Abstract
Galectin 3 (Gal-3), a member of the β-galactoside binding lectin family, exhibits antiapoptotic functions, and its aberrant expression is involved in various aspects of tumor progression. Here we show that p53-induced apoptosis is associated with transcriptional repression of Gal-3. Previously, it has been reported that phosphorylation of p53 at Ser46 is important for transcription of proapoptotic genes and induction of apoptosis and that homeodomain-interacting protein kinase 2 (HIPK2) is specifically involved in these functions. We show that HIPK2 cooperates with p53 in Gal-3 repression and that this cooperation requires HIPK2 kinase activity. Gene-specific RNA interference demonstrates that HIPK2 is essential for repression of Gal-3 upon induction of p53-dependent apoptosis. Furthermore, expression of a nonrepressible Gal-3 prevents HIPK2- and p53-induced apoptosis. These results reveal a new apoptotic pathway induced by HIPK2-activated p53 and requiring repression of the antiapoptotic factor Gal-3.
Gal-3 is a β-galactoside-specific lectin belonging to a large family of carbohydrate-binding proteins, the galectins, characterized by the presence of at least one carbohydrate recognition domain. Gal-3 possesses a chimeric structure that together with the carbohydrate recognition domain comprises a repetitive collagen-like sequence and a short N-terminal domain regulated by posttranslational modifications (5, 23, 56). Gal-3 is widely expressed in epithelial and immune cells while an aberrant expression is present in a large number of human cancers (reviewed in references 37 and 38). Indeed, in the case of thyroid tumors, Gal-3 overexpression is regarded as a marker of malignancy, and it is currently used in the clinical practice for the preoperative characterization of thyroid nodules (6, 39). At the functional level, Gal-3 is involved in different biological events including pre-mRNA processing, cell adhesion, cell cycle regulation, and apoptosis (14, 36, 53, 54). The role in apoptosis is the most characterized and distinguishes Gal-3 from the other galectins that, when involved in this process, possess proapoptotic activities (37, 38). Two sites in the N-terminal tail of Gal-3 were shown to be relevant for its antiapoptotic activity: the Bcl-2 homology domain 1 with a functional NWGR (Asp-Trp-Gly-Arg) antideath motif (1) is responsible for the inhibition of cytochrome c release from mitochondria (58), and the phosphorylation of the casein kinase 1 target Ser6 promotes Gal-3 nuclear export and antiapoptotic function (50). Considerable evidence has accumulated showing that Gal-3 inhibits apoptosis induced by different stresses such as anti-Fas antibody, DNA damaging drugs, and anoikis (1, 34, 53). In mice, targeted disruption of the gal-3 gene sensitizes macrophages to apoptotic stimuli (27, 38), strongly supporting the antiapoptotic role of this galectin. Surprisingly, it is still unclear whether apoptosis requires Gal-3 inactivation to take place.
The wild-type p53 (wtp53) protein is the product of a tumor suppressor gene and it functions as a master regulator of apoptosis. It is a sequence-specific transcription factor that regulates the expression of genes involved in cell cycle arrest or apoptosis in response to a variety of genotoxic damage or cell stress (51), including the apoptotic stimuli that are counteracted by Gal-3. The apoptotic activity of p53 can be regulated independently of the cell cycle arrest function, and several proteins have been identified that are necessary for p53 to mediate the apoptotic response (2). Among such proteins, we have recently identified homeodomain-interacting protein kinase 2 (HIPK2), a Ser/Thr kinase that binds to and activates p53 by phosphorylating it at Ser46 (18, 26). This specific posttranslational modification was shown to be relevant for the induction of p53-mediated apoptosis (43). Indeed, it has been proposed that severe, irreparable DNA damage, which would require cell suicide rather than growth arrest, promotes phosphorylation at Ser46 and a following change in the p53 affinity for different promoters, with a shift from growth arrest-related genes to apoptosis-related ones (9, 43).
HIPK2 was originally discovered as a member of a novel family of Ser/Thr kinases that act as corepressor for homeodomain transcription factors (13, 35). HIPK2 interacts with the carboxy terminus of p53, colocalizes with p53 and PML-3 into the nuclear bodies, and cooperates in the activation of p53-dependent transcription (18, 26). HIPK2 phosphorylates p53 at Ser46 upon induction of severe DNA damage by UV irradiation, doxorubicin, or cisplatin treatments (16, 18, 26, 42). HIPK2 was also shown to induce apoptosis by targeting factors other than p53, such as the CtBP transcriptional corepressor (59), and to modulate the activity of other proteins directly or indirectly related to apoptosis, such as the p53 family members p73 and p63 (33) and the p53 inhibitor MDM2 (15, 52). Thus, increasing evidence points to HIPK2 as an important regulator of apoptosis.
Here, we questioned whether inactivation of the antiapoptotic factor Gal-3 is associated with and is relevant for p53-induced apoptosis. Our present results show that (i) Gal-3 expression is repressed during UV-induced p53-mediated apoptosis, (ii) p53 inhibits Gal-3 expression by repressing its transcription, (iii) HIPK2-induced activation of p53 is required for Gal-3 repression, and (iv) Gal-3 repression is required for p53-induced apoptosis. Altogether, these results indicate that Gal-3, like Bcl-2, belongs to the p53-target genes that are transcriptionally repressed and that this repression strongly contributes to induction of p53-mediated apoptosis.
MATERIALS AND METHODS
Cell culture and treatments.
RKO, H1299, HEK293, and HCT116 human cell lines were maintained and transfected as previously described (18). Human HCT116 colorectal adenocarcinoma cells and the p53−/− derivatives were kindly provided by B. Vogelstein (10). Mouse fibroblasts (MEFs) were obtained and maintained as previously described (45).
The following expression vectors were employed: pCAG3.1, pCAG3.1-wtp53, pCAG3.1-p53S46A (47) (kindly provided by E. Appella), pRSV-p53Val135 (41) (kindly provided by M. Oren), pLp53H175SP, pLp53H273SP, and pLp53S220SP (11), pLXSP, pLHIPK2SP, pLK221RSP, pCMVFlag2B, pCMV-HIPK2Flag, pCMV-HIPK2-K221Rflag, pEGFP-C2, pEGFP-HIPK2, pEGFP-HIPK2-K221R (18), pcDNA3.1/zeo(+) (Invitrogen), and pcDNA3.1/galectin-3 (57) (kindly provided by S. Nakahara and H. Inohara).
The recombinant adenovirus dl70.3 and its Adp53 derivative (4) (kindly provided by S. Bacchetti and F. Graham) were amplified, titrated, and used for infection of adherent cells at the indicated multiplicity of infection (MOI), as previously described (45).
For DNA damage, subconfluent cells were irradiated with 50 or 100 J/m2 of UV light and collected at the indicated time points for further analyses. Fibroblasts were shifted to low-serum medium (1% fetal calf serum) upon irradiation to facilitate apoptosis.
Cytofluorimetric analysis of cellular DNA content was performed on propidium iodide (PI)-stained cells by an Epics XL analyzer (Coulter Corp.).
Western blot analysis.
Total cell extracts (TCEs) were prepared and analyzed as previously described (18). The following antibodies were used in immunoblotting: rabbit anti-p53 antiserum (FL-393; Santa Cruz Biotechnology), rabbit anti-HIPK2 antiserum (kindly provided by L. Schmitz), purified rat monoclonal antibody (MAb) anti-galectin-3 (Mabtech), mouse anti-PARP-1 [where PARP is poly(ADP-ribose) polymerase] MAb (PharMingen International), mouse anti-FLAG and mouse antiactin MAbs (Sigma), mouse anti-α-tubulin MAb (Immunological Sciences), and horseradish peroxidase-conjugated goat anti-mouse, anti-rabbit, or anti-rat antibodies (Cappel). Immunoreactivity was detected by an ECL kit (Amersham Corp.).
RNA extraction and real-time reverse transcription-PCR (real-time RT-PCR).
RNA was extracted from cells using the SV Total RNA isolation system (Promega), following the manufacturer's instruction. cDNA was synthesized in a total volume of 20 μl containing 250 ng of total RNA, using Omniscript reverse transcriptase (QIAGEN). The SYBR Green DNA Master mix (Applied Biosystems) was employed for real-time PCR analysis with the Applied Biosystems 7500 system SDS software. The 18S RNA was used to normalize the amount of total RNA present in each reaction with the following primers for both human and mouse cDNA: 18S-431 forward, 5′-GGAGAGGGAGCCTGAGAAA-3′, and 18S-544 reverse, 5′-CGAAAGAGTCCTGTATTGTTATTTT-3′, as previously described (40). The primer set for Gal-3 was as follows: human Gal-3 forward, 5′-TCCACTTTAACCCACGCTTC-3′; human Gal-3 reverse, 5′-TCTTCCCTTCCCCAGTTATT-3′; mouse Gal-3 forward, 5′-GCTGGAGTTACAGGTGGTTG-3′; and mouse Gal-3 reverse, 5′-GGAGGCATCAGTGGACCT-3′. Each target amplification was performed in duplicate on two different RNA preparations.
