Abstract
The cellular response to DNA damage requires not only direct repair of the damage but also changes in the DNA replication machinery, chromatin, and transcription that facilitate survival. Here, we describe Saccharomyces cerevisiae Doa1, which helps to control the damage response by channeling ubiquitin from the proteosomal degradation pathway into pathways that mediate altered DNA replication and chromatin modification. DOA1 interacts with genes involved in PCNA ubiquitination, including RAD6, RAD18, RAD5, UBC13, and MMS2, as well as genes involved in histone H2B ubiquitination or deubiquitination, including RAD6, BRE1, LGE1, CDC73, UBP8, UBP10, and HTB2. In the absence of DOA1, damage-induced ubiquitination of PCNA does not occur. In addition, the level of ubiquitinated H2B is decreased under normal conditions and completely absent in the presence of DNA damage. In the case of PCNA, the defect associated with the doa1Δ mutant is alleviated by overexpression of ubiquitin, but in the case of H2B, it is not. The data suggest that Doa1 is the major source of ubiquitin for the DNA damage response and that Doa1 also plays an additional essential and more specific role in the monoubiquitination of histone H2B.
To maintain the genome, cells have evolved multiple pathways to detect and respond to DNA damage. The cellular response to DNA damage has been particularly well characterized in the yeast Saccharomyces cerevisiae. An important way in which this organism coordinates different facets of the DNA damage response is the posttranslational modification of proteins. While phosphorylation has received a great deal of attention, it has become increasingly clear that other types of posttranslational modifications, such as ubiquitination, also play critical roles. Protein ubiquitination is catalyzed by ubiquitin (Ub)-conjugating enzymes, which facilitate the formation of an isopeptide bond between the C terminus of Ub and a lysine side chain of a substrate protein (41). Proteins may be monoubiquitinated, or the Ub monomer may act as a point of attachment for additional Ub monomers, resulting in polyubiquitination. The specific biological signal mediated by a polyubiquitin chain is determined, in part, by chain topology, which is differentiated by the Ub lysine residue used for chain extension (22, 32). K48-linked chains appear to target proteins for proteasomal degradation, whereas K63-linked chains appear to regulate proteins involved in a wide range of processes, including DNA repair, mRNA translation, and endocytosis (10, 40). Ub modification of proteins is reversible; Ub may be removed from proteins by deubiquitinating enzymes, which hydrolyze the isopeptide bond between Ub and the substrate protein, or by ubiquitin proteases, which remove Ub monomers from a polyubiquitin chain (2).
Under normal cellular conditions, Ub is produced as a fusion with ribosomal proteins; proteolytic cleavage from these carrier proteins generates a pool of free Ub (31), much of which is used to form K48-linked Ub chains that target proteins for proteosomal degradation. Under conditions of stress, it is thought that increased levels of Ub are supplied by the expression of UBI4, which encodes a head-to-tail pentamer of Ub (9). However, no relationship between UBI4 and any facet of the DNA damage response has been established. While its source remains to be more rigorously characterized, several functions for Ub in the DNA damage response have been identified. For example, to effect changes in DNA synthesis, PCNA is monoubiquitinated by Rad6, with assistance from Rad18; it then may be K63 polyubiquitinated by Rad5 and the Ubc13/Mms2 heterodimer (21, 40). These PCNA modifications are thought to help control various pathways involved in the repair of replication forks that have stalled or collapsed due to damaged DNA (21, 40). In association with Bre1 and Lge1, Rad6 also monoubiquitinates H2B (25, 34, 45), which plays a critical role in both the initiation and elongation of RNA polymerase (Pol) II transcription (6, 19, 27, 48). While H2B ubiquitination is required under normal cellular conditions for transcription of certain genes, maintenance of H2B ubiquitination levels is also critical for the transcriptional and cell cycle response to DNA damage (15).
The doa1Δ mutant was originally identified in screens for mutants that stabilize several normally short-lived proteins (14, 20, 26). The observation that overexpression of Ub can complement the phenotypes led to the suggestion that Doa1 is a regulatory component of the proteasome. Doa1 contains five WD repeats, which are present in a number of eukaryotic proteins involved in cell cycle control, cell fate determination, transcription, transmembrane signaling, RNA metabolism, and vesicular trafficking (29). The N terminus of Doa1 is homologous to PLAP (phospholipase A2-activating protein) (5) and Lis-1 (a subunit of the platelet-activating factor acetylhydrolase) (18), while the C terminus is not homologous to any known protein. High-throughput affinity precipitation studies identified a complex composed of Doa1, Cdc48 (AAA-ATPase), Rpa190 (RNA Pol I subunit), Rpa135 (RNA Pol I subunit), Rpo31 (RNA Pol III subunit), and Shp1 (regulator of Glc7 phosphatase activity) (12, 14). Other studies showed that Doa1 binds proteins containing UBX (Ub regulatory X) domains, such as Ubx4, Ubx6, and Ubx7 (7, 43).
Doa1 was first implicated as being part of the DNA damage response in a genome-wide screen for genes involved in repairing UV- and MMS-induced DNA damage (17). More recently, it was shown that LUB1, the Schizosaccharomyces pombe homologue of DOA1, plays a role in mediating the stress response (30). The temperature sensitivity of a lub1Δ mutant was ameliorated by expression of Ub or the K63R Ub mutant but not by the expression of the K48R Ub mutant. The lub1Δ mutant also exhibited decreased cellular levels of both Ub and polyubiquitin chains but no change in the mRNA levels of ubi4. An increased rate of Ub turnover was also observed. Based on these observations, it was suggested that Lub1 is a negative regulator of Ub degradation that acts to provide Ub specifically for proteasome-mediated protein degradation.
In this report, we characterize the role of Doa1 in the S. cerevisiae DNA damage response. Interestingly, despite the fact that Doa1 and Lub1 are homologous proteins, they appear to play different roles in the stress responses of their respective organisms. We present evidence that in S. cerevisiae, Doa1 provides the majority of Ub used in the damage response. We demonstrate that DOA1 interacts with genes involved in both PCNA and H2B ubiquitination and that in the absence of Doa1, virtually no PCNA ubiquitination is induced in response to DNA damage and levels of ubiquitinated H2B are not maintained. Consistent with the localization of Doa1 to both the cytoplasm and the nucleus (13, 24), the data suggest that Doa1 is involved in the rapid recycling of Ub from the proteasomal degradation pathway into transcriptional and damage response pathways.
MATERIALS AND METHODS
Media and general procedures.
Yeast was cultured at 30°C in yeast extract-peptone-dextrose (YPD), synthetic complete, or Hartwell's complete (HC) medium as described previously (39). Methyl methane sulfonate (MMS; Aldrich), hydroxyurea (HU; U.S. Biological), X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; Amersham), 6-azauracil (6-AU; Fisher), Complete mini protease inhibitor cocktail (Roche), antiubiquitin antibody (Sigma), anti-Flag antibody (M2 mouse monoclonal; Sigma), anti-H3 trimethyllysine 4 antibody (ab8580; Abcam), antihemagglutinin (anti-HA) antibody (12CA5; Roche), antiphosphoglycerokinase (anti-PGK) antibody (22C5; Molecular Probes), anti-myc antibody (9E10; Santa Cruz), anti-mouse antibody-horseradish peroxidase (HRP) conjugate (Upstate), and anti-rabbit antibody-HRP conjugate (Upstate) were also used. Sequences of primers used in this study are available upon request.
Yeast two-hybrid screening.
The LexA-based yeast two-hybrid system (DupLEX-A; Origene Technologies, Inc., Rockville, MD) was employed. DOA1 was PCR amplified from S288C genomic DNA. The 5′ primer contained an NcoI restriction site and an polylinker encoding Gly-Gly-Ser-Gly-Gly. The 3′ primer contained an XhoI restriction site. The PCR product was digested and ligated into NcoI/XhoI-digested pEG202 to generate a Doa1-LexA fusion. The Doa1-LexA-encoding plasmid, the reporter plasmid pSH18-34, and a genomic library in pJG4-5 (obtained from R. Brent, Molecular Sciences Institute, Berkeley, CA) were transformed into yeast EGY48; 1.5 × 106 transformants were screened. Activation of the LEU2 and lacZ reporter genes (located in EGY48 and pSH18-34, respectively) was used to identify physically interacting clones. Library plasmids were isolated and sequenced to reveal the identities of the interactors. The specificity of the interactors was determined using pLexA-Max according to the manufacturer's instructions. For direct yeast two-hybrid studies of Doa1-LexA with Ub chains, DNA encoding one to five ubiquitins was amplified from S288C genomic DNA, using primers specific for UBI4. Isolated fragments were cloned into the prey vector containing the B42 DNA activation domain. Interactions were determined using the lacZ reporter as described above.
Strain construction and sensitivity to DNA damage.
