Abstract
PEA3 is a member of a subfamily of ETS domain transcription factors which is regulated by a number of signaling cascades, including the mitogen-activated protein (MAP) kinase pathways. PEA3 activates gene expression and is thought to play an important role in promoting tumor metastasis and also in neuronal development. Here, we have identified the LIM domain protein LPP as a novel coregulatory binding partner for PEA3. LPP has intrinsic transactivation capacity, forms a complex with PEA3, and is found associated with PEA3-regulated promoters. By manipulating LPP levels, we show that it acts to upregulate the transactivation capacity of PEA3. LPP can also functionally interact in a similar manner with the related family member ER81. Thus, we have uncovered a novel nuclear function for the LIM domain protein LPP as a transcriptional coactivator. As LPP continually shuttles between the cell periphery and the nucleus, it represents a potential novel link between cell surface events and changes in gene expression.
PEA3, ER81, and ERM comprise the PEA3 subfamily of ETS domain transcription factors (reviewed in references 9 and 10). These proteins show a high degree of sequence conservation within their ETS DNA-binding domains and also in an N-terminal acidic domain and the region C-terminal to the ETS domain. These proteins also exhibit a high level of evolutionary conservation with homologues of human PEA3 and ERM, having been identified in other vertebrates such as zebra fish (4, 26). Biologically, PEA3 subfamily members have been shown to be involved in a number of processes including neuronal pathfinding (1, 24) and to play an important role in HER2/Neu-mediated mammary oncogenesis (37). Developmentally, members of the PEA3 subfamily are important recipients of fibroblast growth factor signaling (11, 27, 33, 34). Fibroblast growth factor signaling acts via activating the extracellular signal-regulated kinase/mitogen-activated protein (ERK/MAP) kinase pathway and leads to the upregulation of the expression of PEA3 subfamily members. In addition, the transcriptional activity of several family members is enhanced in response to ERK pathway activation (18, 19, 28). A number of target genes have been identified (reviewed in reference 9 and 10), including genes with important roles in tumor growth and metastasis such as COX-2 (16, 42) and MMP-1 (3).
The LPP (lipoma-preferred partner) protein and zyxin, ajuba, LIMD1, and TRIP6 form a subfamily of LIM domain proteins that are characterized by the presence of three tandem C-terminal LIM domains (44). LPP was first isolated as part of a fusion protein created by chromosomal translocations, in which the C-terminal part of LPP is fused to the N terminus of HMGA2/HMGIC (32). This suggests an important role for LPP in tumorigenesis and, in particular, the C-terminal region containing the LIM domains. LPP is usually localized at the cell periphery in focal adhesions and cell-cell contacts, where it associates with proteins such as α-actinin (23) and Scrib (31). However, in common with other family members (reviewed in reference 44), LPP has been shown to continually shuttle through the nucleus and to possess a Crm1-dependent nuclear export signal (29, 30). TRIP6 has been shown to possess transactivation properties and to act as both a coactivator and corepressor protein in the context of AP-1- and NF-κB-mediated gene regulation under different conditions (20, 43). LPP has also been shown to possess two domains harboring transcriptional activation capacity, which coincide with the LIM domains and the proline-rich region preceding these (29). These observations suggest that LPP might have a role in regulating gene expression in the nucleus.
Our current knowledge of coregulatory partners for PEA3 is limited. To identify such proteins, we previously used a yeast one-hybrid screen and identified USF-1 as an interaction partner for PEA3 (14). Here we identify a clone containing the C-terminal part of LPP from this screen and demonstrate that it is a novel coregulatory protein that affects PEA3 function. LPP is recruited to PEA3-dependent promoters in vivo and acts to potentiate the transactivation potential of PEA3. Functional interactions are also seen with the PEA3 subfamily member ER81. Thus, we have uncovered a novel function for the LIM domain protein LPP as a transcriptional coactivator in the nucleus.
