Abstract
The genetic diversity among globally circulating human immunodeficiency virus type 1 (HIV-1) strains is a serious challenge for HIV-1 vaccine design. We have generated a synthetic group M consensus env gene (CON6) for induction of cross-subtype immune responses and report here a comparative study of T-cell responses to this and natural strain env immunogens in a murine model. Three different strains of mice were immunized with CON6 as well as subtype A, B, or C env immunogens, using a DNA prime-recombinant vaccinia virus boost strategy. T-cell epitopes were mapped by gamma interferon enzyme-linked immunospot analysis using five overlapping Env peptide sets from heterologous subtype A, B, and C viruses. The CON6-derived vaccine was immunogenic and induced a greater number of T-cell epitope responses than any single wild-type subtype A, B, and C env immunogen and similar T-cell responses to a polyvalent vaccine. The responses were comparable to within-clade responses but significantly more than between-clade responses. The magnitude of the T-cell responses induced by CON6 (measured by individual epitope peptides) was also greater than the magnitude of responses induced by individual wild-type env immunogens. Though the limited major histocompatibility complex repertoire in inbred mice does not necessarily predict responses in nonhuman primates and humans, these results suggest that synthetic centralized env immunogens represent a promising approach for HIV-1 vaccine design that merits further characterization.
The high level of genetic variation among human immunodeficiency virus type 1 (HIV-1) isolates poses a major obstacle for HIV-1 vaccine development. The HIV-1 “main” (M) group viruses are primarily responsible for the AIDS pandemic and are further classified into nine genetically distinct subtypes (A to D, F to H, J, and K) (26). With protein sequence differences between Env proteins from various subtypes as high as 35%, wild-type (WT) HIV-1 genes from any one subtype may not induce immune responses that will provide consistent cross-reactive protection against other subtypes. Countries and even cities often have multiple subtypes cocirculating (26, 38). Therefore, a vaccine based on only one strain or subtype is unlikely to prevent or control HIV-1 infection in most parts of the world.
T-cell responses play an important role in the control of viremia in natural infection and vaccinated animals (3, 4, 19, 33, 36). Several candidate vaccines have been shown to induce T-cell responses against HIV-1, but to date, levels of cross-subtype responses have remained suboptimal, highly variable, and epitope specific (12, 13, 20, 22, 34). To maximize the genetic similarity between the candidate vaccine and contemporary HIV-1 strains, to simplify immunogen production, and to provide a baseline from which to build an optimized vaccine, we and others have proposed to use centralized HIV-1 gene sequences (consensus, ancestor, and center of the tree) for vaccine development (10, 11, 14-16, 24, 30, 31).
Because of the star-like phylogeny of HIV-1, a “central sequence” will reduce the amino acid divergence between immunogens and field virus strains (14); however, the three kinds of centralized sequences can differ from each other by 1% to 5%, and their precise sequence depends upon the input data set of natural strains used to generate them. Comparing experimentally defined cytotoxic T-lymphocyte (CTL) epitopes, we have found that consensus and ancestral sequences preserve many known HIV-1 CTL epitopes from different viral strains (14, 16, 39). These observations would predict that centralized HIV-1 genes induce more broadly cross-reactive T-cell responses than WT genes.
We previously generated a group M consensus env gene (CON6) based on the sequences available in the Los Alamos HIV Sequence Database in 1999. CON6 Env-containing pseudovirions are infectious for CD4/CCR5-bearing target cells, and CON6 Env protein trimers induce neutralizing antibodies against select HIV-1 primary isolates (15). Here, we report a comparative study of T-cell responses induced by env gene-based DNA and recombinant vaccinia virus (rVV) vaccines derived from CON6 and three WT subtype A, B, and C HIV-1 strains, tested alone as well as in a polyvalent combination in mice.
MATERIALS AND METHODS
Env gp140CF immunogens.
To generate DNA and rVV vectors expressing secreted forms of envelope immunogens, CON6 (group M consensus), JRFL (subtype B), and 96ZM651 (subtype C) gp140CF plasmids were constructed by introduction of a stop codon before the membrane-spanning domain (YIKIFIMIVGGLIGLRIVFAVLSIVN) and deletion of the gp120/gp41 cleavage site and fusion domain of gp41, as previously described (15). Parts of variable regions (V1, V2, V4, and V5) in CON6 represent the sequences of a contemporary subtype C strain (98CN006) as described previously (15). 92RW020 (subtype A) [pVRC5304 (R5 gp140ΔCFI-Clade-A)] contained an additional deletion between HR1 and HR2 in gp41 (22). All env genes were cloned into a DNA expression vector, pCMV/R, at XbaI and BamHI sites as DNA immunogens. The same genes were also cloned into the shuttle plasmid pSC65 at SalI and KpnI sites to generate rVV as previously described (29). The env gene inserts in both plasmid and rVV DNA were confirmed by PCR and nucleotide sequence analysis. Plasmid DNA was prepared using GenElute HP Plasmid Maxiprep kits as described by the supplier (Sigma, St. Louis, MO). Recombinant vaccinia viruses were purified from infected 293T cell lysates by centrifugation through a 36% sucrose cushion at 32,900 × g for 2 h at 4°C. Purified viruses were titrated on HuTK− 143B cells.
Immunizations.
Female BALB/c, C57BL/6, and C3H-NeJ mice (6 to 8 weeks old) were purchased from Charles River Laboratories (Wilmington, Mass.) and housed in the Duke University Animal Facility under the Association for Assessment and Accreditation of Laboratory Animal Care (AALAC) guidelines with animal use protocols approved by the Duke University Animal Use and Care Committee. Five mice per group were immunized intramuscularly (i.m.) in the quadriceps with gp140CF or gp140ΔCFI and empty vector control DNA (50 μg) three times at 3-week intervals. Three weeks after the last DNA immunization, mice were boosted with rVVs and wild-type VV (107 PFU). For the polyvalent group (a mixture of A_92RW020, B_JRFL, and C_96ZM651), one-third doses of DNA (17 μg) or rVV (0.33 × 107 PFU) were used for each immunogen. Two weeks after rVV boost, mice were euthanized and spleens were collected.
ELISpot assay.
For the enzyme-linked immunospot (ELISpot) assay, a total of five overlapping Env peptide sets from subtypes A, B, and C were used for T-cell epitope mapping. The Envs used for reagent design were from heterologous viruses relative to the vaccine strains. Subtype A (92UG037) gp140 overlapping peptides were synthesized through SynPep Corporation (Dublin, CA) and consisted of 168 peptides that were 15 amino acids long with an 11-amino-acid overlap. Subtype B (MN, SF162P3, and 89.6) and subtype C (Chn19) peptides were obtained from the NIH AIDS Research and Reference Reagent Program (Bethesda, MD). The MN and SF162P3 peptide sets consisted of 174 and 182 peptides, respectively. They were 15 amino acids long, with an 11-amino-acid overlap. The 89.6 and Chn19 peptide sets consisted of 72 peptides that were 20 amino acids long with a 10-amino-acid overlap. Amino acid distances between the Env immunogen and Env peptide sequences are summarized in Table 1. As expected, each subtype immunogen was less divergent from peptide sequences from the same subtype than from peptide sequences from other subtypes. The distances between CON6 and any subtype peptide sequence were comparable to within-subtype distances.
TABLE 1.