Yeast strains used in this study are listed in Table 1. Double deletion mutants were constructed by transformation of the kanMX4 null deletion strains (Open Biosystems) with a double-stranded DNA deletion cassette specific for DOA1. Cassettes were generated by PCR amplification of the Kluyveromyces lactis LEU2 cassette from pUG73 or the kanMX6 cassette from pUG6, using gene-specific primers (16). Gene disruptions were confirmed by PCR, as well as by phenotypic screening of multiple transformants. A ubp10Δ doa1Δ mutant was constructed by mating a MATa ubp10Δ (kindly provided by M. Hochstrasser, Yale University) strain with a MATα doa1Δ strain; the resulting diploid strain was sporulated and dissected. The MMS and HU sensitivity of the mutants was characterized by growing cultures to mid-log phase, normalizing them by cell density, and serially diluting them in fivefold increments. Five-microliter drops were plated on YPD containing different concentrations of MMS or HU, grown for 2 to 3 days at 30°C, and then photographed. Epitope tagging was performed as previously described (28). Briefly, the genomic copy of POL30 was C terminally tagged with the 3-HA epitope by transformation of a double-stranded DNA cassette containing the 3-HA hisMX6 cassette targeted to replace the stop codon of POL30. The genomic copy of DOA1 was C terminally tagged with the 13-myc epitope by transformation of a double-stranded 13-myc kanMX6 cassette targeted to replace the stop codon of DOA1. The correct insertion of the tags was verified by PCR and by anti-HA or anti-myc immunoblotting.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | ATCC |
| orfΔ | BY4741 orfΔ::kanMX4 | Open Biosystems |
| FR1025 | BY4741 doa1Δ::LEU2 | This study |
| FR381 | BY4741 ubi4Δ::kanMX4 doa1Δ::LEU2 | This study |
| FR684 | BY4741 ubx5Δ::kanMX4 doa1Δ::LEU2 | This study |
| FR682 | BY4741 ubx6Δ::kanMX4 doa1Δ::LEU2 | This study |
| FR683 | BY4741 ubx7Δ::kanMX4 doa1Δ::LEU2 | This study |
| FR432 | BY4741 shp1Δ::kanMX4 doa1Δ::LEU2 | This study |
| FR561 | BY4741 ubp7Δ::kanMX4 doa1Δ::LEU2 | This study |
| FR550 | BY4741 rad6Δ::kanMX4 doa1Δ::LEU2 | This study |
| FR552 | BY4741 rad5Δ::kanMX4 doa1Δ::LEU2 | This study |
| FR553 | BY4741 rad18Δ::kanMX4 doa1Δ::LEU2 | This study |
| FR1021 | BY4741 mms2Δ::kanMX4 doa1Δ::LEU2 | This study |
| FR1024 | BY4741 ubc13Δ::kanMX4 doa1Δ::LEU2 | This study |
| FR376 | BY4741 srs2Δ::kanMX4 doa1Δ::LEU2 | This study |
| FR663 | BY4741 bre1Δ::kanMX4 doa1Δ::LEU2 | This study |
| FR1026 | BY4741 lge1Δ::kanMX4 doa1Δ::LEU2 | This study |
| FR685 | BY4741 cdc73Δ::kanMX4 doa1Δ::LEU2 | This study |
| FR664 | BY4741 ubp8Δ::kanMX4 doa1Δ::LEU2 | This study |
| FR686 | BY4741 htb2Δ::kanMX4 doa1Δ::LEU2 | This study |
| FR687 | BY4741 hda1Δ::kanMX4 doa1Δ::LEU2 | This study |
| FR688 | BY4741 htz1Δ::kanMX4 doa1Δ::LEU2 | This study |
| FR555 | BY4741 rad9Δ::kanMX4 doa1Δ::LEU2 | This study |
| FR1218 | BY4741 ubp4Δ::kanMX4 doa1Δ::LEU2 | This study |
| FR1223 | BY4741 rad12Δ::kanMX4 doa1Δ::LEU2 | This study |
| FR1225 | BY4741 rad14Δ::kanMX4 doa1Δ::LEU2 | This study |
| FR521 | BY4741 DOA1-13myc | This study |
| MHY501 | MATalys2-801 leu2-3,112 ura3-52 his3-Δ200 trp1-1 gal2 | M. Hochstrasser |
| MHY1227 | MATalys2-801 leu2-3,112 ura3-52 his3-Δ200 trp1-1 ubp10::HIS3 | M. Hochstrasser |
| FR1231 | MHY501 MATa | This study |
| FR1229 | MHY501 MATadoa1Δ::LEU2 | This study |
| FR1232 | MHY501 MATaubp10Δ::HIS3 | This study |
| FR1230 | MHY501 MATaubp10Δ::HIS3 doa1Δ::LEU2 | This study |
| W1588-4C | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | R. Rothstein |
| FR657 | W1588-4C doa1Δ::LEU2 | This study |
| SUB280 | MATalys2-801 leu2-3,112 ura3-52 his3-Δ200 trp1-1 [am] ubi1-Δ1::TRP1 ubi2-Δ2::ura3 ubi3-Δub-2 ubi4-Δ2::LEU2 (pUB39, pUB100) | D. Finley |
| FR512 | SUB280 doa1Δ::kanMX6 | This study |
| SUB413 | MATa lys2-801 leu2-3,112 ura3-52 his3-Δ200 trp1-1 [am] ubi1-Δ1::TRP1 ubi2-Δ2::ura3 ubi3-Δub-2 ubi4-Δ2::LEU2 [Ub K63R] [pUB100] | D. Finley |
| FR514 | SUB413 doa1Δ::kanMX6 | This study |
| Y0003 | MATa his3-Δ200 leu2-3,112 lys2-801 trp1-1 [am] ura3-52 | S. Jentsch |
| FR1023 | Y0003 doa1Δ::LEU2 | This study |
| FR1088 | Y0003 POL30-3HA | This study |
| FR1090 | Y0003 POL30-3HA doa1Δ::LEU2 | This study |
| Y1192 | Y0003 pol30-K164R | S. Jentsch |
| FR1022 | Y1192 doa1Δ::LEU2 | This study |
| FR1105 | Y1192 pol30-3HA | This study |
| DOA1-TAP | BY4741 DOA1-TAP | Open Biosystems |
| EGY48 | MATaura3 trp l his3 6lexAop-LEU2 | Origene |
Ubiquitin complementation.
Plasmids YEp96, YEp110 (kindly provided by M. Hochstrasser, Yale University), pES12, and pTER103 (kindly provided by M. Ellison, University of Alberta) were transformed into strains W1588-4C (wild type) and FR657 (doa1Δ mutant). Transformants were grown to mid-log phase, normalized by cell density, and serially diluted in fivefold increments. Seven-microliter drops were plated on synthetic complete-Trp media containing different concentrations of MMS, grown for 2 to 3 days at 30°C, and then photographed. Complementation of the H2B ubiquitination and histone H3 K4 methylation was performed by transforming strains BY4741 (wild type) and FR1025 (doa1Δ mutant) with plasmids YEplac195 and pCUP-Ub-195 (kindly provided by E. Johnson, Thomas Jefferson University).
Immunoprecipitation.
Cells were grown to mid-log phase, pelleted, washed once with water, and lysed in buffer A (50 mM Tris, 150 mM NaCl, 5 mM MgCl2 0.5% Triton X-100, 0.1% deoxycholic acid, 1× Complete mini protease inhibitor cocktail). Lysates were clarified by centrifugation. Total protein (2 mg) was incubated with either 200 μg bovine serum albumin (BSA; Fisher) or 200 μg ubiquitin (Fisher) for 2 h at 4°C. Ubiquitin agarose (25 μl; Boston Biochem) was then added, followed by incubation with rocking for 3 h at 4°C. Beads were washed three times with buffer A, resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, and boiled for 5 min. Proteins were separated by 10% SDS-PAGE, transferred to polyvinylidene difluoride (PVDF), detected with anti-myc antibody (1:2,000, 12 h), followed by anti-mouse HRP-conjugated antibody (1:5,000, 1 h), and visualized with ECL Plus (GE Biosciences).
Immunoblotting.
