Abstract
The transcription factor serum response factor (SRF) interacts with its cofactor, MAL/MKL1, a member of the myocardin-related transcription factor (MRTF) family, through its DNA-binding domain. We define a seven-residue sequence within the conserved MAL B1 region essential and sufficient for complex formation. The neighboring Q-box sequence facilitates this interaction. The B1 and Q-box regions also have antagonistic effects on MAL nuclear import, but the residues involved are largely distinct. Both MAL and the ternary complex factor (TCF) family of SRF cofactors interact with a hydrophobic groove and pocket on the SRF DNA-binding domain. Unlike the TCFs, however, interaction of MAL with SRF is impaired by SRF αI-helix mutations that reduce DNA bending in the SRF-DNA complex. A clustered SRF αI-helix mutation strongly impairs MAL-SRF complex formation but does not affect DNA distortion in the MAL-SRF complex. MAL-SRF complex formation is facilitated by DNA binding. DNase I footprinting indicates that in the SRF-MAL complex MAL directly contacts DNA. These contacts, which flank the DNA sequences protected from DNase I by SRF, are required for effective MAL-SRF complex formation in gel mobility shift assays. We propose a model of MAL-SRF complex formation in which MAL interacts with SRF by the addition of a β-strand to the SRF DNA-binding domain β-sheet region, while SRF-induced DNA bending facilitates MAL-DNA contact.
Serum response factor (SRF) is a prototype of the MADS (Mcm1, Agamous, Deficiens, SRF) family of eukaryotic transcription factors, which play important roles in the specification of cell identity during development and differentiation (22, 31). SRF interacts directly with at least two families of signal-regulated cofactors. The ternary complex factor (TCF) family of Ets domain proteins, Elk-1, SAP-1, and Net, which respond to mitogen-activated protein kinase signaling, bind to both SRF and adjacent DNA sequences (34). In contrast, members of the myocardin-related transcription factor (MRTF) coactivator family (MAL/MKL1/MRTFa, MAL16/MKL2/MRTFb, and myocardin) apparently bind SRF without making specific DNA contacts (17, 21, 27, 36, 37). MAL and MAL16 respond to Rho-actin signaling, while activation by myocardin is apparently constitutive (5, 23, 28, 36).
The best-characterized MADS box regulatory complexes are those formed by SRF and its Saccharomyces cerevisiae relative, Mcm1. In each case, the MADS protein forms a three-layered structure comprising the 56-residue MADS box motif and its C-terminal flanking sequences (8, 26, 32). A long coiled-coil at the core of the MADS box dimer straddles the DNA minor groove, making sequence-specific major groove contacts at each side, and inserting its N-terminal extension into the minor groove. Above lies a four-stranded antiparallel β-sheet platform derived from the C-terminal sequences of each MADS motif. The third layer is structurally variable, being formed from subfamily-specific sequences C terminal to the MADS box and is required for high-affinity dimerization. Interaction of TCF with SRF is mediated by the 20-residue B-box sequence (6, 9, 15, 30), while the MATα2-Mcm1 interaction involves an unrelated eight-residue sequence (20, 35). Each cofactor adds an extra β-strand to the MADS box β-sheet platform, albeit in opposite polarity in the two complexes, inserting an aromatic side chain into a hydrophobic pocket in the third layer of the DNA binding structure (8, 16, 32).
The mechanism of MRTF-SRF interaction is poorly understood. Complex formation requires the basic B1 region, also important for nuclear import, and the Q-box, a glutamine-rich sequence C terminal to it (23, 36, 39). Functional and biochemical assays suggest that the SRF surfaces contacted by the MRTFs and TCFs overlap (23, 24, 39), and it has been proposed that the Q box mediates myocardin-SRF interaction via the same mechanism as TCF (39). Two observations suggest that MRTF-SRF and TCF-SRF interactions differ significantly, however: an altered DNA binding specificity SRF derivative can recruit TCF, but not MAL, to DNA (9, 23), and functional studies suggest that the SRF DNA-binding domain must contact DNA to respond to Rho signaling (10). In this paper we analyze MAL-SRF complex formation in detail and identify a short sequence within the B1 region necessary and sufficient for specific interaction with SRF. We show that DNA binding and distortion is necessary for optimal interaction between SRF and MAL, which directly contacts DNA flanking the SRF binding site in the complex.
MATERIALS AND METHODS
Plasmids and proteins.
Flag- or hemagglutinin (HA)-tagged MAL derivatives MAL(fl), MAL(met), MALΔN, MALΔQ, and MALΔB1 were described previously (23). MAL residue numbering takes the first in-frame methionine as residue +1 [MAL(met) isoform (23)]. Mutations were introduced by PCR with MALΔN as template, replacing the BamHI-EcoRI fragment with the altered form. N-terminally myc-tagged SRF.DBD (DNA-binding domain) (residues 120 to 265) derivatives were expressed in vitro from pFTX5 (11) using TNT-T7-coupled reticulocyte lysate (Promega). SRF(120-265), SRF.M2(120-265), and SRF.M1(120-265) were derived from MLV.SRF, MLV.SRF-M2, and MLV.SRF-M1, respectively (9); other derivatives were created by PCR or recloned from the appropriate pAS series plasmids (16). SRF.DBD (residues 133 to 265) was produced by recombinant baculovirus (18). Plasmids MLV-Elk1, MLV-SRF.VP16, 3D.Aluc, MLV.lacZ, c-fosWT, c-fosΔTCF were as described previously (18, 23, 24). SRF.DBD (residues 132 to 223) was expressed in Escherichia coli from pET3a and purified as previously described (8, 26).
Transfections, immunoprecipitations, and reporter assays.
NIH 3T3 cells were transfected using Lipofectamine (Invitrogen). Whole-cell extracts from cells transfected with 1 μg Elk-1 or MAL plasmids were prepared as described previously (23). Protein expression was assessed by immunoblotting. For immunoprecipitations, cells were lysed in radioimmunoprecipitation assay buffer and immunoprecipitated using anti-myc agarose beads in 1% TX buffer in the presence of wild-type (WT) c-fos SRE or c-fos SRE.M DNA (24), generated by PCR as described below. For reporter assays, transfection mixes contained 50 ng MALΔN, 40 ng 3D.Aluc reporter, and 150 ng MLV-lacZ reference plasmids; cells were kept in 0.5% fetal bovine serum for 24 h prior to luciferase assay. Cytochalasin D or serum stimulation was for 7 h. Normalized data were expressed relative to reporter activation by 50 ng SRF.VP16, performed in parallel. Figures show results from three independent experiments ± standard errors of the means.
Pulldown assays.
Purified GST.MAL (glutathione S-transferase-MAL) proteins (10 μg) were incubated with 100 μl in vitro-translated SRF.DBD and 30 μl glutathione Sepharose beads (Amersham) in 500 μl buffer (50 mM Tris, pH 7.9, 150 mM NaCl, 0.1% Triton X-100, and protease inhibitors) for 4 h at 4°C. After two washings, the proteins were fractionated on 16% gels. GST.MAL input was visualized by Ponceau staining, and SRF was visualized by immunoblotting with 9E10 primary (1:1,000, Cancer Research UK) and horseradish peroxidase-conjugated anti-mouse secondary antibody (DAKO).
Immunofluorescence.
Cells transfected with 50 ng MAL expression plasmid were processed for immunofluorescence as described previously (23) and stained with rabbit anti-Flag (1:300; Sigma) primary antibody and fluorescein isothiocyanate (FITC)-conjugated anti-rabbit (1:200; Jackson Laboratories) secondary antibody or with tetramethyl rhodamine isocyanate (TRITC)-conjugated phalloidin (1:100; Molecular Probes). MAL subcellular localization was scored as predominantly nuclear, predominantly cytoplasmic, or evenly distributed in 100 to 200 cells. Percentages represent the averages of two independent experiments. For induction of endogenous genes, cells transfected with 150 ng MALΔN derivatives were kept in Dulbecco’s modified Eagle medium with 1 mg/ml bovine serum albumin for 48 h before staining with mouse anti-smooth muscle α-actin (anti-sm α-actin) (1:200; Sigma) and rabbit anti-Flag or rabbit anti-Egr1 (1:50; Santa Cruz) and mouse anti-Flag M2 (1:200; Santa Cruz) primary antibodies and Cy2-labeled anti-mouse or anti-rabbit (1:200; Jackson Laboratories) and Cy3-labeled anti-mouse or anti-rabbit (1:500; Jackson Laboratories) secondary antibodies. Expression of sm α-actin or Egr-1 was assessed in 100 to 200 cells expressing Flag-MAL. Mock-transfected, serum-starved, and stimulated (30 min) cells were processed in parallel. Percentages are the averages of two independent experiments.