Amino acid distances between Env immunogens and peptides
| Immunogen | Peptide | % Distance |
|---|---|---|
| A_92RW020 | 92UG037 (subtype A) | 20.10 |
| Other subtypes | 24.20 | |
| B_JRFL | MN, 89.6 and SF162P3 (subtype B) | 14.50 |
| Other subtypes | 23.70 | |
| C_96ZM651 | Chn19 (subtype C) | 18.70 |
| Other subtypes | 27.00 | |
| CON6 | Subtypes | 18.50 |
| Among subtypes | 25.00 |
Spleens from individual mice were minced and forced through a 70-μm nylon cell strainer (BD Labware, Franklin Lakes, NJ). Single-cell suspensions of splenocytes were plated in 96-well polyvinylidene difluoride-backed plates (MultiScreen-IP; Millipore, Billerica, MA) coated with 50 μl of anti-mouse gamma interferon (IFN-γ) monoclonal antibody (MAb), AN18 (5 μg/ml; Mabtech, Stockholm, Sweden), overnight at 4°C. The plates were blocked with HEPES-buffered complete RPMI medium at 37°C for 2 h. Equal volumes (50 μl) of each peptide pool and splenocytes (107 cells/ml) were added to the wells in duplicate. All peptides from 89.6 or Chn19 were used in one pool. Peptides from 92UG037, MN, and SF162P3 were equally divided into two pools to keep the peptide numbers similar as for 89.6 and Chn19. The final concentration of each peptide was 1 μg/ml. Plates were incubated overnight (14 to 16 h) at 37°C with 5% CO2. After the plates were washed six times with phosphate-buffered saline (PBS), 50 μl of 1:1,000-diluted biotinylated anti-mouse IFN-γ MAb (Mabtech, Stockholm, Sweden) was added to each well. Plates were incubated at room temperature (RT) for 2 h and then washed with PBS (three times), and 50 μl of streptavidin-alkaline phosphatase conjugate (1:1,000 dilution; Mabtech, Stockholm, Sweden) was added to each well. After incubation (RT for 1 h), the plates were washed with PBS plus 0.05% Tween 20 (five times). Finally, 100 μl of BCIP/NBT (Plus) (5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium) alkaline phosphatase substrate (Moss, Pasadena, MD) was added to each well. The plates were incubated at RT for 10 min. After washing with water, plates were air dried. Spots were counted using an automated ELISpot plate reader (Immunospot counting system; CTL Analyzers, Cleveland, OH) and expressed as spot-forming cells (SFC) per 106 splenocytes. Responses were considered as positive if the number of the spots was fourfold higher than that of the negative control and at least 50 SFC/106 cells. CD4+ and CD8+ T-cell depletion and enrichment were performed using immunomagnetic depletion beads according to the manufacturer's instructions (DYNAL BIOTECH ASA, Oslo, Norway). ELISpot assays were performed with depleted CD4+ or CD8+ T cells to determine whether the T-cell epitopes were either major histocompatibility complex (MHC) class I or II restricted.
Statistical analysis.
The logistic regression models (2, 8) used in this study to investigate the breadth of induced responses were constructed using the statistics package R (www.r-project.org) (32) to fit saturated models and then progressively to eliminate all statistically insignificant interactions: a similar analysis is detailed in, for example, reference 2. One of the models was overdispersed, so we applied the methods described by McCullagh and Nelder (28) to model the overdispersion. In addition, we performed Poisson regressions to model the magnitude of the responses (in SFC per 106 splenocytes), but as these models did not fit very well, we will not discuss them below. The poor fit in these Poisson models is not unexpected: given the considerable variation in immune response between individual mice, it would be surprising if the immunogen and epitope types provided sufficient explanatory power to yield an accurate numerical prediction of the strength of the response. To accommodate this strong biological variation, we have concentrated on more robust statistical procedures. To explore the breadth of the responses, we used the logistic regressions described above in which each response was treated as a Boolean “yes or no” according to whether the response exceeded the threshold of detection and we analyzed contingency tables with Fisher's exact test. Finally, we used rank-based statistics when comparing the magnitudes of responses. These included Kruskal-Wallis tests to compare multiple categories of vaccine groups, coupled with Wilcoxon rank sum tests for further refinement of relationships between pairs of categories. ELISpot tests were done in duplicate; the single highest score for each duplicate test performed was used to compare magnitudes of responses between vaccine groups.
RESULTS
Breadth of cross-subtype T-cell responses induced by CON6.
We determined T-cell responses induced by CON6 and compared these with responses induced by WT Env immunogens administered via a DNA prime/rVV boost immunization strategy. Six groups of mice were immunized with A_92RW020 gp140ΔCFI (subtype A), B_JRFL gp140CF (subtype B), C_96ZM651 gp140CF (subtype C), a polyvalent mixture (A_92RW020 gp140ΔCFI, B_JRFL gp140CF, and C_96ZM651 gp140CF), CON6 gp140CF, or empty plasmid control DNA (three times) at 3-week intervals and boosted once with corresponding rVVs. To evaluate the cross-reactive T-cell responses induced by the CON6 immunogen in different genetic backgrounds with alternative MHC haplotypes, we determined T-cell responses in three mouse strains (BALB/c, C57BL/6, and C3H-NeJ). Splenocytes from mice immunized with each immunogen were used according to the numbers of INF-γ spot-forming cells with five overlapping WT Env peptide sets: 92UG37 (subtype A), SF162P3, MN and 89.6 (subtype B), and Chn19 (subtype C).
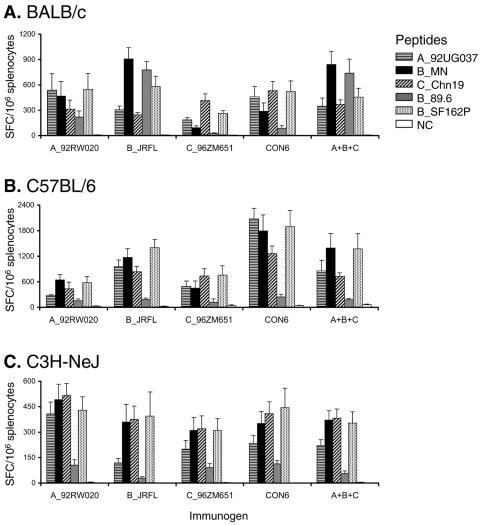
First, we determined the overall T-cell responses to peptide pools containing all Env peptides for each subtype peptide set with splenocytes from individual mice. In general, CON6 induced a similar magnitude of T-cell responses compared to those induced by WT or polyvalent immunogens (Fig. 1). In some instances, the T-cell responses induced by CON6 or the polyvalent vaccine were more potent than those induced by the various subtype immunogens. For example, CON6 induced more potent T-cell responses than A_92RW020, C_96ZM651, and the polyvalent immunogen in C57BL/6 mice (Student's t test: P = 0.001, P = 0.023, and P = 0.040, respectively), although these differences were not seen in C3H and BALB/c strains.
FIG. 1.
T-cell immune responses induced by CON6 and subtype Env immunogens in BALB/c, C57BL/6, and C3H-NeJ mice. Splenocytes were isolated from individual immunized mice (5 mice/group) and stimulated in vitro with five overlapping Env peptide pools. Two pools were used for A_92UG037 (84 peptides in each pool), B_MN (87 peptides in each pool), and SF162P3 (92 peptides in each pool), and one pool was used for B_89.6 (72 peptides) and Chn19 (72 peptides). INF-γ-producing cells were determined by ELISpot analysis. Total responses for each immunogen and peptide pool are expressed as SFC per million splenocytes. The values for each column are the mean ± standard error IFN-γ SFC (n = 5).
To determine the breadth of the T-cell responses, we mapped T-cell epitopes for each immunogen. We combined splenocytes from all five mice in each group for the initial screening using peptide pool matrices (1, 20) because the number of splenocytes from each mouse was insufficient for epitope mapping with all five overlapping peptide sets. When potential positive peptides were identified, we then determined which peptides contained the T-cell epitopes using individual peptides.
