Abstract
The yeast TREX complex physically couples elongating RNA polymerase II with RNA processing and nuclear RNA export factors to facilitate regulated gene expression. Hpr1p is an essential component of TREX, and loss of Hpr1p compromises transcriptional elongation, RNA export, and genome stability. Despite these defects, HPR1 is not essential for viability in yeast. A functional orthologue of Hpr1p has been identified in metazoan species and is variously known as Thoc1, Hpr1, or p84. However, the physiological functions of this protein have not been determined. Here, we describe the generation and phenotypic characterization of mice containing a null allele of the Thoc1 gene. Heterozygous null Thoc1 mice are born at the expected Mendelian frequency with no phenotype distinguishable from the wild type. In contrast, homozygous null mice are not recovered, indicating that Thoc1 is required for embryonic development. Embryonic development is arrested around the time of implantation, as blastocysts exhibit hatching and blastocyst outgrowth defects upon in vitro culture. Cells of the inner cell mass are particularly dependent on Thoc1, as these cells rapidly lose viability coincident with Thoc1 protein loss. While Hpr1p is not essential for the viability of unicellular yeasts, the orthologous Thoc1 protein is required for viability of the early mouse embryo.
Cotranscriptional loading of RNA-processing and export factors onto nascent RNA supports efficient and regulated gene expression (15, 17). Assembly of the TREX complex is one molecular mechanism utilized in yeast to physically couple factors involved in transcriptional elongation with those important for RNA splicing and export. TREX is composed of the THO subcomplex containing four proteins (Hpr1p, Tho2p, Mft1p, and Thp2p) that are essential for normal transcriptional elongation of some yeast genes (3). THO associates with two proteins implicated in nuclear RNA export (Sub2p and Yra1) to form the larger TREX complex (23). Sub2p is the yeast orthologue of human UAP56, which has also been implicated in RNA splicing (12). Hpr1p genetically and physically interacts with RNA polymerase II (1, 3, 6, 23) and is essential for recruitment of Sub2p to genes regulated by TREX (27). Loss of Hpr1p impairs both transcriptional elongation and nuclear RNA export (2, 4, 6, 9, 19, 23). Hpr1p, therefore, is an essential component of the THO and TREX complexes. Hpr1p has also been associated with the Paf1 complex, which influences transcriptional elongation through alterations in histone methylation (1, 10, 16, 26). Despite the defects in gene expression associated with loss of Hpr1p, it is not essential for viability in yeast. However, hpr1Δ yeasts are temperature sensitive for growth (7).
Metazoan species with long, intron-containing genes are likely even more dependent on the physical coupling of transcriptional elongation and RNA processing for efficient gene expression. The presence of metazoan structural homologues for the yeast TREX proteins Tho2p (Thoc2), Sub2p (UAP56), and Yra1p (Aly) suggests that multicellular organisms utilize an analogous mechanism to link transcriptional elongation and RNA processing (12, 22, 24, 25). Although lacking statistically significant similarity at the primary amino acid level, functional orthologues of yeast Hpr1p have also been identified in both human and insect cells (alternatively known as Thoc1, hHpr1, or p84). Depletion of the Hpr1p orthologue from human cancer cell lines or insect cells compromises transcriptional elongation, nuclear RNA export, cell viability, and cell proliferation (11, 18). However, the subunit compositions of the yeast and metazoan TREX complexes are not identical, suggesting the possibility of functional or regulatory differences (13, 18). The physiological requirements for the various TREX proteins have yet to be assessed genetically in a metazoan species, so it is currently unknown whether TREX function is required for the normal growth and development of multicellular organisms. To address this limitation, we have generated a null allele of the murine Thoc1 gene. We describe the phenotypic characterization of mice containing this Thoc1 null allele.
MATERIALS AND METHODS
Generation and genotyping of a Thoc1 null allele in the mouse.
A bacterial artificial chromosome clone spanning the Thoc1 gene was isolated from a 129/SV mouse strain genomic library and used as a template to amplify the homology arms for construction of the targeting vector. The 5′ homology arm is 4 kbp in length and includes most of exon 1 and upstream sequences. The 4.5-kbp 3′ homology arm includes exon 9 and parts of the flanking introns. The neomycin selection cassette was derived from BS524 (21). Fragments were cloned into the BSK cloning vector (Promega, Madison, WI), and all exon and flanking intron sequences were verified by DNA sequencing. The targeting construct was electroporated into a 129/SV-derived embryonic stem (ES) cell line, and G418-resistant colonies were screened by Southern blotting of HindIII-restricted genomic DNA using 5′ and 3′ flanking probes.
