Abstract
The majority of global human immunodeficiency virus infections are caused by viruses characterized by a GPGQ motif at the tip of the V3 loop. Characterization of anti-V3 monoclonal antibodies (MAbs) that neutralize isolates with the GPGQ V3 motif is an important step in designing vaccines that will induce such Abs. Consequently, seven human anti-V3 MAbs derived from the cells of individuals infected with non-B-subtype viruses (anti-V3non-B MAbs) were generated from the cells of individuals from Africa infected with circulating recombinant forms CRF02_AG, CRF09_cpx, and CRF13_cpx, each of which contains a subtype A env gene. Sequence analysis of plasma viruses revealed a GPGQ motif at the apex of the V3 loop from six of the seven subjects and a GPGR motif from one subject. The MAbs were selected with fusion proteins (FP) containing V392UG037.8 or V3JR-CSF from subtype A or B, respectively. In virus binding assays, five of the seven (71%) anti-V3non-B MAbs bound to V3-FPs from both subtype A and subtype B, while only four of the nine (44%) anti-V3B MAbs recognized both V3-FPs. Using two neutralization assays, both the anti-V3non-B and the anti-V3B MAbs neutralized subtype B viruses with similar activities, while the anti-V3non-B MAbs exhibited a tendency toward both increased potency and breadth of neutralization against non-B viruses compared to anti-V3B MAbs. Statistical significance was not achieved, due in large measure to the sizes of the MAb panels, but the overall pattern of data strongly suggests that viruses with the GPGQ motif at the tip of the V3 loop induce anti-V3 Abs with broader cross-neutralizing activity than do viruses with the GPGR motif.
During the past two decades, several epitopes that induce neutralizing antibodies (Abs) have been identified in the human immunodeficiency virus (HIV) envelope through studies of polyclonal and monoclonal Abs (MAbs). These epitopes include the V3 region defined with polyclonal Abs (30, 33) and several MAbs, such as 447-52D (16); the membrane-proximal external region in gp41 defined by MAbs 2F5 and 4E10 (6); the CD4-binding site on gp120 defined by MAb immunoglobulin G1b12 (IgG1b12) (7); and a glycan-rich region on gp120 defined by MAb 2G12 (37). With the exception of V3, none of these epitopes induce neutralizing Abs in the majority of infected humans. Thus, Abs to the membrane-proximal external region of gp41 (G. Shaw, H. Li, J. Decker, S. Allen, E. Hunter, E. Delaporte, M. Peters, B. Hahn, and F. Bibollet-Ruche, Abstr. AIDS Vaccine 2005, abstr. 29, 2005) (45), the CD4 binding site defined by IgG1b12 (25), and the designated carbohydrate moieties on gp120 (23, 37) are rare or absent from the sera of most HIV-infected individuals, and the epitope recognized by 2F5 (9, 11, 29) and the peptide mimotope for IgG1b12 (44) have failed to induce neutralizing Abs when used as experimental immunogens. Moreover, the recently described auto-reactive character of MAbs 2F5, 4E10, and IgG1b12, which recognize cardiolipin and/or double-stranded DNA, indicates that these epitopes may be problematic for the design of an anti-HIV vaccine (22). In contrast, the immunogenicity of the V3 region is reflected by the presence of anti-V3 Abs in the sera of essentially all HIV-infected individuals (38).
Opinions about the V3 loop as an antigen for the induction of neutralizing Abs have changed over time. Early optimism related to the ability of anti-V3 MAbs to neutralize T-cell-line-adapted viruses was replaced by skepticism when it was suggested that the V3 of primary isolate JR-FL was “cryptic” (5). More recent data suggest that V3 is accessible on the surfaces of most virions (31) and that anti-V3 MAbs, such as 447-52D, can neutralize 62 to 92% of primary isolates that carry the epitope for which V3 is specific (3, 43). Nonetheless, recent studies have shown that V3 is masked in many viruses by the V1/V2 region (32) and/or by carbohydrate moieties on the envelope (39), both of which may contribute to the resistance of primary isolates (26, 28). Moreover, it has been demonstrated in several studies that, despite the sequence variation in the V3 loop, many human anti-V3 Abs are cross-reactive (3, 17, 19, 21, 26, 42). For example, recent data show that anti-V3 MAbs derived from the cells of subtype B virus-infected individuals (anti-V3B MAbs) can neutralize various primary isolates from subtype B as well as additional viruses from subtypes A and F if they bear V3 loops containing the GPGR motif (3, 17, 19). This cross-neutralization may be explained by the biologic constraints placed on the V3 loop by the need for it to bind to chemokine receptors in order to mediate infectivity.
The vast majority (>85%) of HIV-1 infections worldwide are due to non-B-subtype viruses, the majority of which bear the GPGQ motif in their V3 loops, while <15% of HIV-1 infections are due to subtype B viruses bearing the GPGR motif (19, 26). However, most anti-V3 MAbs studied have been derived from subtype B virus-infected individuals. Analysis of these MAbs and anti-V3 Abs in the sera of patients infected with subtype B and non-B-subtype viruses suggests that there are differences between the binding and neutralizing activities of different viruses. It has been noted that anti-V3B MAbs poorly neutralize viruses with the GPGQ motifs; in contrast, anti-V3 Abs in the sera of patients infected with non-B-subtype viruses exhibit broader cross-reactivities to B and non-B V3-fusion proteins (FPs) than anti-V3 Abs in the sera of subtype B virus-infected subjects (19, 26). These observations suggest differences between the immunogenic characteristics of V3 loops carrying GPGQ and those carrying GPGR motifs.
Identifying immunogens that induce broad cross-reactivities across different subtype viruses or viruses that infect the majority of individuals worldwide is critical for vaccine design. To better understand the immunologic roles of the GPGQ and GPGR motifs on different viruses and the Abs they induce in infected individuals, we have generated anti-V3 MAbs induced by non-B subtype virus infections and studied their immunochemical properties in binding and neutralization assays.