Gel mobility shift assays.
Gel mobility shift probes (138 bp) were generated by PCR from the wild-type c-fos promoter or its ΔTCF derivative (24). The ΔTCF probe was used to reduce background complex formation by endogenous TCF in the cell extracts. Circular permutation analysis used 138-bp probes with SRF binding sites centered 16, 28, 39, 60, 78, 89, 101, 111, and 122 bp from the fragment end; electrophoresis was on 5% gels. Data were fitted to a cosine function and apparent bend angle (α) estimated using Rf(middle binding site)/Rf(end binding site) = cos (α/2) (29). For nested probe analysis, appropriate PCR was used to generate truncated c-fos probes with ends at positions −27, −25, −22, −19, −16, +28, +25, +22, +19, and +16 from the SRE dyad. Binding assays (10 μl) were as described previously (24). For analysis of MAL derivatives and peptide competitions, reaction mixtures included 0.2 μg MAL extract, recombinant SRF.DBD (residues 133 to 265), 0.1 ng probe, and 0 to 34 μM peptide. For analysis of altered SRF DNA-binding domains, reaction mixtures contained SRF.DBD (residues 120 to 265), MAL or Elk-1 extract, 1 ng probe, and 250 ng/μl poly(dI-dC) · poly(dI-dC). Peptide-binding reaction mixtures contained 0 to 5 μM peptide and SRF.DBD (residues 120 to 265), and the peptides were fractionated on 8% acrylamide-bisacrylamide gels. Peptide concentrations were determined by absorbance at 214 nm. GST.MAL and SRF.DBD reactions contained 0.6 ng GST.MAL proteins and either recombinant SRF.DBD (residues 132 to 223) or in vitro-translated SRF(120-265). Quantitation of binding assays was by phosphorimage analysis or densitometric analysis of scanned autoradiographs using ImageQuant software.
DNase I footprinting.
Probes were generated by EcoRI digestion of the Δ363 c-fos promoter derivative (33), labeling either the 5′ end with [γ-32P]ATP and T4 polynucleotide kinase (NEB) or the 3′ end by filling in with [α-32P]dATP and DNA polymerase I Klenow fragment (NEB), and redigestion with NotI to generate probes of 245 bp and 251 bp, respectively. For footprinting, 20-μl reaction mixtures contained 1 ng (10,000 cpm) probe, 0.85 ng SRF(132-223), and either 0.85 to 22.8 ng GST.MAL(214-298) (WT or H239A) or 1.7 to 85 μM MAL peptides, in 20 mM HEPES, pH 7.5, 50 mM NaCl, 3 mM MgCl2, 1 mM CaCl2, 1 μM ZnCl2, 2% glycerol, 2 mM dithiothreitol, 25 ng poly(dI-dC) · poly(dI-dC), 100 ng bovine serum albumin. After incubation at room temperature for 30 min, 0.25 units DNase I (Sigma) were added for 5 min on ice. Reactions were brought to 25 mM EDTA, 0.1 mg proteinase K, and 1% sodium dodecyl sulfate and incubated at 50°C for 1 h, and the DNA was precipitated. Analysis was on 8% polyacrylamide-8 M urea sequencing gels; AG marker ladders were generated by partial depurination of probes by formic acid, followed by cleavage with piperidine.
RESULTS
Hydrophobic residues within the B1 region are essential for SRF binding.
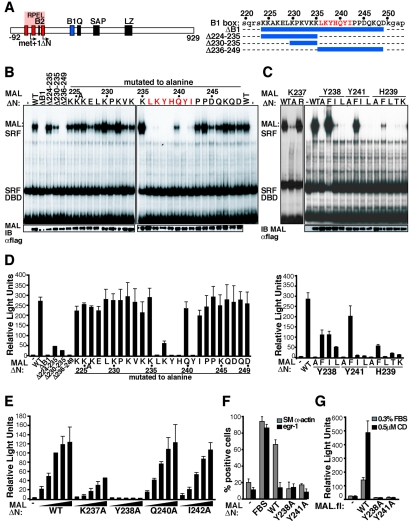
We previously showed the conserved B1 box region is essential for MAL-SRF complex formation (Fig. 1A) (23). To identify residues within this region involved in the MAL-SRF interaction, mutations were introduced into MALΔN, a truncated constitutively nuclear MAL derivative. The altered proteins were expressed by transient transfection in NIH 3T3 cells, and complex formation was assessed in gel mobility shift assays with recombinant SRF DNA-binding domain and a c-fos DNA probe (23).
FIG. 1.
Identification of MAL B1 region residues critical for SRF binding. (A) Conserved motifs in MAL are shown as bars, with RPEL motifs in red. Amino acids are numbered, with the first in-frame methionine (MALmet isoform) as +1; intact NIH 3T3 MAL initiates translation at Leu-92 (23). The MALΔN truncation to position +81 is indicated. The B1 region is shown in blue, and its sequence and deletion mutations are shown. The seven-residue critical sequence identified by the alanine scan is highlighted in red. (B) Deletion and alanine-scanning mutations in the MAL B1 region affect complex formation. Gel mobility shift assays contained Flag-tagged MALΔN B1 region derivatives, SRF.DBD (residues 133 to 265), and the c-fos ΔTCF SRE probe (-, no MALΔN). At the bottom, quantitation of MALΔN expression by immunoblotting (IB) is shown. αflag, anti-Flag antibody. (C) Analysis of interactions by residues K237, Y238, H239, and Y241. Gel mobility shift assays and immunoblotting as in panel B. (D) Activation of the SRF-dependent reporter 3DA.luc by B1 region derivatives. Deletion and alanine scan derivatives (left) and other MALΔN derivatives (right) are shown. (E) Titration of MALΔN derivatives. Reporter assays as in panel D with 6 ng, 18 ng, 55 ng, 166 ng, and 500 ng MALΔN plasmid inputs are shown. (F) Activation of endogenous sm α-actin and egr-1 expression by MALΔN derivatives. Gene expression was scored by immunofluorescence in 100 to 200 transfected cells per experiment (two independent experiments). FBS, fetal bovine serum. (G) Potentiation of cytochalasin D-induced SRF reporter activity by intact MAL requires integrity of the B1 region. Reporter assays were as in panel D. Cells were treated with 0.5 μM cytochalasin D (CD) as indicated.
MALΔN-SRF complex formation was entirely dependent on the B1 region. Small deletions removing the basic N-terminal sequences of the B1 region either had no effect or reduced complex formation slightly, while deletions removing its C-terminal sequences or deletion of the entire region (MALΔB1) completely abolished complex formation (Fig. 1B, left). Next, we conducted an alanine scan across the B1 region (Fig. 1B, right). Substitutions L236A, Y238A, H239A, and Y241A effectively abolished complex formation (<5% wild-type activity), while K237A and I242A significantly reduced it (∼25% wild-type activity). Alanine substitutions elsewhere in the B1 region had only small effects: Q240A and E228A decreased complex formation by 30 to 50%, while K230A, K232A, and K235A increased it by the same amount.
We next investigated the side chain interactions mediating complex formation (Fig. 1C). Substitution K237R did not affect it, suggesting that at this position a basic residue is important. A Y238F substitution slightly increased complex formation, while Y238I and Y238L significantly reduced but did not abolish it; similarly, only the Y241F substitution left complex formation unaffected. The H239F substitution significantly reduced complex formation, while H239L, H239T, or H239K virtually abolished it. The aromatic and/or planar character of Y238, H239, and Y241 is therefore critical for interaction with SRF.
We next tested whether the integrity of the B1 region was required for activation of the SRF-controlled reporter gene 3D.Aluc by MALΔN derivatives (Fig. 1D). Proteins that did not interact detectably with SRF in the gel mobility shift assay did not activate the reporter. The K237A and I242A substitutions had relatively small effects on reporter activation, which were more pronounced at lower plasmid inputs, indicating that the reporter assay appears easily saturated (Fig. 1E). The disproportionately large effects on reporter activation of the K237A substitution and deletions Δ224-235 and Δ230-235 probably arise because these changes also impair nuclear accumulation (see below). The effects on SRF reporter activity of substitutions at residues Y238, H239, and Y241 were broadly similar to their effects on complex formation, although Y238F impaired, rather than enhanced, reporter activation (Fig. 1D). All MALΔN derivatives tested activated the reporter gene independently of functional Rho, indicating that they do not promote activation of endogenous MAL (23).
