Abstract
Gli3 is a zinc finger transcription factor proteolytically processed into a truncated repressor lacking C-terminal activation domains. Gli3 processing is stimulated by protein kinase A (PKA) and inhibited by Hedgehog signaling, a major signaling pathway in vertebrate development and disease. We show here that multisite glycogen synthase kinase 3β (GSK3β) phosphorylation and ubiquitination by SCFβTrCP are required for Gli3 processing. We identified multiple βTrCP-binding sites related to the DSGX2-4S motif in Gli3, which are intertwined with PKA and GSK3β sites, and SCFβTrCP target lysines that are essential for processing. Our results support a simple model whereby PKA triggers a cascade of Gli3 phosphorylation by GSK3β and CK1 that leads to direct βTrCP binding and ubiquitination by SCFβTrCP. Binding of βTrCP to Gli3 N- and C-terminal domains lacking DSGX2-4S-related motifs was also observed, which could reflect indirect interaction via other components of Hedgehog signaling, such as the tumor suppressor Sufu. Gli3 therefore joins a small set of transcription factors whose processing is regulated by the ubiquitin-proteasome pathway. Our study sheds light on the role of PKA phosphorylation in Gli3 processing and will help to analyze how dose-dependent tuning of Gli3 processing is achieved by Hedgehog signaling.
Hedgehog signaling is a major signaling pathway in animal development whose dysregulation is involved in many diseases in humans, including malformation syndromes and several types of cancers (23). In vertebrates, the transcriptional response to Hedgehog factors is mediated by Gli1, Gli2, and Gli3 zinc finger proteins. At the molecular level, Gli3 is translated into a 190-kDa transcriptional activator (Gli3-190) that undergoes proteolytic processing into a truncated 83-kDa repressor (Gli3-83) lacking C-terminal activation domains (7, 34). Hedgehog signaling inhibits Gli3 processing and stimulates transactivation by the resulting full-length protein. Genetic analyses in the mouse support the view that Gli3 acts as a transcription switch in signaling by Sonic Hedgehog (Shh). Alleviating repression by Gli3 is a major step of Shh signaling, and Gli3 activator function is also required for Shh patterning in the neural tube (2). During limb bud development, Gli3-83 exhibits an anteroposterior gradient inversely proportional to Shh levels, which suggests that regulation of Gli3 processing into Gli3-83 is a direct readout of dose-dependent signaling by Shh (34). The importance of fine control of Gli3-83 levels in the limb bud is suggested by molecular findings in patients with Pallister Hall syndrome; stop mutations in Gli3 causing heterozygous expression of constitutively truncated Gli3 and polydactyly (31).
Gli3 processing is known to be stimulated by protein kinase A (PKA) phosphorylation (34), but further molecular events are not characterized and the level at which regulation by Hedgehog proteins operates is unknown. In Drosophila melanogaster, proteolytic processing of Ci, the transcription factor homologous to Gli proteins, is known to require multisite phosphorylation by PKA (5) as well as by glycogen synthase kinase 3β (GSK3β) and CK1, the latter kinases being primed by PKA phosphorylation (16, 29). As described for Ci, there are GSK3β and CK1 sites adjacent to PKA sites in Gli3 (29), but their role has not been tested yet. Importantly, Jiang and Struhl showed that Ci processing is abolished in slimb mutant cells and therefore proposed that processing could result from Ci ubiquitination by the SCFSlimb ubiquitin ligase and subsequent partial degradation by the proteasome (18). SCF ubiquitin ligase complexes contain an F-box protein, such as Slimb, which determines substrate specificity and general components Skp1, Cullin1, and Rbx1, which are needed for ubiquitination per se (11). In agreement with this model, Cullin1 and Rbx1 were recently shown to be required for Ci processing (25, 27). A direct implication of SCFSlimb in Ci processing has, however, remained elusive (22), and the genetic evidence in flies could also be compatible with indirect regulation of Ci processing, with SCFSlimb regulating the stability of another key protein in the pathway as suggested by Chen et al. (4).
In vertebrates, substrates of the Slimb homologue βTrCP contain a consensus DSGX2-4S motif whose phosphorylation is required for βTrCP binding (11). The strict requirement for serine phosphorylation in the DSGX2-4S motif was demonstrated in vitro, using phosphorylated versus nonphosphorylated substrates such as β-catenin (15, 36). Its molecular basis was revealed by solving the structure of a βTrCP-β-catenin complex (37).
In this report, we examine the molecular mechanisms of Gli3 processing stimulated by PKA in a cell culture model. As previously found for Ci in flies, we show that GSK3β and the ubiquitin ligase component βTrCP are required for Gli3 processing. Our results further demonstrate a direct role of SCFβTrCP in Gli3 processing, and we discuss its potential regulation in the context of Hedgehog signaling.
MATERIALS AND METHODS
Plasmids.
Human Gli3 cDNA was cloned into p3×Flag plasmid (Sigma). For transcription shutoff experiments, 3×Flag-Gli3 cDNA was inserted into the pBI-G vector (Clontech). Tet-Off plasmid was from Clontech. Gli3 discrete mutants were obtained in p3×Flag using a QuikChange mutagenesis kit (Stratagene) and were checked by automated sequencing. Other constructs were obtained by standard DNA manipulations.
Cell culture and transfection.
Cell lines were grown in Dulbecco's modified Eagle medium with 10% fetal calf serum. For analysis of cell extracts by immunoblotting, cells were plated in 12-well plates and transfected with a total of 500 ng of plasmid using Lipofectamine 2000 (Invitrogen). Fifty nanograms of Flag-Gli3 expression vector was transfected in each well and, when indicated, 50 ng PKA, 50 ng GSK3β, 50 ng GSK3βR85, or 250 ng hemagglutinin-βTrCP (HA-βTrCP) expression vector was cotransfected. Empty expression vector plasmid was added to complete to a total of 500 ng. When mentioned, cells were treated with the following: 20 μM MG132, 20 mM LiCl, 50 μM forskolin, or 100 ng/ml doxycycline. Cells were harvested 24 h after transfection in phosphate-buffered saline (PBS) and resuspended in lysis buffer (50 mM Tris, pH 8, 150 mM NaCl, 1% NP-40, 1 mM EDTA, and protease inhibitor cocktail [Roche]). Whole-cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting and ECL detection (Amersham). Signals corresponding to truncated and full-length Flag-Gli3 were quantified by Chemigenius2 (Syngene). Relative levels of truncated Gli3-83 were expressed as the percentage of total Gli3 detected. Experiments were repeated at least twice with similar results, and representative experiments including quantification are shown.
Silencing by small interfering RNA.
HEK-293T or HeLa human cells seeded onto 12-well plates at 40 to 50% confluence were transfected with 10 nM small interfering RNA (siRNA) duplexes using Lipofectamine 2000. After 24 h, cells were subjected to a second transfection with a mixture of siRNA and plasmids using Lipofectamine 2000. The siRNA duplexes used for βTrCP silencing targets both βTrCP and the highly homologous βTrCP2 mRNAs (13). siRNA knock-down studies in human cells and knockout of the βTrCP gene in mouse embryonic stem cells have shown that βTrCP and βTrCP2 exert redundant activity towards substrates such as β-catenin and IκB (13). Controls were 21-nucleotide duplexes targeting luciferase.
Immunoprecipitations.
For interaction between exogenously expressed proteins, NIH 3T3 cells were transfected with 3 μg plasmid DNA/well using Lipofectamine 2000 in 6-well plates. For interaction between exogenous βTrCP and endogenous Gli3, C3H-10T1/2 cells were used. At 24 h after transfection, cells were harvested in PBS and resuspended in lysis buffer. Protein extracts were subjected to 1 h of incubation at 4°C with protein G-agarose beads (Roche) for preclearing, 2 h at 4°C with either anti-Flag or anti-HA antibodies, and 1 h at 4°C with protein G-agarose beads for immunoprecipitation. Extracts were washed three times with lysis buffer and twice with lysis buffer supplemented with 0.3 M NaCl and eluted in 30 μl Laemmli buffer. Immunoprecipitated proteins were separated by SDS-PAGE and revealed by Western blotting using horseradish peroxidase-coupled anti-Flag or anti-HA antibodies.
