Abstract
Posttranslational histone modifications and histone variants form a unique epigenetic landscape on mammalian chromosomes where the principal epigenetic heterochromatin markers, trimethylated histone H3(K9) and the histone H2A.Z, are inversely localized in relation to each other. Trimethylated H3(K9) marks pericentromeric constitutive heterochromatin and the male Y chromosome, while H2A.Z is dramatically reduced at these chromosomal locations. Inactivation of a lysosomal and nuclear protease, cathepsin L, causes a global redistribution of epigenetic markers. In cathepsin L knockout cells, the levels of trimethylated H3(K9) decrease dramatically, concomitant with its relocation away from heterochromatin, and H2A.Z becomes enriched at pericentromeric heterochromatin and the Y chromosome. This change is also associated with global relocation of heterochromatin protein HP1 and histone H3 methyltransferase Suv39h1 away from constitutive heterochromatin; however, it does not affect DNA methylation or chromosome segregation, phenotypes commonly associated with impaired histone H3(K9) methylation. Therefore, the key constitutive heterochromatin determinants can dynamically redistribute depending on physiological context but still maintain the essential function(s) of chromosomes. Thus, our data show that cathepsin L stabilizes epigenetic heterochromatin markers on pericentromeric heterochromatin and the Y chromosome through a novel mechanism that does not involve DNA methylation or affect heterochromatin structure and operates on both somatic and sex chromosomes.
In eukaryotic cells, pericentromeric chromosomal regions do not completely decondense in the interphase and form heterochromatin, a more condensed and transcriptionally repressed type of chromatin morphologically distinct from decondensed and transcriptionally active euchromatin (17, 21, 55). Previous molecular and genetic studies had established a set of epigenetic mechanisms such as a special set of histone modifications also referred to as “histone code” (24) and DNA methylation (26) that demarcate silent heterochromatin and transcriptionally active euchromatin, separating one from the other. A key regulatory mechanism that controls heterochromatin formation is the positive feedback loop by which histone H3 trimethylated at lysine 9 (H3me3K9) recruits heterochromatin protein 1 (HP1) through direct interaction with the HP1 chromodomain (2, 27, 30). HP1, in turn, recruits histone H3(K9) methyltransferase Suv39h (24, 54) to methylate adjacent H3(K9). In addition to H3me3K9, other factors can contribute to the stable association of HP1 with chromatin (10, 59). The presence of HP1 at chromosomal loci is sufficient to induce chromatin condensation and gene repression (64). The highest concentration of HP1 and H3me3K9 is found at simple repeats in the vicinity of centromeric regions forming the largest blocks of heterochromatin, also known as pericentromeric constitutive heterochromatin. In addition, H3me3K9 is localized on chromatin clusters associated with certain types of DNA repeats at chromosomal arms (34) and late replicating DNA (67). Dose variations of the Drosophila melanogaster Suv39h homologue Su(var)3-9 (as well as of HP1 and a number of other chromatin-modifying factors) causes a remarkable position-dependent but DNA sequence-independent clonal variegation of expression of certain genes, resulting in mosaic phenotypes long known as position effect variegation (57). Double knockout of Suv39h1/h2 genes in mice causes a major depletion of trimethylated H3(K9) and another important heterochromatin marker, DNA methylation on centromeres, and affects chromosome segregation, indicating a direct role of Suv39h and histone H3 methylation in maintaining heterochromatin integrity and proper chromosomal cohesion (31, 46, 47). Interestingly, the Suv39h double knockout does not affect sex chromosomes, indicating a possible role of another histone methyltransferase(s) in establishing histone methylation on sex chromosomes (47).
The ability of heterochromatin to spread in cis and inactivate normally active genes in a position-specific and sequence-independent manner requires special mechanisms regulating histone H3 methylation on chromosomes. Interestingly, one essential histone H2A variant, H2A.Z, has been previously found to serve as a heterochromatin barrier in Saccharomyces (35) and as a heterochromatin- and H3 trimethylation-promoting protein in Drosophila (63). In vertebrate cells, its localization and role in heterochromatin spreading is ambiguous. H2A.Z is a core but minor component of pericentromeric heterochromatin in mouse cells (I. K. Greaves and D. J. Tremethick, submitted). Depletion of H2A.Z leads to the loss of sister chromatid cohesion and defects in chromosome segregation in mouse and human cells (53). The amount of H2A.Z at constitutive heterochromatin can vary depending on the differentiation state. Pericentromeric heterochromatin of extraembryonic tissue cells becomes highly enriched in H2A.Z during early mouse development, while the inactive X chromosome remains devoid of this histone variant (52). Interestingly, pericentromeric heterochromatin in these cells is depleted of H3me3K9 (34). The significance of this inverse correlation between H2A.Z and H3(K9) methylation and whether it is specific for these cells remains to be determined. Consistent with its heterochromatin functioning, H2A.Z induces the formation of the 30-nm chromatin fiber in vitro and confers a slightly (twofold) higher affinity for HP1α than unmodified H2A-containing nucleosomes (10). Whether this histone variant is linked to histone H3 methylation and heterochromatin formation in vivo remains to be understood.
To determine the spatial relationship between H3me3K9 and H2A.Z in chromatin, we conducted a microscopic immunofluorescence analysis of these two markers' distribution on human and mouse chromosomes. Our studies revealed a global alternate distribution of H2A.Z and H3me3K9 on both mouse and human chromosomes, indicating that such a spatial relationship has a universal nature. The domains defined by this alternating pattern on somatic chromosomes and the unique epigenetic landscape of the Y chromosome provide characteristic landmarks against which the topographic changes in epigenetic chromatin markers and their effect on chromatin structure and activity can be easily analyzed. Using these landmarks, we found that either partial inhibition or a complete knockout of a lysosomal and nuclear protease cathepsin L leads to a dramatic reduction of H3me3K9 on centromeric chromatin and a global repositioning of H2A.Z to the sites vacated by trimethylated H3(K9). Cathepsin L knockout also causes complete reversal of the epigenetic landscape on the male Y chromosome, including loss of H3me3K9 and intrusion of H2A.Z. However, it does not damage DNA methylation or chromosome segregation, the processes affected by Suv39h knockout (31, 47). Thus, our data uncover a novel function of cathepsin L that inversely regulates chromosomal positioning of the principle epigenetic markers H3me3K9 and H2A.Z on mammalian chromosomes in a pathway clearly distinct from that affected by Suv39h inactivation. Our findings are also consistent with the compensatory role of H2A.Z in maintaining constitutive heterochromatin in the absence of cathepsin L and upon the deficiency in histone H3(K9) trimethylation.
MATERIALS AND METHODS
Antibodies and chemicals.
All chemicals were purchased from Fisher Scientific (Pittsburgh, PA), unless otherwise noted. Antibodies against dimethyl-H3(K9), HP1α, HP1β, acetyl-H3(K9, 14), and acetyl-H4 (K12) were described previously (23), as were anti-H2A.Z antibodies (52) and anti-trimethyl-H4 (Lys20) antibodies (28). Antibodies against phosphorylated H3 (Ser10), trimethyl-H3(K9), and mouse Suv39h1 protein were purchased from Upstate (Lake Placid, NY), antibodies against Xpress tag were purchased from Invitrogen (Carlsbad, CA), 5-methylcytosine was purchased from Calbiochem (San Diego, CA), actin beta was purchased from Sigma (St. Louis, MO), and cathepsin L was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Secondary AlexaFluor-conjugated antibodies from Molecular Probes, Inc. (Molecular Probes, Eugene, OR) were used for immunofluorescence, and horseradish peroxidase-conjugated antibodies from Jackson ImmunoResearch (West Grove, PA) were used for Western blot analysis. See Table S1 in the supplemental material for additional information on antibodies and dilutions.
Cell lines and transfection.
NIH 3T3 mouse fibroblasts (ATCC CRL-1658) were cultured in Dulbecco's minimal essential medium containing 10% newborn calf serum (Gibco) as described previously (22). MENT-expressing cells (MENT-63, MENT-ov16) and control G418-resistant cells were described previously (23). Cathepsin L knockout cells, control CatL-expressing cells, and primary mouse fibroblasts (20) were cultured in HEPES-buffered Dulbecco's minimal essential medium supplied with 1 mM sodium pyruvate (VWR), 10% newborn calf serum, 1× penicillin/streptomycin/glutamine (Gibco), and 50 μM 2-mercaptoethanol (Sigma). Transfection experiments were performed using GenePORTER 2 reagent (Gene Therapy Systems) according to the manufacturer's instructions. Cells were analyzed for expression of target protein 48 to 60 h after transfection. In Suv39h1 expression experiments, 1 μM nocodazole was added 12 h after transfection start to prevent transport of tagged Suv39h1 protein to aggresomes. Since transfected cells do not enter mitosis within the time scale of the experiment, this treatment mostly targets aggresomal protein transfer and not mitotic spindle assembly (13, 25). Nocodazole treatment did not alter the nuclear localization of tagged Suv39h1 per se in transfected cells.
Isolation of metaphase chromosomes and chromosome count.