MATERIALS AND METHODS
Monoclonal antibodies.
Seven human anti-V3 MAbs were developed from individuals infected with non-B-subtype HIV-1 using previously described cellular techniques (15, 20). Briefly, Epstein-Barr virus-transformed peripheral blood mononuclear cells reactive with V3-FPs were fused with the heteromyeloma SHM-D33 (36) and the resulting hybridomas cloned to monoclonality. These MAbs were selected using V3-FPs containing the fully glycosylated and folded V3 domains representing the sequence of either the subtype B primary isolate JR-CSF (V3B-FP) (24) or the subtype A primary virus 92UG037.8 (V3A-FP) (26). In addition, nine human anti-V3B MAbs produced in our laboratory from the cells of subtype B virus-infected individuals, 2191, 2219, 2412, 2442, 2456, 2483, 2497, 2580, and 447-52D, were used (16, 19). These MAbs, with the exception of 447-52D, were selected with V3JR-CSF-FP. MAb 447-52D was selected with the V3MN peptide. MAb IgG1b12 (7) and MAb 4E10 (6) were used as positive controls, while MAb 837 (anti-C2) (31) and human anti-parvovirus B19 MAb 1418 (14) served as negative controls.
Viruses.
Eleven primary HIV-1 isolates were studied. Seven primary viruses, Bx08, SF162, BaL (subtype B), 93MW960, 98CN006, 98CN009, and IN98022 (subtype C), were obtained from the NIH AIDS Research and Reference Reagent Program (ARRRP). One isolate, DJ263 (CRF02_AG), was obtained from the Vaccine Research Center, NIAID, NIH (provided by John R. Mascola). Three primary isolates, 03USVA36 and 02USNYU2775 from two individuals infected with subtype B viruses in the United States (1) and NYU129 from one individual infected in Cameroon with CRF02_AG, were isolated in our laboratory. Two viruses pseudotyped with the env genes of SF162 (subtype B) and MW965 (subtype C) were produced in our laboratory as previously described (19). The plasmid containing the env gene of MW965 was obtained from the Vaccine Research Center, NIAID, NIH (provided by John R. Mascola).
RNA extraction, RT-PCR, and phylogenetic analysis.
RNA extraction was performed on 100 μl of plasma as described by Boom et al. (4), followed by a single-tube reverse transcriptase (RT)-PCR (Access RT-PCR system, Promega, Madison, WI) to amplify the C2V5 region of env (40). The amplified products were sequenced and phylogenetically analyzed with reference subtype sequences to determine the virus subtypes as previously described (10, 41).
Binding assay.
A standard enzyme-linked immunosorbent assay (ELISA) was used to determine binding of MAbs to V3-FPs (18). Briefly, ELISA plates were coated overnight at 4°C with V3-FPs at 1.0 μg/ml, blocked with 2% bovine serum albumin in phosphate-buffered saline, and then incubated with human MAbs at a concentration 10.0 μg/ml for 1.5 h at 37°C. The plates were washed, and the bound MAbs were detected by incubation with alkaline phosphatase-conjugated goat anti-human IgG (γ specific) (Zymed, San Francisco, Calif.) for 1.5 h at 37°C. After the plates were washed, substrate was added for 30 min to develop color, and the plates were read at 410 nm.
The relative affinities of MAbs binding to V3-FPs were determined with ELISA by measuring the concentration of each MAb required to achieve 50% maximal binding at saturation, and relative affinities were assessed using MAbs at 0.003 to 10.0 μg/ml. The 50% maximal binding was obtained by linear interpolation.
Neutralization assays.
Two neutralization assays, the GHOST cell and luciferase neutralization assays, were used to test the abilities of the MAbs to neutralize viruses. The GHOST cell neutralization assay was performed as described previously, with minor modifications (8). Briefly, equal volumes of primary virus, at a dilution predetermined in earlier experiments to yield ∼1,000 infected cells per 15,000 cells, and MAb, diluted to yield a final concentration of 25 μg/ml, were incubated for 1 h at 37°C. Each virus/MAb mixture was added to GHOST-CD4+CCR5+ cells in the presence of 8.0 μg/ml DEAE-dextran, and incubation was continued for 3 days, after which the cells were harvested, fixed, and analyzed by flow cytometry (Becton Dickinson). Percent neutralization was calculated using the following formula: (1 − number of infected cells in the presence of MAb/number of infected cells in the absence of MAb) × 100. MAbs IgG1b12 and 4E10 were used in each experiment as positive controls, while MAb 847 served as a negative control. Significant neutralizing activity was determined as any value of >29%, the cutoff based on the 95% confidence level calculated from the fitted normal (Gaussian) distribution of data from 60 experiments using the nonneutralizing MAb 847.
A single-cycle infectivity assay was used to measure the neutralization of luciferase-encoding virions pseudotyped with the desired HIV-1 Env proteins, as previously described (19). Briefly, appropriate dilutions of the virion-containing culture supernatants were preincubated at 37°C for 1.5 h with MAbs at various concentrations. The virus-MAb mixtures were added to HOS-CD4+CCR5+ cells (NIH ARRRP and contributed by Dan Littman) and incubated for 3 days at 37°C. After the cells were washed, the relative light units in the cell lysates were determined on a Lumimark Plus System microplate reader (Bio-Rad Laboratories, Hercules, CA) with luciferase substrate (Promega, Madison, Wis.). The reduction of infectivity was determined by comparing the relative light units in the presence and absence of MAbs and expressed as percent neutralization.
Statistical analyses.
t tests and Fisher's exact test were performed with GraphPad, version 3.00, for Windows (GraphPad Software, San Diego, Calif.). Significance of difference in neutralizing activity against pseudotyped viruses between two panels of MAbs was determined by a new method borrowed from survival analysis. The Kaplan-Meier algorithm was used to estimate the cumulative distribution of neutralizing doses, where the information inherent in the number of MAbs that failed is accounted for. The algorithm then applies the Mantel-Haenszel test to compute the P value for the comparison. This calculation was done by function “survdiff” in the statistical language S-PLUS 7.0 (Insightful Corp.)
