Abstract
Holoprosencephaly (HPE) is the most common structural anomaly of the human brain, resulting from incomplete cleavage of the developing forebrain during embryogenesis. Haploinsufficient mutations in the TG-interacting factor (TGIF) gene were previously identified in a subset of HPE families and sporadic patients, and this gene is located within a region of chromosome 18 that is associated with nonrandom chromosomal aberrations in HPE patients. TGIF is a three-amino-acid loop extension (TALE) homeodomain-containing transcription factor that functions both as a corepressor of the transforming growth factor beta (TGF-β) pathway and as a competitor of the retinoic acid pathway. Here we describe mice deficient in Tgif that exhibited laterality defects and growth retardation and developed kinked tails. Cellular analysis of mutant mouse embryonic fibroblasts (MEFs) demonstrated for the first time that Tgif regulates proliferation and progression through the G1 cell cycle phase. Additionally, wild-type human TGIF was able to rescue this proliferative defect in MEFs. In contrast, a subset of human Tgif mutations detected in HPE patients was unable to rescue the proliferative defect. However, an absence of Tgif did not alter the normal inhibition of proliferation caused by treatment with TGF-β or retinoic acid. Developmental control of proliferation by Tgif may play a role in the pathogenesis of HPE.
Holoprosencephaly (HPE) is the most common birth defect affecting prosencephalic development, resulting in the loss of forebrain midline structures (35). More than 12 chromosomal regions have been mapped in sporadic and familial HPE cases. One region, mapping at 18p11.3 in humans, contains the gene TGIF. Deletions and mutations of TGIF have been associated with HPE (1, 5, 12, 16, 41). Intriguingly, HPE demonstrates considerable variability and incomplete penetrance; the penetrance for TGIF mutations or deletions is only 10% (1, 38).
TGIF is a three-amino-acid loop extension (TALE) homeobox-containing transcription factor. Members of the TALE family, which includes PBC and MEIS class proteins, form heteromeric complexes with each other to regulate transcription of developmentally important genes, such as the Hox and Pax6 genes (31). Members of this family can also function as cofactors to provide specificity and cooperativity for Hox genes. Although TGIF has not been shown to bind PBC proteins (11, 23), TGIF can mutually compete and inhibit downstream target genes with MEIS (58).
TGIF was first implicated as an antagonist in the retinoic acid (RA) pathway because it competed for DNA binding with the RXRα retinoic acid receptor (6). In addition, TGIF has been shown to repress downstream transcriptional activity of RXR (4). Subsequently, TGIF was also shown to be a transcriptional corepressor, interacting with Smad2 to regulate transforming growth factor beta (TGF-β)-responsive gene transcription (55). TGF-β-dependent transcriptional repression by TGIF is mediated through multiple domains, by direct competition with the coactivator p300/CBP for Smad2 interaction and through recruitment of the histone deacetylase complex, the carboxyl terminal binding protein (CtBP) complex, and the Sin3 complex, all of which modify chromatin configuration (33, 54, 56). Finally, TGIF protein stability can also be regulated by the Ras/mitogen-activated protein kinase (MAPK) pathway (28).
Interestingly, both the retinoic acid and the TGF-β/Nodal pathways have been implicated in the development of HPE. Mutations in FOXH1, a transcription factor in the TGF-β pathway, have been identified in HPE patients (34). Similarly, mice deficient for genes in the TGF-β pathway, such as the compound mutant Smad2+/− Nodal+/−, developed HPE (39). In addition, prenatal exposure to retinoic acid causes susceptibility to HPE in humans and model organisms (34, 44).
To investigate the function of Tgif during mouse development, we generated Tgif-null mice. The loss of Tgif resulted in a spectrum of abnormal phenotypes, including laterality defects, all of which occurred at low penetrance. In addition, fibroblasts derived from mutant embryos displayed a cell cycle progression defect. We demonstrated that expression of wild-type human TGIF proteins rescued the proliferative defect in mouse embryonic fibroblasts (MEFs) while expression of proteins containing a subset of mutations within the homeodomain identified in HPE patients did not. Unexpectedly, the absence of Tgif did not alter the normal antiproliferative response to stimulation of the TGF-β, retinoic acid, or Ras/MAPK pathways.
MATERIALS AND METHODS
Targeting construct and generation of Tgif−/− mice.
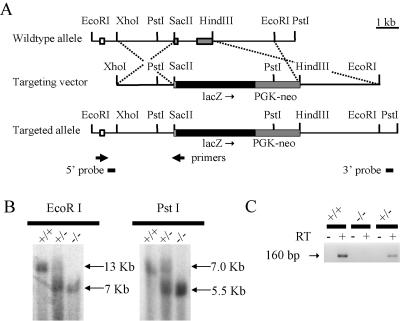
Construction of the targeting vector was as follows. First, a 7.0-kb EcoRI-XhoI fragment was used for homologous arms. Next, exons 2 and 3, which are contained within the 2.3-kb SacII-HindIII fragment, were excised and replaced with lacZ and PGK-neo. lacZ was inserted in frame downstream of amino acid residue 7, leucine, which is the second amino acid of the remaining exon 2. This vector was linearized and electroporated into CCE embryonic stem (ES) cells. ES cells were selected with G418 and screened for homologous recombination events by Southern blot analysis (21). 5′ homologous recombination was screened using EcoRI-digested genomic DNA and a 300-bp XhoI external probe, while 3′ homologous recombination was screened using PstI-digested genomic DNA and a 0.5-kb XbaI external probe. Two independent, correctly targeted ES cell clones were injected into C57BL/6J host blastocysts to generate chimeric mice. The phenotypes observed in mice derived from both ES cell lines were similar and their combined results are included here. All mice and embryos were of a mixed 129/Sv/CD1, 129/Sv/C57BL/6J, or C57BL/6J genetic background. A PCR screening strategy was also used for ear-punch and visceral yolk sac DNA to determine the genotypes of mice and embryos.
Reverse transcriptase PCR (RT-PCR) was performed using primers TGIF-RT (5′-ATGAAAAGCAAGAAGGGTCT-3′) and TGIF 207 (5′-TGTTCATACAGCCAGTCTCG-3′). They are located within exons 1 and 2, respectively (Fig. 1A).
FIG. 1.
Gene targeting of the mouse Tgif locus. (A) Gene targeting strategy utilizing homologous recombination in ES cells. The wild-type allele, targeting vector, and targeted allele are shown with relevant restriction sites and exons (noncoding sequence, open boxes; coding exons, gray boxes). The dashed lines depict regions of homologous recombination. (B) Southern blot analysis of mouse MEF cells demonstrated correct targeting of the locus. EcoRI-digested genomic DNA generated a 13-kb band for the wild-type allele and a 7-kb band for the targeted allele using the 5′ probe indicated in panel A, while PstI-digested genomic DNA generated a 7-kb band for the wild-type allele and a 5.5-kb band for the targeted allele using the indicated 3′ probe. Genotypes are indicated above the lanes. (C) RT-PCR using primers indicated in panel A demonstrated that Tgif transcripts were expressed in Tgif+/+ and Tgif+/−, but not Tgif−/−, MEFs. RT is present (+) or absent (−) in each sample.
Mice were maintained at the British Columbia Cancer Research Centre animal facility. Experiments complied with all relevant federal guidelines and were approved by the Institutional Animal Care and Use Committee.
Generation of Tgif−/− MEFs.