We used an immunofluorescence assay to test whether the B1 region is required for activation of an endogenous MAL-dependent SRF target gene, smooth muscle α-actin, by MALΔN. Expression of MALΔN, but not its Y238A or Y241A derivatives, induced high-level expression of sm α-actin; this effect was specific, since MALΔN remained unable to activate the TCF-controlled Egr-1 gene, which is insensitive to Rho-actin signaling (Fig. 1F) (7). To confirm that the B1 region is also important for SRF activation by intact MAL in vivo, we exploited the observation that overexpression of MAL sensitizes SRF reporter genes to activation by low levels of cytochalasin D (G. Posern, unpublished observation). Intact MAL, but not its Y238A or Y241A derivatives, activated the reporter (Fig. 1G).
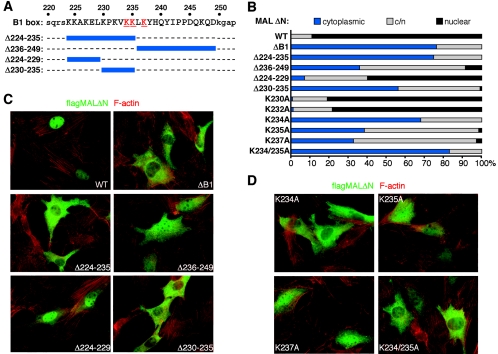
SRF-binding and nuclear import functions of the B1 box are separable.
The B1 region is also required for nuclear accumulation of MALΔN (23). We used an immunofluorescence assay to test whether the B1 sequences involved are distinct from those mediating complex formation with SRF (Fig. 2). Deletion of the entire B1 region (residues 224 to 249), residues 224 to 235, or residues 230 to 235 alone caused MALΔN to become predominantly cytoplasmic; removal of the entire C-terminal part of the B1 region also substantially reduced MALΔN nuclear localization, while MALΔN lacking residues 224 to 229 remained predominantly nuclear (Fig. 2B and C). Alanine replacements at K234, K235, or K237 greatly impaired nuclear localization of MALΔN (Fig. 2B and D). However, activation of the SRF reporter by these mutants did not require functional Rho, indicating that they act directly, rather than by activation of endogenous MAL (activity relative to wild-type MALΔN activity in the presence of C3 transferases was as follows: 103% ± 18% for MALΔN (K234A), 96.4% ± 22% for MALΔN (K235A), and 104% ± 43% for MALΔN (K237A); serum stimulation, 30.4% ± 5%). Other substitutions, including those that completely block complex formation, had no effect on MALΔN localization, and neither the Y238A nor the Y241A mutations affected signal-regulated nuclear accumulation of intact MAL (data not shown). Taken together, these observations render unlikely the possibility that nuclear accumulation results from occlusion of a nuclear export signal upon interaction with SRF. The nuclear localization and SRF binding activities of the B1 box are thus separable, although at least residue K237 must play a role in both activities.
FIG. 2.
Identification of MAL B1 region sequences involved in nuclear import. (A) B1 region sequence, with deletions in blue and residues K234, K235, and K237 highlighted in red. (B) MALΔN B1 mutations affect nuclear import. Subcellular localization was scored as predominantly cytoplasmic, evenly distributed (c/n), or nuclear. Results are averages of two independent experiments (100 to 200 cells each). (C) Subcellular localization of the MALΔN B1 region deletion derivatives. F-actin (red) and transfected MALΔN (green) are shown. (D) Subcellular localization of MALΔN B1 region derivatives, as in panel C.
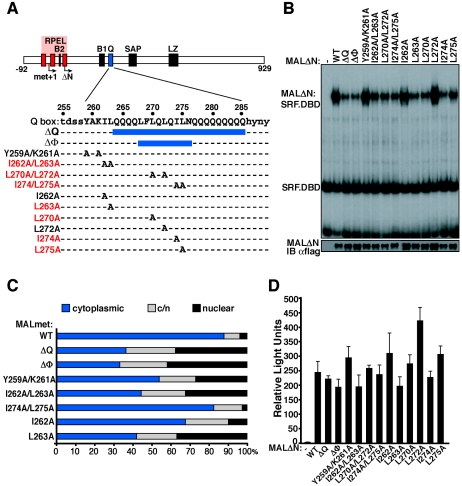
The Q-box enhances SRF interaction but inhibits MAL nuclear accumulation.
The Q-box, a glutamine-rich region located C terminal to the B1 region, is required for optimal MAL-SRF interaction and appears essential for myocardin-SRF interaction (23, 36, 39). We examined MALΔN derivatives containing alterations to the Q-box and its evolutionarily conserved N-terminal flanking sequences (Fig. 3A). Deletion of the Q-box hydrophobic core (MALΔNΔΦ) reduced complex formation by 50%, similar to removal of the entire Q-box (MALΔNΔQ), while alanine substitutions at L270, I274, or L275, but not L272, reduced complex formation by 20 to 40% (Fig. 3B). Alanine substitutions N terminal to the Q-box had no effect on complex formation, apart from L263A, which also reduced it some 25% (Fig. 3B).
FIG. 3.
Effects of the Q-box mutations in the MAL-SRF interaction and the subcellular localization of MAL. (A) Q-box sequence and mutations. Point mutations that affect SRF binding are shown in red. (B) Mutations in the Q-box decrease but do not abolish MAL-SRF complex formation. Gel mobility shift assays were performed using Flag-tagged MALΔN Q-box derivatives, SRF.DBD (residues 133 to 265), and the c-fos ΔTCF SRE probe (-, no MALΔN). Expression levels of the MAL derivatives were confirmed by immunoblotting (IB). αflag, anti-Flag antibody. (C) MAL(met) Q-box mutations affect nuclear import. MAL subcellular localization was scored as described in the legend to Fig. 2. (D) Q-box mutations do not affect SRF reporter activation.
Deletion of the Q-box also causes nuclear accumulation of MAL in unstimulated cells (23). We therefore also examined the effects of Q-box mutations on subcellular localization of intact MAL. Although deletion of the entire Q-box hydrophobic core increased the proportion of cells exhibiting predominantly nuclear MAL, the core mutation I274A/L275A had no effect (Fig. 3C), although both changes impaired complex formation (Fig. 3B). Mutation Y259A/K261A promoted MAL nuclear accumulation (Fig. 3C) but did not affect complex formation (Fig. 3B), while mutations I262A/L263A and L263A both promoted MAL nuclear accumulation (Fig. 3C) and reduced MAL-SRF complex formation (Fig. 3B). The effects of Q-box mutations on subcellular localization and MAL-SRF complex formation show no strict correlation (see Discussion). No Q-box mutations significantly impaired activation of the SRF reporter by MALΔN, consistent with the notion that the Q-box is not required for interaction with SRF in vivo (Fig. 3D) (23).
The hydrophobic core of the B1 basic region is sufficient for SRF interaction.
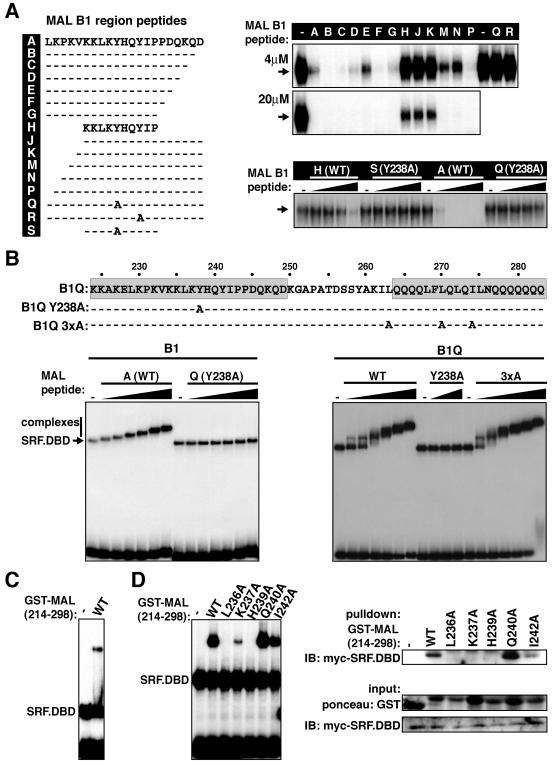
The results presented in the preceding sections are consistent with the view that B1 region residues L236 to I242 represent the primary interaction surface with SRF. To test this directly, we performed peptide competition studies. A 21-residue peptide comprising the hydrophobic core region flanked by seven N- and C-terminal residues (peptide A; residues 229 to 249), but not its Y238A or Y241A derivatives, effectively inhibited complex formation between MALΔN and SRF (Fig. 4A, compare peptides A, Q, and R). Derivatives of peptide A lacking sequences C terminal to position 243 competed for MALΔN complex formation equally effectively, indicating that these sequences are not required for effective complex formation (Fig. 4A, peptides A to G). In contrast, N-terminal truncations of peptide A reduced its effectiveness in the competition assay, coincident with removal of residues K230 and K232, suggesting that at least these two basic residues contribute to the affinity of complex formation (Fig. 4A, compare peptides A, M/N, and J/K; see below). A minimal decapeptide containing the critical residues defined by alanine scanning, N234-KKLKYHQYIP-C243 also competed for MALΔN/SRF complex formation, albeit some 20-fold less effectively than peptide A; again the Y238A substitution abolished binding, indicating the interaction was specific (Fig. 4A, bottom right).
