Abstract
p14ARF is a tumor suppressor that controls a well-described p53/Mdm2-dependent checkpoint in response to oncogenic signals. Here, new insights into the tumor-suppressive function of p14ARF are provided. We previously showed that p14ARF can induce a p53-independent G2 cell cycle arrest. In this study, we demonstrate that the activation of ATM/ATR/CHK signaling pathways contributes to this G2 checkpoint and highlight the interrelated roles of p14ARF and the Tip60 protein in the initiation of this DNA damage-signaling cascade. We show that Tip60 is a new direct p14ARF binding partner and that its expression is upregulated and required for ATM/CHK2 activation in response to p14ARF. Strikingly, both p14ARF and Tip60 products accumulate following a cell treatment with alkylating agents and are absolutely required for ATM/CHK2 activation in this setting. Moreover, and consistent with p14ARF being a determinant of CHK2 phosphorylation in lung carcinogenesis, a strong correlation between p14ARF and phospho-CHK2 (Thr68) protein expression is observed in human lung tumors (P < 0.00006). Overall, these data point to a novel regulatory pathway that mediates the p53-independent negative-cell-growth control of p14ARF. Inactivation of this pathway is likely to contribute to lung carcinogenesis.
ARF (known as p14ARF in humans and p19ARF in mice) was originally identified as an alternative transcript of the Ink4a/ARF tumor suppressor locus, a gene that encodes the p16Ink4a inhibitor of cyclin-dependent kinases (39). By virtue of its unique exon, the ARF transcript encodes a protein that is unrelated to p16Ink4a (35). Nevertheless, like p16Ink4a, ARF exhibits tumor suppressor functions, as demonstrated by the tumor susceptibility phenotype of p14ARF-deficient mice (22). It is well known that the ARF-p53-Mdm2 pathway serves as a checkpoint that protects cells from oncogene-induced transformation (21, 34). However, an increasing number of studies now points to the fact that not all ARF tumor suppressor functions are elicited through the p53-Mdm2 pathway. These new aspects of p53-independent ARF functions have been supported mainly by the characterization of a wide range of new ARF binding partners, such as the B23/nucleophosmin protein involved in ribosome biogenesis (7, 20), the E2F-1 transcription factor (14) and its cofactor DP1 (12) involved in S phase progression, and the recently identified E3 ubiquitin ligase ARF-BP1/Mule (10). In contrast, the cellular signaling pathways involved in these new functions of ARF remain largely unknown.
The DNA damage-signaling pathway is an essential process that is activated following exposure to numerous types of genotoxic stresses and serves to influence or achieve DNA repair, cell cycle delay, and/or apoptosis in order to maintain genomic stability (46). Two members of the phosphatidylinositol 3 (PI-3)-kinase-related kinase family, ATM (for ataxia telangiectasia mutated) and ATR (for ataxia telangiectasia and Rad3 related), play a central role in DNA damage recognition and initial related phosphorylation events (for a review, see reference 36). It is presently well known that DNA single- or double-strand breaks that occur after cellular treatment with radiation or chemotherapeutic drugs activate both ATM and ATR kinases, leading to the recruitment and phosphorylation of a set of cellular proteins involved in the DNA damage response (for a review, see reference 40). Recently, the exposure of cells with a chromatin-modifying drug was also reported to activate ATM, leading to the proposal that ATM activation may result from changes in the structure of chromatin instead of direct binding to DNA strand breaks (2).
Initially discovered as a Tat-interacting protein, Tip60 is a chromatin-modifying enzyme which appears to be a key component in the antiproliferative cellular response. Indeed, Tip60 is required for p19ARF/p53-mediated proliferation arrest (6). Furthermore, its suppression abolishes the G1 cell cycle arrest induced by ionizing radiation, thereby highlighting the involvement of Tip60 in genotoxic signaling networks (6). In agreement with such a role, Tip60 accumulates in cells exposed to UV radiation (26), is required for DNA repair and apoptosis following gamma radiation (19), and has been involved in the exchange of drosophila phospho-H2Av with a nonmodified H2Av at DNA lesions (25). Interestingly, Sun and colleagues recently reported that Tip60 activates ATM following genotoxic stresses, demonstrating that Tip60 is a critical component of the signal transduction pathway that links the detection of DNA breaks to the DNA damage response (42).
We have previously reported that ARF can induce cell growth arrest in tumor cells that lack a functional p53 gene (15). In this study, we demonstrate that the ATM/ATR DNA damage-signaling pathways contribute to the p53-independent tumor suppressor function of p14ARF and provide evidence that Tip60 and p14ARF display interrelated roles in this setting. Moreover, we show that p14ARF is a determinant of CHK2 phosphorylation in lung carcinogenesis. Overall, these data point to a novel regulatory pathway that mediates the p53-independent tumor suppressor function of ARF. Inactivation of this pathway is likely to contribute to lung carcinogenesis.
MATERIALS AND METHODS
Cell lines, cell growth assays, plasmids, and transfection.
H358 and H1299 human lung carcinoma cell lines were cultured in 5% CO2 at 37°C in RPMI 1640 medium (GIBCO, Cergy Pontoise, France) supplemented with 10% (vol/vol) heat-inactivated fetal calf serum. Normal human bronchial epithelial (NHBE; BioWhittaker) cells were cultured in bronchial epithelial cell growth medium. 293 and COS cells as well as MRC5 human fibroblasts were cultured in Dulbecco modified Eagle medium (GIBCO) supplemented with 10% (vol/vol) heat-inactivated fetal calf serum. H358/Tet-On control cells and H358/Tet-On/p14ARF-inducible clones were obtained as previously described (15). Expression of p14ARF was induced when cells were cultured in the presence of 1 μg/ml doxycycline. Clonogenic assays were performed as previously described (15). Transient transfections were carried out using Fugene 6 (Roche Diagnostic). Plasmids used in transient transfections were pcDNA3, pcDNA3-p14ARF, pcDNA3-hemagglutinin (HA)-tagged actinin 4, and pcDNA3-HA-tagged Tip60. The plasmid encoding glutathione S-transferase (GST)-Cdc25C was obtained from Helen Piwinica-Worms (St. Louis, MO).
Cell treatment.
Caffeine, cisplatin, cyclophosphamide monohydrate, doxorubicin, etoposide, and methanesulfonic acid methyl ester were all purchased from Sigma (Saint Quentin Fallavier, France). Cigarette smoke condensate (CSC) was obtained by mechanically smoking cigarettes. The particulate phase of smoke was collected on glass fiber filters and the amount obtained determined by the weight increase of the filter. CSC was prepared by dissolving the collected smoke particles in dimethyl sulfoxide, and aliquots were kept at −80°C. Stable p14ARF-inducible clones were cotreated for 72 h with or without doxycycline (1 μg/ml) in the presence or absence of caffeine (2 to 4 mM). Treatment of H358 cells with cyclophosphamide (10 μM), doxorubicin (10 μM), etoposide (10 μM), methanesulfonic acid methyl ester (10 μM), or CSC (3 to 30 μg/ml) was performed for 24 h. Protein translation inhibition was achieved by treating cells with 30 μg/ml of cycloheximide (CX; Sigma) 24 h posttransfection.
Antibodies.
The anti-Cdc2 p34 (sc-54), anti-Chk1 (G-4), anti-Chk2 (H-300), anti-HA (Y-11), and anti-p14ARF (C-18) antibodies were purchased from Santa Cruz; the anti-p14ARF (Ab-2), anti-ATM (Ab-1), anti-ATR (Ab-2), anti-p14ARF (Ab-1), and anti-Hsp70 (clone JG1; ABR) antibodies were from Oncogene Research; the anti-phospho-ATM (Ser1981), anti-phospho-(Ser/Thr) ATM/ATR substrate, anti-phospho-Cdc2 (Tyr15), anti-phospho-Cdc25c (Ser216), anti-phospho-Chk1 (Ser345), anti-phospho-Chk2 (Thr68), anti-phospho-Rad17 (Ser645), and anti-Rad17 antibodies were from Cell Signaling; the anti-Cdc25c (67211A) antibody was from Pharmingen; the anti-phospho-histone H2AX (Ser139) (clone JBW301) antibody was from Upstate; and the anti-actin (20-33) antibody was from Sigma. Anti-Tip60 antibody was kindly provided by B Amati.
