Abstract
Cystathionine γ-synthase (CGS) is a key enzyme of Met biosynthesis in bacteria and plants. Aligning the amino acid sequences revealed that the plant enzyme has an extended N-terminal region that is not found in the bacterial enzyme. However, this region is not essential for the catalytic activity of this enzyme, as deduced from the complementation test of an Escherichia coli CGS mutant. To determine the function of this N-terminal region, we overexpressed full-length Arabidopsis CGS and its truncated version that lacks the N-terminal region in transgenic tobacco (Nicotiana tabacum) plants. Transgenic plants expressing both types of CGS had a significant higher level of Met, S-methyl-Met, and Met content in their proteins. However, although plants expressing full-length CGS showed the same phenotype and developmental pattern as wild-type plants, those expressing the truncated CGS showed a severely abnormal phenotype. These abnormal plants also emitted high levels of Met catabolic products, dimethyl sulfide and carbon disulfide. The level of ethylene, the Met-derived hormone, was 40 times higher than in wild-type plants. Since the alien CGS was expressed at comparable levels in both types of transgenic plants, we further suggest that post-translational modification(s) occurs in this N-terminal region, which regulate CGS and/or Met metabolism. More specifically, since the absence of the N-terminal region leads to an impaired Met metabolism, the results further suggest that this region plays a role in protecting plants from a high level of Met catabolic products such as ethylene.
The sulfur-containing amino acid Met is an important essential amino acid in animal nutrition. Apart from its role as a protein constituent and its central function in initiating mRNA translation, Met indirectly regulates a variety of cellular processes as the precursor of S-adenosyl-Met (SAM), the primary biological methyl group donor. SAM is also the precursor of plant metabolites such as ethylene, polyamines, vitamin B1, and the Fe-chelator mugineic acid (Anderson, 1990; Ma et al., 1995; Sun, 1998). In addition, Met also serves as a donor for secondary metabolites through S-methyl-Met (SMM; Mudd and Datko, 1990). As can be expected of its cellular importance, Met biosynthesis is subject to complex regulatory control whose mechanism is only now being gradually clarified. Two main elements of this complex regulation have recently been elucidated in plants. In the first, the Met level is controlled by competition between its first specific enzyme, cystathionine γ-synthase (CGS), and Thr synthase, for their common substrate, O-phosphohomo-Ser. Evidence of this competition and its role in Met synthesis was recently obtained by analyzing a mto2-1 mutant of Arabidopsis. This mutant, in which the gene encoding Thr synthase is impaired, demonstrated a approximately 22-fold higher accumulation of soluble Met in rosette leaves than wild-type Arabidopsis (Bartlem et al., 2000).
A second regulatory mechanism of Met synthesis in plants occurs at the level of CGS mRNA and/or protein. Studies conducted with Lemna paucicostata suggest that Met regulates its own synthesis through feedback control of cystathionine synthesis (Datko and Mudd, 1982; Thompson et al., 1983). However, this feedback control is most likely due to the repression of CGS synthesis rather than to the sensitivity of this enzyme to feedback inhibition by Met or its metabolites (Thompson et al., 1983). A mechanism by which Met regulates the CGS level was recently reported through the analysis of mto1 mutants of Arabidopsis, which accumulate up to 40 times more free Met than wild-type plants (Inba et al., 1994). In the mto1 mutant, the steady-state levels of CGS mRNA, protein, and hence enzyme activity are three to five times higher than in wild-type plants (Chiba et al., 1999). The application of Met to wild-type plants reduced the amount of CGS mRNA, whereas no such effect was observed in the mto1-1 mutant. This suggests that the wild-type plant down-regulates the CGS mRNA level in response to exogenous Met or to one of its metabolites, and that this regulation is impaired in the mutant (Chiba et al., 1999).
Plant CGSs possess an N-terminal region that is not present in bacterial CGSs. To elucidate the function of this “plant-specific” region, we overexpressed full-length Arabidopsis CGS in transgenic tobacco (Nicotiana tabacum) plants and its deleted version lacking the N-terminal region of this enzyme. We found that transgenic plants overexpressing the deleted version, but not the wild-type CGS, possess a severe abnormal phenotype and significantly overaccumulate ethylene and other volatile Met catabolic products, suggesting that the N-terminal region of CGS plays an important regulatory role in Met metabolism. Moreover, since no differences were observed in the CGS protein levels between plants expressing these two types of CGS constructs, we further suggest that this function of the N-terminal region of CGS operates at the post-translational level.