MATERIALS AND METHODS
Plasmid constructs.
phPES2 (nucleotides −327 to +59 of human COX-2 promoter [17]), and p5xPEA3 RE-Luc (five PEA3 binding sites and an adenovirus major late promoter [2]) luciferase reporter genes were described previously. Plasmid pCH110 (Pharmacia) contains a simian virus 40-driven β-galactosidase (lacZ) gene and is used to monitor transfection efficiency. pSG5-ERM (encoding full-length human ERM), pSG5-ER81 (encoding full-length human ER81 [25]), and pAS1801 (encoding full-length mouse PEA3) (21) have been described previously. pCMV-MEK1 encodes constitutively active MEK-1(ΔN-S218E/S222D). pAS838 encodes full-length human LPP with C-terminal His and Flag tags and was constructed by ligating a HindIII/NotI fragment from pAS837 into HindIII/NotI-cleaved pCDNA3. The intermediary clone pAS837 was constructed by ligating a HindIII/XhoI fragment from pAS836 into HindIII/XhoI-cleaved pAS728. pAS836 was constructed by replacing the BamHI/XbaI fragment in pAS835 with the same fragment from pNA-C10. pAS835 was constructed by ligating a EcoRI/HindIII-cleaved PCR fragment (primers ADS664/ADS666) into EcoRI/HindIII-cleaved pUC19.
For the yeast one-hybrid experiments, pGAD10 (Clontech), the empty expression vector pMSe4 (40), and pMSe4 containing PEA3(1-494) (amino acids 1 to 494 of PEA3) (pAS497), PEA3(258-494) (pAS613), and PEA3(341-494) (pAS468) (14) have been described before. pAS954 encoding amino acids 254 to 612 of mouse LPP [LPP(254-612)] was isolated during the yeast one-hybrid screen and was initially created by ligating an EcoRI-cut fragment into pGAD10.
The following plasmids were used to create glutathione transferase (GST) fusion proteins. pAS992 (encoding full-length zebra fish PEA3) (14) has been described previously. Plasmid pAS831 encoding human LPP(253-612) was constructed by ligating an EcoRI/SacI fragment from pAS827 into EcoRI/SacI-cleaved pGEX-KG (15). pAS827 encoding LPP(253-612) was constructed by ligating a BglII/SacI-cleaved PCR product, made with the primers ADS660/272, into BamHI/SacI-cleaved pAS1068 (12).
For in vitro transcription/translation, the following plasmids were used. Plasmids pAS477 (encoding full-length zebra fish PEA3 [4]), pSG5-ERM (full-length human ERM), and pSG5-ER81 (full-length human ER81 [25]) have been described previously. Plasmid pAS837 (full-length human LPP) is described above. Plasmids pAS984 and pAS985, encoding LPP(254-612) and LPP(412-612), were constructed by ligating EcoRI/XhoI-cleaved PCR products, made with the primers ADS721/725 and ADS722/725, respectively, into the same sites in pCDNA3. pAS1505 and pAS1501, encoding human LPP(1-285) and LPP(1-418), were constructed by ligating HindIII/BamHI-cleaved PCR products, made with the primers ADS238/828 and ADS238/824, respectively, into the same sites in pAS837.
Protein production and GST pull-down assays.
GST fusion proteins and in vitro translated proteins were made, and subsequent GST pull-down assays were carried out as described previously (38).
Tissue culture, cell transfection, immunofluorescence, RNA interference (RNAi), and reporter gene assays.
MDA-MB-231 cells were grown in RPMI 1640 medium containing 10% fetal calf serum, and 293 cells were maintained as described previously (14). Where indicated, cells were grown in 0.5% fetal calf serum for 24 h and then stimulated with 10 nM phorbol myristate acetate (PMA), and where required, the inhibitor U0126 (10 μM) was added 30 min prior to stimulation.
Transient transfection experiments were carried out in 12-well plates using Polyfect (QIAGEN) for HEK293 cells and Lipofectamine 2000 for MDA-MB-231 cells. Luciferase assays were carried out using a dual light reporter gene assay system (Tropix) as described previously, using pCH110 as an internal control (45).
