Abstract
Isoxaben is a pre-emergence herbicide that inhibits cellulose biosynthesis in higher plants. Two loci identified by isoxaben-resistant mutants (ixr1-1, ixr1-2, and ixr2-1) in Arabidopsis have been reported previously. IXR1 was recently shown to encode the cellulose synthase catalytic subunit CESA3 (W.-R. Scheible, R. Eshed, T. Richmond, D. Delmer, and C. Somerville [2001] Proc Natl Acad Sci USA 98: 10079–10084). Here, we report on the cloning of IXR2, and show that it encodes another cellulose synthase isoform, CESA6. ixr2-1 carries a mutation substituting an amino acid close to the C terminus of CESA6 that is highly conserved among CESA family members. Transformation of wild-type plants with the mutated gene and not with the wild-type gene conferred increased resistance against the herbicide. The simplest interpretation for the existence of these two isoxaben-resistant loci is that CESA3 and CESA6 have redundant functions. However, loss of function procuste1 alleles of CESA6 were previously shown to have a strong growth defect and reduced cellulose content in roots and dark-grown hypocotyls. This indicates that in these mutants, the presence of CESA3 does not compensate for the absence of CESA6 in roots and dark-grown hypocotyls, which argues against redundant functions for CESA3 and CESA6. Together, these observations are compatible with a model in which CESA6 and CESA3 are active as a protein complex.
Cellulose synthesis remains a poorly understood process (for review, see Delmer, 1999). This linear 1,4-β-linked glucan is synthesized in terrestrial plants by a hexameric protein complex referred to as the terminal complex or rosette embedded in the plasma membrane (Kimura et al., 1999). The composition of this complex is unknown. Each of the six particles in the complex is thought to contain several catalytic subunits with an unknown stoichiometry. The relationship between the rosette and the crystalline microfibril structure remains controversial. It is commonly held that one rosette produces one microfibril. This idea was based on calculations based upon the spacing of 1,4-β-glucan chains in algal celluloses and it assumed the presence of 36 glucan chains in a single microfibril (Herth, 1983). Each rosette particle would then contribute six glucan chains to the microfibril. More recent studies using solid-state nuclear magnetic resonance lead to the conclusion that microfibrils in the primary wall consist of 2-nm crystalline units or elementary fibrils, each of which would not contain more than 10 to 15 glucan chains (Ha et al., 1998). The authors propose a scenario in which each of the six particles in the rosette produce the glucan chains of an elementary fibril, which in turn associate into a hexagonal 8- to 10-nm microfibril observed in most primary cell walls.
Genes encoding the cellulose synthase catalytic subunits have been identified in bacteria and plants (Pear et al., 1996). The Arabidopsis genome encodes 10 isoforms of the cellulose synthase catalytic subunit, CESA (http://cellwall.stanford.edu). The strong cellulose-deficient phenotypes observed for mutants in CESA1 (Arioli et al., 1998), CESA7 (Taylor et al., 1999), CESA8 (Taylor et al., 2000), CESA4 (S. Turner, personal communication), or CESA6 (Fagard et al., 2000) indicate specific nonredundant functions for each of these genes. It is interesting that the mutant phenotypes also suggest that CESA isoforms have specialized roles in primary or secondary wall synthesis. Mutants for CESA4, 7, and 8 specifically show a cellulose defect in the secondary wall of the xylem (Taylor et al., 1999, 2000; N. Taylor and S. Turner, personal communication), whereas mutants for CESA1 and 6 have defects in the primary wall (Arioli et al., 1998; Fagard et al., 2000). Furthermore, evidence is accumulating indicating the requirement for more than one isoform in the same cell (Fagard et al., 2000; Taylor et al., 2000), and pull-down experiments suggested that at least CESA7 and CESA8 physically interact and may operate as a heterodimer or multimer (Taylor et al., 2000). Genetic studies finally also indicate a crucial role for a membrane-bound endo-1,4-β-glucanase, KORRIGAN, in the synthesis of cellulose (Nicol et al., 1998; Lane et al., 2001; Sato et al., 2001).
In addition to mutants, chemical inhibitors can be powerful tools for the molecular dissection of biological processes. Several herbicides inhibiting cellulose synthesis have been described such as dichloro-benzonitrile (DCB), isoxaben, and CGA325′615 (Sabba and Vaughn, 1999; Peng et al., 2001). Isoxaben, N-[3(1-ethyl-1-methylpropyl)-5-isoxazolyl] is a pre-emergence, broad leaf herbicide used primarily on small grains, turf, and ornamentals (Huggenberger et al., 1982; Technical report on EL-107 [1987] Lilly Research Laboratories, Indianapolis). It is selectively phytotoxic to dicotyledonous plants, whereas most monocotyledonous species are tolerant. This herbicide is extremely active, with IC50 values in the nanomolar range (Heim et al., 1989). Isoxaben specifically inhibits radioactive Glc incorporation into the acid insoluble cellulosic cell wall fraction (Heim et al., 1990a). Two isoxaben-resistance loci (IXR1 and IXR2) have been described in Arabidopsis (Heim et al., 1989, 1990b).
IXR1 was recently cloned and shown to encode a cellulose synthase catalytic subunit isoform CESA3 (Scheible et al., 2001). Here, we show that isoxaben resistance in ixr2-1 is caused by a mutation in another cellulose synthase isoform, CESA6. This gene was previously identified by knockout mutations (prc1) causing a cellulose-deficient short hypocotyl phenotype (Fagard et al., 2000). These and other data provide new insights into the complexity of the cellulose synthesis machinery in plants.