Primary fibroblasts were isolated from Tgif+/+ and Tgif−/− day 13.5 embryos (37) and cultured in Dulbecco's minimum essential medium supplemented with 10% fetal bovine serum (FBS) and antibiotics (Stem Cell Technologies). Experiments were carried out with early passage (<8) cells from Tgif+/+ and Tgif−/− littermates. MEFs were generated from embryos from 129/Sv/CD1, 129/Sv, and C57BL/6J backgrounds. Unless stated otherwise, all experiments were conducted using MEFs derived from the 129/Sv/CD1 genetic background.
Growth curve analysis.
A total of 2 × 104 cells of each genotype were plated in 12-well culture plates and counted on a daily basis using a Coulter Counter.
BrdU labeling and cell cycle analyses.
A total of 1 × 106 cells at log phase were labeled with bromodeoxyuridine (BrdU) and 7-amino-actinomycin D (a DNA binding dye) following the manufacturer's instructions (BD Biosciences Pharmingen) and fluorescence-activated cell sorter (FACS) analyzed on a Cytopeia Influx. BrdU is incorporated only by cells undergoing DNA synthesis.
[3H]thymidine incorporation into cells.
MEFs derived from embryos of 129/Sv/CD1, 129/Sv and C57BL/6J genetic backgrounds were seeded at 5,000 cells/well in 96-well plates in triplicate. A total of 1 μCi/well [3H]thymidine was added to the cells, and the cells were incubated for 2 h, harvested, and [3H]thymidine incorporation quantified using a β-counter (LKB Wallac, Turku, Finland).
Growth factor and UO126 treatment.
Cells were treated with TGF-β, all trans-RA, 9-cis-RA, or U0126 at the indicated concentrations for the indicated times. TGF-β was a gift from Stem Cell Technologies. All trans-RA, 9-cis-RA, and U0126 were purchased from Sigma.
Serum starvation.
A total of 5,000 cells/well was grown in Dulbecco's minimum essential medium lacking serum for 2 days to induce quiescence; the addition of 10% FBS allowed synchronous reentry into G1 and S phase.
Expression of retroviral constructs in primary MEFs.
The human TGIF cDNA was subcloned upstream of an internal ribosome entry site (IRES)-YFP cassette in the murine stem cell virus (MSCV) retroviral vector (10) to generate MSCV-TGIF-IRES-YFP. Site-directed mutagenesis using a modified restriction site protocol generated the seven-point mutations identified in patients (49). All mutations were sequence verified. Tgif was flag-tagged at the N terminus to allow for Western blot analyses. High-titer helper-free recombinant MSCV-TGIF-IRES-YFP retroviruses were generated and tested according to protocols developed by G. Nolan (www.stanford.edu/group/nolan). Briefly, Phoenix Eco cell lines were transfected using FuGENE (Roche). Supernatant from the Phoenix viral producer cells supplemented with 6 μg of ammonium sulfate per milliliter was used for infection of the MEFs. Yellow fluorescent protein (YFP)-expressing cells were enriched through a Cytopeia Influx sorter, and proliferation was measured as described above.
Embryo harvest and staging.
Embryos were generated by timed matings of congenic C57BL/6J Tgif+/− mice. At E8.0 to E8.25, embryos and yolk sacs were collected and the embryos were staged according to their somite numbers. Subsequently, yolk sacs were used for PCR genotyping.
BrdU incorporation and analysis.
Congenic C57BL/6J Tgif+/− females pregnant with E8.0 to E8.25 embryos received an intraperitoneal injection of 400 μg BrdU per gram of body weight and were euthanized 1 h later, as described previously (18). Embryos were collected and staged as described above, fixed in 4% paraformaldehyde overnight, and frozen in optimum-cutting-temperature embedding compound. Frozen 10-μm sections were stained with anti-BrdU-conjugated Alexa 549 (Molecular Probes) and Hoechst to stain DNA. The fraction of BrdU-incorporated cells was determined by counting the number of positive cells and dividing by the total number of Hoechst-positive cells. Only neuroepithelial cells were counted. Numbers were also compared in dorsal one-third, ventral one-third, and mid-section one-third sections. At least three adjacent sections were counted, and three sets of matched mutant and control embryos were analyzed.
RESULTS
Tgif−/− mice exhibited laterality defects and other pleiotropic phenotypes.
A targeted deletion of Tgif exons 2 and 3, eliminating nearly all coding sequences, was performed in ES cells (Fig. 1A and B). Mice heterozygous for Tgif on the 129/Sv/CD1 background were viable and fertile. A x2 analysis of 80 F2 offspring from these intercrosses at 3 weeks of age demonstrated a normal Mendelian ratio (25 Tgif+/+ [31%], 36 Tgif+/− [45%], and 19 Tgif−/− [24%] [P = 0.43]), showing that Tgif was not essential for viability. To confirm that Tgif expression had been abolished, MEFs were generated from wild-type, heterozygous, and mutant embryos and tested for expression of Tgif by RT-PCR (Fig. 1C). Tgif transcript was robustly detected in wild-type and heterozygous MEFs, while no transcript was observed in mutant MEFs.
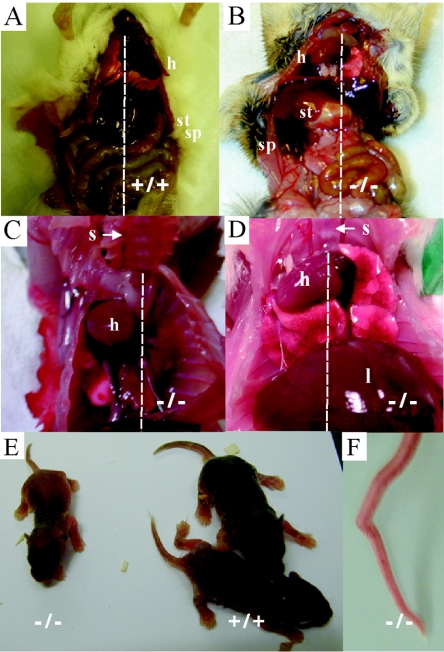
Although the Tgif−/− mice were viable and fertile, a variety of defects were uncovered in this mixed genetic background. Significantly, 13% of Tgif−/− mice developed laterality defects, with both situs inversus and situs ambiguus being observed (Fig. 2A to D; Table 1) after examining 67 mutant mice. Situs ambiguus most often involved the incorrect positioning of the heart and occasionally involved abnormal lung lobe numbers (three cases) or polysplenia (one case). These defects were not observed in control littermates (Table 1). In addition, growth retardation and kinked tails were observed (Fig. 2E and F; Table 1). Taken together, 43% of mutant mice displayed at least one of the defects listed above (Table 1). However, each individual defect was observed in only a limited number of mutant mice, indicating that the penetrance of each defect was low.
FIG. 2.
Tgif mutant mice on the 129/Sv/CD1 genetic background developed pleiotropic phenotypes. (A to D) Adult mice, placed on their backs, were dissected to determine the sidedness of their visceral organs. The dashed line indicates the sternum (s) and the midline of the body axis. (A) In Tgif+/+ mice, the heart (h), stomach (st), and spleen (sp) were on the left side of the body cavity. (B) A Tgif−/− mouse exhibiting situs inversus where the heart, stomach, and spleen were located on the right side of the body cavity. (C and D) Tgif−/− mice exhibiting situs ambiguus where the heart laid within the right chest cavity (liver [l]) (not shown are the stomach and spleen laid on the left side). (E) Some mutant mice were already visibly smaller than littermates 1 week after birth. (F) Tgif mutant mice exhibited kinked tails.
TABLE 1.