For anti-Ub immunoblotting, cells were grown to mid-log phase, pelleted, washed once with water, resuspended in buffer B (50 mM Tris, 100 mM NaCl, 0.5% Triton X-100, 0.1% deoxycholic acid, 1× Complete mini protease inhibitor cocktail), and lysed by mechanical disruption. After concentration was normalized by the Bradford assay (4a), proteins were boiled for 5 min in Tricine loading buffer (0.1 M Tris, 24% glycerol, 8% SDS, 0.2 M dithiothreitol, 0.02% Coomassie blue G-250). Proteins were separated by 11% SDS-PAGE (Tricine), transferred to PVDF, detected with anti-Ub antibody (1:800, 2 to 3 h) and anti-rabbit HRP-conjugated antibody (1:5,000, 1 h), and visualized with ECL Plus. For histone H2B ubiquitination assays, cells were transformed with Flag-HTB1 and Flag-htb1-K123R plasmids (kindly provided by M. A. Osley, University of New Mexico HSC). Cells were grown to early log phase, normalized by cell density, pelleted, washed once with water, resuspended in SDS-PAGE sample buffer, and either immediately frozen or boiled for 10 min. For MMS damage, cells were treated with MMS (0.1%) for 30 min. Proteins were separated by 14% SDS-PAGE, transferred to PVDF, detected with anti-Flag antibody (1:2,000, 2.5 h) and anti-mouse HRP-conjugated antibody (1:5,000, 1 h), and visualized with ECL Plus. For anti-H3 K4 trimethylation immunoblotting, cells were grown to mid-log phase, normalized by cell density, and lysed as previously described (11). Proteins were separated by 16% SDS-PAGE, transferred to PVDF, detected with anti-H3 trimethyllysine 4 antibody (1:5,000, 12 h) and anti-rabbit HRP-conjugated antibody (1:5,000, 1 h), and visualized with ECL Plus. Membranes used for anti-Flag and anti-H3 trimethyllysine 4 immunoblotting were stripped using Restore Western blotting stripping buffer (Pierce) according to the manufacturer's instructions, reprobed with anti-PGK antibody (0.4 μg/ml, 12 h) and anti-mouse HRP-conjugated antibody (1:10,000, 1 h), and visualized with ECL Plus. For PCNA ubiquitination assays, cells expressing Pol30-3HA were grown to mid-log phase, treated with MMS (0.02%, 1 h), pelleted, washed once with water, and flash frozen. Lysates were prepared by mechanical disruption in buffer B. Equal amounts of proteins were separated by 11% SDS-PAGE, transferred to PVDF, detected with anti-HA antibody (1:1,000, 12 h) and anti-mouse horseradish peroxidase-conjugated antibody (1:10,000, 1 h), and visualized with ECL Plus.
Ubiquitin protease assay.
BY4741 (wild type) and Doa1-TAP cells were grown to mid-log phase. Lysates were prepared from 1 liter of cells by mechanical disruption in lysis buffer (50 mM Tris, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 0.5% Triton X-100, 0.1% deoxycholic acid, 1× Complete mini protease inhibitor cocktail). The NaCl concentration of the lysates was increased to 300 mM. Total protein (300 mg) was incubated with 100 μl immunoglobulin G (IgG)-Sepharose 6 fast flow resin (Amersham) at 4°C for 5 h. The resin was washed five times with lysis buffer containing 300 mM NaCl, two times with lysis buffer containing 500 mM NaCl, and three times with ubiquitin protease buffer (50 mM Tris, 5 mM MgCl2, 2 mM DTT). Resin was resuspended in 1 ml of ubiquitin protease buffer. K48-Ub3 (2 μg; Boston Biochem) was added to a 300-μl aliquot of the resuspended resin, and the mixture was incubated at 30°C. The reactions were quenched by the addition of Tricine loading buffer and flash frozen. The reactions were analyzed by 11% SDS-PAGE (Tricine), transferred to PVDF, detected with anti-Ub antibody (1:700, 1 h), followed by anti-rabbit HRP-conjugated antibody (1:5,000, 1 h), and visualized with ECL Plus.
Real-time PCR.
Total RNA from mid-log-phase cultures of strains BY4741 (wild type) and FR1025 (doa1Δ mutant) was prepared using the MasterPure yeast RNA purification kit (Epicentre) according to the manufacturer's instructions. RNA (1 μg) was subjected to cDNA synthesis using the iScript cDNA synthesis kit (Bio-Rad). Quantitative real-time reverse transcription-PCR was carried out on 0.1 μl cDNA in a total reaction volume of 20 μl. Product accumulation was monitored by SYBR green fluorescence. Relative expression levels were determined by comparing the RAD6 or POL30 transcript level to that of the housekeeping gene TCM1. Primer sequences are available upon request.
RESULTS
Doa1 interacts with ubiquitin, ubiquitin chains, and ubiquitin-like proteins.
To identify proteins that may interact with Doa1, a yeast two-hybrid screen was carried out against an S. cerevisiae genomic library using Doa1 fused to LexA as bait. A genomic library, rather than a cDNA library, was used to ensure the representation of genes that might not be expressed under nondamage conditions. Approximately 1.5 × 106 transformants were screened, giving rise to about 200 colonies on selective media. Thirty-nine positive clones were identified by galactose-dependent growth on Leu-deficient medium and lacZ expression. The library plasmids were recovered and sequenced. Four proteins were found to interact with Doa1: Ubi4 (29 inserts recovered), Nfi1 (7), Ubx5 (4), and Ubx7 (1). The same screen was performed in the presence of MMS. Positive interactors were selected on galactose-containing, leucine-deficient media supplemented with 0.02% MMS. All other steps were carried out in a manner identical to that for the nondamage screen. In the presence of MMS, two proteins were found to interact with Doa1: Ubi4 (1) and Ubp7 (1).
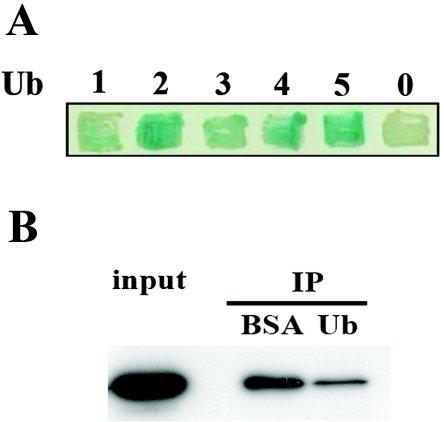
The most frequently isolated library insert in the yeast two-hybrid screens corresponded to UBI4, which encodes a stress-inducible polyubiquitin chain of five head-to-tail-linked Ub monomers. All of the isolated clones began at the N terminus of Ubi4 and contained two to four ubiquitins. To examine whether Doa1 binds Ub monomers and/or polymers, we employed a direct yeast two-hybrid approach to detect interactions between Doa1 and single Ub or Ub chains comprised of two to five head-to-tail-linked Ub moieties. Full-length and truncated constructs were amplified from the genomic UBI4 gene, cloned into pJG4-5, and then tested for interaction with Doa1-LexA. We observed that Doa1 binds both Ub monomers and Ub polymers but has a preference for binding polyubiquitin chains of two to five Ub moieties (Fig. 1A). These results agree with those reported previously by Russell and Wilkinson, who showed that Doa1 interacts with K29-linked polyubiquitin chains (35).
FIG. 1.
Doa1 binds Ub and polyubiquitin chains. (A) Direct yeast two-hybrid interaction assay between Doa1 and Ub chains. Yeast EGY48 was transformed with plasmids expressing Doa1-LexA (DNA binding domain), Ub1-5 (transcriptional activation domain), and a lacZ reporter plasmid, patched onto HC Gal-X-Gal medium, and incubated for 2 days at 30°C. The number of linked Ub monomers, 0 to 5, is indicated. (B) Doa1 binds Ub in vitro. Cell lysate (2 mg) derived from strain Doa1-myc (FR521) was incubated in the presence of ubiquitin (200 μg) or BSA (200 μg) for 2 h at 4°C. Ubiquitin agarose was then added, followed by incubation at 4°C for 3 h. Bound proteins were separated by SDS-PAGE and subjected to anti-myc Western blotting. IP, immunoprecipitation.
To confirm that Doa1 binds Ub, we incubated Ub agarose with whole-cell lysate from a DOA1-myc strain in the presence of either free Ub or BSA. Proteins that bound to the Ub agarose were analyzed by anti-myc Western blotting. Relative to the BSA control, we observed that free Ub effectively competed with the immobilized Ub and significantly reduced the level of Doa1 retained on the Ub agarose (Fig. 1B), confirming that Doa1 binds Ub.
DOA1 interacts with UBI4 and genes that encode UBX domains.
Both doa1Δ and ubi4Δ mutants are sensitive to heat, canavanine (data not shown), and DNA damage, although the damage sensitivity of the doa1Δ mutants is significantly greater (Fig. 2). The similar phenotypes of doa1Δ and ubi4Δ, as well as the observation that doa1Δ mutants have lower levels of Ub, suggested that the two proteins are at least partially functionally redundant. To gain more insight into the relationship between the two genes, we examined the DNA damage sensitivity of a doa1Δ ubi4Δ mutant. The doa1Δ ubi4Δ mutant is synergistically more sensitive to both MMS and HU than either single mutant (Fig. 2), confirming that these genes perform at least partially redundant roles. A similar genetic interaction between LUB1 and UBI4 in S. pombe was also observed (30).