Transfection of siRNA oligonucleotides.
The sequences designed to specifically target human p14ARF, chk1, chk2, atm, atr, and tip60 RNAs were as follows: for p14ARF, 5′-GAACAUGGUGCGCAGGUUCTT-3′; for chk1, 5′-GAAGCAGUCGCAGUGAAGATT-3′; for chk2, 5′-GAACCUGAGGACCAAGAACTT-3′; for atm, 5′-GCGCCUGAUUCGAGAUCCUTT-3′; for atr, 5′-CCUCCGUGAUGUUGCUUGATT-3′; and for tip60, 5′-AAGAAGAUCCAGUUCCCCAAGTT-3′. The scrambled small interfering RNA (siRNA) oligonucleotides used as controls for all RNA interference experiments were as follows: 5′-AAAGGUGACGCUGACGAAGTT-3′ and 5′-CAAGAAAGGCCAGUCCAAGTT-3′. Cells were transfected with siRNA oligonucleotide duplexes using jetSI reagent (Polyplus Transfection, Illkirch, France). Doxycycline (1 μg/ml) was added or not added to the culture medium 4 h after transfection. The cells were analyzed 72 h posttransfection. For experiments with alkylating agents, cells were transfected for 48 h with mismatch, p14ARF, or tip60 siRNAs; then, alkylating agents were added for 24 additional hours.
Cell cycle analysis.
Cells were washed twice in phosphate-buffered saline (PBS) and fixed in ice-cold ethanol for 10 min at −20°C. After two 1× PBS washes, cells were incubated at 37°C for 10 min with 200 U/ml of RNase A (Sigma) and stained with propidium iodide (10 μg/ml in PBS). Cell cycle distribution was determined by flow cytometry using the Cellfit software (Becton Dickinson, Grenoble, France).
Immunoblotting and immunoprecipitation.
Immunoblotting and coimmunoprecipitation experiments were carried out as previously described (15, 26). CHK1/2 kinase assays were performed according to the protocol of Ahn and Prives (1). Briefly, CHK1 or CHK2 was immunoprecipitated and incubated for 30 min at 30°C in the presence of 1 to 2 μg GST or GST-Cdc25C fusion protein prepared according to the manufacturer's protocol (bulk GST purification module; Pharmacia Biotech). Immunoblotting was performed on a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel, and Cdc25C phosphorylation at serine 216 was detected using a specific phospho-antibody. GST fusion proteins were detected by immunoblotting using an anti-GST antibody. For coprecipitation of endogenous Tip60 and p14ARF proteins, nuclear extracts of H1299 cells were prepared by lysing the cells in a buffer containing 15 mM NaCl, 60 mM KCl, 12% sucrose, 2 mM EDTA, 0.5 mM EGTA, 0.65 mM spermidine, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, and 0.1% Triton X-100. Nuclei were pelleted by centrifugation and further lysed for 30 min at 4°C in 1 ml of 300 mM NaCl, 50 mM Tris (pH 8.0), 0.4% NP-40, 10 mM MgCl2, and 2.5 mM CaCl2, supplemented just before use with protease and phosphatase inhibitors. After a 15-min centrifugation step at 10,000 rpm, supernatants containing nuclear proteins were recovered and diluted in 1 ml of 50 mM Tris-HCl (pH 8.0) and 0.4% NP-40. One milligram of total nuclear extracts was then subjected to immunoprecipitation using the anti-p14ARF antibody (C-18; Santa Cruz).
Indirect immunofluorescence.
Cells were fixed in 2% paraformaldehyde-PBS for 5 min at room temperature, washed one time in PBS, and permeabilized in 100% methanol for 20 min at −20°C. After extended washes, nonspecific binding sites were saturated for 45 min at room temperature in the presence of 1% bovine serum albumin and 5% goat serum in PBS, and incubation was carried out with appropriate primary antibody. Cells were then incubated with Alexa 568 or Alexa 488 (Interchim), counterstained with Hoechst 33352, and observed using an Olympus microscope (×40 to ×60 magnification). Images were captured with a Coolview charge-coupled-device camera (Photonic Science) and digitally saved using Visilog software.
GST pull-down assay.
GST pull-down assays were performed as previously described (14), using beads coated with either GST or GST-p14ARF fusion proteins and equivalent amounts of in vitro-translated, wild-type Tip60 protein.
Tissue samples and immunohistochemistry.
One hundred six lung tumor samples were obtained at surgical resections of primary lung tumors or diagnostic lymph node mediastinoscopy. The tumors were classified according to the 1999 World Health Organization (WHO) histological classification of lung tumors (43) and comprised 58 non-small-cell lung carcinomas and 48 neuroendocrine (NE) lung tumors. p14ARF immunostaining was carried out on cryosections of frozen samples using the anti-p14ARF (C-20) antibody as previously described (16). CHK2 and P-CHK2 (Thr68) immunostaining was performed on 3-μm-thick serial paraffin sections using a Ventana Discovery autostainer (Ventana Medical International, Inc., Tucson, AZ). Nonimmune mouse or rabbit sera were used as negative controls for all experiments. Immunostaining was interpreted independently by two observers. Immunostaining scores were calculated by multiplying the percentage of labeled cells (from 0 to 100) by the intensity of staining (1+ to 3+). Tumors with scores of ≥20 were considered positive for p14ARF, in accordance with our previous report (16). Nonneoplastic fibroblasts and endothelial cells, as well as normal adjacent lung tissue showing nuclear reactivity, were used as positive internal controls. Tumor samples exhibiting total scores of ≥10 were considered positive for P-CHK2 (Thr 68) reactivity. As no internal positive staining was available for P-CHK2, its detection in some tumors of the same analysis set was used as a positive control. Tumors were considered positive for CHK2 expression when their overall scores were ≥40. In this case, basal cells of normal bronchi showing nuclear staining were used as positive internal controls.
RESULTS
Caffeine treatment prevents the G2 arrest induced by p14ARF.
We have previously demonstrated the ability of p14ARF to induce a G2 arrest in stably transfected p14ARF-inducible clones derived from the p53-deficient H358 human lung adenocarcinoma cell line (H358/Tet-On-p14ARF) (15). In this setting, we aimed to investigate the cellular signaling pathway(s) involved in this process. To do this, we first assessed the effects of various specific pharmacological inhibitors on the antiproliferative capacity of p14ARF. Interestingly, we observed that treating cells with caffeine significantly prevented both the cell growth inhibition (Fig. 1A) and the G2 arrest (Fig. 1B) induced by p14ARF. To more fully examine the caffeine effect on the p14ARF-induced G2 arrest, immunoblotting experiments were carried out. We previously showed that inactive phospho-Cdc2 (Tyr15) and phospho-Cdc25c (Ser216) proteins accumulated in cells arrested in G2 upon p14ARF expression (15). We observed here that caffeine prevented the accumulation of both phosphorylated products (Fig. 1C), indicating that inhibition of G2 arrest results from modifications upstream of Cdc2 and Cdc25c.
FIG. 1.