FIG. 4.
The B1 region of MAL is necessary and sufficient for SRF interaction. (A) Peptide competition studies. (Left) MAL peptide sequences. (Right) Competition assays. (Top) Binding reaction mixtures contained MALΔN extract, SRF.DBD (residues 133 to 265), and c-fos wild-type SRE with 4 μM or 20 μM MAL B1 peptide as indicated (-, no MAL B1 peptide). (Bottom) Peptide titrations using 1.3, 3.8, 11.3, and 34 μM peptide. Only the SRF.DBD-MAL complex band is shown. (B) Complex formation between MAL B1 and B1Q peptides and SRF.DBD (residues 120 to 265). The sequences of B1Q peptides are shown at the top. Binding reactions with SRF.DBD and c-fos ΔTCF probe with B1 peptides (0.16, 0.32, 0.63, 1.25, 2.5, and 5 μM) or B1Q peptides (wild-type or 3×A peptide, 0.014, 0.04, 0.12, 0.37, 1.11, and 3.33 μM; Y238A peptide, 0.37, 1.11, and 3.33 μM) are shown. (C) Complex formation between purified GST.MAL(214-298) and recombinant SRF.DBD (residues 132 to 223). (D) The B1 region mediates protein-protein interaction with SRF. Mutations in GST.MAL(214-298) have similar effects on complex formation with SRF.DBD (residues 120 to 265) in mobility shift assays (left) and in pulldown assays (right). IB, immunoblotting.
We next used high-density native gels to visualize peptide-SRF interactions directly. Increasing amounts of the B1 peptide, but not its Y238A derivative, decreased the mobility of the SRF.DBD-DNA complex (Fig. 4B, left). The concentration-dependent decrease in mobility might reflect peptide oligomerization or the possibility that although the peptide-SRF complex can dissociate during its passage through the gel, the caging effect of the gel matrix does not allow the peptide to escape from the SRF-DNA complex. Similar phenomena have been seen with other systems (13). In contrast, a longer peptide including the Q box generated a discrete complex of decreased mobility compared to the SRF.DBD-DNA complex at low concentrations, while at higher peptide inputs, an additional complex of yet lower mobility was detectable (Fig. 4B, right). Formation of these complexes was abolished by the Y238A mutation but was unaffected by alanine substitutions at important Q-box hydrophobic residues (Fig. 4B, right). The integrity of the Q-box appears required for complex formation only in the context of the intact protein. Similar results were obtained with a myocardin B1Q peptide (A.-I. Zaromytidou, unpublished data).
Having defined B1 region residues essential for complex formation, we sought to detect direct protein interactions between this region and SRF. A GST fusion protein containing the entire region, GST.MAL(214-298), effectively formed complexes with purified bacterially expressed SRF DNA-binding domain (Fig. 4C). The GST.MAL(214-298) protein could recover the wild-type SRF DNA-binding domain in pulldown experiments; although recovery was very poor, it was also abolished by the critical point changes (Fig. 4D). Taken together, the results in this section show that the core of the B1 region makes direct protein-protein contacts in the MAL-SRF complex.
MAL and TCF interact with a common surface on SRF.
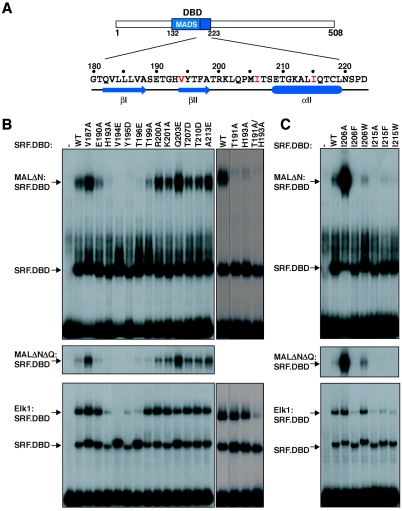
Functional and biochemical studies have shown that the TCF and MAL/myocardin families of SRF cofactors compete for SRF binding (23, 24, 39). In the TCF-SRF and MATα2-Mcm1 complexes, an added β-strand from the cofactor contacts a hydrophobic groove and pocket on the MADS protein (Fig. 5A) (8, 16, 20, 32). We therefore compared the role of the hydrophobic groove in MAL-SRF and TCF-SRF complex formation, using previously characterized SRF point derivatives (16, 40), and also new derivatives designed to alter the dimensions of the pocket.
FIG. 5.
MAL and TCF contact the same hydrophobic groove on the SRF DNA-binding domain. (A) Schematic representation of SRF, with the DNA-binding domain in blue. Mutations and secondary structure elements in the C-terminal half of the DBD are shown below, with hydrophobic pocket residues in red. (B) SRF hydrophobic groove mutations affect complex formation. MALΔN (top), MALΔNΔQ (middle) or Elk-1 (bottom) were used in gel mobility shift assays with SRF.DBD (residues 120 to 265), and either ΔTCF SRE probe (top and middle) or WT SRE probe (bottom) (-, no SRF.DBD). (C) Hydrophobic pocket mutations differentially affect the MAL-SRF and Elk-1-SRF interactions.
All the SRF derivatives bound DNA with comparable affinity, apart from those with substitutions V194E and T196E whose binding activity was somewhat enhanced (Fig. 5B; also data not shown). The effects of these SRF βII-strand substitutions on complex formation with MAL and the Elk-1 TCF differed. While the V194E and T196E substitutions, respectively, abolished or significantly reduced formation of both complexes, T191A, H193A and Y195D significantly reduced complex formation with MAL but not with Elk-1; T199A impaired complex formation by MAL but not Elk-1, while Q203E had small but opposite effects on the two complexes (Fig. 5B, compare top and bottom panels). The changes had similar effects on binding of a MAL derivative lacking the Q-box (Fig. 5B, middle panel). Subtle differences between the interactions of MAL and Elk-1 with SRF were also detected at residues I206 and I215, which line the SRF hydrophobic pocket. At I206, an I206A substitution strongly increased complex formation with MAL but not Elk-1, suggesting this side chain specifically inhibits MAL interaction, while I206F abolished complex formation with both proteins and I206W had no effect (Fig. 5C). At residue I215, complex formation with both MAL and Elk-1 was impaired by the I215A and I215W substitutions, while I215F had a lesser effect (Fig. 5C). Again, similar results were obtained with a MAL derivative lacking the Q-box (Fig. 5C, middle). Taken together, these results show that although both MAL and Elk-1 make contacts with the SRF hydrophobic groove, these contacts are subtly different in the two complexes.
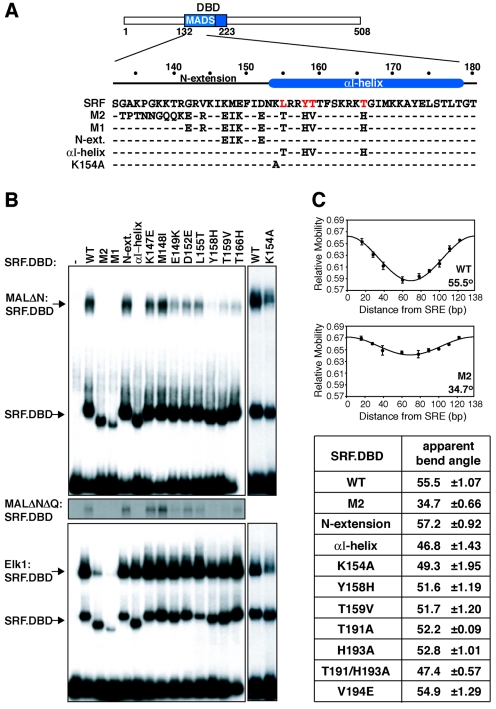
Mutations in the SRF αI-helix specifically block MAL interaction.