FIG. 2.
DOA1 interacts with the genes encoding the proteins it is predicted to bind. Fivefold serial dilutions of 105 cells were plated on YPD plates as well as YPD plates containing MMS or HU and incubated for 3 days at 30°C. The following strains were used: wild-type (WT) (BY4741), doa1Δ (FR1025), ubi4Δ, ubp7Δ, ubx5Δ, ubx6Δ, ubx7Δ, shp1Δ, doa1Δ ubi4Δ (FR381), doa1Δ ubp7Δ (FR561), doa1Δ ubx5Δ (FR684), doa1Δ ubx6Δ (FR682), doa1Δ ubx7Δ (FR683), and doa1Δ shp1Δ (FR432) strains.
Five proteins predicted to bind to Doa1 (Ubx4, Ubx5, Ubx6, Ubx7, and Shp1) contain a UBX domain (7, 12, 43). Although the UBX domain has no significant sequence homology to Ub, it adopts a similar tertiary structure (49). Recently, it was shown that all seven S. cerevisiae UBX domain-containing proteins bind to Cdc48 (which has been shown to bind Doa1 [12]) and act to provide substrate specificity (36). We examined the genetic relationship between DOA1 and genes encoding UBX proteins, including UBX4, UBX5, UBX6, UBX7, and SHP1. Deletion of UBX5, UBX7, or SHP1 suppresses the MMS sensitivity of the doa1Δ mutant, while the ubx6Δ doa1Δ mutant is synergistically sensitive to HU (Fig. 2). Deletion of UBX4 has no effect on the sensitivity of the doa1Δ mutant. These data suggest that the UBX proteins form a complex network of interactions with Doa1 and that at least part of the function of Doa1 involves these UBX proteins.
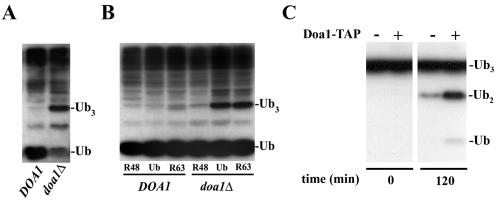
Doa1 is required to process K48-linked Ub trimers.
We next examined the endogenous levels of Ub in the absence of DOA1 (Fig. 3A). As reported previously (14), a reduced level of free Ub is observed in doa1Δ mutants. In addition, we also observed an accumulation of trimeric Ub (Ub3) in the doa1Δ mutant. The assignment of the band as Ub3 was based on both its molecular weight and comparison to a ubp14Δ mutant which is known to accumulate Ub3 (3). In order to determine the linkage topology of Ub3, the K48R and K63R mutants of Ub were overexpressed in DOA1 and doa1Δ cells (Fig. 3B). K48R Ub overexpression reduced the level of Ub3 in doa1Δ cells to that observed in DOA1 cells. However, overexpression of the K63R mutant did not affect the level of Ub3 in doa1Δ cells. Thus, it appears that the K48-linked Ub3 (K48-Ub3) accumulates in the absence of Doa1, suggesting that the activity of Doa1 is directly or indirectly related to the consumption of K48-Ub3.
FIG. 3.
Doa1 is required to process K48-linked polyubiquitin trimers (Ub3) into free Ub. (A) The doa1Δ mutant exhibited low levels of free Ub and accumulation of Ub3. Cell lysates derived from the DOA1 (W1588-4C) and doa1Δ (FR657) strains were subjected to anti-Ub Western blotting. (B) Ub3 is K48 linked. Expression of K48R Ub, but not K63R Ub, suppressed the accumulation of Ub3. The DOA1 (W1588-4C) and doa1Δ (FR657) strains were transformed with plasmids expressing Ub (YEp96), K48R Ub (YEp110), or K63R Ub (pTER103). Whole-cell lysates were prepared from mid-log-phase cells treated with CuSO4 (100 μM) for 2 h and subjected to anti-Ub Western blotting. (C) The Doa1 protein complex possesses in vitro Ub protease activity. Cell lysates derived from the wild-type (BY4741) and Doa1-TAP strains were incubated with IgG-Sepharose at 4°C for 5 h. After being washed extensively, beads were resuspended and incubated with K48-Ub3 at 30°C. Proteins were resolved by SDS-PAGE and subjected to anti-Ub Western blotting.
To further test the hypothesis that Doa1 is involved in the degradation of K48-Ub3, we immobilized Doa1-TAP from whole-cell lysates on IgG-Sepharose beads (Fig. 3C). After extensively washing the beads, we added purified K48-Ub3 and incubated the resulting mixture at 30°C. Analysis by Western blotting with anti-Ub antibody showed a significant accumulation of Ub2 and Ub monomer. In contrast, control reactions using whole-cell lysates from wild-type cells with untagged Doa1 showed virtually no degradation of the K48-Ub3. This result suggests that either Doa1 itself is a Ub protease or Doa1 copurifies with a Ub protease.
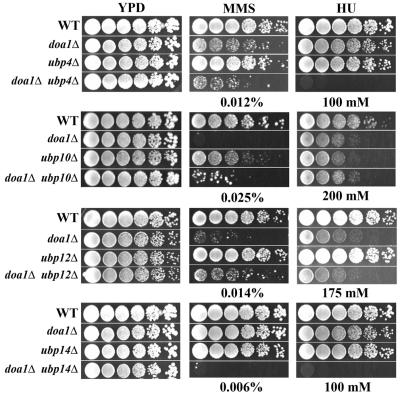
To examine whether the K48-Ub3 protease activity might be provided by an associated protein, we made and characterized double deletion mutants of DOA1 and genes that encode the known Ub proteases. Six of the 17 genes that encode Ub proteases showed genetic interactions with DOA1 (Fig. 4). Deletion of UBP7 suppressed the damage sensitivity of the doa1Δ mutant (Fig. 2). We also found that the doa1Δ ubp14Δ mutant was synergistically more sensitive to MMS and HU than the single mutants, and the doa1Δ ubp4Δ double mutant was synergistically more sensitive to HU. Deletion of UBP8, UBP10, or UBP12 suppressed the MMS sensitivity of the doa1Δ mutant. We next determined whether the deletion of any of the known Ub protease genes results in accumulation of Ub3, as seen in the doa1Δ mutant. Anti-Ub Western blots showed that in the BY4741 background, only ubp1Δ and ubp14Δ mutants accumulated Ub3 (see the supplemental material). We then examined the levels of Ub3 in doa1Δ ubp1Δ and doa1Δ ubp14Δ mutants. We anticipated that if either Ubp1 or Ubp14 provided the Doa1-dependent Ub protease activity, then deletion of the corresponding gene would not increase the accumulation of Ub3 in doa1Δ cells. However, in both cases, the double mutants accumulated more Ub3 than either single mutant (see the supplemental material), suggesting that Doa1-dependent Ub protease activity does not require either UBP1 or UBP14, at least not when deleted alone. In addition, because Doa1 and Ubp7 are predicted to physically interact and because deletion of UBP7 suppressed the MMS and HU sensitivity of the doa1Δ mutant (Fig. 2), we examined the Ub profile in the doa1Δ ubp7Δ double mutant. Deletion of UBP7 did not affect the accumulation of Ub3 in wild-type (see the supplemental material) or doa1Δ cells (data not shown). These data suggest that either Doa1 is itself a Ub protease or Doa1-dependent protease activity is provided by one of several different Ub proteases that act redundantly.
FIG. 4.
DOA1 interacts with Ub proteases. Fivefold serial dilutions of 105 cells were plated on YPD plates as well as YPD plates containing MMS or HU and incubated for 3 days at 30°C. The following strains were used: wild-type (WT) (BY4741 except for ubp10 panel, where FR1231 was used), doa1Δ (FR1025 except for ubp10 panel, where FR1229 was used), ubp4Δ, ubp10Δ (FR1232), ubp12Δ, ubp14Δ, doa1Δ ubp4Δ (FR1218), doa1Δ ubp10Δ (FR1230), doa1Δ ubp12Δ (FR1223), and doa1Δ ubp14Δ (FR1225) strains.
Doa1 provides Ub for a damage response pathway that utilizes K63-linked ubiquitin chains.