Caffeine prevents the antiproliferative effect of p14ARF. H358/Tet-On/p14ARF cells were incubated for 72 h in the presence (+) or absence (−) of 1 μg/ml doxycycline (Dox) with or without caffeine as indicated. (A) Cell survival was evaluated after methylene blue staining. In each case, the growth of cells cultured without doxycycline in the presence or absence of caffeine was normalized to 100%. Results are the means ± standard deviations of three independent experiments performed in duplicate. The P value was calculated by comparing the growth statuses of doxycycline-treated cells cultured in the presence or absence of caffeine. **, P < 0.01 (Student's t test). (B and C) Cell cycle and Western blot analyses. Representative data of at least three independent experiments are shown.
CHK1/2 activation is implicated in the G2 arrest mediated by p14ARF.
As caffeine is a known inhibitor of CHK1/2 kinase signaling pathways, our data suggested that the CHK kinases were involved in the antiproliferative effect of p14ARF. In response to various types of DNA damage, CHK1 and CHK2 are activated by phosphorylation at serines 345 and 317 and at threonine 68, respectively (45). By using Western blot experiments, we reproducibly observed that p14ARF induced a mobility shift for both proteins, highly suggestive of their phosphorylation (Fig. 2A, left panel). Accordingly, when specific phospho-antibodies were used, accumulation of P-CHK1 (Ser345) and P-CHK2 (Thr68) products was detected upon p14ARF expression, although the total CHK1/2 expression levels did not vary. These effects were not observed in an H358/Tet-On control clone cultured in the presence of doxycycline (Fig. 2A, right panel), ensuring the specificity of the p14ARF-dependent responses. Importantly, phosphorylation of CHK1 and CHK2 was also detected in MRC5 human fibroblasts transfected with a plasmid encoding p14ARF compared to cells transfected with a control vector (Fig. 2B). To further confirm the activation of CHK1/2 by p14ARF, kinase assays in which CHK1 or CHK2 immunoprecipitates from H358/Tet-On/p14ARF cells were tested for their capacity to phosphorylate a recombinant GST-Cdc25C fusion protein, a known direct substrate of CHK1/2, were performed. In these experiments, GST-Cdc25C was efficiently phosphorylated by either CHK1 or CHK2 following p14ARF induction (Fig. 2C). Taken together, these results demonstrate the ability of p14ARF to activate CHK1/2 kinases in tumors as well as in primary cells.
FIG. 2.
p14ARF activates CHK1/2 kinases to arrest cells in G2. (A) Expression of CHK1 and CHK2 proteins was studied by Western blotting in H358/Tet-On/p14ARF cells cultured for 72 h in the presence or absence of doxycycline (Dox) as indicated. Phosphorylation of CHK1 and CHK2 proteins was confirmed using specific phospho-antibodies. (B) The same analysis was performed with MRC5 human fibroblasts transiently transfected with control pcDNA3.1 or pcDNA3.1/p14ARF vector. (C) Equal amounts of CHK1 or CHK2 protein were immunoprecipitated (IP) from H358/Tet-On/p14ARF cells and tested for their ability to phosphorylate a GST-Cdc25C fusion protein. Irrelevant mouse or rabbit immunoglobulins (IgG) were used as negative controls. Phosphorylation of GST-Cdc25C at the serine 216 residue was assessed by immunoblotting using a specific phospho-antibody. GST or GST-Cdc25C recombinant proteins were detected by immunoblotting using an anti-GST antibody (arrows). (D) H358/Tet-On/p14ARF cells were transfected for 72 h with either mismatch or specific chk1 and chk2 siRNAs as indicated and subjected to Western blot (left panel) and cell cycle (right panel) analyses. Apoptosis was detected by immunoblotting with an anti-active caspase 3 antibody.
We then asked whether CHK1/2 activation was involved in p14ARF-mediated G2 arrest. To test this hypothesis, H358/Tet-On/p14ARF cells were transfected with siRNAs specifically targeting chk1 or chk2, and the cell cycle profile was analyzed in the absence or presence of p14ARF induction. We repeatedly observed that neutralization of either CHK1 or CHK2 partially prevented the G2 arrest mediated by p14ARF, suggesting that each kinase plays a role in this process (data not shown). Accordingly, when the expressions of both kinases were simultaneously knocked down (Fig. 2D, left panel), the proportion of p14ARF-expressing cells arrested in G2 strongly decreased (Fig. 2D, right panel). Moreover, we also observed, under these conditions, the appearance of a population of cells with a sub-G1 DNA content, in which apoptosis was confirmed by the detection of an active caspase 3 fragment (Fig. 2D, left panel). Therefore, it can be assumed that in the absence of the CHK1/2 kinases, the impaired ability of p14ARF to sustain G2 arrest leads to the induction of apoptosis. Interestingly, a faint sub-G1 peak was also detected in the absence of p14ARF induction, suggesting that inactivation of both kinases might be deleterious in proliferating cells.
p14ARF activates ATM/ATR pathways to mediate G2 arrest.
The pathway involved most frequently in the activation of the CHK proteins is the well-known ATM/ATR DNA damage response pathway, in which CHK1 and CHK2 act as downstream transducers (3). Interestingly, ATM activity was previously reported to contribute to the tumor-suppressive functions of p14ARF through the phosphorylation and accumulation of p53 protein (29). We therefore asked whether activation of ATM could be also involved in some aspects of the p53-independent functions of p14ARF. To test this hypothesis, we first used an affinity-purified antibody recognizing an ATM/ATR/DNA-PK/ATX-phosphorylated consensus target sequence (phosphorylated pS/TQ). As observed in Fig. 3A, p14ARF increased the expression level or led to the appearance of several phosphorylated products (upper panel) which accumulated into numerous nuclear foci (lower panel). Interestingly, these proteins disappeared upon a λ-phosphatase treatment (upper panel), confirming that they were specific phosphorylated products. Taken together, these results were consistent with the activation of some of these ATM/ATR/DNA-PK/ATX signaling pathways in response to p14ARF. As ATM kinase is activated by autophosphorylation at serine 1981 (2), we then used a specific phospho-antibody to look at ATM status. As shown in Fig. 3A (upper panel), the accumulation of a P-ATM (Ser1981) product which disappeared following a λ-phosphatase treatment prior to electrophoresis was observed upon p14ARF expression. Interestingly, we also repeatedly noticed that p14ARF caused the appearance of a higher-migrating ATR band that shifted to a faster-migrating band following the treatment of the cellular extracts with λ-phosphatase, thereby suggesting ATR phosphorylation. To strengthen the notion that p14ARF activates the ATM/ATR signaling pathways, we investigated whether p14ARF could induce the activation of two known ATM/ATR targets, namely, the Rad17 and histone H2AX proteins. In response to DNA damage, these two proteins are rapidly phosphorylated at the Ser645 and Ser139 residues, respectively, and accumulate into nuclear foci (9, 48). Using immunoblotting and immunofluorescence experiments, we clearly observed that phospho-H2AX (Ser139) (γ-H2AX) and P-Rad17 (Ser645) products accumulated into nuclear foci upon p14ARF induction (Fig. 3B). Interestingly, we noticed that these nuclear foci did not colocalize with p14ARF staining, which was observed predominantly in the nucleoli as previously described (15). Furthermore, and in agreement with a previous study (41), accumulation of P-CHK1 (Ser345) or P-CHK2 (Thr68) proteins was never detected within nuclear foci under the same conditions (data not shown). Of note, we also detected a faint γ-H2AX signal in the noninduced cells, which could be consistent with recent data demonstrating that phosphorylation of H2AX can also occur in normally growing cells (18, 31). Collectively, our results indicate that p14ARF activates ATM/ATR signaling pathways.