Metaphase chromosomes were isolated essentially as described previously (61). Briefly, cells were arrested in metaphase by incubation with 100 ng/ml Colcemid (Invitrogen) for various times dependent on cell culture and collected by trypsinization, followed by treatment with hypotonic solution (0.0375 M KCl, 0.4% sodium butyrate) for 13 min at a concentration of 5 · 105 cells/ml. Cell suspensions were then spun down onto poly-l-lysine-coated (Sigma) microscope slides by centrifugation at 1,900 rpm for 10 min in a cytocentrifuge (Cytopro 7620; Wescor). The microscope slides were then incubated in KCM buffer (120 mM KCl, 20 mM NaCl, 10 mM Tris-HCl, pH 7.7, 0.1% Triton X-100) for 10 min at room temperature (RT) and fixed with 2% paraformaldehyde (Electron Microscopy Science, Hatfield, PA) in phosphate-buffered saline (PBS) to preserve the histone association with chromatin. Resulting metaphase spreads were stained with antibodies against modified histone tails and analyzed using an immunofluorescence microscope as described below. For chromosome count, 100 metaphase spreads were randomly selected from each cell type preparation, and the number of chromosomes per karyotype was assessed using Image Pro Plus 4.1 software (Media Cybernetics, Inc., Silver Spring, MD).
Immunofluorescence microscopy and image analysis.
For analysis of interphase nuclei, cells were grown on cover glasses to about 90 to 95% confluence and then fixed with a 1:1 acetone-methanol mixture for 30 min at −20°C. Cover glasses with whole cells or microscope slides with metaphase chromosome spreads were stained with primary and fluorescent secondary antibodies as described previously (22, 23) (see Table S1 in the supplemental material).
The protocol for staining with antibodies against 5-methylcytosine was optimized from several published protocols (4, 37) as follows. Cytospin preparations with metaphase spreads (either unstained or stained with antibodies against histone N tail modifications) were washed twice with double-distilled water, fixed in a 3:1 methanol-acetic acid mixture for 15 min, and dried overnight at RT in the dark. Dry slides were rehydrated for 30 min at 37°C in PBS containing 0.05% Tween 20 and subjected to acid hydrolysis in the preheated 2 N HCl for 20 min at 37°C. Acid was neutralized by washing slides three times for 10 min in 0.1 M sodium borate buffer, pH 8.4, followed by a double wash in PBS with 0.1% Tween 20. Slides were then blocked with 1% bovine serum albumin (Jackson ImmunoResearch) in PBS-0.1% Tween for 30 min at RT, incubated with 1 μg/ml primary mouse anti-5-methylcytosine antibodies for 1 h at 37°C in a humid chamber, washed three times for 5 min in PBS, and then incubated with secondary anti-mouse Alexa Fluor-conjugated antibodies for 1 h at RT, followed by another triple PBS wash (see Table S1 in the supplemental material for antibody dilutions). All samples were counterstained with Hoechst 33258 DNA stain as described previously (22).
Fluorescence microscopy was performed using a Nikon Eclipse microscope with a 60× or 100× plan apo lens. For deconvolution, 16-bit z-axis slices were captured at 0.4-μm steps with a cooled charge-coupled device camera and iteratively deconvolved (usually 20 cycles) using AutoDeblur software (AutoQuant Imaging, Watervliet, NY) as described (22). Deconvolved images of nuclei in the figures represent a single z-axis slice through the center of the nucleus. Image intensity analysis was performed with Image-Pro Plus software.
FISH on metaphase chromosomes.
Fluorescent in situ hybridization (FISH) on metaphase chromosomes was performed essentially as described previously (36, 62), with minor modifications. See supplementary methods in the supplemental material for a detailed description.
Isolation of cell nuclei, total cell extracts, and Western analysis.
Purification of cell nuclei from mouse fibroblasts, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and Western blotting were conducted essentially as described previously (16). Briefly, attached cells were washed once in PBS and lysed in RSB-HEPES buffer (10 mM NaCl, 3 mM MgCl2, 10 mM HEPES, pH 7.5) containing 0.5% Igepal CA-630 (Sigma), followed by centrifugation for 10 min at 7,600 × g. The supernatant was discarded, and the nuclear pellet was used for Western blotting. Detection of HP1 proteins and histones was carried out as described previously (23). For analysis of Suv39h1 protein, total Laemmli cell lysates were prepared by resuspending cells in 1× Laemmli buffer supplemented with 1 mM phenylmethylsulfonyl fluoride, 5 mM dithiothreitol, 1× proteinase inhibitor cocktail, 5 mM sodium vanadate, and 25 mM sodium pyruvate.
Isolation of total RNA, reverse transcription, and semiquantitative PCR.
Total RNA from CLI and CLO cells was isolated using Trizol reagent (Invitrogen) according to the manufacturer's instructions, and cDNA was prepared using the First Strand cDNA synthesis kit (Fermentas, Hanover, MD). Per 25 μl of PCR mixture, 1.5 μl of cDNA (corresponding to ∼0.4 μg of starting total RNA) was used with oligonucleotides specific for Suv39h1 mRNA (forward, 5′-CAACCTTGATGAGCGACTACC-3′; reverse, 5′-TTAAGTGCAATGCCTGATGG-3′). To ensure quantitative PCR, serial dilutions of cDNA samples were prepared, ranging from 1:5 to 1:125, and all samples were analyzed in parallel duplicate experiments. Reverse transcription-PCR (RT-PCR) analysis of housekeeping genes RPS9 (ribosomal protein S9) and beta-actin was used as a control to adjust the amount of cDNA. PCR conditions were as follows: for Suv39h1, 35 cycles, annealing at 56°C; for RPS9, 18 cycles, annealing at 56°C (forward oligonucleotide, 5′-CCTAGCGAGGTCAACATGC-3′; reverse oligonucleotide, 5′-TCTTGGCCAGGGTAAACTTG-3′); for beta-actin, 18 cycles, annealing at 57°C (forward 5′ oligonucleotide, 5′,CAACTGGGACGACATGGAG-3′; reverse oligonucleotide, 5′-TAGATGGGCACAGTGTGGG-3′). PCR products were analyzed by electrophoresis on a 2% agarose gel stained with ethidium bromide.
RESULTS
Alternate distribution of trimethylated H3 and H2A.Z at the Y chromosome and pericentromeric heterochromatin.
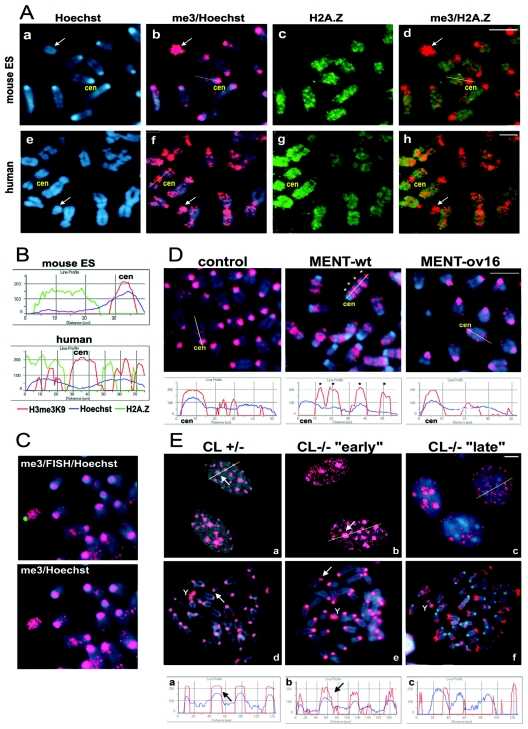
On mouse chromosomes, preferential staining of AT-rich major satellite DNA with Hoechst 33258 provides bright foci at pericentromeric heterochromatin (18). We employed these foci as clear visible landmarks for analysis of localization of H3me3K9 and H2A.Z using immunofluorescent staining of metaphase chromosomes. Chromosomes from mouse embryonic stem (ES)-derived fibroblasts showed a strong mostly centromeric staining by anti-H3me3K9 overlapping with the Hoechst-positive foci (Fig. 1A, subpanel b), consistent with the pattern of this histone modification observed in other studies (34). In sharp contrast, H2A.Z was preferentially localized on noncentromeric chromatin without any detectable overlap with histone H3 methylation on chromosome arms or Hoechst foci (Fig. 1A, subpanels c and d, and Fig. 1B). We also observed a very similar nonoverlapping H3me3K9/H2A.Z pattern on chromosomes from cultured NIH 3T3 mouse fibroblasts (see Fig. S1 in the supplemental material). Human centromeres differ from those of the mouse in the sequence of centromeric DNA repeats, are not preferentially stained by Hoechst, and in a sharp contrast to mouse acrocentric chromosomes, mostly occupy metacentric positions (Fig. 1A, subpanel e). However, despite such sequence and morphology differences, trimethylated H3(K9) again occupied pericentromeric heterochromatin as well as certain domains on chromosome arms, while H2A.Z was mostly excluded from these locations on human chromosomes (Fig. 1A, subpanels f to h, and Fig. 1B). Thus, both human and mouse chromosomes contain a mutually alternating pattern of H3me3K9/H2A.Z distribution, indicating that this spatial chromosomal relationship between the two markers is universal for mammalian cells.
FIG. 1.