Chromatin immunoprecipitation (ChIP).
Proliferating or UV-irradiated, subconfluent cells were cross-linked by adding formaldehyde directly to culture medium. DNA preparation, immunoprecipitation, and amplification were performed as previously described (7). Immunoprecipitation was carried out with anti-p53 sheep polyclonal antibody, and its relative normal sheep serum was used as a negative control (Ab-7 and NSS, respectively; Calbiochem), or anti-NF-YA rabbit polyclonal antibody (item 200-401-100; Rockland). Two primer pairs that amplify 297- and 154-bp-long DNA fragments of the human LGALS3 promoter were selected for PCR amplification of the immunoprecipitated chromatin (see Fig. S1A in the supplemental material). The primers for the control promoters were previously described elsewhere (28). DNA amplification was achieved by employing AmpliTaq Gold DNA polymerase (Perkin Elmer). PCR products were resolved onto a 3% agarose gel composed of NuSieve and SeaKem (FMC Bioproducts) (3:1, wt/wt) and ethidium bromide (0.5 μg/ml) and visualized under UV light.
Cloning and functional analysis of the 5′ flanking region of the human LGALS3 gene.
A 977-bp DNA fragment including 836 bp of the 5′-flanking region, exon 1, and 58 bp of intron 1 of the human LGALS3 gene (see Fig. S1A in the supplemental material) was amplified from genomic DNA of human thyroid cancer cells by PCR in the presence of specific chimeric primers containing extensions of either KpnI or BglII restriction sites. PCRs were performed using Herculase DNA polymerase (Stratagene) in the presence of 250 ng of template genomic DNA. PCR products were gel purified by QIAquick gel extraction kit (QIAGEN), digested with KpnI and BglII, and ligated into the promoterless luciferase (Luc) reporter plasmid pGL-3 basic (Promega). The p(−836/+141)LGALS3-Luc construct generated was analyzed by direct sequencing and compared with the sequence reported in the GenBank database (accession no. AF031421) (30). A series of primers (see Fig. S1A in the supplemental material) was used for PCR amplification of 5′ progressively deleted DNA fragments of the LGALS3 promoter; the products were subcloned into the same pGL-3 basic vector as described for the larger fragment to obtain the following vectors: p(−672/+141)LGALS3-Luc, p(−472/+141)LGALS3-Luc, p(−272/+141)LGALS3-Luc, and p(−13/+141)LGALS3-Luc, each carrying the indicated regions relative to the +1 transcription start site (TSS) described by Kadrofske et al. (30).
Determination of gene reporter activity.
Proliferating H1299 cells plated on 24-well plates were transiently transfected with different expression and reporter vectors by Superfect reagent (QIAGEN). For each experiment, at least three independent transfections were performed. The pRL plasmid (Promega) coding for the Renilla luciferase under the control of a thymidine kinase promoter was cotransfected with the experimental vectors at a 1:200 molar ratio and used as an internal control for both transfection and the luciferase assay. After 24 h, TCEs were prepared with passive lysis buffer (Promega), and luciferase activity was measured by a dual-Luc reporter assay system (Promega) using a TD20/20 double injector luminometer (Turner Designs), as previously described (49). Results were expressed as relative Luc activity, calculated as follows: relative light units = luminescence of experimental vector/luminescence of empty vector. Each luminescence value reported in the formula was normalized with the Renilla luciferase activity as follows: luminescence = firefly Luc activity/Renilla Luc activity.
For stable integration of the reporter vectors, H1299 cells were transfected by Superfect with each of the reporter vectors together with the vector carrying the puromycin resistance gene pLXSP at a 1:20 molar ratio. After 10 days of selection in the presence of 2 μg/ml puromycin (Sigma), mixed populations were induced to express wtp53 by Adp53 infection. The dl70.3 virus was used as a negative control. Equal amounts of TCEs were analyzed for luciferase activity that was calculated as follows: Luc activity = luminescence of experimental vector/microgram of TCE.
Plasmid construction for RNA interference and stable cell transfection.
The pSUPER-LacZ, pSUPER-p53, and pSUPER-HIPK2 plasmids were constructed as previously reported (8, 16). Western blot analysis for STAT-1 protein was used to exclude stimulation of interferon production in each stable transfected cell line. The p53 sequence tested by Brummelkamp et al. (8) was employed. For HIPK2 interference, six different sequences were originally tested, three of which deplete HIPK2 and induce resistance to UV-induced apoptosis to similar extents. Here, the following sequences were used without regard to cell type, with the exception of murine F9 cells, where only the first sequence can be used because it recognizes both human and mouse HIPK2 (conserved motifs are underlined): 5′-GATCCCCGAAAGTACATTTTCAACTGTTCAAGAGACAGTTGAAAATGTACTTTCTTTTTGGAAA-3′ (HIPK2-1376 sense), 5′-AGCTTTTCCAAAAAGAAAAGTACATTTTCAACGTCTCTTGAACAGTTGAAAATGTACTTTCGGG-3′ (HIPK2-1376 antisense), 5′-GATCCCCGAACCACACGTGCTTGGTCTTCAAGAGAGACCAAGCACGTGTGGTTCTTTTTGGAAA-3′ (HIpk2-789 sense), and 5′-AGCTTTTCCAAAAAGAACCACACGTGCTTGGTCTCTCTTGAAGACCAAGCACGTGTGGTTCGGG-3′ (HIPK2-789 antisense).
For Gal-3 interference three different sequences were tested. The two following sequences, which strongly and similarly downregulated Gal-3 expression, were subcloned into pSUPER vector and used interchangeably (conserved motifs are underlined): 5′-GATCCCCCAACAGGAGAGTCATTGTTTTCAAGAGAAACAATGACTCTCCTGTTGTTTTTGGAAA-3′ (Gal3-551, sense), 5′-AGCTTTTCCAAAAACAACAGGAGAGTCATTGTTTCTCTTGAAAACAATGACTCTCCTGTTGGGG-3′ (Gal3-551, antisense), 5′-GATCCCCACCTTACATGTGTAAAGGTTTCAAGAGAACCTTTACACATGTAAGGTTTTTTGGAAA-3′ (Gal3-845, sense), and 5′-AGCTTTTCCAAAAAACCTTACATGTGTAAAGGTTCTCTTGAAACCTTTACACATGTAAGGTGGG-3′ (Gal3-845, antisense).
The pSUPER-LacZ vector for the bacterial lacZ gene was used as control and the cells transfected with this vector were named ctr.
RESULTS
p53-induced apoptosis is associated with repression of Gal-3.
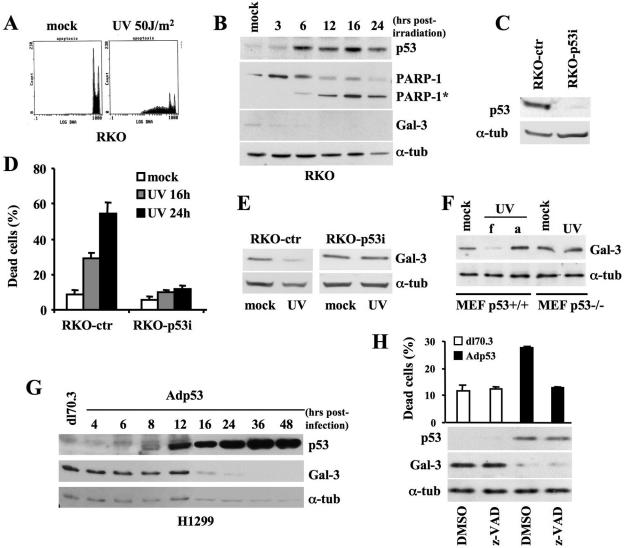
To investigate whether induction of apoptosis is associated with inhibition of the antiapoptotic factor Gal-3, we treated the wtp53-expressing RKO cells with an apoptotic dose of UV irradiation (50 J/m2) (18). As expected, cell cycle analysis of PI-stained cells showed the appearance of a substantial hypo-diploid peak, indicative of cell death (Fig. 1A). Furthermore, Western blot analysis performed at five different time points after UV irradiation (Fig. 1B) revealed the cleavage of PARP-1, a diagnostic marker of apoptosis (22), and the stabilization of p53, a mediator of UV-induced apoptosis (9). Reduction of Gal-3 expression coincided with p53 induction and preceded the PARP-1 cleavage, suggesting that downregulation of Gal-3 is associated with UV-induced apoptosis.
FIG. 1.
