Abstract
Activation-induced cytidine deaminase (AID) is a single-stranded DNA deaminase required for somatic hypermutation of immunoglobulin (Ig) genes, a key process in the development of adaptive immunity. Transcription provides a single-stranded DNA substrate for AID, both in vivo and in vitro. We present here an assay which can faithfully replicate all of the molecular features of the initiation of hypermutation of Ig genes in vivo. In this assay, which detects AID-mediated deamination in the context of transcription by Escherichia coli RNA polymerase, deamination targets either strand and declines in efficiency as the distance from the promoter increases. We show that AID binds DNA exposed by the transcribing polymerase, implicating the polymerase itself as the vehicle which distributes AID on DNA as it moves away from the promoter.
Antibody gene diversification in the secondary lymphoid organs is accomplished by two distinct genetic modifications. Somatic hypermutation is initiated by antigen binding and results in the introduction of point mutations in the variable regions of immunoglobulin (Ig) genes, thereby allowing for selection of high-affinity antibody variants. Class switch recombination, on the other hand, is a deletional recombination reaction between immunoglobulin heavy chain switch regions. This allows a B cell to switch from the production of IgM to the production of IgG, IgE, or IgA.
Even though somatic hypermutation and class switch recombination are independent reactions, they both require transcription through the Ig locus in the presence of a protein named activation-induced cytidine deaminase (AID) (23, 24). AID belongs to a family of enzymes which share certain features with the metabolic cytidine deaminases but differs from them in that AID cannot utilize free nucleotide as a substrate (6, 15). Instead, AID deaminates single-stranded DNA (9, 13, 15, 42). Other enzymes of the family also act on single-stranded nucleic acid (RNA or DNA) (14).
It has been postulated that AID acts directly on the immunoglobulin locus to introduce base changes which are then processed into mutations (20), and compelling genetic evidence in support of this hypothesis exists (16, 27, 29). What process acts on the immunoglobulin locus to create a single-stranded substrate for AID? Single-stranded DNA can be generated through at least three processes: transcription, replication, or DNA breakage (which can be related to the first two or could be the result of the action of a specific endonuclease). Several groups have shown DNA breaks specific to the Ig locus but independent of AID activity (10, 25, 43), though the direct relevance of these to the outcome of the reaction has not been established. Here we consider only the mechanistic involvement of transcription in AID-mediated deamination events.
Several laboratories have shown a strong link between transcription and somatic hypermutation both in vivo (2, 41) and in vitro (8, 13, 31, 35). Here we describe an assay where recombinant AID is able to mutagenize DNA in vitro in the context of transcription by Escherichia coli RNA polymerase (RNAP). We find that AID does not display strand bias: cytidines on the nontemplate (NT) strand can be deaminated just as well as cytidines on the template (T) strand, in a manner similar to Ig gene hypermutation in vivo. In addition, the spectrum of mutations created in the course of in vitro transcription is remarkably similar to that created in the course of the hypermutation reaction on the Ig locus in vivo, and the frequency of these mutations decreases rapidly as a function of distance from the promoter. Finally, we show that AID interacts directly with the elongation complex in vitro, binding through residues that are distinct from the deaminase active site.
MATERIALS AND METHODS
Plasmids and strains.
The ptac-KanL94P plasmid was obtained from M. Nussenzweig, The Rockefeller University. The ptac-CcdB plasmid was obtained by replacing KanL94P with ccdB (amplified from pZero; Invitrogen), using two restriction sites (BamHI and HindIII) which flank the target gene.
Protein purification.
Constructs of human AID were expressed in an E. coli Rosetta strain (Novagen) by using a pET28b vector (Novagen) with a C-terminal hexahistidine tag. Protein expression was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 16°C for 12 to 16 h after the optical density at 600 nm had reached 0.8. Bacterial pellets were directly resuspended in buffer A (50 mM MES [morpholineethanesulfonic acid] [pH 6.5], 500 mM KCl, 1 mM phenylmethylsulfonyl fluoride), frozen, and kept at −80°C. Cells were thawed and lysed with a cell disruptor (French press). The lysate was centrifuged, and the resulting supernatant was loaded onto a Talon resin charged with Co2+ ions (BD Biosciences). The resin was washed first in buffer A and then in buffer B (buffer A plus 30 mM imidazole) and eluted with buffer C (buffer A plus 150 mM imidazole). The eluted protein was immediately dialyzed against buffer A to remove imidazole. The protein was then injected onto a Superdex 200 10/300 GL size exclusion column (Amersham-Pharmacia) by using buffer D (50 mM MES [pH 6.5], 500 mM KCl, 5 mM dithiothreitol). The protein eluted from the gel filtration column as a single peak.
Recombinant human AID purified from E. coli was also obtained from Enzymax, LLC (www.enzymax.net), as was the mutant AID we describe. Human replication protein A (RPA) was a generous gift from Marc Wold. Aliquots of human RPA and Ustilago maydis RPA previously characterized in recombination assays were kindly provided by Haijuan Yang and Nikola Pavletich (39, 40).
Transcription-based assay of AID activity.