Mutually alternating distribution of H3me3K9 and H2A.Z on mammalian chromosomes is altered upon cathepsin L inactivation. (A) Immunofluorescence of metaphase chromosome spreads from mouse ES-derived fibroblasts (mouse ES) and human foreskin fibroblasts (human). Metaphase chromosomes were stained with antibodies against H3me3K9 (me3, red) and H2A.Z (green) as well as Hoechst 33258 DNA stain (blue). cen, centromeres. Arrows indicate the Y chromosome. Bar, 5 μm. (B) Fluorescence intensity profiles of H3me3K9, H2A.Z, and Hoechst plotted along the chromosome paths (indicated by yellow lines in panel A, subpanels b, d, f, and h). Note the “out-of-phase” chromosomal distribution of H2A.Z and H3me3K9 on both mouse and human chromosomes as well as accumulation of H3me3K9 on centromeric regions. (C) FISH of mouse metaphase chromosomes coupled with H3me3K9 fluorescence. Chromosomes were first stained with anti-H3me3K9 antibodies (red) and then subjected to FISH with Y-specific probe (green) and counterstained with Hoechst (blue). (D) H3me3K9 immunofluorescence on metaphase chromosomes from control cells (control) and cells expressing wild-type MENT (MENT-wt) and ov-swap MENT (MENT-ov16). (E) H3me3K9 immunofluorescence of interphase nuclei (subpanels a to c) and metaphase chromosomes (subpanels d to f) from cathepsin L heterozygous (CL+/−) and cathepsin L knockout (CL−/−) mouse embryonic fibroblasts. “Early” and “late” indicate passages of CL−/− cells (see text). Red, H3me3K9; blue, Hoechst. Asterisks indicate bands of H3me3K9 accumulation on MENT-wt chromosome arms. Cen, centromeres; Y, Y chromosome. Arrows on panel E point to pericentromeric chromatin. Profiles illustrate the spatial fluorescence intensity changes of H3me3K9 and Hoechst plotted along the paths shown by yellow lines. Note the accumulation of H3me3K9 in centromeric regions of control, MENT-ov16, CL+/−, and CL−/− “early” fibroblasts but not in MENT-wt or CL−/− “late” fibroblasts. Bar, 5 μm.
We noticed that within the whole-chromosome sets from either human or mouse male cells, one chromosome was intensely stained by H3me3K9 throughout the whole chromosome and contained almost no H2A.Z (Fig. 1A). The morphology and the uniqueness of this chromosome in the metaphase spreads indicated that this is the Y chromosome. We confirmed the identity of the mouse Y chromosome using FISH (Fig. 1C) with the mouse Y chromosome-specific probe pErs5 (5). Thus, the Y chromosome, the most heterochromatic chromosome in the mammalian male cells (44), shares the enrichment in histone H3 trimethylation and the absence of H2A.Z with the pericentromeric heterochromatin and facultative heterochromatin on the inactive female X chromosome in mammalian cells (7). This indicates that there is a general molecular mechanism that establishes the chromosome-wide histone modification landscape on pericentromeric regions and sex chromosomes in mammals.
Chromosomal repositioning of H3me3K9 and H2A.Z by a nuclear cathepsin L inhibitor, MENT.
We then sought to determine whether the alternate global H3me3K9/H2A.Z distribution could be coordinately controlled on chromosomes. Previously, we showed that MENT, a nuclear serpin (serine and cysteine protease inhibitor) involved in chromatin condensation during cell differentiation, causes large-scale remodeling of histone H3 methylation in the interphase nuclei, characterized by formation of new noncentromeric H3me3K9 foci (23). Studies of interphase nuclei did not allow us to assign these foci to the known chromosomal locations; therefore, it was impossible to convincingly demonstrate whether MENT indeed physically changed the position of H3(K9) trimethylation along the DNA. To investigate this issue, we conducted chromosome immunofluorescence experiments with chromosomes from NIH 3T3 cells stably expressing MENT (MENT-wt) as well as with control cells transfected with empty vector (23). The pattern of H3me3K9 on chromosomes from the control cells was similar to the one found on normal mouse chromosomes described above (Fig. 1D, control). In sharp contrast, chromosomes from MENT-expressing cells exhibited a dramatic rearrangement of this marker (Fig. 1D, MENT-wt). H3me3K9 relocated from the pericentromeric regions to the chromosome arms. These chromosomal studies support our previous conclusions that MENT physically rearranges H3me3K9 along DNA.
Previous studies indicated that the MENT interaction with modified histones in the cells involves its reactive center loop (RCL) domain (23). Our in vitro experiments revealed that the RCL is directly involved in MENT's ability to condense chromatin (58). The RCL is also indispensable for direct interaction with and specific inhibition of the protease cathepsin L in vitro (22). To determine if this bifunctional MENT domain is important for the observed rearrangement of epigenetic markers by MENT, we constructed a cell line that stably expressed MENT with RCL inactivated by an “ovalbumin swap” mutation (MENT-ov16), which disrupts protease inhibition and alters chromatin binding by MENT (23, 58). Metaphase chromosomes from the MENT-ov16 cell line showed a similar pattern of H3me3K9 distribution to the control cells (Fig. 1D). Thus, the studies of metaphase chromosomes confirmed the importance of the MENT RCL not only for its association with chromatin but also for specific rearrangement of H3me3K9. The requirement for an intact inhibitory RCL domain of MENT to alter chromosomal distribution of H3me3K9 raised an interesting possibility that this effect may be mediated through cathepsin L inhibition. Cathepsin L is a well-recognized lysosomal protease (3); however, recent studies revealed that some cathepsin L isoforms could enter the cell nucleus and target nuclear proteins (14). MENT acts as a potent and very specific irreversible cathepsin L inhibitor in vitro (22), and using radioactive in vivo cathepsin active site labeling by a specific radioactive peptide (65), we found that stable MENT expression in NIH 3T3 cells interferes with direct cathepsin L labeling (see Fig. S2 in the supplemental material). This and previous findings indicated that MENT can interact with cathepsin L in live cells and further supported the hypothesis that chromatin remodeling by MENT, particularly redistribution of H3me3K9, may involve cathepsin L inhibition.
Primary cathepsin L knockout mouse fibroblasts exhibit destabilized levels of H3 trimethylation.
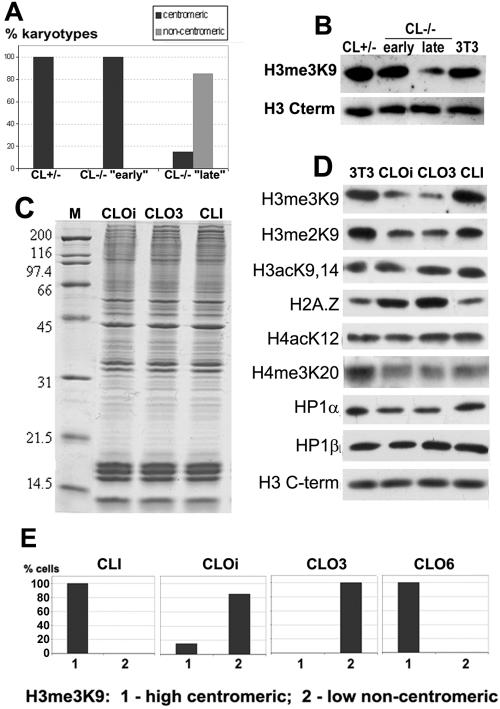
We then sought to determine whether cathepsin L inactivation by itself could affect the topography of chromosomal epigenetic markers without being mediated by MENT. Cathepsin L knockout mice are viable; however, they exhibit a variety of mild abnormalities in different tissues (40, 56, 66). Some of these abnormalities, such as male fertility problems in cathepsin L-deficient mice (66; A. Rudensky, personal communication) could be attributed to the changes in chromatin regulation (see below). We therefore wondered whether the cathepsin L knockout affects the stability and/or localization of the epigenetic chromatin markers. We analyzed the levels and distribution of H3me3K9 in primary mouse embryonic cathepsin L knockout fibroblasts (CL−/−) as well as in heterozygous fibroblasts (CL+/−) as a control. The two cell cultures initially exhibited similar H3me3K9 pericentromeric nuclear patterns (Fig. 1E, subpanels a and b) and protein levels (Fig. 2B, compare CL+/− and CL−/− “early”). However, after several (less than 10) passages, the H3me3K9 phenotype dramatically changed in CL−/− cells: the H3 trimethylation levels dropped (Fig. 2B, CL−/− “late”), and the majority of the cells revealed noncentromeric distribution of H3me3K9 (Fig. 1E, subpanel c). Immunofluorescence analysis of chromosomes from CL+/− and CL−/− cells revealed that, while heterozygous CL+/− and “early” CL−/− cells contained all chromosomes with high-level pericentromeric H3me3K9 (Fig. 1E, subpanels d and e), the majority (80 to 90%) of chromosomes from “late” CL−/− cells had a striking depletion of these markers from the centromeres (Fig. 1E, subpanel f and Fig. 2A, histogram), similar to that observed with MENT-expressing cells, and the remaining H3me3K9 was relocated mainly to telomeric chromosome ends. The Y chromosome has also lost its characteristic trimethylated H3 topography in these later passages of CL−/− cells (Fig. 1E). The observed epigenetic switch was maintained during further CL−/− cell divisions (data not shown). The CL+/− cells maintained 100% high-level pericentromeric H3me3K9 at all times. These findings indicate that cathepsin L stabilizes the cellular levels and localization of H3me3K9. When the cathepsin L-deficient cells proliferate in vitro, the destabilization of cellular epigenetic pathways normally controlled by cathepsin L rapidly leads to clonal epigenetic instability and loss of H3 trimethylation after several rounds of cell division.
FIG. 2.