In BALB/c mice immunized with the polyvalent immunogen containing A, B, and C Envs, we found broader cross-subtype T-cell responses (six, three, and five epitopes for subtypes A, B, and C, respectively) compared to individual WT subtype immunogens (Fig. 2A and see Fig. S1 in the supplemental material). Importantly, we observed comparable broad cross-reactive T-cell responses with the single CON6 env immunogen compared to the polyvalent vaccine (seven, three, and six epitopes for subtypes A, B, and C, respectively). Three epitopes (B1, B6, and B7) that were not detected in CON6-immunized mice were only recognized by one subtype immunogen. Overall, among 12 T-cell epitopes identified in BALB/c mice, individual subtype A, B, and C immunogens failed to elicit T-cell responses to four, eight, and seven epitopes, respectively, while CON6 and the polyvalent immunogens only missed three and two epitopes, respectively. However, eight epitopes were only detected in one of the two repeats in mice immunized with the polyvalent immunogen (Fig. 2A).
FIG. 2.
T-cell epitope mapping of CON6 and subtype Env immunogens. T-cell responses were determined in three mouse strains with five overlapping Env peptide sets from subtypes A, B, and C (A_92UG37, B_SF162P3, B_89.6, B_MN, and C_Chn19). For direct comparison to subtype A and C peptide sets, results from only one (B_SF162P3) of the three subtype B peptide sets are shown in the figure. The T-cell epitopes were first screened with Env overlapping peptide matrices and then confirmed with individual peptides by ELISpot analysis. Positive subtype A, B, and C epitopes are indicated by blue, red, and green boxes, respectively. Epitopes that were positive once in two repeats are indicted with boxes composed of colored and white triangles. White boxes indicate the epitopes that were expected to be detected but did not induce >50 SFC. Epitopes starting with the letters B, C, and H represent the epitopes from BALB/c (A), C57BL/6 (B), and C3H-NeJ (C) mice, respectively. CD8 epitopes are indicated by asterisks, and all others are CD4 epitopes. A deletion containing the C8 epitope between HR1 and HR2 in the gp41 region of 92RW020 (22) is indicated.
In C57BL/6 mice, we identified a total of nine T-cell epitopes (Fig. 2B). CON6 Env performed well as an immunogen, with immunized animals failing to recognize only one epitope (C3) among all epitopes identified in C57BL/6 mice. Two epitopes (C1 and C7) were only detected in CON6-immunized mice. The WT subtype immunogens and the polyvalent immunogen induced similar T-cell responses in C57BL mice, with each missing three or four T-cell epitopes.
Finally, in C3H-NeJ mice, we detected a total of seven epitopes. Broader T-cell responses were induced by CON6 and the polyvalent immunogen than single WT env genes (Fig. 2C). All epitopes induced by individual subtype immunogens were detected in the polyvalent group. The T-cell responses induced by CON6 env missed two epitopes (H4 and H7) which were subtype B or C specific. One additional epitope (H3) induced in CON6-immunized mice was not detected with WT subtype or polyvalent immunogens. B_JRFL induced similar T-cell responses to the polyvalent immunogen. Immunogen and peptide sequences corresponding to all potential T-cell epitopes are summarized in Fig. 3 (and see Table S1 in the supplemental material).
FIG. 3.
Potential T-cell epitope sequence alignment of the immunogens and screening peptides. All sequences were compared to the group M consensus Env sequence (CON6). Sequences of potential T-cell epitopes from immunogens are shown at the top, and sequences of the peptides used for epitope mapping are shown at the bottom for each mouse strain. Epitopes starting with the letters B, C, and H stand for the epitopes from BALB/c (A), C57BL/6 (B), and C3H-NeJ (C) mice, respectively. Peptides that are positive for any one of the three subtypes are underlined. Mutations in the immunogens that might be responsible for failure to induce T-cell responses are indicated by bold letters.
To determine whether responding T cells were either CD8+ or CD4+, we performed ELISpot assays with either depleted CD4+ or CD8+ T cells for all positive peptides in three mouse strains. Four epitopes (B6, C1, C7, and C8) were recognized by CD8+ T cells in BALB/c or C57BL/6 mice. All other epitopes were recognized by CD4+ T cells (Fig. 2).
Since relatively small numbers of T-cell epitopes were identified for each immunogen in each mouse strain, we statistically analyzed the breadth of T-cell responses among CON6, the three subtype A, B, or C monovalent immunogens, and the polyvalent immunogens by combining all T-cell epitopes from three mouse strains with models that include the mouse strain as a variable. To most reliably reveal the relationship among the T-cell responses induced by each immunogen, we took the following variables into consideration in our statistical model: immunogens (n = 5), epitopes (n = 28), peptide sets (n = 5), mouse strains (n = 3), and positive responses. Logistic regression (2, 8) is a natural strategy to use when asking a question of this type: how the probability of a positive response depends on the properties of the immunogen and the peptide. Such regression models yield a set of predicted probabilities as well as a list of coefficients that permit one to compute odds ratios and assign P values. For comparison, we classified the responses into four classes: (i) CON6 (responses induced by CON6 determined by peptides from subtypes A, B, and C); (ii) polyvalent (responses induced by the polyvalent immunogen determined by peptides from subtypes A, B, and C); (iii) intraclade (responses induced by a single subtype immunogen determined by peptides from the same subtype); and (iv) interclade (responses induced by a single subtype immunogen determined by peptides from the other subtypes). We also considered two definitions of whether a given peptide induced a positive response when tested against a given immunogen: in the first, we treated each reaction, including repeats, separately, classifying a response as positive if we saw 50 or more SFC per million cells. For the second definition, we based our judgment on all the repeats of a single peptide-epitope combination, declaring the response to be positive if any one of the repeats showed 50 or more SFC per million cells. Our analysis showed that the immunogen type significantly influenced the probability of positive response. Under the first definition, the CON6 immunogen was most likely to produce a positive response, followed by polyvalent = intraclade > interclade. In this sense, CON6 was significantly more likely to induce a positive response than the polyvalent immunogen (odds ratio, >1.89; P < 0.005) and interclade responses were significantly diminished compared to the polyvalent responses (odds ratio, < 0.56; P < 0.002). With the alternative definition (any single positive reaction implies positivity), the differences between the immunogens were less pronounced: the CON6, polyvalent, and intraclade responses were all comparable, and the only statistically significant effect was the markedly reduced probability of a positive response induced by interclade immunogens (odds ratio, <0.4; P < 0.0004). The modeling results from this second definition are in good qualitative agreement with the broad picture of induced response, in which T-cell responses to the CON6 and polyvalent immunogens were similar, with 22 total epitopes recognized by the polyvalent immunogen-immunized mice and 22 total epitopes recognized by CON6-immunized mice.
Finally, we also explored the dependence of breadth of T-cell response on mouse strain, including this as an extra explanatory factor. With the first, reaction-by-reaction, definition of a positive response, the strongest effect is that the C3H mice are much more likely to produce a positive response than the others (odds ratio, >9.25; P < 0.004). This model was, however, the only one of those presented here that was overdispersed (estimated deviance of 1.8 per degree of freedom), suggesting that this model failed to capture some important source of variation. In contrast, when we fit a similar model, but used the second sense of positivity (any positive response among the duplicate reactions implies positive), a more consistent story emerged: the C3H mice still proved more likely to exhibit a positive response, but the effect was less significant (odds ratio, >14.1; P < 0.015). The only other significant effects were a modestly significant enhancement in the response of C57BL/6 mice to epitopes of subtype B (odds ratio, >3.4; P < 0.017) and a highly significant decrease in probability of a response for interclade epitopes (odds ratio, <0.37; P < 0.0002).
Taken together, we believe that the most appropriate conclusion from our statistical analyses is that the group M consensus env immunogen induced similar breadth to the polyvalent immunogen and was superior to individual WT env immunogens.
Magnitude of cross-subtype T-cell immune responses induced by CON6.
We measured the intensity of the T-cell responses induced by each immunogen with pooled peptides (over 72 peptides in each reaction) (Fig. 1). In these analyses, a pooled response could be dominated by the immunodominant response and responses might also be subjected to interference among peptides in the pool. We obtained the T-cell responses to individual peptides. This allowed us to perform a comparison of distributions of the potencies of T-cell responses induced by each epitope to the different vaccines. All SFC numbers for peptides for each immunogen were plotted in four groups as classified earlier: CON6, polyvalent, intraclade, and interclade (Fig. 4). When multiple counts were available for an epitope (because overlapping peptides were considered to represent a single response and repeats for the same epitope), the highest SFC numbers were used systematically as the single data point included in the analysis. Comparable ELISpot numbers were induced by different immunogens for the shared epitopes, suggesting all immunogens had similar capabilities to induce the T-cell responses.
