Abstract
Using a novel thiol affinity chromatography approach to purify macroH2A1-containing chromatin fragments, we examined the distribution of macroH2A1 histone variants in mouse liver chromatin. We found that macroH2A1 was depleted on the transcribed regions of active genes. This depletion was observed on all of the 20 active genes that we probed, with only one site showing a small amount of enrichment. In contrast, macroH2A1 was concentrated on the inactive X chromosome, consistent with our previous immunofluorescence studies. This preferential localization was seen on genes that are active in liver, genes that are inactive in liver, and intergenic regions but was absent from four regions that escape X inactivation. These results support the hypothesis that macroH2As function as transcriptional repressors. Also consistent with this hypothesis is our finding that the heterochromatin protein HP1β copurifies with the macroH2A1-containing chromatin fragments. This study presents the first detailed examination of the distribution of macroH2A1 variants on specific sequences. Our results indicate that macroH2As have complex distribution patterns that are influenced by both local factors and long-range mechanisms.
As the primary structural unit of eukaryotic chromatin, the nucleosome represents an important target for modifying chromatin function. One mechanism for modifying nucleosome structure and function is the substitution of variant histones for the major or canonical histones. Histone variants of different primary structures have been described in many organisms (38, 53). Genetic studies showed that histone variants H2A.X, H2A.Z, CENPA, and H3.3 have important functional properties that cannot be provided by their conventional counterparts (reviewed in reference 38).
The macroH2A histone variants have an unusual structure consisting of a full-length H2A domain linked to a large nonhistone domain, producing a protein that is nearly three times the size of conventional core histones (40). The H2A domain of macroH2A histones is ∼65% identical to conventional H2As. Most of the nonhistone region appears to be derived from a domain that is found in many contexts in addition to macroH2A including as a domain in proteins involved in the replication of RNA viruses, as domains in some poly(ADP-ribose) polymerases, and as a separate protein in bacteria (4, 41). Recent studies showed that some “macrodomains,” including the one from macroH2A1.1, bind ADP ribose (27). The significance of this binding for macroH2A function is not known.
There are three macroH2A variants. macroH2A1.1 and -1.2 are formed by alternate splicing of macroH2A1, and macroH2A2 is encoded by a separate gene (12, 15, 40, 44). The distribution of these three macroH2As is different in different cell types and changes during development (15, 39). The macroH2A1 (-1.1 and -1.2) content of rat liver chromatin, a relatively rich source of macroH2A1, was estimated to be one molecule for every 30 nucleosomes (40). macroH2As are highly conserved among vertebrates. They appear to be absent from most invertebrates (1, 41) but are in the sea urchin genomic database.
Several lines of evidence indicate that macroH2As have a role in transcriptional repression. Immunofluorescence studies and studies with green fluorescent fusion proteins indicated that macroH2As are preferentially associated with the inactive X chromosome (12, 14-16), and a recent study indicates that macroH2A1 plays a role in the maintenance of X inactivation (23). Immunofluorescence studies indicate that macroH2As are preferentially associated with other large domains of heterochromatin, including pericentromeric heterochromatin in some cell types (16, 21), the XY body of spermatocytes (24), and transcriptionally silent senescence-associated heterochromatic foci (54). Molecular biological and biochemical studies indicate that both the H2A and the nonhistone domains of macroH2A can contribute to transcriptional repression (5, 13, 42).
While immunofluorescence studies indicate that macroH2As are widely but nonrandomly distributed in the chromatin (14), technical limitations of this method make it difficult to probe the distribution of macroH2As on specific sequences. In addition, artifacts associated with antibody accessibility or chromatin density could potentially give a misleading indication of macroH2A distribution. Indeed, it has been asserted that the apparent preferential localization of macroH2A to the inactive X chromosome is due to the increased chromatin concentration of the inactive X and does not reflect a true preferential association with macroH2A (42). In order to directly examine the DNA sequences associated with macroH2A, we developed a biochemical method for purifying macroH2A1-containing chromatin fragments. We applied this technique to mouse liver chromatin and analyzed the distribution of macroH2A1 on active genes, on inactive genes, and on the X chromosome. We also identified some of the proteins that were associated with the purified macroH2A1-containing chromatin fragments.
MATERIALS AND METHODS
Chromatin preparation.
Soluble mouse liver chromatin fragments were prepared from H1-stripped S2 chromatin fractions using standard procedures (see the supplemental material for details).
Thiol affinity chromatography of macroH2A1-containing chromatin.
All chromatography buffers contained 1:100 Complete medium (Roche Applied Science), 1 μg/ml pepstatin, and 5 mM sodium butyrate. Starting with ∼15 mg of DNA in isolated nuclei, we obtained ∼15 ml of solubilized chromatin. The chromatin was applied to a 0.7- × 20-cm column of activated thiol Sepharose (Amersham Biosciences) that was previously equilibrated with column buffer (50 mM NaCl, 1 mM EDTA, 10 mM triethanolamine, pH 7.5). The initial flow rate was 3 ml/h. The activated thiol Sepharose column was connected in series to a 0.7- × 28-cm column of thiopropyl Sepharose (Amersham Biosciences) equilibrated with column buffer. After 3 h we carefully added column buffer to the top of the sample, bringing the total volume to 20 ml, and lowered the flow rate to 1.5 ml/h. The sample was then applied overnight (11 to 12 h) at 1.5 ml/h (slow application appears to increase binding to the thiopropyl column). The following morning the flow rate was increased to 3 ml/h for 1 h, and then the two columns were disconnected and washed separately with 40 ml of column buffer at a high flow rate. The activated thiol Sepharose column was eluted with column buffer containing 100 mM mercaptoethanol. The thiopropyl Sepharose column was first eluted with column buffer containing 0.5 M NaCl (30 ml), reequilibrated with column buffer, and then eluted overnight at a flow rate of 2.5 ml/h with column buffer containing 100 mM mercaptoethanol. We collected 2.5-ml fractions. Washes and the elution were monitored by A260. Mercaptoethanol and thiopyridone released from the beads by mercaptoethanol interfere with the use of A260 to estimate the DNA content of the eluted fractions.
Preparation and electrophoresis of DNA and proteins.
DNA was prepared by digesting the samples with proteinase K (100 μg/ml) in 1% sodium dodecyl sulfate (SDS) and then extracting twice with phenol-chloroform-isoamyl alcohol (25:24:1) followed by precipitation with isopropanol in the presence of a glycogen carrier. The pellet was washed with 70% ethanol, briefly air dried, and dissolved in 10 mM Tris, pH 7.5, 1 mM EDTA. DNA was separated by electrophoresis in agarose gels containing 50 mM Tris base, 50 mM boric acid, and 1 mM EDTA. The DNA was visualized with ethidium bromide.
Proteins were analyzed by electrophoresis in 15% polyacrylamide gels (37.5:1 acrylamide/bisacrylamide) containing SDS (31). Samples were prepared by mixing them with an equal volume of SDS sample buffer (10% glycerol, 3% SDS, 62.5 mM Tris, pH 6.8) containing 100 mM dithiothreitol. Dilute samples, such as the mercaptoethanol-eluted thiopropyl Sepharose fractions, were precipitated by adding trichloroacetic acid to 20%. The pellet was washed once with ice-cold ethanol containing 2% sulfuric acid and twice with ice-cold ethanol, briefly air dried, and then dissolved in SDS sample buffer containing 100 mM dithiothreitol. When needed, Tris base was added to the samples to neutralize acidity. Gels were stained with Coomassie brilliant blue R-250 (Bio-Rad). Western blot assays were performed as described previously (16) with electrophoresis in low-bisacrylamide SDS gels (39) and detection by chemiluminescence.
Subtractive hybridization.
We used the PCR Select subtractive hybridization system (Clontech Laboratories, Palo Alto, Calif.) to enrich for macroH2A1-containing DNA sequences. Forward-subtracted DNA was made by subtraction of macroH2A1-depleted DNA (DNA from thiopropyl flowthrough chromatin) from macroH2A1-enriched DNA (DNA from the mercaptoethanol-eluted thiopropyl chromatin). Reverse-subtracted DNA was made by subtracting thiopropyl-eluted DNA from thiopropyl flowthrough DNA. Following the manufacturer's instructions, clones of forward-subtracted DNA were separately hybridized with forward-subtracted DNA and reverse-subtracted DNA. Clones that preferentially hybridized to forward-subtracted DNA were sequenced and analyzed as described in Results.
Real-time PCR.
