Abstract
Prohibitin is a growth regulatory gene that has pleiotropic functions in the nucleus, mitochondria, and cytoplasmic compartments. Earlier studies had proposed a role for prohibitin in modulating cellular senescence, but the underlying mechanisms remain unknown. Here we show that senescence induced by DNA-damaging agents causes the localization of prohibitin to specific heterochromatic foci. Prohibitin could bind to heterochromatin protein 1 (HP1) family proteins and colocalized with HP1γ in senescence-associated heterochromatic foci. Further, HP1γ could synergize with prohibitin to repress E2F1-mediated transcriptional activity. The depletion of prohibitin by small interfering RNA or antisense techniques led to a reduction in the senescent phenotype, correlating with a reduced expression of senescence-associated β-galactosidase and fewer numbers of senescence-associated heterochromatic foci. Chromatin immunoprecipitation assays showed that prohibitin is needed for the recruitment of HP1γ to E2F1-regulated proliferative promoters, leading to their repression. The ablation of prohibitin prevented the recruitment of HPIγ, but not Suv39H, to the promoters upon senescence. Prohibitin-mediated recruitment of HP1γ occurred in only senescent cells, not in quiescent cells; thus, there is a dichotomy in the recruitment of different corepressors by prohibitin, depending on the type of growth arrest. These studies show that prohibitin plays a vital role in inducing cellular senescence.
Primary mammalian cells in culture undergo a period of rapid proliferation; however, cell growth eventually decelerates and the cells enter a form of permanent cell cycle arrest termed senescence (20). While normal senescence limits the replicative potential of cells by telomere shortening (41), cells can undergo a similar permanent G1 growth arrest in response to treatment with drugs like etoposide or adriamycin or in response to oncogenes like Ras (15, 45, 52, 54, 55). This growth arrest, also called STASIS (stress- or aberrant signaling- induced senescence), is similar to replicative senescence in that cells are unable to divide under mitotic stimulation even though they remain metabolically active. They also show changes in morphology that are typical of senescence (12, 14, 27). It has been shown that cultures of immortal cell lines can also undergo permanent cell cycle arrest in response to DNA-damaging agents, suggesting that senescence may act as a natural barrier to cancer progression (6, 7, 17, 46, 59, 67).
A senescent cell typically has a flattened morphology and increased granularity. At the biochemical level, senescence is accompanied by changes in metabolism and characterized by the induction of senescence-associated β-galactosidase activity (5, 13, 53), while at the genetic level, alterations in chromatin structure and gene expression patterns are observed (2, 25, 37, 49, 57). In senescing cells, the chromatin undergoes remodeling and senescence-associated heterochromatic foci (SAHF) can be detected (37). Heterochromatin is responsible for epigenetic silencing by the sequestration of genes into transcriptionally inactive nuclear neighborhoods, and heterochromatin protein 1 (HP1) has been shown to play a major role in this process (8, 16). Elegant studies from the Lowe and Campisi labs have shown that Rb is hypophosphorylated in senescent cells, and its function is necessary for inducing the senescent phenotype (5, 31). The activation of the Rb and/or p53 protein and the expression of their regulators, such as p16INK4a, p21, and ARF, are required for the senescence process (2, 25, 31, 57).
Rb family members have been shown to interact with various transcriptional corepressors, including histone deacetylase 1 (HDAC1), DNA methyltransferase, and Polycomb proteins as well as chromatin-remodeling complexes like Brg and Brm, to repress the transcriptional activity of E2F (4, 32, 33, 35, 48, 58, 61, 62, 66). Rb has been shown to associate with HP1 and histone methyltransferase Suv39H to facilitate senescence (28, 37, 60). This facilitation is achieved by accumulating on E2F-responsive proliferative promoters, repressing their activity, and inducing the formation of SAHF (36, 37).
Prohibitin, a potential tumor suppressor gene, is implicated in cell cycle control (22, 34, 40, 47, 63-65) and has potent antiproliferative activity (23, 24, 51). Studies from our lab have shown that like Rb, prohibitin physically interacts with and represses E2F family members. This interaction is necessary for the growth-suppressive activity of prohibitin (63, 64). Prohibitin-mediated repression of E2F is involved the recruitment of corepressors like HDAC1 and N-CoR (62). In addition to its effects on proliferation, prohibitin has been shown to influence the replicative capacity of yeast (Saccharomyces cerevisiae) cells as well as human diploid fibroblasts (3, 9-11, 29, 43, 56) but the underlying mechanisms are unknown. The results presented here show that prohibitin physically interacts with the heterochromatin proteins as well as the histone methyltransferase Suv39H (1, 38). Interestingly, prohibitin was detected in senescence-associated heterochromatic foci, where it colocalized with HP1γ; the depletion of prohibitin led to a loss of the senescent phenotype. Prohibitin, Suv39H, and HP1γ were detected on E2F-responsive proliferative promoters upon the induction of senescence, leading to their repression; the ablation of prohibitin prevented the recruitment of HP1γ but not Suv39H. These results suggest that prohibitin, like Rb, contributes to cellular senescence by repressing E2F-responsive promoters utilizing specific corepressors.
MATERIALS AND METHODS
Cell lines.
Breast carcinoma cell lines MCF-7 and T47D were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). MCF-7 cells expressing tetracycline-inducible prohibitin were grown in DMEM supplemented with 10% tetracycline free fetal bovine serum (Clontech), 25 μg/ml zeocin (Invitrogen), and 0.25 μg/ml blasticidin (Invitrogen). Prohibitin expression was induced by incubation of the cells in 1 μg/ml tetracycline for 24 h. T47D cells stably expressing an antisense prohibitin construct were cultured in DMEM supplemented with 10% fetal bovine serum.
Transient transfections.
Transient transfections of MCF-7 cells were performed by electroporation using a Bio-Rad gene pulser at 400 mV. Cells were transfected with 3 μg E2CAT, 3 μg E2F1, and 2 μg prohibitin or 2 μg HP1 family members. Each reaction mixture contained 2 μg of β-galactosidase to serve as an internal control for transfection efficiency. Assays for β-galactosidase activity and chloramphenicol acetyltransferase activity were performed 72 h after transfection using standard protocols (50).
Drug treatment, antibodies, and immunostaining.
Prohibitin monoclonal and polyclonal antibodies were purchased from NeoMarkers, Inc. (Freemont, CA). The HP1γ polyclonal, Suv39H monoclonal, and histone 3-lysine 9 methylation-specific monoclonal antibodies were purchased from Upstate Biotechnologies. Cells were plated onto poly-d-lysine (Sigma)-coated eight-well glass chamber slides (10,000 cells per well) for immunostaining. Twenty-four hours after plating, the cells were treated with 2 μM adriamycin for 24 h (Sigma); subsequently, cells were washed with phosphate-buffered saline (PBS) and incubated for 48 h in fresh medium containing 10% FBS. Cells were fixed in 3.5% paraformaldehyde for 25 min, permeabilized in 0.2% Triton X-100-PBS for 5 min, and blocked in 5% normal goat serum in PBS at room temperature for 1 h. Primary antibody incubations were performed overnight at 4°C. After washing the cells, secondary antibody incubation was performed with goat anti-mouse immunoglobulin G (IgG)-Alexa Fluor-488 and goat anti-rabbit IgG-Alexa Fluor-546 for 30 min at room temperature. DAPI (4′,6′-diamidino-2-phenylindole) was detected using Vectashield mounting medium with DAPI (Vector Laboratories, Inc.). Control experiments demonstrated no cross-reactivity between anti-mouse secondary and anti-rabbit primary antibodies and vice versa; nor was there any detectable staining by secondary antibodies only (data not shown). Cells were visualized with a Zeiss LSM510 (Zeiss, Thornwood, N.Y.) confocal microscope, and areas of colocalization were determined using LSM510 software (Zeiss) or slides were observed by fluorescence microscopy using a Leica DM LB2 microscope (40× lens objective and 0.75 numerical aperture) with a Qimaging Retiga 1300 camera.
Histochemical and quantitative assessment of SA-β-gal activity.
Histochemical detection of senescence-associated β-galactosidase (SA-β-gal) was performed using the method described by Dimri et al. (13). Briefly, cultured cells were treated with adriamycin as described above. After treatment, the cells were washed with PBS and fixed with a 3.5% paraformaldehyde solution for 30 min at room temperature. The cells were washed again in PBS, and SA-β-gal stain solution (pH 6.0) was added and incubated for 16 h at 37°C. A control assay in which the SA-β-gal stain solution was adjusted to pH 7.4 was also performed. The experiment was performed twice, and three fields per treatment in each slide were analyzed for positively stained cells.