In vivo ubiquitination assay.
NIH 3T3 cells were transfected in 6-well plates with 250 ng Flag-Gli3 (or mutants thereof), 1 μg HA-ubiquitin, 125 ng PKA, 125 ng GSK3β expression vectors, and, when indicated, 1 μg myc-βTrCP or control expression vectors. At 24 h after transfection, cells were harvested in PBS, lysed at 95°C for 10 min in 100 μl lysis buffer supplemented with 5% SDS, 10 mM N-ethylmaleimide, and diluted in 500 μl cell lysis buffer supplemented with 10 mM N-ethylmaleimide. Protein extracts were subjected to anti-Flag immunoprecipitation, washed four times with lysis buffer supplemented with 1% SDS, and eluted in Laemmli buffer. Immunoprecipitated proteins were separated by SDS-8% PAGE and revealed by Western blotting using horseradish peroxidase-coupled anti-Flag or anti-HA antibodies. A fraction of eluate (1/20) was used to detect flag-Gli3 and derivatives, and the remainder was used to detect HA-ubiquitin.
Materials.
Antibodies and their manufacturers were the following: anti-FLAG (M2; Sigma), anti-HA (3F10; Roche), anti-Gli3 (N-19; Santa Cruz), and anti-FGFR4 (Santa Cruz). Synthetic siRNAs were from Eurogentech (Belgium). Doxycycline, forskolin, and MG132 were from Sigma.
RESULTS
GSK3β phosphorylation is required for Gli3 proteolytic processing.
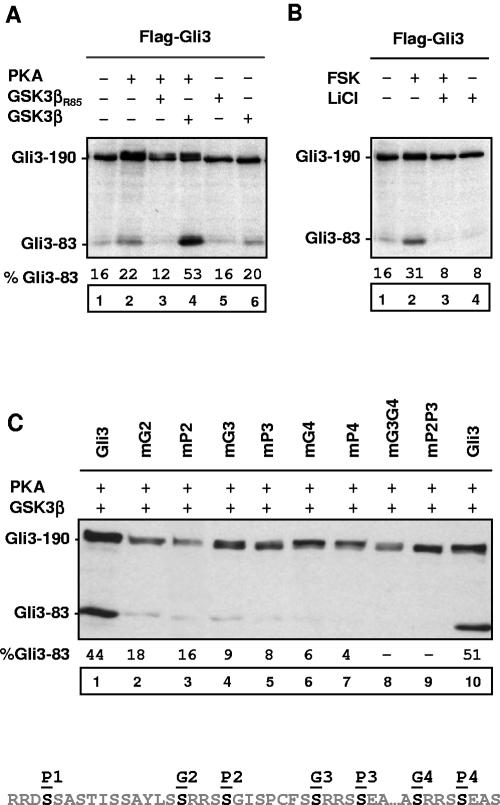
In order to study the role of GSK3β in Gli3 proteolytic processing, we performed transient transfection of a human Gli3 expression vector into NIH 3T3 cells and manipulated GSK3β activity. As previously described (7, 34), exogenous Gli3 was inefficiently processed, and stimulation of PKA by expression of the catalytic subunit of PKA or by treatment of cells with forskolin was necessary to detect significant levels of truncated Gli3 (Fig. 1A and B, lanes 1 and 2). We found that coexpression of GSK3β together with PKA allowed us to reach the highest levels of truncated Gli3, corresponding to 53% total Gli3 (Fig. 1A, lane 4). Conversely, inhibition of GSK3β by coexpression of a dominant-negative mutant of GSK3β (GSK3βR85) (28) inhibited the positive effect of PKA overexpression (Fig. 1A, lane 3), and treatment of cells with LiCl, a pharmacological inhibitor of GSK3β, decreased the effect of PKA stimulation by forskolin (Fig. 1B, lane 3). We next constructed a series of mutants in the Gli3 GSK3β sites and tested their effect on the stimulation of Gli3 processing by GSK3β. PKA sites in Gli3 are numbered from P1 to P4 depending on their relative position in Gli3, and putative GSK3β sites previously located adjacent to sites P2 to P4 by Price and Kalderon (29) were numbered G2 to G4, respectively. Mutations of single GSK3β sites to alanine impaired Gli3 processing as strongly as mutations of the corresponding PKA sites (Fig. 1C, lanes 1 to 7). Furthermore, as observed for PKA sites, simultaneous mutation of multiple GSK3β sites drastically reduced Gli3 processing (Fig. 1C, lanes 8 and 9). Taken together, these findings strongly suggest that regulation of Gli3 processing by direct GSK3β phosphorylation is conserved between D. melanogaster Ci and Gli3. Importantly, these findings provided us with an experimental model in which to analyze molecular events taking place after PKA and GSK3β phosphorylation in Gli3 processing.
FIG. 1.
Stimulation of truncated Gli3 synthesis by PKA and GSK3β. (A) Stimulation of truncated Gli3 synthesis by PKA and GSK3β stimulation. NIH 3T3 cells were transfected with expression vectors for Flag epitope-tagged Gli3 (Flag-Gli3), human PKA catalytic subunit (PKA), human wild-type GSK3β, or human dominant-negative GSK3β (GSK3βR85), and cell extracts were analyzed by immunoblotting with an anti-Flag antibody. (B) Inhibition of endogenous GSK3β by LiCl inhibits forskolin-induced synthesis of truncated Gli3. NIH 3T3 cells were transfected with expression vectors for Flag-Gli3. Where indicated, cells were treated either with 50 μM forskolin (FSK) alone or with 50 μM FSK and 20 mM LiCl for 12 h. Identical quantities of FSK vehicle (ethanol) were added to control cells. (C) Mutation of GSK3β sites adjacent to PKA sites inhibits synthesis of truncated Gli3. GSK3β phosphorylates serine or threonine residues that lie four residues N terminal to a phosphoserine. GSK3β sites SXXXPS, labeled G2 to G4, were found adjacent to PKA sites P2 to P4, respectively (29). Serine-to-alanine mutants at indicated sites of Flag-Gli3 were transfected into NIH 3T3 cells together with PKA and GSK3β expression vectors, and cell extracts were analyzed by immunoblotting with anti-Flag antibody. Autoradiograms were scanned to measure the signals corresponding to truncated and total Gli3 signals. The relative levels of truncated Gli3 are given as percentages of total (truncated + full length) Gli3 signal measured in each condition. A minus sign indicates lanes where truncated products were not quantified. Upon long exposure of blots, low-level processing could be detected and was roughly estimated to be inferior to 3% total Gli3. mG2, mP2, and mP2P3 indicate mutant Flag-Gli3 at site G2, site P2, and both sites P2 and P3, respectively.
βTrCP is required for Gli3 proteolytic processing.
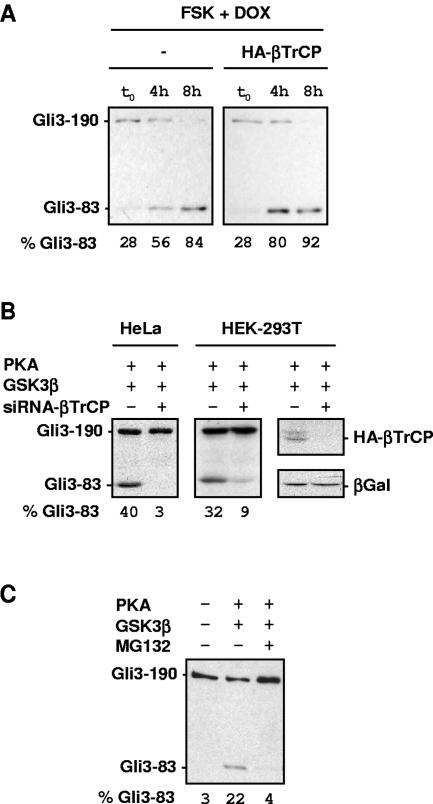
We chose to test the role of βTrCP in Gli3 processing stimulated by PKA and GSK3β. First we modulated SCFβTrCP activity by transfecting an expression vector for wild-type βTrCP (15). In order to follow Gli3 processing, we performed transcription shutoff experiments using a tetracycline-dependent Flag-Gli3 expression vector. Following addition of the tetracycline analog doxycycline, a coexpressed Tet-Off transcription factor inhibits transcription of the Flag-Gli3 vector. When forskolin was simultaneously added to doxycycline, we could observe a gradual increase in the relative levels of truncated Gli3 after transcription shutoff, reaching 56% of total Gli3 within 4 h (Fig. 2A, left panel). Upon βTrCP overexpression, levels of truncated Gli3 increased more rapidly and reached 80% total Gli3 at 4 h (Fig. 2A, compare right and left panels). In order to inhibit SCFβTrCP, we next used RNA interference directed against human βTrCP mRNA (13). Figure 2B shows that specific siRNA directed against human βTrCP (13) led to a strong decrease in the levels of truncated Gli3 stimulated by PKA and GSK3β overexpression, as detected by Western blotting after transfection into human 293T or HeLa cells. We have therefore found that stimulating SCFβTrCP leads to stimulation of Gli3 processing, while conversely, interfering with endogenous SCFβTrCP leads to its inhibition. Taken together, these results show that SCFβTrCP is required for Gli3 proteolytic processing. Furthermore, in agreement with a role of the proteasome after ubiquitination, treatment of cells with the proteasome inhibitor MG132 inhibited Gli3 processing (Fig. 2C), similar to a recent report with a series of proteasome inhibitors (12).