FIG. 3.
p14ARF activates ATM/ATR pathways to induce G2 arrest. (A, B) H358/Tet-On/p14ARF cells were incubated for 72 h in the presence (+) or absence (−) of doxycycline (Dox) and subjected to Western blot and immunofluorescence analyses. pS/TQ antibody recognizes an ATM/ATR/DNA-PK/ATX-phosphorylated consensus target sequence. Phosphorylations of ATM, Rad17, and H2AX were revealed using specific anti-P-ATM (Ser1981), anti-P-Rad17 (Ser645), and anti-γ-H2AX (Ser139) antibodies. Treatment with λ-phosphatase (λ ppase) before loading confirmed the phosphorylation of ATR, ATM, and ATM/ATR substrates upon p14ARF expression. Immunolocalization of p14ARF (green), pS/TQ, γ-H2AX, and P-Rad17 (Ser645) (red) proteins was visualized by coimmunofluorescence in the same cells. Note that the p14ARF staining does not colocalize with pS/TQ ATM/ATR, γ-H2AX, or P-Rad17 (Ser645) nuclear foci (Merge, yellow). (C, D) H358/Tet-On/p14ARF cells were transfected with either mismatch or atm or atr siRNAs for 72 h and subjected to cell cycle (C) and Western blot (D) analyses. The effect of siRNA transfection on ATM or ATR expression is shown in the panel above the histogram. Cell cycle data are the means ± standard deviations of three independent experiments.
To assess the role of ATM/ATR activation in the antiproliferative capacity of p14ARF, we performed siRNA experiments specifically targeting atm or atr in H358/Tet-On/p14ARF cells. Our data showed that neutralization of either ATM or ATR prevented the G2 arrest (Fig. 3C) as well as the accumulation of P-CHK1 (Ser345) and P-Rad17 (Ser645) proteins induced by p14ARF (Fig. 3D). Interestingly, we noticed that the phosphorylation of CHK2 at the Thr68 residue was specifically abrogated following ATM neutralization while the phosphorylation status of histone H2AX was unchanged, suggesting that other PI-3 kinases, such as DNA-PK, could be required for p14ARF to mediate the activation of H2AX. Taken together, these data demonstrate that both ATM/ATR pathways are involved in the G2 checkpoint mediated by p14ARF in a p53-independent context.
p14ARF interacts physically with the Tip60 protein.
A series of experiments was planned to investigate how p14ARF could activate ATM/ATR signaling pathways. Changes in chromatin structure have emerged as an additional mechanism that contributes to the activation of ATM (2), and the chromatin-modifying enzyme Tip60 was recently reported to stimulate ATM autophosphorylation (42). As Tip60 was previously ascribed to an ARF/p53 network (6), we hypothesized that both Tip60 and p14ARF proteins might also be closely connected on p53-independent pathways. To test this hypothesis, we first asked whether p14ARF could interact with Tip60. H1299 cells were transfected with expression vectors encoding p14ARF and/or HA-tagged Tip60 proteins, and coimmunoprecipitation experiments were performed. As Fig. 4A illustrates, HA-Tip60 was detected in p14ARF immunoprecipitates, revealing that both proteins interact. Importantly, after the immunoprecipitation of the endogenous p14ARF, we also observed the coimmunoprecipitation of a fraction of endogenous Tip60 in H1299 nuclear extracts, thus indicating that both proteins can interact in vivo (Fig. 4B). It should be noted that reciprocal immunoprecipitation-Western blot analyses could not be performed, owing to ineffective Tip60 antibodies for immunoprecipitation experiments. Since Tip60 and p14ARF have already been physically linked to Mdm2 (26, 34), their coprecipitation could be the result of Mdm2 binding. To test whether Tip60 and p14ARF could directly complex, an in vitro GST pull-down assay was set up using recombinant GST-p14ARF as bait. As shown in Fig. 4C, a fraction of in vitro-translated HA-Tip60 was able to bind GST-p14ARF, therefore identifying Tip60 as a new direct p14ARF binding partner.
FIG. 4.
p14ARF interacts physically with Tip60. (A) COS cells were transfected with pcDNA3.1-HA-tagged Tip60 (3 μg) and/or pcDNA3.1-p14ARF (1 μg) vector. Whole-cell extracts were prepared and subjected to immunoprecipitation (IP) with anti-p14ARF antibody (C-18) or an irrelevant goat serum as a negative control (Ctl). Immunoprecipitates were tested for the presence of HA-Tip60 and p14ARF using anti-HA and anti-p14ARF antibodies, respectively (left panel). In the right panel, whole-cell extracts were loaded directly and subjected to a Western blotting with both the anti-HA and the anti-p14ARF antibodies. (B) Endogenous p14ARF was immunoprecipitated from H1299 nuclear extracts with anti-p14ARF (C-18) antibody or an irrelevant goat serum as a negative control (Ctl). p14ARF and Tip60 were revealed by Western blots using anti-p14ARF and anti-Tip60 antibodies, respectively. (C) Beads harboring bacterially produced GST-p14ARF or control GST proteins were incubated with an in vitro-translated HA-tagged Tip60 in a GST pull-down experiment and analyzed by Western blot analysis using anti-HA and anti-GST antibodies.
p14ARF stabilizes the Tip60 protein.
When performing the experiments described above, we consistently observed that the level of Tip60 protein was upregulated when p14ARF was transfected (Fig. 4A, right panel). To more fully investigate whether p14ARF could affect Tip60 expression, we cotransfected various cell lines with cytomegalovirus-driven expression vectors for HA-tagged Tip60 and HA-tagged actinin 4 as an internal control, either in the presence or in the absence of p14ARF. Our results showed that the amount of HA-Tip60 significantly increased in the presence of exogenous p14ARF whereas the amount of control HA-actinin 4 was unaffected (Fig. 5A). Similar results were also observed for the endogenous Tip60 protein in stable p14ARF-inducible clones cultured in the presence of doxycycline (Fig. 6A and data not shown). These data revealed that p14ARF induces Tip60 accumulation. To evaluate whether p14ARF modulates the stability of the Tip60 protein, H1299 cells were cotransfected with HA-Tip60 and HA-actinin 4 expression vectors either in the presence or in the absence of p14ARF and incubated with CX, an inhibitor of protein translation. The amounts of HA-Tip60 and HA-actinin 4 were then analyzed by immunoblotting at different times during cycloheximide treatment. As shown in Fig. 5B, p14ARF expression was associated with a significant decrease in Tip60 decay. Taken together, these data indicate that p14ARF stabilizes the Tip60 protein.
FIG. 5.
p14ARF stabilizes the Tip60 protein. (A) H358 and H1299 cells were cotransfected with pcDNA3.1-HA-tagged Tip60 (HA-Tip60; 1 μg) and pcDNA3.1-HA-tagged actinin 4 (HA-Act 4; 1 μg) as an internal control, in the presence (+) or absence (−) of pcDNA3.1-p14ARF (3 μg). Tip60 and actinin 4 (Act 4) were detected by Western blotting using anti-HA antibody. Anti-p14ARF antibody was used to detect p14ARF. Actin was used as a loading control. (B) H1299 cells were transfected with pcDNA3.1-HA-Tip60 (1 μg) and pcDNA3.1-HA-actinin 4 (1 μg) in the presence (+p14ARF) or absence (−p14ARF) of pcDNA3.1-p14ARF (3 μg) and treated with CX for the indicated durations. Whole-cell extracts were then subjected to Western blotting using antiactin, anti-HA, and anti-p14ARF antibodies. Tip60 densitometric signals were normalized to actin as a loading control. A 100% value was arbitrarily assigned to the signal obtained at zero time of cycloheximide treatment. Results are the means ± standard deviations of three independent experiments.