Real-time PCR was performed using the LightCycler System (Roche Applied Science). We used titanium Taq polymerase (BD Biosciences) with its standard buffer. Denaturation was for 1 s at 95°C, annealing was for 3 s (variable temperatures; see Table S1 in the supplemental material), and elongation was for 5 s at 72°C. Detection was with SYBR green I. We used the standard LightCycler data analysis software to quantify the results, using the second derivative maximum to compare different samples. Using three standard primer pairs, we estimated a difference of approximately 1.9-fold per cycle and used this value for all primer pairs. We checked product by melting curves and gel electrophoresis.
Mass spectrometry.
Selected protein bands were excised from the gel, destained, reduced with dithiothreitol, alkylated with iodoacetamide, and digested with trypsin (47). Tryptic peptide mass maps were obtained by matrix-assisted laser desorption ionization-time of flight mass spectrometry in positive ion mode (Bruker Reflex IV mass spectrometer; Bruker Daltonics, Billerica, MA) and used to search publicly available protein sequence databases for the corresponding protein using the MASCOT database search engine (Matrix Science, London, United Kingdom). Trypsin autolysis peaks were used as internal mass calibrants, and monoisotopic m/z values were determined using the SNAP algorithm (Bruker Daltonics).
RESULTS
Purification of macroH2A1-containing chromatin fragments.
Attempts to immunoprecipitate macroH2A1-containing chromatin from mouse liver using several polyclonal antibodies against macroH2A1 gave us unsatisfactory results. As an alternative, we developed a biochemical approach for purifying macroH2A1-containing chromatin that relies on thiol affinity chromatography. This approach was inspired by Vincent Allfrey and his colleagues, who used organomercury chromatography to purify specific nucleosome fractions (3). The nonhistone regions of macroH2A1.1 and -1.2 each contain three cysteine residues that could potentially bind to thiol-specific affinity supports. The crystal structures of the nonhistone regions of macroH2A1.1 and -1.2 show that one cysteine, 282 in 1.1, is exposed on the surface (13, 30). The other two cysteines have very limited surface exposure. We found that macroH2A1-containing nucleosomes bind to thiopropyl Sepharose (Amersham) with relatively high efficiency but show virtually no affinity for the related matrix-activated thiol Sepharose (Amersham). This difference forms the basis for our approach.
We used mouse liver as our source of chromatin because it is relatively rich in macroH2A1 proteins and immunofluorescence indicates that one or both macroH2A1 proteins are preferentially localized to the inactive X chromosome in mouse hepatocytes (14). We prepared an S2 chromatin fraction from micrococcal nuclease-digested nuclei. This fraction contains the majority of the chromatin (18). The S2 chromatin was treated with CM Sephadex to remove most of the H1 (32), and the stripped chromatin was digested with micrococcal nuclease to produce mono- and oligonucleosomes. Using Western blot assays, we found that the macroH2A1 content of this solubilized fraction (relative to the other core histones) was similar to that of whole nuclei.
The solubilized chromatin fragments were first passed through activated thiol Sepharose to remove most proteins and nucleosomes that bind to thiol affinity resins. The unbound chromatin was then passed through thiopropyl Sepharose. Following a buffer wash, the thiopropyl column was washed with buffer containing 0.5 M NaCl to remove nucleosomes that were bound to the beads by cysteine-containing nonhistone proteins. This fraction contains core histones and a variable amount of H1 but very little macroH2A. We then eluted the thiopropyl Sepharose column with mercaptoethanol, releasing a fraction that is highly enriched for macroH2A1 (Fig. 1). Although nucleosomes were bound to the activated thiol resin, Western blots showed that very little macroH2A1 was associated with these nucleosomes (Fig. 1).
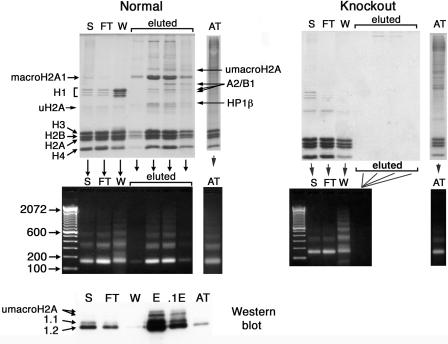
FIG. 1.
Thiol affinity purification of macroH2A1-containing nucleosomes. macroH2A1-containing chromatin fragments were purified by selective thiol affinity chromatography; see Materials and Methods. Normal panels (left) show results obtained with normal mouse liver; knockout panels (right) show results obtained with macroH2A1 knockout mouse liver. The top panels are SDS-polyacrylamide gels showing the distribution of proteins in different fractions. The middle panels are agarose gels showing the distribution of DNA fragments in the same fractions. The bottom panel is a Western blot with antibodies against macroH2A1.1 and -1.2. S, starting material; FT, flowthrough fraction; W, 0.5 M NaCl wash of thiopropyl Sepharose; eluted, fractions eluted from thiopropyl Sepharose with mercaptoethanol; AT, pool of material eluted from activated thiol Sepharose with mercaptoethanol; E, pool of the four peak fractions eluted from thiopropyl Sepharose with mercaptoethanol. The lanes on the Western blot were loaded for equal amounts of H3 and H4 except 0.1E, which was diluted 10-fold. HP1β, chromobox homolog 1 (HP1 beta homolog Drosophila melanogaster), gi:6671696; A2/B1, heterogeneous nuclear ribonucleoprotein A2/B1, gi:7949053 and gi:32880197; uH2A, ubiquitinated H2A; umacroH2A, ubiquitinated macroH2A1. Numbers at left of the middle left panel are molecular sizes in base pairs.
In order to assess the relative macroH2A content of the mercaptoethanol-eluted fractions, we scanned a stained gel of these samples. In the analysis of a typical experiment we found that the average area of the macroH2A peaks from the two peak fractions was 83% of the average of the peaks for H3, H2B, and H4. Assuming that the Coomassie blue staining of these proteins is equivalent, this translates to ∼0.3 mol of macroH2A for each mole of these other core histones. We also examined the depletion of conventional H2A in these fractions. This analysis yielded an average of a 38% reduction of H2A in the two peak fractions in comparison to the starting material.
In order to assess whether the mercaptoethanol-eluted thiopropyl Sepharose fractions contain appreciable amounts of chromatin fragments that do not contain macroH2A1, we ran the purification procedure with liver chromatin fragments prepared from macroH2A1 knockout mice. These mice do not contain macroH2A1.1 or -1.2 (unpublished data), and we did not detect histones or DNA in the mercaptoethanol-eluted thiopropyl fractions when we used macroH2A1 knockout mouse liver (Fig. 1). This indicates that nearly all of the chromatin fragments in the mercaptoethanol-eluted fractions contain at least one molecule of macroH2A1.
In contrast to macroH2A1, the macroH2A2 content of mouse liver is low, which made it difficult to determine where macroH2A2 was distributed during our thiol affinity procedure. We did not detect macroH2A2 in the mercaptoethanol-eluted thiopropyl fractions, but the high concentration of macroH2A1 in these fractions made it difficult to detect a small amount of macroH2A2.
Distribution of macroH2A1-containing nucleosomes in mouse liver chromatin; X-linked genes.
In order to examine the distribution of macroH2A1 on the inactive X chromosome, we purified macroH2A1-containing nucleosomes from female and male mouse livers. Comparison of male samples, containing one active X chromosome, to female samples, containing one active X and one inactive X, should allow us to assess whether macroH2A1 is preferentially concentrated on the inactive X. We used real-time PCR to examine the macroH2A1 distribution on six X-linked genes that are active in liver, four genes that are inactive in liver, and two intergenic regions that are more than 100 kb from any known gene (Table 1). We equalized the male and female DNA concentrations by using UV absorbance and gel electrophoresis in ethidium bromide-containing gels. We obtained similar results from two independent purifications. Results presented in the tables are the averages and standard deviations for the two preparations. Each preparation was tested twice by real-time PCR for each primer pair.
TABLE 1.