The 100-μl in vitro transcription reaction mixtures (20 mM Tris-HCl, pH 7.9, 20 mM KCl, 5 mM MgCl2, 2 nM substrate plasmid, 10 nM RNAP [from Epicenter Technologies], 100 μM ribonucleoside triphosphates [rNTPs], and 20 nM AID) were incubated for 1 h at 37°C. Plasmid DNA was purified by QIAGEN MinElute to yield 10 μl of DNA in H2O, which was introduced into ung-deficient E. coli by electroporation using a Bio-Rad GenePulser. Bacteria were plated on spectinomycin (Spec) and spectinomycin-kanamycin (Kan)-IPTG-containing plates (for ptac-KanL94P assays), on IPTG-spectinomycin (for ptac-CcdB assays), and on kanamycin (for lacZ-ccdB assays).
Assembling the transcription elongation complex (TEC).
The RNAP transcription complex was assembled in transcription buffer (40 mM Tris-HCl, pH 7.9, 40 mM KCl, 10 mM MgCl) following the procedure described by Artsimovitch and Landick (1). Briefly, we assembled the RNA template DNA hybrid by using equimolar quantities of RNA and DNA oligonucleotides. To this we added His-tagged RNAP (a gift from Seth Darst, The Rockefeller University) and finally equimolar amounts of the nontemplate DNA strand. The assembled complex was bound to nickel-agarose beads for 10 min on ice and then washed in 1 M NaCl (to remove single-stranded nucleic acid and incomplete complexes), followed by incubation in transcription buffer.
The TEC oligonucleotide sequences used were as follows: for the nontemplate strand, 5′ CACCACCACGCGGGCTGTAGCGTGCTTTTTTCGATCTTCCAGTG 3′; for the template strand, 5′ CACTGGAAGATCGAAAAAAGCACGCTACAGCCCGCGTGGTGGTG 3′; and for RNA, 5′ GCGGGCUGUAGCGU 3′.
UV cross-linking of AID to the transcription elongation complex.
A transcription complex was assembled with one 32P-labeled oligonucleotide, bound to Ni-agarose beads, and washed with 1 M NaCl. AID with or without competitor tRNA was bound to the beads for 20 min at room temperature. The reaction mixture was then put on ice and irradiated for 10 min at a distance of 2.5 cm from the bulb of a Stratalinker. Sodium dodecyl sulfate sample buffer was added, and the samples were heated to 95°C for 2 min and loaded on a 4 to 20% acrylamide Tris-glycine gel.
RESULTS
In vitro transcription by E. coli RNAP allows AID access to plasmid DNA.
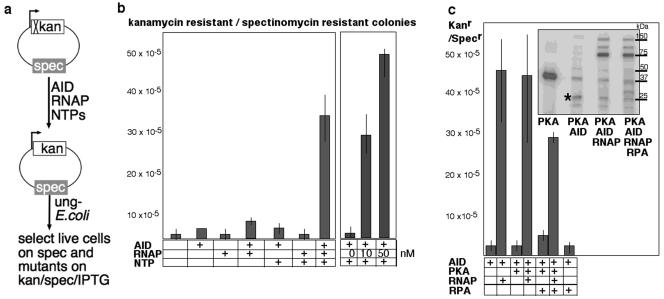
Previous experiments have shown that induction of AID expression in E. coli containing transcribing plasmid ptac-KanL94P can lead to reversion of the L94P mutation and kanamycin resistance (31, 35). To show that transcription is sufficient to target deamination by AID, we incubated the ptac-KanL94P plasmid with recombinant AID, E. coli RNAP, and nucleoside triphosphates in vitro. After deproteinization, we electroporated the plasmid into ung-deficient E. coli cells which were plated both on counting plates (containing spectinomycin) and on selection plates (containing spectinomycin, kanamycin, and IPTG). The mutation frequency was then calculated as Kanr Specr colonies (selected)/Specr colonies (total transfected) (Fig. 1a).
FIG. 1.
In vitro transcription by E. coli RNAP allows AID access to plasmid DNA. (a) In vitro transcription/deamination assay. (b) Transcription limits deamination by AID. (c) Phosphorylation does not affect transcription-dependent deamination by AID with or without RPA in the reaction. PKA (1,000 U) was added as indicated to 2 nM plasmid, 20 nM AID, 40 nM RNAP, and approximately 40 nM RPA with 250 μM nucleoside triphosphates and incubated for 1 h at 37°C. The inset shows sodium dodecyl sulfate-polyacrylamide gel electrophoresis of parallel 10-μl reaction mixtures incubated with 10 nCi [γ-32P]ATP. Molecular sizes indicated were derived from coelectrophoresed standards. *, AID, 24 kDa.
Incubation in vitro of ptac-KanL94P with AID alone did not appreciably increase the frequency of mutation of the Kanr gene (Fig. 1b, left panel). The frequency increased slightly (from 10−5 to 4 × 10−5) with the addition of RNAP to the reaction mixture and reached in vivo levels (from 10−5 to 30 × 10−5) in the context of transcriptional elongation after the addition of ribonucleoside triphosphates. This is an underestimate of the actual frequency of AID-mediated mutations, as, in order to be selected, clones carrying deaminated plasmids must be able to grow in the presence of kanamycin. The mutation frequency was proportional to the amount of RNAP added (Fig. 1b, right panel).