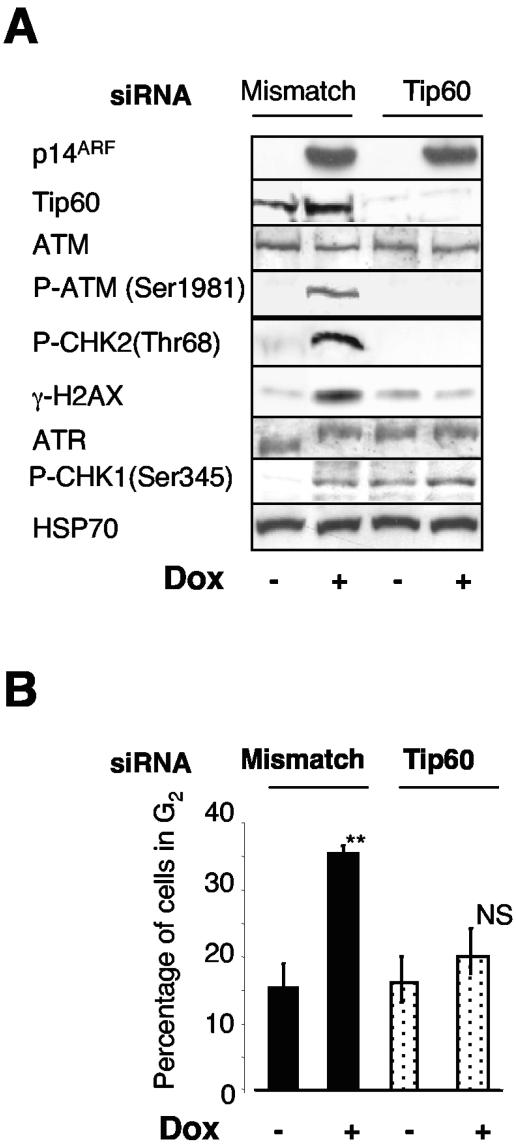
FIG. 6.
Tip60 is required for p14ARF-mediated ATM/CHK2 activation. H358/Tet-On/p14ARF cells cultured in the presence (+) or absence (−) of doxycycline (Dox) were transfected for 72 h with either mismatch or Tip60 siRNA. (A) ATM, ATR, CHK1, CHK2, and H2AX activations were studied by immunoblotting using specific phospho-antibodies. Neutralization of Tip60 was revealed by Western blots using an anti-Tip60 antibody. HSP70 was used as a loading control. Results are representative of three independent experiments. (B) The cell cycle profile of the same cells was analyzed by fluorescence-activated cell sorter cytometry. Results are the means ± standard deviations of three independent experiments.
Tip60 is required for p14ARF-mediated ATM/CHK2 activation.
Having provided evidence that Tip60 is targeted by p14ARF, we then investigated whether it was involved in the activation of the DNA damage-signaling pathways mediated by p14ARF. The H358/Tet-On/p14ARF cells were transfected with siRNAs against tip60, and ATM/ATR/CHK activation and the cell cycle profile were analyzed. As Fig. 6 illustrates, neutralization of Tip60 strongly impaired the ability of p14ARF to induce the accumulation of phosphorylated ATM, CHK2, and H2AX proteins as well as to stop the cells in G2. These data therefore indicated that Tip60 is absolutely required for the activation of ATM/CHK2 by p14ARF in a p53-independent context. Interestingly, and consistent with Tip60 acting upstream of CHK2, we noted that Tip60 neutralization prevented the p14ARF-mediated G2 arrest to the same extent as CHK2 knockdown (data not shown). Furthermore, we observed that p14ARF-induced H2AX phosphorylation was abrogated in Tip60-depleted cells (Fig. 6A) while it was unaffected by ATM or ATR knockdown (Fig. 3D). This suggests that Tip60 might also control the activation of other PI-3 kinases, such as DNA-PK, to regulate the phosphorylation of H2AX in response to p14ARF. Also interesting was the stimulation of CHK1 phosphorylation as well as the shift of ATR when Tip60 was knocked down in the absence of p14ARF induction (Fig. 6A, compare lanes 1 and 3). These data support the notion that Tip60 plays a role in ATR/CHK1 activation independently of p14ARF, which could be consistent with its involvement in G2/M transition (28). Therefore, in order to distinguish between the intrinsic effect of Tip60 on the cell cycle and its role in the p14ARF-mediated activation of the DNA damage-signaling pathway, we decided to focus further studies on the ATM/CHK2 pathway, which was not affected by Tip60 neutralization alone.
p14ARF and Tip60 are upregulated and required for ATM/CHK2 activation in response to DNA-damaging agents.
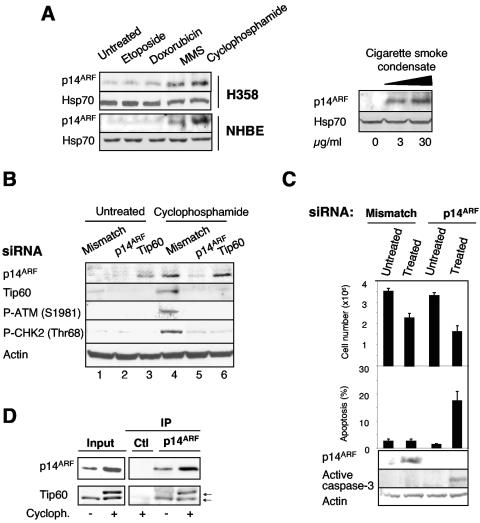
Our results so far demonstrated the ability of p14ARF to stimulate ATM/ATR/CHK signaling pathways in a model of overexpression. Therefore, we attempted to identify some of the upstream signals that could trigger the physiological activation of these pathways by p14ARF. Anticancer agents are among the numerous genotoxic insults which activate the DNA damage response pathways (45). Thus, we first tested the effects of several of these agents on p14ARF expression levels. As shown in Fig. 7A (left panel), treating H358 cells with cyclophosphamide and methyl methanesulfonate, two alkylating agents, clearly increased p14ARF expression levels. In contrast, the topoisomerase inhibitors, etoposide and doxorubicin, had no effect. Similar results were obtained with NHBE cells (Fig. 7A, left panel). Interestingly, p14ARF expression levels were also upregulated in the presence of CSC (Fig. 7A, right panel). Taken together, these data reveal that p14ARF is a target of genotoxic agents. To investigate the functional significance of this p14ARF induction, the protein was neutralized using appropriate siRNA and the activation of the ATM/CHK2 pathway was analyzed by Western blotting with H358 cells cultured in the presence or absence of cyclophosphamide. As expected, accumulation of activated P-ATM and P-CHK2 proteins was observed in response to cyclophosphamide, although total ATM and CHK2 protein levels were similar (Fig. 7B, compare lanes 1 and 4; also data not shown), and was correlated with cell growth inhibition (Fig. 7C, upper panel). Strikingly, the accumulation of both P-ATM and P-CHK2 proteins was prevented by the use of p14ARF siRNA (Fig. 7B, compare lanes 4 and 5), therefore revealing a direct involvement of p14ARF in the activation of the ATM/CHK2 pathway in response to this genotoxic stress. In these settings, we observed the appearance of an obvious population of apoptotic cells, as demonstrated by Hoechst staining and caspase 3 activation, which was not detected in mismatch-transfected, treated cells (Fig. 7C, lower panels). Therefore, it can be assumed that the impaired ability of p14ARF-deprived cells to activate the DNA damage checkpoint in response to alkylating agents ultimately leads to cell death. These results are reminiscent of our previous data showing the induction of apoptosis upon p14ARF expression in cells deprived of CHK1 and CHK2 kinases (Fig. 2D). Then, to complete our investigations, we studied the role of Tip60. By using an anti-Tip60 antibody, we clearly observed an upregulation of Tip60 expression in cells treated with cyclophosphamide (Fig. 7B, compare lanes 1 and 4), which was in agreement with previous data reporting that Tip60 accumulates following DNA damage, in that case from UV radiation (26). Under these conditions, neutralization of Tip60 with siRNA abolished the accumulation of P-ATM/CHK2 despite p14ARF induction (Fig. 7B, compare lanes 4 and 6), providing evidence that Tip60 is crucial for a proper p14ARF-dependent response to cyclophosphamide. Of note, we observed that Tip60 protein levels decreased following p14ARF neutralization in both treated (Fig. 7B, compare lanes 4 and 5) and untreated (Fig. 7B, compare lanes 1 and 2) cells, confirming that p14ARF is a critical regulator of Tip60 expression (Fig. 5 and 7B). Furthermore, we detected a slight accumulation of p14ARF following Tip60 knockdown (Fig. 7B, compare lanes 1 and 3), suggesting the existence of a Tip60-controlled negative feedback loop affecting p14ARF expression levels. Finally, when p14ARF was immunoprecipitated from cyclophosphamide-treated cells, we clearly observed the coimmunoprecipitation of a longer form of Tip60 which was not detected in untreated cells, indicating that p14ARF could interact with a posttranslationally modified form of the protein in response to cyclophosphamide (Fig. 7D). However, it should be noted that p14ARF did not interact preferentially with this slower-migrating form of Tip60 in treated cells. Collectively, these data provide evidence that p14ARF and Tip60 cooperate to activate the DNA damage checkpoint in response to alkylating agents.