RESULTS
Generation of anti-V3non-B MAbs from individuals infected with non-B-subtype viruses.
Seven heterohybridomas producing anti-V3 MAbs were generated from Epstein-Barr virus-transformed peripheral blood mononuclear cells derived from seven different individuals infected with non-B-subtype viruses. Two V3-FPs were used for selection, each containing the V3 region of either subtype A HIV-192UG037.8 or subtype B HIV-1JR-CSF. The utility of this approach was shown previously by demonstrating that MAbs selected with V3-FPs are conformation sensitive and, in terms of neutralizing activity, are more efficient than those selected with linear V3 peptides (17, 19). Six of these MAbs were derived from individuals living in Cameroon, and one, MAb 2182 (previously described [19]), was derived from an individual infected in Côte d'Ivoire (Table 1). All MAbs belong to the IgG1 subclass with lambda light chains (Table 1).
TABLE 1.
Human anti-V3 MAbs generated from individuals infected with non-B-subtype HIV-1
| MAb | Isotype | Subtype of infecting virus | Country of origin |
|---|---|---|---|
| 2182 | IgG1λ | CRF02_AG | Côte d'Ivoire |
| 2557 | IgG1λ | CRF02_AG | Cameroon |
| 2558 | IgG1λ | CRF02_AG | Cameroon |
| 2601 | IgG1λ | CRF13_cpx | Cameroon |
| 3019 | IgG1λ | CRF02_AG | Cameroon |
| 3074 | IgG1λ | CRF02_AG | Cameroon |
| 3224 | IgG1λ | CRF09_cpx | Cameroon |
HIV-1 subtype analysis.
For phylogenetic analysis, viral C2V5 sequences that clustered with reference subtype sequences with bootstrap values of >70% were considered significant. The phylogenetic analysis of the C2V5 sequences revealed that five of the seven heterohybridomas were derived from the cells of HIV-1-positive subjects infected with CRF02_AG viruses. The remaining two were derived from subjects infected with CRF13_cpx and CRF09_cpx (Table 1). Each of these viruses contained the env gene of subtype A.
V3 sequence analysis.
The V3 amino acid sequence of each of the seven primary viruses was aligned with the respective subtype consensus sequence obtained from the Los Alamos Database (Fig. 1). Six V3 sequences contain the GPGQ motif at the tip of the V3 loop, while plasma virus from patient 00USNYU2182 (from whom MAb 2182 was derived) contains the GPGR motif. This GPGR motif appears in ∼4% of subtype A viruses (13). All seven V3 sequences have the same length, 35 amino acids, with no insertions or deletions.
FIG. 1.
V3 sequences of HIV-1 isolates obtained from donors of the anti-V3non-B MAbs. The amino acids were aligned with their CRF02_AG consensus sequence or with individual sequences of viruses from CRF09_cpx and CRF13_cpx (shown in bold) obtained from the Los Alamos Database. Dashes represent identity with the reference sequences.
Cross-reactivities of anti-V3non-B MAbs tested by ELISA.
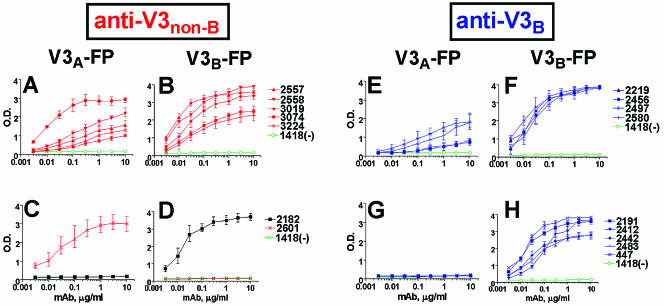
Two panels of MAbs, including seven anti-V3non-B and nine anti-V3B MAbs, were tested by ELISA to determine the binding patterns with V3non-B-FP and V3B-FP containing the GPGQ and GPGR motifs, respectively. Five of the seven (71%) anti-V3non-B MAbs reacted with both V3A-FP and V3B-FP (Fig. 2A and B), while only four of the nine (44%) anti-V3B MAbs showed binding to both V3-FPs (Fig. 2E and F). Given the small sample size, this difference in reactivity was not found to be statistically significant by the Fisher exact test. Subtype-specific reactivities were observed with only two of the seven (29%) anti-V3non-B MAbs (Fig. 2C and D), while five of the nine (56%) anti-V3B MAbs reacted only with V3B-FP (Fig. 2G and H). Interestingly, we note that the anti-V3non-B MAb 2182 generated from the cells of the HIV-1 CRF02_AG-infected patient whose virus carried the GPGR V3 motif (Fig. 1) reacted only with the V3B-FP containing the GPGR motif (Fig. 2C and D). The overall pattern of reactivity displayed in Fig. 2 suggests strongly that the V3 loop carrying the GPGR motif induces a less cross-reactive Ab response than does the V3 loop carrying the GPGQ motif.
FIG. 2.
Binding patterns of V3-fusion proteins with anti-V3 MAbs derived from individuals infected with non-B-subtype (panels A to D) and subtype B (panels E to H) viruses. Seven human anti-V3non-B MAbs generated from the cells of individuals infected with non-B-subtype viruses (A to D) and nine human anti-V3B MAbs from the cells of subtype B virus-infected subjects (E to H) were tested for their abilities to bind to V3-fusion proteins containing a subtype A V3 sequence (V3A-FP) (A, C, E, G) and to a subtype B V3 sequence (V3B-FP) (B, D, F, H). Human anti-parvovirus B19 MAb 1418 was used as negative control. The curves represent the mean binding activities from three separate experiments, and error bars indicate the standard deviations. The binding curves for anti-V3non-B MAbs are shown in red, with the exception of the curves generated with MAb 2182, which are shown in black (C and D); curves generated with anti-V3B MAbs are shown in blue.