Germ line-transmitting chimeras were generated by blastocyst injection of two independent, successfully targeted embryonic stem cell lines. Routine genotyping of genomic DNA extracted from tail biopsy specimens was performed with a PCR assay using the primers 5′TGCCGTAGAAAAATGCACAG3′, 5′AACCACCCCTAATATTCTCCATC3′, and 5′TAAAGCGCATGCTCCAGA3′. Genotyping of preimplantation embryos was performed using a nested-PCR assay. Individual embryos or cell outgrowths were lysed by incubation at 55°C for 4 h in 20 μl of PCR lysis buffer (10 mM Tris-HCl, pH 8.8, 50 mM KCl, 0.08% NP-40, 0.2 mM deoxynucleoside triphosphate, 2.5 mM MgCl2) containing proteinase K (60 μg/ml). After being boiled, an aliquot of the lysates was subjected to nested-PCR amplification using the first-round primers 5′TGCCGTAGAAAAATGCACAG3′, 5′TAAGGTAACTAGAGAGGGAAAGTGTT3′, and 5′TAAAGCGCATGCTCCAGA3′. The second-round primers were 5′GGATCCACTAGTTCTAGAGCGG3′, 5′GTCTTCCCTTGTCACTGG3′, and 5′AACCACCCCTAATATTCTCCATC3′. The null allele generates a 230-bp PCR fragment, while the wild-type allele produces a 527-bp fragment.
All animal work was approved by the Roswell Park Cancer Institute Institutional Animal Care and Use Committee and met federal guidelines.
RT-PCR.
Total RNA was extracted from 30 embryos at each embryonic stage. The RNA was extracted with TRIzol according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). Reverse transcription (RT) was carried out using the SuperScript first-strand synthesis system for RT-PCR (Invitrogen, Carlsbad, CA). The cDNA was used as a template for PCR. PCR primers for mouse Thoc1 were N5-3 (5′-CTCACTTCTTCCAGCCAACC-3′) and N5-4 (5′-AGGGAGCCAGAATCTTCCAT-3′). PCR of β-actin cDNA using primers actin-1 (5′-GGCATCCTCACCCTGAAGTA-3′) and actin-2 (5′-AGAGGCGTACAGGGATAGCA-3′) was used as an input control. These primer sets gave rise to Thoc1 and β-actin PCR fragments of 263 and 248 bp, respectively. The PCR products were resolved by 2% agarose gel electrophoresis and stained with ethidium bromide.
Preimplantation embryo culture.
Timed pregnancies were used to generate preimplantation embryos for analysis. Thoc1 heterozygous females were given an intraperitoneal injection of pregnant mare's serum gonadotropin (5 IU per animal; Sigma-Aldrich, St. Louis, MO), followed 47 h later by an injection of human chorionic gonadotropin (5 IU per animal; Sigma-Aldrich, St. Louis, MO). Treated females were bred with heterozygous Thoc1 males, and the morning of detection of the vaginal plug was designated embryonic day 0.5 (E0.5). Preimplantation embryos (E1.5 to E4.0) were collected by flushing the oviduct or uterus with HEPES-buffered medium 2 (M2; Sigma-Aldrich, St. Louis, MO). For in vitro culture, embryos were cultured in M16 medium without leukemia inhibitory factor.
Immunostaining.