In vitro binding assays.
Glutathione S-transferase (GST) fusions of Rb and prohibitin were prepared as described earlier (63). GST fusions of HP1γ, the HP1γ chromodomain (CD) and chromoshadow domain (CSD), were prepared after the culture was induced with IPTG (isopropyl-β-d-thiogalactopyranoside) (250 mM) for 2 h at 37°C. 35S-labeled HP1α, -β, -γ, and prohibitin proteins were generated by in vitro transcription translation in rabbit reticulocyte lysates by using standard protocols. First, 8 to 10 μl of synthesized polypeptide was incubated with glutathione beads carrying equal amounts of GST fusion proteins in 200 μl of protein binding buffer (20 mM Tris [pH 7.5], 50 mM KCl, 0.5 mM EDTA, 1 mM dithiothreitol, 0.5% NP-40, 3 mg of bovine serum albumin per ml) at 4°C for 2 h. The beads were then washed six times with 1 ml of protein binding buffer. Beads were boiled in 2× loading dye, and proteins were separated on a 12% sodium dodecyl sulfate (SDS)-polyacrylamide gel and visualized by autoradiography. The protein amounts in control input lanes were approximately one-fifth of the total used in the binding assay.
GST pull-down assays in cell lysates.
GST fusions of HP1γ, the HP1γ chromodomain and chromoshadow domain, were immobilized on glutathione beads (Amersham). MCF-7 cell lysate (200 μg protein) was incubated with glutathione beads carrying equal amounts of GST fusion proteins in 200 μl of protein binding buffer (20 mM Tris [pH 7.5], 50 mM KCl, 0.5 mM EDTA, 1 mM dithiothreitol, 0.5% NP-40, 3 mg of bovine serum albumin per ml) at 4°C for 2 h. The beads were then washed six times with 1 ml of protein binding buffer. Beads were boiled in 2× loading dye and proteins were separated on a 12% SDS-polyacrylamide gel. The blots were probed with the appropriate antibody after semidry transfer to supported nitrocellulose membranes. The proteins were detected by using an enhanced chemiluminescence assay system from Amersham.
Immunoprecipitation and Western blotting.
Cell lysates (50 to 200 μg) were treated with 1 μg of the appropriate primary antibody in a volume of 100 μl at 4°C for 1 h. A 3-mg aliquot of protein G-Sepharose in a 100-μl volume was added to each sample and incubated for an additional hour. The binding was performed in a buffer containing 20 mM HEPES [pH 7.9], 40 mM KCl, 1 mM MgCl2, 0.1 mM EGTA, 0.1 mM EDTA, 0.1 mM dithiothreitol, 0.1 mM NaF, 0.1 mM Na3VO4, 0.5% NP-40, and 3 mg of bovine serum albumin per ml. The beads were washed six times with 600 μl of the same buffer, boiled in 20 μl of SDS sample buffer, and separated on 8 or 10% polyacrylamide gels. The blots were probed with the appropriate antibody after semidry transfer to supported nitrocellulose membranes. The proteins were detected by using an enhanced chemiluminescence assay system from Amersham.
Chromatin immunoprecipitation (ChIP) assays.
One confluent plate of T47D or MCF-7 cells (about 3 × 106 cells per plate) was used for each immunoprecipitation reaction as described previously (65). The prohibitin monoclonal, HP1γ polyclonal, Suv39H monoclonal, Rb monoclonal, histone H3 modified at lysine 9 (K9MeH3) monoclonal, and E2F1 monoclonal antibodies were used for immunoprecipitations. The prohibitin monoclonal antibody was purchased from NeoMarkers, Inc. (Freemont, CA). PCRs were then performed using 5 μl of the DNA from the immunoprecipitation reactions or 1 μl of DNA from the input reaction as a template. PCR cycling conditions were as follows: 94°C for 2 min; then 35 cycles of 94°C for 30 s, 56°C for 30 s, and 68°C for 30 s; followed by 68°C for 2 min. The sequences of the PCR primers used in the PCRs were as follows: 5′-TCT GCT GGG AGT TTT CAT TGA CCT C-3′ (cdc25A promoter forward primer), 5′-TTG GCG CCA AAC GGA ATC CAC CAA TC-3′ (cdc25A promoter reverse primer), 5′-TGT TGG CTG CAG CCC GCG AGC AGT TC-3′ (fos promoter forward primer), 5′-GGC GCG TGT CCT AAT CTC GTG AGC AT-3′ (fos promoter reverse primer), 5′-TGG CGC ACG CTC TCTAGA GC-3′ (TS promoter forward primer), and 5′-GAC GGA GGC AGG CCA AGT G-3′ (TS promoter reverse primer).
siRNA transfections.
Small interfering RNA (siRNA) for prohibitin was prepared from prohibitin cDNA which was PCR amplified using the primer set 5′-GGA TCC ATG GCT GCC AAA GTG TT-3′ (forward primer) and 5′-CTC GAG TTA CTG GGG GAG CTG GAG-3′ (reverse primer) and cloned into pCR2.1 (Invitrogen). Sequence and orientation were verified by sequencing. A set of plasmids containing the cis and trans orientations of the inserted fragment with regard to the T7 promoter were used for transcription using T7 RNA polymerase. RNA products were annealed and subjected to dicer reaction for 17 h. The purification was performed according to manufacturer's instructions (BLOCK-iT RNA interference kit; Invitrogen). siRNA oligonucleotides (19 to 21 bp) were purified and checked for integrity and quality on 4% agarose gels. A second set of siRNAs for prohibitin and control nonhomologous siRNA oligonucleotides were obtained from Santa Cruz Biotechnology. The transfections were performed in MCF-7 cells using Oligofectamine (Invitrogen). The treatment of cells with drugs was started 24 h after transfections.
RNA isolation and real-time PCR.
MCF-7 cells were subjected to serum starvation or treatment with adriamycin. Unstimulated asynchronous cells were used as a control. Total RNA was isolated by an RNeasy miniprep kit from QIAGEN following the manufacturer's protocol. One microgram of RNA was DNase treated using RQ1 DNase (Promega), followed by first-strand cDNA synthesis using the iScript cDNA synthesis kit (Bio-Rad). A fraction (1/20) of the final cDNA reaction volume was used in each PCR. Primers sequences are as follows: 5′-CTG CCA GCT GTA CCA GAG AT-3′ (TS forward primer), 5′-ATG TGC ATC TCC CAA AGT GT-3′ (TS reverse primer), 5′-AAC CTG ACC GTC ACT ATG GA-3′ (Cdc25a forward primer), 5′-GAA TCT GTT GAC TCG GAG GA-3′ (Cdc25a reverse primer), 5′-ATC CTC ACC CTG AAG TAC CC-3′ (β-actin forward primer), and 5′-TAG AAG GTG TGG TGC CAG AT-3′ (β-actin reverse primer).
All primers were run at an annealing temperature of 58°C. Real-time PCRs were performed in a volume of 25 μl, which included 200 nM of each forward and reverse primer and iQ SYBR green Supermix (Bio-Rad). Reactions were run in duplicate using an iCycler and iQ software (Bio-Rad). Average threshold cycles (Ct) for TS and Cdc25a were normalized to the average β-actin Ct values for each cDNA sample, and relative levels of TS and Cdc25a were calculated by the ΔΔCt method (30) as follows: 2−TS − β − actin for TS expression or 2−Cdc25a − β − actin for Cdc25a expression.
RESULTS
Distribution of prohibitin in the nucleus is altered upon cellular senescence.