Epigenetic chromatin markers in cathepsin L knockout cells. (A) Frequency of H3me3K9 localization phenotypes on CL+/− (CL+/−) and CL−/− (CL−/− “early” and “late,” see text) chromosomes. H3me3K9 chromosomal distribution in randomly picked metaphase spreads (100 from each cell line) was assessed by immunofluorescence. The percentage of karyotypes with either centromeric (e.g., Fig. 1E, subpanels d and e) or noncentromeric (e.g., Fig. 1E, subpanel f) H3me3K9 in each culture is represented as a bar. Note the 100% centromeric H3me3K9 in CL+/− and CL−/− “early” and the prevalence of noncentromeric H3me3K9 in CL−/− “late.” (B to D) Nuclear proteins of CL−/− and CL+/− fibroblasts as well as NIH 3T3 (3T3), CLO-initial (CLOi), CLO3, and CLI cells were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (C) and Western blotting (B and D) with antibodies against H3me3K9, H3me2K9, H3acK9,14, H2A.Z, H4acK12, H4me3K20, HP1α, and HP1β as indicated. Equal amounts of nuclear protein in each well were confirmed by Coomassie blue R-250 staining (e.g., see panel C) and by Western blotting with antibody against H3 C terminus (H3 C-term). (E) Frequency of H3me3K9 level/localization phenotypes in CLI, CLOi, CLO3, and CLO6 cultures. H3me3K9 nuclear levels and distribution were assessed by immunofluorescence. Bars represent percentages of cells with either high-level centromeric or low-level noncentromeric H3me3K9. More than 200 nuclei were scored in each culture. Note that there was only one H3me3K9 phenotype in CLI, CLO3, and CLO6 cultures and the mixed population of CLOi.
Cathepsin L knockout cell lines exhibit an altered pattern of histone H3 methylation that is restored by cathepsin L expression.
Since the primary cathepsin L knockout fibroblasts displayed epigenetic instability and primary cells tend to enter quiescence, we decided to use immortalized cell lines to further characterize cellular epigenetic phenotypes associated with cathepsin L deficiency. We obtained mouse embryonic cathepsin L knockout fibroblast cells immortalized with large T antigen (further called CLO, “cathepsin L out”) as well as control cathepsin L-expressing cells (CLI, “cathepsin L in”) generated by transducing CLO with vector containing cathepsin L cDNA (20). First, we thoroughly characterized the two cell lines and found that, despite the complete absence of both the cathepsin L protein and cathepsin L-specific activity as determined by Western blotting (see Fig. S3A in the supplemental material) and protease active site labeling (see Fig. S3B in the supplemental material), the two cell lines shared morphological similarity, equal growth rates (see Fig. S3C in the supplemental material), similar overall protein composition (Fig. 2C, compare CLI and CLOi [i stands for “initial”; see also below]), and similar composition of major histones analyzed by reverse-phase high-performance liquid chromatography (see Fig. S4 in the supplemental material). This similarity between the cathepsin L knockout and cathepsin L-expressing cells is consistent with the mild phenotypic changes inflicted by cathepsin L knockout (40, 56, 66).
Further immunofluorescence analysis revealed a strikingly variegating level of H3me3K9 in CLOi cells that precisely correlated with altered nuclear distribution of this marker (Fig. 2E, histograms) as well as other chromatin proteins associated with it. Similar to the primary CL−/− “late” fibroblasts, the majority (85%) of CLOi cells had significantly diminished levels of H3me3K9 (Fig. 2D), which coincided with localization of the remaining H3me3K9 to noncentromeric regions in the cell nuclei (Fig. 2E and Fig. 3A, subpanel f). The rest (15%) of the cells in the CLOi population exhibited H3me3K9 localization and levels similar to those of the CLI fibroblasts (Fig. 2E). We then asked whether this heterogeneity results from a clonal instability of epigenetic pattern in CLOi. Therefore, we isolated a total of 14 single-cell-derived colonies from the initial CLO cell population (CLOi) and analyzed these clones for the levels and localization of H3me3K9. Two types of colonies with a dramatically different H3me3K9 pattern were detected, consistent with the two phenotypes in the initial CLOi cell population: one type (e.g., CLO clone 6, CLO6) with a high-level centromeric H3me3K9 and the other type (e.g., CLO clone 3, CLO3) with a low-level noncentromeric H3me3K9 (Fig. 2E). Detailed immunofluorescence analysis of CLO3 nuclei further confirmed that this clone contained 100% cells with noncentromeric H3me3K9 and thus represented the majority of the CLOi cells.
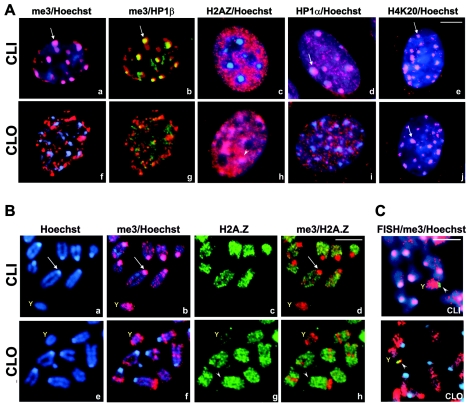
FIG. 3.
Nuclear and chromosomal localization of epigenetic chromatin markers in CLI and CLO (CLO3) cells. (A) Immunofluorescence of chromatin markers in CLI (subpanels a to e) and CLO (subpanels f to j) cell nuclei. Cells were fixed and stained with antibodies against H3me3K9 (red, subpanels a, b, f, and g), HP1β (green, subpanels b and g), HP1α (red, subpanels d and i), H2A.Z (red, subpanels c and h), H4me3K20 (red, subpanels e and j) and counterstained with Hoechst 33258 (blue). Arrows point to the colocalization of epigenetic chromatin markers with pericentromeric heterochromatin (chromocenters) in CLI. Arrowheads indicate accumulation of H2A.Z on heterochromatin of CLO cells. (B) Immunofluorescence of chromosomes from CLI (a to d) and CLO (e to h) cells. Red, H3me3K9; green, H2A.Z; blue, Hoechst 33258; Y, Y chromosome. (C) FISH (green) coupled with H3me3K9 staining (red) of CLI and CLO chromosomes. Arrowheads point at the double FISH signals. Bars, 5 μm.
In a remarkable contrast, all CLI cells always contained uniformly high levels of completely centromeric H3me3K9 (Fig. 2E and Fig. 3A, subpanel a), typical for the majority of mouse cells characterized earlier. We thus concluded that cathepsin L expression was sufficient to prevent the variegation of trimethyl-H3(K9) caused by the protease knockout.
Cathepsin L knockout affects the nuclear localization of epigenetic chromatin markers functionally linked to histone H3 methylation.
We next analyzed levels and localization of several important chromatin epigenetic markers linked to histone H3 methylation. Figure 2D shows the results of Western blot analyses of total nuclear protein isolated from CLI, CLOi, CLO3, and NIH 3T3 cells. Most chromatin markers in CLO3 exhibited levels similar to those observed in the initial CLO population (Fig. 2D, compare CLOi and CLO3). This includes a dramatic decrease in the levels of H3me3K9 (10- to 12-fold as estimated by antibody titration) and also a significant decrease in H3me2K9. Notably, levels of the majority of the rest of the chromatin markers remained unchanged.
Concomitant with down-regulation of H3 trimethylation, we observed an increase in the essential H2A.Z histone isoform (three- to fourfold by titration) in CLOi and CLO3 cells (Fig. 2D). Moreover, while H2A.Z localized mostly to euchromatin and avoided pericentromeric heterochromatin in CLI nuclei (Fig. 3A, subpanel c), CLO3 cells revealed a dramatically altered pattern, with H2A.Z occupying the Hoechst-positive heterochromatin (Fig. 3A, subpanel h). Earlier in this paper we showed that H3me3K9 and H2A.Z exhibit a conserved alternate distribution on mouse and human chromosomes. The increase in H2A.Z that coincides with the decrease in H3me3K9 levels together with their inverse relocation with respect to pericentromeric heterochromatin in CLO cells (H3me3K9 out, H2A.Z in) and very little changes in chromatin morphology further support possible compensating functions of these two chromatin markers.
We also analyzed the CLOi, CLO3, and CLI cell cultures for several other factors correlated with H3 methylation, HP1α, HP1β, and trimethyl-H4 Lys20 (H4me3K20), as well as anticorrelating factors, acetyl-H3 Lys9,14 (H3acK9,14), and acetyl-H4 Lys12 (H4acK12). We found that, like H3me3K9, the levels (Fig. 2D) and localization (Fig. 3A) of HP1 proteins were stable in CLI fibroblasts, with precise colocalization of H3me3K9 and HP1β at the pericentromeric regions (Fig. 3A, subpanel b). HP1α was also localized to the pericentromeric Hoechst-positive heterochromatin in CLI cells (Fig. 3A, subpanel d). In contrast, in CLOi cells, the HP1 nuclear distribution varied dramatically but in a strikingly precise correlation with H3me3K9 (see Fig. S5A in the supplemental material). A smaller fraction (15%) of CLOi cells exhibited patterns of H3me3K9 and HP1 similar to those in CLI nuclei. However, the majority of CLOi cells and all CLO3 cells revealed noncentromeric localization of both HP1α and β proteins and H3me3K9, with almost no overlap between HP1 and H3me3K9 (Fig. 3A, subpanels g and i; see also profiles on Fig. S5B in the supplemental material). CLO3 thus represented the majority of CLOi cells and was used for further studies. In contrast, H4me3K20 did not lose its centromeric association in cathepsin L knockout cells (Fig. 3A, compare subpanels e and j). Neither this marker nor histone H3(K9,14) acetylation or H3(S10) phosphorylation showed any positive or negative correlation with H3me3K9 (Fig. 2D and data not shown), indicating that separate mechanisms control these epigenetic markers and that cathepsin L inactivation specifically affects histone H3(K9) methylation.
Cathepsin L knockout alters the distribution pattern of histone H3(K9) methylation and H2A.Z on centromeres and reverses the epigenetic landscape of the Y chromosome.