RPA has recently been described to bind B-cell-derived, phosphorylated AID and to increase ung-dependent cleavage of a double-stranded DNA fragment transcribed by phage T7 polymerase (5, 12) (see Fig. S1b in the supplemental material). AID protein derived from cultured mammalian cells can also interact with RPA in this assay but only after phosphorylation by protein kinase A (PKA) (4). In our plasmid-based assay, in vitro PKA-phosphorylated, human recombinant AID expressed in bacteria did not deaminate transcribed plasmid DNA with higher efficiency when RPA was included in the reaction (Fig. 1c). Phosphorylation of AID was confirmed by γ-ATP labeling (Fig. 1c, inset). The recombinant human AID (expressed in E. coli) that we used in these experiments is active in an ung cleavage assay (see Fig. S1b in the supplemental material), and the tripartite RPA protein that we used is active in a number of different assays (39, 40). We conclude that, in our assay, the state of the DNA after transcription by RNAP (or T7 [see Fig. S1a in the supplemental material]) does not require RPA as a cofactor for efficient deamination by AID.
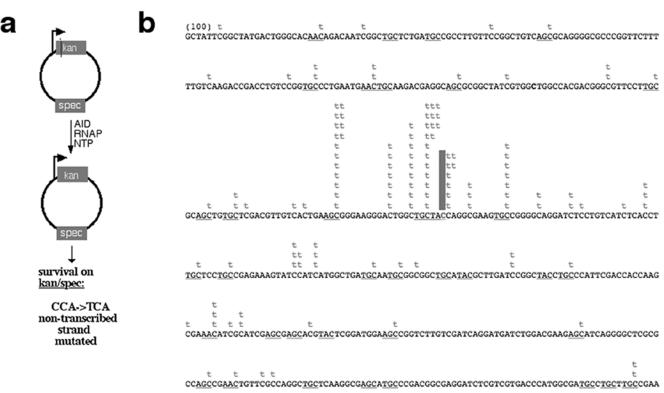
AID has been shown to preferentially deaminate specific sequences known as “hotspot” motifs (WRC motif, where W equals A or T and R equals A or G [33]) and to target only the NT strand in the context of transcription (8, 13, 31, 35). To determine the sequence and strand preferences of AID in our system, we cloned and sequenced the Kanr gene from kanamycin-resistant E. coli clones transformed with ptac-KanL94P plasmid previously treated with AID during transcription by E. coli RNAP. In contrast to previous reports which observed only the C-to-T mutation required for kanamycin resistance and survival (31, 35), 80% of our clones with the reversion mutation had additional mutations (these additional mutations are shown in Fig. 2). Furthermore, mutations exhibited preference for the WRC motif; hence, as noted in many other reports, the mutational bias for C in the context of a WRC motif may be an inherent feature of the AID molecule (28, 42).
FIG. 2.
AID deaminates only the NT strand of the ptac-KanL94P plasmid. (a) In vitro transcription/deamination assay. (b) Compilation of all mutations (from a total of 58 unique sequences). The cytidine residue marked with a solid vertical bar denotes the location of the reversion mutation (CCA to TCA) which was present in every clone. The WRC motif is underlined.
It is interesting to note, however, that the WRC motif found at Ig hotspots for the somatic hypermutation is not strand specific (21); therefore, in vivo, both strands of the V(D)J region should be deaminated equally by AID. In contrast, all ptac-KanL94P mutations were NT-strand C-to-T transitions (Fig. 2). This may indicate that the hypermutation process is initiated in a strand-biased manner and that AID-independent processes then spread the mutations to the T strand as previously hypothesized (31, 35). Alternatively, strand preference in the context of the ptac-KanL94P plasmid may simply reflect that mutation on the NT strand is required for drug selection.
AID can deaminate both DNA strands.
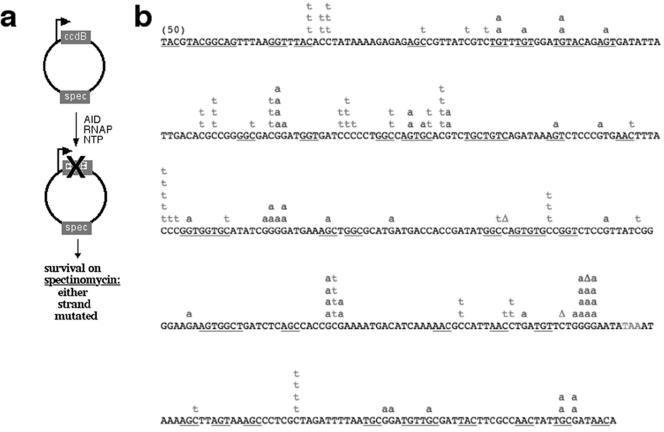
To explore the possibility that, on a different sequence, without selective pressure on specific residues, mutations are generated on either strand, we generated the ptac-CcdB plasmid by simply replacing the kanamycin resistance gene in ptac-KanL94P with ccdB and leaving the rest of the plasmid intact. The CcdB protein kills bacteria by poisoning topoisomerase II complexes (7), so loss-of-function mutants on either strand of the ccdB gene survive. This selection scheme allows for detection of any deleterious mutation that leads to loss of function and would therefore permit the observation of a broader spectrum of mutations than in the gain-of-function system requiring kanamycin resistance. Forty percent of the ptac-CcdB clones had only C-to-T transition mutations in the NT strand, whereas the rest had only G-to-A transitions, which reflect a C-to-T mutation on the T strand (Fig. 3).