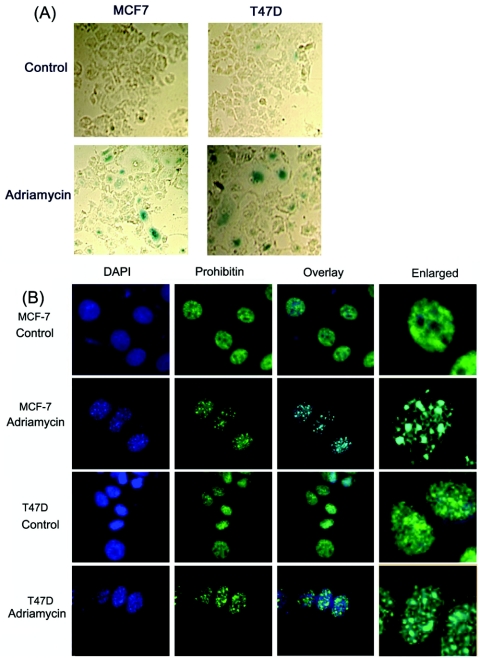
Attempts were made to examine the role of prohibitin in cellular senescence induced by anticancer agents. Previous studies have shown that exposure to DNA-damaging agents like adriamycin, etoposide, SN-38, or VM-26, followed by the removal of the drug, could induce senescence in mammalian cell lines (46, 59). To examine whether senescence induced by drugs affected prohibitin, human breast carcinoma cell lines MCF-7 and T47D were plated on eight-chamber slides and treated with 2 μM adriamycin for 24 h; subsequently, cells were washed with PBS and incubated for 48 h in fresh medium containing 10% FBS. It was found that the drug treatment induced a flattened cell morphology and increased granularity, characteristic of senescence (14). The cells were stained for SA-β-gal activity to confirm that the observed changes were due to the onset of senescence. It was observed that the drug-treated cells showed staining characteristic of senescence-associated β-galactosidase (Fig. 1A), confirming that the observed changes correlated with senescence.
FIG. 1.
(A) Senescence-associated β-galactosidase staining in MCF-7 and T47D cells. The cells were cultured in eight-well chamber slides and treated with adriamycin (2 μM) for 24 h; cells were washed and grown in fresh medium for 48 h. While untreated cells show little or no staining for β-galactosidase, the adriamycin-treated cells show β-galactosidase staining characteristic of senescence. (B) The treatment of cells with adriamycin leads to the appearance of heterochromatic DNA foci. MCF-7 and T47D cells were treated with adriamycin (2 μM) for 24 h and cultured for 48 h in regular growth medium. Cells were stained with DAPI, and heterochromatic foci were visualized by fluorescence microscopy. Immunostaining for prohibitin showed its localization in discrete foci in adriamycin-treated cells; an overlay of the images shows the localization of prohibitin on SAHF.
Narita et al. have shown that senescence induced by the Ras oncogene led to characteristic nuclear staining, where distinct punctuated regions of DNA could be observed by DAPI staining (37); identical foci were induced by the DNA-damaging agents in both cell lines (Fig. 1B). The same cells were immunostained with a monoclonal antibody against prohibitin. It was found that prohibitin was uniformly distributed in the nucleus in the untreated cells; upon treatment with the DNA-damaging agent adriamycin, prohibitin localized to discrete foci. These corresponded to the heterochromatic foci observed with DAPI staining, as seen by an overlay of the images. This result clearly shows that the induction of senescence led to the redistribution of prohibitin to specific heterochromatic foci.
Prohibitin associates with HP1 proteins in vitro.
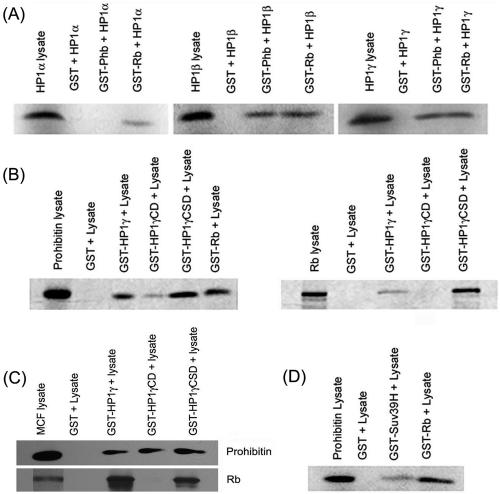
SAHF are related to heterochromatic regions (37), and HP1 is known to be a major component of heterochromatin (16). Since prohibitin localized to discrete foci upon senescence, we assessed whether prohibitin could associate with HP1 by an in vitro GST binding assay. HP1 isoforms α, β, and γ were synthesized by in vitro transcription translation, and their binding to GST fusions of prohibitin or Rb was tested. As shown in the left panel of Fig. 2A, there was no binding of HP1α to unprimed GST or prohibitin-GST beads, while it bound efficiently to Rb-GST beads. HP1β could bind equally well to both GST-Rb and GST-prohibitin beads, but not unprimed GST beads (Fig. 2A, middle panel). Similarly, HP1γ could bind to both GST-Rb and GST-prohibitin beads (Fig. 2A, right panel). These results suggest that prohibitin can directly associate with HP1β and γ proteins.
FIG. 2.
(A) HP1 family members interact with prohibitin in vitro. 35S-labeled HP1α, -β, and -γ were incubated with GST-prohibitin or unprimed GST beads as a control. GST-Rb was used as a positive control. (B) 35S-labeled prohibitin binds to the chromodomain and chromoshadow domain of HP1γ (left panel), while Rb binds preferentially to the chromoshadow domain. (C) Prohibitin in the MCF-7 cell lysates binds to the chromodomain and chromoshadow domain of HP1γ while Rb binds only to the chromoshadow domain. (D) 35S-labeled prohibitin binds to GST-Suv39H in vitro.
Additional GST binding assays were carried out to identify the domains of HP1γ involved in binding to prohibitin. The conserved amino-terminal region of HP1-like proteins features a CD, which plays a role in gene regulation. The conserved carboxyl-terminal region, termed the CSD, is related to the CD in primary amino acid sequence. The binding of 35S-labeled prohibitin to GST fusions of full-length HP1γ, the CD and CSD of HP1γ, was tested. As shown in Fig. 2B, left panel, prohibitin bound to both the CSD and the CD. This was in contrast to the binding of 35S-labeled Rb, which bound with high efficiency to the HP1γ CSD, while it bound only weakly to CD (Fig. 2B, right panel). We confirmed these results by performing GST pull-down assays in a different fashion (Fig. 2C). MCF-7 cell lysates were incubated with the indicated GST beads; after extensive washing, the bound proteins were eluted and the presence of Rb or prohibitin was examined by Western blotting. As shown in Fig. 2C, only GST-HP1γ CSD beads could pull down Rb from MCF-7 cell lysates, while prohibitin was pulled down by both GST-HP1γ CD and GST-HP1γ CSD. Prohibitin thus appears to contact both domains of HP1γ, while Rb interacts preferentially with the chromoshadow domain.
Histone methyltransferases have been implicated in altering chromatin structure in association with Rb and HP1 (39). Hence, a GST pull-down experiment was performed to examine whether prohibitin could bind to Suv39H. As shown in Fig. 2D, 35S-labeled prohibitin synthesized in rabbit reticulocyte lysates could bind to GST-Suv39H, suggesting that it could interact with multiple proteins involved in heterochromatin formation.
HP1γ cooperates with prohibitin to repress E2F-mediated transcription.
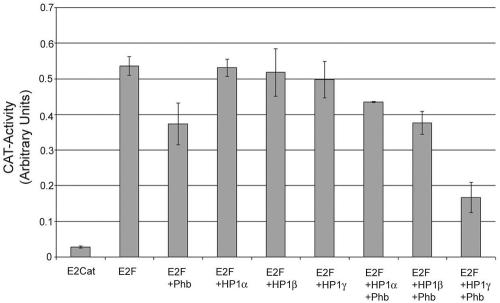
The growth-suppressive effects of prohibitin strongly correlate with its ability to repress E2F-mediated transcription, and prohibitin recruits various transcriptional corepressors like HDAC1, N-CoR, and Sin3A as well as chromatin-remodeling proteins like Brg1 and Brm to repress transcription. Since prohibitin was found to bind to HP1 family members in vitro, whether these proteins could cooperate with prohibitin to repress E2F activity was examined. A transient transfection assay was used for this purpose. MCF-7 cells were transiently transfected with 3 μg each of E2CAT and E2F1. A total of 2 μg of prohibitin, HP1α, HP1β, or HP1γ was cotransfected with E2F1 (Fig. 3). Prohibitin as well as the HP1 family members had negligible repressive effects on prohibitin at these concentrations. To assess the abilities of the three isoforms to cooperate with prohibitin, E2CAT-plus-E2F-plus-Phb-transfected cells were also cotransfected with 2 μg of either HP1α, -β, or -γ. It was found that neither HP1α nor HP1β could increase the repression achieved by prohibitin alone; in contrast, HP1γ could significantly enhance the repressive effects of prohibitin. These results suggest that HP1γ can physically and functionally interact with prohibitin to suppress E2F-mediated transcription.