Gal-3 expression is repressed in p53-mediated apoptosis. (A) RKO cells were exposed to UV (50 J/m2) and collected 24 h later for cell cycle analysis upon PI staining. DNA-content analysis by flow cytometry of one indicative experiment is reported. (B) Immunoblotting kinetics of p53, PARP-1 and its cleaved form (PARP-1*), and Gal-3 at the indicated times after UV irradiation (50 J/m2). α-Tubulin expression is used as a loading control. (C) Immunoblots with antibody to p53 on RKO cells stably transfected with pSUPER-ctr and pSUPER-p53 vectors. α-Tubulin was used as a loading control. (D) Percentage of dead cells determined by trypan blue exclusion in the indicated cells at the indicated times after UV irradiation. Means ± standard deviations of three independent experiments are reported. (E) Immunoblot of Gal-3 on the indicated cells after UV irradiation. α-Tubulin was used as a loading control. (F) Immunoblot of Gal-3 48 h after UV irradiation of MEF from p53+/+ and p53−/− mice. Due to the small amount of apoptosis in these primary cells, the UV-irradiated MEFs were harvested, with the dead, floating cells (f) kept separate from the adherent ones (a). In the case of p53−/− MEFs, only adherent cells were present. (G) Immunoblotting kinetics of p53 and Gal-3 after infection of p53-null H1299 cells with Adp53 recombinant adenovirus or dl70.3 control virus at an MOI of 50. α-Tubulin was used as a loading control. (H) H1299 cells were infected as described in panel G in the presence of the pan-caspase inhibitor z-VAD or its solvent. Percentage of dead cells was calculated as described for panel D at 24 h postinfection when cells were harvested for immunoblot analyses, as described for panel G. α-tub, α-tubulin.
To verify whether p53 is directly involved in the reduction of Gal-3 expression, RKO cells were depleted of p53 by stable transfection with the pSUPER-p53 vector (8) (Fig. 1C). As expected UV irradiation caused significant death in control cells (RKO-ctr), while the p53 depleted RKO-p53i cells were resistant, as shown by the low rate of cell death (Fig. 1D) and by the cell cycle profiles (data not shown). Western blot analysis showed no reduction of Gal-3 expression in the cells lacking p53 proteins (Fig. 1E), indicating that UV irradiation per se is not sufficient to downregulate Gal-3 and that p53 might be a mediator of Gal-3 downregulation in apoptotic conditions.
To confirm this hypothesis, p53+/+ and p53−/− MEFs were UV irradiated in the presence of low serum. A strong downregulation of Gal-3 was associated only with the presence of dead cells in the p53+/+ populations (Fig. 1F), supporting the p53 involvement in Gal-3 repression. Interestingly, the surviving p53+/+ MEFs that remain adherent to the dishes had no reduction of Gal-3 (compare lanes f and a in Fig. 1F), further sustaining the hypothesis that Gal-3 repression is associated to apoptosis. As a further control, p53-null H1299 cells were induced to express apoptotic levels of wtp53 (see Fig. 4C for apoptosis assessment) by infection with recombinant adenoviruses (i.e., dl70.3 control virus or wtp53-carrying Adp53). Time course analysis showed that increased accumulation of wtp53 was followed by Gal-3 downregulation starting, in these experimental conditions, 16 h postinfection (Fig. 1G).
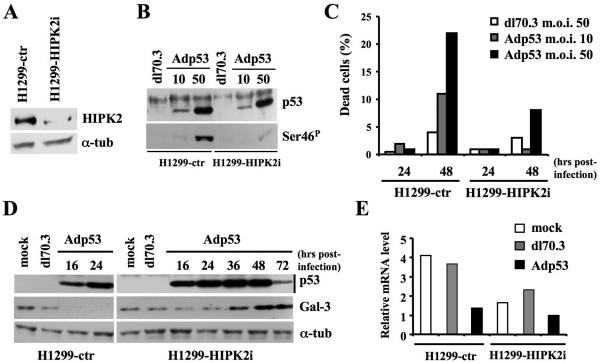
FIG. 4.
p53 requires the presence of HIPK2 to repress Gal-3 expression. (A) Immunoblots with Ab to HIPK2 on H1299 cells stably transfected with pSUPER-ctr or pSUPER-HIPK2 vectors. α-Tubulin was used as a loading control. (B) The cells described in panel A were infected with Adp53 at the indicated MOIs or with dl70.3 at an MOI of 50. TCEs were prepared 24 h postinfection and analyzed by Western blotting for total p53 expression and p53 phosphorylation at Ser46 (Ser-46P). (C) Percentage of dead cells determined by trypan blue exclusion after infection with Adp53 or dl70.3 adenoviruses at the indicated doses and times. One representative of four independent experiments is reported. (D) Immunoblotting kinetics of p53 and Gal-3 after infection of the indicated cells at the indicated times with Adp53 recombinant adenovirus or dl70.3 control virus at an MOI of 50. α-Tubulin was used as a loading control. (E) gal-3 mRNA expression by real-time RT-PCR was analyzed on the indicated cells. Infection was performed as described for panel D, and RNA collected at 24 h postinfection. α-tub, α-tubulin; NSS, normal sheep serum.
To rule out the possibility that Gal-3 repression is a secondary effect of cell death, H1299 cells were infected as described above and incubated in the presence or absence of the pan-caspase inhibitor z-VAD to prevent cell death. As shown in Fig. 1H, z-VAD abolished the p53-induced apoptosis but did not rescue Gal-3 repression, indicating that p53 still represses Gal-3 even when cell death is prevented.
Altogether, these results show that induction of p53-mediated apoptosis correlates with a strong repression of the antiapoptotic factor Gal-3.
p53 represses Gal-3 at the transcription level.
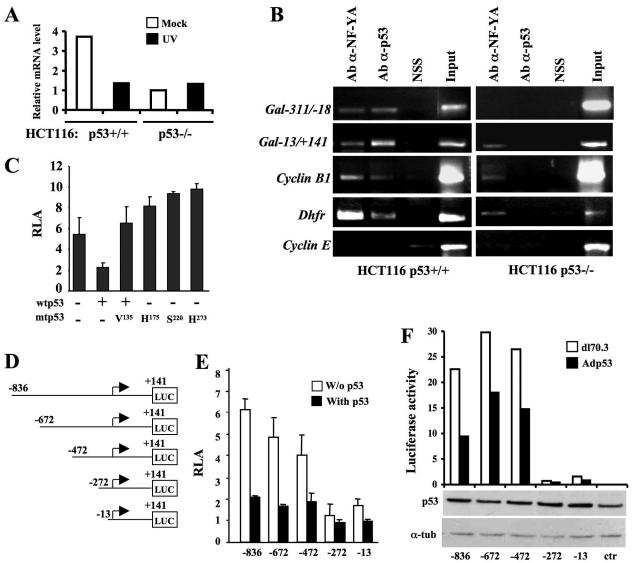
p53 promotes apoptosis through transcriptional and nontranscriptional mechanisms (51). To discriminate between these two mechanisms in Gal-3 regulation, RNA was extracted from isogenic p53+/+ and the p53−/− HCT116 cells (10) in mock conditions and 16 h postirradiation. Different levels of Gal-3 mRNA were present in the two cell lines in mock conditions for reasons that are presently unknown. Nevertheless, after UV irradiation, a strong reduction in the Gal-3 mRNA level was observed only in the p53+/+ HCT116 cells (Fig. 2A), indicating the existence of a p55-mediated regulation at the level of transcription or RNA stability of the LGALS3 gene (i.e., the Gal-3 protein encoding gene).
FIG. 2.
p53 represses Gal-3 at the transcriptional level. (A) Analysis of Gal-3 mRNA expression. UV irradiation was performed as described in the legend of Fig. 1A, and RNA was collected from mock-infected cells and at 16 h postirradiation. Real-time RT-PCR was performed using the 18S RNA as a control. (B) Cross-linked chromatin from the indicated cells was immunoprecipitated with sheep antibody to p53 (Ab-α-p53), normal sheep serum, or rabbit antibody to NF-YA (Ab-α-NF-YA) and analyzed by PCR with primers specific for the indicated promoters. Input corresponds to nonimmunoprecipitated cross-linked chromatin. (C) LGALS3 promoter activity. Coexpression studies were performed in human H1299 cells using the p(−836/+141)LGALS3-Luc reporter construct, together with wtp53, dnp53, or the indicated mutant p53. Means ± standard deviations of at least seven independent experiments are reported. (D) Schematic representation of the human LGALS3 promoter fragments cloned upstream of the firefly luciferase reporter gene in the pGL-3-Basic vector. (E) Luciferase activity of the reporter vectors shown in panel D and indicated with the most-5′ base of the LGALS3 promoter sequence cloned into the relative vectors, in the presence or absence of wtp53. Means ± standard deviations of at least seven independent experiments are reported. (F) Luciferase activity of the same reporter vectors stably transfected into H1299 cells. Each mixed population, indicated as in panel E was infected at an MOI of 50 with dl70.3 control or Adp53 virus. The expression of exogenous p53 in each mixed population is shown by the immunoblot. One indicative experiment out of three performed is reported. α-tub, α-tubulin; NSS, normal sheep serum.