FIG. 2.
Truncated Gli3 synthesis requires βTrCP. (A) Overexpression of wild-type βTrCP stimulates synthesis of truncated Gli3 in transcription shutoff experiments. NIH 3T3 cells were transfected with pBI-G-Flag-Gli3, pTet-Off, and GSK3β expression vectors together with control (−) or HA epitope-tagged βTrCP (HA-βTrCP) expression vectors. Cells were treated with 50 μM forskolin (FSK) and 100 ng/ml doxycycline (DOX) and collected after 0 h, 4 h, and 8 h to analyze the relative levels of truncated versus full-length Flag-Gli3. (B) Downregulation of βTrCP inhibits Gli3 processing. HeLa and 293T cells were transfected with siRNA against human βTrCP (+) or luciferase (−) together with Flag-Gli3, PKA, and GSK3β expression vectors. The right panel shows specific downregulation of HA-βTrCP by siRNA against βTrCP. 293T cells were transfected with siRNA against βTrCP (+) or luciferase (−) together with an expression vector for HA-βTrCP. Equal amounts of cell lysates were probed with anti-HA antibody to detect HA-βTrCP and anti-β-galactosidase (βGal) as a control of transfection and cytomegalovirus expression levels. (C) Gli3 processing is inhibited by MG132 proteasome inhibitor. NIH 3T3 cells were transfected with Flag-Gli3, PKAc, and GSK3β expression vectors. Cells were treated with 20 μM MG132 (+) or vehicle (−) for 6 h, and equal amounts of cell lysates were analyzed by anti-Flag immunoblotting.
βTrCP interacts with multiple Gli3 domains.
In order to test whether βTrCP directly connects Gli3 to the ubiquitin-proteasome system, we examined interaction between Gli3 and βTrCP in coimmunoprecipitation experiments.
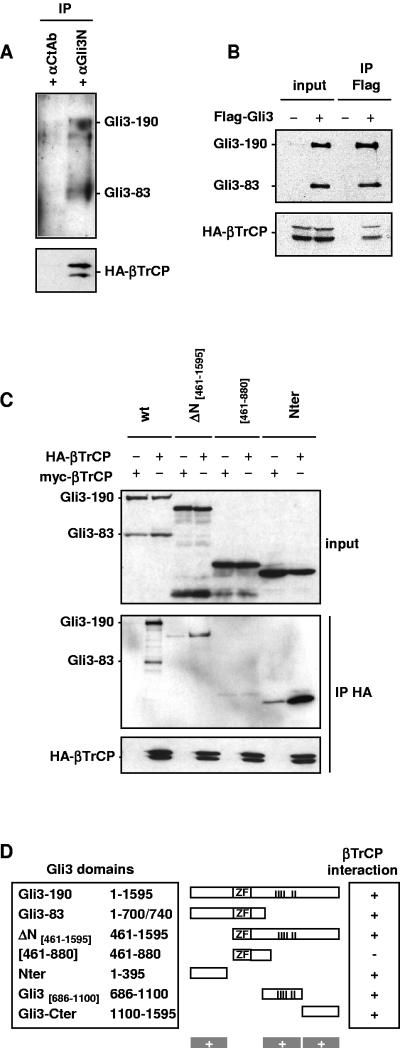
First, endogenous Gli3 was immunoprecipitated from C3H-10T1/2 (previously reported to express Gli3) using a goat anti-Gli3 antibody (Fig. 3A). Endogenous βTrCP could be detected in neither total protein extracts nor in immunopurified fractions due to the lack of an efficient anti-βTrCP antibody. However, HA-tagged βTrCP was specifically coimmunoprecipitated by an anti-Gli3 antibody and not by goat anti-FGFR4 control antibody. Furthermore, HA-βTrCP could be specifically coimmunoprecipitated by Flag-Gli3 (Fig. 3B). These results are consistent with Gli3 and βTrCP being physically associated in vivo.
FIG. 3.
Gli3 interacts with βTrCP. (A) Endogenous Gli3 interact with HA-βTrCP. C3H-10T1/2 cells were transfected with HA-βTrCP expression vector. The cell lysate from four plates of C3H-10T1/2 cells was split in half and subjected to parallel immunoprecipitations using control (αCtAb, control FGF4R antibody) or anti-Gli3N antibody. Immunoprecipitates were analyzed by immunoblotting with anti-HA antibody to detect HA-βTrCP or with anti-Gli3 antibody to detect endogenous Gli3. IP, immunoprecipitation. (B) HA-βTrCP coimmunoprecipitates with Flag-Gli3. NIH 3T3 cells were transfected with HA-βTrCP expression vector and control Flag or Flag-Gli3 expression vectors as indicated together with PKA and GSK3β expression vectors to stimulate synthesis of truncated Gli3. Equal amounts of cell lysates were subjected to anti-Flag immunoprecipitation and analyzed by immunoblotting. (C) Three different regions of Gli3 coimmunoprecipitate with HA-βTrCP. NIH 3T3 cells were transfected with expression vectors as indicated together with PKA and GSK3β expression vectors to stimulate synthesis of truncated Gli3. Plasmid mixtures contained either HA-βTrCP or βTrCP-myc expression constructs, the latter serving as negative controls in coimmunoprecipitation experiments. Cell lysates were subjected to βTrCP immunoprecipitation using anti-HA antibody followed by anti-Flag or anti-HA immunoblotting. In the bottom panel, the different constructs are represented. The position of PKA sites P1 to P6 essential for synthesis of truncated Gli3 (vertical bars) and the zinc finger region responsible for DNA binding (ZF) are indicated. βTrCP-binding sites inferred from analysis by coimmunoprecipitation with HA-βTrCP are indicated by plus signs. Experiments demonstrating that Gli3ΔN contains two independent binding sites to βTrCP are not shown (for Gli3 central 686-1100 domain binding to βTrCP, however, see Fig. 5). Gli3ΔN generated a truncated form upon stimulation by PKA and GSK3β that did not bind to βTrCP, most likely due to absence of the N-terminal βTrCP interaction domain. In the lane corresponding to Gli3 positions 461 to 880, we detected very low levels of truncated products, which could be due to low-level constitutive processing and which, accordingly, were not modulated by PKA, GSK3β, or βTrCP overexpression (data not shown). wt, wild type; Nter, N terminal; Cter, C terminal.