FIG. 3.
HP1γ cooperates with prohibitin to repress E2F-mediated transcription. MCF-7 cells were transiently transfected with 3 μg of E2CAT, E2CAT+E2F, and E2CAT+E2F+Phb, alone or in combination with HP1α, -β, and -γ. A total of 2 μg prohibitin or HP1 isoforms alone had little repressive effect on E2F1; the combination of prohibitin and HP1γ synergized to repress E2F1 activity significantly. Error bars indicate standard deviations.
Prohibitin associates with HP1 proteins in vivo.
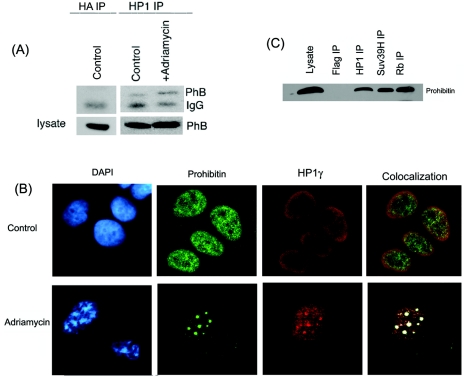
We examined whether the prohibitin-HP1 interaction occurs in vivo; an immunoprecipitation-Western blot analysis was performed to address this. Cell lysates prepared from untreated MCF-7 cells or those treated with adriamycin were immunoprecipitated with a polyclonal antibody to HP1 which recognizes all three isoforms; the presence of prohibitin in the immunoprecipitates was examined by Western blotting using a monoclonal prohibitin antibody. As shown in Fig. 4A, prohibitin was associated with HP1 in control as well as adriamycin-treated cell lysates; however, the binding was more pronounced in the drug-treated lysate, suggesting that the induction of senescence increased the interaction of HP1 with prohibitin in vivo. There was no prohibitin in an immunoprecipitation performed with an antihemagglutinin antibody, which was the negative control.
FIG. 4.
Prohibitin interacts with HP1γ in vivo. (A) Lysates from untreated MCF-7 cells or those treated with adriamycin were immunoprecipitated with a pan-HP1 antibody and Western blotted for prohibitin; immunoprecipitation (IP) with hemagglutinin (HA) antibody was used as negative control. The induction of senescence increased the binding of prohibitin to HP1. (B) A double immunofluorescence experiment on senescent and control MCF-7 cells using a monoclonal antibody against prohibitin and polyclonal antibody against HP1γ. MCF-7 cells were treated with adriamycin (2 μM) for 24 h, washed, and incubated for 48 h in regular growth medium. The slides were fixed and probed with prohibitin and HP1γ antibodies and visualized after staining with goat anti-mouse IgG-Alexa Fluor-488 and goat anti-rabbit IgG-Alexa Fluor-546. It was found that prohibitin colocalizes with HP1γ in senescent cells in discrete SAHF (white and yellow spots), as observed by confocal microsopy. (C) An IP Western blot experiment as described for panel A, showing the interaction of prohibitin with Suv39H in vivo.
Since HP1 could associate with prohibitin in vitro and in vivo, attempts were made to assess whether HP1γ colocalizes with prohibitin in cells. A double immunofluorescence experiment was carried out for this purpose. MCF-7 cells were plated on chamber slides and treated with adriamycin; immunofluorescence was performed using a monoclonal antibody against prohibitin and a polyclonal antibody against HP1γ. The binding of these antibodies was visualized by confocal microscopy following staining with goat anti-mouse IgG-Alexa Fluor-488 and goat anti-rabbit IgG-Alexa Fluor-546 secondary antibodies. Prohibitin and HP1γ (Fig. 4B) were uniformly distributed in the nuclei of control MCF-7 cells; upon the induction of senescence by adriamycin, both proteins localize into specific heterochromatic foci. Interestingly, both proteins were found to colocalize in discrete foci in the senescent cells, as observed by confocal microscopy (Fig. 4B, right panel). It thus appears that HP1γ and prohibitin can physically interact in vitro and in vivo, and they colocalize in specific heterochromatic foci.
Since Suv39H could also bind to prohibitin in vitro, an immunoprecipitation and Western blotting experiment was performed to examine whether Suv39H and prohibitin also associated in vivo. Lysates from MCF-7 cells were immunoprecipitated with Suv39H antibody; HP1 and Rb antibodies were used as positive controls, while a Flag antibody was used as a negative control. Western blotting showed that prohibitin could be detected in Suv39H immunoprecipitate, suggesting that it could indeed bind to Suv39H (Fig. 4C).
Prohibitin and transcriptional corepressors associate with E2F-responsive promoters in senescent cells.
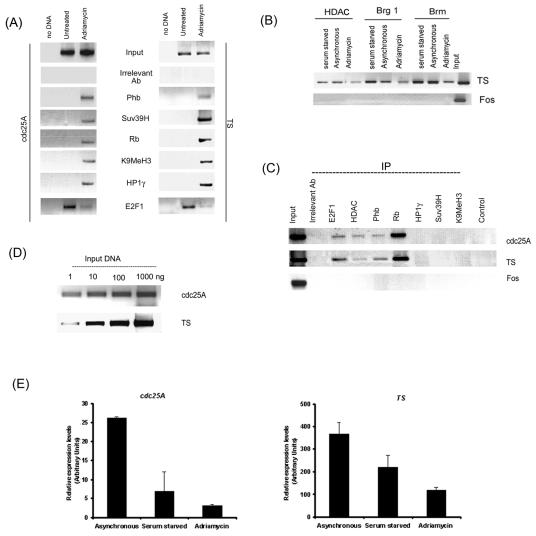
Our lab has shown prohibitin to be recruited to E2F-responsive promoters, and this association correlated with the transcriptional activity of the promoter. Attempts were made to evaluate whether the induction of senescence by DNA-damaging agents also caused its recruitment to E2F-regulated promoters. Chromatin immunoprecipitation assays were used for this purpose. ChIP assay lysates from control MCF-7 cells or those treated with adriamycin were immunoprecipitated using antibodies to HP1γ, Suv39H, K9MeH3, Rb, and prohibitin or an irrelevant antibody as control; their association with proliferative promoters like cdc25A and thymidylate synthase was examined by PCR. The c-fos promoter was used as a control. As shown in Fig. 5A, the binding of Rb, prohibitin, HP1γ, Suv39H, and K9MeH3 to cdc25A and thymidylate synthase promoters was increased upon the induction of senescence. Concurrent with the association of Suv39H, increased methylation of lysine 9 of histone 3 could be observed in the senescent cells; this particular methylation coincides with transcriptional repression. E2F1 was associated with the promoters in asynchronous MCF-7 cells, but this association was lost upon the induction of senescence.
FIG. 5.
(A) ChIP assays showing the recruitment of the indicated proteins as well as K9 methylation of histone H3 on cdc25A and TS promoters upon induction of senescence. The binding of E2F1 was reduced in senescent cells. (B) A ChIP assay showing the recruitment of HDAC1, Brg1, and Brm on cdc25A and TS promoters in asynchronous, quiescent, or senescent cells. (C) A ChIP assay as described for panel B, showing the recruitment of the indicated proteins in serum-starved, quiescent MCF-7 cells. IP, immunoprecipitation. (D) PCR for cdc25A and TS promoters using different amounts of input DNA showing the linear range of the signal. (E) Real-time PCR showing repression of cdc25A and TS in senescent and quiescent cells. TS and Cdc25a were normalized to the average β-actin values for each cDNA sample. Error bars indicate standard deviations.
The association of HDAC1, Brg1, and Brm with the TS promoter was examined in a parallel experiment (Fig. 5B). It was found that a certain amount of HDAC1 was associated with the TS promoter in serum-starved as well as asynchronous cells, but this association was reduced in senescent cells. Brg1 and Brm were associated with the promoter in serum-starved and asynchronous as well as adriamycin-treated cells. Taken together, it appears that the induction of senescence leads to the association of prohibitin, Rb, HP1γ, and Suv39H with the promoters; HDAC1 and Brg/Brm proteins appear to play only a limited role in the senescence process.