RESULTS
Phenotype of Isoxaben-Treated Wild-Type and ixr2-1 Seedlings
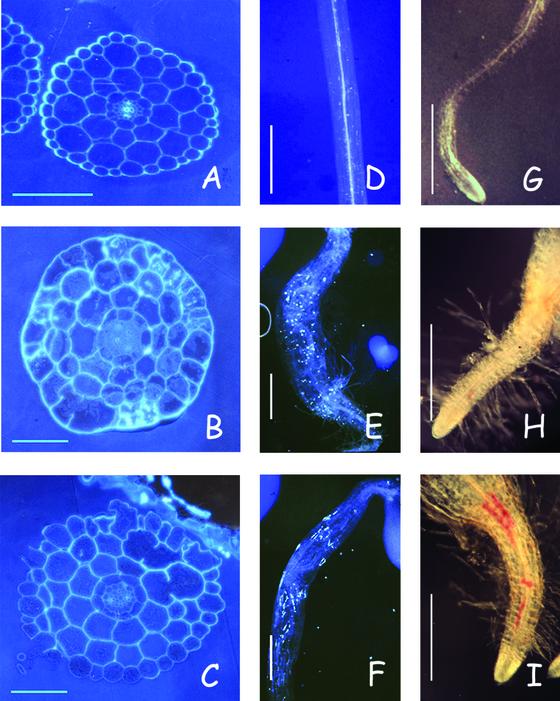
Isoxaben is a potent inhibitor of cellulose synthesis in dicots, including Arabidopsis seedlings (Heim et al., 1990b; unpublished data). Figures 1 and 2 show that isoxaben strongly inhibits hypocotyl and root elongation of dark-grown seedlings. In addition, cells expand radially, callose, lignin, and other phenolic compounds (Fig. 2) accumulate, and seedlings eventually die at concentrations above the IC50 value. In the light, seedlings germinate in the presence of the herbicide, but hypocotyl growth and the development of the root and shoot meristem is blocked (IC50 around 5 nm, data not shown), and seedlings become necrotic and die. The accumulation of callose and lignin is frequently associated with the inhibition of cellulose synthesis as shown for seedlings treated with DCB, another cellulose biosynthesis inhibitor (Fig. 2) or cellulose-deficient mutants (Lukowitz et al., 2001; G. Refrégier, S. Pagant, and H. Höfte, unpublished data). Transverse sections through the hypocotyl of dark-grown seedlings showed the presence of gapped cell walls in isoxaben- or DCB-treated seedlings (Fig. 2). Similar phenotypic characteristics have been described previously for cellulose-deficient mutants (Arioli, et al., 1998; Nicol et al., 1998; Fagard et al., 2000; Lane et al., 2001).
Figure 1.
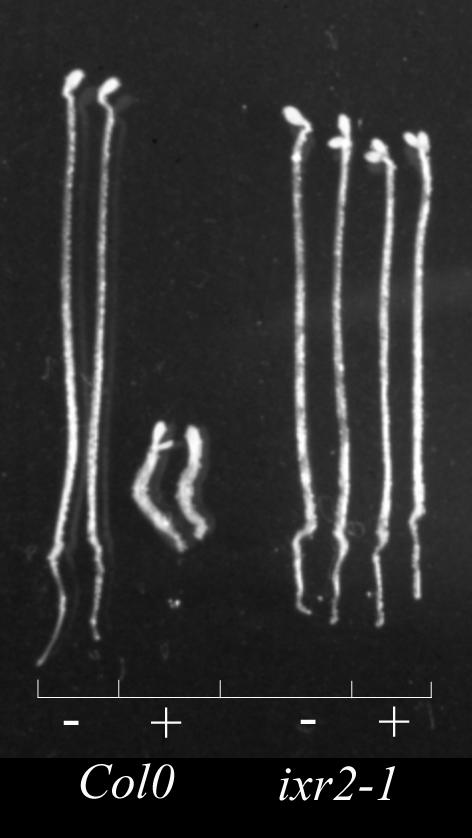
The effect of isoxaben on hypocotyl growth in wild type and ixr2-1. Wild-type Col0 and ixr2-1 seedlings were grown for 4 d in the dark in the absence (−) or presence (+) of 5 nm isoxaben.
Figure 2.
Similar effects of isoxaben and DCB are displayed on hypocotyl and root of wild-type seedlings. A–C, Transverse sections halfway through hypocotyls show gapped walls in seedlings treated with either herbicide. D--F, Hypocotyls stained with sirofluor present an accumulation of callose for both herbicides. G–I, Roots stained with phloroglucinol show an accumulation of lignin for both herbicides, with a stronger staining for DCB. A, D, and G, Control: 4-d-old dark-grown seedlings. B, E, and H, In the presence of 5 nm or (B) 7 nm isoxaben. C, F, and I, In the presence of 0.5 μm or (C) 1.5 μm DCB. Bars in A through C = 100 μm; D through I = 250 μm.