Phenotypes resulting from Tgif heterozygote matings on 129/Sv/CD1 genetic background at 3 weeks of age
| Parameter | Result for mice with indicated genotype
|
||
|---|---|---|---|
| Tgif+/+ | Tgif+/− | Tgif−/− | |
| % with defects | 0 | 0 | 43 |
| % with no apparent defects | 100 | 100 | 57 |
| No. with indicated phenotype/total (%) | |||
| Growth retardationa | 0/35 | 0/73 | 6/31 (19) |
| Kinked tail | 0/124 | 0/243 | 10/92 (11) |
| Situs ambiguusb | 0/72 | 0/145 | 6/67 (9) |
| Situs inversusb | 0/72 | 0/145 | 2/67 (3) |
| Kinked tail and situs ambiguus | 0/72 | 0/145 | 1/67 (1) |
Thirty-one mutant mice were weighed with littermate controls. A mouse was considered growth retarded if its weight was 20% less than the mean weight value of its litter.
Sixty-seven mutant mice were dissected along with littermate controls; 92 mutant mice were physically examined along with littermate controls.
The same chimeras used to generate the 129/Sv/CD1 colony were also mated to C57BL/6J female mice in an attempt to increase the penetrance of the observed phenotypes. A congenic C57BL/6J strain was produced after 10 generations of breeding Tgif+/− mice to C57BL/6J mice. However, no abnormalities were detected after the examination of 26 mutant mice; all mice displayed normal tails, situs, and growth. From this result, we conclude that the phenotypes observed were dependent on the genetic background.
Tgif regulates proliferation in primary MEFs.
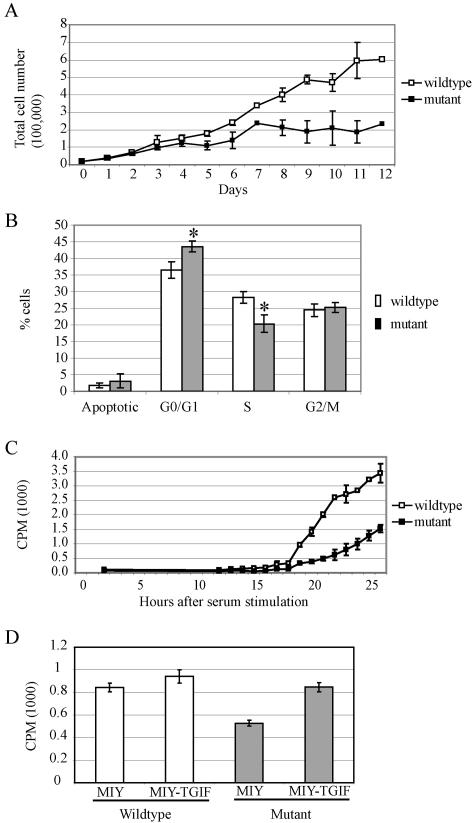
To investigate the function of Tgif, primary MEF cell lines were generated from Tgif+/+ and Tgif−/− embryos on the mixed 129/Sv/CD1 genetic background. RT-PCR confirmed that Tgif was expressed in wild-type and heterozygous MEFs but not in mutant MEFs (Fig. 1C). Unexpectedly, cell lines derived from Tgif−/− embryos failed to proliferate as rapidly as the controls. To examine the proliferative capability of the cells, equal numbers of early passage primary MEFs were plated and counted on a daily basis for 12 days. Mutant cells proliferated at a slower rate and accumulated to a lower density than control cells (Fig. 3A).
FIG. 3.
Tgif−/− MEFs exhibited reduced growth due to a delay in G1 phase progression. (A) Equal numbers of MEFs were plated and counted daily for 12 days. Tgif−/− cells proliferated at a lower rate and accumulated to a lower density than Tgif+/+ cells. Data are pooled from two experiments using independently derived MEFs. (B) BrdU labeling and FACS analysis indicated Tgif−/− cells accumulated in G0/G1 phase and showed delayed entry into S phase. P values were equal to 0.003 and 0.002 between Tgif+/+ (n = 8) and Tgif−/− (n = 8) cells with regard to the percentage of cells in G0/G1 and S phase, respectively, using single-factor analysis of variance. A representative result is shown from five experiments using independently derived MEFs. (C) Tgif+/+ and Tgif−/− cells were growth arrested by serum starvation, and then serum was added to stimulate synchronous reentry into the cell cycle. DNA synthesis was measured by [3H]thymidine incorporation as described in the text. Fewer mutant MEFs reentered the cell cycle at all time points. Consequently, the total number of mutant MEFs entering into the cell cycle was lower. Representative results from two experiments are shown, each performed in triplicate, using cells derived from independent MEFs. (D) Tgif+/+ and Tgif−/− MEFs were infected with MSCV-retroviruses expressing human TGIF and YFP (MIY-TGIF) or YFP alone (MIY). Infected cells were enriched by FACS, and DNA synthesis was measured by [3H]thymidine incorporation as described in the text. Expression of human TGIF restored the proliferation rate of mutant MEFs to normal levels. Error bars indicate standard deviations. CPM, counts per minute. Data shown here were obtained with MEFs derived from embryos on the 129/Sv/CD1 genetic background.
Next, cell cycle progression was monitored by labeling cells with BrdU and 7AAD to distinguish cells in G0/G1, S, and G2/M phases of the cell cycle using flow cytometric analysis. We observed a higher percentage of Tgif−/− MEFs in the G0/G1 phase of the cell cycle (36% Tgif+/+ versus 44% Tgif−/>−; P = 0.003, n = 8) (Fig. 3B); a similarly reduced percentage of Tgif−/− cells was detected in the S phase (28% Tgif+/+ versus 20% Tgif−/−; P = 0.002, n = 8). The percentages of cells undergoing apoptosis or in G2/M phase were comparable between Tgif+/+ and Tgif−/− cells. These results suggest that Tgif−/− MEFs were impaired in G1 progression.
To determine if Tgif−/− MEFs were impaired in their ability to respond to mitogens that regulate the restriction point, proliferation was assessed by [3H]thymidine incorporation by MEFs following stimulation with increasing levels of mitogen (0.1% to 10% FBS). We found that the proliferation rate was directly proportional to the concentration of FBS for both Tgif+/+ and Tgif−/− cells, suggesting that the loss of Tgif does not affect the ability of cells to monitor increasing levels of mitogen (data not shown). We next focused on the kinetics of G1 phase in these cells. Quiescent cells, starved of serum, were stimulated to synchronously enter G1 phase of the cell cycle using serum-supplemented media (25). Approximately twofold more quiescent Tgif+/+ MEFs entered S phase than Tgif−/− MEFs at the indicated times after serum stimulation (Fig. 3C). Consequently, the total number of mutant cells that reentered the cell cycle over time was lower. Taken together, these results suggest that Tgif−/− MEFs do not progress as rapidly through G1 as Tgif+/+ MEFs.