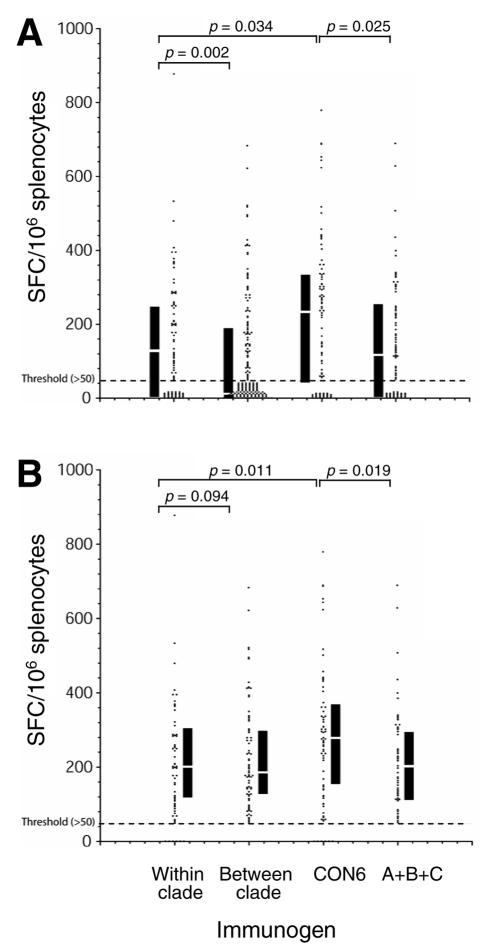
FIG. 4.
Potency of T-cell immune responses among consensus and subtype Env immunogens. T-cell responses (SFC/106 cells) are plotted for each peptide that yielded at least one positive response (>50) to at least one of the five immunogens (A_92UG037, B_JRFL, C_96ZM651, CON6, and polyvalent). Four groups of responses were compared: (i) within clade (responses induced by WT subtype immunogen determined by peptides from the same subtype), (ii) between clade (responses induced by WT subtype immunogen determined by peptides from the other subtypes), (iii) CON6 (responses induced by CON6 determined by peptides from subtypes A, B, and C), and (iv) polyvalent (responses induced by the polyvalent immunogen determined by peptides from subtypes A, B, and C). Each dot represents an SFC count. The dashed line indicates the threshold (50 SFC) below which responses were considered negative. The interquartile ranges and medians are indicated by the black vertical bars and white lines in the bars. The statistical comparison was performed using the Kruskal-Wallis and the Wilcoxon rank tests. The Wilcoxon rank P values are shown for the compared groups.
The Kruskal-Wallis rank sum test was used to compare distributions of response levels to peptides found in the four groups. The null hypothesis—that all four sets of values were drawn from the same distribution—was found to be very unlikely (P = 0.0000025), so we went on to do pairwise comparisons between groups using a Wilcoxon rank sum statistic. This potency comparison analysis showed CON6-immunized animals had slightly better responses than polyvalent immunogen-immunized animals, with polyvalent = intraclade > interclade. About the same number of peptides that induced no response (nonstimulators; <50 SFC) were found in the CON6, polyvalent, and intraclade groups (Fig. 4A), but many more nonstimulators were found in the interclade group. When only positive ELISpot counts were used for the comparison, the potency of T-cell responses induced by CON6 was still stronger than those of all other groups (Fig. 4B). We then reevaluated this comparison for each mouse strain independently. With the smaller number of positive scores available when comparing the results of each strain, the only comparisons that remained significant were the generally higher magnitude of CON6 responses in C57BL/6 mice compared to the polyvalent vaccine (P = 0.0047), intraclade (P = 0.011), and interclade (P = 0.000027) responses.
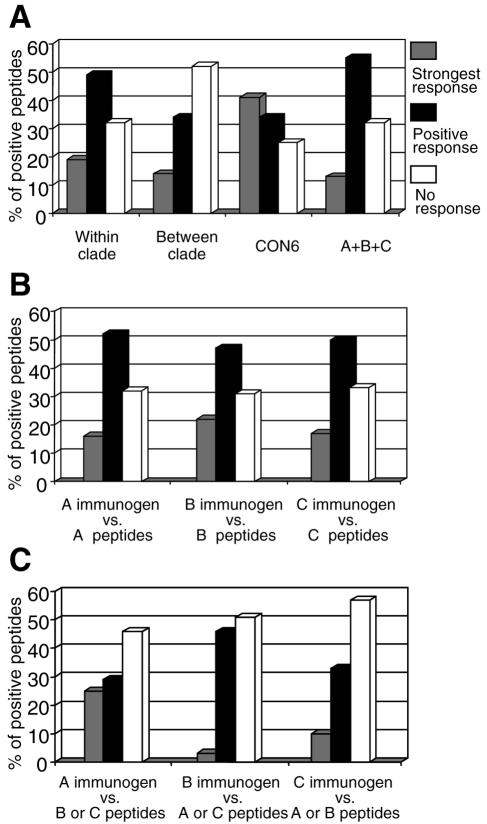
We then performed a second analysis of the data, to directly compare the magnitudes of response to specific peptides rather than the distributions of levels of response. We classified the responses elicited by immunogens against each of the 88 peptides that had detectable responses in at least one vaccinated animal by ranking the level of response to that peptide elicited by each immunogen. The immunogens were categorized three ways for each peptide to ascertain which immunogen tended to give the strongest responses. The three categories were (i) the immunogen with the strongest score for a given peptide (strongest response), (ii) the immunogens that gave a positive score but not the strongest score (positive response), or (iii) the immunogens that had no detectable response to the peptide (no reaction; <50 SFC). In this analysis, CON6 had the highest frequency of the strongest score responses and the lowest frequency of no-reaction responses (Fig. 5A). In contrast, the lower frequencies of strongest score responses were observed for polyvalent, intraclade, and interclade groups. A 3-by-4 contingency table showed that the frequencies were unlikely to be observed by chance (P = 0.0000066), and separate 2-by-3 comparisons showed that the distributions were distinct for CON6 versus polyvalent (P = 0.00008), CON6 versus intraclade (P = 0.0072), and intraclade versus interclade (P = 0.0086). No significant differences were observed between polyvalent and intraclade groups (P = 0.44).
FIG. 5.
Intensity of T-cell immune responses among consensus and subtype Env immunogens by rank. The same data in Fig. 4 were used for comparison by classifying the responses in three categories: (i) the immunogen with the strongest score for a given peptide (strongest response); (ii) the immunogens that gave a positive score, but not the strongest score (positive response); and (iii) the immunogens that had no detectable response to the peptide (no reaction; <50 SFC). The same four groups (within clade, between clade, CON6, and polyvalent) as in Fig. 4 were compared (A). The T-cell responses within the subtype (B) and between subtypes (C) for each WT subtype immunogen were also compared. The 3-by-4 and 2-by-3 contingency table tests were performed for statistical comparisons.
We next compared the potencies of T-cell responses for each subtype immunogen. Single subtype A, B, and C natural antigens gave comparable levels of intraclade responses to the heterologous antigens. No significant differences were observed when T-cell responses induced by subtype immunogens were measured with heterologous peptides from the same subtype (Fig. 5B). However, when heterologous peptides from different subtypes were used to measure T-cell responses, the subtype A immunogen (A_92RW020) had more strongest responses than subtype B or C immunogens (Fig. 5C).
Given the heterogeneity of responses seen between mouse strains, we believe the appropriate interpretation of our analyses is that the group M consensus immunogen induced responses that were comparable in potency to those of the polyvalent immunogen and superior to those of the individual WT immunogens.