Seedlings homozygous for ixr2-1 showed an increased tolerance to the herbicide as shown by the lack of inhibition of hypocotyl growth of dark-grown seedlings by 5 nm isoxaben (Fig. 1). The same was observed for ixr1-2 homozygotes (Scheible et al., 2001 and data not shown).
ixr2-1 Carries a Missense Mutation in CESA6
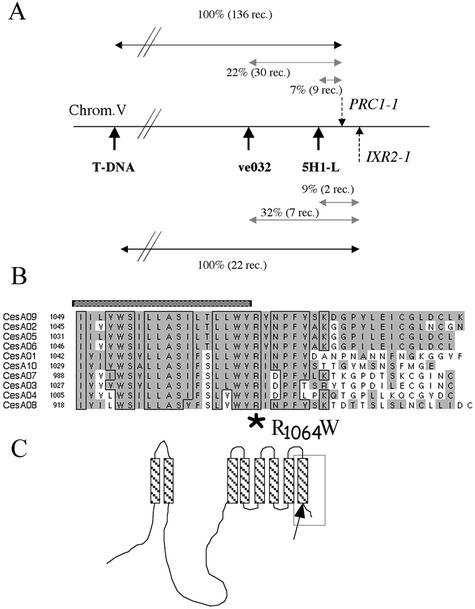
IXR2 was previously shown to map at 0.5 cM from the visual marker yi on the bottom of chromosome 5 (Heim et al., 1990a). Using a cross between a C-24 accession containing a T-DNA insertion north of yi (Van Lijsebettens et al., 1996; Fagard et al., 2000) and ixr2-1, the IXR2 locus was fine mapped (Fig. 3A). It is interesting that IXR2 mapped to the same position as CESA6, a gene encoding a cellulose synthase isoform previously identified by loss of function mutations (prc1) causing a growth defect and cellulose deficiency specifically in roots and etiolated seedlings. Given the fact that low concentrations of isoxaben phenocopy the Prc1− phenotype (Fig. 5; Fagard et al., 2000), the hypothesis that IXR2 is a prc1 allele was tested by sequencing the CESA6 gene in ixr2-1. A single point mutation was detected toward the 3′ end of the coding sequence, causing the amino acid change R1064W. This region of the predicted amino acid sequence is highly conserved between all 10 Arabidopsis CESA genes (Fig. 3B), and the charged residue R1064 delimits the C-terminal end of the 8th predicted transmembrane anchor of CESA6.
Figure 3.
The ixr2-1 mutation causes a R1064W change in a residue conserved for all CESA isoforms in Arabidopsis. A, Fine mapping of IXR2. Twenty-two recombinants were isolated between a T-DNA insertion in a C-24 background, and ixr2-1 in Col0 and recombination breakpoints were mapped using the PCR markers ve032 and 5H1-L. PRC1 was mapped using the same markers on 136 recombinants between prc1-1 in Col0 and the same T-DNA insertion in C-24 (Fagard et al., 2000). The comparison of the maps showed a very close proximity between IXR2 and PRC1. B, Multiple alignment of the C-terminal amino acid sequences of the 10 Arabidopsis CESAs. The numbers indicate the positions of the first amino acid presented in the alignment. Hatched bar indicates the last predicted transmembrane domain. Shaded residues represent a high consensus value of 85%, specified in the program. Boxed residues are identical for at least seven isoforms. The position of the R1064W mutation in the IXR2-1 allele of CESA6 is indicated by the star below the alignment. C, Hypothetical diagram of the membrane topology of CESA proteins. Area in rectangle refers to the part of the sequence shown in B. Predicted transmembrane domains are hatched. The arrow indicates the position of the ixr2-1 mutation.
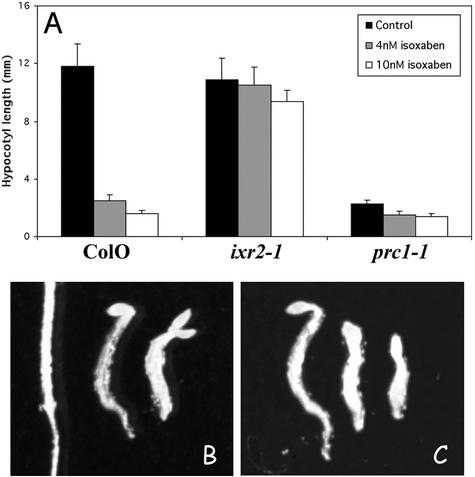
Figure 5.
Evidence for additional isoxaben targets. A, Dark-grown hypocotyl length of 4-d-old seedlings of Col0, ixr2-1 homozygotes, and prc1-1 homozygotes in the absence or presence of 5 or 10 nm isoxaben. B, Seedling phenotype of Col0 grown without (left) and with (middle) 5 nm or (right) 10 nm isoxaben. C, Same conditions as in B, but for prc1-1 seedlings. Note the stronger phenotype for wild-type seedlings grown in the presence of 10 nm isoxaben than that of prc1-1 homozygotes.