RESULTS
Plant CGSs Contained an Extended N-Terminal Region in Comparison with the Bacterial Enzyme
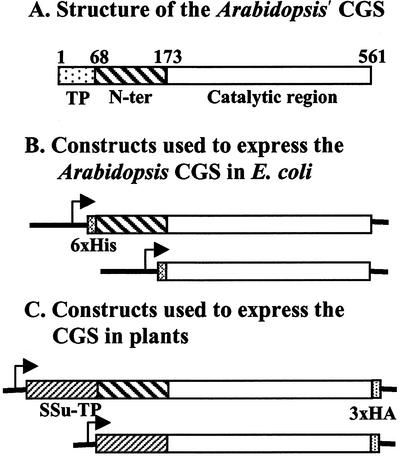
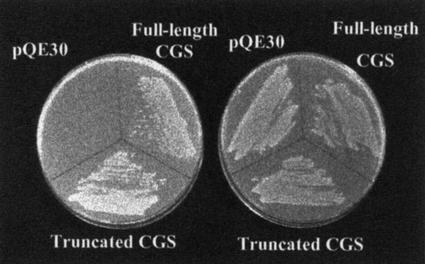
A computer alignment of bacterial and plant CGS protein sequences showed that mature plant CGS enzymes (after removing the plastid transit peptide) contain an N-terminal region of approximately 105 amino acids that is not present in bacterial enzymes. Bacterial CGSs share a high homology only with approximately 390 amino acids of the C terminus of the plant CGSs (Fig. 1A). Since the bacterial and plant CGS genes encode active enzymes, we first wanted to confirm that the N-terminal domain of mature plant CGSs is not essential for their catalytic CGS activity. To this end, we cloned Arabidopsis CGS without its transit peptide and its truncated version lacking the N terminus region (Fig. 1B) into a bacterial expression vector. The resulted constructs were introduced into the Escherichia coli strain, metB, lacking CGS activity (Fig. 2). Both CGS constructs complemented this mutant (Fig. 2), showing that both possess CGS activity, confirming that the N-terminal domain of the mature plant CGS is not essential for its catalytic activity. These results extend the data of a previous report showing that deletion of part of the plant-specific N-terminal region of the mature Arabidopsis CGS enables complementation of an E. coli metB mutant (Kim and Leustek, 1996).
Figure 1.
A, Schematic presentation of Arabidopsis CGS protein. The amino acids are numbered from the first Met of the protein in the transit peptide. TP, the chloroplast-targeting transit peptide, which directs the protein into the chloroplast and is then removed. N-ter, the N-terminal region of CGS (following removal of the transit peptide), which shares no homology to CGSs of bacteria. Catalytic region, the part of CGS sharing a high homology to bacteria CGSs and harboring the catalytic site. B, The constructs used to express the full-length or truncated Arabidopsis CGS in bacteria. A six-His tag was added at the N terminus to the constructs of the bacteria expressed proteins. C, The constructs used to express the full-length or truncated Arabidopsis CGS in plants. The transit peptide of the Rubisco small subunit 3A of pea (Ssu-TP) was used instead of the endogenous one. The sequence encoding three hemagglutinins (3xHA) was added in frame at the C terminus of the protein. Transcription promoters are symbolized by arrows.
Figure 2.
Functional complementation of CGS-deficient E. coli mutant LE392 with Arabidopsis CGS cDNAs. The plasmid pQE30 (as control), the same plasmid containing the full-length Arabidopsis CGS, and the plasmid containing the truncated version (without the N-terminal region) of CGS were transformed into the E. coli mutant. The transformed bacteria were plated onto an M9 minimal medium with (right) or without (left) Met (40 μg mL−1). Plates were incubated at 37°C for 36 h.
Expression of Arabidopsis CGS in Transgenic Tobacco Plants
Since the plant-specific N-terminal region of Arabidopsis CGS is not essential for its catalytic activity, we hypothesized that this region might possess regulatory functions such as correct modulation of the Met level in cells. Thus, we expect that transgenic plants expressing a truncated form of CGS will have an unbalanced Met metabolism. To address this issue, we first transformed Arabidopsis plants with full-length and truncated CGS constructs. A DNA-encoded plastid transit peptide of the pea (Pisum sativum) rbcS-3A was fused in-frame to both constructs to localize the proteins in the chloroplast. A short DNA encoding three copies of the hemagglutinin (HA) epitope tag was also fused in-frame at the 3′ region of the CGS open reading frames of both constructs to enable immunological detection of the proteins in the transgenic plants (Fig. 1C).
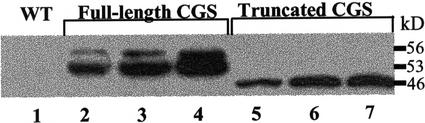
The transgenic Arabidopsis plants grew very poorly and possessed very low levels of wild-type and transgenic CGS gene expression, possibly due to cosuppression (data not shown). To overcome the cosuppression phenomenon, we switched to a heterologous system of transgenic tobacco in which both constructs were well expressed (see below). Thirty independent T0 transgenic tobacco lines expressing each of the Arabidopsis CGS constructs were selected and transferred to the greenhouse for further growth. The expression of CGS constructs in vegetative tissues of the T0 plants was tested by western-blot analysis using anti-HA monoclonal antibodies. Figure 3 shows the results with three representative transgenic plants expressing relatively high levels of each of the Arabidopsis CGS constructs. As expected, no HA-reacting protein bands appeared in the untransformed plants (lane 1). Plants expressing full-length Arabidopsis CGS (lanes 2–4) exhibited two HA cross-reacting bands. The upper band migrated with the expected size of the natural mature Arabidopsis CGS (53 kD; Ravanel et al., 1998) plus the 3-kD HA tag. The second band migrated more rapidly, with an estimated size of 53 kD. This rapidly migrating polypeptide may result from degradation of the full-length mature enzyme. Plants expressing the truncated Arabidopsis CGS (Fig. 3, lanes 5–7) show a single band with expected size of 46 kD. The immunological analysis also revealed that the protein amounts of the full-length and truncated forms of CGS were about equal (Fig. 3). Therefore, eliminating the N-terminal region of Arabidopsis CGS did not change the accumulation amounts of this protein when expressed in tobacco.
Figure 3.