A similar experiment was performed with serum-starved MCF-7 cells to ascertain whether the association of Rb, prohibitin, HP1γ, Suv39H, and K9MeH3 is senescence associated and does not occur in quiescent cells (Fig. 5C). The PCR for TS and cdc25A promoters revealed that prohibitin, Rb, and HDAC were bound to the promoters in quiescent cells; at the same time, HP1γ, Suv39H, and K9MeH3 could not be detected. The recruitment of Rb was significantly higher in quiescent cells than in senescent cells, while prohibitin amounts were comparable in both. It thus appears that different transcriptional corepressors are recruited on proliferative promoters upon senescence and quiescence. The PCR conditions that were used amplified the DNA in a linear, dose-dependent manner (Fig. 5D).
Since the ChIP results demonstrated that Rb, prohibitin, HP1, and Suv39H bind to E2F-responsive proliferative promoters cdc25A and TS, we examined whether the binding correlated with the silencing of these genes. Real-time PCRs were conducted to assess their expression levels in the senescent, serum-starved, and asynchronous cells.
The results of real-time PCR for cdc25A and TS levels in RNA from serum-starved, asynchronous, and senescent cells showed that the treatment of cells with adriamycin led to decreased expression of both genes compared to that of asynchronously growing cells, which is in agreement with our ChIP results (Fig. 5E). Though there was a decline in the expression of both genes in serum-starved cells, the regulation of expression in serum-starved and adriamycin-treated cells appears to involve different repressors. These data suggest that the binding of Rb, prohibitin, HP1, and Suv39H on these promoters upon senescence correlates with their repression.
Prohibitin levels influence SA-β-gal activity.
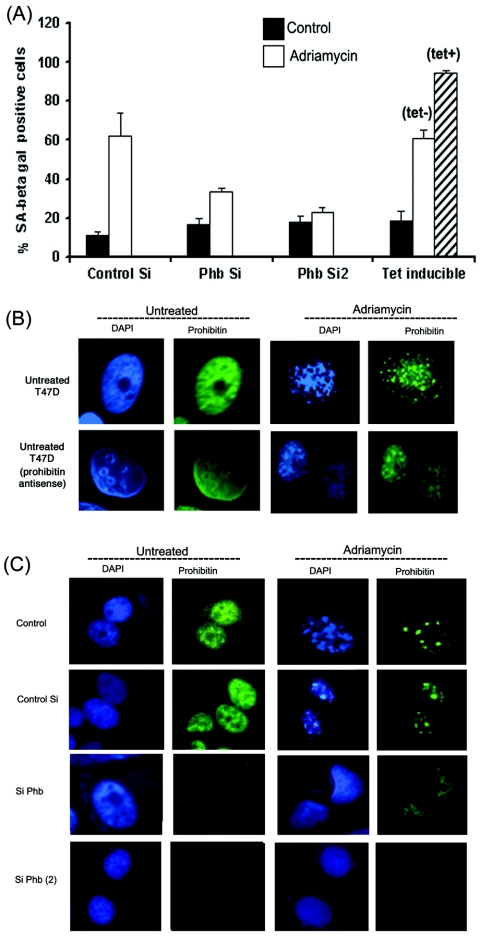
Since the quantitation of foci is difficult, a quantitative assay was used to assess the relative amount of senescing cells. Towards this purpose, control MCF-7 cells, MCF-7 cells expressing a tetracycline-inducible prohibitin, or MCF-7 cells transfected with 1 μg prohibitin siRNA (dicer) or prohibitin siRNA (Santa Cruz Biotechnology) were treated with adriamycin for 24 h. The cells were washed with PBS, fresh medium with 10% FBS was added, and cells were further incubated for 48 h. Subsequently, cells were stained for SA-β-gal activity and positively stained cells were counted under a microscope (Fig. 6A). About 10% of the control siRNA-transfected MCF-7 cells showed positive staining to SA-β-gal; however, treatment with drugs increased it to about 60%. At the same time, only 30% of cells transfected with diced prohibitin siRNA showed SA-β-gal staining upon drug treatment, while only 20% of those transfected with a second Phb siRNA stained positive. Interestingly, almost 100% of cells overexpressing prohibitin showed positive staining for SA-β-gal, while the number of positively stained cells in the absence of tetracycline was comparable to that of control siRNA-transfected cells. Similar results were obtained when senescence was assessed by a colorimetric assay for SA-β-gal (data not shown). These results show that prohibitin can indeed induce senescence and that the absence of prohibitin impairs the ability of the cells to undergo senescence.
FIG. 6.
Prohibitin levels affect senescence in MCF-7 cells. (A) MCF-7 cells transfected with control siRNA, prohibitin siRNA, and MCF-7 cells stably overexpressing tet-inducible prohibitin were treated with adriamycin (2 μM) for 24 h; cells were cultured for 48 h in regular growth medium, fixed, and stained for SA-β-gal. The number of SA-β-gal-positive cells were counted in all the treatments to assess senescence. Error bars indicate standard deviations. (B) Induction of SAHF in control T47D cells (left panels) or those stably expressing antisense prohibitin. The numbers of foci are greatly reduced when prohibitin levels are low, as seen by DAPI staining. (C) An experiment similar to that described for panel B, where SAHF was assessed in MCF-7 cells transfected with siRNA (Si) for prohibitin. SAHF are greatly reduced in cells lacking prohibitin.
Prohibitin levels influence the appearance of heterochromatic foci.
Experiments were done to evaluate whether a reduction in prohibitin levels affects the formation of SAHF. T47D cells stably expressing an antisense prohibitin (19) and MCF-7 cells transfected with siRNA to prohibitin were stained for prohibitin and DNA content. T47D cells stably expressing antisense prohibitin were treated with adriamycin, and cells were immunostained with a monoclonal antibody against prohibitin. Untreated T47D cells stably expressing an antisense prohibitin construct showed reduced expression of prohibitin compared to that of control T47D cells (Fig. 6B). The nuclear staining of DAPI appears to stain whole nucleus evenly in these cells. The drug-treated cells, as shown in Fig. 6B, showed less intense staining for prohibitin, with fewer numbers of foci when stained with DAPI.
The effect of prohibitin levels on senescence was verified using prohibitin siRNA generated by the dicer system and prohibitin siRNA from Santa Cruz Biotechnology (see Fig. 8A). MCF-7 cells were transfected with siRNA; drug treatment was started after 24 h of transfection. As shown in Fig. 6C, a reduction in prohibitin levels led to a significant decrease in foci formation. On an average, about 6.4 ± 2.2 foci were observed per cell on one plane of confocal microscopy; this number was reduced to 1.97 ± 1.47 in cells transfected with Phb siRNA. It thus appears that prohibitin facilitates the formation of SAHF, and the depletion of prohibitin can impair the ability of cells to undergo senescence.
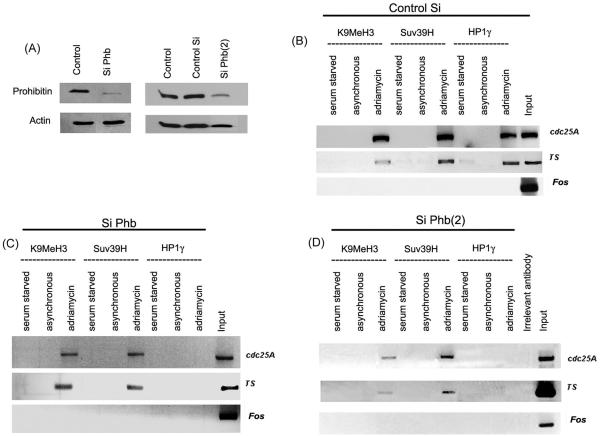
FIG. 8.
(A) Depletion of prohibitin in MCF-7 cells by siRNA, as seen by Western blotting. (B) A ChIP assay showing the recruitment of HP1γ and Suv39H as well as K9 methylation on Cdc25A and TS promoters in MCF-7 cells transfected with control siRNA. (C and D) Depletion of prohibitin by siRNA did not affect the recruitment of Suv39H or K9 methylation upon senescence; the recruitment of HP1γ is completely inhibited. Si, siRNA.
Ablation of prohibitin prevents recruitment of HP1 to SAHF.