Similar T-cell responses induced by subtype B and CON6 Env immunogens to three heterologous subtype B overlapping Env peptide sets.
We used three available heterologous subtype B Env overlapping peptide sets (MN, 89.6, and SF162P3) to determine cross-reactive T-cell responses induced by the subtype B, other subtype, or consensus immunogens. Table 2 shows that more T-cell epitopes (16) were detected in mice immunized with subtype immunogen B_JRFL than in those immunized with A_92RW020 (11) or C_96ZM651 (10). In B_JRFL-immunized mice, only a few epitopes were detected with all three subtype B peptides in either BALB/c mice (two out of five) or C3H mice (one out of six), although all five T-cell epitopes were identified with peptides from all three subtype B sets in C57BL/6 mice. CON6 induced similar T-cell responses to three subtype B peptide sets, as did B_JRFL in terms of total numbers of T-cell epitopes or numbers of cross-reactive epitopes (Table 2 and see Fig. S2 in the supplemental material).
TABLE 2.
Cross-reactive T-cell epitopes identified with three subtype B peptide sets (MN, 89.6, and SF162P3)
| Immunogen | Subtype B peptides T-cell epitopes respond toa:
|
Total no. of epitopes | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BALB/c
|
C57BL/6
|
C3H
|
||||||||
| 3 | 2 | 1 | 3 | 2 | 1 | 3 | 2 | 1 | ||
| B_JRFL | B5 | B1 | B4 | C2 | H5 | H1 | H2 | 16 | ||
| B6 | B2 | C4 | H4 | H6 | ||||||
| C5 | H7 | |||||||||
| C6 | ||||||||||
| C8 | ||||||||||
| A_92RW020 | B4 | B7 | C4 | H5 | H2 | 11 | ||||
| B5 | C5 | H6 | ||||||||
| B9 | C6 | |||||||||
| B11 | ||||||||||
| C_96ZM651 | B9 | B2 | C6 | C2 | H5 | H1 | H2 | 10 | ||
| C4 | H6 | |||||||||
| C5 | ||||||||||
| CON6 | B2 | B4 | B10 | C2 | C7 | H5 | H1 | H2 | 15 | |
| B9 | C4 | H6 | H3 | |||||||
| C5 | ||||||||||
| C6 | ||||||||||
| C8 | ||||||||||
Numbers 3, 2, and 1 represent the number of peptides responded to.
Failure to detect T-cell responses due to mutations in T-cell epitopes.
In the above analyses, we observed that T-cell responses were not always detected among all immunogens or by corresponding peptides. Failures to detect T-cell responses were of two types that were readily understood: those due to amino acid substitutions in screening peptides and those due to amino acid substitutions in immunogens. In the first type, the T-cell response was induced by the immunogen but was not detected when screened with the peptides carrying one or more mutations (Fig. 3). For example, an E-to-D substitution was found in the Chn19 peptide in the C4 epitope in C57BL/6 mice (Fig. 3B). No T cells from any group of immunized mice recognized this peptide (Fig. 2 and see Fig. S1 in the supplemental material). A similar phenomenon has been observed by others for the Th epitope (FEPIPIHYC), in which the E-to-D substitution abrogated T-helper hybridomas' ability to recognize the mutant peptide (40). Some peptides with amino acid substitutions did not completely fail to induce T-cell responses, but the substitutions significantly decreased the peptides' ability to stimulate SFC in vitro. For example, the SFC numbers induced by SF162P3 P18 equivalent peptide with an R-to-K substitution (B6) were at least fivefold lower than those induced by corresponding MN or 89.6 peptides (data not shown).
In the second case, the immunogens containing an amino acid substitution or substitutions in the T-cell epitope regions did not induce T-cell responses for the corresponding epitope responses. For example, in BALB/c and C57BL/6 mice, highly conserved and cross-reactive B2 and C2 epitopes were not detected when A_92RW020 was used as the immunogen, while both epitopes were induced by other immunogens (Fig. 2 and 3). The A-to-E substitution in either the B2 or C2 epitope in the A_92RW020 immunogen sequence might be responsible for the failure to elicit the corresponding T-cell response (Fig. 3A and B).
To determine the potential T-cell epitope coverage of each immunogen, all possible 9-mers in the proteins were compared between the vaccines and the test peptides. CON6 and intraclade single sequences had a similar coverage (27% and 28%, respectively) of the 9-mers in test peptide sequences. The single sequences in the interclade group had the poorest coverage (15%). However, when three sequences were included in the polyvalent immunogen, substantially more 9-mers (37%) were matched among the test peptides (Table 3). Similar results were obtained using 11-mer peptide sequences (Table 3). Given the analysis of 9-mer and 11-mer coverage, it was expected that the polyvalent immunogen would elicit more detectable responses than CON6, but this was not what we observed. This might be explained by the generally diminished magnitude of responses to the polyvalent vaccine reducing our ability to discern responses near the threshold of detection. In the polyvalent group, a few epitopes were not detected (white boxes in Fig. 2) that were induced by individual subtype immunogens. Failure to detect T-cell responses to these peptides in the polyvalent group could not be explained by amino acid substitutions in either immunogens or peptides used for screening, since the same subtype immunogens and peptides were positive when they were tested as individual subtype immunogens (Fig. 2 and 3). Also, often only one of two ELISpot repeats was positive for the polyvalent vaccine. This lack of reactivity may be due to the lower dose of each immunogen (1/3 of individual subtype immunogen doses) in the mixture, interference among epitopes from three subtype immunogens in the mixture (18, 21), or other mechanisms.
TABLE 3.
Amino acid similarity of all possible 9- or 11-mer peptide sequences between immunogens and peptides used in this study
| No. of mismatches in peptide | Relative amino acid similaritye
|
|||
|---|---|---|---|---|
| CON6a | A+B+Cb | Intracladec | Intercladed | |
| Peptide match length of 9 | ||||
| 0 | 0.2674 ± 0.0147 | 0.3703 ± 0.0234 | 0.2822 ± 0.0356 | 0.1532 ± 0.0100 |
| 1 | 0.5423 ± 0.0144 | 0.6262 ± 0.0203 | 0.5356 ± 0.0293 | 0.3709 ± 0.0126 |
| 2 | 0.7177 ± 0.0232 | 0.7679 ± 0.0120 | 0.6886 ± 0.0245 | 0.5592 ± 0.0078 |
| 3 | 0.8310 ± 0.0224 | 0.8435 ± 0.0094 | 0.7743 ± 0.0215 | 0.7133 ± 0.0070 |
| Peptide match length of 11 | ||||
| 0 | 0.2078 ± 0.0159 | 0.3076 ± 0.0229 | 0.2291 ± 0.0355 | 0.1080 ± 0.0101 |
| 1 | 0.4682 ± 0.0134 | 0.5538 ± 0.0270 | 0.4686 ± 0.0342 | 0.2945 ± 0.0137 |
| 2 | 0.6430 ± 0.0224 | 0.7103 ± 0.0154 | 0.6263 ± 0.0269 | 0.4730 ± 0.0078 |
| 3 | 0.7665 ± 0.0277 | 0.7927 ± 0.0117 | 0.7190 ± 0.0267 | 0.6238 ± 0.0087 |
CON6, the CON6 immunogen sequence was compared to subtype A, B, and C peptide sequences.
A+B+C, all three sequences in the polyvalent immunogen were compared to subtype A, B, and C peptide sequences.
Intraclade, the individual subtype immunogen sequence was compared to subtype matching peptide sequences.
Interclade, the individual subtype immunogen sequence was compared to subtype mismatching peptide sequences.
The data are shown as mean ± standard error relative to 1 = 100%.
Most of the T-cell epitopes of HIV-1 envelope proteins were in conserved regions.