We next decided to examine whether the changes in the histone tail modifications observed in the interphase nuclei persist on metaphase chromosomes. Consistent with the findings with normal mouse cells described earlier, the trimethylated H3(K9) resided mostly in pericentromeric regions on chromosomes from CLI cells (Fig. 3B, subpanel b). In contrast, chromosomes from CLO3 cells (also called CLO) revealed a dramatic reduction of H3me3K9 over the centromeric regions and its relocation to chromosome arms, mainly to distal telomeric ends of chromosomes (Fig. 3B, subpanel f), similar to the pattern observed on “late” CL−/− chromosomes (Fig. 1E, subpanel f). Concomitant with this, the histone variant H2A.Z exhibited a strikingly opposite relocation. In all chromosomes from CLO cells, H2A.Z was present at the centromeres as well as chromosome arms (Fig. 3B, subpanels g and h), whereas on CLI chromosomes, it was completely excluded from pericentromeric chromatin (subpanels c and d). CLI chromosomes were thus no different from other mouse chromosomes described previously in this work. Chromosomes from CLO cells, in contrast, present a novel pattern of H3me3K9 distribution reminiscent of that observed in cells expressing a cathepsin L inhibitor, MENT. However, in contrast to cathepsin L knockout, MENT-expressing cells did not exhibit a significant loss of histone H3 methylation (23; also data not shown). Thus, the cathepsin L knockout system presented much stronger alterations in localization of H3me3K9 (as well as its levels) and H2A.Z. This increased effect apparently reflects the complete inactivation of cathepsin L in the knockout cells (see Fig. S3B in the supplemental material) in contrast to partial inhibition of cathepsin L by MENT. Thus, our studies for the first time reveal the effect of cathepsin L inactivation on chromatin epigenetic markers per se and provide a new way of fine regulation of cellular epigenetics through cathepsin L enzymatic activity and its nuclear inhibitors.
The CLI and CLO cell lines were derived from a male mouse embryo, and in the CLI cells, one chromosome per metaphase set contained high levels of H3me3K9 and almost no H2A.Z (Fig. 3B, subpanels a to d), consistent with the staining of the Y chromosome from normal mouse and human cells (Fig. 1). Morphology and FISH analysis with the Y-chromosome-specific probe confirmed the identity of this chromosome (Fig. 3C) and further indicated that CLI cells are no different than other mouse fibroblasts. In the CLO cells, in contrast, the Y chromosome lost its signature H3me3K9/H2AZ pattern (Fig. 3B, subpanels e to h). It became depleted in H3me3K9 and enriched in H2A.Z and, therefore, often indistinguishable from somatic chromosomes. FISH studies confirmed the identity of the Y chromosome in CLO cells (Fig. 3C). Interestingly, while the FISH probe was localized to the only region on the Y chromosome devoid of H3me3K9 in CLI cells, on the Y chromosome from CLO cells, the FISH signal perfectly colocalized with an island of H3(K9) trimethylation (Fig. 3C). These findings confirmed that cathepsin L knockout not only destabilized total levels of H3 trimethylation but also altered its overall chromosomal location. Thus, our findings indicate that the chromosome-wide histone N-tail modifications on the male Y chromosome can be positionally regulated and that this regulation is mediated by cathepsin L.
Nuclear localization of Suv39h1 is altered by cathepsin L knockout without affecting chromosome segregation and DNA methylation.
Most of histone H3 trimethylation in mice is mediated by Suv39h histone methyltransferases (HMTases), which are responsible for almost 100% of centromeric methylation and are encoded by two genes, Suv39h1 and Suv39h2 (43). While both HMTases are expressed during embryogenesis, Suv39h1 functions as the main H3(K9) methylating enzyme in the majority of adult tissues and Suv39h2 expression is restricted to adult testes, indicating a more universal role of Suv39h1 (43). We analyzed the levels of Suv39h1 transcription by semiquantitative RT-PCR (using primers specific for Suv39h1 mRNA) and Suv39h1 protein levels using specific antibodies. We did not find any difference in Suv39h1 mRNA (Fig. 4A) or protein (Fig. 4B) levels between CLO and CLI cells. In addition, we did not observe any severe phenotypes in CLO cells described for Suv39h gene double knockout, such as chromosome instability and aberrant segregation or down-regulation of H3acK9 and H4acK12 (47). We next decided to analyze Suv39h1 nuclear localization using expression of tagged Suv39h1, since the available antibodies against endogenous Suv39h1 were not suitable for immunofluorescence. Previous studies indicated that the presence of the N-terminal tag does not inhibit Suv39h1 activity or association with chromatin (11, 29). Figure 5A shows immunofluorescence analysis of nuclear localization of the full-length Xpress-tagged Suv39h1 in CLO and CLI cells. In agreement with other studies, Suv39h1 revealed nuclear, preferably heterochromatic localization in CLI cells (Fig. 5A, upper row), where the majority of tagged protein colocalized with pericentromeric Hoechst- and H3me3K9-positive foci (Fig. 5B, profiles). Double immunofluorescence analysis using antibodies against HP1β also revealed significant colocalization of this heterochromatin marker with tagged Suv39h1 (Fig. 5C, upper row). In contrast, in CLO cells, Xpress-Suv39h1 was not targeted to pericentromeric heterochromatin. Instead, it formed bright foci in other nuclear locations, where it significantly colocalized with H3me3K9 but not with Hoechst (Fig. 5A, lower row, and Fig. 5B). Interestingly, while Suv39h1 colocalized with H3me3K9 in both cell cultures (albeit at different nuclear locations), no significant correlation with HP1β was found in CLO cells (Fig. 5C). Green fluorescent protein (GFP)-tagged Suv39h1 exhibited similar nuclear localization patterns (Fig. 5C), indicating that the altered chromatin location of this HMTase in CLO cells is a property of the Suv39h1 itself, independent of the tag fusion. Furthermore, the observed localization pattern of Suv39h1 in CLO and CLI cells was not dependent on the methyltransferase activity, since deletion of the catalytic SET domain of Suv39h1 did not change its nuclear distribution in either cell culture (data not shown). Thus, in cathepsin L knockout cells, H3me3K9, Suv39h1, and HP1 can be completely depleted from pericentromeric heterochromatin without causing its decondensation or any effect on general protein composition, cell morphology, growth rates, and cell survival. Importantly, expression and localization of euchromatic H3(K9) methyltransferase, G9a was not altered in CLO cells compared to CLI cells (data not shown), which indicates a very specific effect of cathepsin L knockout on Suv39h1. Interestingly, DNA methylation was also not affected by cathepsin L knockout, as judged by immunofluorescence analysis of chromosomes from CLI and CLO cells using antibodies against methylated cytosine (Fig. 5D). In addition, cathepsin L knockout did not alter chromosome segregation, since CLI and CLO cells exhibited similar distribution of nondiploid chromosome numbers per karyotype (Fig. 5E). These observations clearly distinguish the effect of cathepsin L knockout from the phenotype of Suv39h1/h2 double knockout cells, where the morphology of centromeres, DNA methylation, and chromosome segregation were substantially damaged as a result of the HMTase depletion (46, 47). Thus, cathepsin L knockout provides an opportunity to modulate the global pattern of histone H3 methylation and related heterochromatin markers without compromising overall heterochromatin structure and genome integrity.
FIG. 4.

Suv39h1 expression and protein levels are similar in CLI and CLO cells. (A) RT-PCR analysis of total RNA from CLI and CLOi cells. Total RNA from logarithmically growing CLI and CLOi cells was isolated and analyzed by RT-PCR with oligonucleotides specific for Suv39h1 (upper panel) or beta-actin (lower panel) mRNA, as described in Materials and Methods. 1:5, cDNA from CLOi cells was diluted 5 times in a parallel experiment to ensure the sensitivity of PCR analysis. (B) Upper panel, Western blot analysis of total protein from CLI and CLOi cells with antibodies against mouse Suv39h1 protein. Numbers indicate molecular weight markers. Lower panel, the same membrane was stripped and stained with antibodies against beta-actin to confirm equal protein loading.
FIG. 5.
Suv39h1 relocation does not affect DNA methylation or chromosome segregation. (A) Immunofluorescence of CLI (upper row) and CLO (lower row) cells transfected with Xpress-Suv39h1 and stained with antibodies against Xpress (green), H3me3K9 (red), and Hoechst 33258 DNA stain (blue) 48 h after transfection. (B) Fluorescence intensity profiles illustrate spatial distribution of Xpress, me3, and Hoechst in the nuclei along the paths indicated by yellow lines on panel A. Note the colocalization of Xpress and H3me3K9 foci with Hoechst-positive chromocenters in CLI cells (arrows) but not CLO cells (arrowheads). Bar, 5 μm. (C) Cathepsin L knock-in (CLI) or knockout (CLO) cells were transfected with GFP-tagged Suv39h1, fixed, and stained with antibodies against HP1β 48 h after transfection (CLI, upper row; CLO, lower row). Blue, Hoechst 33258 DNA stain; green, GFP fluorescence; red, HP1β. Profiles indicate fluorescence intensity changes along the yellow lines. Note the almost complete correlation of GFP-Suv39h1 and HP1β signals with Hoechst foci in CLI cells (arrow) and the absence of any correlation between these markers in CLO cells. (D) 5-Methylcytosine (5-meC) immunofluorescence of metaphase chromosomes from CLI and CLOi cells. Chromosomes were stained with antibodies against 5-meCy (red) and counterstained with Hoechst (blue) as described in Materials and Methods. Note similar distribution of 5-meCy on centromeres (arrows) and the Y chromosome (Y). Bar, 5 μm. (E) Nondiploid chromosome number distribution in metaphase spreads from CLI (black) and CLOi (gray) cells. One hundred metaphase sets from each culture were randomly selected, and chromosomes were counted using Image Pro Plus software.