Altered DNA binding specificity SRF derivatives, in which the MADS box N-terminal sequences are substituted with those from yeast Mcm1, fail to form complexes with MAL on their cognate binding sites but remain competent for TCF binding (23). We therefore investigated the role of the MADS box N-terminal sequence in complex formation. Altered-specificity derivatives SRF-M1 and SRF-M2 (9) weakly bound the c-fos SRF binding site but did not form complexes with MAL and exhibited reduced interaction with Elk-1 (Fig. 6B, compare top and bottom panels). The authentic DNA binding specificity of SRF is determined by the extreme N-terminal sequences of the MADS box (Fig. 6A) (25). Leaving these specificity-determining sequences intact, we constructed two SRF derivatives in which only sequences in the MADS box N-extension or αI-helix were substituted by Mcm1 sequences (Fig. 6A). Both proteins bound the c-fos SRE efficiently; however, the αI-helix substitution reduced complex formation with MAL to 5 to 10% wild-type activity, while the N-extension substitution had no effect (Fig. 6B, top). Substitution of individual αI-helix residues had less pronounced effects, with the Y158H, T159V, and T166H substitutions all reducing MAL-SRF complex yield (Fig. 6B, top). None of these mutations affected complex formation with Elk-1, although a further αI-helix alanine substitution, at K154, reduced complex formation with both cofactors (Fig. 6B, bottom blot).
FIG. 6.
Mutations in the SRF.DBD αI-helix inhibit MAL-SRF complex formation through their effect on DNA bending. (A) Secondary structure elements and mutations in the N-terminal half of the SRF DBD. Critical αI-helix residues are highlighted in red. N-ext., N-extension. (B) SRF αI-helix mutations disrupt MAL-SRF complex formation. Extracts from cells expressing MALΔN (top), MALΔNΔQ (middle), or Elk-1 (bottom) were used in gel mobility shift assays with the indicated SRF.DBD derivatives (residues 120 to 265) and either ΔTCF SRE probe (top and middle) or WT SRE probe (bottom) (-, no SRF.DBD). (C) Circular permutation analysis. (Top) Representative SRF mobility plots versus distance of the center of the SRF binding site from the probe end (for probes, see Materials and Methods). (Bottom) Estimated apparent bend angles for the different derivatives (two independent experiments).
The MADS box N-terminal changes that impair MAL complex formation also affect the mobility of the SRF-DNA complex, suggesting that they may affect DNA bending (Fig. 6B). Indeed, the SRF αI-helix was previously implicated in DNA bending by a relaxed-specificity SRF derivative, metCORE (40, 41). To compare DNA bending by the different SRF derivatives, we performed circular permutation analysis, using a set of DNA probes in which SRF binds at different distances from the probe end (Fig. 6C). The wild-type SRF DNA-binding domain induced an apparent bend of 55.5° in this assay (this estimate is less than the 72° observed in crystallographic analysis, probably because of the different method used). The SRF-M2 and αI-helix mutations, which greatly impair interaction with MAL, reduced the apparent SRF-DNA bend angle to 34.7° and 46.8°, respectively; in contrast, the N-extension mutation, which did not affect MAL complex formation, did not affect DNA bending (Fig. 6C). Individual αI-helix changes that impaired complex formation had lesser effects, in parallel with their effect on complex formation (Fig. 6C). SRF αI-helix mutations that impair MAL interaction thus also reduce its ability to bend DNA. In contrast, the SRF βII-strand/pocket region, changes T191A and H193A, which reduced MAL complex formation, had only marginal effects on bending, while SRF V194E, which abolished complex formation, distorted DNA similarly to the wild-type protein (Fig. 6C).
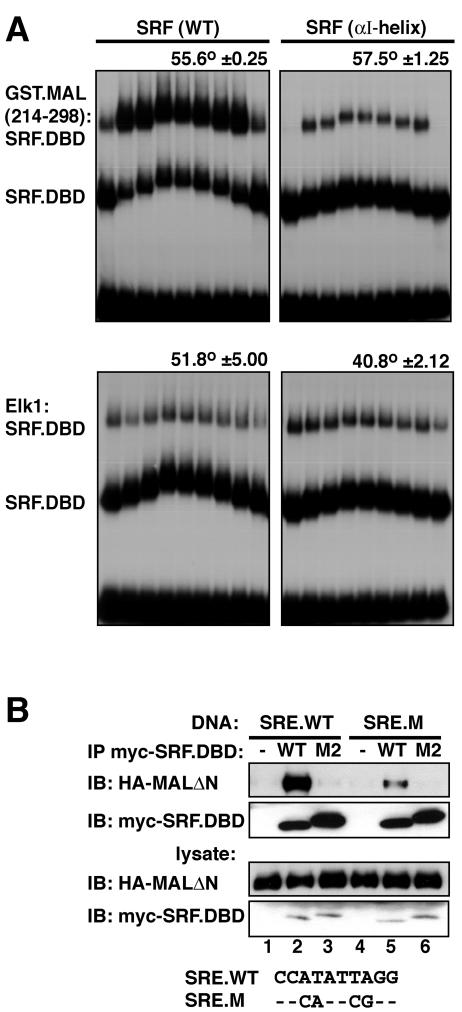
We next used the circular permutation assay to test whether the αI-helix mutation also affects DNA bending in the MAL-SRF complex. Although the different protein content of the MAL-SRF-DNA complex precludes a quantitative comparison of its apparent DNA bend angle with those of SRF-DNA complexes, the assay can be used to compare bending by complexes containing different SRF derivatives. Complexes between the SRF DNA-binding domain and the MAL fusion protein GST.MAL(214-298) distorted DNA with an apparent bend of 55.6° ± 0.25°; however, in contrast to its effect on DNA bending in the SRF-DNA complex, the SRF αI-helix mutation left DNA bending in the MAL-SRF-DNA complex unaffected at 57.5 ± 1.25° (Fig. 7A, top). This result implies that interaction of MAL with the mutant SRF-DNA complex must increase the extent of DNA bending, either through direct MAL-DNA contacts or through allosteric effects. The Elk-1-SRF-DNA complex exhibited a smaller apparent DNA bend than SRF-DNA alone (51.8 ° ± 5°) (29), and this was not affected by the αI-helix mutation (40.8 ° ± 2° [Fig. 7A, bottom]).
FIG. 7.
Optimal MAL-SRF interaction requires DNA binding and distortion. (A) The SRF αI-helix mutation does not affect DNA bending in the MAL-SRF-DNA complex. Circular permutation assays were performed using wild-type SRF.DBD or its αI-helix derivative (residues 120 to 265) and either GST.MAL(214-298) or transiently expressed Elk-1 as indicated. The apparent bend angles for the ternary complexes are indicated above each panel. Note that MAL complex formation is specifically impaired on probes in which the SRF binding site is centered 16 base pairs from the fragment end (see Fig. 8). (B) Cognate DNA enhances MAL-SRF complex formation. Extracts containing HA-tagged MALΔN were incubated with myc-tagged SRF.DBD (residues 120 to 265) from wild-type SRF or SRF-M2 in the presence of either the wild-type c-fos SRE or its derivative SRE.M (sequences shown below). Following immunoprecipitation (IP) with anti-myc beads, MAL was detected by immunoblotting (IB) with anti-HA. (-, no SRF).
MAL-SRF complex formation is facilitated by direct MAL-DNA contact.
The observations presented in the preceding section led us to examine the role of DNA in MAL-SRF complex formation in more detail. To test whether DNA binding facilitates SRF-MAL interaction, we used a coimmunoprecipitation assay. Myc-tagged wild-type or mutant SRF was coexpressed with HA-tagged MALΔN and immunoprecipitated in the presence of DNA containing either wild-type or mutated SRF binding sites. MAL was efficiently recovered in SRF immunoprecipitates in the presence of the wild-type SRF binding site, but its recovery was greatly reduced in the presence of the mutant binding site SRE.M, which cannot bind wild-type SRF (Fig. 7B, compare lanes 2 and 5) (9). In contrast, MAL was not recovered in SRF-M2 immunoprecipitates, regardless of which SRF binding site was present, consistent with the electrophoretic mobility shift assay results (Fig. 7B, lanes 3 and 6, and 6B) (23, 24, 39). Thus, MAL recognizes SRF more effectively in the context of the wild-type SRF-DNA complex.
