. M. Ernst, and R. J. Gorelick. 1994. Evidence that a central domain of nucleocapsid protein is required for RNA packaging in murine leukemia virus. J. Virol. 68:6124-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roldan, A., R. S. Russell, B. Marchand, M. Götte, C. Liang, and M. A. Wainberg. 2004. In vitro identification and characterization of an early complex linking HIV-1 genomic RNA recognition and Pr55Gag multimerization. J. Biol. Chem. 279:39886-39894. [DOI] [PubMed] [Google Scholar]
- 69.Rumlova-Klikova, M., E. Hunter, M. V. Nermut, I. Pichova, and T. Ruml. 2000. Analysis of Mason-Pfizer monkey virus Gag domains required for capsid assembly in bacteria: role of the N-terminal proline residue of CA in directing particle shape. J. Virol. 74:8452-8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rumlova, M., T. Ruml, J. Pohl, and I. Pichova. 2003. Specific in vitro cleavage of Mason-Pfizer monkey virus capsid protein: evidence for a potential role of retroviral protease in early stages of infection. Virology 310:310-318. [DOI] [PubMed] [Google Scholar]
- 71.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 72.Schmalzbauer, E., B. Strack, J. Dannull, S. Guehmann, and K. Moelling. 1996. Mutations of basic amino acids of NCp7 of human immunodeficiency virus type 1 affect RNA binding in vitro. J. Virol. 70:771-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Urbaneja, M. A., B. P. Kane, D. G. Johnson, R. J. Gorelick, L. E. Henderson, and J. Casas-Finet. 1999. Binding properties of the human immunodeficiency virus type 1 nucleocapsid protein p7 to a model RNA: elucidation of the structural determinants for function. J. Mol. Biol. 287:59-75. [DOI] [PubMed] [Google Scholar]
- 74.Urbaneja, M. A., C. F. McGrath, B. P. Kane, L. E. Henderson, and J. Casas-Finet. 2000. Nucleic acid binding properties of the simian immunodeficiency virus nucleocapsid protein NCp8. J. Biol. Chem. 275:10394-10404. [DOI] [PubMed] [Google Scholar]
- 75.Vogt, M. V., and M. Simon. 1999. Mass determination of Rous sarcoma virus virions by scanning transmission electron microscopy. J. Virol. 73:7050-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.von Schwedler, U. K., T. L. Stemmler, V. Y. Klishko, S. Li, K. H. Albertine, D. R. Davis, and W. I. Sundquist. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 17:1555-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.von Schwedler, U. K., K. M. Stray, J. E. Garrus, and W. I. Sundquist. 2003. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J. Virol. 77:5439-5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang, S. W., and A. Aldovini. 2002. RNA incorporation is critical for retroviral particle integrity after cell membrane assembly of Gag complexes. J. Virol. 76:11853-11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang, S. W., K. Noonan, and A. Aldovini. 2004. Nucleocapsid-RNA interactions are essential to structural stability but not to assembly of retroviruses. J. Virol. 78:716-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.You, J. C., and C. S. McHenry. 1993. HIV nucleocapsid protein: expression in Escherichia coli, purification, and characterization. J. Biol. Chem. 268:16519-16527. [PubMed] [Google Scholar]
- 81.Yu, F., S. M. Joshi, Y. M. Ma, R. L. Kingston, M. N. Simon, and V. M. Vogt. 2001. Characterization of Rous sarcoma virus Gag particles assembled in vitro. J. Virol. 75:2753-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang, Y., H. Qian, Z. Love, and E. Barklis. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J. Virol. 72:1782-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zlotnick, A. 2003. Are weak protein-protein interactions the general rule in capsid assembly? Virology 315:269-274. [DOI] [PubMed] [Google Scholar]