Distribution of macroH2A1 on X-linked genesa
| Gene | Male | Female | Gene | Male | Female | |
|---|---|---|---|---|---|---|
| Active genes | Testis-expressed gene 11 (Tex11) | |||||
| Coagulation factor VIII(F8) | Sixth intron | 0.9 ± 0.18 | 3.0 ± 0.4 | |||
| 2,300 bp before start | 0.2 ± 0.06 | 3.9 ± 0.6 | Twenty-first exon | 0.8 ± 0.17 | 2.8 ± 0.05 | |
| 1,250 bp before start | 0.4 ± 0.01 | 4.3 ± 1.5 | ||||
| First exon | 0.5 ± 0.02 | 4.0 ± 0.8 | Testis-specific X-linked gene (Tsx) | |||
| Last intron | 0.7 ± 0.03 | 2.6 ± 0.3 | Second intron | 1.2 ± 0.02 | 3.4 ± 0.4 | |
| Sixth intron | 0.7 ± 0.08 | 2.7 ± 0.3 | ||||
| Coagulation factor IX (F9) | ||||||
| First intron | 0.5 ± 0.02 | 4 ± 0.4 | Intergenic region, band A6 | 0.3 ± 0.06 | 2.3 ± 0.2 | |
| Third intron | 0.5 ± 0.05 | 3.4 ± 0.4 | ||||
| Intergenic region, band D | 0.7 ± 0.09 | 2.5 ± 0.3 | ||||
| Glucose-6-phosphate dehydrogenase (G6pdx) | ||||||
| First exon | 0.2 ± 0.01 | 3.1 ± 0.3 | Escape inactivation | |||
| Second intron | 0.3 ± 0.01 | 2.8 ± 0.4 | Eukaryotic translation initiation factor 2, subunit 3 structural gene X linked (Eif2s3x) | |||
| Glycerol kinase (Gyk) | ||||||
| 1,650 bp before start | 0.7 ± 0.06 | 4.0 ± 0.7 | 1,840 bp before start | 0.9 ± 0.05 | 1.5 ± 0.01 | |
| 600 bp before start | 0.2 ± 0.03 | 5.5 ± 0.1 | First intron | 0.1 ± 0.02 | 0.2 ± 0.01 | |
| First intron | 0.2 ± 0.02 | 3.0 ± 0.6 | Tenth intron | 0.1 ± 0.01 | 0.2 ± 0.01 | |
| Fourteenth exon | 0.2 ± 0.02 | 2.6 ± 0.3 | ||||
| Jumonji, AT-rich interactive domain 1C (Rbp2) | ||||||
| Hypoxanthine guanine phosphoribosyltransferase 1 (Hprt1) | ||||||
| 2,505 bp before start | 0.7 ± 0.01 | 1.6 ± 0.1 | ||||
| First intron | 0.1 ± 0.05 | 3.6 ± 0.4 | 680 bp before start | 0.3 ± 0.02 | 0.9 ± 0.01 | |
| Eighth intron | 0.2 ± 0.01 | 2.5 ± 0.3 | First intron | 0.1 ± 0.04 | 0.2 ± 0.01 | |
| Seventh intron | 0.1 ± 0.02 | 0.2 ± 0.02 | ||||
| Phosphoglycerate kinase 1 (Pgk1) | ||||||
| 1,260 bp before start | 1.7 ± 0.02 | 3.1 ± 0.4 | Xist | |||
| First intron | 0.4 ± 0.01 | 4.1 ± 1.0 | 360 bp before start | 1.5 ± 0.2 | 1.1 ± 0.03 | |
| Eighth intron | 0.2 ± 0.02 | 2.3 ± 0.3 | 2.5 kb into first exon | 1.0 ± 0.2 | 0.7 ± 0.16 | |
| 560 bp before 3′ end | 0.9 ± 0.12 | 0.5 ± 0.16 | ||||
| Inactive genes | ||||||
| Gamma-aminobutyric acid (GABA) A receptor, theta (Gabrq) | Pseudoautosomal boundary | |||||
| Midline 1 (Mid1) | ||||||
| Second intron | 0.8 ± 0.08 | 2.9 ± 0.4 | First intron | 0.3 ± 0.02 | 1.7 ± 0.2 | |
| Eighth intron | 0.6 ± 0.02 | 2.8 ± 0.4 | Second exon | 0.3 ± 0.05 | 1.6 ± 0.2 | |
| Third intron | 1.1 ± 0.02 | 1.3 ± 0.2 | ||||
| Opsin 1 (Opn1mw) | Fourth exon | 2.4 ± 0.03 | 2.8 ± 0.1 | |||
| 500 bp before start | 1.1 ± 0.2 | 2.6 ± 0.03 | Eighth exon | 3.1 ± 0.1 | 3.2 ± 0.2 | |
| First intron | 1.1 ± 0.2 | 2.5 ± 0.2 | 900 bp beyond end | 7.4 ± 0.2 | 9.3 ± 0.6 | |
| Last intron | 1.2 ± 0.2 | 3.3 ± 0.5 | 15 kb to 34 kb beyond end | 0.4 ± 0.05 | 0.4 ± 0.06 |
Values are the ratios of the concentrations in the macroH2A1-enriched thiopropyl Sepharose eluted fractions to the concentrations in the starting materials, determined by real-time PCR.
macroH2A1 was consistently depleted on the transcribed regions of the active genes (male samples of active genes). It was also depleted on the upstream regions of factor VIII and glycerol kinase but moderately enriched in a region approximately 1 kb upstream of phosphoglycerate kinase 1 (Table 1). In contrast, female samples showed significant enrichment for macroH2A1 on all probed regions of these genes, consistent with our previous conclusion based on immunofluorescence that macroH2A is preferentially concentrated on the inactive X (14). For the inactive genes, male samples gave a neutral macroH2A1 distribution on Tex11 and opsin1 and a small depletion for Tsx and Gabrq. macroH2A1 was significantly enriched on all four of these genes with the female samples, indicating that the preferential enrichment for macroH2A1 on the inactive X occurs for genes that are inactive in liver as well as ones that are active. Similar results were obtained for two X-linked intergenic regions, one in chromosome band A6 and the other in band D (Table 1).
We examined four regions that escape X inactivation. Xist had less macroH2A1 in the female samples than in the male samples (Table 1), which is consistent with it being active on the inactive X and inactive on the active X (7-9). Jarid1c and Eif2s3x escape X inactivation and are expressed in liver (19, 46). macroH2A1 was depleted on the transcribed regions of both genes in males and females. The upstream regions of Jarid1c and Eif2s3x had higher concentrations of macroH2A1 in the female samples than in the male samples (Table 1), indicating that macroH2A1 localization in these regions is shifting toward the pattern seen in regions of the X chromosome that do not escape inactivation.
The pseudoautosomal region of the X chromosome is generally believed to escape inactivation (43), so we examined the boundary of this region. The 5′ end of Mid1 is present only on the X chromosome, while its 3′ end is part of the pseudoautosomal region, being present on both the X and Y chromosomes. The transition to the pseudoautosomal region occurs in the third intron (37). Probes in the first and second introns showed depletion of macroH2A1 in male samples and a moderate enrichment in female samples. In contrast, probes in the third, fourth, and eighth introns gave nearly equivalent results for male and female samples. The 16-kb region immediately downstream of Mid1 is composed primarily of a 31-bp tandem repeat. We probed a small area of unique sequences approximately 900 bp downstream of Mid1 that is located between clusters of this repeat. This probe showed strong enrichment for macroH2A1 that was similar in males and females. A different 31-bp repeat is present in the region extending from ∼16 kb downstream of Mid1 to ∼34 kb downstream. This area is almost entirely composed of this tandem repeat, so we used primers to the repeated sequence. Because this repeat appears to be present only in this region of the X chromosome and probably the equivalent region of the Y chromosome, our primers should be specific for this domain. This distal repeat domain was equally depleted of macroH2A1 in males and females (Table 1).
Active and inactive autosomal genes.
In order to investigate whether the depletion of macroH2A1 from active genes is a more general phenomenon, we probed 12 active autosomal genes (Table 2). macroH2A1 was depleted on the transcribed regions of all of these genes except for one site on glucokinase that showed a small amount of enrichment. Probes in the near-upstream regions of albumin, fibrinogen gamma, and transferrin were also depleted of macroH2A1, while regions further upstream of these genes showed enrichment.
TABLE 2.