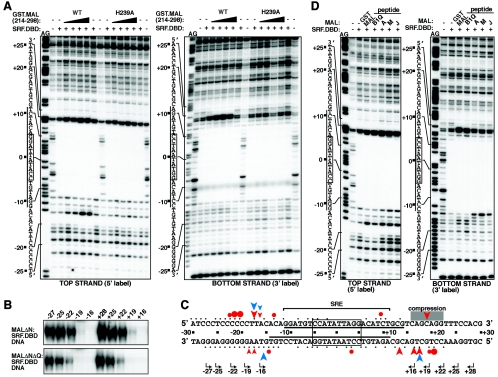
To test whether MAL makes contacts with DNA, we performed DNase I footprinting. End-labeled c-fos DNA probes were incubated with recombinant SRF DNA-binding domain, either alone or with increasing amounts of GST.MAL(214-298) and treated with limiting amounts of DNase I. The SRF DNA-binding domain protected probe sequences spanning the classical SRE element (33) (Fig. 8A). Addition of increasing amounts of wild-type GST.MAL(214-298), but not its nonbinding H239A derivative, induced additional marked changes in DNase I accessibility symmetrically around the SRE dyad (Fig. 8A; summarized in Fig. 8C). On the top strand, the most prominent of these changes included protections 5′ to positions −22, −21, and −20 and enhancements 5′ to −17 and −16 and on the bottom strand, protections 5′ to positions +20 and +19 and enhancements 5′ to +17, +16, and +13 (Fig. 8C). At least at the 5′ side of the SRE, the major perturbations in DNase I accessibility were restricted to the top DNA strand (Fig. 8A and C); a gel compression prevented conclusive interpretation of the pattern 3′ to the SRE.
FIG.8.
MAL makes direct DNA contacts that are required for complex formation. (A) DNase I footprinting of the MAL-SRF complex. Reaction mixtures contained c-fos DNA, SRF.DBD (residues 132 to 223) (+) as indicated, and increasing amounts (0.8, 2.5, 7.6, and 22.8 ng) of WT or H239A GST.MAL(214-298) proteins. The probe DNA sequences are aligned with the AG marker ladders (line, SRE; open box, SRF core consensus binding site). (B) Complex formation between recombinant SRF.DBD (residues 132 to 223) and transiently expressed MALΔN (top) or MALΔNΔQ (bottom) on nested DNA probes with ends at the indicated positions relative to the SRE dyad (see panel C). Only the MAL-SRF-DNA complex is shown. Similar results were obtained with GST.MAL (214-298) (data not shown). (C) Summary of the DNase I footprinting data. The c-fos SRE region is shown, with black dots representing phosphodiester bonds where DNase I cleavage was readily detectable on naked DNA. For simplicity, the footprint with SRF alone is indicated by the solid lines. The colored circles and arrowheads show protection from and enhancement of DNase I cleavage, respectively. Red symbols, GST.MAL(214-298); blue symbols, peptide J. The red arrowhead within a gray bar indicates where a sequencing compression precludes conclusive interpretation of top strand changes at the 3′ side of the SRE. Within the SRE region, changes were visible only after prolonged exposure of the autoradiogram. The start positions and direction of the nested band shift probes used in panel B are shown at the bottom. (D) Footprint analysis of MAL B1 region peptides (Fig. 4A), presented as in panel A. Reaction mixtures contained 7.6 ng GST.MAL(214-298), 1.7 μM peptide B1Q, 8.5 μM peptides A and M, and 85 μM peptide J.
These results strongly suggest that MAL-SRF complex formation involves direct contacts between MAL and DNA. This observation was intriguing in light of the circular permutation experiment presented in Fig. 7A, which showed that MAL-SRF-DNA complex formation, but not SRF binding, was impaired when the SRF binding site was located 16 base pairs from the fragment end. To investigate the role of MAL-DNA contact in complex formation directly, we performed gel mobility shift assays with nested probes in which the SRF binding site was brought progressively closer to the fragment end in 3-base-pair increments. Complex formation between transiently expressed MALΔN and the SRF DNA-binding domain was relatively efficient on probes in which the binding site is located 22 base pairs from the fragment end; however, further truncations which impinged on or deleted the footprinted region (+19, −19, +16, and −16 probes) substantially impaired complex formation (Fig. 8B). These results show that MAL makes direct DNA contacts in the MAL-SRF-DNA complex. These contacts are not mediated by the Q-box region, since similar results were obtained with MALΔNΔQ, which lacks these sequences (Fig. 8B).
To gain further insight into the MAL sequences involved in the flanking DNA contacts, we investigated footprinting by MAL peptide-SRF complexes. The B1Q peptide, residues MAL224-283 (Fig. 4B) in which the entire B1 region is intact generated a footprint essentially identical to that produced by GST.MAL(214-298) (Fig. 8D). In contrast, peptide A (Fig. 4A), which lacks the N-terminal residues of the B1 region, generated a modified footprint. At the 5′ side of the SRE, the peptide A-SRF footprint lacked the top strand protections 5′ to positions −22/−21/−20, while an additional enhanced cleavage was seen on the bottom strand 5′ to position −16; similar results were seen at the 3′ side of the SRE, although in this case the top strand enhancements were not resolved (Fig. 8D, summarized in Fig. 8C). Shorter peptides gave similar results (Fig. 8D). The altered pattern of MAL-DNA interaction seen with peptides lacking the N-terminal part of the MAL basic region suggests that these sequences are required for, but do not necessarily mediate, authentic MAL-DNA contacts.
DISCUSSION
In this study we have characterized the interaction between the SRF transcription factor and its cofactor MAL/MKL1, a member of the MRTF family of SRF coactivators. Formation of the MAL-SRF complex is absolutely dependent on a short predominantly hydrophobic core sequence located within the conserved MAL B1 region, which is also sufficient for specific interaction. Hydrophobic residues in the neighboring Q-box region of MAL contribute toward affinity but are unlikely to contact SRF directly. Sequences within both the B1 and Q-box regions that control MAL nuclear accumulation are largely distinct from those required for complex formation. The MAL and TCF families of SRF coactivators both interact with a deep hydrophobic pocket on the SRF DNA-binding domain. SRF αI-helix mutations that reduce DNA bending by SRF reduce complex formation with MAL but do not affect DNA distortion in the complex. Efficient MAL-SRF interaction requires that SRF be bound to DNA. In the complex MAL makes symmetric DNA contacts or close approaches at positions +16 through +22 and −16 through −22 with respect to the SRF dyad, and these interactions are required for efficient complex formation. Taken together, the data suggest a model in which efficient MAL-SRF complex formation is dependent on SRF-induced DNA bending, which facilitates MAL-DNA interaction.
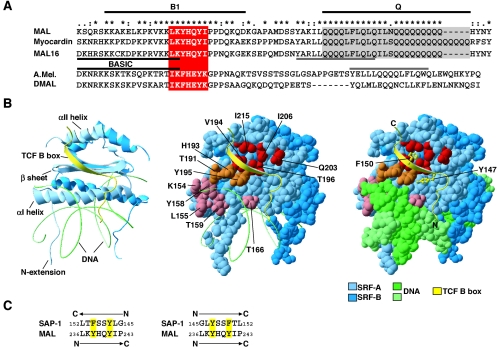
Our previous studies showed that the conserved MAL B1 basic region is essential for formation of the MAL-SRF complex, while the neighboring Q-box contributes to affinity (23). Here we used alanine scanning to identify a seven-residue sequence within the B1 region that is critical for interaction with SRF (Fig. 9A, red box). A short peptide including this sequence is sufficient for interaction, albeit with substantially reduced affinity. Within the critical region, basic, aromatic, and planar conserved residues play important roles in complex formation. The core sequence is conserved in insect MAL homologs, and that from drosophila can replace the mouse sequence (Fig. 9A; A.-I. Zaromytidou, unpublished data). While individual basic residues in the N-terminal part of the B1 region are dispensable for specific interaction with SRF, peptide competition studies suggest that this region contributes to the affinity of complex formation. Consistent with this, the integrity of the basic sequences is required to generate the authentic MAL-SRF footprint. The strand asymmetry of the DNase I footprint suggests that the flanking footprints result from interactions with the DNA phosphate backbone, but we cannot rule out additional base-specific contacts. The symmetric DNA contacts made by MAL provides at least a partial explanation for previous observations that, in the context of the c-fos promoter, both the TCF and AP1/ATF sites, located on either side of the SRF binding site, contribute to occlusion of SRF from MAL interaction (24, 38).
FIG. 9.
MAL and SRF residues involved in complex formation. (A) Alignment of the sequences of B1 and Q regions of mouse MAL, myocardin, and MAL16, with identities and conservative replacements indicated by asterisks, colons, and dots above the sequence. Conservation of the critical seven-residue MAL sequence (red) and Q-box-related sequence (gray) in bee (A.Mel) and fruit fly MAL (DMAL) is shown. (B) The MAL-binding surface of SRF. The SAP-1-SRF ternary complex (8) is shown: blue, SRF; green, DNA; yellow, SAP-1 B-box. (Left) Ribbon model, with secondary structure elements indicated. (Center) SRF and SAP-1 are shown in van der Waals and ribbon representations, respectively. SRF residues implicated in MAL interaction are shown in red (hydrophobic pocket), orange (β-strand residues also contacting DNA), and pink (critical αI-helix residues). (Right) Same as center but with DNA shown as van der Waals representation and SAP-1 aromatic side chains Y147 and F150 in backbone representation. Images were produced using Deep View/Swiss-Pdb Viewer version 3.7. (C) Potential sequence relationships between the critical MAL B1 region sequence and the β-strand segment of the SAP-1 B-box. For discussion, see text.