Next, we attempted in vivo detection of p53 on the LGALS3 promoter by performing ChIP assays with anti-p53 Ab. Cross-linked chromatin from p53+/+ and p53−/− HCT116 cells (see Fig. S1B in the supplemental material) was employed to ensure the specificity of the immunoprecipitation. Two primer pairs (see Fig. S1A in the supplemental material) that amplify the −311 to −18 (−311/−18) and the −13/+141 regions of the LGALS3 promoter, relative to the TSS, were employed. The presence of two NF-Y consensus sequences proximal to the aforementioned amplified regions (see Fig. S1A in the supplemental material) led us to use an anti-NF-YA Ab as a positive control (28). As a further control, we employed the cyclin B1 and Dhfr promoters that are bound independently by NF-Y and p53 and the cyclin E promoter that is not bound by either protein (28). As shown in Fig. 2B, the 297- and the 154-bp fragments of the LGALS3 promoter were present in the p53 immunoprecipitations from p53+/+ HCT116 cells but not from their p53−/− counterparts, indicating that p53 is present in vivo in the complexes bound to this promoter and suggesting that regulation occurs at the level of transcription.
To directly test p53 activity on the LGALS3 promoter, we cloned the genomic fragment of the human LGALS3 gene encompassing nucleotides −836 to + 141, previously shown to possess significant promoter activity (30), into the pGL-3 basic reporter vector. Transient transfection experiments and dual luciferase assays were performed in p53-null H1299 cells. As expected, this genomic fragment retains clear promoter activity when transfected alone into the cells (Fig. 2C). We then analyzed the effects of p53 on this promoter by expressing wtp53 with or without the p53Val135 mutant that possesses p53 dominant-negative (dnp53) function (41). Expression of wtp53 markedly inhibited LGALS3 promoter activity (Fig. 2C) in a dose-dependent manner (see Fig. S1C in the supplemental material) while coexpression of wtp53 and dnp53 completely abolished the wtp53 inhibitory effect (Fig. 2C). Furthermore, three different p53 tumor-derived mutants that lost the wild-type transcriptional activity (24) were unable to repress the LGALS3 promoter (Fig. 2C), strongly indicating that wtp53 is specifically involved in this repression.
To try to identify the promoter region responsible for p53 repression, we examined a series of 5′ progressively deleted LGALS3 promoter constructs (Fig. 2D) upon their transient or stable transfection into H1299 cells. Comparable results were obtained with the two transfection systems. Indeed, in agreement with previous studies (30), a dramatic reduction of luciferase activity, independently of p53, was observed in the p(−272/+141)LGALS3-Luc and p(−13/+141)LGALS3-Luc constructs (Fig. 2E and F; see Fig. S1D in the supplemental material for the absolute counts), indicating the presence of relevant positive transcription responsive elements in the −472/−272 region. In addition, all vectors were repressed by wtp53 expression including the p(−272/+141)LGALS3-Luc and p(−13/+141)LGALS3-Luc that, despite their low basal activity, were reproducibly and differentially repressed by wtp53, with the p(−272/+141)LGALS3-Luc vector being less repressed than the shorter one (Fig. 2E and F; see Fig. S1D in the supplemental material for the absolute counts). This suggests the existence of both positive and negative regulation in the same region. Nevertheless, altogether these results show that p53 downregulates Gal-3 by repressing its promoter activity.
HIPK2-mediated phosphorylation of p53 at Ser46 promotes Gal-3 downregulation.
Transcriptional regulation of p53-target genes depends on p53 posttranslational modifications including the apoptosis-specific phosphorylation at Ser46 (9, 43). To test whether Gal-3 downregulation depends on phosphorylation of p53 at Ser46, p53-null H1299 cells were transiently transfected with expression vectors encoding wtp53 or the p53(S46A) mutant that cannot be phosphorylated (9). Transient expression of p53 mutants with a single change of Ser to Ala is frequently reported to induce biological outcomes similar to those of wtp53 (3). Thus, a small amount of expression vectors was transfected. Although under this experimental condition the effect on Gal-3 downregulation was less dramatic than that occurring upon recombinant adenovirus infection, a significant difference was observed between wtp53 and p53(S46A) mutant expression (Fig. 3A), suggesting that Ser46 phosphorylation is functionally involved in Gal-3 downregulation.
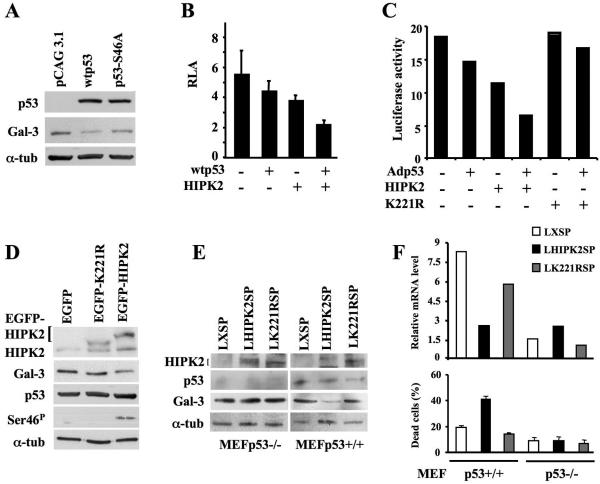
FIG. 3.
p53 phosphorylation at Ser46 by HIPK2 is required for p53-mediated repression of Gal-3. (A) Immunoblots with antibodies to p53 and Gal-3 on H1299 cells transfected with pCAG3.1 control, pCAG3.1-wtp53, or pCAG3.1-p53S46A mutant vector. α-Tubulin was used as a loading control. (B) Coexpression studies were performed in human H1299 cells by transfecting the p(−836/+141)LGALS3-Luc reporter vector with wtp53 and/or HIPK2 expression vectors. Means ± standard deviations of at least seven independent experiments are reported. (C) Luciferase activity of the same p(−836/+141)LGALS3-Luc reporter vector stably transfected into H1299 cells. The polyclonal population was infected with Adp53 at MOI of 10 and transiently transfected with HIPK2- or K221R mutant-expressing vectors. The results of one indicative experiment out of three performed are reported. (D) Immunoblot of total p53 and p53 phosphorylation at Ser46 (Ser-46P), HIPK2, and Gal-3 in p53-positive HEK293 cells transiently transfected with pEGFP vectors encoding EGFP-tagged wild-type HIPK2 or its KD K221R mutant. α-Tubulin was used as a loading control. (E and F) MEFs from p53+/+ and p53−/− mice were infected with recombinant LHIPK2SP, LK221RSP, or control LXSP retroviruses. At 36 h postinfection, cells were harvested to perform Western blotting on the indicated proteins (E) and real-time RT-PCR on the gal-3 transcript (F, upper panel) and to measure cell death (F, lower panel). α-tub, α-tubulin.
It was previously shown that the Ser/Thr kinase HIPK2 phosphorylates p53 at Ser46 and stimulates p53-mediated transcription of proapoptotic genes such as Bax, PIG-3, Noxa, and p53AIP-1 (15, 17, 18, 26). Therefore, we investigated the role of HIPK2 in p53-mediated repression of the LGALS3 promoter. H1299 cells were cotransfected with the p(−836/+141)LGALS3-Luc vector in the presence or absence of a low repressing dose of wtp53 (e.g., 25 ng of plasmid DNA per sample, at a 1:200 molar ratio with the reporter vector) and HIPK2 expression vectors (200 ng of plasmid DNA per sample). Evaluation of luciferase activity showed significant cooperation between wtp53 and HIPK2 in LGALS3 promoter repression (Fig. 3B). To confirm and extend these results, H1299 cells stably transfected with p(−836/+141)LGALS3-Luc vector were infected with Adp53 at an MOI of 10 in the presence or absence of HIPK2 or its kinase-dead (KD) K221R mutant. The strong cooperation between p53 and HIPK2 in Gal-3 repression was confirmed by this experiment. In addition, no effect was induced by the KD mutant, supporting a role for p53 phosphorylation (Fig. 3C).
To characterize the HIPK2-p53 cooperation, p53-expressing HEK293 cells were induced to express increasing doses of Flag-tagged wild-type HIPK2 protein or enhanced green fluorescent protein (EGFP)-tagged wild-type HIPK2 or the KD K221R mutant by transient transfection. Western blot analysis of endogenous Gal-3 protein showed that HIPK2 overexpression reduced Gal-3 levels in a dose-dependent manner (see Fig. S2A in the supplemental material) and that this reduction requires the HIPK2 kinase activity (Fig 3D). Furthermore, this HIPK2 effect was mediated by p53 since EGFP-HIPK2 overexpression in p53-null H1299 cells did not modify Gal-3 protein levels (see Fig. S2B in the supplemental material). Since we have recently identified in the murine p53 a site homologous to Ser46 (12), similar experiments were performed with mouse cells. Results comparable to those with human cells were obtained when MEFs from p53+/+ and p53−/− mice were infected with recombinant retroviruses carrying the wild-type HIPK2 (pLHIPK2SP) or the KD mutant (LK221RSP). A reduction of Gal-3 protein (Fig. 3E) and mRNA levels (Fig. 3F, top graph) was present only in the p53+/+ cells infected with the wild-type HIPK2-carrying virus. In addition, only MEFs from p53+/+ mice, overexpressing wild-type HIPK2, underwent apoptosis (Fig. 3F, bottom graph).
Altogether, these results indicate that the HIPK2-induced activation of p53 contributes to p53-mediated repression of Gal-3.
HIPK2 depletion impairs p53-mediated downregulation of Gal-3.