Wild-Type Plants Transgenic for IXR2-1 Show Reduced Sensitivity to Isoxaben
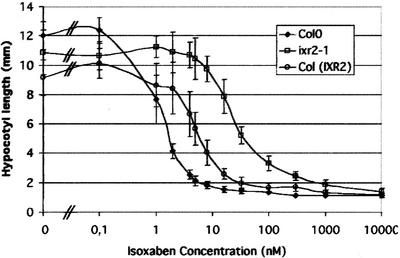
To confirm that the R1064W change is responsible for the isoxaben-resistant phenotype, transgenic Arabidopsis plants were constructed carrying a 12.74-kb genomic clone containing CESA6 with or without the R1064W mutation. This fragment carries a functional PRC1 gene as shown by its ability to complement the prc1 phenotype (data not shown). T2 progenies of four of four transformants carrying the wild-type CESA6 fragment did not show an increased resistance against 5 nm isoxaben (e.g. The length of dark-grown Columbia [Col0] hypocotyls was 11.8 ± 1.6 mm and 2.1 ± 0.3 mm, respectively, in the absence and presence of 5 nm isoxaben. For a T2 family of Col0 transformed with the wild-type CESA6 fragment, hypocotyls measured 12.9 ± 0.9 mm and 2.3 ± 0.3 mm, respectively, in the absence and presence of 5 nm isoxaben. The three other T2 families behaved in the same way). In contrast, T2 progenies of five of five independent transformants carrying the R1064W mutation segregated for an increased isoxaben-resistance phenotype (data not shown). A homozygous T3 line (line 201.3) containing CESA6-R1064W was selected and analyzed more in detail. Measuring the inhibition of hypocotyl growth of dark-grown seedlings is a sensitive way to quantify isoxaben activity. Wild-type seedlings showed a steep dose-response curve with an IC50 of around 1.5 nm (Fig. 4). This result was confirmed by the inhibition of the incorporation of 14C-Glc into the acid-insoluble cellulosic fraction (IC50 around 1 nm, data not shown). The line homozygous for ixr2-1 showed an increased resistance against isoxaben with IC50values 15 times higher than those for the wild-type control (Fig. 4). In the absence of isoxaben, hypocotyls of the line carrying CESA6-R1064W (line 201.3) were, for unknown reasons, slightly shorter than those of wild-type plants. The isoxaben dose-response curve for 201.3 (Fig. 4) revealed a resistance intermediate between that of the wild-type and ixr2 homozygotes (IC50 of 6 nm). Together, these results demonstrate that the R1064W point mutation in CESA6 is sufficient to confer increased isoxaben resistance.
Figure 4.
Transgenic Col0 homozygous for an insertion carrying CESA6-R1064W shows increased resistance to isoxaben. Isoxaben dose-response curve for hypocotyl length of 4-d-old dark-grown seedlings. Curves for wild-type Col0, ixr2-1 homozygotes, and Col0 plants homozygote for CESA6-R1064W [Col(IXR2)] are shown.
Other Functionally Redundant Isoforms of CESA6 Also May Be Isoxaben Targets
All six prc1 alleles sequenced so far contain premature stop codons and are complete loss-of-function alleles of CESA6 (Fagard et al., 2000). The results reported here show that CESA6 is a target for isoxaben. As a result, the Prc1− phenotype is expected to be comparable with that of the wild-type treated with isoxaben. This is only in part true. Dark-grown hypocotyls of prc1-1 were as long as those of the wild type grown on 4 nm isoxaben (Fig. 5). However, higher concentrations of isoxaben further inhibited hypocotyl growth. Isoxaben also had a strong effect on light-grown seedlings (data not shown), whereas aerial parts of prc1-1 grown in the light did not show a detectable phenotype (Fagard et al., 2000). Paradoxically, despite the nonessential role for CESA6 in these conditions, ixr2-1 seedlings were also resistant against isoxaben when grown in the light. These observations indicate that isoxaben recognizes other targets besides CESA6, which are partially or totally redundant with CESA6. This was confirmed by the observation that despite the absence of a growth defect in the light, hypocotyl growth of prc1 was hypersensitive to isoxaben. Hypocotyls of 7-d-old wild-type and prc1-1 seedlings grown in the absence of isoxaben showed comparable lengths (0.67 ± 0.03 cm and 0.63 ± 0.04 cm, respectively, n > 20). In contrast, the hypocotyls of prc1-1 seedlings grown for 7 d on 3 nm isoxaben were significantly shorter than those of the wild-type controls (0.32 ± 0.02 cm versus 0.62 ± 0.03 cm, respectively, n > 20).
DISCUSSION
IXR2 encodes CESA6
The following evidence shows that resistance against isoxaben in the ixr2-1 mutant is caused by a mutation in CESA6. First, fine mapping showed a colocalization of IXR2-1 with PRC1, which encodes CESA6. Second, CESA6 in ixr2-1 carried an amino acid change, R1064W. Third, transformation of a genomic fragment carrying the CESA6 gene with this mutation, but not with the wild-type control, conferred increased isoxaben-resistance to wild-type plants.
The observed reduced resistance level of homozygotes for the mutant transgene compared with ixr2-1 homozygotes is not surprising. The semidominant nature of the mutation has been reported previously, with ixr2-1 heterozygotes showing a 10-fold higher IC50 value than homozygotes (Heim et al., 1990a). The same observation was reported for ixr1 mutants. In addition, ixr1 homozygotes transformed with the wild-type gene showed a lower resistance to isoxaben than nontransformed ixr1 controls (Scheible et al., 2001). This observation is consistent with the idea that multiple copies of CESA6 and/or CESA3 constitute the rosette complex. The different catalytic subunits within a rosette would coordinately synthesize the different glucan chains that constitute the microfibril. It is not clear how the activity of the subunits is coordinated, but it can be expected that the presence of even a few isoxaben-sensitive subunits within a, for the rest resistant, complex may lead to the poisoning of the entire complex in the presence of the herbicide.