FIG. 3.
AID deaminates either strand of the ptac-CcdB plasmid. (a) In vitro transcription/deamination assay. (b) Compilation of all unique mutations (from a total of 50 sequences). Triangles mark the positions of short deletions. The WRC motif is underlined.
Since the ptac-CcdB and ptac-KanL94P plasmid backbones are identical, bidirectional transcription from a cryptic promoter cannot account for the observation that both strands are targets for AID-mediated deamination in ptac-CcdB but only the top strand is targeted in ptac-KanL94P. Furthermore, RNase protection experiments do not reveal bidirectional transcription of this locus (not shown).
As with the ptac-KanL94P substrate, the majority of mutations we recovered were at a distance from one another, leaving a number of intervening CG base pairs untouched (see Fig. S2b in the supplemental material, aligned raw sequence data). In contrast to the ptac-KanL94P plasmid, ptac-CcdB mutations did not appear to exhibit preference for the WRC motif as defined by Rogozin and Diaz (33). This can be explained by the inability of the ptac-CcdB-carrying bacteria to tolerate silent mutations on the ccdB gene (such mutations would allow toxin production and would be lethal). This, combined with the fact that AID-targeted deamination of 28% of WRC hotspots found in the ccdB coding sequence would resolve into silent mutations (see Fig. S2c in the supplemental material), results in the perceived absence of hotspot targeting (Fig. 3).
The distance between elongation complex and promoter determines the efficiency of AID-mediated deamination.
It is well known that mutability in the Ig locus decreases as the distance from the promoter increases, so that mutations peak between 150 nucleotides (nt) and 450 nt from the promoter and decline exponentially thereafter (30). It has been proposed that lack of mutation in the first 100 to 150 nt may be related to nucleosome positioning in vivo; alternatively, it may reflect an inherent feature of transcriptional elongation (19).
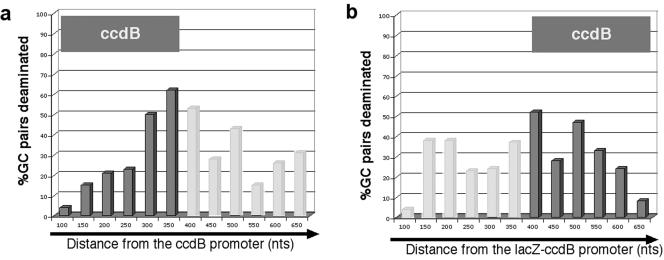
To analyze whether ptac-CcdB mutations followed this pattern, we catalogued mutated CG pairs in each clone per 50-nt interval so that the position of each mutated pair is represented in the catalog only once. We then compared this catalog to the total number of GC pairs found in each 50-nt interval (compiled sequence data are shown in Fig. 3) and plotted the percentage of mutated pairs as a function of distance from the promoter. Since the ccdB gene is only 350 nt long (its coding sequence is represented in Fig. 4), mutations extend into the plasmid sequence past it. Surprisingly, we found that ptac-CcdB mutations in vitro began ∼80 nt from the transcription start and occurred at various distances from the ptac promoter but peaked from 300 nt to 500 nt downstream from the transcription start site (Fig. 4a). A very similar distribution of mutations (not shown) with respect to distance from the promoter was also recovered from the kanamycin revertants (this is readily apparent even in the uncatalogued deamination sites presented in aggregate in Fig. 2). Therefore, for two different substrates, mutations decline with distance from the promoter, strongly arguing that the distribution of mutations is not a feature of a particular sequence (for example, it cannot reflect the distribution of pause/arrest sites, since it is unlikely that the ccdB and Kanr genes, whose sequences are quite different, would happen to share this highly sequence-specific feature).
FIG. 4.
AID deamination efficiency decreases as the elongation complex travels away from the promoter. (a) Histogram showing the percentage of deaminated GC pairs in 50-nt intervals from the transcription start of the ccdB gene, with the area spanning the coding region for ccdB depicted by dark gray bars (data from 50 unique sequences). (b) Histogram showing the percentage of deaminated GC pairs in 50-nt intervals from the transcription start of the lacZ-ccdB fusion protein. The coding region of the ccdB gene is depicted by dark gray bars (data from 31 unique sequences).
This observation predicts that the addition of irrelevant “filler” sequence between the promoter and the target gene would not alter the distribution of mutations with respect to their distance from the promoter; therefore, the target gene itself would be less mutated (3). To test this prediction, we used a vector encoding a lacZ-ccdB fusion protein (pZero), in which the ccdB sequence begins 350 nt from the promoter. Transcription of this new plasmid in the presence of AID led to similar numbers of recovered mutants, because mutations in the lacZ sequence itself lead to inactivation of the entire lacZ-ccdB fusion protein and allow survival. When we sequenced the mutants, we again found that mutations initiated ∼100 nt from the transcription start and declined 600 nt from the promoter (Fig. 4b). However, over half of the mutations were focused on the lacZ sequence and mutations were already at their peak even at the start of the ccdB coding sequence (Fig. 4b). Thus, by introducing the lacZ “filler” sequence in front of the ccdB coding sequence we shifted the mutation spectrum on the ccdB sequence (compare Fig. 4a and b).