DISCUSSION
Since the discovery of the role of trimethylated histone H3(K9) and the histone methyltransferases in establishing and spreading heterochromatin (24), numerous studies were performed which were aimed toward understanding their regulation and interaction with other factors important for chromatin structure and activity. Here, we describe for the first time the chromosome-wide topographic relationship between H3me3K9 and histone isoform H2A.Z and show that this relationship is coordinately regulated by the nuclear and lysosomal protease cathepsin L (catL). These data, together with that of previously published studies, provide new clues for understanding the role(s) of these epigenetic factors in heterochromatin establishment and function.
While H3me3K9 has a well-established role in promoting heterochromatin formation, H2A.Z may fulfill more complex functions in chromatin regulation, related to its ability to assemble unique higher-order chromatin structures that can be utilized in different ways (9, 10). For example, it can act as a heterochromatin-promoting protein in Drosophila (63) but as a heterochromatin barrier in Saccharomycescerevisiae (35). H2A.Z is associated with the 5′ ends of both active and inactive genes in yeast euchromatin (51) and with pericentromeric heterochromatin as well as euchromatic regions in mammalian cells (52). While the bulk of H2A.Z is localized to chromosome arms, a small portion of it found at pericentromeric heterochromatin (Greaves and Tremethick, submitted) is crucial for chromosome segregation in mammalian cells (53). Consistent with these studies, we found that H2A.Z mainly accumulates on chromosome arms in normal mouse and human fibroblasts (Fig. 1A); however, it is translocated to heterochromatin in catL knockout cells (Fig. 3B).
Remarkably, the removal of catL also causes dramatic variegation in total H3 trimethylation levels (Fig. 2D). These changes are accompanied by redistribution of the HP1 isoforms but, surprisingly, do not involve alteration in heterochromatin compaction or chromosome stability, previously linked to the decreased H3 methylation (8, 45, 46). The inactivation of Suv39h HMTases by double gene knockout causes down-regulation of H3 methylation (47) as well as severe aberrations in DNA methylation at pericentromeric satellite repeats (31), which is consistent with the functional link between histone H3(K9) and DNA methylation. In contrast, catL-deficient cells exhibit a normal DNA methylation pattern and morphology of pericentromeric Hoechst-positive chromatin (Fig. 5D) and show no alterations in chromosome number compared to catL-expressing cells (Fig. 5E). This indicates that the CLO cells manage to uncouple histone and DNA methylation and overcome the negative effect of H3me3K9 deficiency on heterochromatin. In addition to heterochromatin, Suv39h and HP1 are known to act at several (or many) euchromatic loci (for a review, see reference 21), where they may not necessarily be affected by cathepsin L, thus explaining the relatively mild phenotype of catL knockout compared to Suv39h1/h2 double knockout. Also, other epigenetic factors may be responsible for compensating for the effect of Suv39h and HP1 relocation in cathepsin L knockout cells. Suv39h1/2 double knockout cells were shown to have increased H3K9 monomethylation and H3K27 trimethylation at pericentric heterochromatin (45). The authors proposed (45) that this change may constitute a mechanism for heterochromatin “rescue” involving substitution of H3me3K9 by H3me3K27 known to mark the facultative heterochromatin in normal cells (32, 48). The exact structural role of H3me3K27 in the formation of facultative or constitutive heterochromatin needs further investigation.
The distinguishing feature of the catL knockout system is that here the down-regulation of H3 trimethylation is accompanied by a significant (fourfold) up-regulation of H2A.Z and its accumulation in pericentromeric heterochromatin (Fig. 2D and Fig. 3, respectively). Although H2A.Z is generally believed to be a euchromatic protein, recent studies have shown that this histone variant binds HP1 and promotes compaction of reconstituted chromatin fibers, indicating that it can be involved in heterochromatin formation (10, 52) and chromosome segregation (53). Therefore, we suggest that H2A.Z functionally replaces H3me3K9 in catL knockout cells, thus allowing them to survive without suffering from the major defects in constitutive heterochromatin and chromosome segregation caused by H3me3K9 deficiency.
The chromosome-wide enrichment in H3me3K9 and depletion of H2A.Z has been previously reported only for the inactive X (Xi) chromosome in female mammalian cells (7). Our work reveals that all mouse and human chromosomes share the alternate epigenetic landscape of these two epigenetic markers (Fig. 1). We also detected the enrichment in H3me3K9 and absence of H2A.Z on the Y chromosome of male fibroblasts, thus revealing the similarity of chromatin composition between the two sex chromosomes that are mostly transcriptionally inactive in somatic mammalian tissues. Interestingly, the condensed Y chromosome in plants also reveals enrichment in H3(K9) methylation (39). These findings indicate that the formation of a signature epigenetic landscape on the X and Y chromosomes may reflect a molecular mechanism of heterochromatin maintenance common for both sex chromosomes. Consistent with this, very recent data suggest that dimethyl-H3(K9) and H2A.Z may assemble the X and Y chromosomes into facultative heterochromatin in round spermatid bodies during meiosis (D. Tremethick, personal communication).
One of the phenotypes associated with catL inactivation is the abnormal spermatogenesis in furless mice that express catalytically inactive catL (66; A. Rudensky, personal communication). Interestingly, a similar defect is observed in Suv39h double knockout mice, where the impaired spermatogenesis leads to complete spermatogenic failure that is attributed to the defects in spermatocyte differentiation and chromosome missegregation. The importance of histone modifications and their controlled regulation on sex chromosomes for mammalian spermatogenesis was noted in several studies (reviewed in reference 15). In this light, our findings that catL knockout destabilizes Y chromosome epigenetics provide a potential link between altered histone modification patterns and abnormal germ cell development in catL-deficient mice and emphasize the importance of further studies of the catL role in epigenetic control of germ cell differentiation.
Besides the dramatic variegation in histone H3(K9) methylation, we did not observe any other significant alterations in histone tail modifications in catL knockout cells, either at lysine 9 [e.g., H3(K9) acetylation] (Fig. 2D) or at neighboring amino acids, such as phosphorylation of Ser10 (data not shown), which has recently been found to influence the H3(K9) methylation and HP1 binding to chromatin (12, 19). Other heterochromatin-associated methylated sites, such as H4(K20) and H3(K27), were also unaffected in CLO cells (Fig. 2D, Fig. 3A, and data not shown). This indicates that the effect of catL knockout is specific for histone H3(K9) methylation and that the function of Suv39h1, the major HMTase in pericentromeric heterochromatin, is affected in CLO cells. Indeed, consistent with the role of Suv39h1 in the feedback mechanism of epigenetic regulation of heterochromatin spreading (24), we observed almost absolute colocalization of H3me3K9, HP1, and expressed tagged Suv39h1 in catL-positive cells and complete segregation of HP1 from Suv39h and H3me3K9 in CLO cells (Fig. 5A and C). At the same time, the mRNA and protein levels of endogenous Suv39h1 were similar between CLI and CLO cells, indicating that the impaired H3 methylation in catL-deficient cells is not due to the altered HMTase levels. The requirement of several cell passages for the redistribution of H3(K9) trimethylation in primary CL−/− cells indicates that this is a result of the gradual decrease of histone methylation during replication rather than active demethylation. This is consistent with the absence of histone demethylase activity specific for histone H3(K9) in mouse cells.
Interestingly, Suv39h1 still partially colocalized with trimethyl-H3(K9) in CLO cells (although at altered nuclear locations), and this localization was independent of the HMTase catalytic SET domain (data not shown). This indicates that other Suv39h1 domain(s) are responsible for its altered chromatin targeting in CLO cells. Thus, our work clearly demonstrates the functional relationship between catL activity and Suv39h1 recruitment, although the exact epigenetic mechanism(s) directly affected by cathepsin L remains to be discovered.
The apparent stability of Suv39h1 protein level raises a question about which chromatin factors could be affected by cathepsin L proteolytic activity and responsible for the altered function and targeting of Suv39h1 in CLO cells. Previous studies by Goulet and colleagues and Moon et al. indicated that, during S phase, the small (24 kDa) catL isoform localizes to the cell nucleus where it proteolytically cleaves the essential CDP/cux transcription factor, causing alteration of its DNA-binding activity and sequence specificity (14, 38). CDP/cux is an important regulator of many housekeeping genes, including several histone genes (reviewed in reference 41), and it has been shown to directly recruit G9a HMTase to the target genes and also to affect Suv39h1 (42). However, we found that transient overexpression of either the full-length or truncated (imitation of cleaved) isoform of CDP/cux in CLO cells did not restore total levels of histone H3 methylation and that the nuclear localization and mRNA levels of G9a HMTase were not different between CLO and CLI cells (data not shown). This indicates that the global epigenetic changes observed in CLO cells cannot be readily explained by altered CDP/cux processing. Furthermore, our studies using ectopically expressed catL (not shown here) indicate that the truncated catL isoform (identical to that of Goulet et al.) goes very efficiently into the nuclei of mouse fibroblasts but, after prolonged expression, forms nuclear aggregates, indicating that the protease is most probably misfolded and, hence, inactive. Also, the expression of a full-length, but not a truncated, catL affects MENT localization and chromatin binding inside the cell nuclei (unpublished observations). Therefore, the mechanism of catL nuclear transport and the activity of the truncated catL form in the cell nucleus remain to be determined.