Redundancy within the CESA Gene Family
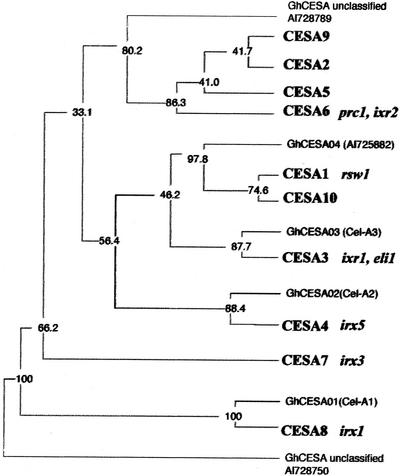
It is important to know the function of all 10 CESA isoforms in Arabidopsis (Fig. 6). Most isoforms appear to have an evolutionary conserved, specialized function, as shown by the presence of orthologs in unrelated plant species such as cotton (Fig. 6). Mutants rsw1 (Arioli et al., 1998) and prc1 (Fagard et al., 2000) have a strong seedling phenotype, indicating that the encoded isoforms (CESA1 and CESA6, respectively) are required for cellulose synthesis in the primary wall. Mutants for CESA7 (irx3), CESA8 (irx1), and CESA4 (irx5; Taylor et al., 1999, 2000; N. Taylor and S. Turner, personal communication) are characterized by their collapsed xylem phenotype, and the isoforms appear to be specialized for cellulose synthesis during secondary wall formation. The very similar dark-grown seedling phenotype for rsw1 and prc1 (Fagard et al., 2000) suggests that at least in the hypocotyl, the two encoded isoforms are required in the same cell types and have nonredundant functions. The same was observed for irx1, 3, and 5 in xylem cells (Taylor et al., 1999, 2000; Taylor, 2001). Expression in the same cell types was further suggested for CESA1 and CESA3 using promoter-β-glucuronidase fusion constructs (Scheible et al., 2001). The requirement for several isoforms in the same cell type may indicate the coexistence of several specialized cellulose synthase complexes or the presence of one or more isoforms in the same complex. The latter idea was supported by pull-down experiments showing that CESA7 and CESA8 physically interact in vitro.
Figure 6.
Members of the CESA family are functionally specialized as shown by loss-of-function and isoxaben-resistant alleles. A phylogenetic tree based on a hypervariable region among CESA protein sequences is shown (Fagard et al., 2000). Sequences from Arabidopsis (in bold) were aligned with those from cotton (Gossypium hirsutum), and a parsimonious consensus tree was constructed. Numbers are bootstrapped values (n = 100). Corresponding mutant alleles are indicated in italics and mutant phenotypes are discussed in the text. Clustering of one or more specific Arabidopsis sequences with cotton homologs suggests a functional specialization of the corresponding isoforms. CESA6 clusters with three other Arabidopsis isoforms, suggesting that they have arisen by more recent gene duplications.
The cloning of ixr1 and ixr2 provides additional functional information for the encoded isoforms. The existence of two resistance loci on first sight would suggest redundant roles for the corresponding proteins. However, the strong phenotype observed for dark-grown hypocotyls of prc1 suggests that CESA3 is absent in this organ or that CESA3 is present but unable to compensate for the absence of CESA6 in the mutant. Although CESA3 protein has not yet been detected directly, two lines of evidence strongly suggest that CESA3 is present in dark-grown hypocotyls. First, CESA3 transcripts are constitutively expressed (Scheible et al., 2001), including in dark-grown and light-grown seedlings (A. Van Tuinen, T. Desprez, and H. Höfte, unpublished data), and second, ixr1 hypocotyl growth in the dark was also resistant against isoxaben. Given the fact that ixr1 mutations do not alter the expression pattern of CESA3 transcripts (Scheible et al., 2001), it is reasonable to conclude that in dark-grown hypocotyls, the function of CESA3 is not redundant with that of CESA6. The absence of redundancy is further suggested by the strong phenotype observed for dark-grown seedlings carrying recessive loss of function mutations in CESA3 (M. Bevan, personal communication), This leads to the interesting possibility that at least in dark-grown hypocotyls, CESA3 and CESA6 are active as a complex. This is similar to the complex reported for CESA7 and CESA8 in cells synthesizing secondary walls. In this scenario, isoxaben may bind directly, or via another protein, at the interface between the two monomers or may recognize a conformation requiring the presence of both wild-type isoforms.
A closer examination of the Prc1− phenotype, however, shows that in the absence of CESA6, other targets for isoxaben exist in dark-grown hypocotyls. Complete knockout mutants for CESA6 showed a milder phenotype than wild-type plants treated with 10 nm isoxaben, a concentration that did not significantly affect growth in ixr2-1. This residual isoxaben-sensitive CESA activity may correspond to CESA3, which in that case would only partially replace CESA6 in the prc1 mutant, but it may also correspond to other CESA isoforms. More strikingly, aerial parts of prc1 mutants grown in the light did not show a growth phenotype, but were nevertheless hypersensitive to isoxaben action. Therefore, other CESA isoforms must have redundant roles with CESA6 in these conditions. In theory, this may be CESA3, but this would imply that the specificity of CESA3 is altered in light because CESA3 was unable to completely replace CESA6 in dark-grown prc1 seedlings. In an alternate manner, isoforms CESA2, 5, and 9, which are highly similar to CESA6, may be redundant with CESA6 and constitute other isoxaben targets. As shown by expressed sequence tag data, CESA2 and CESA5 mRNAs are also expressed in dark- and light-grown seedlings, but at lower levels than CESA6 (http://cellwall.stanford.edu). CESA9 has a very low expression level as shown by the single expressed sequence tag that was found in a library from rosette leaves. The exact function of these three isoforms remains to be determined and will require the isolation of the corresponding mutants.