Distribution of macroH2A1 on active and inactive genesa
| Gene | Male | Female | Gene | Male | Female | |
|---|---|---|---|---|---|---|
| Active genes | ||||||
| Albumin 1 (Alb1), chromosome 5 | TATA box binding protein (Tbp), chromosome 17 | |||||
| 1,800 bp before start | 3.8 ± 0.69 | 4.0 ± 0.31 | ||||
| 570 bp before start | 1.2 ± 0.42 | 1.6 ± 0.11 | Second exon | 0.1 ± 0.01 | 0.2 ± 0.01 | |
| 70 bp before start | 0.8 ± 0.09 | 0.8 ± 0.22 | Fifth exon | 0.2 ± 0.01 | 0.2 ± 0.01 | |
| First intron | 0.4 ± 0.07 | 0.4 ± 0.02 | ||||
| Twelfth intron | 0.6 ± 0.08 | 0.7 ± 0.11 | Transferrin (Trf), chromosome 9 | |||
| 2,110 bp beyond end | 0.6 ± 0.06 | 0.7 ± 0.05 | 1,500 bp before start | 2.8 ± 0.47 | 3.5 ± 0.33 | |
| 290 bp before start | 0.2 ± 0.05 | 0.3 ± 0.04 | ||||
| ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 (Atp5c1), chromosome 2 | First intron | 0.5 ± 0.05 | 0.5 ± 0.04 | |||
| Sixteenth exon | 0.3 ± 0.01 | 0.3 ± 0.01 | ||||
| 1,240 bp beyond end | 0.5 ± 0.03 | 0.6 ± 0.02 | ||||
| First intron | 0.2 ± 0.01 | 0.2 ± 0.01 | ||||
| Third intron | 0.1 ± 0.02 | 0.2 ± 0.01 | Ubiquinol-cytochrome c reductase hinge protein (Uqcrh), chromosome 4 | |||
| Citrate synthase (Cs), chromosome 10 | First intron | 0.1 ± 0.01 | 0.1 ± 0.01 | |||
| First intron | 0.2 ± 0.03 | 0.3 ± 0.01 | Second intron | 0.1 ± 0.02 | 0.2 ± 0.01 | |
| Third intron | 0.2 ± 0.06 | 0.2 ± 0.01 | ||||
| Cytochrome c, somatic (Cycs), chromosome 6 | Potentially active genes | |||||
| First intron | 0.2 ± 0.01 | 0.1 ± 0.01 | Alpha fetoprotein (Afp), chromosome 5 | |||
| 580 bp before start | 1.5 ± 0.18 | 2.0 ± 0.12 | ||||
| Enoyl coenzyme A hydratase, short chain, 1, mitochondrial (Echs1), chromosome 7 | Junction of third exon | 0.6 ± 0.16 | 0.7 ± 0.1 | |||
| Twelfth intron | 0.5 ± 0.08 | 0.5 ± 0.05 | ||||
| First intron | 0.2 ± 0.02 | 0.2 ± 0.04 | ||||
| Fourth exon | 0.3 ± 0.01 | 0.3 ± 0.01 | ||||
| Inactive genes | ||||||
| Fibrinogen gamma (Fgg), chromosome 3 | Myosin, heavy polypeptide 6, cardiac muscle, alpha (Myh6), chromosome 14 | |||||
| 2,115 bp before start | 1.6 ± 0.11 | 2.2 ± 0.09 | ||||
| 620 bp before start | 0.5 ± 0.03 | 0.7 ± 0.09 | 1,750 bp before start | 2.2 ± 0.25 | 2.0 ± 0.09 | |
| Third exon | 0.3 ± 0.04 | 0.4 ± 0.05 | Third intron | 1.5 ± 0.21 | 1.5 ± 0.18 | |
| Seventh intron | 0.3 ± 0.11 | 0.4 ± 0.09 | Fourteenth intron | 4.1 ± 0.12 | 4.5 ± 1.03 | |
| 340 bp beyond end | 0.4 ± 0.02 | 0.4 ± 0.04 | Thirty-seventh intron | 1.9 ± 0.1 | 1.7 ± 0.04 | |
| Glucokinase (Gck), chromosome 11 | Glial fibrillary acidic protein (Gfap), chromosome 11 | |||||
| First intron | 0.6 ± 0.05 | 0.8 ± 0.03 | ||||
| Second intron | 1.1 ± 0.1 | 1.5 ± 0.2 | First intron | 1.6 ± 0.25 | 1.7 ± 0.04 | |
| Sixth intron | 0.8 ± 0.01 | 1 ± 0.1 | Seventh intron | 2.9 ± 0.32 | 2.8 ± 0.37 | |
| Last exon | 0.6 ± 0.01 | 0.8 ± 0.01 | ||||
| Insulin I (Ins1), chromosome 19 | ||||||
| Liver glycogen phosphorylase (Pyg1), chromosome 12 | 3′ exonb | 0.7 ± 0.06 | 0.8 ± 0.06 | |||
| 1,240 bp before start | 0.9 ± 0.1 | 1.5 ± 0.08 | Keratin (Krt2-19), chromosome 15 | |||
| First intron | 0.4 ± 0.03 | 0.4 ± 0.03 | First intron | 1.6 ± 0.17 | 1.8 ± 0.12 | |
| Next to second exon | 0.5 ± 0.02 | 0.7 ± 0.03 | Eighth intron, 1 | 1.7 ± 0.12 | 1.8 ± 0.16 | |
| Eighth intron, 2 | 1.3 ± 0.07 | 1.2 ± 0.13 | ||||
| Spot 14 (Thrsp), chromosome 7 | ||||||
| 5′ exon | 0.5 ± 0.06 | 0.6 ± 0.06 | Trypsinogen (Try4), chromosome 6 | |||
| Intron | 0.5 ± 0.07 | 0.6 ± 0.1 | First intron | 0.5 ± 0.11 | 0.4 ± 0.07 | |
| 3′ exon | 0.3 ± 0.03 | 0.3 ± 0.03 | Second intron | 0.5 ± 0.1 | 0.5 ± 0.03 | |
| 845 bp beyond end | 0.3 ± 0.03 | 0.3 ± 0.04 |
Values are the ratios of the concentrations in the macroH2A1-enriched thiopropyl Sepharose eluted fractions to the concentrations in the starting materials, determined by real-time PCR.
This probe may also detect insulin II.
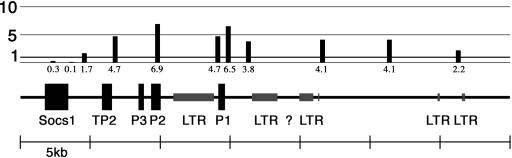
We probed five genes that should be inactive in liver and found enrichment on some and depletion on others (Table 2). We investigated the distribution of macroH2A1 in a region around a cluster of spermatogenesis genes that contains protamine 1, protamine 2, protamine 3, and transition protein 2 (Fig. 2). macroH2A1 was significantly enriched at all sites probed across this region extending until Socs1, at which point there was a sharp transition to being depleted of macroH2A1. One notable feature of this gene cluster is the presence of large “mammalian apparent LTR-retrotransposon” long terminal repeat (MaLR-LTR) elements (49) as identified by RepeatMasker (50). Large blocks of these elements are present in the upstream regions of protamine 1 and in the region between protamine 1 and protamine 2. macroH2A1 was present in high concentrations around these LTR clusters and in regions extending well beyond them in both directions (Fig. 2).
FIG. 2.
Distribution of macroH2A1 around protamine gene cluster. Real-time PCR was used to estimate the relative concentrations of macroH2A1 at various sites along a segment of mouse chromosome 16. The relative macroH2A1 concentration is expressed as the concentration in the macroH2A1-enriched thiopropyl Sepharose fractions divided by the concentration in the chromatin applied to the columns. Genes: Socs1, suppressor of cytokine signaling 1; TP2, transition protein 2; P3, protamine 3; P2, protamine 2; P1, protamine 1. Segments marked LTR were identified by RepeatMasker (50) and are primarily MaLR-LTR elements. The question mark indicates a region of unknown sequence in the genomic database.
Analysis of mononucleosomal DNA.
Active and inactive genes may have different nucleosome repeat lengths (51) that could potentially affect our results. It is also possible that there are small amounts of nonnucleosomal DNA in our preparations. In order to examine whether these factors affected our results, we gel purified mononucleosomal DNA from the macroH2A1-enriched thiopropyl-eluted fraction and from the starting material of one male and one female experiment. This DNA was used to probe a subset of active genes and X-linked genes (Table 3). The results with mononucleosomal DNA were virtually identical to what we obtained with the total DNA isolated from these fractions.
TABLE 3.
Results with mononucleosomesa
| Gene | Male | Female |
|---|---|---|
| Active genes | ||
| Albumin 1, first intron | 0.5 | 0.4 |
| ATP5c1, first intron | 0.1 | 0.1 |
| Citrate synthase, third intron | 0.1 | 0.1 |
| Cytochrome c, first intron | 0.1 | 0.1 |
| TBP, fifth exon | 0.1 | 0.1 |
| X-linked genes, active | ||
| Factor VIII, first exon | 0.6 | 3.6 |
| Glycerol kinase, 600 bp before start | 0.1 | 4.7 |
| HPRT, first intron | 0.1 | 3.7 |
| X-linked genes, inactive | ||
| Gabrq, second intron | 0.9 | 3.3 |
| Tex11, sixth intron | 1.8 | 3.1 |
| Tsx, sixth intron | 0.8 | 3.5 |
Values are ratios of the concentrations in mononucleosomal DNA isolated from the macroH2A1-enriched thiopropyl Sepharose eluted fractions to the concentrations in mononucleosomal DNA isolated from the starting materials, determined by real-time PCR.
Use of subtraction hybridization to identify regions enriched for macroH2A1.