A series of Gli3 protein fragments were next tested for their ability to bind βTrCP in vivo, as shown in Fig. 3C and summarized in Fig. 3D. Both full-length and truncated Gli3 could be detected in HA-βTrCP immunoprecipitates, which suggested that truncated Gli3, corresponding roughly to a fragment of position 1 to roughly position 700/740, was sufficient for βTrCP binding. A fragment of positions 461 to 880 containing the zinc finger region responsible for DNA binding and a DSGSHS sequence at positions 665 to 670 was not sufficient for βTrCP binding. This indicated that the DSGX2-4S motif at positions 665 to 670 in Gli3 was not sufficient for βTrCP binding and was in agreement with our finding that mutation of this motif does not affect Gli3 processing (data not shown). Interestingly, we found that an N-terminal domain, which lacks a DSGX2-4S motif, was sufficient for βTrCP binding and was likely responsible for the interaction observed between truncated Gli3 and βTrCP. However, ΔN[461-1595], lacking this N-terminal domain, could still bind βTrCP. Finally, in further coimmunoprecipitation assays, we could locate two fragments from ΔN[461-1595] that were each able to bind βTrCP independently: a central fragment of positions 686 to 1100 (designated the central 686-1100 domain) containing PKA and GSK3β sites and a C-terminal domain of positions 1100 to 1595 (data not shown) (schematic representation in Fig. 3D). We therefore conclude that βTrCP interacts with at least three independent domains in Gli3. In order to further analyze the role of βTrCP-binding domains in Gli3 processing, we next chose (i) to examine if the N- and C-terminal binding domains are necessary for processing; (ii) to test the importance of PKA sites in binding of βTrCP to the central 686-1100 domain; and (iii) to investigate if βTrCP interaction with Gli3 domains is direct.
βTrCP-binding domains are necessary for efficient Gli3 processing.
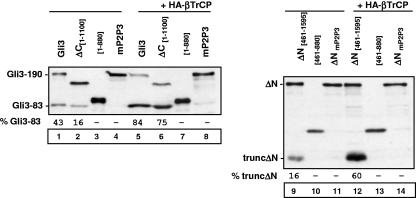
We first examined the role of N- and C-terminal βTrCP-binding domains in Gli3 processing. Deletion of the N- or C-terminal domain significantly diminished the levels of truncated Gli3 expressed from the corresponding ΔN[461-1595] and ΔC[1-1100] fragments (Fig. 4, compare lanes 9 and 2, respectively, to lane 1). As expected, the deletion of the Gli3 domain containing PKA phosphorylation sites in the latter constructs (giving, respectively, fragments spanning positions 461 to 880 and 1 to 880) diminished processing as markedly as observed upon mutation of multiple PKA sites (Fig. 4, lanes 10 and 11 as well as lanes 3 and 4, respectively). Because our studies in coimmunoprecipitation assays showed that βTrCP could still bind to ΔN[461-1595] and ΔC[1-1100] fragments, we tested whether βTrCP overexpression could still efficiently stimulate processing. We found that processing of mutants with deletions of N- and C-terminal domains was strongly induced upon βTrCP overexpression and observed that the levels of truncated protein were close to those obtained with wild-type Gli3 (Fig. 4, lanes 12 and 6, respectively). Strikingly, the multiple PKA site mutants were still unable to produce truncated protein (Fig. 4, lanes 14 and 8, respectively). These findings suggest that binding of βTrCP to the N- and C-terminal domains is necessary for efficient proteolytic processing but is dispensable in the context of βTrCP overexpression.
FIG. 4.
Gli3 N- and C-terminal domains are necessary for efficient processing. The indicated constructs were transfected into NIH 3T3 cells with PKA and GSK3β expression vectors, and the relative levels of truncated (trunc) and full-length products were analyzed by immunoblotting as described in the legend to Fig. 1. mP2P3 indicates serine-to-alanine mutations at PKA sites P2 and P3.
βTrCP binding to the central 686-1100 domain of Gli3 is dependent on PKA phosphorylation.
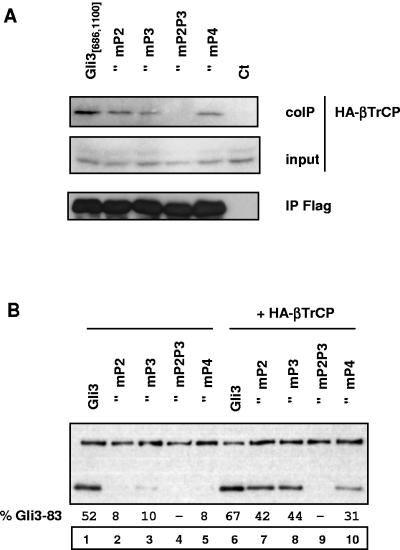
We next examined the consequence of mutations in PKA sites on βTrCP binding to the central 686-1100 domain of Gli3. Mutation in single site P2, P3, or P4 did not significantly impair the ability of βTrCP to bind to the central 686-1100 domain, whereas a mutant bearing mutations in sites P2 and P3 was no longer able to interact with βTrCP (Fig. 5A). In agreement with these results, we found that processing of Gli3 mutants at single PKA sites was significantly stimulated by βTrCP overexpression, whereas that of Gli3 mutants in multiple phosphorylation sites was not (Fig. 5B, compare lanes 1 to 5 to lanes 6 to 10). It therefore appears that multisite PKA phosphorylation of Gli3 is required for βTrCP binding to the Gli3 central 686-1100 domain and processing. In contrast, we did not detect any effect of single or multiple PKA site mutations on the interaction between βTrCP and full-length Gli3 in coimmunoprecipitation assays (data not shown). Furthermore, we found that binding of βTrCP to the N- and C-terminal domains was not modified upon PKA stimulation (data not shown). Taken together, these results suggests that in our experimental system, βTrCP interacts with Gli3 independently of PKA phosphorylation by means of its N- and C-terminal domains and that additional binding to the central 686-1100 domain is induced upon PKA phosphorylation.
FIG. 5.
Effect of PKA site mutations on binding of βTrCP to Gli3 central 686-1100 domain. (A) Cells were transfected with HA-βTrCP, PKA, and GSK3β expression vectors and either wild-type Gli3 central 686-1100 domain, mutant Gli3 central 686-1100 domain, or control (ct) expression vectors as indicated. Cell lysates were subjected to anti-Flag immunoprecipitation (IP) and analyzed by immunoblotting. (B) The indicated constructs were transfected into NIH 3T3 cells with PKA and GSK3β expression vectors, and the relative levels of truncated and full-length products were analyzed by immunoblotting as described in the legend to Fig. 1. mG2, mP2, and mP2P3 indicate mutant Flag-Gli3 at site G2, site P2, and both sites P2 and P3, respectively.
Identification of direct βTrCP-binding sites in the central 686-1100 domain of Gli3.
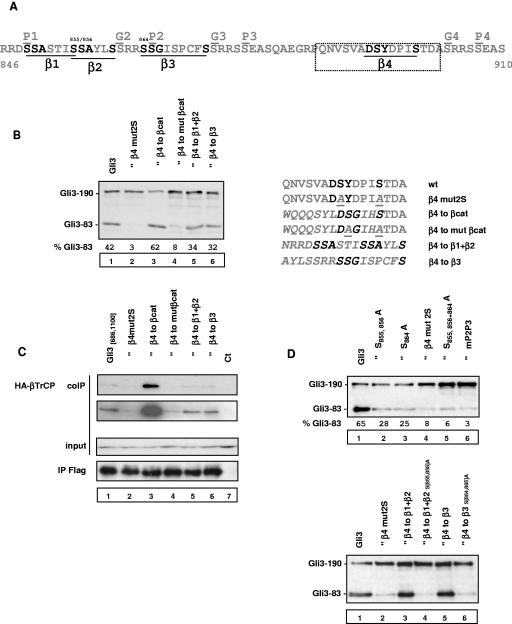
Direct binding of βTrCP is usually mediated by a phosphorylated DSGX2-4S motif in ubiquitination substrates (11, 36). The domains of Gli3 found to interact with βTrCP in coimmunoprecipitation assays do not have a DSGX2-4S motif. However, in the central 686-1100 domain, we could identify four sequence motifs, designated β1 to β4, that are related to the DSGX2-4S motif and could, therefore, be involved in direct binding of βTrCP (Fig. 6A). One of these motifs does not overlap with PKA or GSK3β sites previously identified. We therefore focused on testing its role in Gli3 processing and direct interaction with βTrCP. Figures 6B and C show that motif β4 is indeed required for both Gli3 processing and binding of βTrCP to the central 686-1100 domain of Gli3 (Fig. 6B and D, compare lanes 1 and 2). In order to confirm that the motifs identified in Gli3 are βTrCP-binding sites, we performed peptide swapping experiments and replaced motif β4 with either the βTrCP-binding motif from β-catenin, an inactive mutant thereof, the tandem motif β1+β2, or motif β3. When motif β4 was replaced by the βTrCP-binding motif from β-catenin, Gli3 processing was slightly enhanced (Fig. 6B, lane 3) and βTrCP binding to the central domain was concomitantly strongly reinforced (Fig. 6C, lane 3), while conversely, replacement of β4 by an inactive mutant motif from β-catenin did not allow significant processing and binding of βTrCP (Fig. 6B and C, lanes 4). Furthermore, we found that motif β4 could be replaced by the tandem β1+β2 or β3 motifs (Fig. 6B, lanes 5 and 6), and Fig. 6C strongly suggests that motifs β1+β2, β3, and β4 are direct βTrCP-binding sites (lanes 5 and 6). Importantly, these data establish a tight correlation between direct βTrCP binding and proteolytic processing of Gli3.