The 50% maximal binding concentrations (half-max) for the five anti-V3non-B and four anti-V3B MAbs that bound both V3-FPs were not statistically different. Calculated from data shown in Fig. 2A and E, the average half-max for anti-V3non-B MAbs binding to V3A-FP was 0.16 ± 0.11 μg/ml and the average half-max for anti-V3B MAbs binding to V3A-FP was 0.27 ± 0.12 μg/ml (P = 0.27). Similarly, calculated from data shown in Fig. 2 B and F, the average half-max for anti-V3non-B MAbs reacting with V3B-FP was 0.022 ± 0.01 μg/ml and the average half-max for anti-V3B MAbs was 0.018 ± 0.006 μg/ml (P = 0.64). Moreover, there was no significant difference in half-max values for binding to V3B-FP between the four anti-V3B MAbs that react with both V3-FPs (0.018 ± 0.006 μg/ml) (Fig. 2F) and the five V3B MAbs that bind to V3B- but not V3A-FP (0.039 ± 0.03 μg/ml; P = 0.25) (Fig. 2H). These results suggest that cross-reactivity among anti-V3 MAbs is due to specificity differences and is independent of their binding affinities.
Neutralization of pseudotyped viruses.
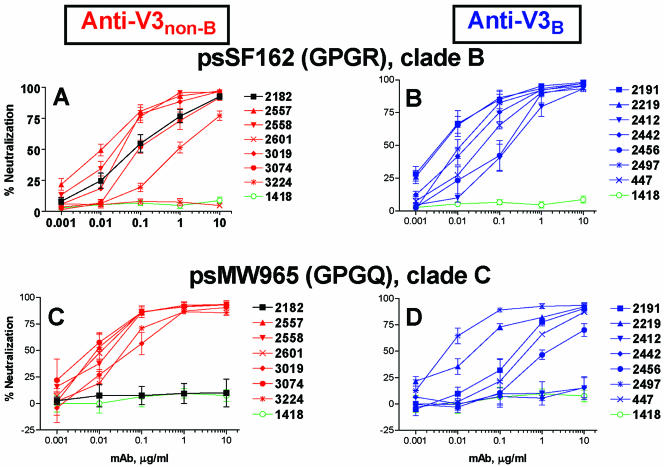
The neutralizing activities of seven anti-V3non-B MAbs were compared with those of seven anti-V3B MAbs in assays using luciferase-expressing pseudotyped viruses carrying Env from either subtype B SF162 (psSF162) or subtype C MW965 (psMW965). All MAbs were tested at concentrations ranging from 0.001 to 10 μg/ml. Overall, both anti-V3B and anti-V3non-B MAbs displayed similar neutralizing activities against psSF162, which bears the V3 GPGR motif (Fig. 3A and B). Thirteen of the 14 MAbs exhibited 50% neutralizing doses (ND50s) against psSF162 at <1.0 μg/ml; however, MAb 2601 (anti-V3non-B) did not neutralize psSF162 (Fig. 3A).
FIG. 3.
Neutralization patterns of pseudotype viruses with anti-V3 MAbs derived from subjects infected with non-B-subtype (panels A and C) and subtype B (panels B and D) viruses. The neutralization capacities of seven anti-V3non-B MAbs and seven anti-V3B MAbs with psSF162 (bearing the GPGR motif) and psMW965 (bearing the GPGQ motif) were tested in a single-cycle infectivity assay. Human anti-parvovirus B19 MAb 1418 was used as a negative control. The neutralizing curves represent the means from three experiments, and error bars indicate standard deviations. The curves representing neutralizing activities for anti-V3non-B MAbs are shown in red, and those for anti-V3B MAbs are shown in blue. Data for the negative control MAb 1418 are shown in green, and data for the MAb 2182 are shown in black.
In contrast, psMW965, which bears the GPGQ V3 motif, was neutralized more efficiently by anti-V3non-B than by anti-V3B MAbs (Fig. 3C and D). Six of the seven anti-V3non-B MAbs achieved an ND50 at <0.1 μg/ml against psMW965, while only two of the seven anti-V3B MAbs reached this level of activity (Fig. 3C and D). There was a clear difference found in this pattern of neutralization when these two groups of MAbs were analyzed for ND85 values (P = 0.04), but the difference was not found to be significant when they were analyzed for ND50 values. Statistical significance was evaluated by the Mantel-Haenszel test applied to Kaplan-Meier estimates of the distributions (see Materials and Methods).
Of note again is MAb 2182, the anti-V3non-B MAb generated from cells of the CRF02_AG-infected patient whose virus bears the GPGR motif. The neutralization pattern of this MAb resembled those of MAbs induced by subtype B viruses bearing the GPGR motif in that MAb 2182 could neutralize subtype B psSF162 (Fig. 3A) but not subtype C psMW965 (Fig. 3C).
Neutralization of primary isolates.