The small interfering RNA (siRNA) constructs against LPP were synthesized by in vitro transcription according to the manufacturer's instructions (Ambion) using the following primers: for Si-LPP-A, ADS1287 (5′-AACAAGGTCACCCAAATACCTCCTGTCTC-3′) and ADS1288 (5′-AAAGGTATTTGGGTGACCTTGCCTGTCTC-3′); for Si-LPP-B, ADS1289 (5′-AAATGACTCTGACCCTACCTACCTGTCTC-3′) and ADS1290 (5′-AATAGGTAGGGTCAGAGTCATCCTGTCTC-3′); for Si-LPP-C, ADS1291 (5′-AAGAAGACCTATATCACAGATCCTGTCTC-3′) and ADS1292 (5′-AAATCTGTGATATAGGTCTTCCCTGTCTC-3′). Antisense reverse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) oligonucleotides were used as controls (Ambion).
Cells in 12-well plates were transfected with 2.5 μl (for HEK293) or 0.5 μl (for MDA-MB-231) of 20 μM LPP-siRNA. siPORT Amine (Ambion) was used for the transfection of HEK293 cells, and siPORTLipid (Ambion) was used for the transfection of MDA-MB-231 cells. Control siRNAs were antisense GAPDH duplexes. A two-step transfection protocol was followed, with the first carried out 24 h before reporter constructs were added, and a second cotransfection of the siRNA construct was carried out using Lipofectamine 2000 (Invitrogen).
Isolation of RNA and reverse transcription-PCR.
Total cellular RNA was extracted from cells cultured in six-well plates according to the QIAGEN RNeasy Mini Protocol. Reverse transcription was carried out using a kit provided by Roche. Subsequent PCR was performed using ImmoMix Red (Bioline). The primers used were MMP-1 (ADS1283, 5′-GTTCAGGGACAGAATGTGCTA-3′, and ADS1284, 5′-CTGCAGTTGAACCAGCTATTAG-3′) and GAPDH (ADS1285, 5′-TGATGACATCAAGAAGGTGGTGAAG-3′, and ADS1286, 5′-TCCTTGGAGGCCATGTAGGCCAT-3′).
Yeast one-hybrid analysis.
Yeast one-hybrid analysis was carried out using the ETS-driven LacZ reporter system as described previously (14, 40).
Western blot analysis.
Western blotting was carried out using Supersignal West Dura Extended Duration Substrate (Pierce) and the following primary antibodies: anti-PEA3, anti-ER81 (Santa Cruz), anti-LPP (MP2 [29]), anti-GAPDH antibody (Abcam), and mouse or rabbit anti-immunoglobulin G (IgG)-horseradish peroxidase (BD-Pharminogen). Data were visualized using a Bio-Rad Fluor-S MultiImager and Quantity One software.
ChIP.
Chromatin immunoprecipitation (ChIP) assays were carried out essentially as described previously (41). Cells grown in 100-mm-diameter dishes were cross-linked with 1% formaldehyde for 5 min. Immunoprecipitations were carried out using the antibodies anti-PEA3 (Santa Cruz), anti-LPP (MP2 [29]), or nonspecific IgG (Upstate). The following specific primers were used to amplify the precipitated DNA by PCR. Primers used were for the promoter region of (PEA3)x5-Luc (with five PEA3 binding sites) (ADS1277, 5′-AGTGCAGGTGCCAGAACATT-3′, and ADS1278, 5′-GCCTTATGCAGTTGCTCTCC-3′), the luciferase coding region (ADS1279, 5′-CGTCGCCAGTCAAGTAACAA-3′, and ADS1280, 5′-AATTACACGGCGATCTTTCC-3′), the human MMP-1 promoter (nucleotides −4013 to −3834; ADS1281, 5′-CTTGAGGCCAGGAGTTTGAG-3′, and ADS1282, 5′-CGCTTAGGCTGGAGTGTAGG-3′), and the GAPDH coding sequence (ADS1285/ADS1286).
Immunohistochemistry and immunofluorescence.
To construct tissue microarrays (TMA), the following procedure was used. Following ethical approval, formalin-fixed, paraffin-embedded tissue blocks from 70 breast cancer patients were identified. Using a hematoxylin- and eosin-stained slide as a template, representative areas of carcinoma were identified and marked. A TMA was constructed by sampling four cores (0.6 mm in diameter). Four cores were sampled from different tumor areas to account for heterogeneity in any one tumor and to minimize the number of lost cases during subsequent processing of the microarray. Cores were introduced into a recipient block, and sections of the microarray were sectioned for immunohistochemical and immunofluorescence staining.