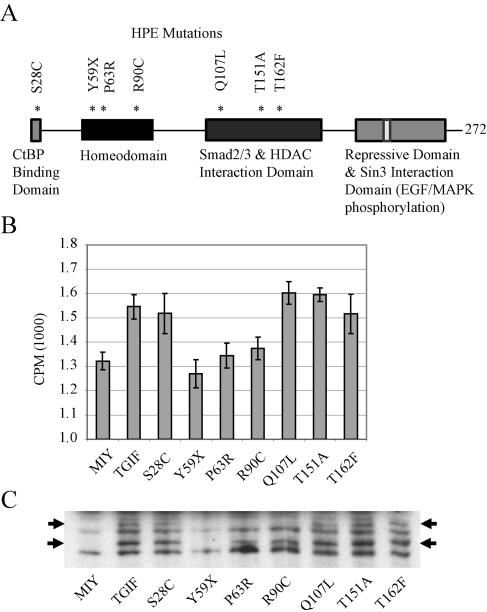
To determine whether the proliferative defect observed in MEFs could be rescued, a cDNA for human TGIF was expressed in mutant MEFs, which were then tested for proliferative capability. Retroviral constructs were generated to express human TGIF cDNAs upstream of an IRES-YFP sequence. The infection efficiency of MEFs was 50 to 60% and cells expressing TGIF were enriched by fluorescence-activated cell sorting for YFP expression. Expression of human TGIF was able to rescue the proliferation defect in mutant MEFs (Fig. 3D). Overexpression of TGIF in wild-type MEFs resulted in a small, but consistent, increase in proliferation.
A proliferation defect was also present in MEFs derived from congenic 129/Sv and C57BL/6J embryos (see Fig. S1 in the supplemental material), indicating that the loss of Tgif caused a proliferation defect in multiple genetic backgrounds.
Growth retardation was observed in vivo as early as embryonic day 8.
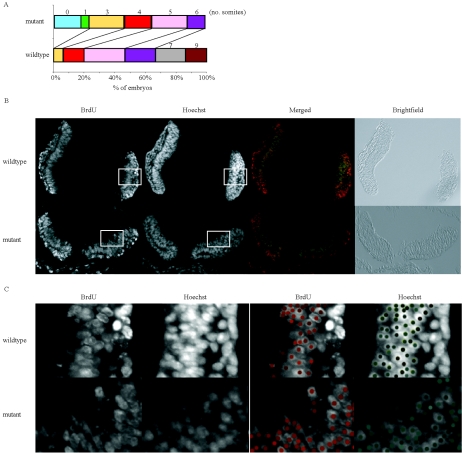
To assess whether proliferation defects were present in vivo, embryos were dissected between embryonic days 8.0 and 8.25, the stage at which dorsal-ventral patterning is initiated in the forebrain. Embryos were precisely staged with respect to their somite number. Upon examination of 15 wild-type and 17 mutant embryos from seven litters, mutant embryos were determined to be developmentally delayed by 2 to 3 somites (0.001 < P < 0.002 using the Mann-Whitney test) (Fig. 4A). Consistent with this observation, some neonates and adult mutant mice were growth retarded (Fig. 2D and Table 1). Together, these observations suggest that reduced proliferation in the absence of Tgif led to a growth retardation phenotype in vivo as early as embryonic day 8.
FIG. 4.
Tgif−/− mutant embryos exhibited growth retardation. (A) Examination of the developmental stage of Tgif+/+ and Tgif−/− embryos (15 and 17 embryos, respectively, from seven litters) at E8.0 to E8.25 according to their somite numbers. Mutant embryos were determined to be developmentally delayed by two or three somites (0.001 < P < 0.002 by the Mann-Whitney test). (B and C) In vivo BrdU incorporation rates of wild-type and mutant forebrains at E8.0 to E8.25 were similar. Embryos labeled with BrdU were used to determine the percentage of cells in S phase in vivo. The fraction of BrdU-incorporated cells was determined by counting the number of BrdU-positive cells (red) and dividing by the total number of Hoechst-positive cells (green). At least three adjacent sections were counted, and three sets of matched mutant and control embryos were analyzed. The boxed regions in panel B were enlarged in panel C, and spots were placed over positively stained cells to determine the percentage of BrdU-positive cells. The embryos shown here were on the C57BL/6J genetic background.
To determine whether embryonic cells in vivo showed a reduced rate of proliferation, in vivo BrdU incorporation experiments were conducted to compare the proportion of cells in S phase between the wild-type and mutant embryos at embryonic day 8. We did not detect a significant reduction in the proportion of cells that incorporated BrdU between wild-type and mutant forebrains (Fig. 4B and C). The sensitivity of in vivo BrdU incorporation may be too low to detect the small difference in proliferation expected from the in vitro MEF assays (Fig. 3B). Alternatively, it is also possible that in vivo proliferation was not significantly different between wild-type and mutant embryos at the developmental stage examined, as cell cycle is normally tightly regulated during in vivo development.
The proliferative function of TGIF requires the homeodomain.
To test whether mutations identified in HPE patients (Fig. 5A) (1, 12, 16) were able to rescue the proliferation defect, TGIF point mutations were recreated in the retroviral constructs described above. Not surprisingly, the Y59X mutation did not rescue the proliferative defect, presumably because the protein preterminated within the homeodomain (Fig. 5B). Notably, the other point mutations within the homeodomain, P63R and R90C, were also unable to compensate for the loss of Tgif. In contrast, mutations outside of the homeodomain, S28C, Q107L, T151A, and T162F, within the CtBP and SMAD2 and histone deacetylase interaction domains, rescued the proliferative defect in mutant MEFs. The levels of protein expression for wild-type and mutant TGIF proteins were similar, with the exception of Y59X, which did not produce a detectable protein at the expected size (Fig. 5C). Thus, these results indicate a novel function for Tgif that requires the homeodomain. Furthermore, the association between proliferation defects and human mutations suggest that cell cycle regulation may be disrupted in some HPE patients.
FIG. 5.
Expression of a subset of mutated human TGIF rescued the proliferative defect of MEFs. (A) Diagram of HPE patient mutations and their positions relative to the functional domains of TGIF, which were generated in MSCV retroviral constructs. (B) Tgif−/− MEFs were infected with MSCV retroviruses expressing human TGIF and YFP (MIY-TGIF), YFP alone (MIY), or human TGIF containing the patient mutations shown in panel A. Infected cells were enriched by FACS, and DNA synthesis was measured by [3H]thymidine incorporation as described in the text. Expression of human TGIF restored the proliferation rate of mutant MEFs to normal levels. Mutations affecting the homeodomain were unable to rescue the proliferative defect. Representative results from five experiments are shown; each experiment was performed in sextuplicate using cells derived from independent Tgif−/− embryos. Error bars represent standard deviations of the means. CPM, counts per minute. (C) Tgif−/− MEFs were infected with MSCV retroviral constructs containing flag-tagged TGIF as described in the text. Western blot analysis of MEF cell extracts showed similar expression levels for all mutant proteins. TGIF generally migrated as a doublet of 30 to 32 kDa; the upper band likely represents the phosphorylated form.
Tgif−/− MEFs retain the ability to respond to TGF-β and retinoic acid.