FIG. 6.
Direct binding of βTrCP is required for Gli3 processing. (A) Identification of four sequence motifs related to the DSGX2-4S βTrCP-binding site in between PKA sites P1 and P4. SCFβTrCP substrates previously identified contain a DSGX2-4S sequence whose phosphorylation is necessary for βTrCP binding. The sequence motifs β1 to β4 underlined in the figure are related to the DSGX2-4S motif by alignment of the residues indicated in boldface. The 16-amino-acid sequence indicated by the box was mutated to test the role of motif β4 in processing and binding of βTrCP. (B) Effect of mutations in motif β4 on Gli3 processing. The indicated constructs were transfected into NIH 3T3 cells with PKA and GSK3β expression vectors, and the relative levels of truncated and full-length Gli3 were analyzed by immunoblotting. The constructs tested contained mutations of the 16-amino-acid box containing motif β4 as indicated in italics. The β-catenin (βcat) βTrCP-binding site was positioned such that the key serines are expected to be phosphorylated by sequential GSK3β activity after phosphorylation of site P4 by PKA (i.e., in a context mimicking their normal phosphorylation [1]). The mutant β-catenin motif does not bind βTrCP (15, 36). (C) Effect of mutations in motif β4 on binding of βTrCP to Gli3 central 686-1100 domain. NIH 3T3 cells were transfected with HA-βTrCP, PKA, and GSK3β expression vectors and wild-type (wt) or mutant (mut) Gli3 central 686-1100 domain expression vectors or control expression vector (ct) as indicated. Cell lysates were subjected to anti-Flag immunoprecipitation (IP) and analyzed by immunoblotting. Short and long exposures of immunoblots of coimmunoprecipitated (coIP) HA-βTrCP are shown. (D) Effect of mutations in motifs β1+β2 and β3 on Gli3 processing in their natural context (upper panel) or when replacing motif β4 (lower panel). The indicated constructs were transfected into NIH 3T3 cells with PKA and GSK3β expression vectors, and the relative levels of truncated and full-length Gli3 were analyzed by immunoblotting. mP2P3 indicates mutant Flag-Gli3 at PKA sites P2 and P3.
In order to further test the importance of β1+β2 and β3 motifs, we chose to mutate serines 855 and 856 in β1+β2, which are separate from phosphorylation sites P1 and P2, and serine S864, whose mutation to alanine does not modify the RRXS PKA site and should, therefore, preserve the ability of PKA to phosphorylate S865 (site P2). The upper panel in Fig. 6D shows that each mutation impaired Gli3 processing (lanes 2 and 3) and that a mutant bearing S855A, S856A, and S864A mutations was processed as inefficiently as a multisite PKA mutant (lanes 5 and 6). In addition, we checked that mutant β1+β2 and β3 motifs could not replace motif β4 (Fig. 6D, lower panel, lanes 4 and 6). We conclude that the multiple βTrCP-binding sites identified are all required for full efficiency of Gli3 processing upon PKA and GSK3β stimulation.
Identification of lysines necessary for Gli3 processing that are ubiquitinated by SCFβTrCP.
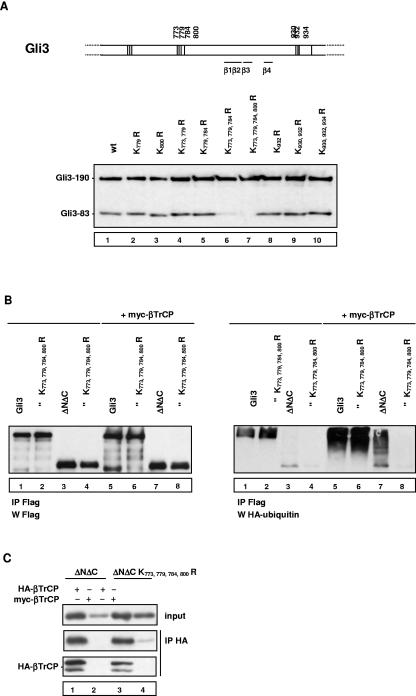
In order to confirm that direct binding of βTrCP to Gli3 results in Gli3 ubiquitination by SCFβTrCP, we sought to locate the corresponding target lysines. We considered the possibility that lysines located near the βTrCP-binding sites could be ubiquitination targets necessary for processing and therefore introduced a series of single or multiple lysine-to-arginine mutations into Gli3 and tested their effect on processing (Fig. 7A). Mutation of the four lysines located N terminal to the Gli3 βTrCP-binding sites, K773, K779, K784, and K800, abolished Gli3 processing (Fig. 7A, lane 7). A strong inhibition was also observed with mutation of K773, K779, and K784 (Fig. 7, lane 6), while single or double mutations of lysines 773 to 800 had no effect (Fig. 7, lanes 2 to 5). In contrast, single or multiple mutations of the three lysines located C-terminal to βTrCP-binding sites had no effect (Fig. 7, lanes 8 to 10).
FIG. 7.
Identification of lysines essential for Gli3 processing that are ubiquitinated by SCFβTrCP. (A) Effect of single or multiple lysine-to-arginine mutations on Gli3 processing. The positions of lysines tested for their potential role in processing and ubiquitination are indicated. The position 846 to 910 domain containing βTrCP-binding motifs β1 to β4 does not contain any lysine. Lysines near this domain are represented by a vertical line. The indicated constructs were transfected into NIH 3T3 cells with PKA and GSK3β expression vectors, and the relative levels of truncated and full-length Gli3 were analyzed by immunoblotting. (B) Lysines 773, 779, 784, and 800 are essential for ubiquitination of Gli3ΔNΔC[461,1100] and its stimulation by βTrCP overexpression. The indicated Gli3 or Gli3ΔNΔC[461,1100] constructs were transfected into NIH 3T3 cells together with PKA, GSK3β, HA-ubiquitin, and control or myc-βTrCP expression vectors. Cells were treated with the proteasome inhibitor MG132 for 4 h at 20 μM, which inhibited proteolytic processing as described in Fig. 2C and favored detection of ubiquitinated proteins, and cells were lysed in lysis buffer containing 5% SDS at 95°C for 10 min. The resulting extracts were subjected to immunoprecipitation with anti-Flag antibody and analyzed by immunoblotting with anti-HA antibody to detect ubiquitinated species (right panel) or with anti-Flag antibody (left panel, labeled “IP Flag, W Flag”). (C) Mutation of lysines 773, 779, 784, and 800 to arginine does not impair binding of Gli3ΔNΔC[461,1100] to HA-βTrCP. NIH 3T3 cells were transfected with expression vectors as indicated together with PKA and GSK3β expression vectors. Cell lysates were subjected to βTrCP immunoprecipitation (IP) using anti-HA antibody followed by anti-Flag or anti-HA immunoblotting. wt, wild type.