To directly evaluate whether p53 requires HIPK2 to repress Gal-3, we interfered with HIPK2 expression in p53-null H1299 cells by stable transfection with a pSUPER-HIPK2 vector (Fig. 4A). Control (H1299-ctr) cells and HIPK2 cells subjected to interference (H1299-HIPK2i) were infected at two different MOIs with Adp53 or at the highest MOI with dl70.3 control virus. In agreement with previous results obtained with HIPK2-specific antisense oligonucleotides (18, 26), the TP53 gene transfer resulted in phosphorylation of p53 at Ser46 (Fig. 4B) and cell death (Fig. 4C) much more efficiently in control than in H1299-HIPK2i cells. Consistent with these data, Western blot analysis showed that wtp53 overexpression drastically reduced Gal-3 levels in H1299-ctr cells (Fig. 4D, left). However, no reduction of Gal-3 protein was detectable in H1299-HIPK2i cells even at later time points (Fig. 4D, right). Comparable results were obtained at the mRNA level (Fig. 4E) indicating that HIPK2 depletion strongly weakens p53-induced repression of Gal-3 and apoptosis.
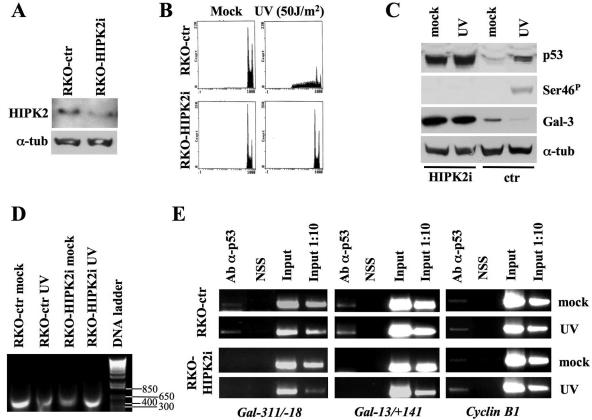
To verify this effect in a more physiological context, we activated the endogenous p53 protein in HIPK2-depleted wtp53-carrying RKO cells with an apoptotic dose of UV irradiation. Interference of HIPK2 was obtained by stable transfection of RKO cells with pSUPER-HIPK2 vectors (Fig. 5A). After UV irradiation, significant death was observed in control cells, while RKO-HIPK2i cells were completely resistant, as shown by cell cycle profiles (Fig. 5B). Consistent with the results of H1299 cells upon infection, the UV-induced phosphorylation of p53 at Ser46 and Gal-3 repression detectable in the RKO-ctr cells were absent in the RKO-HIPK2i cells (Fig. 5C). Interestingly, a significant and reproducible increase in Gal-3 expression was observed in RKO-HIPK2i cells even under basal conditions, suggesting that HIPK2 contributes to the regulation of Gal-3 levels also independently from apoptosis.
FIG. 5.
HIPK2 depletion blocks Gal-3 repression upon DNA damage and in basal conditions. (A) Immunoblots with antibody to HIPK2 on RKO cells stably transfected with pSUPER-ctr or pSUPER-HIPK2 vectors. α-Tubulin was used as a loading control. (B) The indicated cells were treated with UV light at 50 J/m2 and collected 24 h postirradiation for cell cycle analysis upon PI staining. DNA content analysis by flow cytometry of one indicative experiment is reported. (C) Immunoblotting kinetics of total p53, p53 phosphorylation at Ser46, and Gal-3 on the indicated cells before and after UV irradiation. α-Tubulin was used as a loading control. (D and E) RKO-ctr and RKO-HIPK2i were irradiated as described above. At 16 h postirradiation cells were cross-linked. The size and quality of sonicated DNA were controlled on agarose gel (D). ChIP assays were performed with anti-p53 Ab on two regions (i.e., −311/−18 and −13/+141 relative to the TSS) of the LGALS3 promoters and one of the cyclin B1 promoters, as described in the legend of Fig. 2B. α-tub, α-tubulin.
To evaluate whether this latter effect was reproducible in other cells and whether it was dependent on the presence of p53, stable depletion of HIPK2 was induced in four different cell lines (e.g., mouse F9, human MCF7, and HCT116 cells, all carrying wtp53, and HCT116 p53−/− cells). As observed in the RKO-HIPK2i cells, upregulation of Gal-3 expression was observed in p53 expressing cells but not in the p53−/− ones (see Fig. S2C and D in the supplemental material) in basal conditions (i.e., in the absence of DNA damage).
To begin to characterize this phenomenon at the molecular level, cross-linked DNA extracted from RKO-ctr and RKO-HIPK2i cells before and after UV-irradiation (Fig. 5D) was employed in ChIP experiments. PCR amplification of the LGALS3 and cyclin B1 promoters was performed on chromatin immunoprecipitated with the anti-p53 Ab. In the RKO-ctr cells, p53 bound the LGALS3 promoter before and after DNA damage (Fig. 5E, left and middle panels), and, as expected (28), similar results were obtained with the cyclin B1 promoter (Fig. 5E, right panel). Interestingly, HIPK2 depletion abolished the binding of p53 to the LGALS3 promoter while not affecting its presence on the cyclin B1 promoter, strongly supporting a role for HIPK2 in the p53-mediated repression of Gal-3.
Gal-3 is a mediator of p53-dependent apoptosis.
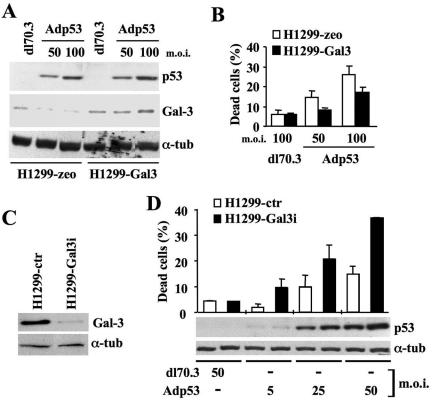
To investigate whether the HIPK2-dependent, p53-mediated repression of Gal-3 we observed in apoptotic conditions plays a causal role in this type of cell death, we employed a nonrepressible Gal-3. H1299 cells were stably transfected with an expression vector carrying an exogenous gal-3 cDNA driven by a heterologous promoter that cannot be transcriptionally repressed by p53. To avoid the effects of excessive Gal-3 expression, cells were transfected with a small amount of plasmid DNA. After selection, polyclonal populations of H1299-Gal-3 and H1299-zeo control cells were tested for Gal-3 expression; the amount of Gal-3 was only slightly more than the endogenous levels in the two Gal-3-transfected mixed populations analyzed (Fig. 6A, compare dl70.3 infected cells; see Fig. S3A in the supplemental material). These cells were subsequently infected with Adp53 or dl70.3 control virus and again analyzed by Western blotting. No repression of Gal-3 was detectable in the TCEs from H1299-Gal-3 upon overexpression of p53 (Fig. 6A). Most importantly, Gal-3-transfected cells survived better than their relative controls (Fig. 6B), indicating that a nonrepressible Gal-3 protects from p53-induced apoptosis. Interestingly, a slight but consistent increase in Gal-3 expression was detected in both H1299-Gal-3 populations upon p53 overexpression (Fig. 6A; see Fig. S3B in the supplemental material). Since a comparable Gal-3 induction was also observed in the adherent fraction of the p53+/+ MEFs upon UV irradiation, it can be postulated that increased levels of Gal-3 might associate with p53-induced growth arrest. To begin testing this hypothesis, cell cycle profiles of the adenovirus-infected H1299-zeo and H1299-Gal-3 cells were analyzed by flow cytometry. We found that the increased levels of Gal-3 were associated with cell accumulation in the G2/M phases of the cell cycle (see Fig. S3D in the supplemental material). Whether this correlation is functional remains to be addressed.
FIG. 6.
Gal-3 repression is required for p53-mediated apoptosis. (A) Immunoblotting kinetics of p53 and Gal-3 on H1299 cells stably transfected with pcDNA3.1/zeo or pcDNA3.1/Gal-3 vectors and maintained as mixed populations. The cells were infected at the indicated MOIs with dl70.3 control or Adp53 adenoviruses. α-Tubulin was used as a loading control. (B) At 24 h postinfection, the percentage of dead cells was determined by trypan blue exclusion on the same cells as in panel A. Means ± standard deviations of two independent experiments performed in duplicate are reported. (C) Immunoblots for Gal-3 on H1299 cells stably transfected with pSUPER-ctr or pSUPER-Gal3 vectors. α-Tubulin was used as a loading control. (D) The same cells analyzed in panel C were infected with the indicated viruses at the indicated MOIs. At 36 h postinfection, cells were harvested to measure the percentage of death and the levels of p53 expression by Western blotting. α-Tubulin was used as a loading control. α-tub, α-tubulin.