For immunohistochemistry studies, 5-μm-thick tissue sections were cut from paraffin-embedded breast tumor tissue blocks and mounted on Superfrost Plus slides (BDH, Poole, United Kingdom). Sections were dewaxed, rehydrated, and washed in phosphate-buffered saline (PBS). Endogenous peroxidase was blocked using 3% hydrogen peroxidase in PBS for 10 min. Antigen retrieval was performed by immersing sections in 0.6 M citrate buffer and microwaving on high power for 7 min. Antigens were detected using the Vectastain Elite kit (Vector Labs, Burlingame, Calif.) according to the manufacturer's instructions. Briefly, sections were blocked in serum for 90 min. Sections were incubated with the primary antibodies rabbit anti-human LPP (1:1,000) and mouse anti-human PEA3 (5 μg/ml) (Santa Cruz) for 60 min at room temperature. Subsequently, sections were incubated in the corresponding biotin-labeled secondary antibody (1 in 2,000) for 30 min, followed by peroxidase-labeled avidin biotin complex. Sections were developed in 3,3-diaminobenzidine tetrahydrochloride and counterstained with hematoxylin. Negative controls were performed using matched IgG controls (Dako, Denmark). Sections were examined under a light microscope. Immunostained slides were scored for LPP and PEA3. Independent observers, without knowledge of prognostic factors, scored slides.
For immunofluorescence microscopy studies, breast cancer sections were prepared as above and incubated in goat serum for 60 min. Rabbit anti-human LPP (1:100 dilution in 10% human serum) was placed on each slide for 90 min. The sections were incubated with goat anti-rabbit Oregon Green 488 (1 in 500) (Molecular Probes) for 60 min. Subsequently, the slides were blocked in goat serum for 90 min. Each slide was incubated with anti-PEA3 (50 μg/ml in 10% human serum) for 90 min, and sections were subsequently incubated with goat anti-mouse Alexa Fluor 594 (Molecular Probes) for 60 min and were counterstained with DAPI (4′,6′-diamidino-2-phenylindole) (Sigma-Aldrich). Sections were mounted using fluorescent mounting media (DAKO, Denmark). Confocal microscopy was performed using a confocal microscope (Zeiss LSM 510 UV META system), and images were captured using laser capture software. Negative controls were performed using matched IgG. Similarly, for investigating LPP expression in MDA-MB-231 cells, a similar staining procedure was used. However, cells were grown on coverslips, washed in PBS, and fixed with 4% paraformaldehyde in PBS. Cells were washed three times in PBS, made permeable in 0.5% Triton X-100 in PBS for 4 min, and blocked with 3% bovine serum albumin in PBS overnight. Cells were washed three times with PBS and then incubated with rabbit anti-human LPP (1:300 dilution in 1.5% bovine serum albumin) for 1 h. Staining was performed as above, but cells were visualized by confocal microscopy on a Leica system.
RESULTS
The identification of LPP as an interaction partner for PEA3.
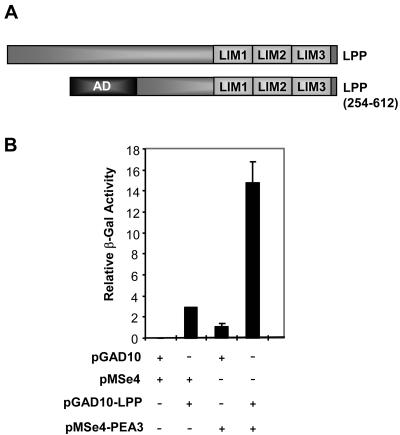
A yeast one-hybrid screen was used to identify potential coactivators for PEA3. In this screen we used a strain containing an integrated lacZ reporter driven by five ETS binding sites (40), full-length zebra fish PEA3, and a cDNA library encoding Gal4 activation domain fusion proteins derived from day 11 mouse embryos. We reasoned that by using zebra fish PEA3, we would identify evolutionarily conserved and hence functionally relevant binding partners. Among the clones isolated were USF-1 (14) and the C-terminal part of the LIM domain protein LPP (Fig. 1A). The reporter strain was retransformed with plasmids encoding PEA3, LPP, or both together, demonstrating that cotransformation was required for high levels of reporter gene activity (Fig. 1B).