We next tested the effects of mutating the four lysines N-terminal to βTrCP-binding sites on ubiquitination of Gli3 and Gli3ΔNΔC, a fragment of positions 461 to 1100 lacking N- and C-terminal βTrCP-binding domains. For this purpose, we performed in vivo ubiquitination assays: cells were lysed under strong denaturation conditions favoring preservation of ubiquitinated proteins, and HA-ubiquitin was detected in immunoprecipitates of the Flag-tagged protein studied. Figure 7B shows that Gli3 and Gli3-K[773, 779, 784, 800]R were ubiquitinated and that ubiquitination was strongly stimulated by βTrCP overexpression (Fig. 7B, right panel, compare lanes 1 and 2 to lanes 5 and 6). Ubiquitination of Gli3ΔNΔC was weaker than that of Gli3, possibly due to the lack of N- and C-terminal βTrCP-binding domains, but it was still significantly stimulated by βTrCP overexpression, while in contrast, Gli3ΔNΔCK[773, 779, 784, 800]R was only moderately ubiquitinated, and its ubiquitination could not be stimulated by βTrCP (Fig. 7B, right panel, compare lanes 3 and 4 to lanes 7 and 8). Furthermore, we checked that βTrCP interacts with Gli3ΔNΔCK[773, 779, 784, 800]R as efficiently as its wild-type counterpart in coimmunoprecipitation assays (Fig. 7C, compare lanes 1 and 3). Taken together, these findings indicate that Gli3ΔNΔC is ubiquitinated at lysines 773, 778, 784, and 800 by SCFβTrCP and strongly suggest that ubiquitination of these residues is essential for Gli3 processing.
DISCUSSION
The data presented here demonstrate that phosphorylation-dependent processing of Gli3 occurs through SCFβTrCP-mediated ubiquitination. As expected from studies of Ci (16, 29), the fly homologue of Gli3, we first showed that GSK3β phosphorylation is required for Gli3 processing, GSK3β stimulation having been found to enhance processing while, conversely, downregulation of GSK3β or mutation of GSK3β site S861, S873, or S903 inhibited it. Modulating the ubiquitin ligase receptor βTrCP in overexpression and RNA interference experiments showed that Gli3 processing is tightly correlated to βTrCP levels. We then showed that multisite phosphorylation by PKA and GSK3β is required for direct binding of βTrCP at multiple motifs related to the DSGX2-4S consensus. Moreover, we identified lysines necessary for Gli3 processing that are ubiquitinated by SCFβTrCP. We discuss potential molecular mechanisms involved in this unusual regulation of transcription factor function and its control in the context of Hedgehog signaling.
Processing by the ubiquitin-proteasome system.
Our study shows that Gli3 belongs to a small family of transcription factors regulated by ubiquitin-proteasome-dependent processing (30). In the NF-κB pathway, p100 and p105 are processed into C-terminally truncated proteins that translocate to the nucleus and activate transcription (6, 10). Ubiquitination directs targeting of these transcription factors to the proteasome, but the molecular mechanisms that direct processing rather than degradation are not well understood. In vitro studies have recently confirmed that the proteasome can perform processing of model substrates, proteolysis being stopped upon reaching resistant protein domains (21). The cleavage site in Gli3 appears to be near the end of the zinc finger domain. It will be interesting to study whether proteasomal processing is blocked in cis by an unusual structure near the cleavage site of Gli3 or in trans, possibly by dimerization (20).
A revised consensus βTrCP-binding motif.
We uncovered multiple βTrCP-binding sites necessary for Gli3 processing which depart from the DSGX2-4S motif found in most βTrCP substrates. By replacing motif β4 with the β-catenin motif, we found that binding of βTrCP and processing were restored, which formally demonstrated that βTrCP binding is required for Gli3 processing and strongly suggested that motif β4 is a bona fide βTrCP-binding site (Fig. 6C). Motifs β1+β2 and β3 could also replace motif β4 and are also likely direct βTrCP-binding sites (Fig. 6B and C). Non-DSGX2-4S βTrCP-binding motifs have recently been found in several other SCFβTrCP substrates. We aligned them with βTrCP-binding motifs in Gli3 and propose a revised consensus βTrCP-binding motif (Fig. 8). Molecular modeling and in vitro interaction studies will be important to address how βTrCP binds to the multiple motifs in Gli3 compared to the DSGX2-4S motif.
FIG. 8.

Alignment of βTrCP-binding motifs in known SCFβTrCP substrates. The first SCFβTrCP substrates identified allowed defining a common DSGX2-4S sequence whose phosphorylation is necessary for βTrCP binding (11). Alignment of βTrCP-binding sites in recently identified substrates allows proposing a revised consensus βTrCP-binding motif. For hCDC25A, Per2, p100, xCDC25A, and Wee1A substrates, see references 3, 8, 10, 19, and 35, respectively.
PKA triggers a cascade of Gli3 phosphorylation by GSK3β and CK1.
Interestingly, in the case of Wee1A and xCDC25A, one or both serines in the DSGX2-4S motif are replaced by the serine phosphomimetics aspartic or glutamic acid (Fig. 8). In contrast, the motifs in Gli3 have serines that fit the DSGX2-4S motif, and binding of βTrCP is therefore expected to be strictly phosphorylation dependent. Indeed, S865 in motif β3 corresponds to PKA site P2. For other serines in βTrCP-binding motifs of Gli3, we propose that they are phosphorylated by sequential GSK3β and CK1 activity after initial priming by PKA (Fig. 9). S855 and S864 occupy the position of aspartic acid in the alignment of motifs β2 and β3 with the DSGX2-4S motif, and one could anticipate that phosphorylation of these serines through the GSK3β/CK1 cascade will contribute to efficient βTrCP binding. PKA therefore likely triggers a cascade of Gli3 phosphorylation by GSK3β and CK1 that provokes direct βTrCP binding and ubiquitination. This simple model nicely explains the importance of PKA and GSK3β sites for βTrCP binding and Gli3 processing (Fig. 1 and 5). Studies published while this paper was in review demonstrated that GSK3β-primed CK1 phosphorylation, and not only PKA-primed CK1 phosphorylation as previously described, is indeed necessary for Ci processing in D. melanogaster, most likely by leading to direct Slimb binding (17).
FIG. 9.
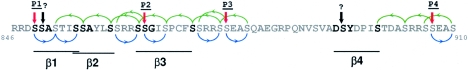
Phosphorylation of βTrCP-binding sites by a putative cascade of GSK3β, CK1, and PKA phosphorylations. PKA phosphorylates serines in consensus RRXS sites indicated by red arrows. GSK3β phosphorylates serines four residues N terminal to a phosphoserine, while CK1 phosphorylates serines three residues C terminal to a phosphoserine; both can sequentially multiphosphorylate substrates after priming (1, 9, 14). S855 in motif β1 could, therefore, be phosphorylated as follows: S849 (P1) phosphorylation by PKA priming sequential phosphorylation of S852 and S855 by CK1. S856 phosphorylation in motif β2 could be as follows: S865 (P2) by PKA priming S868 by CK1 and then S864, S860, and S856 by GSK3β. Similar phosphorylation pathways can easily be proposed for all serines in β1 to β4 motifs (blue and green arrows representing phosphorylations by CK1 and GSK3β, respectively), except S850 and S894 (indicated by black arrows). S850 and S894 lack serines at n + 4 or n − 3 positions for phosphorylation priming, and their sequence context is not similar to that in unprimed CK1 sites (14). An alternative candidate kinase is Fused. S850 phosphorylation may not be required if βTrCP could bind to the overlapping DSS850ASTIS motif (with S850 aligned to G/A in the consensus) rather than the motif proposed in Fig. 8. In any case, it appears that 19 serines in the 65-amino-acid segment from P1 to P4, including most serines in βTrCP-binding motifs β1 to β4, are likely phosphorylated by GSK3β and CK1 after priming by PKA.
The cascade of PKA/GSK3β/CK1 phosphorylations, however, may not be the only phosphorylation events involved in Gli3 processing and potentially regulated by Hedgehog signaling. S850 and S894 in motifs β1 and β4 are unlikely to be phosphorylated by GSK3β and CK1 (Fig. 9). An alternative candidate kinase could be the Fused serine/threonine kinase (24). In addition, PKA sites P5 and P6 are essential for Gli3 processing (34), but they are not adjacent to GSK3β, CK1, or potential βTrCP-binding motifs and probably act by a different mechanism.
Gli3, a noncanonical substrate with multiple βTrCP-binding sites.