This work confirms the efficiency of MENT ectopically expressed in NIH 3T3 cells as a nuclear inhibitor of catL. Interestingly, in terminally differentiated chicken erythrocytes that express relatively low MENT (less than in ectopic MENT-expressing NIH 3T3), histone H3(K9) trimethylation is focal and heterochromatic (23), while in granulocytes that express high levels of MENT (chicken) or a related human serpin MNEI, H3me3K9 is redistributed and blocked from immunofluorescence detection (49). Thus, catL inhibition by high doses of a nuclear serpin (MENT or MNEI) may contribute to chromatin rearrangement in mature granulocytes. It is plausible to think that other closely related intracellular serpins, such as SCCA1, SQN-5, and Spi2A (1, 33), that inhibit papain-like cysteine proteases may participate in controlling catL-mediated chromatin regulation in other cell types.
In addition to the variegation of chromatin markers described in the current work, other catL-linked phenotypes include defective skin and bone cell differentiation (50), dilated cardiomyopathy (60), and male fertility defects (66), some of which have been attributed to the misbalance between cell proliferation, differentiation, and death. Furthermore, cells expressing a cathepsin L knockdown construct show a significant increase in senescence (68), a phenotype that provides an interesting parallel to that of Suv39h1-dependent senescence (6). However, the primary CL−/− fibroblasts showed growth rates similar to those of CL+/− cells and no signs of senescence (data not shown). Future studies should clarify whether the above-mentioned phenotypes resulting from cathepsin L deficiency involve the epigenetic heterochromatin changes reported here.
Supplementary Material
Acknowledgments
We thank A. Rudensky for providing the cathepsin L knockout cells and valuable comments, L. Carrel for advice in chromosome work, M. Miljkovic for help with high-performance liquid chromatography, S. Bronson for help with the cytospin used in chromosome spreads, and E. Popova for critical discussion and valuable comments. We thank P. Singh for antibodies against HP1 and methylated histone H4, T. Kouzarides for the plasmid expressing Suv39h1, A. Roopra for the plasmid expressing G9a, and A. Nepveu for the plasmid expressing CDP/cut.
This work was supported by an award from the American Heart Association (Y.B.), and in part by NIH COBRE grant 5P20RR020173 (R.W.M.). This project was funded, in part, under a grant with the Pennsylvania Department of Health using Tobacco Settlement Funds. The Department specifically disclaims responsibility for any analyses, interpretations, or conclusions.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Al-Khunaizi, M., C. J. Luke, Y. S. Askew, S. C. Pak, D. J. Askew, S. Cataltepe, D. Miller, D. R. Mills, C. Tsu, D. Bromme, J. A. Irving, J. C. Whisstock, and G. A. Silverman. 2002. The serpin SQN-5 is a dual mechanistic-class inhibitor of serine and cysteine proteinases. Biochemistry 41:3189-3199. [DOI] [PubMed] [Google Scholar]
- 2.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 3.Barrett, A. J., and H. Kirschke. 1981. Cathepsin B, cathepsin H, and cathepsin L. Methods Enzymol. 80(Pt C):535-561. [DOI] [PubMed] [Google Scholar]
- 4.Barton, S. C., K. L. Arney, W. Shi, A. Niveleau, R. Fundele, M. A. Surani, and T. Haaf. 2001. Genome-wide methylation patterns in normal and uniparental early mouse embryos. Hum. Mol. Genet. 10:2983-2987. [DOI] [PubMed] [Google Scholar]
- 5.Bean, C. J., P. A. Hunt, E. A. Millie, and T. J. Hassold. 2001. Analysis of a malsegregating mouse Y chromosome: evidence that the earliest cleavage divisions of the mammalian embryo are non-disjunction-prone. Hum. Mol. Genet. 10:963-972. [DOI] [PubMed] [Google Scholar]
- 6.Braig, M., S. Lee, C. Loddenkemper, C. Rudolph, A. H. Peters, B. Schlegelberger, H. Stein, B. Dorken, T. Jenuwein, and C. A. Schmitt. 2005. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436:660-665. [DOI] [PubMed] [Google Scholar]
- 7.Chadwick, B. P., and H. F. Willard. 2004. Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc. Natl. Acad. Sci. USA 101:17450-17455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esteller, M. 2005. Aberrant DNA methylation as a cancer-inducing mechanism. Annu. Rev. Pharmacol. Toxicol. 45:629-656. [DOI] [PubMed] [Google Scholar]
- 9.Fan, J. Y., F. Gordon, K. Luger, J. C. Hansen, and D. J. Tremethick. 2002. The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat. Struct. Biol. 9:172-176. [DOI] [PubMed] [Google Scholar]
- 10.Fan, J. Y., D. Rangasamy, K. Luger, and D. J. Tremethick. 2004. H2A.Z alters the nucleosome surface to promote HP1alpha-mediated chromatin fiber folding. Mol. Cell 16:655-661. [DOI] [PubMed] [Google Scholar]
- 11.Firestein, R., X. Cui, P. Huie, and M. L. Cleary. 2000. Set domain-dependent regulation of transcriptional silencing and growth control by SUV39H1, a mammalian ortholog of Drosophila Su(var)3-9. Mol. Cell. Biol. 20:4900-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischle, W., B. S. Tseng, H. L. Dormann, B. M. Ueberheide, B. A. Garcia, J. Shabanowitz, D. F. Hunt, H. Funabiki, and C. D. Allis. 2005. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438:1116-1122. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Mata, R., Z. Bebok, E. J. Sorscher, and E. S. Sztul. 1999. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J. Cell Biol. 146:1239-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goulet, B., A. Baruch, N. S. Moon, M. Poirier, L. L. Sansregret, A. Erickson, M. Bogyo, and A. Nepveu. 2004. A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol. Cell 14:207-219. [DOI] [PubMed] [Google Scholar]
- 15.Govin, J., C. Caron, C. Lestrat, S. Rousseaux, and S. Khochbin. 2004. The role of histones in chromatin remodelling during mammalian spermiogenesis. Eur. J. Biochem. 271:3459-3469. [DOI] [PubMed] [Google Scholar]
- 16.Grigoryev, S. A., and C. L. Woodcock. 1998. Chromatin structure in granulocytes. A link between tight compaction and accumulation of a heterochromatin-associated protein (MENT). J. Biol. Chem. 273:3082-3089. [DOI] [PubMed] [Google Scholar]
- 17.Heitz, E. 1928. Das Heterochromatin der Moose. I. Jb. Wiss. Bot. 69:762-818. [Google Scholar]
- 18.Hilwig, I., and A. Gropp. 1973. Decondensation of constitutive heterochromatin in L cell chromosomes by a benzimidazole compound (“33258 Hoechst”). Exp. Cell Res. 81:474-477. [DOI] [PubMed] [Google Scholar]
- 19.Hirota, T., J. J. Lipp, B. H. Toh, and J. M. Peters. 2005. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 438:1176-1180. [DOI] [PubMed] [Google Scholar]
- 20.Hsieh, C. S., P. deRoos, K. Honey, C. Beers, and A. Y. Rudensky. 2002. A role for cathepsin L and cathepsin S in peptide generation for MHC class II presentation. J. Immunol. 168:2618-2625. [DOI] [PubMed] [Google Scholar]
- 21.Huisinga, K. L., B. Brower-Toland, and S. C. Elgin. 2006. The contradictory definitions of heterochromatin: transcription and silencing. Chromosoma 115:110-122. [DOI] [PubMed] [Google Scholar]
- 22.Irving, J. A., S. S. Shushanov, R. N. Pike, E. Y. Popova, D. Bromme, T. H. Coetzer, S. P. Bottomley, I. A. Boulynko, S. A. Grigoryev, and J. C. Whisstock. 2002. Inhibitory activity of a heterochromatin-associated serpin (MENT) against papain-like cysteine proteinases affects chromatin structure and blocks cell proliferation. J. Biol. Chem. 277:13192-13201. [DOI] [PubMed] [Google Scholar]
- 23.Istomina, N. E., S. S. Shushanov, E. M. Springhetti, V. L. Karpov, I. A. Krasheninnikov, K. Stevens, K. S. Zaret, P. B. Singh, and S. A. Grigoryev. 2003. Insulation of the chicken beta-globin chromosomal domain from a chromatin-condensing protein, MENT. Mol. Cell. Biol. 23:6455-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 25.Johnston, J. A., C. L. Ward, and R. R. Kopito. 1998. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143:1883-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones, P. A., and D. Takai. 2001. The role of DNA methylation in mammalian epigenetics. Science 293:1068-1070. [DOI] [PubMed] [Google Scholar]
- 27.Khorasanizadeh, S. 2004. The nucleosome: from genomic organization to genomic regulation. Cell 116:259-272. [DOI] [PubMed] [Google Scholar]
- 28.Kourmouli, N., P. Jeppesen, S. Mahadevhaiah, P. Burgoyne, R. Wu, D. M. Gilbert, S. Bongiorni, G. Prantera, L. Fanti, S. Pimpinelli, W. Shi, R. Fundele, and P. B. Singh. 2004. Heterochromatin and tri-methylated lysine 20 of histone H4 in animals. J. Cell Sci. 117:2491-2501. [DOI] [PubMed] [Google Scholar]
- 29.Krouwels, I. M., K. Wiesmeijer, T. E. Abraham, C. Molenaar, N. P. Verwoerd, H. J. Tanke, and R. W. Dirks. 2005. A glue for heterochromatin maintenance: stable SUV39H1 binding to heterochromatin is reinforced by the SET domain. J. Cell Biol. 170:537-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 31.Lehnertz, B., Y. Ueda, A. A. Derijck, U. Braunschweig, L. Perez-Burgos, S. Kubicek, T. Chen, E. Li, T. Jenuwein, and A. H. Peters. 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13:1192-1200. [DOI] [PubMed] [Google Scholar]