Transgenic tobacco plants expressing the Arabidopsis truncated and full-length CGSs accumulated similar amounts of the corresponding protein. Western-blot analysis of proteins extracts from transgenic plants expressing full-length CGS (lanes 2–4) and truncated CGS (lanes 5–7). The wild-type plant (lane 1) is marked WT. Crude protein extracts (25 μg) from wild-type and representative 7-week-old transgenic tobacco plants were separated by SDS-PAGE and subjected to immunoblot analysis using antiserum against 3xHA epitope tag. The migration of Mr protein markers is indicated on the right.
Since differences were observed between plants expressing full-length and truncated CGS at the phenotype level (see below), we next analyzed these differences.
Expression of the Truncated CGS Construct Caused a Severe Abnormal Phenotype
The transgenic tobacco plants expressing full-length Arabidopsis CGS grew with an indistinguishable phenotype and at a similar rate as the wild-type plants. In contrast, the transgenic plants expressing the truncated Arabidopsis CGS exhibited a severely abnormal phenotype, which could be easily recognized after 6 weeks of growth. This included stunted growth, a slow developmental rate, loss of apical dominance, and narrow, greener, and curly leaves (Fig. 4, A–D). In some of these plants, the apical meristems and leaf primordium became brown and dry (Fig. 4D). Flower buds were produced in only some of the transgenic plants, but they fell rapidly after an abscission zone formed, causing total sterility. The transgenic plants expressing the truncated CGS construct produced many secondary stems and were able to survive for more than 2 years. All analyses were performed on vegetatively propagated T0 plants due to the issue of sterility. As a result of the phenotype differences between these two transgenic lines, we next wanted to reveal the Met content in these lines.
Figure 4.
Severe abnormalities of tobacco plants expressing truncated CGS. A, Transgenic plants expressing full-length CGS display essentially a wild-type appearance, whereas transgenic plants expressing truncated CGS display severe abnormalities. B, Leaves of transgenic plants expressing truncated CGS. C, Transgenic plants expressing truncated CGS lost their apical dominance and produced several shoots. D, The meristem and primordial leaves were withering in some plants expressing truncated CGS.
Analysis of Met, SMM, and Asp-Related Amino Acids in the Transgenic Plants
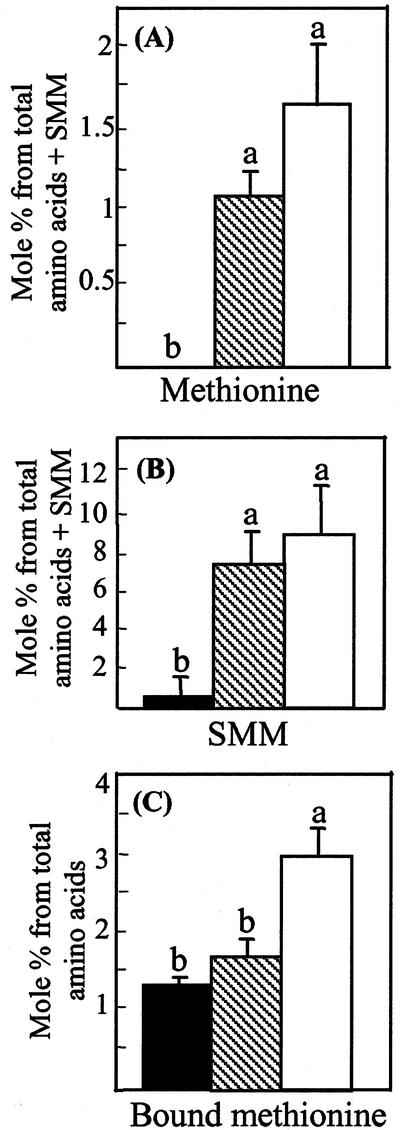
The free amino acid analyses were performed on leaves of 7-week-old wild-type and transgenic plants expressing the two CGS constructs. We first measured the soluble Met content in these plants. Whereas the Met level was less than the detection level in wild-type plants, plants expressing full-length and truncated CGS exhibited an average of 1 and 1.5 mol% Met, respectively (Fig. 5A). Free Met levels differed significantly (P < 0.05) between the two types of transgenic plants and the wild-type plants, but not between the two types of transgenic plants (Fig. 5A).
Figure 5.
A, Met; B, SMM; C, Met incorporated to proteins level, from wild type (▪); transgenic lines expressing full-length (▧); and truncated Arabidopsis CGS (□). The amounts of Met and SMM were calculated from total free amino acids plus SMM as detected by HPLC and is given in mol% of this total. The Met level incorporated into proteins was calculated from phosphate-buffered saline-soluble proteins that were subjected to amino acid analysis following protein hydrolysis by HPLC. The levels of soluble Met, SMM, and bound Met were determined from the extraction of leaves of plants grown for 7 weeks. The data are presented as the mean ± se of eight individual plants per line, one measurement per plant. Statistically significant differences (P < 0.05) are identified by letters.
The SMM level was then determined in these plants because this metabolite had recently been postulated to function as a storage reservoir of labile methyl moieties in the phloem (Mudd and Datko, 1990; Bourgis et al., 1999), and was also found to correlate with the free Met level (Gakiere et al., 2000; Kim and Leustek, 2000). Both types of transgenic plants expressing Arabidopsis CGS contained a similar elevated level of SMM—about 10 times higher than that the wild-type plants (Fig. 5B).
Met belongs to the Asp family of amino acids. Thus, we further studied the effect of Met and SMM elevation on other amino acids belonging to this family. No significant difference was observed between wild-type and transgenic plants (Table I). The levels of other amino acids were also comparable between wild-type and transgenic plants.