We used subtractive hybridization to identify DNA regions that are enriched for macroH2A1. DNA from the flowthrough fractions (depleted in macroH2A1) was subtracted from DNA from the mercaptoethanol-eluted thiopropyl fractions (highly enriched for macroH2A1). Twenty-seven clones were sequenced. Three of the clones contained a closely related tandemly repeated 31-bp sequence. Hybridization of one of these clones with nonsubtracted DNA showed that these repeat sequences were significantly enriched in the macroH2A1-containing thiopropyl fractions (see Fig. S1 in the supplemental material). This tandem repeat was previously shown to be abundant in the pseudoautosomal regions of the X and Y chromosomes and also to be present on chromosomes 9 and 13 (22). The locations of many of these repeats were not mapped in the mouse genomic database. The largest mapped cluster of these repeats is the 31-bp repeats in and around the 3′ end of the mid1 gene discussed above.
Of the remaining 24 subtraction clones, 11 contained a single genomic segment; the other sequences contained two or more genomic segments and were not analyzed further. We used real-time PCR to examine the distribution of macroH2A1 on these 11 sequences. Primers were designed to amplify a unique genomic sequence located in or right next to the cloned sequence; one exception was the primers for clone 28, which amplify a repetitive endogenous retroviral sequence. The PCR results indicated that macroH2A1 is enriched on 10 of these sequences. The amount of enrichment ranged from ∼2-fold to more than 9-fold (Table 4). We used RepeatMasker (50) to probe for repetitive sequences in and around these 10 sequences (Table 4). Two clones contained large GT/CA repeats, a dinucleotide repeat commonly present in the genome. Hybridization with one of these clones suggested that there is no general enrichment of macroH2A1 on GT/CA repeats (not shown).
TABLE 4.
Distribution of macroH2A1 on subtraction clones
| Clone(s) | Malea | Femalea | Repeat(s) presentb | Chromosome(s) | Nearest gene |
|---|---|---|---|---|---|
| Forward subtraction clones | |||||
| 1 | 2.7 ± 1.89 | 5.4 ± 0.65 | MaLR LTR and LINE L1 | 1 | Trpm8, intron |
| 5 | 2.1 ± 0.34 | 2.1 ± 0.2 | Abuts an MaLR LTR | 13 | LOC432934, >100 kb |
| 8 | 4.1 ± 0.49 | 4.1 ± 0.24 | GT repeat | 4 | Bach2, 15 kb |
| 13 | 5.8 ± 0.56 | 6.6 ± 0.84 | MIR | 6 | Atp2b2, intron |
| 24 | 6.0 ± 0.66 | 7.7 ± 0.83 | GT repeat and MIR | 9 | Edg8, 8.3 kb |
| 26 | 5.2 ± 0.6 | 5.3 ± 0.17 | Flanked by MaLR and ERVL LTRs | 11 | 1700019I23Rik, 27 kb |
| 27 | 5.0 ± 1.04 | 5.6 ± 0.3 | MaLR LTR | 2 | Gm356, 14 kb |
| 28 | 3.8 ± 0.36 | 4.2 ± 0.7 | RLTR4-int LTR | Multiple | Reverse transcriptase |
| 15, 18, 21 | 7.4 ± 0.24c | 9.3 ± 0.56c | 31-bp tandem repeat | X, Y, 9, 13 | Mid1 and others |
| 29 | 3.1 ± 0.16 | 3.2 ± 0.01 | MER1 and MIR | 6 | LOC269739, 19 kb |
| 32 | 3.1 ± 0.17 | 3.5 ± 0.28 | MaLR LTR and LINE L1 | 1 | Cnih3, intron |
| Reverse subtraction clones | |||||
| O5 | 0.2 ± 0.02 | 0.8 ± 0.57 | Repeated on Y and X | Y and X | |
| O7 | 0.5 ± 0.11 | 0.7 ± 0.09 | LINE L1 | Multiple | |
| H2 | 0.6 ± 0.07 | 0.7 ± 0.05 | LINE L1 | Multiple |
Values are ratios of the concentrations in the macroH2A1-enriched thiopropyl Sepharose eluted fractions to the concentrations in the starting materials, determined by real-time PCR. All target sequences were nonrepetitive except for clone 28 and the reverse subtraction clones.
Repeats were identified by Repeat Masker (50).
Result for a region 900 bp downstream of Mid1 between two clusters of these repeats.
Three clones contained a MaLR-LTR element, and a fourth clone abutted a MaLR-LTR. MaLR-LTR elements are common in the mouse genome, and it was estimated that they should occur on average once per 30 to 100 kb (49). Since our subtracted clones together contain about 5 kb, the presence of three MaLR-LTR elements suggests that there may be a preferential association of macroH2A1 with these elements. We examined this possibility by real-time PCR using one type of MaLR-LTR consensus sequence (ORR1a, the type of MaLR-LTR that is prominent around the protamine genes) to design primers that should probe these sequences at more than 100 sites. We observed only a small enrichment for macroH2A1with these primers (Table 5). Subtraction clone 28 is a segment of the reverse transcriptase of an endogenous retroviral element. Using primers based on consensus sequences from the LTR, gag, polymerase, and envelope regions of this class of endogenous retroviral elements (RLTR4), we found that these regions were on average enriched for macroH2A1, with the largest enrichment occurring on the clone 28 sequence (Table 5).
TABLE 5.
Distribution of macroH2A1 on some repeated sequences
| Sequence | Repeata | Maleb | Femaleb |
|---|---|---|---|
| Clone O7 | LINE L1 | 0.5 ± 0.11 | 0.7 ± 0.09 |
| Clone H2 | LINE L1 | 0.6 ± 0.07 | 0.7 ± 0.05 |
| LINE 5′ | L1 consensus, 5′ | 1.0 ± 0.13 | 1.3 ± 0.11 |
| LINE 3′ | L1 consensus, 3′ | 0.8 ± 0.27 | 1.5 ± 0.12 |
| MaLR-LTR | LTR, ORR1a consensus | 1.5 ± 0.09 | 1.7 ± 0.07 |
| MaLR-LTR | Internal, ORR1a consensus | 1.2 ± 0.11 | 1.4 ± 0.05 |
| MaLR-LTR | Internal, ORR1a consensus | 1.4 ± 0.15 | 1.8 ± 0.25 |
| Clone 28 | RLTR4, polymerase | 3.9 ± 0.27 | 3.9 ± 0.01 |
| RLTR4 | RLTR4, envelope | 2.0 ± 0.09 | 2.2 ± 0.1 |
| RLTR4 | RLTR4, gag | 2.2 ± 0.22 | 2.7 ± 0.08 |
| RLTR4 | RLTR4, LTR | 1.7 ± 0.13 | 1.6 ± 0.2 |
| γ satellite | Centromeric | 1.9 ± 0.01 | 1.6 ± 0.05 |
Repeats were identified by RepeatMasker (50).
Values are ratios of the concentrations in the macroH2A1-enriched thiopropyl Sepharose eluted fractions to the concentrations in the starting materials, determined by real-time PCR.
Three of the 10 subtraction clones enriched for macroH2A1 contain mammalian-wide interspersed repeat (MIR) elements, with two MIRs present in clone 13. MIRs are short interspersed elements that are widely present in mammals and appear to be derived from tRNA (26, 48). Although their precise prevalence in the mouse genome has not been reported, they probably represent less than 1% of the total genome. Because MIR sequences are not highly conserved and often less than 100 bp, it is difficult to design primers that probe the distribution of macroH2A on more than one MIR sequence.
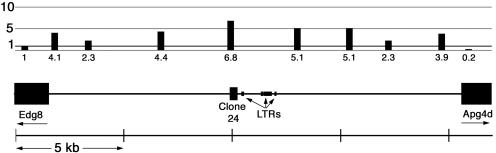
We developed a more complete map of macroH2A1 distribution in a region of about 20 kb around subtraction clone 24 (Fig. 3). macroH2A1 was enriched on all the sequences that we probed in this region until we reached the neighboring genes Apg4d and Edg8. According to the NCBI Expressed Sequence Tag database, both of these genes are expressed in mouse liver.
FIG. 3.
Distribution of macroH2A1 around subtraction clone 24. Real-time PCR was used to estimate the relative concentrations of macroH2A1 at various sites along a segment of mouse chromosome 9. The relative macroH2A1 concentration is expressed as the concentration in the macroH2A1-enriched thiopropyl Sepharose fractions divided by the concentration in the chromatin applied to the columns. Genes: Edg8, endothelial differentiation, sphingolipid G-protein-coupled receptor, 8; Apg4d, APG4 (ATG4) autophagy-related homolog D. Segments marked LTR were identified by RepeatMasker (50) as endogenous retroviral element RMER6A.