The presence of multiple binding sites is unusual in SCFβTrCP substrates. It is likely that βTrCP-binding sites found in Gli3 possess a lower binding affinity than that from β-catenin. When motif β4 was replaced by the βTrCP-binding site from β-catenin, a marked increase of βTrCP binding to Gli3[686,1100] was observed in coimmunoprecipitation assays (Fig. 6C). In contrast, the replacement of β4 by motif β1+β2 or β3 did not change the levels of interaction detected. Differences in phosphorylation levels could also be involved, however, and here again in vitro studies using synthetic peptides will be required to directly examine how βTrCP binding to sites in Gli3 differs from that to the site in β-catenin.
Interestingly, Sic1 from Saccharomyces cerevisiae has been shown to contain multiple low-affinity phosphorylation-dependent binding sites for the F-box protein Cdc4. Seminal structure-function studies have shown that this configuration results in high-affinity binding, as obtained with a single high-affinity site, but allows a tighter control of binding by phosphorylation (26). Gli3 processing, at least in the limb bud, results in graded levels of truncated protein inversely proportional to Shh doses along the anteroposterior axis (34), and such spatially graded response appears different from the tight all-or-none control of Sic1 degradation during the yeast cell cycle. If, as in Sic1, a threshold in multisite phosphorylation were necessary for Gli3 ubiquitination and processing, it would be expected to play a permissive rather than instructive role in dose-dependent regulation by Shh. Alternatively, different levels of Gli3 phosphorylation might result in different levels of Gli3 processing. In our assays, mutations in single phosphorylation sites indeed allowed us to observe intermediate levels of Gli3 processing (Fig. 1C and 6D), and the levels of direct binding of βTrCP to Gli3 may be more finely controlled by phosphorylation than could be detected here in coimmunoprecipitation assays using overexpressed βTrCP (Fig. 5A). Analyzing in vivo the role of the intertwined phosphorylation and βTrCP-binding sites reported here will be an important challenge in understanding the regulation of Gli3 processing by Hedgehog signaling.
Modeling of SCF complexes showed that binding of βTrCP to substrates results in juxtaposition of Cdc34, the enzyme that performs ubiquitin conjugation, to the βTrCP-binding peptide and explained that the lysine residues adjacent to the DSGX2-4S motif, lying between 8 and 20 residues N terminal, are the ubiquitination targets in β-catenin and IκB (37). In Gli3, however, there are no lysines adjacent to the βTrCP-binding motifs, and we showed that lysines lying at least 48 residues N terminal to motif β1 are SCFβTrCP ubiquitination targets necessary for processing (Fig. 7). This result importantly strengthens our demonstration that SCFβTrCP-mediated ubiquitination is required for Gli3 processing. The proximity of lysines 773, 779, 784, and 800 to Cdc34, which is a prerequisite for ubiquitin conjugation, may be intrinsic to Gli3 structure, or alternatively, it may be achieved upon a change in conformation that could be an additional level of regulation of Gli3 processing. Interestingly, in contrast to what was observed in the Gli3ΔNΔC context, mutation of lysines 773, 779, 784, and 800 had no effect on the levels of ubiquitinated Gli3 (Fig. 7C). This suggests that the N- and C-terminal domains contain alternative lysine targets accessible to Cdc34 upon binding of SCFβTrCP to Gli3 but that ubiquitination of such lysines is not sufficient for processing. Ubiquitination of particular lysines (at positions 773, 779, 784, and 800) may therefore be essential to mediate processing rather than destruction by the proteasome.
Gli3 ubiquitination takes place within a multiprotein complex regulated by Hedgehog signaling.
In addition to the central 686-1100 domain, we have found that βTrCP could interact independently with N- and C-terminal domains. N- and C-terminal domains lack βTrCP DSGX2-4S-binding motifs, and interactions detected with βTrCP in coimmunoprecipitation assays are likely indirect, taking place in a multiprotein complex. The indirect interaction of βTrCP and Gli3 may favor direct binding to the central domain of Gli3 upon phosphorylation, and conversely destabilization of the complex by N- or C-terminal deletion of Gli3 might result in less efficient recruitment of βTrCP to Gli3 and consequently less efficient ubiquitination and processing (as observed in Fig. 4 and 7).
In D. melanogaster, Ci belongs to a complex regulated by Hedgehog signaling that can contain Costal, a kinesin-like protein, Fused, and Sufu proteins as well as PKA, GSK3β, and CK1 kinases (22, 38). One attractive possibility is that proteins in the vertebrate complex, such as Sufu or Costal, mediate indirect binding of βTrCP to N- and C-terminal Gli3 domains. For example, Sufu has been shown previously to interact with βTrCP in vitro (32), and we could confirm this interaction in coimmunoprecipitation assays as well as interaction of Sufu with the N-terminal domain of Gli3 (unpublished results). Moreover, Sufu is a tumor suppressor in the Hedgehog pathway, and this likely implies that vertebrate Sufu is required for Gli3 processing (33). These findings were unexpected, since Sufu mutant flies show no overt phenotypic defect, and they may point to important differences in the control of Gli3 and Ci processing. Further studies of the possible interactions of βTrCP with proteins in the Gli3 regulatory complex will be important in understanding how Hedgehog signaling inhibits Gli3 processing and exerts dose-dependent effects during development and in disease.
Acknowledgments
D.T. was supported by doctoral fellowships from MNRT and ARC, and M.C. was supported by a doctoral fellowship from the Ministerio de Planificacion Nacional (Chile). This work was supported by INSERM and ARC.
We are grateful to the Benarous laboratory for βTrCP expression plasmids and discussion. B. Vogelstein, H. Sasaki, D. Bohmann, M. Raymonjean, M. Pap, G. M. Cooper, M. J. Birnbaum, and K. Hattori kindly provided plasmids. We thank F. Letourneur and his colleagues at the DNA Sequencing Facility of Institut Cochin.
REFERENCES
- 1.Amit, S., A. Hatzubai, Y. Birman, J. S. Andersen, E. Ben-Shushan, M. Mann, Y. Ben-Neriah, and I. Alkalay. 2002. Axin-mediated CKI phosphorylation of beta-catenin at Ser 45: a molecular switch for the Wnt pathway. Genes Dev. 16:1066-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai, C. B., D. Stephen, and A. L. Joyner. 2004. All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev. Cell 6:103-115. [DOI] [PubMed] [Google Scholar]
- 3.Busino, L., M. Donzelli, M. Chiesa, D. Guardavaccaro, D. Ganoth, N. V. Dorrello, A. Hershko, M. Pagano, and G. F. Draetta. 2003. Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage. Nature 426:87-91. [DOI] [PubMed] [Google Scholar]
- 4.Chen, C. H., D. P. von Kessler, W. Park, B. Wang, Y. Ma, and P. A. Beachy. 1999. Nuclear trafficking of Cubitus interruptus in the transcriptional regulation of Hedgehog target gene expression. Cell 98:305-316. [DOI] [PubMed] [Google Scholar]
- 5.Chen, Y., N. Gallaher, R. H. Goodman, and S. M. Smolik. 1998. Protein kinase A directly regulates the activity and proteolysis of cubitus interruptus. Proc. Natl. Acad. Sci. USA 95:2349-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciechanover, A., H. Gonen, B. Bercovich, S. Cohen, I. Fajerman, A. Israel, F. Mercurio, C. Kahana, A. L. Schwartz, K. Iwai, A. Orian, and E. Eytan. 2001. Mechanisms of ubiquitin-mediated, limited processing of the NF-kappaB1 precursor protein p105 SCF(beta)(-TrCP) ubiquitin ligase-mediated processing of NF-kappaB p105 requires phosphorylation of its C terminus by IkappaB kinase. Biochimie 83:341-349. [DOI] [PubMed] [Google Scholar]