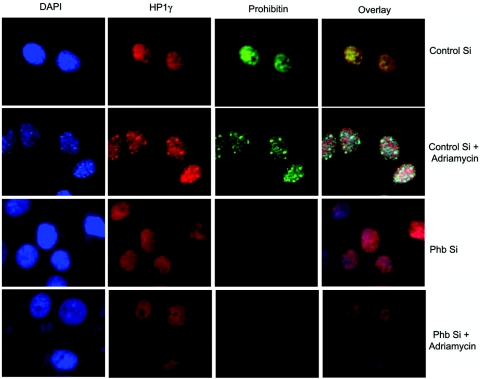
MCF-7 cells transfected with prohibitin siRNA were analyzed to see whether a reduction in the levels of prohibitin affects the recruitment of HP1 to SAHF. Cells were grown on glass chamber slides and transfected with dicer-generated siRNA for prohibitin or control siRNA, and drug treatment was initiated 24 h after transfection. As shown in Fig. 7, the transfection with siRNA to prohibitin led to decreased numbers of foci, as seen by DAPI staining. Interestingly, HP1γ also did not show any punctate staining and was distributed in the nucleus in a granular fashion. This result suggests that that reduced prohibitin levels may be responsible for the nonrecruitment of HP1γ to the SAHF; conversely, it appears likely that prohibitin is necessary for the recruitment of HP1γ to the heterochromatic foci.
FIG. 7.
Depletion of prohibitin by siRNA (Si) reduces SAHF formation and recruitment of HP1γ to heterochromatic regions as seen by double immunofluorescence. The cells were transfected with control siRNA or prohibitin siRNA and treated with adriamycin (2 μM) for 24 h; cells were cultured for 48 h in regular growth medium, fixed, and stained for prohibitin and HP1γ.
Ablation of prohibitin leads to the dissociation of HP1γ from E2F-responsive promoters.
ChIP assays revealed the binding of Rb, prohibitin, HP1γ, Suv39H, and K9MeH3 to the Cdc25A and TS promoters in senescent cells (Fig. 5A). Since the binding of HP1γ, Suv39H, and K9MeH3 is considered to be a senescence-associated event, we examined whether the reduction in prohibitin levels affected the recruitment of these proteins. Towards this purpose, MCF-7 cells were transfected with control siRNA or siRNA to prohibitin and either treated with drugs to induce senescence or serum starved to induce quiescence. ChIP lysates were immunoprecipitated with K9MeH3, Suv39H, and HP1γ antibodies, and their association with cdc25A and TS promoters was assessed by PCR. As shown in Fig. 8C and D, the recruitment of both K9MeH3 and Suv39H upon drug treatment was not affected by Phb siRNA transfection; in stark contrast, HP1γ was not recruited to either of these promoters upon drug treatment, while in control siRNA-transfected cells, HP1γ was recruited to both promoters (Fig. 8B), suggesting that prohibitin is necessary for the recruitment of HP1γ to these promoters upon the induction of senescence. These results strongly suggest that prohibitin facilitates the recruitment of HP1γ to proliferative promoters as well as senescence-associated heterochromatic foci, promoting senescence.
DISCUSSION
Prohibitin was initially cloned on the basis of its ability to induce G1/S arrest (40) and later studies have correlated its growth suppressive properties with its ability to repress E2F transcriptional activity (62-65). While several nuclear and mitochondrial functions have been attributed to prohibitin in the past (21, 62), recent studies also show that it might regulate cell surface functions, acting as a receptor for hitherto unknown ligands (26), and might regulate cell migration in collaboration with Raf-1 (44). A role for prohibitin in cellular senescence has been proposed for human fibroblasts and yeast (29, 43). Studies of human fibroblasts correlated the decreased levels of prohibitin and passage numbers, leading to the hypothesis that prohibitin might be affecting senescence (10); no mechanistic studies were carried out on this observation. Studies described in this paper provide in-depth analyses of the molecular mechanisms underlying senescence induced by prohibitin in mammalian cells.
It has been demonstrated that various environmental stresses and DNA-damaging agents induce cell cycle arrest, leading to STASIS. This growth arrest is similar to replicative senescence since cells remain metabolically and synthetically active and show characteristic changes in morphology and physiology. Our study shows that in addition to SA-β-gal staining, the cells show unique staining patterns for prohibitin as well as DAPI, relative to the cells in which senescence was not induced. This distinct staining pattern was observed in breast cancer cell lines. It was recently reported that senescent cells accumulate a distinct type of heterochromatin, which stain brightly with DAPI and are termed senescent-associated heterochromatic foci (37); the similarity in staining pattern for DAPI in drug-treated cells suggests that the foci formation can occur in cell lines other than fibroblasts. However, an interesting difference in DAPI staining was that the MCF-7 cells did not show single large nucleoli as described by Narita et al. but almost all the drug-treated cells showed irregular nuclear envelopes (37). The cells transfected with siRNA to prohibitin showed decreased foci formation, while those overexpressing prohibitin had higher numbers of foci. While these observations contradict the report from another group that prohibitin might inhibit senescence since its levels are reduced (10), the reduction in protein levels cannot be indicative of a repression of senescence.
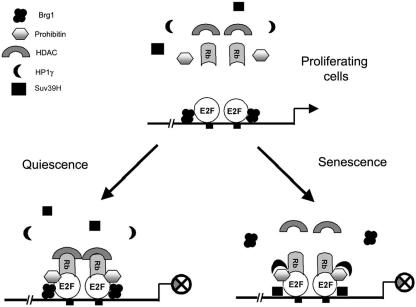
An elegant study from the laboratory of Lowe and coworkers has shown that SAHF are enriched in HP1 family proteins (37), and other studies have shown that HP1 contributes to heterochromatin formation (16). We find that prohibitin interacts with HP1 family members in vitro and in vivo. It has been shown that the association of HP1, Rb, and Suv39H with the cyclin E promoter correlates with gene silencing (39). Our in vitro binding assay reveals that HP1β and -γ show robust binding to prohibitin; at the same time, it appears that the interaction of HP1γ with prohibitin has more profound effects on E2F transcriptional activity than other HP1 family members. It was recently reported that, in senescent cells, Rb acts directly on E2F target promoters to nucleate regions of heterochromatin, leading to the silencing of E2F target genes (37). In addition, Rb was found to interact with HP1 and the histone methyltransferase Suv39H1, which have been shown to be required for maintenance of heterochromatin in mammalian cells (42). Our findings on prohibitin and its association with HP1 suggest that prohibitin might be involved in recruiting HP1γ to the heterochromatic foci and E2F-regulated genes. Based on our findings, we propose the model depicted in Fig. 9. E2F-regulated proliferative promoters are occupied by E2F family members in asynchronous cells; repressors like Rb or prohibitin are not associated. Upon induction of quiescence by serum withdrawal, repressors like Rb, prohibitin, and corepressors like HDAC1 are recruited; corepressors like HP1γ and Suv39H, which are associated with long-term transcriptional repression, are not recruited. In contrast, upon the induction of senescence, the promoters are occupied by Rb, prohibitin, HP1γ, and Suv39H; further, the methylation of lysine 9 of histone 3, a hallmark of transcriptional repression (18), is observed. The ChIP assays also reveal that the depletion of prohibitin prevents the recruitment of HP1γ to the promoters, even though Rb is present. This observation, in conjunction with the finding that depletion of prohibitin prevents the formation of SAHF and the recruitment of HP1γ, leads us to propose that prohibitin plays a major role in recruiting HP1γ to chromatin and inducing senescence.
FIG. 9.
A model showing the recruitment of different corepressors on E2F-regulated promoters in proliferating, quiescent, and senescent cells. In proliferating cells, E2F is bound to its target sites; Brg1/Brm may also be present, but other corepressors are not, and the promoter is transcribed (top panel). Upon induction of quiescence, Rb, prohibitin, HDAC1, and Brg1 are recruited; Suv39H and HP1γ are not (lower left panel). Induction of senescence (lower right panel) leads to the recruitment of Rb, prohibitin, HP1γ, and Suv39H; K9 methylation occurs on histone 3. The depletion of prohibitin prevents the recruitment of HP1γ to the promoter.
These results throw new light on prohibitin function and reveal a new facet in the process of cellular senescence and STASIS. It appears reasonable to imagine that the ability of prohibitin to induce and promote senescence contributes to the proposed tumor suppressive functions of this protein.
Acknowledgments
Thanks go to Scott Lowe and Gideon Grafi for the generous gift of HP1 constructs. The help and suggestions of Sophie Bolick and Jian Wu regarding real-time PCR are appreciated. Support of the Analytical Microscopy Core at Moffitt is gratefully acknowledged.