- 32.Lewis, A., K. Mitsuya, D. Umlauf, P. Smith, W. Dean, J. Walter, M. Higgins, R. Feil, and W. Reik. 2004. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat. Genet. 36:1291-1295. [DOI] [PubMed] [Google Scholar]
- 33.Liu, N., T. Phillips, M. Zhang, Y. Wang, J. T. Opferman, R. Shah, and P. G. Ashton-Rickardt. 2004. Serine protease inhibitor 2A is a protective factor for memory T cell development. Nat. Immunol. 5:919-926. [DOI] [PubMed] [Google Scholar]
- 34.Martens, J. H., R. J. O'Sullivan, U. Braunschweig, S. Opravil, M. Radolf, P. Steinlein, and T. Jenuwein. 2005. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 24:800-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meneghini, M. D., M. Wu, and H. D. Madhani. 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112:725-736. [DOI] [PubMed] [Google Scholar]
- 36.Miller, A. P., K. Gustashaw, D. J. Wolff, S. H. Rider, A. P. Monaco, B. Eble, D. Schlessinger, J. L. Gorski, G. J. van Ommen, J. Weissenbach, et al. 1995. Three genes that escape X chromosome inactivation are clustered within a 6 Mb YAC contig and STS map in Xp11.21-p11.22. Hum. Mol. Genet. 4:731-739. [DOI] [PubMed] [Google Scholar]
- 37.Miller, O. J., W. Schnedl, J. Allen, and B. F. Erlanger. 1974. 5-Methylcytosine localised in mammalian constitutive heterochromatin. Nature 251:636-637. [DOI] [PubMed] [Google Scholar]
- 38.Moon, N. S., P. Premdas, M. Truscott, L. Leduy, G. Berube, and A. Nepveu. 2001. S phase-specific proteolytic cleavage is required to activate stable DNA binding by the CDP/Cut homeodomain protein. Mol. Cell. Biol. 21:6332-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mosiolek, M., P. Pasierbek, J. Malarz, M. Mos, and A. J. Joachimiak. 2005. Rumex acetosa Y chromosomes: constitutive or facultative heterochromatin? Folia Histochem. Cytobiol. 43:161-167. [PubMed] [Google Scholar]
- 40.Nakagawa, T., W. Roth, P. Wong, A. Nelson, A. Farr, J. Deussing, J. A. Villadangos, H. Ploegh, C. Peters, and A. Y. Rudensky. 1998. Cathepsin L: critical role in Ii degradation and CD4 T cell selection in the thymus. Science 280:450-453. [DOI] [PubMed] [Google Scholar]
- 41.Nepveu, A. 2001. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene 270:1-15. [DOI] [PubMed] [Google Scholar]
- 42.Nishio, H., and M. J. Walsh. 2004. CCAAT displacement protein/cut homolog recruits G9a histone lysine methyltransferase to repress transcription. Proc. Natl. Acad. Sci. USA 101:11257-11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Carroll, D., H. Scherthan, A. H. Peters, S. Opravil, A. R. Haynes, G. Laible, S. Rea, M. Schmid, A. Lebersorger, M. Jerratsch, L. Sattler, M. G. Mattei, P. Denny, S. D. Brown, D. Schweizer, and T. Jenuwein. 2000. Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol. Cell. Biol. 20:9423-9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pardue, M. L., and J. G. Gall. 1970. Chromosomal localization of mouse satellite DNA. Science 168:1356-1358. [DOI] [PubMed] [Google Scholar]
- 45.Peters, A. H., S. Kubicek, K. Mechtler, R. J. O'Sullivan, A. A. Derijck, L. Perez-Burgos, A. Kohlmaier, S. Opravil, M. Tachibana, Y. Shinkai, J. H. Martens, and T. Jenuwein. 2003. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12:1577-1589. [DOI] [PubMed] [Google Scholar]
- 46.Peters, A. H., J. E. Mermoud, D. O'Carroll, M. Pagani, D. Schweizer, N. Brockdorff, and T. Jenuwein. 2002. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat. Genet. 30:77-80. [DOI] [PubMed] [Google Scholar]
- 47.Peters, A. H., D. O'Carroll, H. Scherthan, K. Mechtler, S. Sauer, C. Schofer, K. Weipoltshammer, M. Pagani, M. Lachner, A. Kohlmaier, S. Opravil, M. Doyle, M. Sibilia, and T. Jenuwein. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107:323-337. [DOI] [PubMed] [Google Scholar]
- 48.Plath, K., J. Fang, S. K. Mlynarczyk-Evans, R. Cao, K. A. Worringer, H. Wang, C. C. de la Cruz, A. P. Otte, B. Panning, and Y. Zhang. 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300:131-135. [DOI] [PubMed] [Google Scholar]
- 49.Popova, E. Y., D. Claxton, E. Lukasova, P. Bird, and S. A. Grigoryev. 2006. Epigenetic heterochromatin markers distinguish terminally differentiated leukocytes from incompletely differentiated leukemia cells in human blood. Exp. Hematol. 34:453-462. [DOI] [PubMed] [Google Scholar]
- 50.Potts, W., J. Bowyer, H. Jones, D. Tucker, A. J. Freemont, A. Millest, C. Martin, W. Vernon, D. Neerunjun, G. Slynn, F. Harper, and R. Maciewicz. 2004. Cathepsin L-deficient mice exhibit abnormal skin and bone development and show increased resistance to osteoporosis following ovariectomy. Int. J. Exp. Pathol 85:85-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raisner, R. M., P. D. Hartley, M. D. Meneghini, M. Z. Bao, C. L. Liu, S. L. Schreiber, O. J. Rando, and H. D. Madhani. 2005. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123:233-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rangasamy, D., L. Berven, P. Ridgway, and D. J. Tremethick. 2003. Pericentric heterochromatin becomes enriched with H2A.Z during early mammalian development. EMBO J. 22:1599-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rangasamy, D., I. Greaves, and D. J. Tremethick. 2004. RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nat. Struct. Mol. Biol. 11:650-655. [DOI] [PubMed] [Google Scholar]
- 54.Rea, S., F. Eisenhaber, D. O'Carroll, B. D. Strahl, Z. W. Sun, M. Schmid, S. Opravil, K. Mechtler, C. P. Ponting, C. D. Allis, and T. Jenuwein. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593-599. [DOI] [PubMed] [Google Scholar]
- 55.Richards, E. J., and S. C. Elgin. 2002. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 108:489-500. [DOI] [PubMed] [Google Scholar]
- 56.Roth, W., J. Deussing, V. A. Botchkarev, M. Pauly-Evers, P. Saftig, A. Hafner, P. Schmidt, W. Schmahl, J. Scherer, I. Anton-Lamprecht, K. Von Figura, R. Paus, and C. Peters. 2000. Cathepsin L deficiency as molecular defect of furless: hyperproliferation of keratinocytes and perturbation of hair follicle cycling. FASEB J. 14:2075-2086. [DOI] [PubMed] [Google Scholar]
- 57.Schotta, G., A. Ebert, V. Krauss, A. Fischer, J. Hoffmann, S. Rea, T. Jenuwein, R. Dorn, and G. Reuter. 2002. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 21:1121-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Springhetti, E. M., N. E. Istomina, J. C. Whisstock, T. Nikitina, C. L. Woodcock, and S. A. Grigoryev. 2003. Role of the M-loop and reactive center loop domains in the folding and bridging of nucleosome arrays by MENT. J. Biol. Chem. 278:43384-43393. [DOI] [PubMed] [Google Scholar]
- 59.Stewart, M. D., J. Li, and J. Wong. 2005. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol. Cell. Biol. 25:2525-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stypmann, J., K. Glaser, W. Roth, D. J. Tobin, I. Petermann, R. Matthias, G. Monnig, W. Haverkamp, G. Breithardt, W. Schmahl, C. Peters, and T. Reinheckel. 2002. Dilated cardiomyopathy in mice deficient for the lysosomal cysteine peptidase cathepsin L. Proc. Natl. Acad. Sci. USA 99:6234-6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sullivan, B. A., and S. Schwartz. 1995. Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum. Mol. Genet. 4:2189-2197. [DOI] [PubMed] [Google Scholar]
- 62.Sullivan, B. A., and P. E. Warburton. 1999. Studying the progression of vertebrate chromosomes through mitosis by immunofluorescence and FISH, p. 81-101. In W. A. Bickmore (ed.), Chromosome structural analysis. Oxford University Press, Oxford, United Kingdom.
- 63.Swaminathan, J., E. M. Baxter, and V. G. Corces. 2005. The role of histone H2Av variant replacement and histone H4 acetylation in the establishment of Drosophila heterochromatin. Genes Dev. 19:65-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Verschure, P. J., I. van Der Kraan, E. M. Manders, and R. van Driel. 1999. Spatial relationship between transcription sites and chromosome territories. J. Cell Biol. 147:13-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilcox, D., and R. W. Mason. 1992. Inhibition of cysteine proteinases in lysosomes and whole cells. Biochem. J. 285(Pt 2):495-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wright, W. W., L. Smith, C. Kerr, and M. Charron. 2003. Mice that express enzymatically inactive cathepsin L exhibit abnormal spermatogenesis. Biol. Reprod. 68:680-687. [DOI] [PubMed] [Google Scholar]
- 67.Wu, R., A. V. Terry, P. B. Singh, and D. M. Gilbert. 2005. Differential subnuclear localization and replication timing of histone H3 lysine 9 methylation states. Mol. Biol. Cell 16:2872-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng, X., P. M. Chou, B. L. Mirkin, and A. Rebbaa. 2004. Senescence-initiated reversal of drug resistance: specific role of cathepsin L. Cancer Res. 64:1773-1780. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.