Our assay recapitulates the promoter-dependent distribution exhibited by in vivo mutation in the Ig locus. Hence, the initiation and decay of AID-mediated deamination events are mechanistically linked to transcription. We hypothesize that the initiation of mutation events reflects the transition from the RNAP initiation complex to the fully processive, elongation-competent form of RNAP. This transition is thought to happen ∼70 to 100 nt from the promoter (38), creating a 5′ mutational boundary.
Modes of AID binding to the transcription elongation complex.
Within transcribed DNA, the first 100 transcribed base pairs are essentially mutation free, both in vivo (19) and in vitro (Fig. 4). Since transcription becomes processive only after the polymerase clears the first 70 bp, this lack of access of AID to the 5′ end of the transcribed DNA may reflect a specific interaction between AID and DNA within the TEC.
To assess whether single-stranded DNA exposed by the TEC could be a target for AID, as previously hypothesized (31), we assembled an immobilized TEC in vitro. Because AID does not bind double-stranded DNA (15) but can bind and deaminate any single-stranded DNA, we took care that the assembled TEC was free of single-stranded nucleic acid (by assembling it with hexahistidine-tagged RNAP and purifying it on nickel-agarose beads [see Materials and Methods]). We then confirmed that the assembled TEC was active and able to add templated rNTPs to the nascent RNA molecule (not shown). Moreover, we could readily cross-link AID to the in vitro-assembled TEC (Fig. 5a).
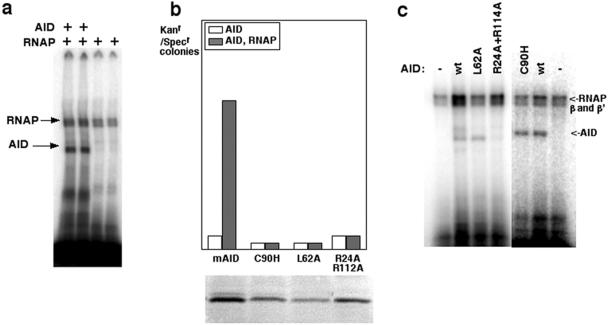
FIG. 5.
AID binds DNA in an RNAP transcription complex. (a) UV cross-linking of AID to a transcription complex purified on nickel-agarose. The RNAP-DNA-RNA complex was assembled as described in Materials and Methods, with the NT strand labeled with 32P. (b) Specific mutations modulate AID activity in vitro. Equal volumes of AID, truncated AID, and single amino acid mutants were assayed with 20 nM RNAP. The mutation frequency was normalized to AID protein concentration as determined from Coomassie-stained protein gels (shown below results for each assay). Mutated amino acids are indicated below the corresponding reactions. mAID, wild-type (wt) mouse AID. (c) Effect of inactivating mutations on binding to the transcription complex.
We then wondered whether we could identify mutants of AID that would disrupt the AID-TEC interaction and would therefore result in loss of AID activity. Loss of AID activity in humans (32) can be due to three types of mutations in the AID molecule: mutations which affect the structural stability of the protein, mutations which directly interfere with catalysis (such as mutations in residues H56, E58, C88, C90, and P87), and, finally, mutations which interfere with substrate recognition.
We concentrated on mutations which did not affect the biophysical behavior of the protein during purification, suggesting that structural stability was not compromised (Fig. 5b, bottom). Specifically, we concentrated on three different mutants: AIDC90H, AIDL62A, and the double mutant AIDR24A R112A. AIDC90H is a catalytic mutant which, based on the structure of metabolic deaminases, is predicted to have at least a 100-fold-lower activity than that of the wild type and which had no activity in our assay (Fig. 5b). The mutation in AIDL62A is proximal to residues H56 and E58, which are mechanistically involved in catalysis. L62 is not a conserved catalytic residue, but the insertion of alanine in that position may destabilize the architecture of the active site: AIDL62A was inactive in our assay (Fig. 5b). Finally, mutations in AIDR24A R112A do not destabilize the protein, and residues R24 and R112 are not part of the known deaminase active site; yet, AIDR24A R112A was unable to catalyze deamination in vitro (Fig. 5b).
To determine whether specific residues could be involved in binding the TEC, we assembled the elongation complex and UV cross-linked the mutant proteins to the assembled TEC. Although catalytically inactive, AIDC90H could readily be cross-linked to the TEC (Fig. 5c). AIDL62A was also cross-linked to the TEC and was hence not deficient in binding. Therefore, although residue L62 is not mechanistically involved with catalysis, its proximity to the active site, as well as the inability of AIDL62A to catalyze deamination in our assay, suggests that this residue is important for the structural stability of the catalytic pocket. Remarkably, AIDR24A R112A was unable to bind the assembled TEC (Fig. 5c) although its protein concentration was comparable to that of the wild type (Fig. 5b, bottom). Since residues R24 and R112 are at a distance from the active site, they should not affect the architecture of the binding pocket. Rather, our data imply that their lack of activity in our assay is due to their inability to bind the TEC. Therefore, our experiments identify AID residues distinct from the active site of the deaminase that promote its interaction with the TEC.