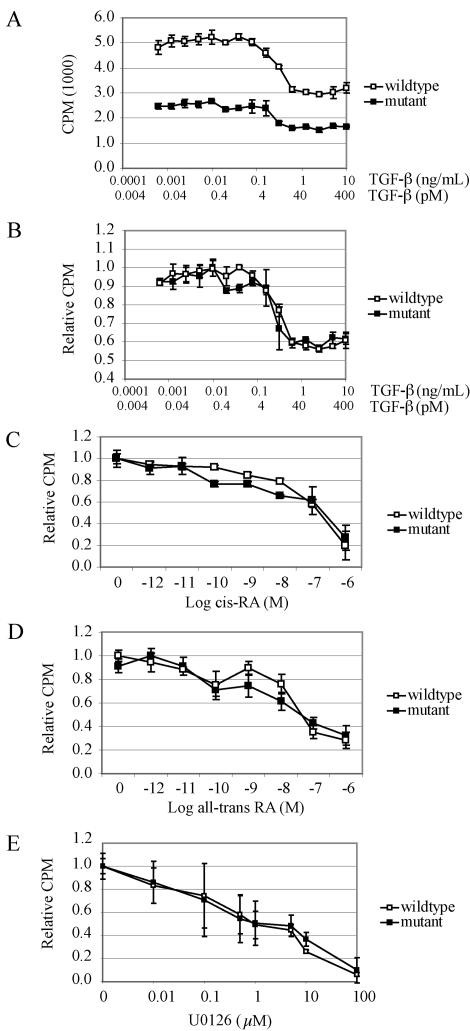
Tgif has been proposed to modulate signaling from both the TGF-β and the RA pathways by repressing the level of transcription (6, 55). In wild-type MEFs, treatment with TGF-β or retinoic acid results in a dose-dependent inhibition of proliferation. To determine whether the loss of Tgif sensitized MEFs to this inhibitory response, we assessed [3H]thymidine incorporation into MEFs after treatment with these compounds. A typical proliferation inhibition response to TGF-β in Tgif+/+ MEFs was observed: growth inhibition was detected at TGF-β concentrations as low as 0.08 ng/ml, or 3 pM, and reached a maximum twofold inhibition at 0.6 ng/ml or 25 pM (46). Tgif−/− MEFs proliferated more slowly compared with Tgif+/+ MEFs in response to increasing concentrations of TGF-β (Fig. 6A). However, since Tgif−/− cells normally proliferated at a lower rate, the level of [3H]thymidine uptake was normalized to the maximum incorporation level. Through this normalization, it became apparent that the degrees of growth inhibition of Tgif+/+ and Tgif−/− MEFs were similar at similar concentrations of TGF-β (Fig. 6B).
FIG. 6.
Analysis of MEF proliferation in response to TGF-β, 9-cis-RA, all-trans-RA, and U0126. (A) Primary Tgif+/+ and Tgif−/− MEF cells were grown in the presence of 0.6 pg/ml to 10 ng/ml TGF-β for 24 h and DNA synthesis was measured by [3H]thymidine incorporation as described in the text. Mutant MEFs displayed lower proliferation at all concentrations of TGF-β relative to wild-type MEFs. (B) Results shown in panel A were normalized to the maximum value of each genotype, since mutant MEFs consistently showed reduced proliferation. The proliferation inhibitory responses of wild-type and mutant MEFs were similar at all concentrations of TGF-β. (C to D) Fresh 9-cis-RA (C) and all-trans-RA (D) were added to MEFs on a daily basis for 5 days, and [3H]thymidine incorporation was performed on day 6. Results were normalized as described for panel B. The proliferative inhibitory responses to 9-cis-RA and all-trans-RA were similar for wild-type and mutant MEFs. (E) Concentrations ranging from 0.01 to 100 μM U0126 were added to MEFs and [3H]thymidine incorporation was performed after 24 h of culture. The proliferative inhibitory responses to U0126 were similar for wild-type and mutant MEFs. All results are from representative experiments. For each, three experiments were performed in triplicate using cells derived from independent Tgif+/+ and Tgif−/− embryos. Error bars represent standard deviations of the means. CPM, counts per minute.
We also characterized the growth arrest response for 9-cis-RA and all-trans-RA in MEFs. Similar to keratinocytes, treatment of Tgif+/+ MEFs with 10−12 M 9-cis-RA caused a reduction in DNA synthesis, with a maximal effect seen at 10−6 M, which caused an 80% reduction in proliferation (15). Using all-trans-RA, 10−11 M was effective at inhibiting proliferation, with a maximal effect at 10−6 M, resulting in a 70% reduction in proliferation. When normalized, these dose-response experiments again showed that the levels of inhibition of proliferation in response to RA were similar between Tgif+/+ and Tgif−/− MEFs (Fig. 6C and D).
TGIF protein is stabilized by phosphorylation of two sites near its C terminus by the Ras/MAP kinase pathway (28). We examined the effect of U0126, an inhibitor of the kinase activity of MEK in the Ras/MAP kinase pathway to determine whether TGIF stability altered MEF proliferation. U0126 inhibited cell proliferation at 0.01 μM and almost completely inhibited cell cycling at 100 μM. The proliferative response to inhibition of Ras/MAPK signaling in Tgif+/+ and Tgif−/− MEFs did not differ significantly (Fig. 6E). Taken together, the pathways known to be modulated by Tgif did not show an increased sensitivity to ligand in Tgif−/− cells, and the mutant MEFs displayed growth inhibition responses that were unaltered relative to Tgif+/+ cells.
DISCUSSION
Deletion, nonsense and missense mutations in TGIF have been identified in a subset of HPE patients (1, 5, 12, 16, 41). Significantly, HPE can develop in patients carrying heterozygous mutations in TGIF, suggesting that the processes being disrupted are exquisitely sensitive to the level of TGIF. Paradoxically, most carriers do not develop HPE, indicating a low level of penetrance for this disorder and that phenotypic manifestation may be dependent on genetic modifiers (1, 38). Here, we show that mice lacking Tgif were viable and fertile without overt evidence of an HPE phenotype. However, some mutant mice did develop multiple defects, including laterality defects, growth retardation, and kinked tails, suggesting that Tgif modulates many processes during development.
Events determining left-right patterning take place early during embryogenesis before the onset of organogenesis. Soon after the breaking of symmetry, expression of the TGF-β family member Nodal becomes enhanced on the left side of the node during gastrulation (17). The left-sided Nodal signal is relayed to the left lateral plate mesoderm where downstream target genes such as Lefty1, Lefty2, and Pitx2 are activated. The left-sided signals and pathways are barred from the right side by midline mesodermal cells. We observed laterality defects in 13% of our null mice, with two mice showing complete situs inversus, indicating that Tgif plays a role in laterality determination. TGIF has been shown to repress the level of TGF-β-activated transcription through interaction with Smad2 and Smad3. Since Smad2 and Smad3 also mediate Nodal signaling, the absence of TGIF may also disrupt Nodal signaling (55). Alternatively, recent reports demonstrated that retinoic acid and Shh are involved in breaking of the left-right symmetry (48, 50). Since TGIF has been shown to compete, and thus inhibit, retinoic acid signaling and repress retinoic acid responsive transcription, the loss of TGIF may disrupt this step of the laterality determination pathway (4, 6). A third possible mechanism by which laterality defects may occur in Tgif mutants is abnormal midline development, which was suggested by the presence of kinked tails (42). Expression analysis by in situ hybridization of embryos between E8.0 to E9.5 revealed that while changes to Nodal and Lefty2 expression were not detected (20 embryos for each marker), 1 mutant embryo (out of 20) did exhibit bilateral Pitx2 expression (data not shown). Bilateral Pitx2 expression suggests that the loss of Tgif may disrupt laterality at a molecular level upstream of Pitx2. The low penetrance of this defect impeded further studies, but this result does suggest that TGIF is a novel candidate gene for laterality disorders in humans. Significantly, many genes involved in the development of the axial midline lead to both laterality and HPE defects when disrupted (43).