We sequenced three clones from a reverse subtraction experiment that were depleted of macroH2A1 (Table 4). Each of these clones was composed entirely of repetitive sequence. Two of them are from different regions of long interspersed nucleotide element (LINE) L1. The third one is a moderately repetitive sequence that appears to be primarily on the Y chromosome and to a lesser extent on the X and unmapped sites. RepeatMasker did not identify this repeat. The depletion of macroH2A1 from this sequence was higher in males than in females, possibly due to the prevalence of this repeat on the Y chromosome and an enrichment of macroH2A on the repeats on the inactive X.
Proteins associated with macroH2A-containing chromatin.
We used mass spectrometry to identify some of the proteins associated with macroH2A1-containing chromatin fragments. We obtained high-quality results on two proteins: heterochromatin protein HP1β and hnRNP A2/B1 (Fig. 1) (see Fig. S2 and S3 in the supplemental material). Western blot analyses confirmed the identities of HP1β and hnRNP A2/B1 and the enrichment of these proteins on the purified macroH2A1-containing chromatin (see Fig. S5 in the supplemental material). When we used macroH2A1 knockout liver, Western blot assays did not detect any A2/B1 in the mercaptoethanol-eluted thiopropyl fraction and only traces of HP1β (not shown). Therefore, we believe that these proteins are associated with macroH2A1-containing chromatin. A2 and B1 are very similar proteins formed by alternate splicing (10), and we were unable to distinguish these proteins in our analyses. On the basis of its position in the gel, the largest A2/B1 band appears be the full-length protein. We also observed multiple A2/B1 proteolytic degradation products (Fig. 1) that were identified as A2/B1 by mass spectrometry.
We examined two closely spaced bands just above the main macroH2A bands. These bands reacted with macroH2A1 antibodies (Fig. 1) and gave peptides with molecular weights that matched those of macroH2A1 and other peptides that matched ubiquitin (see Fig. S4 in the supplemental material). On the basis of these results and the position of these bands in the gel, we believe that they are monoubiquitinated macroH2A1.1 and -1.2 (Fig. 1). macroH2As have a lysine in their H2A region that corresponds to the lysine that is monoubiquitinated in other H2As, and recent studies demonstrated the presence of monoubiquitinated macroH2A in cultured human cells (23).
We identified topoisomerase I as one of the proteins associated with the mercaptoethanol-eluted thiopropyl Sepharose fractions by mass spectrometry. However, we were unable to confirm this identification by Western blot analysis, possibly due to the partial degradation of the topoisomerase I in our preparations.
DISCUSSION
The binding of macroH2A1-containing nucleosomes to thiopropyl Sepharose, but not activated thiol Sepharose, provides a powerful method for enriching macroH2A1-containing nucleosomes. The basic thiol affinity chemistry of these two matrixes is similar, suggesting that the difference lies in the accessibility of their reactive groups to thiols in macroH2A1 proteins. The most likely explanation is the difference in the linker arms. Activated thiol Sepharose has a branched arm with two carboxylate groups, while thiopropyl Sepharose has a shorter arm that is uncharged. As with any procedure used to purify a specific chromatin fraction, our results may not be representative of all macroH2A1-containing chromatin. While we attempted to maximize the representation of chromatin by using a chromatin fraction that contains most of the DNA, some chromatin is lost in our procedure. The enrichment of macroH2A1 that we observed on the inactive X correlates well with what we observe by immunofluorescence of nuclei, indicating that our results are representative of a significant fraction of the chromatin. One limitation of this procedure is that it does not resolve macroH2A1.1 and -1.2, which may have distinct distributions.
Our previous immunofluorescence study showing the preferential localization of macroH2A to the inactive X (14) was challenged on the basis that the higher macroH2A immunofluorescence of the inactive X was due to increased nucleosome density (42). In our current study we have essentially eliminated the complication of nucleosome density by examining the distribution of specific sequences in DNA isolated from macroH2A1-containing nucleosomes. Our study with gel-purified mononucleosomal DNA largely eliminated effects related to differences in nucleosome spacing or possible contributions of nonnucleosomal DNA to the analysis. These analyses confirm our previous conclusion that macroH2A is preferentially concentrated on the inactive X. This preferential localization is not simply the consequence of the transcriptional silence of the inactive X. This can be seen by the distribution of macroH2A1 on X-linked genes or sequences that are not transcribed. We probed four X-linked genes that are inactive in liver and two intergenic regions that are far away from any known gene. None of these regions showed significant enrichment for macroH2A1 on the active X, and all of them showed significant macroH2A1 enrichment on the inactive X. macroH2A1 was preferentially localized to the inactive X at multiple regions of the 10 X-linked genes and two intergenic regions that we examined. In contrast, macroH2A1 was not preferentially localized to four regions that escape X inactivation: Xist, Eif2s3x, Jarid1c, and the pseudoautosomal region around Mid1. Our results provide evidence for boundaries for preferential macroH2A1 localization in the 5′ ends of Eif2s3x and Jarid1 and near the third intron of Mid1. The shift in the pattern of macroH2A1 localization in upstream regions of Eif2s3x and Jarid1 is consistent with a recent study that identified CTCF binding boundary elements in the promoter regions of these genes (20). Taken together our results indicate that macroH2A1 is broadly targeted to the inactive X but is not preferentially localized to regions that escape inactivation.
A very interesting finding of our studies is the depletion of macroH2A1 from the transcribed regions of active genes. This was observed on all but one site probed for 12 active autosomal genes and all sites probed on eight active X-linked genes (Tables 1 and 2). At least in some cases the domain of macroH2A1 depletion on active genes appeared to continue into the promoter region and beyond the last exon, although this is an uncertain conclusion because the start and end for many genes in the mouse genomic database are tentative. We typically observe a depletion of approximately two- to fivefold in transcribed regions. The concentration of macroH2A1 in mouse liver appears to be similar to that of rat liver (39), which we previously estimated to be approximately one macroH2A molecule for every 30 nucleosomes (40). This suggests that macroH2A1 is very sparsely present on most transcribed regions. These results together with our results showing an increased concentration of macroH2A1 on the silent regions of the inactive X are consistent with the hypothesis that macroH2A1 histones are transcriptionally repressive. The presence of the heterochromatin protein HP1β on the purified macroH2A1-containing chromatin fragments is consistent with macroH21 having a repressive role (34). Both macroH2A and HP1β can associate with pericentromeric chromatin in some cell types (16, 21, 34). However, immunofluorescence staining indicates that pericentromeric macroH2A1 is not a prominent feature in hepatocytes (14), and we observed only a small enrichment of a γ-satellite centromeric repeat sequence in the purified macroH2A1-containing chromatin from liver (Table 5). This suggests that some of the HP1β associated with macroH2A1-containing chromatin is in regions other than pericentromeric heterochromatin. We do not know whether this HP1β directly interacts with macroH2A1, but they are likely in close proximity given the small size of the chromatin fragments that we used.
Our results indicate that macroH2A1-containing nucleosomes are interspersed with nucleosomes that contain conventionally sized H2As, even in regions where they are relatively concentrated, such as the inactive X chromosome. On the basis of our experiments with macroH2A1 knockout liver, we believe that nearly all of the chromatin fragments eluted from thiopropyl Sepharose by mercaptoethanol contain at least one molecule of macroH2A1. Even with the relatively short fragments that we used in these studies, we observed as much or more conventionally sized H2A in these purified nucleosomes than macroH2A. This suggests extensive interspersion. Technical limitations make it difficult to accurately estimate the concentration of macroH2A1 on specific sequences. Variability in the macroH2A subtype composition and distribution in different cell types (15) will affect our results. Examination of the inactive X is complicated by the presence of the active X, which will reduce the observed concentration of macroH2A in female cells. The average enrichment that we observed on the X-linked genes that we probed (excluding genes that escape inactivation) was 3.2-fold. The active X seems unlikely to produce more than a twofold reduction in enrichment. While factors such as cell type heterogeneity may also increase the estimated macroH2A concentration, it appears to be well below what we would expect for complete substitution. A few regions, such as the 3′ end of Mid1, gave an enrichment of close to 10-fold, but most regions were less. These results lead us to believe that macroH2A1 is interspersed with conventionally sized H2As in most regions.
In the regions that we examined with multiple probes, macroH2A1 appeared to be arranged in relatively large domains of enrichment or depletion. The most notable domains of depletion are the transcribed regions of active genes. In addition to the inactive X chromosome, we mapped two substantial domains of enrichment. One of these domains occurs around the protamine gene cluster and the other in the region upstream of two genes, Apg4d and Edg8. Interestingly, both of these domains of enrichment showed relatively sharp transitions to macroH2A1 depletion as they extended to neighboring genes. In the case of the protamine cluster this transition occurred just upstream of Socs1. Socs1 is expressed in liver, and its expression in hepatocytes is increased by treatment with lipopolysaccharide (52).