DISCUSSION
Multiple lines of work have established a direct correlation between transcription and somatic hypermutation in vivo. Promoter exchange (17) and promoter duplication (26) experiments have shown that a promoter is absolutely required for hypermutation in vivo. Furthermore, experiments with artificial constructs in transfected cell lines established a direct correlation between the rate of transcription and the frequency of mutation (2, 41). Analyses of the patterns of mutations recovered from Ig genes have suggested a close linkage between hypermutation and the transcription machinery: mutations increased sharply ∼150 nt from the promoter (18, 30) but then decayed exponentially past 400 to 450 nt from the promoter (30). Taken together, these studies have led to a model of hypermutation (37) in which a mutator factor is loaded onto the RNAP (polymerase II [Pol II]) initiation complex and is then carried along the gene by the polymerase during transcription.
Recent genetic and biochemical evidence suggests that AID is the postulated mutator (11). Multiple laboratories have shown that AID deaminates transcribed genes in E. coli (31, 35) and genes transcribed by phage polymerases in vitro (8, 12). These experiments recapitulated some of the features of hypermutation: mutations were C-to-T transitions and preferentially occurred at recognized hotspot motifs (WRC). However, data from these experiments were at odds with many in vivo characteristics of the reaction.
We present here an in vitro assay in which AID mediates deamination in the context of transcription but without marked preference for either strand. Previous assays have used transcription of a Kanr gene in E. coli to show AID targeting the NT strand. When we replicated this gain-of-function assay in vitro, we found deamination exclusively on the NT strand (Fig. 2). We then generated a loss-of-function system and repeated the experiment. Strikingly, deamination by AID, although still transcription dependent, then targeted either the NT strand, creating C-to-T mutations, or the T strand, creating G-to-A mutations on the NT strand (Fig. 3). In our assay, AID did not function processively: the majority of mutations we recovered left a number of intervening CG base pairs untouched (see Fig. S2 in the supplemental material, aligned raw sequence data). Our data provide direct evidence for the ability of AID to target either DNA strand during transcription by RNA polymerase.
One of the intriguing features of in vivo hypermutation is that mutability decreases as the distance from the promoter increases, so that mutations in the Ig locus peak between 150 nt and 450 nt from the promoter and decline precipitously thereafter (30). It has been proposed that the lack of mutation in the first 100 to 150 nt could be related to nucleosome positioning in vivo (19). Alternatively, it could reflect an inherent feature of transcriptional elongation. The latter possibility is strongly supported by our mutational analysis of clones transcribed and deaminated in vitro. Essentially, in our assay we did not recover mutations until 80 nt past the transcription start. In addition, when we looked at all mutated clones in aggregate (Fig. 4) we found that on either strand mutations peaked 200 to 500 nt from the transcription start and decayed rapidly thereafter. Since our in vitro assay recapitulates this in vivo feature of Ig mutation but contains only RNAP, rNTPs, and AID, the distribution of AID-mediated deamination events must be intimately and mechanistically linked to transcription.
The failure of T7 transcription systems to recapitulate this feature of the reaction (see Fig. S3 in the supplemental material) (34) is most likely due to the fact that phage polymerases are significantly different from mammalian RNA Pol II. Phage polymerases are single-subunit enzymes, more similar to eukaryotic DNA polymerases both in their protein structure and in the structure of their transcription bubble (36) as well as in enzyme kinetics, transcribing at a rate of 200 to 400 nt/s (38). In contrast, E. coli RNAP is homologous to core RNA Pol II from eukaryotes (22), generates similar transcription bubbles, and polymerizes RNA at similar rates (15 to 20 nt/s) (38). Our data suggest that the RNAP-based transcription assay faithfully recapitulates features of Ig hypermutation that the phage-based assays cannot.
We hypothesize that concentration of mutation events reflects the transition from the RNAP initiation complex to the fully processive, elongation-competent form of RNAP. This transition involves not only the breaking of initial ties with the promoter but also the shedding of accessory initiation factors (such as σ70 in the case of E. coli RNAP or TATA-binding protein in the case of mammalian Pol II). Conversion of RNAP or Pol II to an elongation-competent form happens ∼70 to 100 nt from the promoter, depending on the gene studied (38); this may set the 5′ boundary of mutation. Assuming the mutator can load only onto the RNAP initiation complex at the promoter but can dissociate as the complex transcribes would sufficiently explain the rapid decline of mutation frequency 400 to 450 nt from the transcriptional start site.
The strong correlation between transcription and AID-mediated deamination prompted us to ask whether AID could directly interact with the transcription complex. We therefore performed cross-linking studies between in vitro-assembled RNAP-DNA complexes and AID. We discovered that AID could be directly cross-linked to DNA in the TEC (Fig. 5a) and that the stability of this interaction required residues R24 and R112, which are distinct from the catalytic core (Fig. 5c). Finally, our data suggest that AID and transcription are both necessary and together are sufficient for hypermutation both in vivo and in vitro. However, not all transcribed genes are hypermutated in vivo, and so the mechanism by which AID is targeted nearly exclusively to the Ig loci remains to be elucidated.