Tgif−/− mice did not develop HPE and were viable. Similar observations were also made recently by others (4, 20, 45). This distinction between humans and mice may reflect the importance of genetic modifiers in HPE (36). In addition, three mammalian Tgif paralogues have been identified, and Tgif2 possesses many biochemical properties similar to those of Tgif (32), potentially compensating for the lack of Tgif. The other reports did not describe any abnormal phenotypes in Tgif mutants, suggesting that some differences were present between the mutant mice described by us and those reported previously (4, 20, 45). Since Tgif is a simple, three-exon-containing gene encompassed within 7 kb of genomic DNA, the targeting constructs from all four groups were similarly designed to remove the majority of coding sequences. The low-penetrant defects reported here were observed in only mutant mice, demonstrating that the phenotypes were associated with the loss of Tgif. The phenotypic differences between this report and others are likely due to differences in genetic background, as we observed pleiotropic defects in 129/Sv/CD1 while none of the four groups observed defects in the C57BL/6 genetic background. Genetic background has been commonly observed to influence the phenotypes of mutant mice (8, 27, 36). We examined 26 C57BL/6J congenic Tgif−/− mice, and all have displayed normal tails, situs, and size. In the future, the function of Tgif may be better studied when an inbred background that increases the penetrance of the intriguing phenotypes we have described is identified. Tgif−/− mice may also become a useful genetic modification to aid in the development of HPE models through the generation of compound mutants.
We provide the first evidence of a functional role for Tgif during proliferation in mammalian cells. MEF cell lines lacking Tgif proliferated at a slower rate and did not reach the same density as normal controls; cell lines derived from 129/Sv/CD1 and C57BL/6J and mixed 129/Sv/CD1 mutant mice all exhibited reduced proliferation. Further cell cycle analysis revealed that the mutants were delayed in G1 phase and that reexpression of human TGIF rescued this proliferation defect. Consistent with our observations, the yeast protein Tos8, a TALE homeodomain transcription factor related to TGIF, regulates multiple genes involved in G1/S transition events (19). Furthermore, expression of human TGIF (CPR1) restored cell cycle progression by overcoming G1 arrest signals in yeast Far-negative mutants (14). Also of relevance, the Drosophila TGIF orthologues achintya and vismay were shown to function during meiosis (3, 52). This suggests that a role for Tgif in the regulation of the cell cycle may be conserved in eukaryotes from yeasts to mammals.
Accordingly, the expression patterns of Tgif support its role in proliferation. In the mouse embryo, Tgif expression was detected in proliferating cell populations, including those within the branchial arches, developing lung, limb buds, and tongue (7). Tgif expression is also widespread in the developing central nervous system while it undergoes dramatic growth and expansion (7, 20, 45). Altogether, the expression analyses currently available indicate that Tgif potentially plays a role in the proliferation of multiple neuronal populations, as well as other tissues. Consistent with this idea, Tgif−/− mice became growth retarded and mutant embryos were often developmentally delayed as early as embryonic day 8 when somite numbers were used to assess developmental progress.
Assays of in vivo proliferation did not demonstrate a significant difference in the number of proliferating cells in the developing forebrains of wild-type and mutant embryos at embryonic day 8. While in vivo BrdU incorporation was useful in mutant mice that displayed dramatic proliferation defects, such as Pax6 (53), it may not be sufficiently sensitive to detect small proliferation changes. Alternatively, in vivo proliferation may not be altered by the loss of Tgif at the stage examined. Cell cycle is tightly regulated in vivo during development, and other studies have failed to detect in vivo proliferation defects after initially observing such defects in vitro. For example, in vivo proliferation defects could not be demonstrated in mice lacking the D-type cyclin-dependent kinase genes Cdk4 and Cdk6 and in mice lacking the cyclin D1, D2, and D3 genes (24, 30).
TGIF possesses many well-characterized properties that indicate its links to many signaling pathways, namely as a direct competitor of RXR for downstream target genes (6), a transcriptional corepressor in the retinoic acid (4) and TGF-β pathways (55), and a factor in the modulation of Tgif protein stability by the Ras/MAP kinase pathway (28). Wotton et al. showed that Tgif can down-regulate TGF-β signaling (55). Bertolino et al. similarly showed that Tgif competed with retinoic acid during transcription activation of retinoid X receptor-responsive elements, such as the cellular retinol-binding protein II promoter, through direct competition for DNA binding elements (6). More recently, Bartholin et al. provided an additional mechanism through which Tgif can function as a competitive antagonist independent of direct DNA binding; TGIF can directly bind to the ligand binding domain of the RXR family of receptors and repress downstream transcription through recruitment of CtBP complexes (4). However, while we expected the loss of Tgif in mutant MEFs to cause hypersensitivity to TGF-β or retinoic acid, we did not observe increased growth arrest in these cells.
Further studies utilizing TGIF mutations identified in HPE patients addressed the mechanism of TGIF in regulating proliferation. These experiments indicated mutations located in the homeodomain were not able to rescue the proliferative defect. These results suggest a novel mechanism by which Tgif may regulate cell cycle genes or downstream targets through DNA binding or protein-protein interactions through the homeodomain. Given the fact that only transcriptional repressive activity has been demonstrated for TGIF, its target genes may be inhibitors of G1 progression.
Proliferation plays an important role during central nervous system development (29, 57). Recent reports showed the formation of a complex between Geminin, a cell cycle suppressor, and Six3 regulates the proliferation of neuronal precursors (13). Accordingly, overexpression of Geminin leads to cyclopia, a severe form of HPE, and the loss of forebrain, caused by a reduction in proliferating cells and premature differentiation of neurons. Correspondingly, some HPE patients carry mutations in Six3 (51). In fact, many genes and pathways associated with HPE, namely, Shh, Ptch, Gli2 in the Shh pathway, megalin, BMP4 in the TGF-β pathway, and the retinoic acid pathway, were shown to affect proliferation in the developing neural tube (9, 22, 26, 40, 44, 47). Mutations in Zic2 have also been identified in HPE patients, and while the molecular functions of Zic2 are poorly understood, Zic1 activity is associated with the expansion and differentiation of neuronal progenitors (2). It is tempting to speculate that deregulated proliferation may give rise to HPE in patients, based on our observation of a proliferative defect in the absence of Tgif activity. Further studies of Tgif mutants may elucidate its role in cell cycle progression and any possible role cell cycle may play in HPE development.
Supplementary Material
Acknowledgments
We thank K. Humphries and P. Rosten for the use of their BAC clone library, E. Robertson for providing the CCE cell line, D. Wotton and J. Massague for human TGIF cDNA, G. Nolan for Phoenix Eco cell lines, R. Grewal for injecting blastocysts, and G. DeJong, L. Laycock, and G. Thornbury for FACS support.