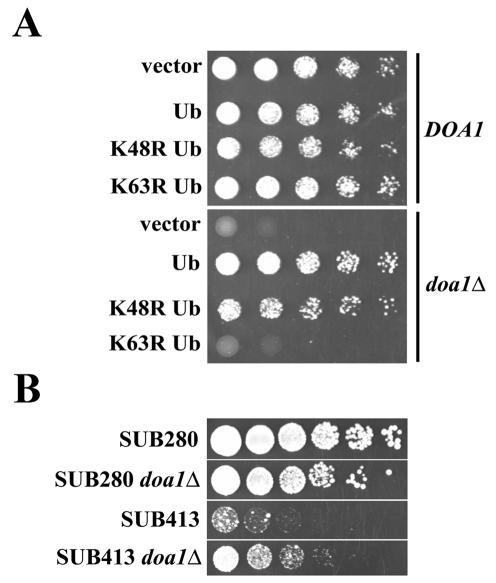
It was previously demonstrated that overexpression of Ub partially suppresses the Ub-proline-β-galactosidase degradation defect of doa1Δ cells (14). In addition, Ub expression suppressed the heat sensitivity of S. pombe lub1Δ cells (30). To examine the effect of Ub expression on the DNA damage sensitivity of doa1Δ cells, exogenous Ub was overexpressed in a doa1Δ mutant. The MMS sensitivity associated with doa1Δ cells was nearly completely complemented by the expression of Ub (Fig. 5A), consistent with the idea that Doa1 provides Ub for the DNA damage response.
FIG. 5.
Doa1 is required to maintain normal levels of K63-linked Ub used in the DNA damage response. (A) Expression of Ub as well as K48R Ub, but not K63R Ub, suppressed the MMS sensitivity associated with the doa1Δ mutant. The DOA1 (W1588-4C) and doa1Δ (FR657) strains were transformed with plasmids expressing Ub (YEp96), K48R Ub (YEp110), K63R Ub (pTER103), or an empty vector (pES12). Fivefold serial dilutions of 105 cells were plated on HC-Trp media containing 0.02% MMS. Plates were incubated for 2 days at 30°C. (B) Deletion of DOA1 in the SUB413 strain (which expresses ubiquitin solely as K63R) suppressed the sensitivity of SUB413 to MMS. Fivefold serial dilutions of 105 cells were plated on YPD media with or without 0.007% MMS and incubated for 3 days at 30°C.
We next determined whether Ub-mediated complementation of the doa1Δ mutant requires K48- or K63-linked Ub polymers (Fig. 5A). Expression of the K48R Ub mutant suppressed the MMS sensitivity of doa1Δ cells to the same extent as the expression of wild-type Ub. However, expression of K63R Ub failed to suppress the MMS sensitivity. To further investigate the relationship of Doa1 and K63-linked Ub chains, we examined the effect of deleting DOA1 in strains engineered to express the K63R mutant as the sole source of Ub (Fig. 5B). Strains SUB280 and SUB413 carry deletions of all chromosomal Ub genes (UBI1, UBI2, UBI3, and UBI4) and express Ub (SUB280) or the K63R mutant (SUB413) from a plasmid. As expected, the SUB413 strain was significantly more sensitive to MMS than the SUB280 strain; however, the sensitivity was significantly suppressed by deletion of DOA1. This suppression is consistent with a mechanism by which Doa1 degrades K48-linked Ub trimers and commits the resulting Ub to a pathway that assembles K63-linked polymers. Perhaps, if the K63-linked polymers cannot be produced (i.e., in the SUB413 strain), then the activity of Doa1 will result in the nonproductive consumption of the K48-linked polymers, and thus, the deletion of DOA1 will be beneficial to the cell.
Doa1 supplies Ub for damage-induced PCNA ubiquitination.
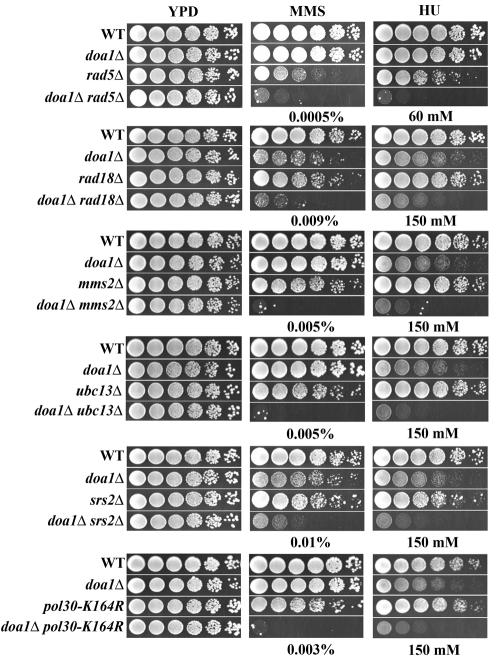
The involvement of Doa1 in providing Ub for a pathway involving K63-linked Ub chains, as well as the sensitivity of doa1Δ cells to DNA-damaging agents, prompted us to investigate the relationship between DOA1 and the genes that encode components of the PCNA ubiquitination machinery. The DNA damage sensitivities of double deletion mutants of DOA1 with RAD6, RAD18, RAD5, UBC13, MMS2, and SRS2 were determined. All of these double mutants are synergistically more sensitive to MMS and HU than the corresponding single mutants (the genetic interaction between RAD18 and DOA1 reported in reference 17 was in error and appeared to result from a suppressor mutation in the rad18Δ library strain). In addition, deletion of DOA1 in a POL30-K164R mutant (in which PCNA cannot be ubiquitinated) results in synergistic sensitivity to MMS and HU (Fig. 6). To rule out the possibility that these sensitivities result from transcriptional defects in the absence of DOA1 (see below), we examined the transcript levels of RAD6 and POL30 in wild-type and doa1Δ cells. Transcript levels for both RAD6 and POL30 were identical in each case, suggesting that the genetic interactions reflect disruption of protein function.
FIG. 6.
DOA1 interacts with genes involved in PCNA ubiquitination. Fivefold serial dilutions of 105 cells were plated on YPD plates as well as YPD plates containing MMS or HU and incubated for 3 days at 30°C. The following strains were used: wild-type (WT) (BY4741), doa1Δ (FR1025), rad5Δ, rad18Δ, mms2Δ, ubc13Δ, srs2Δ, doa1Δ rad5Δ (FR552), doa1Δ rad18Δ (FR553), doa1Δ mms2Δ (FR1021), doa1Δ ubc13Δ (FR1024), doa1Δ srs2Δ (FR376), wild-type (Y0003), doa1Δ (FR1023), pol30-K164R (Y1192), and doa1Δ pol30-K164R (FR1022) strains.
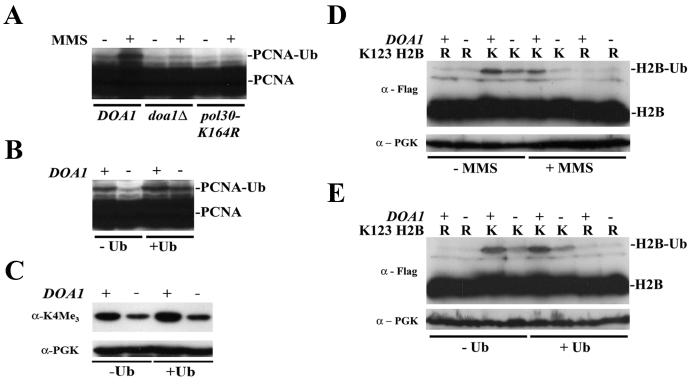
To obtain direct evidence that Doa1 is involved in PCNA ubiquitination, the modification of PCNA-HA was monitored after DNA damage. The introduction of the HA tag results in increased sensitivity to DNA damage; however, the sensitivity was significantly less than that observed in the inactive POL30-K164R mutant (data not shown). After treating cells with MMS to induce ubiquitination of PCNA (21), we observed a slower- migrating species, which we attributed to monoubiquitinated PCNA-HA (Fig. 7A). Under the same conditions, no PCNA modification was observed in a POL30-K164R mutant in which PCNA cannot be ubiquitinated. Ubiquitinated PCNA-HA is also not observed in the doa1Δ mutant, suggesting that Doa1 is required to ubiquitinate PCNA. Indeed, when the experiment was repeated in the presence of exogenously expressed Ub, we observed that PCNA was modified (Fig. 7B). Because we were unable to detect polyubiquitinated forms of PCNA, we were unable to address any role Doa1 might have in PCNA polyubiquitination. Nonetheless, the data clearly demonstrate that Doa1 plays a central role in providing Ub for PCNA modification.
FIG. 7.