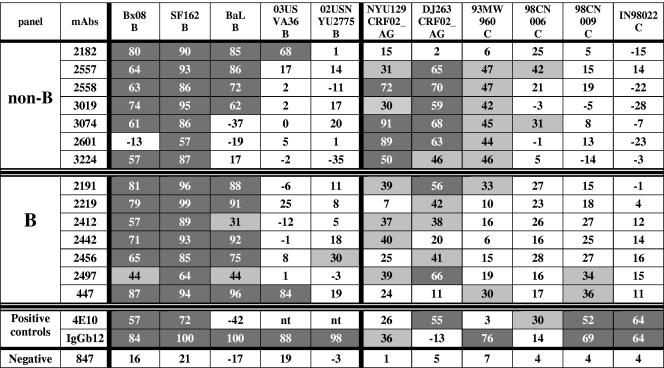
Using the GHOST cell neutralization assay, the neutralizing activity of anti-V3non-B and anti-V3B MAbs were tested at 25 μg/ml against five subtype B primary isolates (bearing the GPGR V3 motif) and against two CRF02_AG and four subtype C isolates (bearing the GPGQ V3 motif). Four anti-V3non-B MAbs, 2182, 2557, 2558, and 3019, neutralized three subtype B viruses (Bx08, SF162, and BaL) with similar potencies, as did the anti-V3B MAbs. The three remaining anti-V3non-B MAbs, 3074, 2601, and 3224, neutralized only one or two of these three subtype B isolates. (Fig. 4). Two subtype B viruses (03USVA36 and 02USNYU2775) were resistant to neutralization by both categories of MAbs, with the exception of 2182 (anti-V3non-B) and 447 (anti-V3B), which neutralized 03USVA36, and MAb 2456 (anti-V3B), which only weakly neutralized 02USNYU2775 (Fig. 4). These data were consistent with those above, suggesting that there was little or no difference between the abilities of anti-V3B and anti-V3non-B MAbs to react with subtype B viruses. Here, the percentage of subtype B virus/MAb combinations which showed neutralizing activities was slightly higher for anti-V3B MAbs (63%) than for anti-V3non-B MAbs (51%), but the difference was not significant as determined by the Fisher exact test.
FIG. 4.
Neutralization of primary isolates by anti-V3 MAbs derived from individuals infected with non-B- and B-subtype viruses. Neutralization of primary isolates was performed in the GHOST cell neutralization assay, with MAbs at a final concentration of 25 μg/ml. The cutoff value of 29% is based on the 95% confidence level obtained with 60 experiments using the nonneutralizing human anti-C2 MAb 847. The percentages represent the means from three separate experiments. Light gray cells indicate neutralization in excess of the cutoff value; dark gray cells indicate virus/MAb combinations giving >50% neutralization. nt, not tested.
The two panels of anti-V3 MAbs were also tested against six low-passage, non-B subtype primary isolates, including four subtype C and two CRF02_AG viruses. Overall, the anti-V3non-B MAbs more efficiently neutralized three non-B viruses (NYU129, DJ263, and 93MW960) than did the anti-V3B MAbs: 20 of the 42 (48%) non-B virus/anti-V3non-B MAb test combinations showed significant neutralization; nine of these combinations neutralized virus at >50%. In contrast, only 13 of the 42 (31%) non-B virus/anti-V3B MAb combinations displayed significant neutralizing activity, of which only two combinations achieved >50% neutralization (Fig. 4). While the difference in neutralizing activity between both panels was not found to be significant when measured by the Fisher exact test, it is again noteworthy that the anti-V3non-B MAb 2182, derived from the individual infected with a CRF02-AG virus bearing the GPGR V3 motif, did not neutralize primary isolates carrying the GPGQ motif but could neutralize the subtype B viruses carrying the GPGR motif (Fig. 4). Thus, the overall pattern of primary isolate neutralization was consistent with the results of pseudovirus neutralization and virus binding, suggesting that viruses bearing the GPGQ V3 motif induce an Ab response of broader cross-reactivity than that induced by viruses bearing the GPGR V3 motif.
DISCUSSION
Three types of experiments were used to study anti-V3 MAbs from subjects infected with subtype B or non-B strains of HIV: (i) binding of MAbs to V3A- versus V3B-FPs, (ii) neutralization of pseudotyped viruses bearing the Env proteins of viruses from either subtype B or subtype C, and (iii) neutralization of primary isolates from subtype B, C, or CRF02_AG. The data from all three sets of experiments suggest that anti-V3non-B MAbs derived from subjects infected with non-B subtype viruses carrying the GPGQ V3 motif exhibit broader reactivities than anti-V3B MAbs from subjects infected with subtype B viruses carrying the GPGR V3 motif. Our present findings corroborate those from studies of polyclonal anti-V3 serum Abs from HIV-infected subjects living in Cameroon, where GPGQ viruses predominate, showing broader cross-reactivities than did polyclonal anti-V3 serum Abs from HIV-infected subjects living in North America, where GPGR viruses predominate (26). Taken together, these studies suggest that the immunogenic properties of V3 loops carrying the GPGQ V3 motif can be different from those bearing the GPGR V3 motif.
The critical immunogenic role of the motif at the tip of the V3 loop is further strengthened by the pattern of reactivity exhibited by the MAb 2182 derived from the cells of a subject from Côte d'Ivoire who was infected with a CRF02_AG virus bearing the GPGR V3 motif (Fig. 1). Thus, while being classified as an “anti-V3non-B MAb” and included as such in all statistical analyses, MAb 2182 was nonetheless induced by a “GPGR virus” and displayed the characteristics of “anti-V3B MAbs” stimulated by GPGR viruses. This MAb bound to V3B-FP but not to V3A-FP (Fig. 2C and D), neutralized a pseudovirus bearing a subtype B envelope with a GPGR V3 motif but not pseudoviruses bearing the GPGQ V3 motif (Fig. 3A and C), and neutralized four of the five subtype B primary isolates but did not neutralize any of the CRF02_AG or subtype C primary isolates (Fig. 4). These results were confirmed recently, when it was shown that MAb 2182 was strongly dependent on the presence of the Arg (R) residue in the GPGR motif and was unable to neutralize chimeric viruses containing the subtype B consensus variants in which the R was replaced by Gln (Q) or Lys (K) (32a). The MAb 2182 therefore serves as an internal control, confirming that the critical feature determining V3 immunogenicity is not the genotype to which the envelope maps but the nature of the V3 motif it possesses.