FIG. 7.
p14ARF and Tip60 are required for ATM/CHK2 activation in response to DNA-damaging agents. (A) H358 and NHBE cells were treated for 24 h with the indicated cytotoxic agents (left panel) or cigarette smoke condensate (right panel) and analyzed for p14ARF expression by Western blotting. Hsp70 was used as a loading control. (B) H358 cells were transfected for 48 h with either mismatch, p14ARF, or tip60 siRNA and treated or not treated for an additional 24 h with cyclophosphamide (10 μM). Western blot analyses were performed with the indicated antibodies. (C) H358 cells were transfected for 48 h with either mismatch or p14ARF siRNA and treated or not treated for 24 additional hours with cyclophosphamide (10 μM). For each condition, cells were counted using trypan blue staining (upper panel). Apoptosis was evaluated after Hoechst staining and immunoblotting with anti-active caspase 3 antibody (lower panels). (D) Endogenous p14ARF was immunoprecipitated from nuclear extracts of H358 cells treated (+) or not treated (−) for 24 h with cyclophosphamide (Cycloph.; 10 μM). Immunoblotting was performed using anti-p14ARF or anti-Tip60 antibody. An irrelevant goat serum was used as a negative control (Ctl). Notice the appearance of a Tip60 doublet (arrows) that coimmunoprecipitates with p14ARF in cyclophosphamide-treated cells.
Direct correlation between p14ARF and P-CHK2 (Thr68) expression in human lung tumors.
We previously reported a frequent inactivation of p14ARF in aggressive human lung tumors (16), and our present data demonstrate that p14ARF is required for activation of DNA damage-signaling pathways in response to genotoxic stresses in cell lines. We therefore asked whether p14ARF expression could correlate with the activation of some of the components of the DNA damage network in human lung tumors. To this end, we studied p14ARF, CHK2, and P-CHK2 (Thr68) expression using immunohistochemistry in 106 resected lung tumors of various histological types (Fig. 8). Compared to results for normal lung structures present at the vicinity of the tumors, p14ARF immunostaining was undetectable in 23/106 (22%) human lung tumors, predominantly in NE lung tumors (16/48; 33%), as previously reported (16). Moreover, CHK2 expression was observed in 86/106 (81%) human lung tumors. Of these 86 CHK2-expressing tumors, 56 (65%) accumulated the active P-CHK2 (Thr68) protein. A direct correlation linking the expression of both p14ARF and P-CHK2 (Thr68) was found in all tumors tested (P = 6 × 10−5), with 65/86 (75%) samples exhibiting either the presence or the absence of the expression of both proteins. Interestingly, this direct correlation was also observed in NE tumors (P = 9 × 10−3), which were predominantly negative for both p14ARF and P-CHK2 (Thr68) staining (10/35; 28%) compared to non-small-cell lung carcinomas (4/51; 8%) (Fig. 8B). Taken together, these data are consistent with p14ARF being a determinant of CHK2 phosphorylation in human lung tumors.
FIG. 8.
Immunohistochemical analysis of p14ARF, CHK2, and P-CHK2 (Thr68) status in human lung tumors. (A) Representative examples are shown. (Upper panels) A small-cell lung carcinoma (SCLC) exhibiting negative p14ARF and P-CHK2 (Thr68) staining and positive CHK2 nuclear expression (magnification, ×40; bar scale = 100 μm). (Lower panels) A squamous lung carcinoma with positive p14ARF, CHK2, and P-CHK2 (Thr68) staining (magnification, ×20; bar scale = 200 μm). (B) Relationships between p14ARF status, CHK2 status, and P-CHK2 (Thr68) status in human lung tumors. Statistical analyses were performed using a chi-square test. NSCLC, non-small-cell lung carcinoma; −, negative immunostaining; +, positive immunostaining; NS, not significant.
DISCUSSION
During the past decade, the p14ARF/MDM2/p53 signaling pathway has been the subject of intense investigations. In contrast, and even if it is now well admitted that p14ARF also has tumor suppressor functions that do not depend on p53 or Mdm2, the cellular signaling pathways associated with these functions remain largely unexplored. Here, we provide evidence that p14ARF triggers a G2 checkpoint independently of p53 through activation of both ATM and ATR kinases and their downstream transducers CHK1 and CHK2. Interestingly, p14ARF was recently shown to activate ATM, leading to the phosphorylation and stabilization of p53 (29, 33). Our data therefore demonstrate that the ATM/ATR DNA damage-signaling pathways also contribute to the p53-independent tumor suppressor function of p14ARF. The contribution of p14ARF to the cellular response induced by treatments with genotoxic agents remains a subject of debate. Although it was first thought that p14ARF was not involved in this process (22, 30, 47), later studies reported that its level increases after exposure to gamma radiation and that its loss results in a defective DNA damage arrest upon exposure to ionizing radiation (23, 24). In addition, p14ARF was also shown to play a role in DNA repair after treatment of cells by UV irradiation (37). In this study, we provide evidence that p14ARF accumulates in cells treated with alkylating agents as well as with cigarette smoke condensate and demonstrate that its expression is crucial for the activation of ATM and CHK2 in these settings. Therefore, our data identify p14ARF as a key component for the initiation of the DNA damage-signaling cascade.
How might p14ARF activate the ATM/ATR signaling pathways? It was recently demonstrated that oncogenic stimuli induce a DNA damage response in human tumors through aberrations in DNA replication (5, 17). Interestingly, the Foxm1b protein, a component of the DNA replication machinery, was recently identified as a p14ARF target (11), and it was also shown that ATM becomes activated during replication in the absence of DNA damage (38). Therefore, it is possible that p14ARF activates ATM/ATR/CHK by acting on replicative processes. Another possibility comes from the recent notion of ATM activation through epigenetic mechanisms (2). Consistent with this hypothesis, we highlight the role of the Tip60 protein in the checkpoint control induced by p14ARF independently of p53. Indeed, we provide evidence that Tip60 is absolutely required for p14ARF-mediated ATM/CHK2 activation and cell cycle arrest. Several data support the notion that Tip60 and p14ARF have interrelated roles in the control of cell growth in a p53-dependent context. First, both proteins are able to inhibit Mdm2-induced degradation of p53 and to stimulate its transcriptional activity (27, 34). Second, the suppression of Tip60 confers resistance to the ARF/p53-dependent proliferation arrest (6). Altogether, these and our results demonstrate that Tip60 is a critical mediator of both the p53-dependent and the p53-independent tumor suppressor functions of p14ARF. Consistent with Tip60 being closely connected to p14ARF, we further show that p14ARF is a positive regulator of Tip60 expression both in normally growing cells and in response to cyclophosphamide treatment. Stabilization of Tip60 following UV irradiation has been previously reported and was correlated with a decrease in its Mdm2-mediated polyubiquitination (26). As p14ARF is a well-known inhibitor of the E3 ligase activity of Mdm2 (34), it would be interesting to determine whether Tip60 stabilization by p14ARF involves the Mdm2 protein.