Doa1-mediated Ub production is required for the ubiquitination or maintenance of monoubiquitinated PCNA and histone H2B and for the efficient trimethylation of histone H3 on K4. (A) Cell lysates from the DOA1 POL30-3HA (FR1088), doa1Δ POL30-3HA (FR1090), and pol30-K164R-3HA (FR1105) strains were subjected to anti-HA Western blotting. The addition of MMS (0.02%, 1 h) caused induction of PCNA ubiquitination in DOA1. PCNA-Ub conjugates were absent in the doa1Δ and pol30-K164R strains. (B) Ectopic expression of Ub restored the PCNA ubiquitination defect in doa1Δ cells. The DOA1 POL30-3HA (FR1088) and doa1Δ POL30-3HA (FR1090) strains were transformed with plasmids YEplac195 (control) and pCUP-Ub-195 (Ub), grown to mid-log phase, and treated with MMS (0.02%, 1 h). Cell lysates were subjected to anti-HA Western blotting. (C) The DOA1 (BY4741) and doa1Δ (FR1025) strains were transformed with plasmid YEplac195 (control) or pCUP-Ub-195 (Ub). Cell lysates were subjected to anti-H3 trimethyllysine 4 Western blotting. Protein loading was verified by reprobing the membranes with anti-PGK antibodies. (D) The DOA1 (BY4741) and doa1Δ (FR1025) strains were transformed with plasmids expressing either Flag-HTB1 or Flag-htb1-K123R, grown to early log phase, and either mock treated or treated with MMS (0.1%, 30 min). Cell lysates were subjected to anti-Flag Western blotting. (E) The DOA1 (BY4741) and doa1Δ (FR1025) strains were transformed with plasmids expressing either Flag-HTB1 or Flag-htb1-K123R and either YEplac195 (control) or pCUP-Ub-195 (Ub). Cell lysates were subjected to anti-Flag Western blotting. K indicates Flag-HTB1; R indicates Flag-htb1-K123R. The presence of H2B (H2B) or ubiquitinated H2B (H2B-Ub) is also indicated. Protein loading was verified by reprobing the membranes with anti-PGK antibodies.
Doa1 is required for histone H2B ubiquitination.
A high-throughput synthetic genetic array screen identified a synthetic lethal interaction between DOA1 and CDC73 (42), which is an essential component of the Paf1 complex that is required for H2B ubiquitination (46). We constructed a doa1Δ cdc73Δ mutant by transforming a haploid cdc73Δ strain with a deletion cassette targeted for DOA1 and observed a slow growth phenotype (Fig. 8). The mutant also exhibited synergistic sensitivity to MMS and HU. Based on these results and the genetic interaction between DOA1 and RAD6, we speculated that Doa1 might be involved in H2B ubiquitination. We thus determined the genetic relationship between DOA1 and other genes involved in H2B ubiquitination (Fig. 8). Deletion of DOA1 in bre1Δ and lge1Δ mutants resulted in synergistic sensitivity to MMS and HU. As mentioned above, deletion of UBP8 or UBP10, both histone H2B deubiquitinases (2), suppresses the sensitivity of doa1Δ cells to DNA damage. Moreover, deletion of HTB2, one of the two genes encoding histone H2B (44), also suppresses the sensitivity of doa1Δ cells to MMS. These genetic data suggest that Doa1 is involved in the ubiquitination of histone H2B.
FIG. 8.
DOA1 interacts with genes involved in the ubiquitination or maintenance of ubiquitinated H2B. Fivefold serial dilutions of 105 cells were plated on YPD plates as well as YPD plates containing MMS or HU and incubated for 3 days at 30°C. The following strains were used: wild-type (WT) (BY4741), doa1Δ (FR1025), rad6Δ, bre1Δ, lge1Δ, cdc73Δ, ubp8Δ, htb2Δ, hda1Δ, htz1Δ, rad9Δ, doa1Δ rad6Δ (FR550), doa1Δ bre1Δ (FR663), doa1Δ lge1Δ (FR1026), doa1Δ cdc73Δ (FR685), doa1Δ ubp8Δ (FR664), doa1Δ htb2Δ (FR686), doa1Δ hda1Δ (FR687), doa1Δ htz1Δ (FR688), and doa1Δ rad9Δ (FR555) strains.
To determine whether Doa1 is directly involved in H2B ubiquitination, the level of ubiquitinated H2B was examined in the absence of DOA1. Flag-HTB1 (encoding Flag-tagged H2B) and Flag-htb1-K123R (encoding Flag-tagged H2B that cannot be ubiquitinated [34]) expression plasmids were transformed into DOA1 and doa1Δ strains. Cell lysates were prepared and subjected to anti-Flag immunoblotting. Deletion of DOA1 results in a decrease in H2B-Ub levels under normal cellular conditions. After treatment with MMS, doa1Δ cells showed a profound H2B ubiquitination defect (Fig. 7D). Because MMS treatment decreases the levels of free Ub in DOA1 and doa1Δ cells to similar extents, this defect is unlikely the result of decreased levels of Ub. Indeed, expression of Ub does not affect the H2B ubiquitination defect observed in a doa1Δ strain (Fig. 7E). The data suggest that Doa1 plays a specific role in histone H2B ubiquitination that is required under normal cellular conditions and especially in the presence of DNA damage.
H2B ubiquitination regulates methylation of histone H3 at K4 by Set1. Mutants that have defects in H2B ubiquitination show reduced levels of H3 K4 methylation. Likewise, DOA1 deletion results in a reduction of H3 methylation at K4 (Fig. 7C). Importantly, as with the H2B ubiquitination defect, the methylation defect is not suppressed by Ub overexpression, suggesting that, at least for H2B ubiquitination, Doa1 does more than simply produce free cellular Ub. Because this assay detects methylation of an unmodified and endogenously expressed protein, it also suggests that the observable defects are not related to ectopic expression of modified H2B. These data support the hypothesis that Doa1 plays a specific and functionally important role that is required for H2B ubiquitination and the subsequent H3 K4 methylation.
DOA1 interacts genetically with other genes involved in transcription.
The mammalian homolog of Doa1, PLAP, associates with HDAC6 (a class II histone deacetylase), p97/VCP/Cdc48, and polyubiquitin (23, 37). In S. cerevisiae, Doa1 has been shown to bind Cdc48 (14), and in this report, we describe its binding to polyubiquitin. The homolog of HDAC6 is also conserved across species, and its S. cerevisiae homologue Hda1 is known to act at the ENA1 promoter and regulate the transcriptional response to DNA damage (47). We examined whether the mammalian PLAP-HDAC6-Cdc48 polyubiquitin complex is conserved in yeast. Deletion of HDA1 suppressed the sensitivity of doa1Δ cells to MMS (Fig. 8). However, efforts to detect a physical interaction between Hda1 and Doa1 by immunoprecipitation were unsuccessful. It seems likely that the genetic interaction reflects a functional interaction, but the physical interaction is too weak to detect or is disrupted in the presence of other proteins. Consistent with this possibility, it has been reported for mammalian cells that the interactions within the PLAP-HDAC6-Cdc48 complex are disrupted by the presence of Ub (37).
HTZ1 encodes a histone H2A variant, is essential for the recruitment of both RNA Pol II and TATA binding protein to the GAL1-10 promoters, and is thought to act in a chromatin-remodeling pathway that is partially redundant with H2B ubiquitination (25). htz1Δ cells are not only sensitive to DNA-damaging agents and heat but are also sensitive to the transcriptional inhibitors 6-azauracil and mycophenolic acid, suggesting that HTZ1 plays a role in transcriptional elongation under DNA damage and/or stress conditions. Interestingly, doa1Δ and htz1Δ cells show synergistic sensitivities to MMS and HU (Fig. 8), suggesting that Doa1 functions in a pathway that functionally overlaps with Htz1. We also determined the genetic interaction of Doa1 with Rad9. In addition to its checkpoint function, Rad9 is involved in the transcriptional induction of several genes involved in multiple DNA metabolism/repair pathways and is itself regulated by H2B ubiquitination (1, 15). We observed a synergistic sensitivity to MMS in a doa1Δ rad9Δ strain (Fig. 8). In all, the data suggest that Doa1 is required to maintain appropriate levels of H2B ubiquitination, possibly to help control the transcriptional response to DNA damage.
The doa1Δ mutant is sensitive to transcriptional inhibitors and shows a Gal defect.
In order to further test the hypothesis that Doa1 plays a role in transcription, we examined the sensitivity of a doa1Δ mutant to the transcriptional inhibitor 6-AU. 6-AU inhibits Pol II elongation and has been used to identify mutants with defective elongation due to their increased sensitivity to 6-AU (38). Consistent with the proposed function of Doa1, the doa1Δ mutant is sensitive to 6-AU (see the supplemental material). In fact, the level of sensitivity is similar to that observed for the K123R mutant of H2B, which cannot be ubiquitinated (48). Histone ubiquitination is required for the proper activation of GAL1 (19, 27). To determine whether Doa1 is involved in transcriptional regulation of the GAL locus, the growth of the doa1Δ mutant was examined on galactose-containing media. Indeed, a significant growth defect was observed in doa1Δ cells relative to wild-type cells (see the supplemental material). In all, the data suggest that Doa1 is required to ubiquitinate or maintain ubiquitinated H2B.