- 7.Dai, P., H. Akimaru, Y. Tanaka, T. Maekawa, M. Nakafuku, and S. Ishii. 1999. Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J. Biol. Chem. 274:8143-8152. [DOI] [PubMed] [Google Scholar]
- 8.Eide, E. J., M. F. Woolf, H. Kang, P. Woolf, W. Hurst, F. Camacho, E. L. Vielhaber, A. Giovanni, and D. M. Virshup. 2005. Control of mammalian circadian rhythm by CKIɛ-regulated proteasome-mediated PER2 degradation. Mol. Cell. Biol. 25:2795-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiol, C. J., A. Wang, R. W. Roeske, and P. J. Roach. 1990. Ordered multisite protein phosphorylation. Analysis of glycogen synthase kinase 3 action using model peptide substrates. J. Biol. Chem. 265:6061-6065. [PubMed] [Google Scholar]
- 10.Fong, A., and S. C. Sun. 2002. Genetic evidence for the essential role of beta-transducin repeat-containing protein in the inducible processing of NF-kappa B2/p100. J. Biol. Chem. 277:22111-22114. [DOI] [PubMed] [Google Scholar]
- 11.Fuchs, S. Y., V. S. Spiegelman, and K. G. Kumar. 2004. The many faces of beta-TrCP E3 ubiquitin ligases: reflections in the magic mirror of cancer. Oncogene 23:2028-2036. [DOI] [PubMed] [Google Scholar]
- 12.Garrett, I. R., D. Chen, G. Gutierrez, M. Zhao, A. Escobedo, G. Rossini, S. E. Harris, W. Gallwitz, K. B. Kim, S. Hu, C. M. Crews, and G. R. Mundy. 2003. Selective inhibitors of the osteoblast proteasome stimulate bone formation in vivo and in vitro. J. Clin. Investig. 111:1771-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guardavaccaro, D., Y. Kudo, J. Boulaire, M. Barchi, L. Busino, M. Donzelli, F. Margottin-Goguet, P. K. Jackson, L. Yamasaki, and M. Pagano. 2003. Control of meiotic and mitotic progression by the F box protein beta-Trcp1 in vivo. Dev. Cell 4:799-812. [DOI] [PubMed] [Google Scholar]
- 14.Ha, N. C., T. Tonozuka, J. L. Stamos, H. J. Choi, and W. I. Weis. 2004. Mechanism of phosphorylation-dependent binding of APC to beta-catenin and its role in beta-catenin degradation. Mol. Cell 15:511-521. [DOI] [PubMed] [Google Scholar]
- 15.Hart, M., J. P. Concordet, I. Lassot, I. Albert, R. del los Santos, H. Durand, C. Perret, B. Rubinfeld, F. Margottin, R. Benarous, and P. Polakis. 1999. The F-box protein beta-TrCP associates with phosphorylated beta-catenin and regulates its activity in the cell. Curr. Biol. 9:207-210. [DOI] [PubMed] [Google Scholar]
- 16.Jia, J., K. Amanai, G. Wang, J. Tang, B. Wang, and J. Jiang. 2002. Shaggy/GSK3 antagonizes Hedgehog signalling by regulating Cubitus interruptus. Nature 416:548-552. [DOI] [PubMed] [Google Scholar]
- 17.Jia, J., L. Zhang, Q. Zhang, C. Tong, B. Wang, F. Hou, K. Amanai, and J. Jiang. 2005. Phosphorylation by double-time/CKIepsilon and CKIalpha targets cubitus interruptus for Slimb/beta-TRCP-mediated proteolytic processing. Dev. Cell 9:819-830. [DOI] [PubMed] [Google Scholar]
- 18.Jiang, J., and G. Struhl. 1998. Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature 391:493-496. [DOI] [PubMed] [Google Scholar]
- 19.Kanemori, Y., K. Uto, and N. Sagata. 2005. Beta-TrCP recognizes a previously undescribed nonphosphorylated destruction motif in Cdc25A and Cdc25B phosphatases. Proc. Natl. Acad. Sci. USA 102:6279-6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, L., G. N. DeMartino, and W. C. Greene. 2000. Cotranslational dimerization of the Rel homology domain of NF-kappaB1 generates p50-p105 heterodimers and is required for effective p50 production. EMBO J. 19:4712-4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, C. W., M. J. Corboy, G. N. DeMartino, and P. J. Thomas. 2003. Endoproteolytic activity of the proteasome. Science 299:408-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lum, L., and P. A. Beachy. 2004. The Hedgehog response network: sensors, switches, and routers. Science 304:1755-1759. [DOI] [PubMed] [Google Scholar]
- 23.McMahon, A. P., P. W. Ingham, and C. J. Tabin. 2003. Developmental roles and clinical significance of hedgehog signaling Hedgehog signaling in animal development: paradigms and principles. Curr. Top. Dev. Biol. 53:1-114. [DOI] [PubMed] [Google Scholar]
- 24.Methot, N., and K. Basler. 2000. Suppressor of fused opposes hedgehog signal transduction by impeding nuclear accumulation of the activator form of Cubitus interruptus. Development 127:4001-4010. [DOI] [PubMed] [Google Scholar]
- 25.Noureddine, M. A., T. D. Donaldson, S. A. Thacker, and R. J. Duronio. 2002. Drosophila Roc1a encodes a RING-H2 protein with a unique function in processing the Hh signal transducer Ci by the SCF E3 ubiquitin ligase. Dev. Cell 2:757-770. [DOI] [PubMed] [Google Scholar]
- 26.Orlicky, S., X. Tang, A. Willems, M. Tyers, and F. Sicheri. 2003. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112:243-256. [DOI] [PubMed] [Google Scholar]
- 27.Ou, C. Y., Y. F. Lin, Y. J. Chen, and C. T. Chien. 2002. Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes Dev. 16:2403-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pap, M., and G. M. Cooper. 1998. Role of glycogen synthase kinase-3 in the phosphatidylinositol 3-kinase/Akt cell survival pathway. J. Biol. Chem. 273:19929-19932. [DOI] [PubMed] [Google Scholar]
- 29.Price, M. A., and D. Kalderon. 2002. Proteolysis of the Hedgehog signaling effector Cubitus interruptus requires phosphorylation by glycogen synthase kinase 3 and casein kinase 1. Cell 108:823-835. [DOI] [PubMed] [Google Scholar]
- 30.Rape, M., and S. Jentsch. 2002. Taking a bite: proteasomal protein processing. Nat. Cell Biol. 4:E113-E116. [DOI] [PubMed] [Google Scholar]
- 31.Shin, S. H., P. Kogerman, E. Lindstrom, R. Toftgard, and L. G. Biesecker. 1999. GLI3 mutations in human disorders mimic Drosophila cubitus interruptus protein functions and localization. Proc. Natl. Acad. Sci. USA 96:2880-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stone, D. M., M. Murone, S. Luoh, W. Ye, M. P. Armanini, A. Gurney, H. Phillips, J. Brush, A. Goddard, F. J. de Sauvage, and A. Rosenthal. 1999. Characterization of the human suppressor of fused, a negative regulator of the zinc-finger transcription factor Gli. J. Cell Sci. 112:4437-4448. [DOI] [PubMed] [Google Scholar]
- 33.Taylor, M. D., L. Liu, C. Raffel, C. C. Hui, T. G. Mainprize, X. Zhang, R. Agatep, S. Chiappa, L. Gao, A. Lowrance, A. Hao, A. M. Goldstein, T. Stavrou, S. W. Scherer, W. T. Dura, B. Wainwright, J. A. Squire, J. T. Rutka, and D. Hogg. 2002. Mutations in SUFU predispose to medulloblastoma. Nat. Genet. 31:306-310. [DOI] [PubMed] [Google Scholar]
- 34.Wang, B., J. F. Fallon, and P. A. Beachy. 2000. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell 100:423-434. [DOI] [PubMed] [Google Scholar]
- 35.Watanabe, N., H. Arai, Y. Nishihara, M. Taniguchi, T. Hunter, and H. Osada. 2004. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFbeta-TrCP. Proc. Natl. Acad. Sci. USA 101:4419-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winston, J. T., P. Strack, P. Beer-Romero, C. Y. Chu, S. J. Elledge, and J. W. Harper. 1999. The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev. 13:270-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu, G., G. Xu, B. A. Schulman, P. D. Jeffrey, J. W. Harper, and N. P. Pavletich. 2003. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol. Cell 11:1445-1456. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, W., Y. Zhao, C. Tong, G. Wang, B. Wang, J. Jia, and J. Jiang. 2005. Hedgehog-regulated Costal2-kinase complexes control phosphorylation and proteolytic processing of Cubitus interruptus. Dev. Cell 8:267-278. [DOI] [PubMed] [Google Scholar]