Supplementary Material
Acknowledgments
We thank Mark Chlenov and Seth Darst for advice and reagents, Marc Wold for human RPA, Haijuan Yang and Nikola Pavletich for their very generous gifts of human RPA and U. maydis RPA, and Paul Hakimpour for help with bioinformatics.
This work has been partially supported by grants from the Keck Foundation, the Searle Trust, and the Irene Diamond Professorships Fund.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Artsimovitch, I., and R. Landick. 2002. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell 109:193-203. [DOI] [PubMed] [Google Scholar]
- 2.Bachl, J., and C. Olsson. 1999. Hypermutation targets a green fluorescent protein-encoding transgene in the presence of immunoglobulin enhancers. Eur. J. Immunol. 29:1383-1389. [DOI] [PubMed] [Google Scholar]
- 3.Bachl, J., C. Steinberg, and M. Wabl. 1997. Critical test of hot spot motifs for immunoglobulin hypermutation. Eur. J. Immunol. 27:3398-3403. [DOI] [PubMed] [Google Scholar]
- 4.Basu, U., J. Chaudhuri, C. Alpert, S. Dutt, S. Ranganath, G. Li, J. P. Schrum, J. P. Manis, and F. W. Alt. 2005. The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature 438:508-511. [DOI] [PubMed] [Google Scholar]
- 5.Basu, U., J. Chaudhuri, and F. W. Alt. 2005. B cell development, function and disease. Presented at the Keystone Meeting, Steamboat Springs, Colo.
- 6.Beale, R. C., S. K. Petersen-Mahrt, I. N. Watt, R. S. Harris, C. Rada, and M. S. Neuberger. 2004. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J. Mol. Biol. 337:585-596. [DOI] [PubMed] [Google Scholar]
- 7.Bernard, P., P. Gabant, E. M. Bahassi, and M. Couturier. 1994. Positive-selection vectors using the F plasmid ccdB killer gene. Gene 148:71-74. [DOI] [PubMed] [Google Scholar]
- 8.Bransteitter, R., P. Pham, P. Calabrese, and M. F. Goodman. 2004. Biochemical analysis of hypermutational targeting by wild type and mutant activation-induced cytidine deaminase. J. Biol. Chem. 279:51612-51621. [DOI] [PubMed] [Google Scholar]
- 9.Bransteitter, R., P. Pham, M. D. Scharff, and M. F. Goodman. 2003. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl. Acad. Sci. USA 100:4102-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bross, L., and H. Jacobs. 2003. DNA double strand breaks occur independent of AID in hypermutating Ig genes. Clin. Dev. Immunol. 10:83-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaudhuri, J., and F. W. Alt. 2004. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat. Rev. Immunol. 4:541-552. [DOI] [PubMed] [Google Scholar]
- 12.Chaudhuri, J., C. Khuong, and F. W. Alt. 2004. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature 430:992-998. [DOI] [PubMed] [Google Scholar]
- 13.Chaudhuri, J., M. Tian, C. Khuong, K. Chua, E. Pinaud, and F. W. Alt. 2003. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature 422:726-730. [DOI] [PubMed] [Google Scholar]
- 14.Conticello, S. G., C. J. Thomas, S. K. Petersen-Mahrt, and M. S. Neuberger. 2005. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol. Biol. Evol. 22:367-377. [DOI] [PubMed] [Google Scholar]
- 15.Dickerson, S. K., E. Market, E. Besmer, and F. N. Papavasiliou. 2003. AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med. 197:1291-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Noia, J. M., and M. S. Neuberger. 2004. Immunoglobulin gene conversion in chicken DT40 cells largely proceeds through an abasic site intermediate generated by excision of the uracil produced by AID-mediated deoxycytidine deamination. Eur. J. Immunol. 34:504-508. [DOI] [PubMed] [Google Scholar]
- 17.Fukita, Y., H. Jacobs, and K. Rajewsky. 1998. Somatic hypermutation in the heavy chain locus correlates with transcription. Immunity 9:105-114. [DOI] [PubMed] [Google Scholar]
- 18.Lebecque, S. G., and P. J. Gearhart. 1990. Boundaries of somatic mutation in rearranged immunoglobulin genes: 5′ boundary is near the promoter, and 3′ boundary is approximately 1 kb from V(D)J gene. J. Exp. Med. 172:1717-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Longerich, S., A. Tanaka, G. Bozek, D. Nicolae, and U. Storb. 2005. The very 5′ end and the constant region of Ig genes are spared from somatic mutation because AID does not access these regions. J. Exp. Med. 202:1443-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin, A., and M. D. Scharff. 2002. AID and mismatch repair in antibody diversification. Nat. Rev. Immunol. 2:605-614. [DOI] [PubMed] [Google Scholar]
- 21.Milstein, C., M. S. Neuberger, and R. Staden. 1998. Both DNA strands of antibody genes are hypermutation targets. Proc. Natl. Acad. Sci. USA 95:8791-8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murakami, K. S., and S. A. Darst. 2003. Bacterial RNA polymerases: the wholo story. Curr. Opin. Struct. Biol. 13:31-39. [DOI] [PubMed] [Google Scholar]