DISCUSSION
During normal growth, a significant portion of Ub is used to target proteins for proteasomal degradation, and it is presumably sequestered within these pathways. However, in the presence of DNA damage, Ub must quickly be made available for posttranslational modification of proteins involved in sensing, repairing, and/or tolerating the damage, such as PCNA and histone H2B. Thus, DNA damage might be expected to induce the expression of UBI4, which has conventionally been thought to be the major source of Ub for various stress responses (9). We have found that doa1Δ and ubi4Δ mutants share several phenotypes, including sensitivity to heat (9), canavanine (9), and DNA-damaging agents (17). Moreover, doa1Δ and ubi4Δ mutants are synergistically sensitive to MMS and HU, suggesting that both proteins might supply Ub for the DNA damage response. However, we observed that only the doa1Δ single mutant showed strong sensitivity to these damaging agents and that the deletion of UBI4 results in a significant sensitivity only if DOA1 is also absent. In addition, as discussed below, doa1Δ mutants show no MMS-induced ubiquitination of either PCNA or histone H2B, despite the presence of functional UBI4. Thus, we conclude that Doa1 plays the dominant role in supplying Ub for the DNA damage response.
Elements of the DNA damage response that appear to rely on Doa1 include the ubiquitination of both PCNA and histone H2B. Several observations support the conclusion that Doa1 is involved in PCNA ubiquitination. First, DOA1 deletion results in synergistic sensitivity to DNA damage when deleted in combination with other genes involved in either the monoubiquitination (RAD6 and RAD18) or polyubiquitination (RAD5, UBC13, and MMS2) of PCNA. Second, doa1Δ mutants show no damage-induced PCNA monoubiquitination. Third, while we were unable to visualize polyubiquitinated forms of PCNA, Ub expression suppresses the sensitivity of the doa1Δ mutant but only if the expressed Ub is capable of forming the K63-linked polymers required for PCNA polyubiquitination. Finally, ectopic expression of Ub suppressed the PCNA monoubiquitination defect. The simplest model consistent with the conclusion is that Doa1 is required to maintain or produce sufficient levels of Ub for PCNA modification and possibly for other facets of the damage response.
Several arguments support a more specific function for Doa1 in the ubiquitination of H2B. First, DOA1 interacts with genes involved in producing or maintaining ubiquitinated H2B (RAD6, BRE1, LGE1, CDC73, UBP8, UBP10, and HTB2) as well as genes involved in chromatin remodeling or the transcriptional response to DNA damage (HTZ1, HDA1, and RAD9). Second, Doa1 is required to maintain ubiquitinated H2B under normal conditions and especially in response to DNA damage. Third, in the absence of DOA1, histone H3 methylation at K4, which is regulated by H2B ubiquitination, is significantly reduced. Importantly, neither H2B ubiquitination nor H3 K4 methylation defects are suppressed by ectopic expression of Ub, suggesting that unlike PCNA modification, Doa1 plays a more specific and essential role in the ubiquitination of histone H2B. Finally, the doa1Δ mutant and mutants of H2B that cannot be ubiquitinated show similarly impaired transcription of the GAL1 gene and sensitivity to transcriptional inhibitors. It is also interesting to note that deletion of the gene that encodes the Doa1 binding partner Shp1 results in MMS sensitivity, and the shp1Δ mutant was also recently shown to confer sensitivity to the transcriptional inhibitor mycophenolic acid (8). These data suggest that Doa1 is required for H2B ubiquitination and/or maintenance of the modified protein, as well as the subsequent methylation of histone H3 and the transcriptional response to DNA damage.
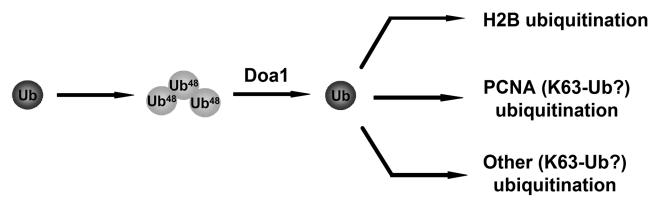
A potential mechanism describing the role of Doa1 is illustrated in Fig. 9. We suggest that when DNA is damaged, Doa1 acts to rapidly recycle Ub from proteosomal degradation pathways into pathways that modify PCNA and histone H2B and possibly other proteins. This mechanism is supported by the physical interaction between Doa1 and Ub polymers, the ability of Doa1, or an associated protein, to catalyze the cleavage of K48-Ub3, and the accumulation in vivo of K48-Ub3 in a doa1Δ mutant. A functional association between Doa1 and the proteasomal degradation pathways is further supported by the previously reported physical interaction between Doa1 and Cdc48, which together with Ufd1 and Npl4 recruit substrates to the 26S proteasome (33). Moreover, the Doa1 binding partner Shp1 has been shown to link the cellular stress response to proteasomal protein degradation (36). While Doa1 may itself provide the Ub protease activity, the genetic and physical data are also consistent with its association with multiple different Ub proteases, such as Ubp7, which may be specific for different protein targets, such as histone H2B and PCNA. In addition, specificity may be provided by other factors, such as the Ubx proteins, which may bind Doa1 and act as regulatory subunits. While further elucidation of these details requires additional experiments, it is clear that the Doa1-associated pathway is the major source of Ub for the cellular response to DNA damage and that it also plays an essential role in H2B modification.
FIG. 9.
Model for Doa1 function. The K48-linked Ub trimer, possibly produced as a peptide fusion after proteolytic degradation of ubiquitinated proteins, is processed by a complex containing Doa1, and the Ub is channeled into the DNA damage response.
Although Doa1 was shown to complement the deletion of LUB1 in S. pombe, the two genes do not appear to be functionally redundant. Lub1 appears to provide Ub specifically for proteasome-mediated protein degradation, whereas we have shown that Doa1 recycles Ub from proteosomal degradation pathways into pathways that control chromatin structure and replication. While these apparent contradictions may reflect differences between S. cerevisiae and S. pombe, the phenotypes are sufficiently distinct to suggest that despite the high sequence homology of Doa1 and Lub1 (53% similarity), their functions may have at least partially diverged. Interestingly, the mammalian homolog of Doa1, PLAP, has been shown to form a complex with HDAC6 and Cdc48, which binds polyubiquitin and copurifies with ubiquitin protease activity (36), suggesting that the functions of Doa1 might be conserved in S. cerevisiae and mammals. Further testing of the proposed model for Doa1 function, as well as its conservation in other eukaryotes, will greatly contribute to our understanding of how cells regulate ubiquitination and respond to DNA damage.
Supplementary Material
Acknowledgments
We gratefully acknowledge M. A. Osley for helpful discussions. Helpful suggestions of a referee are also acknowledged. We thank M. Hochstrasser, M. Ellison, E. Johnson, M. A. Osley, R. Rothstein, D. Finley, and S. Jentsch for providing strains and plasmids.