Preimplantation embryos were washed in 4 mg/ml of bovine serum albumin/phosphate-buffered saline (BSA/PBS) and then fixed for 30 min at 37°C in 3% paraformaldehyde in stabilization buffer [0.l M piperazine-N,N′-bis(2-ethanesulfonic acid), 5 mM MgCl2, and 2.5 mM EGTA]. The embryos were rinsed two times in BSA/PBS, transferred to 0.2% Triton in BSA/PBS at room temperature for 45 min, and then blocked overnight in 3% fetal calf serum, 0.1% Tween-20, 0.02% Na-azide, 0.4% powdered milk. The embryos were then incubated in the primary antibody for 1 h at 37°C, washed, and incubated with secondary antibody for an additional hour at 37°C. Immunostaining was performed using primary antibodies directed against Thoc1 protein (pThoc1) (5E10; Genetex, San Antonio, TX), activated caspase 3 (9661; Cell Signaling, Beverly, MA), Oct4 (Santa Cruz Biotechnology, Santa Cruz, CA), and serine 10 phosphorylated histone H3 (06-570; Upstate, Waltham, MA). Fluorescent secondary antibodies and the DNA counterstain Hoechst 33342 were from Molecular Probes (Carlsbad, CA). Whole E12.5 embryos were frozen, fixed, and sectioned as previously described (8) and then immunostained for pThoc1 and counterstained for DNA as described above. Overlapping fluorescent images were captured using a 10× objective on a Zeiss Axioplan2 microscope. Individual images were merged using Adobe Photoshop to generate the whole-embryo composite image.
RESULTS AND DISCUSSION
A Thoc1 null allele was generated by targeted homologous recombination in murine ES cells. The targeting vector was designed to delete 11.5 kbp of genomic DNA containing exons 2 through 8 of Thoc1 (Fig. 1A). The deleted exons include codons 19 to 173, which span a region of Thoc1 protein (pThoc1) that is highly conserved throughout evolution. The deletion also creates a reading frame shift, causing premature termination of potential readthrough translation. Hence, the targeted allele does not express pThoc1. Two of 128 G418-resistant ES cell clones isolated upon transfection of the targeting vector are heterozygous for the correctly targeted allele as assessed by Southern blotting using 5′ and 3′ flanking probes (Fig. 1B). These two ES cell clones were used to generate germ line-transmitting chimeras that served as strain founders. The phenotypes observed are completely penetrant in each strain, so data from the two strains are not further distinguished. Routine genotyping was performed using a PCR assay designed to detect the wild-type and mutant alleles (Fig. 1C).
FIG. 1.
Generation of a Thoc1 null allele in the mouse. (A) A representation of the exon/intron structure of the targeted region of the Thoc1 gene, the targeting vector, and the expected structure of the successfully targeted mutant allele. Exons are numbered and shown as solid boxes. neo, neomycin selection marker. The positions of HindIII restriction enzyme sites (H), the flanking hybridization probes, and the expected sizes of fragments detected by Southern blotting are indicated. (B) Southern blot analysis of representative mice with the indicated genotypes using the 3′ and 5′ flanking probes and HindIII-restricted genomic DNA. (C) PCR genotyping of mice. The targeted allele generates a 403-bp band, while the wild-type allele amplifies a 550-bp band.
Adult Thoc1 heterozygous (Thoc1+/−) mice are born at the expected Mendelian frequency and are overtly normal. In contrast, no homozygous null (Thoc1−/−) mice were recovered from more than 121 genotyped offspring from intermating of Thoc1+/− mice (Table 1). The lack of Thoc1−/− mice indicates that Thoc1 is required for embryonic development. We investigated the timing of presumptive embryonic mortality by genotyping embryos from heterozygote intercrosses at various stages of gestation. No Thoc1−/− postimplantation embryos were detected among those analyzed at E8.5 to E11.5. However, empty deciduae were often observed. While the empty deciduae may account for the missing Thoc1−/− embryos, pure embryonic tissue sufficient for PCR genotyping could not be recovered to confirm this possibility. In contrast, preimplantation Thoc1−/− blastocysts genotyped at E3.5 were recovered at the expected Mendelian ratio, suggesting that embryonic development ceases around the time of implantation.
TABLE 1.
Genotypes of neonates and embryos from Thoc1+/− intercrossesa
| Stage | No. of embryos/neonates with genotype:
|
No. of empty deciduae | ||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| P14 | 46 | 75 | 0 | |
| E10.5 to E11.5 | 6 | 11 | 0 | 16 |
| E8.5 | 8 | 16 | 0 | 11 |
| E3.5 | 17 | 40 | 15 | |
Embryos from Thoc1+/− heterozygote intercrosses were collected on day 14 postpartum (P14) or at the indicated days of embryonic gestation (E3.5 to E11.5), and the genotypes were determined by PCR.