The structure of a subtype B V3 loop to which anti-V3B MAb 447 binds has been obtained by both nuclear magnetic resonance and crystallographic studies (34, 35). It was shown that the Arg (R) residue present in the GPGR motif plays a critical role in determining Ab specificity. This observation was confirmed by neutralization studies showing that MAb 447 could neutralize GPGR viruses from subtypes A, B, F, and H but not GPGQ viruses or viruses with other “non-GPGR” sequences at the tip of the V3 loop (3, 43). The present data support our previous studies showing that specific motifs at the tip of the V3 loop are critical in inducing Abs with characteristic properties.
Molecular modeling based on the crystallographic studies of the V3/MAb 447 complex also helps to explain the differential immunogenicities of V3 loops characterized by the GPGR and GPGQ motifs. Preliminary modeling of GPGR and GPGQ V3 loops suggest that these two motifs may present different patterns of surface charges, with the GPGR motif concentrating surface charges at the tip of the loop, while in the GPGQ motif, the electrostatic charges are spread over the length of the N-terminal β strand of V3 (T. Cardozo and S. Zolla-Pazner, unpublished data). Since charge often plays a critical role in defining B-cell epitopes, different patterns of surface charge between GPGR and GPGQ viruses could strongly affect the immunogenicities of these two categories of V3 and the resulting anti-V3 Abs they induce. Structural studies and molecular modeling also illuminate the patterns of reactivity of the anti-V3 Abs induced by GPGR and GPGQ viruses. Thus, when the Arg (R) residue of GPGR is replaced with a Gln (Q) residue, Gln fits into the charged pocket of the combining site of MAb 447, which is normally occupied by the Arg residue (M. Schapira and S. Zolla-Pazner, unpublished data). However, the Gln residue cannot form the salt bridge and cation π interactions that the Arg residue forms with critical residues in the binding site of MAb 447 (35).
The present study has examined the specificities and neutralizing activities of anti-V3 MAbs from human volunteers infected with various strains of subtype B and non-B viruses. The data suggest that viruses bearing the GPGQ V3 motif induce anti-V3 Abs with broader immunologic activity than do viruses bearing the GPGR motif. Given that >85% of global HIV infections are caused by non-B subtype viruses, most of which carry a V3 loop with the GPGQ motif at the tip, while the remaining ∼15% HIV infections, caused by subtype B viruses, carry the GPGR motif, and given that anti-V3 Abs may be an important component in protection against HIV infection (2, 12, 27), these data suggest that if a monovalent vaccine is used, it should preferentially include a V3 region containing the GPGQ V3 motif. A polyvalent vaccine would benefit from a combination of immunogens representing viruses with the GPGQ and the GPGR motifs.
Acknowledgments
This study was supported in part by NIH grants HL59725, AI36085, and AI47053, by the Immunology Core of the NYU Center for AIDS Research (NIH grant AI27742), by research funds from Department of Veterans Affairs, and by Fogarty International Center Training grant TW01409.
We are grateful to Dennis R. Burton of the Scripps Research Institute, La Jolla, CA, for providing MAb IgG1b12 and to Hermann Katinger of the Institute of Applied Microbiology, Vienna, Austria, for MAb 4E10.
REFERENCES
- 1.Achkar, J. M., S. Burda, F. Konings, M. Urbanski, C. Williams, D. Seifen, M. N. Kahirimbanyi, M. Vogler, M. Parta, H. C. Lupatkin, S. Zolla-Pazner, and P. N. Nyambi. 2004. Infection with HIV Type 1 group M non-B subtypes in individuals living in New York City. J. Acquir. Immune Defic. Syndr. 36:835-844. [DOI] [PubMed] [Google Scholar]
- 2.Andrus, L., A. M. Prince, I. Bernal, P. McCormack, D. H. Lee, M. K. Gorny, and S. Zolla-Pazner. 1998. Passive immunization with a human immunodeficiency virus type-1 neutralizing monoclonal antibody in Hu-PBL-SCID mice: isolation of a neutralization escape variant. J. Infect. Dis. 177:889-897. [DOI] [PubMed] [Google Scholar]
- 3.Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bou-Habib, D. C., G. Roderiquez, T. Oravecz, P. W. Berman, P. Lusso, and M. A. Norcross. 1994. Cryptic nature of envelope V3 region epitopes protects primary monocytotropic human immunodeficiency virus type 1 from antibody neutralization. J. Virol. 68:6006-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchacher, A., R. Predl, K. Strutzenberger, W. Steinfellner, A. Trkola, M. Purtscher, G. Gruber, C. Tauer, F. Steindl, A. Jungbauer, and H. Katinger. 1994. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retrovir. 10:359-369. [DOI] [PubMed] [Google Scholar]
- 7.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. Parren, L. S. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 8.Cecilia, D., V. N. KewalRamani, J. O'Leary, B. Volsky, P. N. Nyambi, S. Burda, S. Xu, D. R. Littman, and S. Zolla-Pazner. 1998. Neutralization profiles of primary human immunodeficiency virus type 1 isolates in the context of coreceptor usage. J. Virol. 72:6988-6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coeffier, E., J. M. Clement, V. Cussac, N. Khodaei-Boorane, M. Jehanno, M. Rojas, A. Dridi, M. Latour, R. El Habib, F. Barre-Sinoussi, M. Hofnung, and C. Leclerc. 2000. Antigenicity and immunogenicity of the HIV-1 gp41 epitope ELDKWA inserted into permissive sites of the MalE protein. Vaccine 19:684-693. [DOI] [PubMed] [Google Scholar]
- 10.Delwart, E. L., E. G. Shpaer, J. Louwagie, F. E. McCutchan, M. Grez, H. Rubsamen-Waigmann, and J. I. Mullins. 1993. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science 262:1257-1261. [DOI] [PubMed] [Google Scholar]
- 11.Eckhart, L., W. Raffelsberger, B. Ferko, A. Klima, M. Purtscher, H. Katinger, and F. Ruker. 1996. Immunogenic presentation of a conserved gp41 epitope of human immunodeficiency virus type 1 on recombinant surface antigen of hepatitis B virus. J. Gen. Virol. 77:2001-2008. [DOI] [PubMed] [Google Scholar]
- 12.Emini, E. A., W. A. Schleif, J. H. Nunberg, A. J. Conley, Y. Eda, S. Tokiyoshi, S. D. Putney, S. Matsushita, K. E. Cobb, C. M. Jett, J. W. Eichberg, and K. K. Murthy. 1992. Prevention of HIV-1 infection in chimpanzees by gp120 V3 domain-specific monoclonal antibody. Nature 355:728-730. [DOI] [PubMed] [Google Scholar]
- 13.Gaschen, B., B. T. Korber, and D. T. Foley. 1999. Global variation in the HIV-1 V3 region, p. 594-602. In C. Kuiken, B. Foley, B. H. Hahn, P. Marx, F. McCutchan, J. W. Mellors, J. Mullins, S. Wolinsky, and B. Korber (ed.), Human retroviruses and AIDS 1999. Los Alamos National Library, Los Alamos, N. Mex.