S.R. is a recipient of the AHA postdoctoral fellowship. This study was funded by a grant from the NCI to S.C. (CA77301).
REFERENCES
- 1.Ait-Si-Ali, S., V. Guasconi, L. Fritsch, H. Yahi, R. Sekhri, I. Naguibneva, P. Robin, F. Cabon, A. Polesskaya, and A. Harel-Bellan. 2004. A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells. EMBO J. 23:605-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcorta, D. A., Y. Xiong, D. Phelps, G. Hannon, D. Beach, and J. C. Barrett. 1996. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc. Natl. Acad. Sci. USA 93:13742-13747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger, K. H., and M. P. Yaffe. 1998. Prohibitin family members interact genetically with mitochondrial inheritance components in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:4043-4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brehm, A., E. A. Miska, D. J. McCance, J. L. Reid, A. J. Bannister, and T. Kouzarides. 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391:597-601. [DOI] [PubMed] [Google Scholar]
- 5.Campisi, J. 2001. Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol. 11:S27-31. [DOI] [PubMed] [Google Scholar]
- 6.Chang, B. D., E. V. Broude, M. Dokmanovic, H. Zhu, A. Ruth, Y. Xuan, E. S. Kandel, E. Lausch, K. Christov, and I. B. Roninson. 1999. A senescence-like phenotype distinguishes tumor cells that undergo terminal proliferation arrest after exposure to anticancer agents. Cancer Res. 59:3761-3767. [PubMed] [Google Scholar]
- 7.Chang, B. D., M. E. Swift, M. Shen, J. Fang, E. V. Broude, and I. B. Roninson. 2002. Molecular determinants of terminal growth arrest induced in tumor cells by a chemotherapeutic agent. Proc. Natl. Acad. Sci. USA 99:389-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheutin, T., A. J. McNairn, T. Jenuwein, D. M. Gilbert, P. B. Singh, and T. Misteli. 2003. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299:721-725. [DOI] [PubMed] [Google Scholar]
- 9.Coates, P. J., D. J. Jamieson, K. Smart, A. R. Prescott, and P. A. Hall. 1997. The prohibitin family of mitochondrial proteins regulate replicative lifespan. Curr. Biol. 7:607-610. [DOI] [PubMed] [Google Scholar]
- 10.Coates, P. J., R. Nenutil, A. McGregor, S. M. Picksley, D. H. Crouch, P. A. Hall, and E. G. Wright. 2001. Mammalian prohibitin proteins respond to mitochondrial stress and decrease during cellular senescence. Exp. Cell Res. 265:262-273. [DOI] [PubMed] [Google Scholar]
- 11.Dell'Orco, R. T., J. K. McClung, E. R. Jupe, and X. T. Liu. 1996. Prohibitin and the senescent phenotype. Exp. Gerontol. 31:245-252. [DOI] [PubMed] [Google Scholar]
- 12.Dimri, G. P., E. Hara, and J. Campisi. 1994. Regulation of two E2F-related genes in presenescent and senescent human fibroblasts. J. Biol. Chem. 269:16180-16186. [PubMed] [Google Scholar]
- 13.Dimri, G. P., X. Lee, G. Basile, M. Acosta, G. Scott, C. Roskelley, E. E. Medrano, M. Linskens, I. Rubelj, O. Pereira-Smith, et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 92:9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dimri, G. P., A. Testori, M. Acosta, and J. Campisi. 1996. Replicative senescence, aging and growth-regulatory transcription factors. Biol. Signals 5:154-162. [DOI] [PubMed] [Google Scholar]
- 15.Drayton, S., and G. Peters. 2002. Immortalisation and transformation revisited. Curr. Opin. Genet. Dev. 12:98-104. [DOI] [PubMed] [Google Scholar]
- 16.Eissenberg, J. C., and S. C. Elgin. 2000. The HP1 protein family: getting a grip on chromatin. Curr. Opin. Genet. Dev. 10:204-210. [DOI] [PubMed] [Google Scholar]
- 17.Elmore, L. W., C. W. Rehder, X. Di, P. A. McChesney, C. K. Jackson-Cook, D. A. Gewirtz, and S. E. Holt. 2002. Adriamycin-induced senescence in breast tumor cells involves functional p53 and telomere dysfunction. J. Biol. Chem. 277:35509-35515. [DOI] [PubMed] [Google Scholar]
- 18.Fuks, F. 2005. DNA methylation and histone modifications: teaming up to silence genes. Curr. Opin. Genet. Dev. 15:490-495. [DOI] [PubMed] [Google Scholar]
- 19.Fusaro, G., P. Dasgupta, S. Rastogi, B. Joshi, and S. Chellappan. 2003. Prohibitin induces the transcriptional activity of p53 and is exported from the nucleus upon apoptotic signaling. J. Biol. Chem. 278:47853-47861. [DOI] [PubMed] [Google Scholar]
- 20.Hayflick, L., and P. S. Moorhead. 1961. The serial cultivation of human diploid cell strains. Exp. Cell Res. 25:585-621. [DOI] [PubMed] [Google Scholar]
- 21.Ikonen, E., K. Fiedler, R. G. Parton, and K. Simons. 1995. Prohibitin, an antiproliferative protein, is localized to mitochondria. FEBS Lett. 358:273-277. [DOI] [PubMed] [Google Scholar]
- 22.Joshi, B., D. Ko, D. Ordonez-Ercan, and S. P. Chellappan. 2003. A putative coiled-coil domain of prohibitin is sufficient to repress E2F1-mediated transcription and induce apoptosis. Biochem. Biophys. Res. Commun. 312:459-466. [DOI] [PubMed] [Google Scholar]
- 23.Jupe, E. R., X. T. Liu, J. L. Kiehlbauch, J. K. McClung, and R. T. Dell'Orco. 1995. Prohibitin antiproliferative activity and lack of heterozygosity in immortalized cell lines. Exp. Cell Res. 218:577-580. [DOI] [PubMed] [Google Scholar]
- 24.Jupe, E. R., X. T. Liu, J. L. Kiehlbauch, J. K. McClung, and R. T. Dell'Orco. 1996. Prohibitin in breast cancer cell lines: loss of antiproliferative activity is linked to 3′ untranslated region mutations. Cell Growth Differ. 7:871-878. [PubMed] [Google Scholar]
- 25.Kamijo, T., F. Zindy, M. F. Roussel, D. E. Quelle, J. R. Downing, R. A. Ashmun, G. Grosveld, and C. J. Sherr. 1997. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91:649-659. [DOI] [PubMed] [Google Scholar]
- 26.Kolonin, M. G., P. K. Saha, L. Chan, R. Pasqualini, and W. Arap. 2004. Reversal of obesity by targeted ablation of adipose tissue. Nat. Med. 10:625-632. [DOI] [PubMed] [Google Scholar]
- 27.Kwak, I. H., H. S. Kim, O. R. Choi, M. S. Ryu, and I. K. Lim. 2004. Nuclear accumulation of globular actin as a cellular senescence marker. Cancer Res. 64:572-580. [DOI] [PubMed] [Google Scholar]
- 28.Li, Y., D. A. Kirschmann, and L. L. Wallrath. 2002. Does heterochromatin protein 1 always follow code? Proc. Natl. Acad. Sci. USA 99(Suppl. 4):16462-16469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, X. T., C. A. Stewart, R. L. King, D. A. Danner, R. T. Dell'Orco, and J. K. McClung. 1994. Prohibitin expression during cellular senescence of human diploid fibroblasts. Biochem. Biophys. Res. Commun. 201:409-414. [DOI] [PubMed] [Google Scholar]
- 30.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 31.Lowe, S. W., and C. J. Sherr. 2003. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr. Opin. Genet. Dev. 13:77-83. [DOI] [PubMed] [Google Scholar]
- 32.Luo, R. X., A. A. Postigo, and D. C. Dean. 1998. Rb interacts with histone deacetylase to repress transcription. Cell 92:463-473. [DOI] [PubMed] [Google Scholar]
- 33.Magnaghi-Jaulin, L., R. Groisman, I. Naguibneva, P. Robin, S. Lorain, J. P. Le Villain, F. Troalen, D. Trouche, and A. Harel-Bellan. 1998. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature 391:601-605. [DOI] [PubMed] [Google Scholar]