To determine whether preimplantation Thoc1−/− embryos exhibit developmental defects, embryos were flushed from the oviducts or uteri and cultured in vitro. Freshly isolated E3.5 Thoc1−/− embryos had a morphology similar to that of wild-type (Thoc1+/+) embryos but generally failed to hatch from the zona pellucida or form blastocyst outgrowths in culture (Fig. 2A). A small fraction (<5%) of freshly isolated E3.5 Thoc1 null embryos were able to hatch upon in vitro culture, and a few cells with trophoblast morphology were able to attach to the culture dish. However, such embryos never produced viable blastocyst outgrowths, as cells of the inner cell mass (ICM) were lacking (data not shown). While E3.5 Thoc1−/− embryos cultured in vitro failed to hatch normally, the frequency of empty deciduae detected in vivo suggests that development may proceed sufficiently to induce a decidual reaction.
FIG. 2.
Developmental defects in Thoc1 nullizygous embryos cultured in vitro. (A) E3.5 embryos produced by intermating Thoc1+/− mice were collected and cultured in vitro for up to 3 days. Representative phase-contrast images of embryos of the indicated genotypes are shown at various times of in vitro culture. Heterozygous embryos can be seen to hatch from the zona pellucida at E4.5 and form blastocyst outgrowths by E6.5. Blastocyst morphological features apparent in the images are indicated by TE, ICM, and BC (blastocoele). (B) Freshly isolated E1.5 embryos produced as in panel A were cultured in vitro for 6 days. Representative images are shown at each day of in vitro culture. Compaction of both wild-type and homozygous null embryos are observed at E2.5, at which point the zona pellucida (ZP) was removed experimentally by acid treatment. Embryos of both genotypes formed blastocysts by E3.5. Wild-type blastocysts fully expanded by E4.5 and formed blastocyst outgrowths by E7.5. Thoc1 null blastocysts failed to fully expand and began to degenerate by E4.5. (C) Freshly isolated E2.5 embryos were cultured in vitro for 1.5 days and immunostained for pThoc1 and the activated form of caspase 3. Nuclear DNA was counterstained with Hoechst 33342, and representative images were captured under fluorescence microscopy. Apoptotic cells were frequently observed only in the absence of pThoc1. (D) Freshly isolated E2.5 embryos of the indicated genotypes were cultured as in panel C and immunostained for pThoc1 and the phosphorylated form of histone H3 to mark mitotic cells. DNA was counterstained, and images were captured as described above.
Freshly isolated E1.5 Thoc1−/− embryos cultured in vitro compacted normally after 1 day in culture (E2.5) and developed into normal-appearing blastocysts by E3.5 (Fig. 2B). In contrast to Thoc1+/+ embryos, however, these Thoc1−/− embryos typically failed to reach the fully expanded blastocyst stage and did not form blastocyst outgrowths even after experimental removal of the zona pellucida (Fig. 2B). These data suggest that Thoc1−/− embryos suffer from developmental defects at the late blastocyst stage, which compromises hatching, implantation, and subsequent development. Consistent with this possibility, freshly isolated Thoc1−/− E2.5 embryos cultured in vitro for 1.5 days had significantly fewer cells, on average, than Thoc1+/+ or Thoc1+/− embryos (25 versus 36 cells per embryo, respectively; P < 0.0005). Similarly cultured Thoc1−/− embryos exhibited an increase in the number of apoptotic cells as measured by immunostaining for the activated form of caspase 3 (Fig. 2C). Condensed nuclear fragments characteristic of the remnants of apoptotic cells were also apparent in Thoc1−/− embryos as visualized by DNA staining. However, there was no significant difference in the mitotic indexes of Thoc1+/+ and Thoc1−/− E2.5 embryos cultured in vitro for 1.5 days as assayed by immunostaining for the phosphorylated form of histone H3 (Fig. 2D). Further, E2.5 Thoc1−/− and Thoc1+/+ embryos cultured in vitro for only 1 day showed no difference in total cell numbers (24 versus 25; P > 0.8). Hence, cell numbers in Thoc1−/− embryos are relatively normal up to around E3.5 but thereafter fail to accumulate due to a loss of cell viability. This loss of cell viability is the likely cause of the embryonic lethality observed around the time of implantation.