- 14.Gigler, A., S. Dorsch, A. Hemauer, C. Williams, S. Kim, N. S. Young, S. Zolla-Pazner, H. Wolf, M. K. Gorny, and S. Modrow. 1999. Generation of neutralizing human monoclonal antibodies against parvovirus B19 proteins. J. Virol. 73:1974-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorny, M. K. 1994. Production of human monoclonal antibodies via fusion of Epstein-Barr virus-transformed lymphocytes with heteromyeloma, p. 276-281. In J. E. Celis (ed.), Cell biology: a laboratory handbook, vol. 2. Academic Press, San Diego, Calif. [Google Scholar]
- 16.Gorny, M. K., A. J. Conley, S. Karwowska, A. Buchbinder, J. Y. Xu, E. A. Emini, S. Koenig, and S. Zolla-Pazner. 1992. Neutralization of diverse HIV-1 variants by an anti-V3 human monoclonal antibody. J. Virol. 66:7538-7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorny, M. K., K. Revesz, C. Williams, B. Volsky, M. K. Louder, C. A. Anyangwe, C. P. Krachmarov, S. C. Kayman, A. Pinter, A. Nadas, P. N. Nyambi, J. R. Mascola, and S. Zolla-Pazner. 2004. The V3 loop is accessible on the surface of most human immunodeficiency virus type 1 primary isolates and serves as a neutralization epitope. J. Virol. 78:2394-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorny, M. K., T. C. VanCott, C. Hioe, Z. R. Israel, N. L. Michael, A. J. Conley, C. Williams, J. A. Kessler II, P. Chigurupati, S. Burda, and S. Zolla-Pazner. 1997. Human monoclonal antibodies to the V3 loop of HIV-1 with intra- and interclade cross-reactivity. J. Immunol. 159:5114-5122. [PubMed] [Google Scholar]
- 19.Gorny, M. K., C. Williams, B. Volsky, K. Revesz, S. Cohen, V. R. Polonis, W. J. Honnen, S. C. Kayman, C. P. Krachmarov, A. Pinter, and S. Zolla-Pazner. 2002. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize HIV-1 primary isolates from various clades. J. Virol. 76:9035-9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorny, M. K., J.-Y. Xu, V. Gianakakos, S. Karwowska, C. Williams, H. W. Sheppard, C. V. Hanson, and S. Zolla-Pazner. 1991. Production of site-selected neutralizing human monoclonal antibodies against the third variable domain of the HIV-1 envelope glycoprotein. Proc. Natl. Acad. Sci. USA 88:3238-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorny, M. K., J.-Y. Xu, S. Karwowska, A. Buchbinder, and S. Zolla-Pazner. 1993. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J. Immunol. 150:635-643. [PubMed] [Google Scholar]
- 22.Haynes, B. F., J. Fleming, W. E. St. Clair, H. Katinger, G. Stiegler, R. Kunert, J. Robinson, R. M. Scearce, K. Plonk, H. F. Staats, T. L. Ortel, H. X. Liao, and M. S. Alam. 2005. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 308:1906-1908. [DOI] [PubMed] [Google Scholar]
- 23.Jeffs, S. A., C. Shotton, P. Balfe, and J. A. McKeating. 2002. Truncated gp120 envelope glycoprotein of human immunodeficiency virus 1 elicits a broadly reactive neutralizing immune response. J. Gen. Virol. 83:2723-2732. [DOI] [PubMed] [Google Scholar]
- 24.Kayman, S. C., Z. Wu, K. Revesz, H. Chen, R. Kopelman, and A. Pinter. 1994. Presentation of native epitopes in the V1/V2 and V3 regions of human immunodeficiency virus type 1 gp120 by fusion glycoproteins containing isolated gp120 domains. J. Virol. 68:400-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kessler, J. A. II, P. M. McKenna, E. A. Emini, C. P. Chan, M. D. Patel, S. K. Gupta, G. E. Mark III, C. F. Barbas III, D. R. Burton, and A. J. Conley. 1997. Recombinant human monoclonal antibody IgG1b12 neutralizes diverse human immunodeficiency virus type 1 primary isolates. AIDS Res. Hum. Retrovir. 13:575-582. [DOI] [PubMed] [Google Scholar]
- 26.Krachmarov, C., A. Pinter, W. J. Honnen, M. K. Gorny, P. N. Nyambi, S. Zolla-Pazner, and S. C. Kayman. 2005. Antibodies that are cross-reactive for human immunodeficiency virus type 1 clade A and clade B V3 domains are common in patient sera from Cameroon, but their neutralization activity is usually restricted by epitope masking. J. Virol. 79:780-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Letvin, N. L., S. Robinson, D. Rohne, M. K. Axthelm, J. W. Fanton, M. Bilska, T. J. Palker, H. X. Liao, B. F. Haynes, and D. C. Montefiori. 2001. Vaccine-elicited V3 loop-specific antibodies in rhesus monkeys and control of a simian-human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate envelope. J. Virol. 75:4165-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, M., F. Gao, J. R. Mascola, L. Stamatatos, V. R. Polonis, M. Koutsoukos, G. Voss, P. Goepfert, P. Gilbert, K. M. Greene, M. Bilska, D. L. Kothe, J. F. Salazar-Gonzalez, X. Wei, J. M. Decker, B. H. Hahn, and D. C. Montefiori. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108-10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang, X., S. Munshi, J. Shendure, G. Mark III, M. E. Davies, D. C. Freed, D. C. Montefiori, and J. W. Shiver. 1999. Epitope insertion into variable loops of HIV-1 gp120 as a potential means to improve immunogenicity of viral envelope protein. Vaccine 17:2862-2872. [DOI] [PubMed] [Google Scholar]