Recent studies have highlighted a role for Tip60 in the cellular response to DNA damage. Indeed, Tip60 was shown to be essential for the repair of DNA strand breaks following gamma radiation (19) and was also involved in the exchange of drosophila phospho-H2Av with a nonmodified H2Av at DNA lesions (25). In this study, we demonstrate that both Tip60 and p14ARF are absolutely required for a proper activation of the ATM/CHK2 pathway in response to cyclophosphamide, unraveling their interrelated roles in the initiation of the DNA damage-signaling cascade. One critical question that remains to be elucidated is this: in which way do p14ARF and Tip60 cooperate to activate ATM? Tip60 was very recently reported to bind and to acetylate ATM, leading to the activation of ATM kinase activity (42). In this setting, it was also demonstrated that the acetyltransferase activity of the ATM-Tip60 complex is specifically activated by DNA damage. We found that both Tip60 and p14ARF interact. It is thus tempting to speculate that p14ARF could be part of an ATM/Tip60 complex and could contribute to its activation. Moreover, in contrast to the ATM/Tip60 interaction being not modulated by DNA damage (42), we observed that p14ARF bound Tip60 as well as a slower-migrating form of Tip60 in cyclophosphamide-treated cells. Phosphorylation of Tip60 has already been described and was found to stimulate its histone acetyltransferase activity (28). Whether p14ARF interacts with such a posttranslationally modified form of Tip60 to affect its hypoxanthine-aminopterin-thymidine activity requires further investigation. Alternatively, the modulation of Tip60 activity towards other nonhistone proteins (32, 44) and/or the regulation of Tip60 transcriptional functions (8) might also contribute to the activation of the ATM/CHK2 signaling pathway by p14ARF.
The activation of an ATM/CHK2-regulated DNA damage response network was recently identified as a barrier against human cancer and genetic instability (4, 5, 13, 17). In this study, we found a direct correlation between the expression of both p14ARF and phospho-CHK2 (Thr68) proteins in a large series of human lung tumors. In keeping with our data on cell lines, these results confirm that p14ARF is a critical determinant of CHK2 activation. Therefore, based on our data, we propose that p14ARF inactivation would allow tumor cells to dodge the CHK2-dependent checkpoint control that is normally activated in response to cigarette smoke carcinogens, thus favoring lung tumorigenesis. This hypothesis fits perfectly well with the high frequency of p14ARF loss we had previously reported in aggressive small-cell lung cancers, which are tumors strictly related to tobacco smoking (16).
Acknowledgments
We thank B. Amati for providing us with the Tip60 antibody. We thank Celine Lampreia and Pascal Perron for technical assistance and Mary Callanan for help with editing.
This work was supported by a grant to E.B. from La Ligue Nationale Contre le Cancer as an équipe labellisée, by the Region Rhône Alpes (Thématique Prioritaire Cancer and Canceropole LARA [2003 and 2005], Oncocell, Epimed, INACancer), and by INCa (Institut National du Cancer, EpiPro). The laboratory of S.K. was supported by a Sidaction grant, and E.C. was supported by Sidaction (2004) as well as by ANRS (2002 to 2004).
REFERENCES
- 1.Ahn, J., and C. Prives. 2002. Checkpoint kinase 2 (Chk2) monomers or dimers phosphorylate Cdc25C after DNA damage regardless of threonine 68 phosphorylation. J. Biol. Chem. 277:48418-48426. [DOI] [PubMed] [Google Scholar]
- 2.Bakkenist, C. J., and M. B. Kastan. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421:499-506. [DOI] [PubMed] [Google Scholar]
- 3.Bartek, J., and J. Lukas. 2003. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 3:421-429. [DOI] [PubMed] [Google Scholar]
- 4.Bartkova, J., P. Guldberg, K. Gronbaek, K. Koed, H. Primdahl, K. Moller, J. Lukas, T. F. Orntoft, and J. Bartek. 2004. Aberrations of the Chk2 tumour suppressor in advanced urinary bladder cancer. Oncogene 23:8411-8418. [DOI] [PubMed] [Google Scholar]
- 5.Bartkova, J., Z. Horejsi, K. Koed, A. Kramer, F. Tort, K. Zieger, P. Guldberg, M. Sehested, J. M. Nesland, C. Lukas, T. Orntoft, J. Lukas, and J. Bartek. 2005. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434:864-870. [DOI] [PubMed] [Google Scholar]
- 6.Berns, K., E. M. Hijmans, J. Mullenders, T. R. Brummelkamp, A. Velds, M. Heimerikx, R. M. Kerkhoven, M. Madiredjo, W. Nijkamp, B. Weigelt, R. Agami, W. Ge, G. Cavet, P. S. Linsley, R. L. Beijersbergen, and R. Bernards. 2004. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428:431-437. [DOI] [PubMed] [Google Scholar]
- 7.Bertwistle, D., M. Sugimoto, and C. J. Sherr. 2004. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol. Cell. Biol. 24:985-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrozza, M. J., R. T. Utley, J. L. Workman, and J. Cote. 2003. The diverse functions of histone acetyltransferase complexes. Trends Genet. 19:321-329. [DOI] [PubMed] [Google Scholar]
- 9.Celeste, A., O. Fernandez-Capetillo, M. J. Kruhlak, D. R. Pilch, D. W. Staudt, A. Lee, R. F. Bonner, W. M. Bonner, and A. Nussenzweig. 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5:675-679. [DOI] [PubMed] [Google Scholar]
- 10.Chen, D., N. Kon, M. Li, W. Zhang, J. Qin, and W. Gu. 2005. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell 121:1071-1083. [DOI] [PubMed] [Google Scholar]
- 11.Costa, R. H., V. V. Kalinichenko, M. L. Major, and P. Raychaudhuri. 2005. New and unexpected: forkhead meets ARF. Curr. Opin. Genet. Dev. 15:42-48. [DOI] [PubMed] [Google Scholar]
- 12.Datta, A., J. Sen, J. Hagen, C. K. Korgaonkar, M. Caffrey, D. E. Quelle, D. E. Hughes, T. J. Ackerson, R. H. Costa, and P. Raychaudhuri. 2005. ARF directly binds DP1: interaction with DP1 coincides with the G1 arrest function of ARF. Mol. Cell. Biol. 25:8024-8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiTullio, R. A., Jr., T. A. Mochan, M. Venere, J. Bartkova, M. Sehested, J. Bartek, and T. D. Halazonetis. 2002. 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat. Cell Biol. 4:998-1002. [DOI] [PubMed] [Google Scholar]
- 14.Eymin, B., L. Karayan, P. Seite, C. Brambilla, E. Brambilla, C. J. Larsen, and S. Gazzeri. 2001. Human ARF binds E2F1 and inhibits its transcriptional activity. Oncogene 10:1033-1041. [DOI] [PubMed] [Google Scholar]
- 15.Eymin, B., C. Leduc, J. L. Coll, E. Brambilla, and S. Gazzeri. 2003. p14ARF induces G2 arrest and apoptosis independently of p53 leading to regression of tumours established in nude mice. Oncogene 22:1822-1835. [DOI] [PubMed] [Google Scholar]
- 16.Gazzeri, S., V. Della Valle, L. Chaussade, C. Brambilla, C. J. Larsen, and E. Brambilla. 1998. The human p19ARF protein encoded by the beta transcript of the p16INK4a gene is frequently lost in small cell lung cancer. Cancer Res. 58:3926-3931. [PubMed] [Google Scholar]