Consistent with the interrogation of gene expression databases, we find that Thoc1 RNA is present in the fertilized oocyte and throughout preimplantation embryonic development (Fig. 3A). Thoc1 protein is also widely expressed at later stages of development. Sagittal sections of E12.5 wild-type embryos have been immunostained for pThoc1, and nuclear pThoc1 staining is observed in all tissues and cell types that are detectable in such sections (Fig. 3B). Similar results are observed in E13.5 embryos (data not shown). We conclude that Thoc1 is widely expressed during embryonic development. Thoc1 expression has previously been detected in a wide range of adult tissues (5).
FIG. 3.
Thoc1 expression during embryonic development. (A) Total RNA was isolated from wild-type embryos at the indicated developmental stages (C, cell number stage; M, morula; B, blastocyst). RNA was reverse transcribed, and Thoc1 cDNA was amplified by PCR using primers specific for Thoc1. PCR was also performed using primers specific for β-actin to serve as a loading control. The no-RT panel shows the PCR products generated in the absence of reverse transcriptase to control for possible DNA contamination. The PCR products were resolved by agarose gel electrophoresis and stained with ethidium bromide. The images were inverted for clarity. (B) A frozen sagittal section from a wild-type E12.5 embryo was immunostained for pThoc1 and counterstained for DNA using Hoechst 33342. Representative images were captured under fluorescence microscopy. The inset image is magnified from the location of the white square on the whole embryo to show nuclear pThoc1 staining. The head (H) and ventral (V) aspects of the embryo are indicated. (C) Freshly isolated preimplantation embryos of the indicated gestational ages and genotypes were immunostained for pThoc1. Cells were counterstained for DNA, and images were captured as for panel A.
Since pThoc1 may be present in early Thoc1−/− embryos due to translation of maternally supplied mRNA, we determined whether the loss of cell viability observed in Thoc1 null embryos coincided with the disappearance of pThoc1. Immunostaining of E2.5 embryos showed that nuclear pThoc1 staining was barely detectable in E2.5 Thoc1−/− embryos while it was readily apparent in heterozygous embryos (Fig. 3C). By E3.5, pThoc1 nuclear staining was undetectable in Thoc1−/− embryos while strong nuclear pThoc1 staining remained in Thoc1+/− embryos. Hence, pThoc1 levels were declining by E2.5 and undetectable by E3.5 in Thoc1−/− embryos, coinciding with the timing of loss of cell and embryo viability.
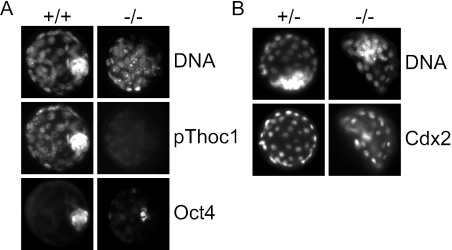
The blastocyst stage marks segregation into the first two cell lineages in the mammalian embryo, the ICM, comprised of undifferentiated embryonic stem cells that ultimately give rise to the embryo proper, and the differentiating trophoectoderm (TE), which contributes to extraembryonic tissues, like the placenta. We immunostained blastocysts for the differentiation markers Oct4 (ICM) and Cdx2 (TE) to determine if cellular differentiation occurs in Thoc1−/− blastocysts and to identify which cell type fails to accumulate. As expected, wild-type late-stage blastocysts showed a well-organized inner cell mass comprised of cells expressing Oct4, but not Cdx2 (Fig. 4). Cells comprising the presumptive TE of Thoc1-expressing blastocysts express Cdx2, but not Oct4. In contrast to wild-type blastocysts, there were very few Oct4-positive cells in age-matched Thoc1−/− embryos, and a well-organized inner cell mass was not apparent (Fig. 4A). However, the few Oct4-positive cells observed were Cdx2 negative. The presumptive TE of Thoc1−/− blastocysts contained approximately normal numbers of Cdx2-positive and Oct4-negative cells in the appropriate spatial organization (Fig. 4B). Proper cellular differentiation of the TE in Thoc1−/− blastocysts is also supported by the appearance of the blastocoel cavity, which requires formation of adherens junctions and tight junctions, the apparent initiation of a decidual reaction (see above), and normal-appearing E-cadherin staining (data not shown). These data suggest that cellular differentiation is properly initiated in Thoc1−/− embryos but that Oct4-positive Cdx2-negative cells of the ICM fail to survive. We conclude that Thoc1 deficiency causes peri-implantation embryonic lethality due initially to the failure of Oct4-positive cells of the ICM to survive.