- 30.Matthews, T. J., A. J. Langlois, W. G. Robey, N. T. Chang, R. C. Gallo, P. J. Fischinger, and D. P. Bolognesi. 1986. Restricted neutralization of divergent human T-lymphotropic virus type III isolates by antibodies to the major envelope glycoprotein. Proc. Natl. Acad. Sci. USA 83:9709-9713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nyambi, P. N., H. A. Mbah, S. Burda, C. Williams, M. K. Gorny, A. Nadas, and S. Zolla-Pazner. 2000. Conserved and exposed epitopes on intact, native, primary human immunodeficiency virus type 1 virions of group M. J. Virol. 74:7096-7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pinter, A., W. J. Honnen, Y. He, M. K. Gorny, S. Zolla-Pazner, and S. C. Kayman. 2004. The V1/V2 domain of gp120 is a global regulator of sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 78:5205-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.Pinter, A., C. P. Krachmarov, W. J. Honnen, S. C. Kayman, M. K. Gorny, and S. Zolla-Pazner. 2006. Factors determining the breadth and potency of neutralization by V3-specific human monoclonal antibodies derived from subjects infected with clade A or clade B strains of human immunodeficiency virus type 1. J. Virol. 80:7273-7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rusche, J. R., K. Javaherian, C. McDanal, J. Petro, D. L. Lynn, R. Grimaila, A. Langlois, R. C. Gallo, L. O. Arthur, P. J. Fischinger, D. P. Bolognesi, S. D. Putney, and T. J. Matthews. 1988. Antibodies that inhibit fusion of human immunodeficiency virus-infected cells bind a 24-amino acid sequence of the viral envelope, gp120. Proc. Natl. Acad. Sci. USA 85:3198-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharon, M., N. Kessler, R. Levy, S. Zolla-Pazner, M. Gorlach, and J. Anglister. 2003. Alternative conformations of HIV-1 V3 loops mimic Β hairpins in chemokines, suggesting a mechanism for coreceptor selectivity. Structure 11:225-236. [DOI] [PubMed] [Google Scholar]
- 35.Stanfield, R. L., M. K. Gorny, C. Williams, S. Zolla-Pazner, and I. A. Wilson. 2004. Structural rationale for the broad neutralization of HIV-1 by human antibody 447-52D. Structure 12:193-204. [DOI] [PubMed] [Google Scholar]
- 36.Teng, N. N., K. S. Lam, F. Calvo Riera, and H. S. Kaplan. 1983. Construction and testing of mouse-human heteromyelomas for human monoclonal antibody production. Proc. Natl. Acad. Sci. USA 80:7308-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogel, T., R. Kurth, and S. Norley. 1994. The majority of neutralizing Abs in HIV-1-infected patients recognize linear V3 loop sequences. Studies using HIV-1MN multiple antigenic peptides. J. Immunol. 153:1895-1904. [PubMed] [Google Scholar]
- 39.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 40.Zhong, P., S. Burda, F. Konings, M. Urbanski, L. Ma, L. Zekeng, L. Ewane, L. Agyingi, M. Agwara, D. Saa, Z. E. Afane, T. Kinge, S. Zolla-Pazner, and P. Nyambi. 2003. Genetic and biologic properties of HIV type 1 isolates prevalent in villagers of the Cameroon equatorial rain forests and grass fields: Further evidence of broad HIV-1 genetic diversity. AIDS Res. Hum. Rertrovir. 19:1167-1178. [DOI] [PubMed] [Google Scholar]
- 41.Zhong, P., S. Burda, M. Urbanski, H. Kenfack, M. Tongo, L. Heyndrickx, A. Nanfack, J. Shang, L. Agyingi, S. Zolla-Pazner, L. Zekeng, and P. Nyambi. 2002. HIV type 1 group M clades infecting subjects from rural villages in equatorial rain forests of Cameroon. J. Acquir. Immune Defic. Syndr. 31:495-505. [DOI] [PubMed] [Google Scholar]
- 42.Zolla-Pazner, S., M. K. Gorny, P. N. Nyambi, T. C. VanCott, and A. Nadas. 1999. Immunotyping of human immunodeficiency virus type 1 (HIV): an approach to immunologic classification of HIV. J. Virol. 73:4042-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zolla-Pazner, S., P. Zhong, K. Revesz, B. Volsky, C. Williams, P. Nyambi, and M. K. Gorny. 2004. The cross-clade neutralizing activity of a human monoclonal antibody is determined by the GPGR V3 motif of HIV Type 1. AIDS Res. Hum. Rertrovir. 20:1254-1258. [DOI] [PubMed] [Google Scholar]
- 44.Zwick, M. B., L. L. Bonnycastle, A. Menendez, M. B. Irving, C. F. Barbas III, P. W. Parren, D. R. Burton, and J. K. Scott. 2001. Identification and characterization of a peptide that specifically binds the human, broadly neutralizing anti-human immunodeficiency virus type 1 antibody b12. J. Virol. 75:6692-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]