This work was funded by the NIH (GM068569).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aboussekhra, A., J. Vialard, D. Morrison, M. de la Torre-Ruiz, L. Cernakova, F. Fabre, and N. Lowndes. 1996. A novel role for the budding yeast RAD9 checkpoint gene in DNA damage-dependent transcription. EMBO J. 15:3912-3922. [PMC free article] [PubMed] [Google Scholar]
- 2.Amerik, A., S. Li, and M. Hochstrasser. 2000. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol. Chem. 381:981-992. [DOI] [PubMed] [Google Scholar]
- 3.Amerik, A., S. Swaminathan, B. A. Krantz, K. D. Wilkinson, and M. Hochstrasser. 1997. In vivo disassembly of free polyubiquitin chains by yeast Ubp14 modulates rates of protein degradation by the proteasome. EMBO J. 16:4826-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnason, T., and M. Ellison. 1994. Stress resistance in Saccharomyces cerevisiae is strongly correlated with assembly of a novel type of multiubiquitin chain. Mol. Cell. Biol. 14:7876-7883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Clark, M., L. Ozgur, T. Conway, J. Dispoto, S. Crooke, and J. Bomalaski. 1991. Cloning of a phospholipase A2-activating protein. Proc. Natl. Acad. Sci. USA 88:5418-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniel, J. A., M. S. Torok, Z.-W. Sun, D. Schieltz, C. D. Allis, J. R. Yates III, and P. A. Grant. 2004. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J. Biol. Chem. 279:1867-1871. [DOI] [PubMed] [Google Scholar]
- 7.Decottignies, A., A. Evain, and M. Ghislain. 2004. Binding of Cdc48p to a ubiquitin-related UBX domain from novel yeast proteins involved in intracellular proteolysis and sporulation. Yeast 21:127-139. [DOI] [PubMed] [Google Scholar]
- 8.Desmoucelles, C., B. Pinson, C. Saint-Marc, and B. Daignan-Fornier. 2002. Screening the yeast “disruptome” for mutants affecting resistance to the immunosuppressive drug, mycophenolic acid. J. Biol. Chem. 277:27036-27044. [DOI] [PubMed] [Google Scholar]
- 9.Finley, D., E. Ozkaynak, and A. Varshavsky. 1987. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell 48:1035-1046. [DOI] [PubMed] [Google Scholar]
- 10.Galan, J.-M., and R. Haguenauer-Tsapis. 1997. Ubiquitin Lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 16:5847-5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardner, R. G., Z. W. Nelson, and D. E. Gottschling. 2005. Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin. Mol. Cell. Biol. 25:6123-6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gavin, A., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. Rick, A. Michon, C. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. Heurtier, R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 13.Ghaemmaghami, S., W. Huh, K. Bower, R. Howson, A. Belle, N. Dephoure, E. O'Shea, and J. Weissman. 2003. Global analysis of protein expression in yeast. Nature 425:737-741. [DOI] [PubMed] [Google Scholar]
- 14.Ghislain, M., R. Dohmen, F. Levy, and A. Varshavsky. 1996. Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 15:4884-4899. [PMC free article] [PubMed] [Google Scholar]
- 15.Giannattasio, M., F. Lazzaro, P. Plevani, and M. Muzi-Falconi. 2005. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J. Biol. Chem. 280:9879-9886. [DOI] [PubMed] [Google Scholar]
- 16.Gueldener, U., J. Heinisch, G. J. Koehler, D. Voss, and J. H. Hegemann. 2002. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 30:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanway, D., J. K. Chin, G. Xia, G. Oshiro, E. A. Winzeler, and F. E. Romesberg. 2002. Previously uncharacterized genes in the UV- and MMS-induced DNA damage response in yeast. Proc. Natl. Acad. Sci. USA 99:10605-10610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hattori, M., H. Adachi, M. Tsujimoto, H. Arai, and K. Inoue. 1994. The catalytic subunit of bovine brain platelet-activating factor acetylhydrolase is a novel type of serine esterase. J. Biol. Chem. 269:23150-23155. [PubMed] [Google Scholar]
- 19.Henry, K. W., A. Wyce, W.-S. Lo, L. J. Duggan, N. C. T. Emre, C.-F. Kao, L. Pillus, A. Shilatifard, M. A. Osley, and S. L. Berger. 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17:2648-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hochstrasser, M., and A. Varshavsky. 1990. In vivo degradation of a transcriptional regulator: the yeast alpha 2 repressor. Cell 61:697-708. [DOI] [PubMed] [Google Scholar]
- 21.Hoege, C., B. Pfander, G. Moldovan, G. Pyrowolakis, and S. Jentsch. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419:135-141. [DOI] [PubMed] [Google Scholar]
- 22.Hofmann, R. M., and C. M. Pickart. 2001. In vitro assembly and recognition of Lys-63 polyubiquitin chains. J. Biol. Chem. 276:27936-27943. [DOI] [PubMed] [Google Scholar]
- 23.Hook, S. S., A. Orian, S. M. Cowley, and R. N. Eisenman. 2002. Histone deacetylase 6 binds polyubiquitin through its zinc finger (PAZ domain) and copurifies with deubiquitinating enzymes. Proc. Natl. Acad. Sci. USA 99:13425-13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huh, W., J. Falvo, L. Gerke, A. Carroll, R. Howson, J. Weissman, and E. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 25.Hwang, W., S. Venkatasubrahmany, A. Ianculescu, A. Tong, C. Boone, and H. Madhani. 2003. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell 11:261-266. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, E. S., P. C. M. Ma, I. M. Ota, and A. Varshavsky. 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270:17442-17456. [DOI] [PubMed] [Google Scholar]
- 27.Kao, C.-F., C. Hillyer, T. Tsukuda, K. Henry, S. Berger, and M. A. Osley. 2004. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. 18:184-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 29.Neer, E., C. Schmidt, R. Nambudripad, and T. Smith. 1994. The ancient regulatory-protein family of WD-repeat proteins. Nature 371:297-300. [DOI] [PubMed] [Google Scholar]
- 30.Ogiso, Y., R. Sugiura, T. Kamo, S. Yanagiya, Y. Lu, K. Okazaki, H. Shuntoh, and T. Kuno. 2004. Lub1 participates in ubiquitin homeostasis and stress response via maintenance of cellular ubiquitin contents in fission yeast. Mol. Cell. Biol. 24:2324-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozkaynak, E., D. Finley, M. Solomon, and A. Varshavsky. 1987. The yeast ubiquitin genes: a family of natural gene fusions. EMBO J. 6:1429-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pickart, C. 1997. Targeting of substrates to the 26S proteasome. FASEB J. 11:1055-1066. [DOI] [PubMed] [Google Scholar]
- 33.Richly, H., M. Rape, S. Braun, S. Rumpf, C. Hoege, and S. Jentsch. 2005. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell 120:73-84. [DOI] [PubMed] [Google Scholar]
- 34.Robzyk, K., J. Recht, and M. A. Osley. 2000. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287:501-504. [DOI] [PubMed] [Google Scholar]
- 35.Russell, N., and K. Wilkinson. 2004. Identification of a novel 29-linked polyubiquitin binding protein, Ufd3, using polyubiquitin chain analogues. Biochemistry 43:4844-4854. [DOI] [PubMed] [Google Scholar]
- 36.Schuberth, C., H. Richly, S. Rumpf, and A. Buchberger. 2004. Shp1 and Ubx2 are adaptors of Cdc48 involved in ubiquitin-dependent protein degradation. EMBO Rep. 5:818-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seigneurin-Berny, D., A. Verdel, S. Curtet, C. Lemercier, J. Garin, S. Rousseaux, and S. Khochbin. 2001. Identification of components of the murine histone deacetylase 6 complex: link between acetylation and ubiquitination signaling pathways. Mol. Cell. Biol. 21:8035-8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaw, R. J., and D. Reines. 2000. Saccharomyces cerevisiae transcription elongation mutants are defective in PUR5 induction in response to nucleotide depletion. Mol. Cell. Biol. 20:7427-7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sherman, F., G. R. Fink, and J. Hicks. 1983. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Stelter, P., and H. Ulrich. 2003. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425:188-191. [DOI] [PubMed] [Google Scholar]
- 41.Sung, P., S. Prakash, and L. Prakash. 1988. The RAD6 protein of Saccharomyces cerevisiae polyubiquitinates histones, and its acidic domain mediates this activity. Genes Dev. 2:1476-1485. [DOI] [PubMed] [Google Scholar]
- 42.Tong, A. H. Y., G. Lesage, G. D. Bader, H. Ding, H. Xu, X. Xin, J. Young, G. F. Berriz, R. L. Brost, M. Chang, Y. Chen, X. Cheng, G. Chua, H. Friesen, D. S. Goldberg, J. Haynes, C. Humphries, G. He, S. Hussein, L. Ke, N. Krogan, Z. Li, J. N. Levinson, H. Lu, P. Menard, C. Munyana, A. B. Parsons, O. Ryan, R. Tonikian, T. Roberts, A.-M. Sdicu, J. Shapiro, B. Sheikh, B. Suter, S. L. Wong, L. V. Zhang, H. Zhu, C. G. Burd, S. Munro, C. Sander, J. Rine, J. Greenblatt, M. Peter, A. Bretscher, G. Bell, F. P. Roth, G. W. Brown, B. Andrews, H. Bussey, and C. Boone. 2004. Global mapping of the yeast genetic interaction network. Science 303:808-813. [DOI] [PubMed] [Google Scholar]
- 43.Uetz, P., L. Giot, G. Cagney, T. Mansfield, R. Judson, J. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 44.Wallis, J., L. Hereford, and M. Grunstein. 1980. Histone H2B genes of yeast encode two different proteins. Cell 22:799-805. [DOI] [PubMed] [Google Scholar]
- 45.Wood, A., N. Krogan, J. Dover, J. Schneider, J. Heidt, M. Boateng, K. Dean, A. Golshani, Y. Zhang, J. Greenblatt, M. Johnston, and A. Shilatifard. 2003. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11:267-274. [DOI] [PubMed] [Google Scholar]
- 46.Wood, A., J. Schneider, J. Dover, M. Johnston, and A. Shilatifard. 2003. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278:34739-34742. [DOI] [PubMed] [Google Scholar]
- 47.Wu, J., N. Suka, M. Carlson, and M. Grunstein. 2001. TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol. Cell 7:117-126. [DOI] [PubMed] [Google Scholar]
- 48.Xiao, T., C.-F. Kao, N. J. Krogan, Z.-W. Sun, J. F. Greenblatt, M. A. Osley, and B. D. Strahl. 2005. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 25:637-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan, X., A. Shaw, X. Zhang, H. Kondo, J. Lally, P. Freemont, and S. Matthews. 2001. Solution structure and interaction surface of the C-terminal domain from p47: a major p97-cofactor involved in SNARE disassembly. J. Mol. Biol. 311:255-263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.