FIG. 4.
Thoc1−/− blastocyst stage embryos lack Oct4-positive cells of the inner cell mass. (A) Freshly isolated E2.5 embryos of the indicated genotypes were cultured in vitro for 1.5 days and then immunostained for pThoc1 and Oct4 protein. DNA was counterstained with Hoechst 33342. Representative images were captured under fluorescence microscopy. (B) Freshly isolated E2.5 embryos were cultured as described above and then immunostained for Cdx2 protein.
The widespread expression of pThoc1 in developing embryos and adults suggests the possibility that pThoc1 may also be required for later stages of embryonic development, as well as for normal homeostasis of adult tissue. Due to the early embryonic lethality observed in Thoc1−/− mice, however, it is currently unclear whether this is the case. Alternatively, undifferentiated stem or progenitor cells may be uniquely dependent on pThoc1. This requirement would block early embryonic development but might not influence later stages of embryonic or adult development. We currently favor the latter hypothesis based on the preliminary observation that conditional ablation of mouse Thoc1 in differentiating mammary epithelial cells or embryonic fibroblasts has little effect on cell viability (X. Wang, Y. Li, and D. Goodrich, unpublished data). Phenotypic characterization of mice containing such conditionally null or hypomorphic alleles of Thoc1 will be necessary to definitively test this hypothesis.
The mammalian TREX component pThoc1 is essential for the viability of at least some cell types in mice. Given that murine pThoc1 has a high degree of primary amino acid sequence similarity to pThoc1 from other metazoan species, the requirement for pThoc1 is likely to extend to other multicellular organisms. For example, Drosophila melanogaster and human cancer cell lines both exhibit reduced viability upon depletion of their respective orthologous Thoc1 proteins (11, 18). In contrast, HPR1 is not essential for the viability of the unicellular yeast S. cerevisiae, although Hpr1p-deficient yeasts grow more slowly, are temperature sensitive for growth, and have a reduced cellular life span (14, 20). The differences in the physiological requirements for HPR1/Thoc1 may reflect TREX-independent functions of pThoc1 or may be due to differences in the functions of the yeast and metazoan TREX complexes. While all detectable pThoc1 is resident within TREX complexes (13), we currently cannot exclude the possibility that TREX independent functions are responsible for the pThoc1 requirement for cell viability. Since the yeast and metazoan TREX complexes differ in subunit composition, we favor the second hypothesis, that metazoan TREX complexes may have functions and regulatory inputs distinct from their yeast counterpart and that these differences account for the different physiological requirements for HPR1/Thoc1. Resolution of this issue will require further comparison of the molecular, cellular, and physiological functions of the yeast and metazoan TREX complexes.
Acknowledgments
We thank members of the Goodrich laboratory for helpful discussions. We acknowledge Deborah Ogden and Jennifer Black for performing the whole-embryo frozen sectioning.
This work was supported by grants to D.W.G. from the National Cancer Institute (CA-70292) and the Ralph C. Wilson Foundation Medical Research Foundation. National Cancer Institute Cancer Center Support Grant CA016056 supported the Cell Analysis and Transgenic Mouse facilities utilized in this work.