The genetic data indicate that CESA1 is also essential for cellulose synthesis, including in dark-grown hypocotyls (Fagard et al., 2000). From this, it can be inferred that CESA1 is still active in the presence of isoxaben in an ixr2 or ixr1 background, which would suggest that CESA1 is not an isoxaben target. However, it is not formally excluded that isoxaben recognizes a conformation that requires the association of not only CESA3 and CESA6, but also of CESA1, and that mutations in any of these proteins may destabilize the complex. These two possibilities can be now distinguished by expressing CESA1 versions carrying their equivalent of the ixr2-1, ixr1-1, or ixr1-2 mutations in transgenic plants and assessing the resistance against isoxaben.
The R1064W mutation in ixr2-1 removes a positive charge at the end of the last predicted membrane spanning domain. This may cause a dramatic conformational change associated with a change in the membrane topology of the protein, which may prevent the binding of isoxaben or isoxaben-associated protein(s). ixr1-1 and ixr1-2 also carry mutations close to the C terminus of CESA3 (Scheible et al., 2001), suggesting again that this region is targeted by the herbicide. It is unlikely, however, that the amino acids mutated in the ixr1 and ixr2 alleles fold together in a single isoxaben-binding site. According to the predicted topology, R1064W in IXR2-1, and G998D in IXR1-1 would be on the opposite side of the membrane compared with T942I in IXR1-2. The prediction of the topology is not correct or one has to assume that the mutations all induce a conformational change of the protein complex, leading to the loss of isoxaben binding. In the absence of a comprehensive three-dimensional structure for the cellulose synthase complex, it is not clear how isoxaben interferes with the synthesis of cellulose. In ixr2 and ixr1 alleles, the mutated amino acid is far downstream from the active site residues, which makes it unlikely that the compound directly interferes with the catalytic site. In an alternate manner, the extrusion of the glucan chain supposedly requires a membrane channel that may be formed by the transmembrane domains. Isoxaben could block such a channel. Isoxaben binding could interfere with the interaction between CESA subunits and in this way destabilize the hexameric rosette or each particle that constitutes the rosette. A destabilizing effect on the rosettes was also suggested for the herbicide CGA325′615 (Peng et al., 2001).
In conclusion, the identification of the isoxaben target CESA6 provides further insights in the complexity of the cellulose synthesis machinery and the role of the 10 cellulose synthase isoforms. Indirect evidence for complex formation between CESA3 and CESA6 is provided and partial redundancy between CESA6 and other CESA isoforms, possibly CESA2, CESA5, or both. With the targets characterized, isoxaben becomes an excellent tool for the study of cellulose synthesis in Arabidopsis, and also in other plant species.
MATERIALS AND METHODS
Plant Material and Growth Conditions
prc1-1(Desnos et al., 1996), ixr1-2 (DH48), ixr2-1 (DH1; kindly provided by the Arabidopsis Biological Resource Center), and wild-type Arabidopsis plants were of the Col0 ecotype. A KanR5 line used for ixr2-1 map-based cloning that bears a T-DNA insertion close to marker LFY3 (Van Lijsebettens et al., 1996) at the bottom of chromosome 5 (Lister and Dean, 1993) was of C24 ecotype. Seedlings were grown in dark conditions as described in Fagard et al. (2000).
Measurement of Hypocotyl Length
Growth of seedlings was arrested by addition of an aqueous solution of 0.4% (w/v) formaldehyde. Seedlings were spread on agar plates and an image was captured using a digital camera. The lengths of hypocotyls were measured using image analysis software (Optimas 5.2; IMASYS, Surennes, France) as described in Gendreau et al. (1997).
Cross Sections and Calcofluor Staining
For light microscopy, seedlings were fixed in 4% (w/v) paraformaldehyde and 0.2% (w/v) glutaraldehyde in phosphate-buffered saline buffer and were embedded in historesin (Technovit 7100; Kulzer, Wehrheim, Germany) following the manufacturer's instructions. Sections 3 μm thick were cut using a microtome (Jung RM2055; Leica, Wetzlar, Germany). The cross sections were stained with a 0.005% (w/v) aqueous solution of Calcofluor (Fluorescent Brightener 28; Sigma, St. Louis) for 2 min and they were visualized under UV light using a microscope (microphot FXA; Nikon, Tokyo).
Callose and Lignin Staining and Microscopic Observations
Plants were grown in vitro for 4 d in dark conditions on a medium containing 1% (w/v) Suc and 5 nm isoxaben or 0.5 μm DCB. Seedlings were directly spread on slides and were gently squashed under the coverslip before adding the staining components between the slide and the coverslip. Callose staining was done with Aniline Blue Fluorochrome or sirofluor (Biosupplies Australia, Parkville, Victoria, Australia) at a concentration of 0.1 mg mL−1. Slides were kept in the dark for 30 to 120 min before observation under UV light.
Pholoroglucinol in hydrochloric solution (Prolabo, Fontenay, France) was used for lignin staining and was observed under white light with Nomarski optics.