Table I.
Contents of aspartate-related amino acids in wild-type and transgenic plants expressing truncated and full-length Arabidopsis CGS
| Amino Acid | Wild Type | Truncated CGS | Full-Length CGS |
|---|---|---|---|
| mol % | |||
| Asp | 7.2 ± 4.3 | 4.9 ± 1.9 | 5.2 ± 2.8 |
| Thr | 2.9 ± 0.8 | 2.3 ± 1.1 | 4.1 ± 1.8 |
| Lys | 0.9 ± 0.4 | 1.0 ± 0.2 | 1.0 ± 0.4 |
| Ile | 0.8 ± 0.3 | 1.2 ± 0.8 | 1.0 ± 0.7 |
The amount of each amino acid was determined by HPLC analysis using external calibration standards. Eight plants of each type were analyzed and the data are presented as the mean ± se.
Expression of Full-Length and Truncated CGS Constructs Resulted in Increased Leaf Protein-Bound Met
We then tested whether increased Met production in the transgenic plants was associated with increased incorporation of this amino acid into leaf proteins. To address this issue, aqueous-soluble proteins were subjected to amino acid analysis following protein hydrolysis. The proportion of Met in the aqueous-soluble proteins was only slightly higher in transgenic plants expressing full-length CGS than in wild-type plants. However, it nearly doubled in transgenic plants expressing the truncated CGS construct (Fig. 5C). This result suggested that more Met was produced in the transgenic plants expressing the truncated CGS and that this Met was incorporated into proteins.
Transgenic Plants Expressing the Truncated CGS Emitted Significant Levels of Volatile Met Catabolic Products
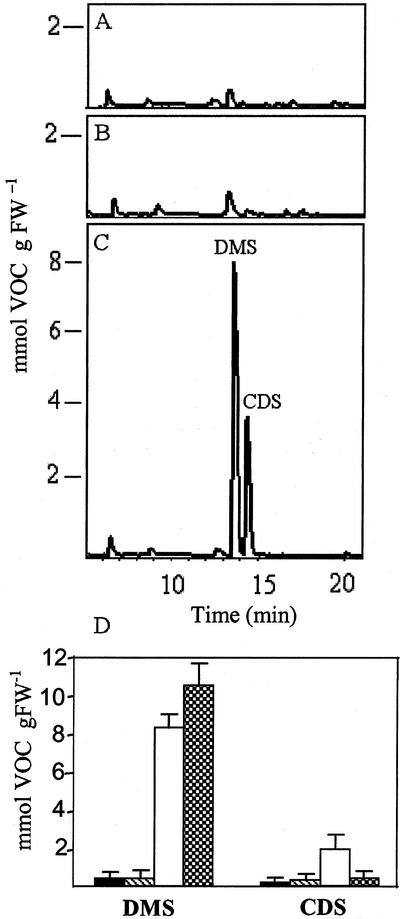
In addition to the severely abnormal phenotype of the transgenic plants expressing truncated Arabidopsis CGS, these plants also possessed a very typical smell, unlike those expressing full-length CGS. To determine what chemical compounds caused this smell, gas chromatography-mass spectrometry (GC-MS) analysis was performed on wild-type plants and on eight independently transformed tobacco lines expressing a relatively similar level of each of the transgenic CGS gene products. This analysis showed that the plants expressing truncated CGS emitted substantially higher levels of the sulfide-containing compounds dimethyl sulfide (DMS) and carbon disulfide (CDS) than wild-type tobacco or transgenic tobacco plants expressing full-length Arabidopsis CGS (Fig. 6). The DMS levels emitted by some of the transgenic plants expressing truncated CGS (lines N46 and N66) were about 45 times higher than in wild-type plants. On average, the levels of DMS emission were 20 and 21 times higher than in transgenic plants expressing full-length CGS and wild-type plants, respectively (Fig. 6D).
Figure 6.
Tobacco plants transformed with truncated Arabidopsis CGS produces volatile organic compounds (VOCs). Patterns of VOC analysis of wild-type plant (A); transgenic plants expressing full-length CGS (B); transgenic plants expressing truncated CGS (C). The two peaks were identified as DMS and CDS. D, Quantitation of the amounts of DMS sulfide and CDS in wild-type (▪); transgenic lines expressing full-length (▧); transgenic lines expressing truncated Arabidopsis CGS (□); and wild-type plants irrigated with 10 mm Met for 10 d ( ). One gram of fresh weight leaf was taken from 7-week-old plants. The amount of these volatile compounds was calculating by determined the area of the corresponding peak in the GC-MS graph as compared with a known standard. The data are presented as the mean + se (black bars) of eight individual plants, one measurement per plant.
A characteristic smell was detected in tobacco transgenic plants in which the SAM synthase was suppressed. Two VOCs were identified: methanethiol and its oxidation product, dimethyl disulfide (Boerjan et al., 1994). DMS and CDS were not previously detected from plants exhibiting a high Met level. Therefore, to study whether these two volatile compounds were Met catabolic products, 7-week-old wild-type plants were irrigated with 10 mm Met or water for 10 d and subject to volatile detection by GC-MS. The results showed a high emission of DMS and a slightly higher (12%) elevation of CDS in plants irrigated with Met (Fig. 6D). These results suggested that at least DMS is a catabolic product of Met and more likely CDS, as well. The fact that DMS and CDS, both containing sulfide, are the major compounds emitted from the plants expressing truncated CGS suggests that the sulfur supply and the Cys level do not limit or regulate the Met content in plants.