REFERENCES
- 1.Chang, M., D. French-Cornay, H. Y. Fan, H. Klein, C. L. Denis, and J. A. Jaehning. 1999. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol. Cell. Biol. 19:1056-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chavez, S., and A. Aguilera. 1997. The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev. 11:3459-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chavez, S., T. Beilharz, A. G. Rondon, H. Erdjument-Bromage, P. Tempst, J. Q. Svejstrup, T. Lithgow, and A. Aguilera. 2000. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J. 19:5824-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chavez, S., M. Garcia-Rubio, F. Prado, and A. Aguilera. 2001. Hpr1 is preferentially required for transcription of either long or G+C-rich DNA sequences in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:7054-7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durfee, T., M. A. Mancini, D. Jones, S. J. Elledge, and W.-H. Lee. 1994. The amino-terminal region of the retinoblastoma gene product binds a novel nuclear matrix protein that co-localizes to centers for RNA processing. J. Cell Biol. 127:609-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan, H. Y., K. K. Cheng, and H. L. Klein. 1996. Mutations in the RNA polymerase II transcription machinery suppress the hyperrecombination mutant hpr1 delta of Saccharomyces cerevisiae. Genetics 142:749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan, H. Y., R. J. Merker, and H. L. Klein. 2001. High-copy-number expression of Sub2p, a member of the RNA helicase superfamily, suppresses hpr1-mediated genomic instability. Mol. Cell. Biol. 21:5459-5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frey, M. R., J. A. Clark, O. Leontieva, J. M. Uronis, A. R. Black, and J. D. Black. 2000. Protein kinase C signaling mediates a program of cell cycle withdrawal in the intestinal epithelium. J. Cell Biol. 151:763-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huertas, P., and A. Aguilera. 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 12:711-721. [DOI] [PubMed] [Google Scholar]
- 10.Krogan, N. J. D. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11:721-729. [DOI] [PubMed] [Google Scholar]
- 11.Li, Y., X. Wang, X. Zhang, and D. W. Goodrich. 2005. Human hHpr1/p84/Thoc1 regulates transcriptional elongation and physically links RNA polymerase II and RNA processing factors. Mol. Cell. Biol. 25:4023-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo, M. L., Z. Zhou, K. Magni, C. Christoforides, J. Rappsilber, M. Mann, and R. Reed. 2001. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413:644-647. [DOI] [PubMed] [Google Scholar]
- 13.Masuda, S., R. Das, H. Cheng, E. Hurt, N. Dorman, and R. Reed. 2005. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 19:1512-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merker, R. J., and H. L. Klein. 2002. hpr1Delta affects ribosomal DNA recombination and cell life span in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:421-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore, M. J. 2005. From birth to death: the complex lives of eukaryotic mRNAs. Science 309:1514-1518. [DOI] [PubMed] [Google Scholar]
- 16.Ng, H. H., S. Dole, and K. Struhl. 2003. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 278:33625-33628. [DOI] [PubMed] [Google Scholar]
- 17.Reed, R. 2003. Coupling transcription, splicing and mRNA export. Curr. Opin. Cell Biol. 15:326-331. [DOI] [PubMed] [Google Scholar]
- 18.Rehwinkel, J., A. Herold, K. Gari, T. Kocher, M. Rode, F. L. Ciccarelli, M. Wilm, and E. Izaurralde. 2004. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat. Struct. Mol. Biol. 11:558-566. [DOI] [PubMed] [Google Scholar]
- 19.Rondon, A. G., S. Jimeno, M. Garcia-Rubio, and A. Aguilera. 2003. Molecular evidence that the eukaryotic THO/TREX complex is required for efficient transcription elongation. J. Biol. Chem. 278:39037-39043. [DOI] [PubMed] [Google Scholar]
- 20.Santos-Rosa, H., and A. Aguilera. 1994. Increase in incidence of chromosome instability and non-conservative recombination between repeats in Saccharomyces cerevisiae hpr1 delta strains. Mol. Gen. Genet. 245:224-236. [DOI] [PubMed] [Google Scholar]
- 21.Soukharev, S., J. L. Miller, and B. Sauer. 1999. Segmental genomic replacement in embryonic stem cells by double lox targeting. Nucleic Acids Res. 27:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strasser, K., and E. Hurt. 2000. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 19:410-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strasser, K., S. Masuda, P. Mason, J. Pfannstiel, M. Oppizzi, S. Rodriguez-Navarro, A. G. Rondon, A. Aguilera, K. Struhl, R. Reed, and E. Hurt. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417:304-308. [DOI] [PubMed] [Google Scholar]
- 24.Stutz, F., A. Bachi, T. Doerks, I. C. Braun, B. Seraphin, M. Wilm, P. Bork, and E. Izaurralde. 2000. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA 6:638-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.West, R. W., Jr., B. Kruger, S. Thomas, J. Ma, and E. Milgrom. 2000. RLR1 (THO2), required for expressing lacZ fusions in yeast, is conserved from yeast to humans and is a suppressor of SIN4. Gene 243:195-205. [DOI] [PubMed] [Google Scholar]
- 26.Wood, A., J. Schneider, J. Dover, M. Johnston, and A. Shilatifard. 2003. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278:34739-34742. [DOI] [PubMed] [Google Scholar]
- 27.Zenklusen, D., P. Vinciguerra, J. C. Wyss, and F. Stutz. 2002. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 22:8241-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]