- 34.McClung, J. K., E. R. Jupe, X. T. Liu, and R. T. Dell'Orco. 1995. Prohibitin: potential role in senescence, development, and tumor suppression. Exp. Gerontol. 30:99-124. [DOI] [PubMed] [Google Scholar]
- 35.Meloni, A. R., E. J. Smith, and J. R. Nevins. 1999. A mechanism for Rb/p130-mediated transcription repression involving recruitment of the CtBP corepressor. Proc. Natl. Acad. Sci. USA 96:9574-9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narita, M., and S. W. Lowe. 2004. Executing cell senescence. Cell Cycle 3:244-246. [PubMed] [Google Scholar]
- 37.Narita, M., S. Nunez, E. Heard, A. W. Lin, S. A. Hearn, D. L. Spector, G. J. Hannon, and S. W. Lowe. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113:703-716. [DOI] [PubMed] [Google Scholar]
- 38.Nielsen, A. L., M. Oulad-Abdelghani, J. A. Ortiz, E. Remboutsika, P. Chambon, and R. Losson. 2001. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell 7:729-739. [DOI] [PubMed] [Google Scholar]
- 39.Nielsen, S. J., R. Schneider, U. M. Bauer, A. J. Bannister, A. Morrison, D. O'Carroll, R. Firestein, M. Cleary, T. Jenuwein, R. E. Herrera, and T. Kouzarides. 2001. Rb targets histone H3 methylation and HP1 to promoters. Nature 412:561-565. [DOI] [PubMed] [Google Scholar]
- 40.Nuell, M. J., D. A. Stewart, L. Walker, V. Friedman, C. M. Wood, G. A. Owens, J. R. Smith, E. L. Schneider, R. Dell'Orco, C. K. Lumpkin, et al. 1991. Prohibitin, an evolutionarily conserved intracellular protein that blocks DNA synthesis in normal fibroblasts and HeLa cells. Mol. Cell. Biol. 11:1372-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pandita, T. K., and J. L. Roti Roti. 2003. Role of telomerase in radiocurability (review). Oncol. Rep. 10:263-270. [PubMed] [Google Scholar]
- 42.Peters, A. H., D. O'Carroll, H. Scherthan, K. Mechtler, S. Sauer, C. Schofer, K. Weipoltshammer, M. Pagani, M. Lachner, A. Kohlmaier, S. Opravil, M. Doyle, M. Sibilia, and T. Jenuwein. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107:323-337. [DOI] [PubMed] [Google Scholar]
- 43.Piper, P. W., and D. Bringloe. 2002. Loss of prohibitins, though it shortens the replicative life span of yeast cells undergoing division, does not shorten the chronological life span of G0-arrested cells. Mech. Ageing Dev. 123:287-295. [DOI] [PubMed] [Google Scholar]
- 44.Rajalingam, K., C. Wunder, V. Brinkmann, Y. Churin, M. Hekman, C. Sievers, U. R. Rapp, and T. Rudel. 2005. Prohibitin is required for Ras-induced Raf-MEK-ERK activation and epithelial cell migration. Nat. Cell Biol. 7:837-843. [DOI] [PubMed] [Google Scholar]
- 45.Ramirez, R. D., C. P. Morales, B. S. Herbert, J. M. Rohde, C. Passons, J. W. Shay, and W. E. Wright. 2001. Putative telomere-independent mechanisms of replicative aging reflect inadequate growth conditions. Genes Dev. 15:398-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rebbaa, A., X. Zheng, P. M. Chou, and B. L. Mirkin. 2003. Caspase inhibition switches doxorubicin-induced apoptosis to senescence. Oncogene 22:2805-2811. [DOI] [PubMed] [Google Scholar]
- 47.Roskams, A. J., V. Friedman, C. M. Wood, L. Walker, G. A. Owens, D. A. Stewart, M. S. Altus, D. B. Danner, X. T. Liu, and J. K. McClung. 1993. Cell cycle activity and expression of prohibitin mRNA. J. Cell. Physiol. 157:289-295. [DOI] [PubMed] [Google Scholar]
- 48.Ross, J. F., X. Liu, and B. D. Dynlacht. 1999. Mechanism of transcriptional repression of E2F by the retinoblastoma tumor suppressor protein. Mol. Cell 3:195-205. [DOI] [PubMed] [Google Scholar]
- 49.Sage, J., A. L. Miller, P. A. Perez-Mancera, J. M. Wysocki, and T. Jacks. 2003. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature 424:223-228. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 51.Sato, T., H. Saito, J. Swensen, A. Olifant, C. Wood, D. Danner, T. Sakamoto, K. Takita, F. Kasumi, Y. Miki, et al. 1992. The human prohibitin gene located on chromosome 17q21 is mutated in sporadic breast cancer. Cancer Res. 52:1643-1646. [PubMed] [Google Scholar]
- 52.Shay, J. W., and I. B. Roninson. 2004. Hallmarks of senescence in carcinogenesis and cancer therapy. Oncogene 23:2919-2933. [DOI] [PubMed] [Google Scholar]
- 53.Shelton, D. N., E. Chang, P. S. Whittier, D. Choi, and W. D. Funk. 1999. Microarray analysis of replicative senescence. Curr. Biol. 9:939-945. [DOI] [PubMed] [Google Scholar]
- 54.Sherr, C. J. 1998. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 12:2984-2991. [DOI] [PubMed] [Google Scholar]
- 55.Sherr, C. J., and R. A. DePinho. 2000. Cellular senescence: mitotic clock or culture shock? Cell 102:407-410. [DOI] [PubMed] [Google Scholar]
- 56.Steglich, G., W. Neupert, and T. Langer. 1999. Prohibitins regulate membrane protein degradation by the m-AAA protease in mitochondria. Mol. Cell. Biol. 19:3435-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stein, G. H., L. F. Drullinger, A. Soulard, and V. Dulic. 1999. Differential roles for cyclin-dependent kinase inhibitors p21 and p16 in the mechanisms of senescence and differentiation in human fibroblasts. Mol. Cell. Biol. 19:2109-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strober, B. E., J. L. Dunaief, Guha, and S. P. Goff. 1996. Functional interactions between the hBRM/hBRG1 transcriptional activators and the pRB family of proteins. Mol. Cell. Biol. 16:1576-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.te Poele, R. H., A. L. Okorokov, L. Jardine, J. Cummings, and S. P. Joel. 2002. DNA damage is able to induce senescence in tumor cells in vitro and in vivo. Cancer Res. 62:1876-1883. [PubMed] [Google Scholar]
- 60.Trimarchi, J. M., and J. A. Lees. 2002. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 3:11-20. [DOI] [PubMed] [Google Scholar]
- 61.Vandel, L., E. Nicolas, O. Vaute, R. Ferreira, S. Ait-Si-Ali, and D. Trouche. 2001. Transcriptional repression by the retinoblastoma protein through the recruitment of a histone methyltransferase. Mol. Cell. Biol. 21:6484-6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, S., G. Fusaro, J. Padmanabhan, and S. P. Chellappan. 2002. Prohibitin colocalizes with Rb in the nucleus and recruits N-CoR and HDAC1 for transcriptional repression. Oncogene 21:8388-8396. [DOI] [PubMed] [Google Scholar]
- 63.Wang, S., N. Nath, M. Adlam, and S. Chellappan. 1999. Prohibitin, a potential tumor suppressor, interacts with RB and regulates E2F function. Oncogene 18:3501-3510. [DOI] [PubMed] [Google Scholar]
- 64.Wang, S., N. Nath, G. Fusaro, and S. Chellappan. 1999. Rb and prohibitin target distinct regions of E2F1 for repression and respond to different upstream signals. Mol. Cell. Biol. 19:7447-7460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang, S., B. Zhang, and D. V. Faller. 2002. Prohibitin requires Brg-1 and Brm for the repression of E2F and cell growth. EMBO J. 21:3019-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang, H. S., M. Gavin, A. Dahiya, A. A. Postigo, D. Ma, R. X. Luo, J. W. Harbour, and D. C. Dean. 2000. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell 101:79-89. [DOI] [PubMed] [Google Scholar]
- 67.Zheng, X., P. M. Chou, B. L. Mirkin, and A. Rebbaa. 2004. Senescence-initiated reversal of drug resistance: specific role of cathepsin L. Cancer Res. 64:1773-1780. [DOI] [PubMed] [Google Scholar]