Based on the results described above and on their similarity to what was originally observed in macrophages from the gal-3 knockout mice following exposure to stresses that induce apoptosis (27, 38), it would be expected that depletion of Gal-3 sensitizes cells to p53-induced apoptosis. Thus, H1299 cells were stably transfected with pSUPER-Gal3 vectors (Fig. 6C). When increasing doses of p53 were expressed by infecting these cells at different MOIs, we observed that infection with Adp53 at an MOI of 5 in the H1299-Gal3i cells induced the same level of apoptosis that requires fivefold more virus in the H1299-ctr cells, supporting this hypothesis (Fig. 6D).
Altogether, these results indicate that repression of Gal-3 expression strongly contributes to p53-induced apoptosis.
DISCUSSION
In this study, we investigated whether inhibition of the multifunctional lectin Gal-3, whose overexpression is frequently observed in human cancer and induces resistance to apoptosis, is associated with and required for p53-dependent apoptosis. We show that p53-induced apoptosis correlates with a consistent downregulation of Gal-3 mRNA and protein levels due to transcriptional repression of the LGALS3 promoter activity by HIPK2-activated p53. Furthermore, expression of a nonrepressible Gal-3, similar to depletion of p53 or HIPK2, is sufficient to block p53-induced apoptosis, while interference of Gal-3 sensitizes the cells to p53. These data reveal the existence of a functional pathway that links the HIPK2 kinase activity to the ability of p53 to suppress the antiapoptotic functions of Gal-3 by silencing its transcription.
The most extensively studied function of Gal-3 is regulation of apoptosis. A large body of evidence had clearly demonstrated the antiapoptotic activity of Gal-3 to diverse apoptotic stimuli in a variety of normal and tumor cells in vitro and in vivo (37, 38). At the molecular level, significant sequence similarity was observed between Gal-3 and the antiapoptotic factor Bcl-2 (1). Current evidence indicates that the mechanism by which Gal-3 inhibits apoptosis depends on its subcellular localization. Upon different apoptotic stimuli, Gal-3 was shown to translocate to the mitochondria blocking changes in membrane potential (58). Gal-3 phosphorylation at Ser6 by casein kinase 1 was shown to be relevant for the export of Gal-3 from the nucleus in response to chemotherapeutic drugs and resistance to apoptosis (50, 56). In addition, gal-3 knockout mice, although viable and fertile, are more sensitive to apoptosis and show an attenuated inflammatory response, in part because of this sensitivity (27, 38). Despite the extensive characterization of the antiapoptotic functions of Gal-3, to our knowledge, this study is the first to show that suppression of Gal-3 is necessary for a cell to execute the apoptotic program. Since the apoptotic pathways inhibited by Gal-3 are usually activated by the tumor suppressor p53, we asked whether p53-induced apoptosis is associated with inhibition of Gal-3 antiapoptotic activity. Here, we show that p53-induced apoptosis by Adp53 infection or UV irradiation is associated with suppression of Gal-3 expression and is facilitated by Gal-3 interference, while enforced expression of Gal-3 inhibits the apoptotic function of p53. Thus, our results strongly indicate that to complete the apoptotic program, at least some types of apoptotic pathways require repression of Gal-3 and that p53 accomplishes this goal at the transcription level only in the presence of a functional HIPK2.
p53 regulates different biological functions by activating or repressing the transcription of many target genes (reviewed in reference 19). The p53 transrepression activity was shown to play a major role in apoptosis by the repression of several antiapoptotic factors such as Bcl-2, survivin, and others. Interestingly, p53-mediated transcriptional repression has been associated with each of the transcriptional repression mechanisms, i.e., interference with the functions of DNA-binding transcriptional activators or with the basal transcriptional machinery and alteration of chromatin structure by recruiting chromatin-remodeling factors such as histone deacetylases (reviewed in reference 25). Thus far, the LGALS3 gene has not been shown to be modified in the gene expression profiles performed on human cells overexpressing p53 by employing cDNA microarray (31, 32, 60). However, by the same technology, LGALS3 overexpression was not consistently detected in tumor samples known to express high levels of Gal-3 mRNA, suggesting that probe design might account for these discrepancies (55). A p53-mediated transcriptional repression of the LGALS3 gene was reported by Raimond and collaborators, who described a promoter region located inside the second intron of the human LGALS3 gene that was downregulated by wtp53 but not by mutant p53 (46). However, the same group subsequently demonstrated that this promoter region drives the transcription of another gene, named the Gal-3 internal gene, totally unrelated to Gal-3 (23a). Since we found a p53-mediated downregulation of Gal-3 at protein and mRNA levels and p53 was shown to repress the rabbit gal-3 promoter (21), we tested whether the most upstream promoter region of the LGALS3 gene encoding for Gal-3 was regulated by p53. No p53 consensus sites were detected in this promoter region; however, in vivo ChIP shows the presence of p53, and experiments of reporter gene expression demonstrate a p53-mediated transcriptional repression. Our results suggest that the promoter region potentially implicated in the p53-induced repression of LGALS3 promoter is comprised of the region between nucleotides −472 and −272. Because this region is also critical for the LGALS3 basal activity, further analysis is required to fully characterize the p53 repressing mechanism. We are currently developing new HIPK2 and phospho-specific antibodies for p53 phosphorylated at Ser 46 to overcome the limits imposed thus far by the poor performances of the available reagents in ChIP assays.
HIPK2 was shown to participate as a positive transcription regulator in p53-mediated transcriptional activation of proapoptotic genes such as Bax, PIG-3, Noxa, and p53AIP-1 (15, 17, 18, 26). Here we observed that HIPK2 can contribute to p53-mediated apoptosis also through the repression of at least one antiapoptotic factor, i.e., Gal-3. The corepression activity of HIPK2 is not surprising, since it works as a transcriptional corepressor with homeobox transcription factors (13, 35). However, in-depth characterization of the transcriptional activity of HIPK2 still awaits antibodies that work in ChIP assays. By an indirect approach, e.g., by performing ChIP with anti-p53 Ab in HIPK2-depleted or control cells, we observed that HIPK2 is required for p53 docking on the LGALS3 promoter. This is not a general mechanism, since HIPK2 is irrelevant for p53 binding to the cyclin B1 promoter; however, it might explain the increased basal levels of Gal-3 expression we found in the wtp53-carrying HIPK2-depleted cells and suggests the existence of a new mechanism of HIPK2-induced, p53-mediated transcriptional repression of a subset of genes.
The stronger effect of HIPK2 depletion rather than p53 depletion or knockout on Gal-3 expression (for example, compare Fig. 1E and Fig. 5C, both coming from the same Western blot) as well as the mild HIPK2 activity in repressing the LGALS3 promoter in the absence of p53 (Fig. 3B and C), suggests the possibility of a broader effect of HIPK2 versus p53. Since HIPK2 interacts also with the p53 family members p73 and p63 (33), it is conceivable that defects of HIPK2 might also be responsible for the functional impairment of these proteins. Interestingly, loss of p73 biological activity in the presence of high levels of the protein was recently observed in thyroid tumor cells; such loss could only partially be explained by the known mechanisms of p73 inactivation (i.e., interaction with ΔNp73, the transcriptionally inactive variants of p73, or with mutant p53) (20). Although we did not observe significant apoptosis upon HIPK2 overexpression in p53 null cells, p53-independent apoptosis by HIPK2 has been reported (59) and might also contribute to the Gal-3 increment induced by HIPK2 depletion.
Defects in apoptosis are though to play a major role in tumorigenesis in addition to tumor response to anticancer treatments (i.e., chemo- and radiotherapy), and p53 alterations are frequent causes of such defects (48). Increasing evidence supports the involvement of Gal-3 in tumorigenesis and resistance to chemotherapeutic drugs through its strong antiapoptotic activity (reviewed in reference 38). Our data describing wtp53-mediated transcriptional repression of Gal-3 in p53-induced apoptosis are fully consistent with these observations. Taken together, these data would predict a mutual exclusion between the presence of wtp53 protein and Gal-3 overexpression in the same tumor cells. Although this is true in several types of human cancers, at least one exception to this prediction is represented by the well-differentiated thyroid carcinomas that almost invariably express wtp53 and high levels of Gal-3 (6, 29). A possible explanation for this paradox might be linked to our further observation that p53 needs to be activated by HIPK2 to repress Gal-3 and induce apoptosis. Impairment of this pathway due to HIPK2 alterations might contribute to the aberrant accumulation of Gal-3 in human cancer even in the presence of wtp53. Indeed, reduced levels of HIPK2 mRNA were found in a few human thyroid cancers (44 and data not shown). We are currently testing this hypothesis by genetic analysis at the HIPK2 loci in biopsies from patients with well-differentiated thyroid cancers.
In summary, our study identifies a new apoptotic pathway triggered by HIPK2-activated p53 and requiring p53-mediated repression of the antiapoptotic molecule Gal-3.
Supplementary Material
Acknowledgments
We thank all the people cited in the text for providing us with cells and/or expression vectors. We are particularly grateful to A. Prodosmo, C. Lazzari, and G. Parsons for providing their unpublished, stably transfected HIPK2-interfered cells; A Prodosmo, F. Moretti, M. Crescenzi, and M. Fanciulli for helpful advice and stimulating discussions; A. Sacchi for support and scientific inspiration; S. Bacchetti for helpful discussion and critical revision of the manuscript; and N. De Stefano and R. Dominici for technical support.