To determine the locations of all identified T-cell epitopes in the Env proteins, we mapped epitopes from three mouse strains (Fig. 6). Most of the epitopes were found in the conservative regions of the envelope protein, with only a few exceptions. One epitope (C1) in C57BL/6 mice immunized with CON6 was found in the signal peptide region. It was only detected with Chn19 peptides and not with any others due to the high level of sequence variation (Fig. 3B). The same epitope was also confirmed with CON6 autologous peptides (data not shown). In BALB/c mice, a well-studied subtype B-specific P18 epitope (B6) was confirmed in the V3 region (35), which was actually not much more variable than other conserved regions in the Env proteins (Fig. 3A) (16). Epitopes B3 and C7 overlapped with the 3′ end of V3 and 5′ end of V4, respectively. However, both overlapping regions were conserved. We did not have autologous peptide sets for each subtype immunogen to map potential T-cell epitopes in the variable regions. However, we mapped the T-cell epitopes for CON6 with autologous overlapping peptides and found only one extra epitope in C57BL/6 mice and one extra epitope in C3H mice. Both were in the conserved regions (data not shown). Parts of variable regions (V1, V2, V4, and V5) in CON6 were from a subtype C Env sequence. However, we did not observe more cross-reactive T-cell responses to subtype C peptides (Chn19) despite greater similarity in the variable regions between CON6 and Chn19 sequences.
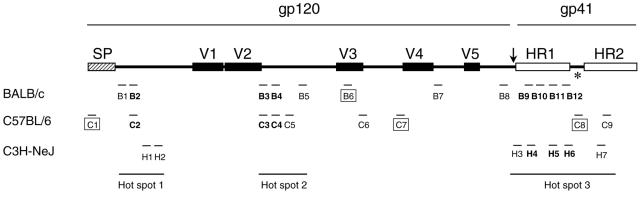
FIG. 6.
The locations of T-cell epitopes in gp140 protein detected in all three mouse strains. The signal peptide (SP), variable regions, and heptad repeats (HR1 and HR2) are indicated by hatched, solid, and empty boxes, respectively. The deletion of the gp120/gp41 cleavage site and the fusion domain of gp41 are indicated by an arrow. The deletion of the spacer between HR1 and HR2 is indicated by an asterisk. The three T-cell epitope hot spots are indicted at the bottom of the figure. CD8 epitopes are indicated by boxed epitopes; all others were CD4 epitopes.
Three murine T-cell epitope hot spots were found in the gp140 protein (Fig. 6). The first one was at the N terminus of gp120 (B1, B2/C2, H1, and H2). The second one was at the first half of the C2 region (B3/C3, B4/C4, C5, and B5). The third one was at the gp41 heptad repeat region (H3, B9/H4, B10/H5, B11/H6, B12, C8, H7, and C9). All epitopes in each hot spot were localized in close proximity to each other (Fig. 3 and 6). These results suggested that these regions may be major T-cell response targets in mice.
DISCUSSION
In this study, we demonstrated that the group M consensus env gene (CON6) induced a higher number of cross-reactive and more potent T-cell responses to subtype A, B, and C viruses than any single subtype env immunogen alone. CON6 induced cross-reactive T-cell responses to all three subtypes, while single WT clade A, B, or C immunogens tended to induce subtype-specific T-cell immune responses and induced fewer responses that were cross-reactive with other subtypes. Interestingly, the polyvalent immunogen induced similar numbers of epitope responses compared to CON6. While we recognize that immunogenicity of an immunogen in mice does not necessarily predict immunogenicity in nonhuman primates or humans, these results provide proof of the concept that a consensus immunogen can induce broader and more potent T-cell responses across clades than individual strain-based immunogens. Thus, the theoretical advantages of the consensus approach as a T-cell immunogen have been experimentally validated in an animal model system. Further testing of centralized gene vaccines in nonhuman primates and humans, alone or in combination with subtype consensus or WT immunogens, are thus warranted.
While it was anticipated that intraclade responses would be more intense and more common than interclade responses, the performance of the CON6 immunogen with respect to both the breadth and magnitude of the elicited immune response was not expected. Previous studies have shown that a polyvalent immunogen induces broader immune responses than single immunogens (6, 7, 22, 34). In two studies, the T-cell responses were analyzed with peptide pools but not individual peptides (22, 34). In such an analysis, a few predominant T-cell responses might overshadow many weaker T-cell responses and the breadth of T-cell responses could not be evaluated on the basis of individual T-cell epitopes. In the current study, we defined T-cell epitopes using individual peptides from five overlapping Env peptide sets of three subtypes (A, B, and C) and found that the majority of the T-cell epitopes were recognized by responses elicited by the polyvalent immunogen. However, some T-cell epitopes were not detected in mice immunized with the polyvalent immunogen, while the same epitopes were detected in the animals immunized with individual subtype immunogens. The failure to induce some T-cell responses might be due to the lower doses of each immunogen in the polyvalent cocktail. Alternatively, there may have been interference between some of the epitopes among the multiple immunogens. Interference has been reported in some (17, 22, 34) but not all (18, 21) studies of multivalent immunogens, thus leaving its potential impact on our results unclear. Still another explanation may be that the ancestral nature of CON6 (group M consensus and ancestor sequences being the same in this case) (13, 14) may have features that make it more immunogenic, while some of these immune responses have been lost in modern strains that may have accumulated immunorefractive mutations as HIV has evolved in people. While HLA and MHC molecules are very different in humans and mice, the processing of T-cell epitopes by enzymes may not be.
A recent study by Doria-Rose et al. showed that an ancestral subtype B env immunogen (An1-envB) induced weak neutralizing activity in rabbits (10). We have observed neutralizing activity induced with CON6 gp120 and gp140 proteins in guinea pigs similar to those induced by An1-envB (15). The study by Doria-Rose et al. did not perform T-cell immune response analysis. In a more recent study, we have studied a second generation of group M consensus Env immunogen (CON-S) with shorter variable loops for its ability to induce neutralizing antibodies and discovered that a newer generation of group M consensus env gene (CON-S) with shorter variable loops can induce broader neutralizing antibody responses to subtype B and C viruses than wild-type Env immunogens (27). This study showed that the consensus gene approach has potential for inducing more broadly reactive neutralizing antibodies than current immunogens and was superior to wild-type subtype A, B, or C primary HIV-1 Envs. Further studies are needed to compare the neutralizing antibody responses induced by the group M consensus env gene with a large panel of polyvalent Env immunogens.
Few studies have been performed to map T-cell epitopes with single overlapping Env peptide sets in mice (5, 9, 37, 40). T-cell epitopes in each subtype WT Env protein used in this study have not been identified before. Our study is the first to systemically map the Env T-cell epitopes induced by consensus and multiple subtype Env immunogens with different subtype overlapping peptide sets. In the Los Alamos HIV Molecular Immunology Database, many CD4 epitopes were registered from the literature but only four CD8 epitopes were reported in mice (23). In this study, we identified three new CD8 epitopes (C1, C7, and C8) in C57BL/6 mice and four new CD4 epitopes (B9/H3, B10/H4, H7, and C9) in three strains of mice, although we did not detect some CD4 epitopes that have been previously reported (23).
The majority of T-cell epitopes identified in this study were in conserved envelope regions (Fig. 6). These data differed from results from other studies in which Th epitopes were identified in envelope variable regions (9, 37). This discrepancy may be partly explained by the use of different immunogens, peptides, and detection methods of Th responses. However, Yusim et al. showed that HIV CTL epitopes were concentrated in relatively conserved regions when heterologous reference peptides were used for detection of CTL activity (39). They also found that variable regions tend to have higher concentrations of amino acids that never serve as C-terminal anchor residues and to have reduced propensity for epitope cleavage and processing. The analysis suggests that variable regions that more readily accommodate changes in HIV-1 genomes tend to accumulate mutations that make them generally less immunogenic as well as less likely to be cross-recognized either within a subtype or among subtypes. Thus, the sequence analysis is in agreement with our observation.