How are macroH2As targeted to certain regions and depleted from others? Multiple mechanisms may be involved, since studies with chimeric proteins indicate that both the H2A domain and the nonhistone domain can direct macroH2As to the inactive X (11). Xist RNA is required to maintain the preferential localization of macroH2A1 to the inactive X (6, 17), but it is not known whether Xist-containing RNPs have a direct role in macroH2A localization. Our finding that the hnRNP protein A2/B1 is present in purified macroH2A1-containing chromatin suggests that RNA may be a component of this chromatin. However, A2/B1 can also bind single-stranded DNA (25, 35), so its presence does not necessarily indicate the presence of RNA. Based on the study of other histone variants (28, 29, 36), it seems reasonable to believe that there is a chromatin-remodeling complex(es) that preferentially targets the assembly or exchange of macroH2A into chromatin. The targeting of such complexes to specific regions by interaction with chromatin-bound proteins or RNPs could lead to the preferential assembly of macroH2As in particular regions. macroH2As would appear to be available for exchange into chromatin as they are expressed in tissues with few dividing cells such as adult mouse liver (44) and continue to be relatively abundant in adult cells that have not undergone cell division for months or years (2). Depletion of macroH2A in transcribed and other regions could occur by the localized use of remodeling complexes that incorporate H2As other than macroH2A. Differential stability of macroH2As in different regions could also be an important contributor to their distribution.
Are there local DNA sequences that promote macroH2A localization to particular regions? In the case of the inactive X, it appears that Xist can promote macroH2A localization over very large distances. However, there could still be local elements that help recruit Xist RNP or macroH2A. On the basis of studies of chromosomal translocations involving the inactive X, it was suggested that there are “way stations” that boost inactivation along the chromosome (45). Mary Lyon hypothesized that interspersed repetitive elements could act as way stations (33). Thus far, our results show a positive correlation with certain LTR elements. MaLR-LTR elements are relatively abundant in the macroH2A1-enriched sequences that we identified by subtractive hybridization and are prominent in a macroH2A1-enriched domain that we identified around the protamine gene cluster. Additionally, one of our subtraction clones corresponded to a segment of a reverse transcriptase from an endogenous retrovirus. However, the correlation between macroH2A1 enrichment and LTR elements is not straightforward. The amount of enrichment that we observed when we made probes to consensus sequences of MaLR-LTR elements was small (Table 5), indicating that substantial macroH2A1 enrichment is not a consistent feature of these elements. Also, when we probed sequences next to two randomly selected endogenous retroviruses of the same class, we found significant macroH2A enrichment next to one and no enrichment next to the other (not shown). Thus, if LTR elements and/or endogenous retroviruses have a role in macroH2A localization, it would appear to be only part of a more complex mechanism. One interesting candidate for a macroH2A1-targeting sequence is the 31-bp tandem repeat that is present around the 3′ end of mid1. Perhaps proteins bound to these sequences or unusual structural features of this DNA could target macroH2A1 to these sequences.
This study substantiates our previous conclusions based on immunofluorescence studies that macroH2As are preferentially associated with the inactive X chromosome and are widely and nonrandomly distributed in the rest of the chromatin. Interestingly, macroH2A1 was depleted from the transcribed genes that we examined, consistent with its apparent function as a transcriptional repressor. Our results indicate that macroH2As have complex distribution patterns that are influenced by local factors as well as long-range targeting mechanisms. Much remains to be learned about how these patterns are established and how they affect transcriptional activity and other chromatin functions.
Supplementary Material
Acknowledgments
We thank Steve Seeholzer for the identification of proteins by mass spectrometry; Chhaya Dharia, Leslie Taylor, Dannee Chen, Carl Costanzi, and Denys Volgin for technical help and suggestions; Mike Atchison and Narayan Avadhani for comments on the manuscript; and Sandra Martin for LINE L1 clones.
This work was supported by Public Health Service grant GM49351 from the NIH.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abbott, D. W., M. Laszczak, J. D. Lewis, H. Su, S. C. Moore, M. Hills, S. Dimitrov, and J. Ausio. 2004. Structural characterization of macroH2A containing chromatin. Biochemistry 43:1352-1359. [DOI] [PubMed] [Google Scholar]
- 2.Akbarian, S., R. Z. Chen, J. Gribnau, T. P. Rasmussen, H. Fong, R. Jaenisch, and E. G. Jones. 2001. Expression pattern of the Rett syndrome gene MeCP2 in primate prefrontal cortex. Neurobiol. Dis. 8:784-791. [DOI] [PubMed] [Google Scholar]
- 3.Allegra, P., R. Sterner, D. F. Clayton, and V. G. Allfrey. 1987. Affinity chromatographic purification of nucleosomes containing transcriptionally active DNA sequences. J. Mol. Biol. 196:379-388. [DOI] [PubMed] [Google Scholar]
- 4.Allen, M. D., A. M. Buckle, S. C. Cordell, J. Lowe, and M. Bycroft. 2003. The crystal structure of AF1521, a protein from Archaeoglobus fulgidus with homology to the non-histone domain of macroH2A. J. Mol. Biol. 330:503-511. [DOI] [PubMed] [Google Scholar]
- 5.Angelov, D., A. Molla, P. Y. Perche, F. Hans, J. Cote, S. Khochbin, P. Bouvet, and S. Dimitrov. 2003. The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol. Cell 11:1033-1041. [DOI] [PubMed] [Google Scholar]
- 6.Beletskii, A., Y.-K. Hong, J. Pehrson, M. Egholm, and W. M. Strauss. 2001. PNA interference mapping demonstrates functional domains in the noncoding RNA Xist. Proc. Natl. Acad. Sci. USA 98:9215-9220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borsani, G., R. Tonlorenzi, M. C. Simmler, L. Dandolo, D. Arnaud, V. Capra, M. Grompe, A. Pizzuti, D. Muzny, C. Lawrence, and A. Ballabio. 1991. Characterization of a murine gene expressed from the inactive X chromosome. Nature 351:325-329. [DOI] [PubMed] [Google Scholar]
- 8.Brockdorff, N., A. Ashworth, G. F. Kay, P. Cooper, S. Smith, V. M. McCabe, D. P. Norris, G. D. Penny, D. Patel, and S. Rastan. 1991. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature 351:329-331. [DOI] [PubMed] [Google Scholar]
- 9.Brown, C. J., A. Ballabio, J. L. Rupert, R. G. Lafreniere, M. Grompe, R. Tonlorenzi, and H. F. Willard. 1991. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349:38-44. [DOI] [PubMed] [Google Scholar]
- 10.Burd, C. G., M. S. Swanson, M. Gorlach, and G. Dreyfuss. 1989. Primary structures of the heterogeneous nuclear ribonucleoprotein A2, B1, and C2 proteins: a diversity of RNA binding proteins is generated by small peptide inserts. Proc. Natl. Acad. Sci. USA 86:9788-9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chadwick, B. P., C. M. Valley, and H. F. Willard. 2001. Histone variant macroH2A contains two distinct macrochromatin domains capable of directing macroH2A to the inactive X chromosome. Nucleic Acids Res. 29:2699-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chadwick, B. P., and H. F. Willard. 2001. Histone H2A variants and the inactive X chromosome: identification of a second macroH2A variant. Hum. Mol. Genet. 10:1101-1113. [DOI] [PubMed] [Google Scholar]
- 13.Chakravarthy, S., S. K. Y. Gundimella, C. Caron, P.-Y. Perche, J. R. Pehrson, S. Khochbin, and K. Luger. 2005. Structural characterization of the histone variant macroH2A. Mol. Cell. Biol. 25:7616-7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costanzi, C., and J. R. Pehrson. 1998. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 393:599-601. [DOI] [PubMed] [Google Scholar]
- 15.Costanzi, C., and J. R. Pehrson. 2001. MACROH2A2, a new member of the MACROH2A core histone family. J. Biol. Chem. 276:21776-21784. [DOI] [PubMed] [Google Scholar]