This work was supported by Associazione Italiana per la Ricerca sul Cancro, Ministero della Salute, Ministero dell'Areonautica, Air Force Medical Institute—Rome, Compagnia San Paolo-Progetto Oncologia, and by EC FP6 funding (contract 503576). C.R. and L.L. are recipients of fellowships from Fondazione Italiana per la Ricerca sul Cancro, and A.U. is the recipient of a fellowship from AFaR.
This article reflects the authors' views and not necessarily those of the European Community. The EC is not liable for any use that may be made of the information contained herein.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Akahani, S., P. Nangia-Makker, H. Inohara, H.-R. Kim, and A. Raz. 1997. Galectin-3: a novel antiapoptotic molecule with a functional BH1 (NWGR) domain of Bcl-2 family. Cancer Res. 57:5272-5276. [PubMed] [Google Scholar]
- 2.Appella, E., and C. W. Anderson. 2000. Signaling to p53: breaking the post-translational modification code. Pathol. Biol. 48:227-245. [PubMed] [Google Scholar]
- 3.Ashcroft, M., M. H. Kubbutat, and K. H. Vousden. 1999. Regulation of p53 function and stability by phosphorylation. Mol. Cell. Biol. 19:1751-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bacchetti, S., and F. Graham. 1993. Inhibition of cell proliferation by an adenovirus vector expressing the human wild type p53 protein. Int. J. Oncol. 3:781-788. [DOI] [PubMed] [Google Scholar]
- 5.Barondes, S. H., V. Castronovo, D. N. Cooper, R. D. Cummings, K. Drickamer, T. Feizi, M. A. Gitt, J. Hirabayashi, C. Hughes, K. Kasai, et al. 1994. Galectins: a family of animal beta-galactoside-binding lectins. Cell 76:597-598. [DOI] [PubMed] [Google Scholar]
- 6.Bartolazzi, A., A. Gasbarri, M. Papotti, G. Bussolati, T. Lucante, A. Khan, H. Inohara, F. Marandino, F. Orlandi, A. Vecchione, R. Tecce, and O. Larsson. 2001. Application of an immunodiagnostic method for improving the preoperative diagnosis of nodular thyroid lesions. Lancet 357:1644-1650. [DOI] [PubMed] [Google Scholar]
- 7.Boyd, K. E., J. Wells, J. Gutman, S. M. Bartley, and P. J. Farnham. 1998. c-Myc target gene specificity is determined by a post-DNA-binding mechanism. Proc. Natl. Acad. Sci. USA 95:13887-13892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 9.Bulavin, D. V., S. Saito, M. C. Hollander, C. W. Anderson, E. Appella, and A. J. Fornace, Jr. 1999. Phosphorylation of human p53 by p38 kinase coordinates N-terminal phosphorylation and apoptosis in response to UV radiation. EMBO J. 18:6845-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bunz, F., A. Dutriaux, C. Lengauer, T. Waldman, S. Zhou, J. P. Brown, J. M. Sedivy, K. W. Kinzler, and B. Vogelstein. 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282:1497-1501. [DOI] [PubMed] [Google Scholar]
- 11.Capponcelli, S., S. Fontanesi, E. Pedrini, M. A. Cerone, V. Corti, M. Alessio, A. Bachi, S. Soddu, D. Ribatti, P. Picci, L. J. Helman, G. Cantelli-Forti, and L. Sangiorgi. 2005. Evaluation of the molecular mechanisms involved in the gain of function of a Li-Fraumeni TP53mutation. Hum. Mut. 26:94-103. [DOI] [PubMed] [Google Scholar]
- 12.Cecchinelli, B., A. Porrello, C. Lazzari, A. Gradi, G. Bossi, M. D'Angelo, A. Sacchi, S. Soddu. Ser58 of mouse p53 is the homologue of human Ser46 and is phosphorylated by HIPK2 in apoptosis. Cell Death Differ., in press. [DOI] [PubMed]
- 13.Choi, C. Y., Y. O. Kim, H. J. Kwon, and Y. Kim. 1999. The homeodomain protein NK-3 recruits Groucho and a histone deacetylase complex to repress transcription. J. Biol. Chem. 274:33194-33197. [DOI] [PubMed] [Google Scholar]
- 14.Dagher, S. F., J. L. Wang, and R. J. Patterson. 1995. Identification of galectin-3 as a factor in pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 92:1213-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Stefano, V., G. Blandino, A. Sacchi, S. Soddu, and G. D'Orazi. 2004. HIPK2 neutralizes MDM2 inhibition rescuing p53 transcriptional activity and apoptotic function. Oncogene 23:5185-5192. [DOI] [PubMed] [Google Scholar]
- 16.Di Stefano, V., C. Rinaldo, A. Sacchi, S. Soddu, and G. D'Orazi. 2004. Homeodomain-interacting protein kinase-2 activity and p53 phosphorylation are critical events for cisplatin-mediated apoptosis. Exp. Cell Res. 293:311-320. [DOI] [PubMed] [Google Scholar]
- 17.Di Stefano, V., S. Soddu, A. Sacchi, and G. D'Orazi. 2005. HIPK2 contributes to PCAF-mediated p53 acetylation and selective transactivation of p21(Waf1) after nonapoptotic DNA damage. Oncogene 24:5431-5442. [DOI] [PubMed] [Google Scholar]
- 18.D'Orazi, G., B. Cecchinelli, T. Bruno, I. Manni, Y. Higashimoto, S. Saito, M. Gostissa, S. Coen, A. Maschetti, G. Del Sal, G. Piaggio, M. Fanciulli, E. Appella, and S. Soddu. 2002. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat. Cell Biol. 4:11-19. [DOI] [PubMed] [Google Scholar]
- 19.el-Deiry, W. S. 1998. Regulation of p53 downstream genes. Semin. Cancer Biol. 8:345-357. [DOI] [PubMed] [Google Scholar]
- 20.Frasca, F., V. Vella, A. Aloisi, A. Mandarino, E. Mazzon, R. Vigneri, and P. Vigneri. 2003. p73 tumor-suppressor activity is impaired in human thyroid cancer. Cancer Res. 63:5829-5837. [PubMed] [Google Scholar]
- 21.Gaudin, J. C., M. Monsigny, and A. Legrand. 1997. Modulation of the expression of the rabbit galectin-3 gene by p53 and c-Ha-ras proteins and PMA. Glycobiology 7:1089-1098. [DOI] [PubMed] [Google Scholar]
- 22.Germain, M., E. B. Affar, D. D'Amours, V. M. Dixit, G. S. Salvesen, and G. G. Poirier. 1999. Cleavage of automodified poly(ADP-ribose) polymerase during apoptosis. Evidence for involvement of caspase-7. J. Biol. Chem. 274:28379-28384. [DOI] [PubMed] [Google Scholar]
- 23.Gong, H. C., Y. Honjo, P. Nangia-Makker, V. Hogan, N. Mazurak, R. S. Bresalier, and A. Raz. 1999. The NH2 terminus of galectin-3 governs cellular compartmentalization and functions in cancer cells. Cancer Res. 59:6239-6245. [PubMed] [Google Scholar]
- 23a.Guittant, M., S. Charpentier, T. Normand, M. Dubois, J. Raimond, and A. Legrand. 2001. Identification of an integral gene to the human galectin-3 gene with two different overlapping reading frames that do not encode galectin-3. J. Biol. Chem. 276:2652-2657. [DOI] [PubMed] [Google Scholar]
- 24.Hainaut, P., and M. Hollstein. 2000. p53 and human cancer: the first ten thousand mutations. Adv. Cancer Res. 77:81-137. [DOI] [PubMed] [Google Scholar]
- 25.Ho, J., and S. Benchimol. 2003. Transcriptional repression mediated by the p53 tumour suppressor. Cell Death Differ. 4:404-408. [DOI] [PubMed] [Google Scholar]
- 26.Hofmann, T. G., A. Moller, H. Sirma, H. Zentgraf, Y. Taya, W. Droge, H. Will, and M. L. Schmitz. 2002. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat. Cell Biol. 4:1-10. [DOI] [PubMed] [Google Scholar]
- 27.Hsu, D. K., R. Y. Yang, Z. Pan, L. Yu, D. R. Salomon, W. P. Fung-Leung, and F. T. Liu. 2000. Targeted disruption of the galectin-3 gene results in attenuated peritoneal inflammatory responses. Am. J. Pathol. 156:1073-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imbriano, C., A. Gurtner, F. Cocchiarella, S. Di Agostino, V. Basile, M. Gostissa, M. Dobbelstein, G. Del Sal, G. Piaggio, and R. Mantovani. 2005. Direct p53 transcriptional repression: in vivo analysis of CCAAT-containing G2/M promoters. Mol. Cell. Biol. 25:3737-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito, T., T. Seyama, T. Mizuno, N. Tsuyama, T. Hayashi, Y. Hayashi, K. Dohi, N. Namamura, and M. Akiyama. 1992. Unique association of p53 mutations with undifferentiated but not with differentiated carcinomas of the thyroid gland. Cancer Res. 52:1369-1371. [PubMed] [Google Scholar]