Transgenic Plants Expressing Truncated CGS Produced a High Level of Ethylene
Ethylene, one of the major phytohormones in plants, is synthesized from Met via SAM. Since the transgenic plants expressing the truncated CGS possess some phenotype that resembles ethylene symptoms, we tested the rate of ethylene emission in 3-week-old shoots regenerated from transgenic and wild-type plants. As shown in Table II, ethylene production was comparable between wild-type and transgenic plants expressing full-length Arabidopsis CGS. However, in the transgenic plants expressing the truncated CGS construct, ethylene production was nearly 40 times higher than in wild-type plants. This high level of ethylene may explain some of the abnormal phenotypes observed in the transgenic plants such as enhanced abscission of flower buds, dryness of the apical meristem and leaf primordium, retarded stem elongation, as well as curled leaves (Salisbury and Ross, 1991).
Table II.
Ethylene production by young shoots
| Strains | Ethylene Production |
|---|---|
| mg kg−1 h−1 fresh wt | |
| Wild type | 0.11 ± 0.04 |
| Transgenic plants expressing full-length CGS | 0.15 ± 0.08 |
| Transgenic plants expressing truncated CGS | 4.35 ± 0.46 |
Ethylene production by shoots (about 10 cm high) regenerated from transgenic and wild-type plants after 3 weeks of growth in soil. The average ± SE of five different shoots is shown.
DISCUSSION
Plant CGS enzymes possess an extended N-terminal region that is not found in the bacterial enzyme and is not essential for its catalytic activity, as we determined using a complementation test in an E. coli mutant. These results are in agreement with a recent crystal structure analysis of tobacco CGS showing that the catalytic residues, the active site, and the substrate binding residues are all localized in the C-terminal part that is conserved with the bacterial CGSs (Clausen et al., 1998, 1999).
Since the N-terminal region is not essential for CGS catalytic activity, we hypothesized that this region might possess regulatory functions such as the correct modulation of Met level in cells. To examine this hypothesis, we overexpressed full-length Arabidopsis CGS and a truncated version (lacking its N-terminal region) in tobacco plants. It was found that both transgenic plants contained higher levels of Met and its metabolites SMM, as expected from CGS overexpression (Gakiere et al., 2000). However, although the plants expressing full-length CGS exhibited the same developmental rate and phenotype as wild-type plants, those expressing truncated CGS showed severely abnormal phenotypes. These abnormal plants also produced higher contents of Met (33%), SMM (17%), and Met in proteins (41%), than plants expressing the full-length CGS, but more extensively, these plants emitted a significantly higher level of Met catabolic products. These products contain methyl and/or sulfide groups (DMS and CDS), as well as the hormone, ethylene. It is assumed that the severe abnormal phenotype characterizing these plants is derived from overproduction of these metabolites and/or from other unknown Met catabolic products.
The high levels of Met and its catabolic products found in plants expressing truncated CGS, but not in those expressing the full-length CGS, strongly suggested that sequences in the N-terminal region are important for the regulation of proper Met metabolism. The absence of this region leads to an impaired Met metabolism that results in an abnormal phenotype. Thus, it is further suggested that this region plays a regulatory role in protecting plants from a high level of Met catabolic products such as ethylene.
How can the N-terminal part of CGS regulate Met metabolism? The differences between the two transgenic lines in the level of catabolic metabolites of Met could be explained by the “Met overflow” hypothesis. According to this hypothesis, the differences between the two lines of transgenic plants arise in Met synthesis and hence in Met content. Thus, in plants expressing truncated CGS, the Met level rises, the reservoirs of soluble and protein-bound Met and the SMM pool fill up, and the catabolic pathway is induced beyond this threshold. Based on this hypothesis, the Met level does not reach this threshold in plants expressing full-length CGS.
This Met elevation in plants expressing truncated CGS may be the result of the high stability of the CGS transcript or the protein, and/or of a higher enzyme activity. Regulation at the level of transcript stability of CGS was suggested by Chiba et al. (1999), who showed that mutations in the MTO1 region located in the N-terminal region led to the loss of regulation of this transcript level and, consequently, to increases in protein content and Met level. Therefore, transgenic plants expressing truncated CGS would be expected to possess a higher transcript level of CGS, and hence a higher protein and Met level as a result of the deletion of the MTO1 regulatory sequence from the CGS sequence. However, we did not find this to be the case. No significant differences were found in the CGS protein levels between plants expressing full-length and truncated CGS. Furthermore, the CGS band intensity (on average) of plants expressing full-length CGS was higher than those expressing truncated CGS (Fig. 3). Thus, our results suggest that this regulation takes place at the post-translation level, driven by as yet unidentified post-translation machinery within the N-terminal region.
An example of post-translation modification that may occur in the N-terminal region is its involvement in feedback inhibition of the enzyme activity. Met metabolite or protein can bind to this region, leading to conformational changes and inhibition of CGS catalytic activity. However, it should be taken into account that a study conducted with purified Arabidopsis CGS showed that metabolites such as Met, SAM, cystathionine, homo-Cys, SMM, S-adenosylhomo-Cys, 5-methylthioadenosine, Thr, and iso-Leu had no significant effect on CGS activity (Ravanel et al., 1998). At the same time, it is possible that some other metabolite or protein could interact with elements in this N-terminal region and alter CGS activity, such as Met catabolic products (ethylene or DMS, for example).