This study was supported by the National Cancer Institute of Canada with funding from the Terry Fox Foundation. P.A.H. is a Michael Smith Foundation for Health Research Scholar and a Canadian Institute of Health Research New Investigator. L.M. acknowledges the Effie I. Lefeaux Scholarship in Mental Retardation and the Albert B. and Mary Steiner award for travel.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aguilella, C., C. Dubourg, J. Attia-Sobol, J. Vigneron, M. Blayau, L. Pasquier, L. Lazaro, S. Odent, and V. David. 2003. Molecular screening of the TGIF gene in holoprosencephaly: identification of two novel mutations. Hum. Genet. 112:131-134. [DOI] [PubMed] [Google Scholar]
- 2.Aruga, J., T. Tohmonda, S. Homma, and K. Mikoshiba. 2002. Zic1 promotes the expansion of dorsal neural progenitors in spinal cord by inhibiting neuronal differentiation. Dev. Biol. 244:329-341. [DOI] [PubMed] [Google Scholar]
- 3.Ayyar, S., J. Jiang, A. Collu, H. White-Cooper, and R. A. White. 2003. Drosophila TGIF is essential for developmentally regulated transcription in spermatogenesis. Development 130:2841-2852. [DOI] [PubMed] [Google Scholar]
- 4.Bartholin, L., S. E. Powers, T. A. Melhuish, S. Lasse, M. Weinstein, and D. Wotton. 2006. TGIF inhibits retinoid signaling. Mol. Cell. Biol. 26:990-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bendavid, C., B. H. Haddad, A. Griffin, M. Huizing, C. Dubourg, I. Gicquel, L. R. Cavalli, L. Pasquier, B. Long, M. Ouspenskaia, S. Odent, F. Lacbawan, V. David, and M. Muenke. 2006. Multicolor FISH and quantitative PCR and detect submicroscopic deletions in holoprosencephaly patients with a normal karyotype. J. Med. Genet. 43 [Epub ahead of print.] [DOI] [PMC free article] [PubMed]
- 6.Bertolino, E., B. Reimund, D. Wildt-Perinic, and R. G. Clerc. 1995. A novel homeobox protein which recognizes a TGT core and functionally interferes with a retinoid-responsive motif. J. Biol. Chem. 270:31178-31188. [DOI] [PubMed] [Google Scholar]
- 7.Bertolino, E., S. Wildt, G. Richards, and R. G. Clerc. 1996. Expression of a novel murine homeobox gene in the developing cerebellar external granular layer during its proliferation. Dev. Dyn. 205:410-420. [DOI] [PubMed] [Google Scholar]
- 8.Bonyadi, M., S. A. Rusholme, F. M. Cousins, H. C. Su, C. A. Biron, M. Farrall, and R. J. Akhurst. 1997. Mapping of a major genetic modifier of embryonic lethality in TGF beta 1 knockout mice. Nat. Genet. 15:207-211. [DOI] [PubMed] [Google Scholar]
- 9.Britto, J., D. Tannahill, and R. Keynes. 2002. A critical role for sonic hedgehog signaling in the early expansion of the developing brain. Nat. Neurosci. 5:103-110. [DOI] [PubMed] [Google Scholar]
- 10.Buske, C., M. Feuring-Buske, J. Antonchuk, P. Rosten, D. E. Hogge, C. J. Eaves, and R. K. Humphries. 2001. Overexpression of HOXA10 perturbs human lymphomyelopoiesis in vitro and in vivo. Blood 97:2286-2292. [DOI] [PubMed] [Google Scholar]
- 11.Chang, C. P., Y. Jacobs, T. Nakamura, N. A. Jenkins, N. G. Copeland, and M. L. Cleary. 1997. Meis proteins are major in vivo DNA binding partners for wild-type but not chimeric Pbx proteins. Mol. Cell. Biol. 17:5679-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, C. P., S. R. Chern, S. H. Du, and W. Wang. 2002. Molecular diagnosis of a novel heterozygous 268C→T (R90C) mutation in TGIF gene in a fetus with holoprosencephaly and premaxillary agenesis. Prenat. Diagn. 22:5-7. [DOI] [PubMed] [Google Scholar]
- 13.Del Bene, F., K. Tessmar-Raible, and J. Wittbrodt. 2004. Direct interaction of geminin and Six3 in eye development. Nature 427:745-749. [DOI] [PubMed] [Google Scholar]
- 14.Edwards, M. C., N. Liegeois, J. Horecka, R. A. DePinho, G. F. Sprague, Jr., M. Tyers, and S. J. Elledge. 1997. Human CPR (cell cycle progression restoration) genes impart a Far− phenotype on yeast cells. Genetics 147:1063-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goyette, P., C. Feng Chen, W. Wang, F. Seguin, and D. Lohnes. 2000. Characterization of retinoic acid receptor-deficient keratinocytes. J. Biol. Chem. 275:16497-16505. [DOI] [PubMed] [Google Scholar]
- 16.Gripp, K. W., D. Wotton, M. C. Edwards, E. Roessler, L. Ades, P. Meinecke, A. Richieri-Costa, E. H. Zackai, J. Massague, M. Muenke, and S. J. Elledge. 2000. Mutations in TGIF cause holoprosencephaly and link NODAL signalling to human neural axis determination. Nat. Genet. 25:205-208. [DOI] [PubMed] [Google Scholar]
- 17.Hamada, H., C. Meno, D. Watanabe, and Y. Saijoh. 2002. Establishment of vertebrate left-right asymmetry. Nat. Rev. Genet. 3:103-113. [DOI] [PubMed] [Google Scholar]
- 18.Hebert, J. M., Y. Mishina, and S. K. McConnell. 2002. BMP signaling is required locally to pattern the dorsal telencephalic midline. Neuron 35:1029-1041. [DOI] [PubMed] [Google Scholar]
- 19.Horak, C. E., N. M. Luscombe, J. Qian, P. Bertone, S. Piccirrillo, M. Gerstein, and M. Snyder. 2002. Complex transcriptional circuitry at the G1/S transition in Saccharomyces cerevisiae. Genes Dev. 16:3017-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin, J. Z., S. Gu, P. McKinney, and J. Ding. 2006. Expression and functional analysis of Tgif during mouse midline development. Dev. Dyn. 235:547-553. [DOI] [PubMed] [Google Scholar]
- 21.Joyner, A. 1993. Gene targeting, a practical approach, vol. 126. Oxford University Press, New York, N.Y.
- 22.Kenney, A. M., M. D. Cole, and D. H. Rowitch. 2003. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development 130:15-28. [DOI] [PubMed] [Google Scholar]
- 23.Knoepfler, P. S., K. R. Calvo, H. Chen, S. E. Antonarakis, and M. P. Kamps. 1997. Meis1 and pKnox1 bind DNA cooperatively with Pbx1 utilizing an interaction surface disrupted in oncoprotein E2a-Pbx1. Proc. Natl. Acad. Sci. USA 94:14553-14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozar, K., M. A. Ciemerych, V. I. Rebel, H. Shigematsu, A. Zagozdzon, E. Sicinska, Y. Geng, Q. Yu, S. Bhattacharya, R. T. Bronson, K. Akashi, and P. Sicinski. 2004. Mouse development and cell proliferation in the absence of D-cyclins. Cell 118:477-491. [DOI] [PubMed] [Google Scholar]
- 25.Krek, W., and J. A. DeCaprio. 1995. Cell synchronization. Methods Enzymol. 254:114-124. [DOI] [PubMed] [Google Scholar]
- 26.Lai, K., B. K. Kaspar, F. H. Gage, and D. V. Schaffer. 2003. Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nat. Neurosci. 6:21-27. [DOI] [PubMed] [Google Scholar]
- 27.LeCouter, J. E., B. Kablar, P. F. Whyte, C. Ying, and M. A. Rudnicki. 1998. Strain-dependent embryonic lethality in mice lacking the retinoblastoma-related p130 gene. Development 125:4669-4679. [DOI] [PubMed] [Google Scholar]