Mapping of the ixr2 Mutation
The ixr2 mutant (Col0 ecotype; Heim et al., 1990a) was crossed to a T-DNA line (C24 ecotype) bearing an insertion close to RI marker LFY3. The F1 hybrid was allowed to self-pollinate and the progeny was collected. To identify seedlings homozygous for the ixr2 mutation, the F2 progeny was plated directly on 6 nm isoxaben-containing growth medium and cultured in the dark for 4 d. In these conditions, it is possible to distinguish homozygous ixr2/ixr2, heterozygous ixr2/IXR2, and homozygous wild-type (IXR2/IXR2) seedlings (data not shown). Homozygous mutant seedlings (ixr2/ixr2) were then transferred to fresh medium containing 50 mg L−1 kanamycin. After 10 d, 22 kanamycin-resistant seedlings were transferred to greenhouse and were considered recombinant between the ixr2-1 mutation and the T-DNA insertion. Genomic DNA was extracted from leaves and flower buds as described previously (Bouchez and Camilleri, 1998). These recombinant seedlings were further analyzed using the markers ve032 and 5H1-L generated during the map-based cloning of the CESA6 gene (Fagard et al., 2000).
DNA Construction and Plant Transformation
Standard molecular cloning techniques were performed as described by Sambrook et al. (1989). A 6.4-kb EcoRI-HindIII DNA fragment of the P1 clone MVP7 (accession no. AB025637) was cloned into the site EcoRI-HindIII of the binary vector pDE 1001 containing a kanamycin resistance marker (Denecke et al., 1992) to obtain the pDE-EHMVP7 clone. A 6.3-kb HindIII DNA fragment of the P1 clone MVP7 was introduced into the HindIII site of pDE-EHMVP7 to obtain the pDE-CESA6 clone.
A PCR reaction was performed using the oligonucleotides TGGTTATGGAGGTGGGTTGA, forward, and ATTTTCAATTTAGAAGACCGCAT, reverse, that surrounds the ixr2-1 mutation. The resulting 2.6-kb DNA fragment was cleaved with SalI-XbaI enzymes, and the digested fragment was cloned into the SalI-XbaI sites of pDE-CESA6, giving rise to the pDE-CESA6-R1064W clone. The binary T-DNA constructs were mobilized into Agrobacterium tumefaciens C58 Pmp90. The pDE-CESA6 and pDE-CESA6-R1064W constructs were introduced into Col0 seedlings using the infiltration protocol described by Bechtold and Pelletier (1998). The T1 transformants were selected on a kanamycin-containing medium (50 mg L−1). The T2 transformants were obtained after selfing T1 kanamycin-resistant seedlings. Four Col0(CESA6) T2 transformants were analyzed on a kanamycin-containing medium, and the line 206 carrying an unique transgene was selected for the analyses.
Five Col0(CESA6-R1064W) = Col(IXR2) T2 transformants were tested for their isoxaben resistance. The T2 transformants were plated on 10−9 m isoxaben-containing medium in dark conditions so that the wild-type seedlings were classified as sensitive. We observed a segregating population representing homozygous and heterozygous ixr2 states. The F3 progeny of the most resistant line (line 201.3) was obtained and it was 100% kanamycin-resistant, confirming the homozygous state of the unique transgene.
DNA Sequencing
Genomic DNA was extracted from adult seedlings grown in normal greenhouse conditions using standard protocol. PCR fragments covering the entire coding sequence were generated and sequenced by Genome Express (Meylan, France).
The ixr2 mutation was detected in the PCR fragment amplified with the specific primers TGGTTATGGAGGTGGGTTGA, forward, and ACGTGGCACAATATGGCTGA, reverse.
Alignments and Phylogenetic Tree
The alignment presented in the Figure 3 was obtained with the Multalign program (http://www.toulouse.inra.fr/multalin.html). For multiple alignment, selected sequences (from the last transmembrane segment through the end of each AtCESA) were combined in SeqVu. A high consensus value of 85% was chosen in the program. For the dendrogram (Fig. 6), sequences from the HVR2 domains corresponding to amino acids 690 and 743 in CESA6 were aligned with the program ClustalW (http://www.infobiogen.fr/services/analyseq/cgi-bin/clustalw_in.pl). An additional sequence, which is more divergent, was inserted as an outlier (AI727450). This alignment was analyzed by PROTPARS to construct a consensus tree from 100 bootstrapped data sets.
ACKNOWLEDGMENTS
We thank Jocelyne Kronenberger for excellent technical assistance and the Ohio Arabidopsis Biological Resource Center for providing ixr1-2 and ixr2-1 seeds. We also thank Michael Bevan and Simon Turner for allowing us to cite their unpublished data.
Footnotes
This work was financed in part by the Ministère de la Recherche et de Technology (grants to M.F., G.R., and T.D.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010822.