Further analysis would be required to reveal whether some protein or metabolite is involved in CGS regulation and/or the Met metabolism in plants, but differences between plants in terms of the regulation mode of the N-terminal region of CGS are to be expected. The computer alignment of plant CGS protein sequences within this region shows that it is not a conserved region except for a small segment of the MTO1. This may lead to the various metabolites or proteins that bind CGS and regulate its activity, which differs from one plant to another. Differences in catabolic product patterns are found between Arabidopsis and tobacco, which contain a high Met level. In mto1 and mto2 mutants of Arabidopsis, whose Met levels are 40 and 22 times higher, respectively, than the wild-type plant, catabolic products were barely detected (Chiba et al., 1999; Bartlem et al., 2000). On the other hand, a rise in Met content was accompanied by a rise in the level of dimethyl-disulfide and methanethiol in transgenic tobacco plants in which SAM synthase was suppressed (Boerjan et al., 1994).
Taken together, due to the major role played by Met and its catabolic products in the metabolism of plant cells, a complex regulation is expected. Previous studies have shown that the level of Thr synthase that compete with CGS for their common substrate, O-phosphohomo-Ser, and the carbon skeleton availability to Met biosynthesis are major factors limiting Met biosynthesis (Bartlem et al., 2000). However, additional studies show that sequences at the CGS gene are important for CGS transcript stability and affect Met synthesis (Chiba et al., 1999). In this study, we add another point to this complexity in Met regulation. We suggest that post-translation modification occurring in the N-terminal region of CGS may affect enzyme activity or protein interaction with unknown elements that regulate Met synthesis and/or Met catabolism. However, further analysis is required to clarify the role of the N-terminal in Met metabolism, its role in CGS activity, and the post-translation modification that apparently occurs in this region.
MATERIALS AND METHODS
Plants and Strains
An Escherichia coli mutant, LE392 (met B1, tryp R55, and P2 lysogen; Stratagene, La Jolla, CA), a Met auxotroph, was used for complementation analysis with Arabidopsis CGS constructs. The complemented mutants were cultured for 36 h at 37°C in M9 medium (Sambrook et al., 1989), which was supplemented with 50 μg mL−1 ampicillin, Trp (40 mg L−1) and, for the positive control, Met (40 mg L−1). The solid medium contained 1.5% (w/v) agarose (Invitrogen, Grand Island, NY).
Arabidopsis (ecotype C24) and tobacco (Nicotiana tabacum cv Samsun NN) were grown in the growth chamber under a light regimen of 16 h of light, and 8 h of dark at 22°C to 25°C.
Constructing the Plasmids for the Expression of Arabidopsis CGS in Bacteria
The Arabidopsis CGS cDNA was PCR amplified from a flower cDNA library, kindly donated by the Arabidopsis Biological Resource Center (Columbus, OH). Fragments of DNA encoding mature CGS (without its plastid transit peptide), starting with Val-68 (Ravanel et al., 1998) and truncated CGS (i.e. without its transit peptide and the N-terminal region) starting with Ser-173, were amplified. Primer 1 (5′-AGGATCCGTCCGTCAGCT GAGCATTAAAGC-3′) was used to amplify the sequence of the mature protein. Primer 2 (5′-AGGATCTTGAGCTCCGATGGGAGCC TCAC-3′) was used to amplify the truncated CGS. For reverse amplification of both cDNAs, the same primer, primer 3 (5′-AAAGCTT GATGGCTTCGAGAGCTTGAAG-3′), was used. The BamHI site located at primers 1 and 2 and the HindIII site located at primer 3 were used to insert the amplified DNA fragments into the pQE30 expression vector (Qiagen, Valencia, CA). The nucleotide sequences of the constructed plasmids were verified by DNA sequencing.
Constructing the Binary Plasmids for the Expression of Arabidopsis CGSs to Transgenic Plants
The two forms of cDNA encoding full-length and truncated CGS were amplified from a flower cDNA library using primers 1, 2, and 3. However, the SphI restriction site containing the ATG translation-initiation codon was replaced by the BamHI site in the forward primers (1 and 2), and the reverse primer (primer 3) contained SmaI site instead of HindIII. The PCR fragments (1,479 and 1,164 bp, respectively) were ligated to a PCR vector, pGMT (Promega, Madison, WI), and were then digested with SphI; one SphI site was located in the primer and the other in the plasmid. The fragments were subcloned to a pCE vector (Shaul and Galili, 1992) digested by the same enzyme. This vector contains the 35S promoter of cauliflower mosaic virus, an Ω DNA sequence from the coat protein gene of tobacco mosaic virus for translation enhancement, and the transit peptide pea (Pisum sativum) rbcS-3A chloroplast (Shaul and Galili, 1992). The pCE vector was then cut with SmaI (present at 5′ in the pCE plasmid) and the fragment was subcloned into the binary Ti plasmid, pZP111 (Hadjukiewicz et al., 1994), digested by the same enzyme. Tang et al. (2000) designed a pZP111 vector with the SmaI site at the end of its polylinker site, and an epitope tag of 3xHA, followed by a TGA stop codon. Using this vector, the DNA fragments containing CGS were fused in frame to 3xHA, replacing its natural TGA stop codon (Fig. 1C). The pZP111 plasmid carries the gene for kanamycin resistance.