- 23.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102:553-563. [DOI] [PubMed] [Google Scholar]
- 24.Muramatsu, M., V. S. Sankaranand, S. Anant, M. Sugai, K. Kinoshita, N. O. Davidson, and T. Honjo. 1999. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 274:18470-18476. [DOI] [PubMed] [Google Scholar]
- 25.Papavasiliou, F. N., and D. G. Schatz. 2002. The activation-induced deaminase functions in a postcleavage step of the somatic hypermutation process. J. Exp. Med. 195:1193-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters, A., and U. Storb. 1996. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity 4:57-65. [DOI] [PubMed] [Google Scholar]
- 27.Petersen-Mahrt, S. K., R. S. Harris, and M. S. Neuberger. 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature 418:99-103. [DOI] [PubMed] [Google Scholar]
- 28.Pham, P., R. Bransteitter, J. Petruska, and M. F. Goodman. 2003. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature 424:103-107. [DOI] [PubMed] [Google Scholar]
- 29.Rada, C., J. M. Di Noia, and M. S. Neuberger. 2004. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell 16:163-171. [DOI] [PubMed] [Google Scholar]
- 30.Rada, C., and C. Milstein. 2001. The intrinsic hypermutability of antibody heavy and light chain genes decays exponentially. EMBO J. 20:4570-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramiro, A. R., P. Stavropoulos, M. Jankovic, and M. C. Nussenzweig. 2003. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat. Immunol. 4:452-456. [DOI] [PubMed] [Google Scholar]
- 32.Revy, P., T. Muto, Y. Levy, F. Geissmann, A. Plebani, O. Sanal, N. Catalan, M. Forveille, R. Dufourcq-Labelouse, A. Gennery, I. Tezcan, F. Ersoy, H. Kayserili, A. G. Ugazio, N. Brousse, M. Muramatsu, L. D. Notarangelo, K. Kinoshita, T. Honjo, A. Fischer, and A. Durandy. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the hyper-IgM syndrome (HIGM2). Cell 102:565-575. [DOI] [PubMed] [Google Scholar]
- 33.Rogozin, I. B., and M. Diaz. 2004. Cutting edge: DGYW/WRCH is a better predictor of mutability at G:C bases in Ig hypermutation than the widely accepted RGYW/WRCY motif and probably reflects a two-step activation-induced cytidine deaminase-triggered process. J. Immunol. 172:3382-3384. [DOI] [PubMed] [Google Scholar]
- 34.Shen, H. M., S. Ratnam, and U. Storb. 2005. Targeting of the activation-induced cytosine deaminase is strongly influenced by the sequence and structure of the targeted DNA. Mol. Cell. Biol. 25:10815-10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sohail, A., J. Klapacz, M. Samaranayake, A. Ullah, and A. S. Bhagwat. 2003. Human activation-induced cytidine deaminase causes transcription-dependent, strand-biased C to U deaminations. Nucleic Acids Res. 31:2990-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steitz, T. A. 2004. The structural basis of the transition from initiation to elongation phases of transcription, as well as translocation and strand separation, by T7 RNA polymerase. Curr. Opin. Struct. Biol. 14:4-9. [DOI] [PubMed] [Google Scholar]
- 37.Storb, U., H. M. Shen, N. Michael, and N. Kim. 2001. Somatic hypermutation of immunoglobulin and non-immunoglobulin genes. Philos. Trans. R. Soc. Lond. B 356:13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uptain, S. M., C. M. Kane, and M. J. Chamberlin. 1997. Basic mechanisms of transcript elongation and its regulation. Annu. Rev. Biochem. 66:117-172. [DOI] [PubMed] [Google Scholar]
- 39.Yang, H., P. D. Jeffrey, J. Miller, E. Kinnucan, Y. Sun, N. H. Thoma, N. Zheng, P. L. Chen, W. H. Lee, and N. P. Pavletich. 2002. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 297:1837-1848. [DOI] [PubMed] [Google Scholar]
- 40.Yang, H., Q. Li, J. Fan, W. K. Holloman, and N. P. Pavletich. 2005. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature 433:653-657. [DOI] [PubMed] [Google Scholar]
- 41.Yoshikawa, K., I. M. Okazaki, T. Eto, K. Kinoshita, M. Muramatsu, H. Nagaoka, and T. Honjo. 2002. AID enzyme-induced hypermutation in an actively transcribed gene in fibroblasts. Science 296:2033-2036. [DOI] [PubMed] [Google Scholar]
- 42.Yu, K., F. T. Huang, and M. R. Lieber. 2004. DNA substrate length and surrounding sequence affect the activation-induced deaminase activity at cytidine. J. Biol. Chem. 279:6496-6500. [DOI] [PubMed] [Google Scholar]
- 43.Zan, H., X. Wu, A. Komori, W. K. Holloman, and P. Casali. 2003. AID-dependent generation of resected double-strand DNA breaks and recruitment of Rad52/Rad51 in somatic hypermutation. Immunity 18:727-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.