LITERATURE CITED
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R et al. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science. 1998;279:717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thalianaplants by vacuum infiltration. Methods Mol Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- Bouchez D, Camilleri C. High molecular weight DNA extraction from Arabidopsis. Methods Mol Biol. 1998;82:61–70. doi: 10.1385/0-89603-391-0:61. [DOI] [PubMed] [Google Scholar]
- Delmer D. Cellulose biosynthesis: exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:245–276. doi: 10.1146/annurev.arplant.50.1.245. [DOI] [PubMed] [Google Scholar]
- Denecke J, De Rycke R, Botterman J. Plant and mammalian sorting signals for protein retention in the endoplasmic reticulum contain a conserved epitope. EMBO J. 1992;11:2345–2355. doi: 10.1002/j.1460-2075.1992.tb05294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnos T, Orbovic V, Bellini C, Kronenberger J, Caboche M, Traas J, Höfte H. Procuste1 mutants identify two distinct genetic pathways controlling hypocotyl cell elongation, respectively in dark- and light-grown Arabidopsisseedlings. Development. 1996;122:683–693. doi: 10.1242/dev.122.2.683. [DOI] [PubMed] [Google Scholar]
- Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, McCann M, Rayon C, Vernhettes S, Höfte H. PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell. 2000;12:2409–2423. doi: 10.1105/tpc.12.12.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau E, Traas J, Desnos T, Grandjean O, Caboche M, Höfte H. Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 1997;114:295–305. doi: 10.1104/pp.114.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M-A, Apperley DC, Evans BW, Huxham IM, Jardine WG, Viëtor RJ, Reis D, Vian B, Jarvis MC. Fine structure in cellulose microfibrils: NMR evidence from onion and quince. Plant J. 1998;16:183–190. doi: 10.1046/j.1365-313x.1998.00291.x. [DOI] [PubMed] [Google Scholar]
- Heim DR, Roberts JL, Pike PD, Larrinua IM. Mutation of a locus of Arabidopsis thalianaconfers resistance to the herbicide isoxaben. Plant Physiol. 1989;90:146–150. doi: 10.1104/pp.90.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim DR, Roberts JL, Pike PD, Larrinua IM. A second locus, Ixr B1 in Arabidopsis thaliana, that confers resistance to herbicide isoxaben. Plant Physiol. 1990a;92:858–861. doi: 10.1104/pp.92.3.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim DR, Skomp JR, Tschabold EE, Larrinua I. Isoxaben inhibits the synthesis of acid-insoluble cell wall materials in Arabidopsis thaliana. Plant Physiol. 1990b;93:695–700. doi: 10.1104/pp.93.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim DR, Skomp JR, Waldron C, Larrinua IM. Differential response to isoxaben of cellulose biosynthesis by wild-type and resistance strains of Arabidopsis thaliana. Pestic Biochem Physiol. 1991;39:93–99. [Google Scholar]
- Herth W. Arrays of plasma-membrane “rosettes” involved in cellulose microfibril formation of Spirogyra. Planta. 1983;159:347–356. doi: 10.1007/BF00393174. [DOI] [PubMed] [Google Scholar]
- Huggenberger F, Jennings EA, Ryan PJ, Burow KW. Isoxaben, a new selective herbicide for use in cereals. Weeds. 1982;1:47. [Google Scholar]
- Kimura S, Laosinchai W, Itoh T, Cui X, Linder R, Brown R. Immunogold labeling of rosette terminal cellulose-synthase complexes in the vascular plant Vigna angularis. Plant Cell. 1999;11:2075–2085. doi: 10.1105/tpc.11.11.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DR, Wiedemeier A, Peng L, Höfte H, Vernhettes S, Desprez T, Hocart CH, Birch RJ, Baskin TI, Burn JE et al. Temperature-sensitive alleles of rsw2 link the korrigan endo-1,4-β-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol. 2001;126:278–288. doi: 10.1104/pp.126.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister C, Dean C. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 1993;4:745–750. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Nickle TC, Meinke DW, Last RL, Conklin PL, Somerville CR. Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. Proc Natl Acad Sci USA. 2001;98:2262–2267. doi: 10.1073/pnas.051625798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol F, His I, Jauneau A, Vernhettes S, Canut H, Höfte H. A plasma membrane-bound putative endo-1,4-β-d-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 1998;17:5563–5576. doi: 10.1093/emboj/17.19.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA. 1996;93:12637–12642. doi: 10.1073/pnas.93.22.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Xiang F, Roberts E, Kawagoe Y, Greve LC, Kreuz K, Delmer DP. The experimental herbicide CGA 325′615 inhibits synthesis of crystalline cellulose and causes accumulation of non-crystalline β-1,4-glucan associated with CesA protein. Plant Physiol. 2001;126:981–992. doi: 10.1104/pp.126.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabba RP, Vaughn KC. Herbicides that inhibit cellulose biosynthesis. Weed Sci. 1999;47:757–763. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sato S, Kato T, Kakegawa K, Ishii T, Liu YG, Awano T, Takabe K, Nishiyama Y, Kuga S, Nakamura Y et al. Role of the putative membrane-bound endo-1,4-β-glucanase KORRIGAN in cell elongation and cellulose synthesis in Arabidopsis thaliana. Plant Cell Physiol. 2001;42:251–263. doi: 10.1093/pcp/pce045. [DOI] [PubMed] [Google Scholar]
- Scheible WR, Eshed R, Richmond T, Delmer D, Somerville C. Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in ArabidopsisIxr1 mutants. Proc Natl Acad Sci USA. 2001;98:10079–10084. doi: 10.1073/pnas.191361598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor N. Molecular genetic analysis of cellulose synthesis in Arabidopsis: Conference proceedings of plant protein club workshop: added value products from plants 2: cell wall components. Presented at University of York, March 21–23, 2001. 2001. p. 29. [Google Scholar]
- Taylor NG, Laurie S, Turner SR. Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell. 2000;12:2529–2540. doi: 10.1105/tpc.12.12.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Scheible WR, Cutler S, Somerville CR, Turner SR. The irregular xylem3 locus of Arabidopsisencodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell. 1999;11:769–780. doi: 10.1105/tpc.11.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lijsebettens M, Wang X, Cnops G, Boerjan W, Desnos T, Höfte H, Van Montagu M. Transgenic Arabidopsistester lines with dominant marker genes. Mol Gen Genet. 1996;251:365–372. doi: 10.1007/BF02172528. [DOI] [PubMed] [Google Scholar]