- 16.Costanzi, C., P. Stein, D. M. Worrad, R. M. Schultz, and J. R. Pehrson. 2000. Histone macroH2A1 is concentrated in the inactive X chromosome of female preimplantation embryos. Development 127:2283-2289. [DOI] [PubMed] [Google Scholar]
- 17.Csankovszki, G., B. Panning, B. Bates, J. R. Pehrson, and R. Jaenisch. 1999. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat. Genet. 22:323-324. [DOI] [PubMed] [Google Scholar]
- 18.Davis, A. H., T. L. Reudelhuber, and W. T. Garrard. 1983. Variegated chromatin structures of mouse ribosomal RNA Genes. J. Mol. Biol. 167:133-155. [DOI] [PubMed] [Google Scholar]
- 19.Ehrmann, I. E., P. S. Ellis, S. Mazeyrat, S. Duthie, N. Brockdorff, M. G. Mattei, M. A. Gavin, N. A. Affara, G. M. Brown, E. Simpson, M. J. Mitchell, and D. M. Scott. 1998. Characterization of genes encoding translation initiation factor eIF-2gamma in mouse and human: sex chromosome localization, escape from X-inactivation and evolution. Hum. Mol. Genet. 7:1725-1737. [DOI] [PubMed] [Google Scholar]
- 20.Filippova, G. N., M. K. Cheng, J. M. Moore, J. P. Truong, Y. J. Hu, D. K. Nguyen, K. D. Tsuchiya, and C. M. Disteche. 2005. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev. Cell 8:31-42. [DOI] [PubMed] [Google Scholar]
- 21.Grigoryev, S. A., T. Nikitina, J. R. Pehrson, P. B. Singh, and C. L. Woodcock. 2004. Dynamic relocation of epigenetic chromatin markers reveals an active role of constitutive heterochromatin in the transition from proliferation to quiescence. J. Cell Sci. 117:6153-6162. [DOI] [PubMed] [Google Scholar]
- 22.Harbers, K., U. Francke, P. Soriano, R. Jaenisch, and U. Muller. 1990. Structure and chromosomal mapping of a highly polymorphic repetitive DNA sequence from the pseudoautosomal region of the mouse sex chromosomes. Cytogenet. Cell Genet. 53:129-133. [DOI] [PubMed] [Google Scholar]
- 23.Hernandez-Munoz, I., A. H. Lund, P. van der Stoop, E. Boutsma, I. Muijrers, E. Verhoeven, D. A. Nusinow, B. Panning, Y. Marahrens, and M. van Lohuizen. 2005. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc. Natl. Acad. Sci. USA 102:7635-7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoyer-Fender, S., C. Costanzi, and J. R. Pehrson. 2000. Histone macroH2A1.2 is concentrated in the XY-body by the early pachytene stage of spermatogenesis. Exp. Cell Res. 258:254-260. [DOI] [PubMed] [Google Scholar]
- 25.Ishikawa, F., M. J. Matunis, G. Dreyfuss, and T. R. Cech. 1993. Nuclear proteins that bind the pre-mRNA 3′ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol. Cell. Biol. 13:4301-4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jurka, J., E. Zietkiewicz, and D. Labuda. 1995. Ubiquitous mammalian-wide interspersed repeats (MIRs) are molecular fossils from the mesozoic era. Nucleic Acids Res. 23:170-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karras, G. I., G. Kustatscher, H. R. Buhecha, M. D. Allen, C. Pugieux, F. Sait, M. Bycroft, and A. G. Ladurner. 2005. The macro domain is an ADP-ribose binding module. EMBO J. 24:1911-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krogan, N. J., M. C. Keogh, N. Datta, C. Sawa, O. W. Ryan, H. Ding, R. A. Haw, J. Pootoolal, A. Tong, V. Canadien, D. P. Richards, X. Wu, A. Emili, T. R. Hughes, S. Buratowski, and J. F. Greenblatt. 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12:1565-1576. [DOI] [PubMed] [Google Scholar]
- 29.Kusch, T., L. Florens, W. H. Macdonald, S. K. Swanson, R. L. Glaser, J. R. Yates III, S. M. Abmayr, M. P. Washburn, and J. L. Workman. 2004. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306:2084-2087. [DOI] [PubMed] [Google Scholar]
- 30.Kustatscher, G., M. Hothorn, C. Pugieux, K. Scheffzek, and A. G. Ladurner. 2005. Splicing regulates NAD metabolite binding to histone macroH2A. Nat. Struct. Mol. Biol. 12:624-625. [DOI] [PubMed] [Google Scholar]
- 31.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 32.Libertini, L. J., and E. W. Small. 1980. Salt induced transitions of chromatin core particles studied by tyrosine fluorescence anisotropy. Nucleic Acids Res. 8:3517-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lyon, M. F. 1998. X-chromosome inactivation: a repeat hypothesis. Cytogenet. Cell Genet. 80:133-137. [DOI] [PubMed] [Google Scholar]
- 34.Maison, C., and G. Almouzni. 2004. HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 5:296-304. [DOI] [PubMed] [Google Scholar]
- 35.McKay, S. J., and H. Cooke. 1992. hnRNP A2/B1 binds specifically to single stranded vertebrate telomeric repeat TTAGGGn. Nucleic Acids Res. 20:6461-6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizuguchi, G., X. Shen, J. Landry, W.-H. Wu, S. Sen, and C. Wu. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303:343-348. [DOI] [PubMed] [Google Scholar]
- 37.Palmer, S., J. Perry, D. Kipling, and A. Ashworth. 1997. A gene spans the pseudoautosomal boundary in mice. Proc. Natl. Acad. Sci. USA 94:12030-12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pehrson, J. R. 2004. Core histone variants, p. 181-204. In J. Zlatanova and S. H. Leuba (ed.), Chromatin structure and dynamics: state of the art, vol. 29. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 39.Pehrson, J. R., C. Costanzi, and C. Dharia. 1997. Developmental and tissue expression patterns of histone macroH2A1 subtypes. J. Cell. Biochem. 65:107-113. [DOI] [PubMed] [Google Scholar]
- 40.Pehrson, J. R., and V. A. Fried. 1992. MacroH2A, a core histone containing a large nonhistone region. Science 257:1398-1400. [DOI] [PubMed] [Google Scholar]
- 41.Pehrson, J. R., and R. N. Fuji. 1998. Evolutionary conservation of macroH2A subtypes and domains. Nucleic Acids Res. 26:2837-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perche, P.-Y., C. Vourc'h, L. Konecny, C. Souchier, M. Robert-Nicoud, S. Dimitrov, and S. Khochbin. 2000. Higher concentrations of histone macroH2A in the Barr body are correlated with higher nucleosome density. Curr. Biol. 10:1531-1534. [DOI] [PubMed] [Google Scholar]
- 43.Rappold, G. A. 1993. The pseudoautosomal regions of the human sex chromosomes. Hum. Genet. 92:315-324. [DOI] [PubMed] [Google Scholar]
- 44.Rasmussen, T. P., T. Huang, M. A. Mastrangelo, J. Loring, B. Panning, and R. Jaenisch. 1999. Messenger RNAs encoding mouse histone macroH2A1 isoforms are expressed at similar levels in male and female cells and result from alternative splicing. Nucleic Acids Res. 27:3685-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riggs, A. D. 1990. Marsupials and the mechanisms of X chromosome inactivation. Aust. J. Zool. 37:419-441. [Google Scholar]
- 46.Sheardown, S., D. Norris, A. Fisher, and N. Brockdorff. 1996. The mouse Smcx gene exhibits developmental and tissue specific variation in degree of escape from X inactivation. Hum. Mol. Genet. 5:1355-1360. [DOI] [PubMed] [Google Scholar]
- 47.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 48.Smit, A. F., and A. D. Riggs. 1995. MIRs are classic, tRNA-derived SINEs that amplified before the mammalian radiation. Nucleic Acids Res. 23:98-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smit, A. F. A. 1993. Identification of a new, abundant superfamily of mammalian LTR-transposons. Nucleic Acids Res. 21:1863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smit, A. F. A., R. Hubley, and P. Green. 1996;-2004. RepeatMasker Open-3.0._1996-2004.
- 51.Smith, R. D., R. L. Seale, and J. Yu. 1983. Transcribed chromatin exhibits an altered nucleosomal spacing. Proc. Natl. Acad. Sci. USA 80:5505-5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ueki, K., T. Kondo, and C. R. Kahn. 2004. Suppressor of cytokine signaling 1 (SOCS-1) and SOCS-3 cause insulin resistance through inhibition of tyrosine phosphorylation of insulin receptor substrate proteins by discrete mechanisms. Mol. Cell. Biol. 24:5434-5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Holde, K. E. 1988. Chromatin. Springer-Verlag, New York, N.Y.
- 54.Zhang, R., M. V. Poustovoitov, X. Ye, H. A. Santos, W. Chen, S. M. Daganzo, J. P. Erzberger, I. G. Serebriiskii, A. A. Canutescu, R. L. Dunbrack, J. R. Pehrson, J. M. Berger, P. D. Kaufman, and P. D. Adams. 2005. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell 8:19-30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.