In addition to its role in contacting SRF and DNA, the B1 region affects nuclear accumulation of MALΔN. It appears to act as a nuclear import signal, as opposed to promoting MAL nuclear retention through SRF interaction or acting as a nuclear export signal occluded upon SRF binding, since its functions in complex formation and nuclear accumulation are separable. While K234, K235, and K237 are critical for import, only K237 is also essential for MAL-SRF complex formation.
In the Q-box region, we identified a number of hydrophobic residues that contribute to complex formation. While our results are similar to the results of a previous study of myocardin, we do not concur with the proposal that the Q-box makes essential interactions with SRF similar to the TCF B-box (39). First, in our hands MAL and myocardin derivatives lacking the Q-box effectively activate SRF reporters (A.-I. Zaromytidou, unpublished data). Second, substitutions in SRF affect complex formation regardless of the presence of the Q-box, suggesting that there is no unique SRF surface contacted by the Q-box. Third, peptides including the entire Q-box region do not interact with SRF in the absence of the intact B1 region. Fourth, the Q-box region is poorly conserved through evolution (Fig. 9A), and its alignment with the TCF B-box cannot easily be reconciled with the SAP-1/SRF crystal structure (8). It is unlikely that the Q-box contributes to the interactions with DNA flanking the SRE, since MAL derivatives lacking the Q-box remain dependent on these sequences for efficient complex formation with SRF. Instead, we suggest that the Q-box contributes to MAL-SRF complex formation indirectly, either by stabilizing the structure of the B1 region in the MAL-SRF complex or by acting as an intramolecular chaperone to structure the B1 region in uncomplexed MAL. The latter mechanism could explain why Q-box mutations also promote MAL nuclear import, since disruption of a B1/Q-box interaction might expose B1 residues critical for nuclear import.
Our analysis of complex formation by mutated SRF derivatives indicates that MAL interaction requires residues along a hydrophobic groove, containing a deep hydrophobic pocket, on the SRF DNA-binding domain. MAL-SRF complex formation is disrupted by SRF substitutions H193A, V194E, Y195D, and T196E, while substitutions at residues I206 and I215 which are predicted to alter the size of the pocket affect complex formation appropriately (Fig. 9B, red and orange residues). Mutations at all these positions also affect TCF binding (16), although their relative effects on the TCF-SRF complex are subtly different. Structural studies have shown that the hydrophobic groove and pocket play important roles in the TCF-SRF and MATα2-Mcm1 complexes (8, 16, 19, 32). Our data support previous functional and biochemical studies demonstrating that binding of MRTFs and TCFs to SRF is mutually exclusive (23, 24, 39).
In the TCF-SRF and MATα2-Mcm1 complexes, the cofactor contributes an additional β-strand to the MADS box β-sheet region (8, 32). Although the β-strand is added in antiparallel and parallel orientations in TCF-SRF and MATα2-Mcm1 complexes, respectively, the side chain interactions are strikingly similar. In each case, an aromatic residue is docked in the hydrophobic pocket, while hydrophobic interactions occur between the strand and the MADS box αI-helix (SAP-1 TCF interactions shown in Fig. 9B). An alanine-scanning mutagenesis of the TCF Elk-1 identified two essential aromatic residues within its seven-residue added β-strand (15), while an in vivo functional analysis of the MATα2-Mcm1 interaction showed that all eight added β-strand residues are sensitive to alanine substitution (20). Intriguingly, the critical seven-residue MAL hydrophobic sequence contains aromatic residues spaced similarly to those in the TCF β-strand (Fig. 9C). This, its length, and the involvement of the SRF hydrophobic groove all suggest that the core hydrophobic sequence contacts SRF directly through a similar “added β-strand” mechanism. Our data do not address the direction of the added β-strand, since in either orientation aromatic residues are available for insertion into the hydrophobic pocket. We note, however, that only if the MAL β-strand is added in opposite polarity to the SAP-1 B-box does MAL L236 align with SAP-1 L152, a residue critical for stability of the SAP-1-SRF ternary complex (Fig. 9C) (8, 15).
Despite the common involvement of the SRF hydrophobic groove and pocket, MAL-SRF interaction differs from that with TCF in that it is critically dependent on residues within the SRF αI-helix which are largely masked by DNA in the SRF-DNA complex (Fig. 9B, pink). SRF αI-helix residues have previously been implicated in DNA contact and bending by SRF and Mcm1 (1, 2, 8, 26, 32, 40, 41), and our results confirm and extend these observations. In contrast to its effect on DNA bending in the SRF-DNA complex, however, the αI-helix mutation does not alter DNA bending in the context of the MAL-SRF-DNA ternary complex, implying that induction of an appropriate DNA bend is a prerequisite for formation of the MAL-SRF complex. According to this view, the impairment of MAL-SRF complex formation by SRF mutations that impair DNA bending, such as the αI-helix mutant, would reflect the use of binding energy to further distort the DNA in the ternary complex. It is likely that the direct contacts between MAL and DNA flanking the SRF binding site are responsible for the maintenance of the DNA bend in such complexes. Our results suggest that MAL-SRF complex formation might also be influenced by the sequence of the SRF binding site itself, according to its capacity for bending, as seen with Mcm1 and the Arabidopsis thaliana squamosa proteins (2, 40, 41). Structural analysis and site selection experiments will address these issues.
In summary, our results support a model in which MAL adds a β-strand composed of the critical B1 core hydrophobic sequence to the SRF β-sheet region, while simultaneously making direct contacts, probably via B1 region basic residues to the DNA flanking the SRF binding site. Functional studies of both SRF and Mcm1 have previously implicated DNA bending in transcriptional activation (3, 4, 10, 14). Since the mechanisms of ternary complex formation by the MRTFs and TCFs are related but distinct, it may be possible to use transgenes expressing SRF derivatives specific for each cofactor family to test to what extent SRF inactivation phenotypes in vivo reflect disruption of Rho-actin or mitogen-activated protein kinase signaling to SRF target genes. The SRF αI-helix residues T159 and S162 are targets for signal-induced phosphorylation (12): such modifications might be involved in controlling cofactor access to SRF, which occurs during the switch from proliferative to differentiated phenotype in smooth muscle (39). The structure of the SRF-MAL complex and the influence of DNA sequence and external signals on complex formation will be exciting topics for future experiments.
Acknowledgments
We thank Paul Fitzjohn and Graham Smith for help with the circular permutation analysis; Sara Ross for advice on DNase I footprinting; Rob Nicolas for advice and encouragement; Nicola O'Reilly for peptide synthesis; Andrew Sharrocks for SRF DNA-binding domain derivatives; Paul Bates, Markus Hassler, Caroline Hill, and Transcription Laboratory members for discussions and comments on the manuscript; and reviewer 2 for forcing us at last to do footprints.
F.M. was in part funded by an E.U. Marie Curie Fellowship. This work was funded by Cancer Research UK.