Based on the crystal structure of gp120 protein (25), we found most of the Th epitope targets (H1, H2, C3/B3, C4/B4, C5, B5, and B8) were in the nonexposed inner domain and only three Th epitope targets (C6, C7, and B7) were in the outer domain. Other Th epitope targets were in either the signal peptide region (C1) or the C1 region (B1 and B2/C2), for which no structural data were available. Therefore, our data do not support the hypothesis that exposed protein fragments are uniquely susceptible to proteolytic activity and are thus preferentially shuttled through the antigen-processing pathway in mice (5, 37).
T-cell responses were sensitive to amino acid substitutions between immunogens and peptides, and nearly all nonrecognized T-cell epitopes had amino acid substitutions in the epitope sequences in the vaccines relative to testing strains. This was also true for the subtype B immunogen (JRFL) when three heterologous subtype B Env overlapping peptide sets were used for epitope mapping. Since vaccinated individuals will not be exposed to, nor infected with, homologous viruses, a successful vaccine must be able to prevent or control heterologous viral infection. Therefore, our study with heterologous immunogens and screening peptides within the same subtype was designed to begin to mimic this viral diversity. The level of cross-reactivity induced by the subtype B immunogen to three heterologous subtype B Env-overlapping peptide sets was similar to what we observed with CON6 immunogen (Table 2). Our sequence analysis showed that the differences in amino acid sequences between group M consensus and subtype Env proteins were the same as those between two viruses within the same subtype (Table 1) (14). Therefore, our experimental results were in agreement with our expectations based on sequence analysis and suggest that the group M consensus immunogens may be as efficient as a natural appropriately selected single subtype immunogen for induction of T-cell immune responses against viruses in regions where one subtype is predominant.
The inbred mouse is not an ideal model with which to study T-cell responses, since mice from the same strain have a common genetic background and mouse MHC molecules are different from HLA molecules in humans. The other major difference is that viruses constantly evolve under the immune selection pressure and accumulate many escape mutations along infection history in humans, while they are completely new in mice when used as immunogens. However, if the mechanisms for T-cell responses to HIV-1 immunogens in general are similar between humans and mice, more cross-reactive T-cell responses after immunization with group M consensus immunogens might be anticipated in humans, although the T-cell epitopes recognized may be different. To address this concern, a similar comparison experiment is warranted to determine if cross-subtype T-cell responses can also be induced in nonhuman primates and, if successful, in human clinical trials.
Supplementary Material
Acknowledgments
We thank Allan Zajac for stimulating discussions, Thomas Kepler for statistical analyses and perspective, Stacie Vanleeuwen for technical assistance, Olga Torres for artwork, and the Los Alamos National Laboratory Advanced Computing Laboratory for providing parallel computing time used to inform the design of the group M consensus sequence (CON6).
This work was supported by NIH, NIAID grants AI85338, AI54497, AI55386, PO1 AI52816, IPCAVD AI35351, and HIVRAD AI61734; Duke Center for Translational Research grant AI51445; and the NIAID AIDS Research and Reference Reagent Program. E.A.W. and Z.T.C. were supported by NIH training grant 5T32 AI07392. B.T.K. was funded through internal Los Alamos National Laboratory LDRD research funds.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 77:2081-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agresti, A. 2002. Categorical data analysis. John Wiley & Sons, Inc., Hoboken, N.J.
- 3.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H. L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69-74. [DOI] [PubMed] [Google Scholar]
- 4.Barouch, D. H., S. Santra, J. E. Schmitz, M. J. Kuroda, T. M. Fu, W. Wagner, M. Bilska, A. Craiu, X. X. Zheng, G. R. Krivulka, K. Beaudry, M. A. Lifton, C. E. Nickerson, W. L. Trigona, K. Punt, D. C. Freed, L. Guan, S. Dubey, D. Casimiro, A. Simon, M. E. Davies, M. Chastain, T. B. Strom, R. S. Gelman, D. C. Montefiori, M. G. Lewis, E. A. Emini, J. W. Shiver, and N. L. Letvin. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290:486-492. [DOI] [PubMed] [Google Scholar]
- 5.Brown, S. A., J. Stambas, X. Zhan, K. S. Slobod, C. Coleclough, A. Zirkel, S. Surman, S. W. White, P. C. Doherty, and J. L. Hurwitz. 2003. Clustering of Th cell epitopes on exposed regions of HIV envelope despite defects in antibody activity. J. Immunol. 171:4140-4148. [DOI] [PubMed] [Google Scholar]
- 6.Chakrabarti, B. K., X. Ling, Z. Y. Yang, D. C. Montefiori, A. Panet, W. P. Kong, B. Welcher, M. K. Louder, J. R. Mascola, and G. J. Nabel. 2005. Expanded breadth of virus neutralization after immunization with a multiclade envelope HIV vaccine candidate. Vaccine 23:3434-3445. [DOI] [PubMed] [Google Scholar]
- 7.Cho, M. W., Y. B. Kim, M. K. Lee, K. C. Gupta, W. Ross, R. Plishka, A. Buckler-White, T. Igarashi, T. Theodore, R. Byrum, C. Kemp, D. C. Montefiori, and M. A. Martin. 2001. Polyvalent envelope glycoprotein vaccine elicits a broader neutralizing antibody response but is unable to provide sterilizing protection against heterologous simian/human immunodeficiency virus infection in pigtailed macaques. J. Virol. 75:2224-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crawly, M. 2002. Statistical computing: an introduction to data analysis using S-Plus. John Wiley & Sons, Inc., Chichester, West Sussex, United Kingdom.
- 9.Dai, G., N. K. Steede, and S. J. Landry. 2001. Allocation of helper T-cell epitope immunodominance according to three-dimensional structure in the human immunodeficiency virus type I envelope glycoprotein gp120. J. Biol. Chem. 276:41913-41920. [DOI] [PubMed] [Google Scholar]
- 10.Doria-Rose, N. A., G. H. Learn, A. G. Rodrigo, D. C. Nickle, F. Li, M. Mahalanabis, M. T. Hensel, S. McLaughlin, P. F. Edmonson, D. Montefiori, S. W. Barnett, N. L. Haigwood, and J. I. Mullins. 2005. Human immunodeficiency virus type 1 subtype B ancestral envelope protein is functional and elicits neutralizing antibodies in rabbits similar to those elicited by a circulating subtype B envelope. J. Virol. 79:11214-11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellenberger, D. L., B. Li, L. D. Lupo, S. M. Owen, J. Nkengasong, M. S. Kadio-Morokro, J. Smith, H. Robinson, M. Ackers, A. Greenberg, T. Folks, and S. Butera. 2002. Generation of a consensus sequence from prevalent and incident HIV-1 infections in West Africa to guide AIDS vaccine development. Virology 302:155-163. [DOI] [PubMed] [Google Scholar]
- 12.Ferrari, G., W. Humphrey, M. J. McElrath, J. L. Excler, A. M. Duliege, M. L. Clements, L. C. Corey, D. P. Bolognesi, and K. J. Weinhold. 1997. Clade B-based HIV-1 vaccines elicit cross-clade cytotoxic T lymphocyte reactivities in uninfected volunteers. Proc. Natl. Acad. Sci. USA 94:1396-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrari, G., D. D. Kostyu, J. Cox, D. V. Dawson, J. Flores, K. J. Weinhold, and S. Osmanov. 2000. Identification of highly conserved and broadly cross-reactive HIV type 1 cytotoxic T lymphocyte epitopes as candidate immunogens for inclusion in Mycobacterium bovis BCG-vectored HIV vaccines. AIDS Res. Hum. Retrovir. 16:1433-1443. [DOI] [PubMed] [Google Scholar]