Plant Transformations
Arabidopsis and tobacco plants were transformed as previously described (Horsch et al., 1985; Clough and Bent, 1998). Transgenic plants were selected on media containing 100 mg L−1 kanamycin.
Western-Blot Analysis
Leaves of transgenic and wild-type plants were homogenized by mortar and pestle in a buffer containing 100 mm Tris-HCl, pH 7.5, 2 mm EDTA, 1 mm dithiothreitol, and 1 mm phenylmethylsulfonyl fluoride (PMSF) at 4°C. After 5 min of centrifugation (16,000g at 4°C), the supernatant was collected. Protein samples (30 μg) were fractionated on 12% (w/v) SDS-PAGE (Laemmli, 1970) and transferred to a PMSF membrane using a Protein Trans-Blot apparatus (Bio-Rad, Hercules, CA). The membrane was blocked overnight at 4°C in a solution of 5% (v/v) nonfat dried milk, and it was then reacted with commercial anti-HA monoclonal antibodies (Roche, Basel) for 2 h at room temperature, followed by incubation with anti-mouse IgG conjugated with horseradish-peroxidase under the same conditions. Immunodetection was conducted with an enhanced chemiluminescence kit (Pierce, Rockford, IL) in accordance with the manufacturer's instructions.
Measurements of Free Amino Acid Levels in Leaves of Transgenic Plants
Five young leaves from 7-week-old transgenic tobacco plants were ground in liquid nitrogen and kept frozen. Free amino acids were extracted from a sample of frozen leaves basically as described by Bieleski and Turner (1966). Approximately 200 mg of tissue was homogenized by mortar and pestle in the presence of 600 μl of water:chloroform:methanol (3:5:12, v/v). After brief centrifugation, the supernatant was collected and the residue was extracted with 600 μl of the same mixture. The two supernatants were combined. Chloroform (300 μl) and water (450 μl) were added, and the resulting mixture was centrifuged again. The upper water-methanol phase was collected, dried, and dissolved in 200 μl of water. The concentration of free amino acids was determined using O-phthalaldehyde reagent, followed by measuring the 335/447 nm fluorescence. The composition of amino acids was determined by loading a 66-nmol sample of total free amino acids on an Amino Quant Liquid Chromatograph (Hewlett-Packard, Palo Alto, CA).
Measurement of Amino Acids Incorporated into Proteins
Leaves were homogenized and extracted at 4°C in a phosphate saline buffer containing 1 mm PMSF and 1 μm leupeptin. Following centrifugation (5 min, 16,000g, 4°C), the supernatant was collected and dialyzed against water. Protein concentration was then determined using the Bradford method, and a batch of 39 mg of protein was hydrolyzed in 0.3 mL of distilled 6 n HCl at 110°C for 22 h under vacuum. A sample of 4 μg of the hydrolyzed protein was analyzed by HPLC (Bio LC Amino Acid Analyzer; Dionex, Sunnyvale, CA).
Determination of the VOCs by GC-MS
The VOCs from leaves of the transgenic plants were determined using the OI 4560 Purge and Trap system connected to a 5890 Series II Gas Chromatograph equipped with a 5972 MS Detector (all Hewlett-Packard). Data were analyzed using HP MS Chemstation software (Hewlett-Packard) according to Environmental Protection Agency method no. 524.2 with 60-m fused silica capillary columns (ID of 0.25 μm and film thickness of 1.4 μm). Ten milliliters of water was added to 10/150-mm glass tubes, and 1 g of fresh young leaves was analyzed. The sample was purged at room temperature with 99.999% helium (40 mL min−1 for 11 min). The helium transferred the VOCs to an ambient temperature micro trap containing silica, Tenax, and charcoal as adsorbents. After purging and trapping, the volatile analytes were thermally desorbed at 180°C and were injected onto the GC column through a heated (100°C) transfer line. The chromatographic separation was applied with a temperature gradient from 35°C to 220°C at a rate of 10°C min−1.
Assay of Ethylene Production in the Transgenic Plants
The procedure described by Guzman and Ecker (1990) was applied. Ethylene production was assayed from young shoots (about 10 cm high) that regenerated from the transgenic and wild-type plants. These shoots were planted in soil for 3 weeks of growth in a growth chamber (25°C, 16 h of light, 8 h of dark). The resulting plants (in their pots) were then incubated for 24 h at 22°C in an airtight 1-L glass jar. The ethylene was assayed by GC (model 5890; Hewlett-Packard), using an Alumina 60/80 column (model 020283; Supleco, Bellefonte, PA). A standard of 640 μl L−1 ethylene (balance N2) was used to calibrate the ethylene concentrations.
ACKNOWLEDGMENTS
We would like to thank Dr. Gadi Schuster for his critical reading of this manuscript. We would also like to thank Adi Nov for her statistical work and Igal Bar-Ilan and Edna Hadar for their help with the GC-MS analysis.
Footnotes
This study was supported by the Israel Science Foundation (grant no. 410/98–2).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010819.