- 28.Lo, R. S., D. Wotton, and J. Massague. 2001. Epidermal growth factor signaling via Ras controls the Smad transcriptional co-repressor TGIF. EMBO J. 20:128-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukaszewicz, A., P. Savatier, V. Cortay, P. Giroud, C. Huissoud, M. Berland, H. Kennedy, and C. Dehay. 2005. G1 phase regulation, area-specific cell cycle control, and cytoarchitectonics in the primate cortex. Neuron 47:353-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malumbres, M., R. Sotillo, D. Santamaria, J. Galan, A. Cerezo, S. Ortega, P. Dubus, and M. Barbacid. 2004. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell 118:493-504. [DOI] [PubMed] [Google Scholar]
- 31.Mann, R. S., and M. Affolter. 1998. Hox proteins meet more partners. Curr. Opin. Genet. Dev. 8:423-429. [DOI] [PubMed] [Google Scholar]
- 32.Melhuish, T. A., C. M. Gallo, and D. Wotton. 2001. TGIF2 interacts with histone deacetylase 1 and represses transcription. J. Biol. Chem. 276:32109-32114. [DOI] [PubMed] [Google Scholar]
- 33.Melhuish, T. A., and D. Wotton. 2000. The interaction of the carboxyl terminus-binding protein with the Smad corepressor TGIF is disrupted by a holoprosencephaly mutation in TGIF. J. Biol. Chem. 275:39762-39766. [DOI] [PubMed] [Google Scholar]
- 34.Ming, J. E., and M. Muenke. 2002. Multiple hits during early embryonic development: digenic diseases and holoprosencephaly. Am. J. Hum. Genet. 71:1017-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muenke, M., and P. A. Beachy. 2000. Genetics of ventral forebrain development and holoprosencephaly. Curr. Opin. Genet. Dev. 10:262-269. [DOI] [PubMed] [Google Scholar]
- 36.Nadeau, J. H. 2003. Modifier genes and protective alleles in humans and mice. Curr. Opin. Genet. Dev. 13:290-295. [DOI] [PubMed] [Google Scholar]
- 37.Nagy, A., M. Gertsenstein, K. Vintersten, and R. R. Behringer. 2003. Manipulating the mouse embryo: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Nanni, L., J. E. Ming, M. Bocian, K. Steinhaus, D. W. Bianchi, C. Die-Smulders, A. Giannotti, K. Imaizumi, K. L. Jones, M. D. Campo, R. A. Martin, P. Meinecke, M. E. Pierpont, N. H. Robin, I. D. Young, E. Roessler, and M. Muenke. 1999. The mutational spectrum of the sonic hedgehog gene in holoprosencephaly: SHH mutations cause a significant proportion of autosomal dominant holoprosencephaly. Hum. Mol. Genet. 8:2479-2488. [DOI] [PubMed] [Google Scholar]
- 39.Nomura, M., and E. Li. 1998. Smad2 role in mesoderm formation, left-right patterning and craniofacial development. Nature 393:786-790. [DOI] [PubMed] [Google Scholar]
- 40.Ohkubo, Y., C. Chiang, and J. L. Rubenstein. 2002. Coordinate regulation and synergistic actions of BMP4, SHH and FGF8 in the rostral prosencephalon regulate morphogenesis of the telencephalic and optic vesicles. Neuroscience 111:1-17. [DOI] [PubMed] [Google Scholar]
- 41.Overhauser, J., H. F. Mitchell, E. H. Zackai, D. B. Tick, K. Rojas, and M. Muenke. 1995. Physical mapping of the holoprosencephaly critical region in 18p11.3. Am. J. Hum Genet. 57:1080-1085. [PMC free article] [PubMed] [Google Scholar]
- 42.Purandare, S. M., S. M. Ware, K. M. Kwan, M. Gebbia, M. T. Bassi, J. M. Deng, H. Vogel, R. R. Behringer, J. W. Belmont, and B. Casey. 2002. A complex syndrome of left-right axis, central nervous system and axial skeleton defects in Zic3 mutant mice. Development 129:2293-2302. [DOI] [PubMed] [Google Scholar]
- 43.Roessler, E., and M. Muenke. 2001. Midline and laterality defects: left and right meet in the middle. Bioessays 23:888-900. [DOI] [PubMed] [Google Scholar]
- 44.Schneider, R. A., D. Hu, J. L. Rubenstein, M. Maden, and J. A. Helms. 2001. Local retinoid signaling coordinates forebrain and facial morphogenesis by maintaining FGF8 and SHH. Development 128:2755-2767. [DOI] [PubMed] [Google Scholar]
- 45.Shen, J., and C. A. Walsh. 2005. Targeted disruption of Tgif, the mouse ortholog of a human holoprosencephaly gene, does not result in holoprosencephaly in mice. Mol. Cell. Biol. 25:3639-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sirard, C., S. Kim, C. Mirtsos, P. Tadich, P. A. Hoodless, A. Itie, R. Maxson, J. L. Wrana, and T. W. Mak. 2000. Targeted disruption in murine cells reveals variable requirement for Smad4 in transforming growth factor beta-related signaling. J. Biol. Chem. 275:2063-2070. [DOI] [PubMed] [Google Scholar]
- 47.Spoelgen, R., A. Hammes, U. Anzenberger, D. Zechner, O. M. Andersen, B. Jerchow, and T. E. Willnow. 2005. LRP2/megalin is required for patterning of the ventral telencephalon. Development 132:405-414. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka, Y., Y. Okada, and N. Hirokawa. 2005. FGF-induced vesicular release of Sonic hedgehog and retinoic acid in leftward nodal flow is critical for left-right determination. Nature 435:172-177. [DOI] [PubMed] [Google Scholar]
- 49.Tao, B. 1994. Mutagenesis by PCR, p. 69-83. In H. Griffin and A. Griffin (ed.), PCR technology. Current innovations, vol. I. CRC Press, Boca Raton, Fla. [Google Scholar]
- 50.Vermot, J., and O. Pourquie. 2005. Retinoic acid coordinates somitogenesis and left-right patterning in vertebrate embryos. Nature 435:215-220. [DOI] [PubMed] [Google Scholar]
- 51.Wallis, D. E., E. Roessler, U. Hehr, L. Nanni, T. Wiltshire, A. Richieri-Costa, G. Gillessen-Kaesbach, E. H. Zackai, J. Rommens, and M. Muenke. 1999. Mutations in the homeodomain of the human SIX3 gene cause holoprosencephaly. Nat. Genet. 22:196-198. [DOI] [PubMed] [Google Scholar]
- 52.Wang, Z., and R. S. Mann. 2003. Requirement for two nearly identical TGIF-related homeobox genes in Drosophila spermatogenesis. Development 130:2853-2865. [DOI] [PubMed] [Google Scholar]
- 53.Warren, N., D. Caric, T. Pratt, J. A. Clausen, P. Asavaritikrai, J. O. Mason, R. E. Hill, and D. J. Price. 1999. The transcription factor, Pax6, is required for cell proliferation and differentiation in the developing cerebral cortex. Cereb. Cortex 9:627-635. [DOI] [PubMed] [Google Scholar]
- 54.Wotton, D., P. S. Knoepfler, C. D. Laherty, R. N. Eisenman, and J. Massague. 2001. The Smad transcriptional corepressor TGIF recruits mSin3. Cell Growth Differ. 12:457-463. [PubMed] [Google Scholar]
- 55.Wotton, D., R. S. Lo, S. Lee, and J. Massague. 1999. A Smad transcriptional corepressor. Cell 97:29-39. [DOI] [PubMed] [Google Scholar]
- 56.Wotton, D., R. S. Lo, L. A. Swaby, and J. Massague. 1999. Multiple modes of repression by the Smad transcriptional corepressor TGIF. J. Biol. Chem. 274:37105-37110. [DOI] [PubMed] [Google Scholar]
- 57.Xuan, S., C. A. Baptista, G. Balas, W. Tao, V. C. Soares, and E. Lai. 1995. Winged helix transcription factor BF-1 is essential for the development of the cerebral hemispheres. Neuron 14:1141-1152. [DOI] [PubMed] [Google Scholar]
- 58.Yang, Y., C. K. Hwang, U. M. D'Souza, S. H. Lee, E. Junn, and M. M. Mouradian. 2000. Three-amino acid extension loop homeodomain proteins Meis2 and TGIF differentially regulate transcription. J. Biol. Chem. 275:20734-20741. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.