- 14.Gao, F., T. Bhattacharya, B. Gaschen, J. Taylor, J. P. Moore, V. Novitsky, K. Yusim, D. Lang, B. Foley, S. Beddows, M. Alam, B. Haynes, B. H. Hahn, and B. Korber. 2003. Consensus and ancestral state HIV vaccines. Science 299:1517-1518. (Letter.) [Google Scholar]
- 15.Gao, F., E. A. Weaver, Z. Lu, Y. Li, H.-X. Liao, B. Ma, S. M. Alam, R. M. Scearce, L. L. Sutherland, J.-S. Yu, J. M. Decker, G. M. Shaw, D. C. Montefiori, B. T. Korber, B. H. Hahn, and B. F. Haynes. 2005. Antigenicity and immunogenicity of a synthetic human immunodeficiency virus type 1 group M consensus envelope glycoprotein. J. Virol. 79:1154-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaschen, B., J. Taylor, K. Yusim, B. Foley, F. Gao, D. Lang, V. Novitsky, B. Haynes, B. H. Hahn, T. Bhattacharya, and B. Korber. 2002. Diversity considerations in HIV-1 vaccine selection. Science 296:2354-2360. [DOI] [PubMed] [Google Scholar]
- 17.Haynes, B. F., M. A. Moody, C. S. Heinley, B. Korber, W. A. Millard, and R. M. Scearce. 1995. HIV type 1 V3 region primer-induced antibody suppression is overcome by administration of C4-V3 peptides as a polyvalent immunogen. AIDS Res. Hum. Retrovir. 11:211-221. [DOI] [PubMed] [Google Scholar]
- 18.Insel, R. A. 1995. Potential alterations in immunogenicity by combining or simultaneously administering vaccine components. Ann. N. Y. Acad. Sci. 754:35-47. [DOI] [PubMed] [Google Scholar]
- 19.Jin, X., D. E. Bauer, S. E. Tuttleton, S. Lewin, A. Gettie, J. Blanchard, C. E. Irwin, J. T. Safrit, J. Mittler, L. Weinberger, L. G. Kostrikis, L. Zhang, A. S. Perelson, and D. D. Ho. 1999. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 189:991-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keating, S. M., R. C. Bollinger, T. C. Quinn, J. B. Jackson, and L. M. Carruth. 2002. Cross-clade T lymphocyte-mediated immunity to HIV type 1: implications for vaccine design and immunodetection assays. AIDS Res. Hum. Retrovir. 18:1067-1079. [DOI] [PubMed] [Google Scholar]
- 21.Kjerrstrom, A., J. Hinkula, G. Engstrom, V. Ovod, K. Krohn, R. Benthin, and B. Wahren. 2001. Interactions of single and combined human immunodeficiency virus type 1 (HIV-1) DNA vaccines. Virology 284:46-61. [DOI] [PubMed] [Google Scholar]
- 22.Kong, W.-P., Y. Huang, Z.-Y. Yang, B. K. Chakrabarti, Z. Moodie, and G. J. Nabel. 2003. Immunogenicity of multiple gene and clade human immunodeficiency virus type 1 DNA vaccines. J. Virol. 77:12764-12772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korber, B., C. Brander, B. Haynes, R. Koup, C. Kuiken, J. Moore, B. Walker, D. Watkins, et al. 2002. HIV molecular immunology 2002. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 24.Korber, B., M. Muldoon, J. Theiler, F. Gao, R. Gupta, A. Lapedes, B. H. Hahn, S. Wolinsky, and T. Bhattacharya. 2000. Timing the ancestor of the HIV-1 pandemic strains. Science 288:1789-1796. [DOI] [PubMed] [Google Scholar]
- 25.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leitner, T., B. Foley, B. H. Hahn, P. A. Marx, F. McCutchan, J. W. Mellors, S. Wolinksy, B. Korber, et al. 2004. HIV sequence compendium 2003. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 27.Liao, H. X., L. L. Sutherland, S.-M. Xia, M. E. Brock, R. M. Scearce, S. Vanleeuwen, M. Alam, S. McAdam, E. Weaver, Z. T. Camacho, B. Ma, Y. Li, J. Decker, G. J. Nabel, D. Montefiori, B. H. Hahn, B. Korber, F. Gao, and B. Haynes. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology, in press. [DOI] [PMC free article] [PubMed]
- 28.McCullagh, P., and J. Nelder. 1989. Generalized linear models. Chapman and Hall, London, United Kingdom.
- 29.Moss, B., and P. Earl. 1998. Expression of proteins in mammalian cells using vaccinia viral vectors, p. 16.15.1-16.19.9. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., Indianapolis, Ind.
- 30.Nickle, D. C., M. A. Jensen, G. S. Gottlieb, D. Shriner, G. H. Learn, A. G. Rodrigo, and J. I. Mullins. 2003. Consensus and ancestral state HIV vaccines. Science 299:1515-1517. (Letter.) [DOI] [PubMed] [Google Scholar]
- 31.Novitsky, V., U. R. Smith, P. Gilbert, M. F. McLane, P. Chigwedere, C. Williamson, T. Ndung'u, I. Klein, S. Y. Chang, T. Peter, I. Thior, B. T. Foley, S. Gaolekwe, N. Rybak, S. Gaseitsiwe, F. Vannberg, R. Marlink, T. H. Lee, and M. Essex. 2002. Human immunodeficiency virus type 1 subtype C molecular phylogeny: consensus sequence for an AIDS vaccine design? J. Virol. 76:5435-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.R Development Core Team. 2005. A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- 33.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857-860. [DOI] [PubMed] [Google Scholar]
- 34.Seaman, M. S., L. Xu, K. Beaudry, K. L. Martin, M. H. Beddall, A. Miura, A. Sambor, B. K. Chakrabarti, Y. Huang, R. Bailer, R. A. Koup, J. R. Mascola, G. J. Nabel, and N. L. Letvin. 2005. Multiclade human immunodeficiency virus type 1 envelope immunogens elicit broad cellular and humoral immunity in rhesus monkeys. J. Virol. 79:2956-2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shirai, M., C. D. Pendleton, and J. A. Berzofsky. 1992. Broad recognition of cytotoxic T cell epitopes from the HIV-1 envelope protein with multiple class I histocompatibility molecules. J. Immunol. 148:1657-1667. [PubMed] [Google Scholar]
- 36.Shiver, J. W., T. M. Fu, L. Chen, D. R. Casimiro, M. E. Davies, R. K. Evans, Z. Q. Zhang, A. J. Simon, W. L. Trigona, S. A. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. S. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, V. R. Fernandez, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. C. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415:331-335. [DOI] [PubMed] [Google Scholar]
- 37.Surman, S., T. D. Lockey, K. S. Slobod, B. Jones, J. M. Riberdy, S. W. White, P. C. Doherty, and J. L. Hurwitz. 2001. Localization of CD4+ T cell epitope hotspots to exposed strands of HIV envelope glycoprotein suggests structural influences on antigen processing. Proc. Natl. Acad. Sci. USA 98:4587-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vidal, N., M. Peeters, C. Mulanga-Kabeya, N. Nzilambi, D. Robertson, W. Ilunga, H. Sema, K. Tshimanga, B. Bongo, and E. Delaporte. 2000. Unprecedented degree of human immunodeficiency virus type 1 (HIV-1) group M genetic diversity in the Democratic Republic of Congo suggests that the HIV-1 pandemic originated in Central Africa. J. Virol. 74:10498-10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yusim, K., C. Kesmir, B. Gaschen, M. M. Addo, M. Altfeld, S. Brunak, A. Chigaev, V. Detours, and B. T. Korber. 2002. Clustering patterns of cytotoxic T-lymphocyte epitopes in human immunodeficiency virus type 1 (HIV-1) proteins reveal imprints of immune evasion on HIV-1 global variation. J. Virol. 76:8757-8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhan, X., K. S. Slobod, S. Surman, S. A. Brown, T. D. Lockey, C. Coleclough, P. C. Doherty, and J. L. Hurwitz. 2003. Limited breadth of a T-helper cell response to a human immunodeficiency virus envelope protein. J. Virol. 77:4231-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.