LITERATURE CITED
- Anderson JW. Sulfur metabolism in plants. In: Miflin BJ, Lea PJ, editors. The Biochemistry of Plants: A Comprehensive Treatise. Vol. 16. London: Academic Press; 1990. pp. 327–381. [Google Scholar]
- Bartlem D, Lambein I, Okamoto T, Itaya A, Uda Y, Kijima F, Tamaki Y, Nambara E, Naito S. Mutation in the threonine synthase gene results in an overaccumulation of soluble methionine in Arabidopsis. Plant Physiol. 2000;123:101–110. doi: 10.1104/pp.123.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieleski RL, Turner NA. Separation and estimation of amino acids in crude plant extracts by thin-layer electrophoresis and chromatography. Anal Biochem. 1966;17:278–293. doi: 10.1016/0003-2697(66)90206-5. [DOI] [PubMed] [Google Scholar]
- Boerjan W, Bauw G, Montagu MV, Inze D. Distinct phenotypes generate by over-expression and suppression of S-adenosyl-l-methionine synthetase reveal developmental patterns of gene silencing in tobacco. Plant Cell. 1994;6:1401–1414. doi: 10.1105/tpc.6.10.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgis F, Roje S, Nuccio ML, Fisher DB, Tarczynski MC, Li C, Herschbach C, Rennenberg H, Pimenta MJ, Shen TL et al. S-Methylmethionine plays a major role in phloem sulfur transport and is synthesized by a novel type of methyltransferase. Plant Cell. 1999;11:1485–1498. doi: 10.1105/tpc.11.8.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba Y, Ishikawa M, Kijima F, Tyson RH, Kim J, Yamamoto A, Nambara E, Leustek T, Wallsgrove RM, Naito S. Evidence for autoregulation of cystathionine γ-synthase mRNA stability in Arabidopsis. Science. 1999;286:1371–1374. doi: 10.1126/science.286.5443.1371. [DOI] [PubMed] [Google Scholar]
- Clausen T, Huber R, Prade L, Wahl MC, Messerschmidt A. Crystal structure of Escherichia colicystathionine γ-synthase at 1.5 A resolution. EMBO J. 1998;17:6827–6838. doi: 10.1093/emboj/17.23.6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T, Wahl MC, Messerschmidt A, Huber R, Fuhrmann JC, Laber B, Streber W, Steegborn C. Cloning, purification and characterization of cystathionine γ-synthase from Nicotiana tabacum. Biol Chem. 1999;380:1237–1242. doi: 10.1515/BC.1999.157. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Datko AH, Mudd SH. Methionine biosynthesis in Lemna: inhibitor studies. Plant Physiol. 1982;69:1070–1076. doi: 10.1104/pp.69.5.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gakiere B, Denis L, Droux M, Ravanel S, Job D. Methionine synthesis in higher plants: sense strategy applied to cystathionine γ-synthase and cystathionine β-lyase in Arabidopsis thaliana. In: Brunold C, editor. Sulfur Nutrition and Sulfur Assimilation in Higher Plants. Bern, Switzerland: Paul Haupt; 2000. pp. 313–315. [Google Scholar]
- Guzman P, Ecker JR. Exploiting the triple response of Arabidopsisto identify ethylene-related mutants. Plant Cell. 1990;2:513–523. doi: 10.1105/tpc.2.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacteriumbinary vectors for plant transformation. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley R. A simple and general method for transferring genes into plants. Science. 1985;227:1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- Inba K, Fujiwara T, Hayashi H, Chino M, Komeda Y, Naito S. Isolation of an Arabidopsis thalianamutant, mto1, that overaccumulates soluble methionine: temporal and spatial patterns of soluble methionine accumulation. Plant Physiol. 1994;104:881–887. doi: 10.1104/pp.104.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Leustek T. Cloning and analysis of the gene for cystathionine γ-synthase from Arabidopsis thaliana. Plant Mol Biol. 1996;32:1117–1124. doi: 10.1007/BF00041395. [DOI] [PubMed] [Google Scholar]
- Kim J, Leustek T. Repression of cystathionine γ-synthase in Arabidopsis thalianaproduces partial methionine auxotrophy and developmental abnormalities. Plant Sci. 2000;151:9–18. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Ma JF, Shinada T, Matsuda C, Nomoto K. Biosynthesis of phytosiderophores, mugineic acids, associated with methionine cycling. J Biol Chem. 1995;270:16549–16554. doi: 10.1074/jbc.270.28.16549. [DOI] [PubMed] [Google Scholar]
- Mudd H, Datko AH. The S-methylmethionine cycle in Lemna paucicostata. Plant Physiol. 1990;93:623–630. doi: 10.1104/pp.93.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanel S, Gakiere B, Job D, Douce R. Cystathionine γ-synthase from Arabidopsis thaliana: purification and biochemical characterization of the recombinant enzyme overexpressed in Escherichia coli. Biochem J. 1998;331:639–648. doi: 10.1042/bj3310639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury FB, Ross CW. Plant Physiology. Belmont, CA: Wadsworth Press; 1991. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shaul O, Galili G. Increased lysine synthesis in tobacco plants that express high levels of bacterial dihydrodipicolinate synthase in their chloroplasts. Plant J. 1992;2:203–209. [Google Scholar]
- Sun S. Methionine enhancement in plants. In: Singh BK, editor. Plant Amino Acids: Biochemistry and Biotechnology. New York: Academic Press; 1998. pp. 205–213. [Google Scholar]
- Tang G, Zhu X, Tang X, Galili G. A novel composite locus of Arabidopsisencoding two polypeptides with metabolically related but distinct functions in lysine catabolism. Plant J. 2000;23:195–203. doi: 10.1046/j.1365-313x.2000.00770.x. [DOI] [PubMed] [Google Scholar]
- Thompson G, Datko AH, Mudd SH. Adaptation of Lemna paucicostatato sublethal methionine deprivation. Plant Physiol. 1983;71:241–247. doi: 10.1104/pp.71.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]