- 17.Gorgoulis, V. G., L. V. Vassiliou, P. Karakaidos, P. Zacharatos, A. Kotsinas, T. Liloglou, M. Venere, R. A. Ditullio, Jr., N. G. Kastrinakis, B. Levy, D. Kletsas, A. Yoneta, M. Herlyn, C. Kittas, and T. D. Halazonetis. 2005. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434:907-913. [DOI] [PubMed] [Google Scholar]
- 18.Ichijima, Y., R. Sakasai, N. Okita, K. Asahina, S. Mizutani, and H. Teraoka. 2005. Phosphorylation of histone H2AX at M phase in human cells without DNA damage response. Biochem. Biophys. Res. Commun. 336:807-812. [DOI] [PubMed] [Google Scholar]
- 19.Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang, M. Horikoshi, R. Scully, J. Qin, and Y. Nakatani. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102:463-473. [DOI] [PubMed] [Google Scholar]
- 20.Itahana, K., K. P. Bhat, A. Jin, Y. Itahana, D. Hawke, R. Kobayashi, and Y. Zhang. 2003. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol. Cell 12:1151-1164. [DOI] [PubMed] [Google Scholar]
- 21.Kamijo, T., J. D. Weber, G. Zambetti, F. Zindy, M. F. Roussel, and C. J. Sherr. 1998. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc. Natl. Acad. Sci. USA 95:8292-8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamijo, T., F. Zindy, M. F. Roussel, D. E. Quelle, J. R. Downing, R. A. Ashmun, G. Grosveld, and C. J. Sherr. 1997. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91:649-659. [DOI] [PubMed] [Google Scholar]
- 23.Khan, S., C. Guevara, G. Fujii, and D. Parry. 2004. p14ARF is a component of the p53 response following ionizing irradiation of normal human fibroblasts. Oncogene 23:6040-6046. [DOI] [PubMed] [Google Scholar]
- 24.Khan, S. H., J. Moritsugu, and G. M. Wahl. 2000. Differential requirement for p19ARF in the p53-dependent arrest induced by DNA damage, microtubule disruption, and ribonucleotide depletion. Proc. Natl. Acad. Sci. USA 97:3266-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kusch, T., L. Florens, W. H. MacDonald, S. K. Swanson, R. L. Glaser, J. R. Yates III, S. M. Abmayr, M. P. Washburn, and J. L. Workman. 2004. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306:2084-2087. [DOI] [PubMed] [Google Scholar]
- 26.Legube, G., L. K. Linares, C. Lemercier, M. Scheffner, S. Khochbin, and D. Trouche. 2002. Tip60 is targeted to proteasome-mediated degradation by Mdm2 and accumulates after UV irradiation. EMBO J. 21:1704-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Legube, G., L. K. Linares, S. Tyteca, C. Caron, M. Scheffner, M. Chevillard-Briet, and D. Trouche. 2004. Role of the histone acetyl transferase Tip60 in the p53 pathway. J. Biol. Chem. 279:44825-44833. [DOI] [PubMed] [Google Scholar]
- 28.Lemercier, C., G. Legube, C. Caron, M. Louwagie, J. Garin, D. Trouche, and S. Khochbin. 2003. Tip60 acetyltransferase activity is controlled by phosphorylation. J. Biol. Chem. 278:4713-4718. [DOI] [PubMed] [Google Scholar]
- 29.Li, Y., D. Wu, B. Chen, A. Ingram, L. He, L. Liu, D. Zhu, A. Kapoor, and D. Tang. 2004. ATM activity contributes to the tumor-suppressing functions of p14ARF. Oncogene 23:7355-7365. [DOI] [PubMed] [Google Scholar]
- 30.Lowe, S. W., E. Cepero, and G. Evan. 2004. Intrinsic tumour suppression. Nature 432:307-315. [DOI] [PubMed] [Google Scholar]
- 31.McManus, K. J., and M. J. Hendzel. 2005. ATM-dependent DNA damage-independent mitotic phosphorylation of H2AX in normally growing mammalian cells. Mol. Biol. Cell 16:5013-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel, J. H., Y. Du, P. G. Ard, C. Phillips, B. Carella, C. J. Chen, C. Rakowski, C. Chatterjee, P. M. Lieberman, W. S. Lane, G. A. Blobel, and S. B. McMahon. 2004. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol. Cell. Biol. 24:10826-10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pauklin, S., A. Kristjuhan, T. Maimets, and V. Jaks. 2005. ARF and ATM/ATR cooperate in p53-mediated apoptosis upon oncogenic stress. Biochem. Biophys. Res. Commun. 334:386-394. [DOI] [PubMed] [Google Scholar]
- 34.Pomerantz, J., N. Schreiber-Agus, N. J. Liegeois, A. Silverman, L. Alland, L. Chin, J. Potes, K. Chen, I. Orlow, H. W. Lee, C. Cordon-Cardo, and R. A. DePinho. 1998. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell 92:713-723. [DOI] [PubMed] [Google Scholar]
- 35.Quelle, D. E., F. Zindy, R. A. Ashmun, and C. J. Sherr. 1995. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell 83:993-1000. [DOI] [PubMed] [Google Scholar]
- 36.Rouse, J., and S. P. Jackson. 2002. Interfaces between the detection, signaling, and repair of DNA damage. Science 297:547-551. [DOI] [PubMed] [Google Scholar]
- 37.Sarkar-Agrawal, P., I. Vergilis, N. E. Sharpless, R. A. DePinho, and T. M. Runger. 2004. Impaired processing of DNA photoproducts and ultraviolet hypermutability with loss of p16INK4a or p19ARF. J. Natl. Cancer Inst. 96:1790-1793. [DOI] [PubMed] [Google Scholar]
- 38.Shechter, D., V. Costanzo, and J. Gautier. 2004. ATR and ATM regulate the timing of DNA replication origin firing. Nat. Cell Biol. 6:648-655. [DOI] [PubMed] [Google Scholar]
- 39.Sherr, C. J. 2001. The INK4a/ARF network in tumour suppression. Nat. Rev. Mol. Cell Biol. 2:731-737. [DOI] [PubMed] [Google Scholar]
- 40.Shiloh, Y. 2001. ATM and ATR: networking cellular responses to DNA damage. Curr. Opin. Genet. Dev. 11:71-77. [DOI] [PubMed] [Google Scholar]
- 41.Sorensen, C. S., R. G. Syljuasen, J. Falck, T. Schroeder, L. Ronnstrand, K. K. Khanna, B. B. Zhou, J. Bartek, and J. Lukas. 2003. Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell 3:247-258. [DOI] [PubMed] [Google Scholar]
- 42.Sun, Y., X. Jiang, S. Chen, N. Fernandes, and B. D. Price. 2005. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl. Acad. Sci. USA 102:13182-13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Travis, W. D., T. V. Colby, B. Corrin, Y. Shimosato, and E. Brambilla. 1999. Histological typing of lung and pleural tumours. World Health Organization international histological classification of tumours, 3rd ed. Springer, Berlin, Germany.
- 44.Yang, X. J. 2004. Lysine acetylation and the bromodomain: a new partnership for signaling. Bioessays 26:1076-1087. [DOI] [PubMed] [Google Scholar]
- 45.Zhou, B. B., and J. Bartek. 2004. Targeting the checkpoint kinases: chemosensitization versus chemoprotection. Nat. Rev. Cancer 4:216-225. [DOI] [PubMed] [Google Scholar]
- 46.Zhou, B. B., and S. J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433-439. [DOI] [PubMed] [Google Scholar]
- 47.Zindy, F., C. M. Eischen, D. H. Randle, T. Kamijo, J. L. Cleveland, C. J. Sherr, and M. F. Roussel. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 12:2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zou, L., D. Cortez, and S. J. Elledge. 2002. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 16:198-208. [DOI] [PMC free article] [PubMed] [Google Scholar]