- 30.Kadrofske, M., K. P. Openo, and J. L. Wang. 1998. The human LGALS3 (galectin-3) gene: determination of the gene structure and functional characterization of the promoter. Arch. Biochem. Biophys. 349:7-20. [DOI] [PubMed] [Google Scholar]
- 31.Kannan, K., N. Amariglio, G. Rechavi, J. Jakob-Hirsch, I. Kela, N. Kaminski, G. Getz, E. Domany, and D. Givol. 2001a. DNA microarrays identification of primary and secondary target genes regulated by p53. Oncogene 20:2225-2234. [DOI] [PubMed] [Google Scholar]
- 32.Kannan, K., N. Kaminski, G. Rechavi, J. Jakob-Hirsch, N. Amariglio, D. Givol. 2001b. DNA microarray analysis of genes involved in p53 mediated apoptosis: activation of Apaf-1. Oncogene 20:3449-3455. [DOI] [PubMed] [Google Scholar]
- 33.Kim, E.-J., J.-S. Park, and S.-J. Um. 2002. Identification and characterization of HIPK2 interacting with p73 and modulating functions of the p53 family in vivo. J. Biol. Chem. 277:32020-32028. [DOI] [PubMed] [Google Scholar]
- 34.Kim, H. R., H. M. Lin, H. Bilirian, and A. Raz. 1999. Cell cycle arrest and inhibition of anoikis by galectin-3 in human breast epithelial cells. Cancer Res. 59:4148-4154. [PubMed] [Google Scholar]
- 35.Kim, Y. H., C. Y. Choi, S. J. Lee, M. A. Conti, and Y. Kim. 1998. Homeodomain-interacting protein kinases, a novel family of co-repressors for homeodomain transcription factors. J. Biol. Chem. 273:25875-25879. [DOI] [PubMed] [Google Scholar]
- 36.Kuwabara, I., and F. T. Liu. 1996. Galectin-3 promotes adhesion of human neutrophils to laminin. J. Immunol. 156:3939-3944. [PubMed] [Google Scholar]
- 37.Liu, F. T., R. J. Patterson, J. L. Wang. 2002. Intracellular functions of galectins. Biochim. Biophys. Acta 1572:263-273. [DOI] [PubMed] [Google Scholar]
- 38.Liu, F. T., and G. A. Rabinovich. 2005. Galectins as modulators of tumour progression. Nat. Rev. Cancer 5:29-41. [DOI] [PubMed] [Google Scholar]
- 39.Lloyd, R. V. 2001. Distinguishing benign from malignant thyroid lesions: galectin-3 as the latest candidate. Endrocr. Pathol. 12:255-257. [DOI] [PubMed] [Google Scholar]
- 40.Marchetti, A., F. Barassi, C. Martella, A. Chella, S. Salvatore, A. Castrataro, F. Mucilli, R. Sacco, and F. Buttitta. 2004. Down-regulation of high in normal-1 (HIN-1) is a frequent event in stage I non-small cell lung cancer and correlates with poor clinical outcome. Clin. Cancer Res. 10:1338-1344. [DOI] [PubMed] [Google Scholar]
- 41.Michalovitz, D., O. Halevy, and M. Oren. 1990. Conditional inhibition of transformation and of cell proliferation by a temperature-sensitive mutant of p53. Cell 62:671-680. [DOI] [PubMed] [Google Scholar]
- 42.Moller, A., H. Sirma, T. G. Hofmann, S. Rueffer, E. Klimczak, W. Droge, H. Will, and M. L. Schmitz. 2003. PML is required for homeodomain-interacting protein kinase 2 (HIPK2)-mediated p53 phosphorylation and cell cycle arrest but is dispensable for the formation of HIPK domains. Cancer Res. 63:4310-4314. [PubMed] [Google Scholar]
- 43.Oda, K., H. Arakawa, T. Tanaka, K. Matsuda, C. Tanikawa, T. Mori, H. Nishimori, K. Tamai, T. Tokino, Y. Nakamura, and Y. Taya. 2000. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 102:849-862. [DOI] [PubMed] [Google Scholar]
- 44.Pierantoni, G. M., A. Bulfone, F. Pentimalli, M. Fedele, R. Iuliano, M. Santoro, L. Chiariotti, A. Ballabio, and A. Fusco. 2002. The homeodomain-interacting protein kinase 2 gene is expressed late in embryogenesis and preferentially in retina, muscle, and neural tissues. Biochem. Biophys. Res. Commun. 290:942-947. [DOI] [PubMed] [Google Scholar]
- 45.Porrello, A., M. A. Cerone, S. Coen, A. Gurtner, G. Fontemaggi, L. Cimino, G. Piaggio, A. Sacchi, and S. Soddu. 2000. p53 regulates myogenesis by triggering the differentiation activity of pRb. J. Cell Biol. 151:1295-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raimond, J., F. Rouleux, M. Monsigny, and A. Legrand. 1995. The second intron of the human galectin-3 gene has a strong promoter activity down-regulated by p53. FEBS Lett. 363:165-169. [DOI] [PubMed] [Google Scholar]
- 47.Saito, S., A. A. Goodarzi, Y. Higashimoto, Y. Noda, S. P. Lees-Miller, E. Appella, and C. W. Anderson. 2002. ATM mediates phosphorylation at multiple p53 sites, including Ser(46), in response to ionizing radiation. J. Biol. Chem. 277:12491-12494. [DOI] [PubMed] [Google Scholar]
- 48.Schmitt, C. A., J. S. Fridman, M. Yang, E. Baranov, R. M. Hoffman, and S. W. Lowe. 2002. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell 1:289. [DOI] [PubMed] [Google Scholar]
- 49.Sciacchitano, S., A. Orecchio, L. Lavra, S. Misiti, A. Giacchini, M. Zani, D. Danese, A. Gurtner, S. Soddu, U. Di Mario, and M. Andreoli. 2002. Cloning of the mouse insulin receptor substrate-3 (mIRS-3) promoter, and its regulation by p53. Mol. Endocrinol. 16:1577-1589. [DOI] [PubMed] [Google Scholar]
- 50.Takenaka, Y., T. Fukumori, T. Yoshii, N. Oka, H. Inohara, H. R. Kim, R. S. Bresalier, and A. Raz. 2004. Nuclear export of phosphorylated galectin-3 regulates its antiapoptotic activity in response to chemotherapeutic drugs. Mol. Cell. Biol. 24:4395-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vousden, K. H. 2000. p53: death star. Cell 103:691-694. [DOI] [PubMed] [Google Scholar]
- 52.Wang, Y., K. M. Debatin, and H. Hug. 2001. HIPK2 overexpression leads to stabilization of p53 protein and increased p53 transcriptional activity by decreasing Mdm2 protein levels. BMC Mol. Biol. 2:8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang, R. Y., D. K. Hsu, and F. T. Liu. 1996. Expression of galectin-3 modulates T-cell growth and apoptosis. Proc. Natl. Acad. Sci. USA 93:6737-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang, R. Y., and F. T. Liu. 2003. Galectins in cell growth and apoptosis. Cell. Mol. Life Sci. 60:267-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yano, Y., N. Uematsu, T. Yashiro, H. Hara, E. Ueno, M. Miwa, G. Tsujimoto, Y. Aiyoshi, and K. Uchida. 2004. Gene expression profiling identifies platelet-derived growth factor as a diagnostic molecular marker for papillary thyroid carcinoma. Clin. Cancer Res. 10:2035-2043. [DOI] [PubMed] [Google Scholar]
- 56.Yoshii, T., T. Fukumori, Y. Honjo, H. Inohara, H. R. Kim, and A. Raz. 2002. Galectin-3 phosphorylation is required for its anti-apoptotic function and cell cycle arrest. J. Biol. Chem. 277:6852-6857. [DOI] [PubMed] [Google Scholar]
- 57.Yoshii, T., H. Inohara, Y. Takenaka, Y. Honjo, S. Akahani, T. Nomura, A. Raz, and T. Kubo. 2001. Galectin-3 maintains the transformed phenotype of thyroid papillary carcinoma cells. Int. J. Oncol. 18:787-792. [DOI] [PubMed] [Google Scholar]
- 58.Yu, F., R. L. Finley, Jr., A. Raz, and H.-R. Kim. 2002. Galectin-3 translocates to the perinuclear membranes and inhibits cytochrome c release from the mitochondria. J. Biol. Chem. 277:15819-15827. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, Q., Y. Yoshimatsu, J. Hildebrand, S. M. Frisch, and R. H. Goodman. 2003. Homeodomain interacting protein kinase 2 promotes apoptosis by down-regulating the transcriptional corepressor CtBP. Cell 115:177-186. [DOI] [PubMed] [Google Scholar]
- 60.Zhao, R., G. Kurt, M. Murphy, Y. Yin, D. Notterman, W. H. Hoffman, E. Tom, D. H. Mack, and A. J. Levine. 2000. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 14:981-993. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.