REFERENCES
- 1.Acton, T. B., J. Mead, A. M. Steiner, and A. K. Vershon. 2000. Scanning mutagenesis of Mcm1: residues required for DNA binding, DNA bending, and transcriptional activation by a MADS-box protein. Mol. Cell. Biol. 20:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acton, T. B., H. Zhong, and A. K. Vershon. 1997. DNA-binding specificity of Mcm1: operator mutations that alter DNA-bending and transcriptional activities by a MADS box protein. Mol. Cell. Biol. 17:1881-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruhn, L., and G. F. Sprague, Jr. 1994. MCM1 point mutants deficient in expression of α-specific genes: residues important for interaction with α1. Mol. Cell. Biol. 14:2534-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carr, E. A., J. Mead, and A. K. Vershon. 2004. Alpha1-induced DNA bending is required for transcriptional activation by the Mcm1-alpha1 complex. Nucleic Acids Res. 32:2298-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cen, B., A. Selvaraj, R. C. Burgess, J. K. Hitzler, Z. Ma, S. W. Morris, and R. Prywes. 2003. Megakaryoblastic leukemia 1, a potent transcriptional coactivator for serum response factor (SRF), is required for serum induction of SRF target genes. Mol. Cell. Biol. 23:6597-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalton, S., and R. Treisman. 1992. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell 68:597-612. [DOI] [PubMed] [Google Scholar]
- 7.Gineitis, D., and R. Treisman. 2001. Differential usage of signal transduction pathways defines two types of serum response factor target gene. J. Biol. Chem. 276:24531-24539. [DOI] [PubMed] [Google Scholar]
- 8.Hassler, M., and T. J. Richmond. 2001. The B-box dominates SAP-1-SRF interactions in the structure of the ternary complex. EMBO J. 20:3018-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hill, C. S., R. Marais, S. John, J. Wynne, S. Dalton, and R. Treisman. 1993. Functional analysis of a growth factor-responsive transcription factor complex. Cell 73:395-406. [DOI] [PubMed] [Google Scholar]
- 10.Hill, C. S., J. Wynne, and R. Treisman. 1994. Serum-regulated transcription by serum response factor (SRF): a novel role for the DNA binding domain. EMBO J. 13:5421-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howell, M., and C. S. Hill. 1997. XSmad2 directly activates the activin-inducible, dorsal mesoderm gene XFKH1 in Xenopus embryos. EMBO J. 16:7411-7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iyer, D., N. Belaguli, M. Fluck, B. G. Rowan, L. Wei, N. L. Weigel, F. W. Booth, H. F. Epstein, R. J. Schwartz, and A. Balasubramanyam. 2003. Novel phosphorylation target in the serum response factor MADS box regulates alpha-actin transcription. Biochemistry 42:7477-7486. [DOI] [PubMed] [Google Scholar]
- 13.Klejman, M. P., L. A. Pereira, H. J. van Zeeburg, S. Gilfillan, M. Meisterernst, and H. T. Timmers. 2004. NC2α interacts with BTAF1 and stimulates its ATP-dependent association with TATA-binding protein. Mol. Cell. Biol. 24:10072-10082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim, F. L., A. Hayes, A. G. West, A. Pic-Taylor, Z. Darieva, B. A. Morgan, S. G. Oliver, and A. D. Sharrocks. 2003. Mcm1p-induced DNA bending regulates the formation of ternary transcription factor complexes. Mol. Cell. Biol. 23:450-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ling, Y., J. H. Lakey, C. E. Roberts, and A. D. Sharrocks. 1997. Molecular characterization of the B-box protein-protein interaction motif of the ETS-domain transcription factor Elk-1. EMBO J. 16:2431-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ling, Y., A. G. West, E. C. Roberts, J. H. Lakey, and A. D. Sharrocks. 1998. Interaction of transcription factors with serum response factor. Identification of the Elk-1 binding surface. J. Biol. Chem. 273:10506-10514. [DOI] [PubMed] [Google Scholar]
- 17.Ma, Z., S. W. Morris, V. Valentine, M. Li, J. A. Herbrick, X. Cui, D. Bouman, Y. Li, P. K. Mehta, D. Nizetic, Y. Kaneko, G. C. Chan, L. C. Chan, J. Squire, S. W. Scherer, and J. K. Hitzler. 2001. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat. Genet. 28:220-221. [DOI] [PubMed] [Google Scholar]
- 18.Marais, R., J. Wynne, and R. Treisman. 1993. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell 73:381-393. [DOI] [PubMed] [Google Scholar]
- 19.Mead, J., A. R. Bruning, M. K. Gill, A. M. Steiner, T. B. Acton, and A. K. Vershon. 2002. Interactions of the Mcm1 MADS box protein with cofactors that regulate mating in yeast. Mol. Cell. Biol. 22:4607-4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mead, J., H. Zhong, T. B. Acton, and A. K. Vershon. 1996. The yeast α2 and Mcm1 proteins interact through a region similar to a motif found in homeodomain proteins of higher eukaryotes. Mol. Cell. Biol. 16:2135-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mercher, T., M. B. Coniat, R. Monni, M. Mauchauffe, F. N. Khac, L. Gressin, F. Mugneret, T. Leblanc, N. Dastugue, R. Berger, and O. A. Bernard. 2001. Involvement of a human gene related to the Drosophila spen gene in the recurrent t(1;22) translocation of acute megakaryocytic leukemia. Proc. Natl. Acad. Sci. USA. 98:5776-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Messenguy, F., and E. Dubois. 2003. Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene 316:1-21. [DOI] [PubMed] [Google Scholar]
- 23.Miralles, F., G. Posern, A. I. Zaromytidou, and R. Treisman. 2003. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 113:329-342. [DOI] [PubMed] [Google Scholar]
- 24.Murai, K., and R. Treisman. 2002. Interaction of serum response factor (SRF) with the Elk-1 B-box inhibits RhoA-actin signalling to SRF and potentiates transcriptional activation by Elk-1. Mol. Cell. Biol. 22:7083-7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nurrish, S. J., and R. Treisman. 1995. DNA binding specificity determinants in MADS-box transcription factors. Mol. Cell. Biol. 15:4076-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pellegrini, L., S. Tan, and T. J. Richmond. 1995. Structure of serum response factor core bound to DNA. Nature 376:490-498. [DOI] [PubMed] [Google Scholar]
- 27.Sasazuki, T., T. Sawada, S. Sakon, T. Kitamura, T. Kishi, T. Okazaki, M. Katano, M. Tanaka, M. Watanabe, H. Yagita, K. Okumura, and H. Nakano. 2002. Identification of a novel transcriptional activator, BSAC, by a functional cloning to inhibit tumor necrosis factor-induced cell death. J. Biol. Chem. 277:28853-28860. [DOI] [PubMed] [Google Scholar]
- 28.Selvaraj, A., and R. Prywes. 2003. Megakaryoblastic leukemia-1/2, a transcriptional co-activator of serum response factor, is required for skeletal myogenic differentiation. J. Biol. Chem. 278:41977-41987. [DOI] [PubMed] [Google Scholar]
- 29.Sharrocks, A. D., and P. Shore. 1995. DNA bending in the ternary nucleoprotein complex at the c-fos promoter. Nucleic Acids Res. 23:2442-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shore, P., and A. D. Sharrocks. 1994. The transcription factors Elk-1 and serum response factor interact by direct protein-protein contacts mediated by a short region of Elk-1. Mol. Cell. Biol. 14:3283-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shore, P., and A. D. Sharrocks. 1995. The MADS-box family of transcription factors. Eur. J. Biochem. 229:1-13. [DOI] [PubMed] [Google Scholar]
- 32.Tan, S., and T. J. Richmond. 1998. Crystal structure of the yeast MATalpha2/MCM1/DNA ternary complex. Nature 391:660-666. [DOI] [PubMed] [Google Scholar]
- 33.Treisman, R. 1986. Identification of a protein-binding site that mediates transcriptional response of the c-fos gene to serum factors. Cell 46:567-574. [DOI] [PubMed] [Google Scholar]
- 34.Treisman, R. 1994. Ternary complex factors: growth factor regulated transcriptional activators. Curr. Opin. Genet. Dev. 4:96-101. [DOI] [PubMed] [Google Scholar]
- 35.Vershon, A. K., and A. D. Johnson. 1993. A short, disordered protein region mediates interactions between the homeodomain of the yeast alpha 2 protein and the MCM1 protein. Cell 72:105-112. [DOI] [PubMed] [Google Scholar]
- 36.Wang, D., P. S. Chang, Z. Wang, L. Sutherland, J. A. Richardson, E. Small, P. A. Krieg, and E. N. Olson. 2001. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell 105:851-862. [DOI] [PubMed] [Google Scholar]
- 37.Wang, D. Z., S. Li, D. Hockemeyer, L. Sutherland, Z. Wang, G. Schratt, J. A. Richardson, A. Nordheim, and E. N. Olson. 2002. Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc. Natl. Acad. Sci. USA 99:14855-14860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, Y., and R. Prywes. 2000. Activation of the c-fos enhancer by the erk MAP kinase pathway through two sequence elements: the c-fos AP-1 and p62TCF sites. Oncogene 19:1379-1385. [DOI] [PubMed] [Google Scholar]
- 39.Wang, Z., D. Z. Wang, D. Hockemeyer, J. McAnally, A. Nordheim, and E. N. Olson. 2004. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature 428:185-189. [DOI] [PubMed] [Google Scholar]
- 40.West, A. G., and A. D. Sharrocks. 1999. MADS-box transcription factors adopt alternative mechanisms for bending DNA. J. Mol. Biol. 286:1311-1323. [DOI] [PubMed] [Google Scholar]
- 41.West, A. G., P. Shore, and A. D. Sharrocks. 1997. DNA binding by MADS-box transcription factors: a molecular mechanism for differential DNA bending. Mol. Cell. Biol. 17:2876-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]