FIG. 1.
One-hybrid identification of LPP as a PEA3 interaction partner. (A) Diagrammatic representation of full-length LPP and the truncated version isolated in a yeast one-hybrid assay. The locations of the LIM domains and the Gal4 fused transcriptional activation domain (AD) are shown. (B) Yeast one-hybrid interactions between PEA3 and LPP(254-612) on an ETS site-driven β-galactosidase reporter gene. The presence of the empty expression plasmids pMSe4 and pGAD10, pMSe4 containing zebra fish PEA3, or pGAD10 containing LPP(254-612) is indicated.
Expression of LPP in normal mammary gland and tumorigenic breast cancer cells.
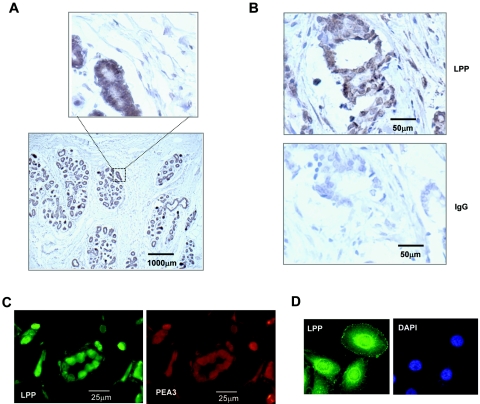
Members of the PEA3 subfamily of ETS domain transcription factors are expressed in a temporal manner in different cells in the mouse mammary gland and are upregulated in many HER2/Neu-positive metastatic human breast cancers (5; reviewed in references 10 and 22). We therefore asked whether LPP was also expressed in normal human mammary gland and metastatic human breast cancer cells. Strong LPP expression could be detected in the luminal epithelial cells in the ducts of the normal mammary gland (Fig. 2A). In contrast, little expression could be seen in the surrounding stromal and myoepithelial cells. PEA3 expression could not be detected in normal adult mammary gland tissue (data not shown). Next, we examined sections containing invasive ductal carcinomas. Here, LPP expression could be seen in cells surrounding the lumen of the ducts and also in the stromal cells and cells invading the surrounding tissue (Fig. 2B). To establish whether PEA3 expression could be detected in the same cells as LPP in the ductal carcinomas, we carried out immunofluorescence assays with two fluorescent probes. Clear colocalization of LPP and PEA3 could be seen in many cells (Fig. 2C). Furthermore, using a TMA, coexpression of PEA3 and LPP could be detected in 71% (P = 0.002) of PEA3-positive ductal carcinoma samples (n = 53), but LPP was only detected in 41% of PEA3-negative samples (n = 17), suggesting a functional importance to their coexpression.
FIG. 2.
LPP expression in normal and cancerous breast tissue. (A) Immunohistochemical analysis of LPP protein expression in normal human breast tissue sections. An enlarged part of the image is shown to illustrate expression in the ductal epithelial cells. (B) LPP expression from human tissue derived from patients with invasive ductal carcinomas. LPP expression is revealed by brown staining. A control IgG-stained adjacent section is shown in the bottom panel. (C) Immunofluorescence analysis of LPP expression (green, fluorescein isothiocyanate) and PEA3 expression (red, tetramethyl rhodamine isothiocyanate) in ductal carcinoma samples. (D) Confocal analysis of LPP expression (fluorescein isothiocyanate; green) in MDA-MB-231 cells. Nuclei are visualized with DAPI staining.
The expression of LPP in ductal carcinomas appeared to be localized at least in part to the nucleus (Fig. 2C). Previous studies indicated that LPP could be detected in the nucleus of keratinocytes, in addition to its localization to focal adhesions (29). We therefore investigated the localization of LPP in MDA-MB-231 breast cancer cells, using confocal microscopy. LPP was detected in the focal adhesions as expected but was also strongly expressed in the perinuclear region and in the nucleus itself (Fig. 2D). PEA3 could also be weakly detected in the nuclear compartment of MDA-MB-231 cells (data not shown).
Thus, the identification of LPP as an interaction partner for PEA3 and the colocalization of LPP and PEA3 suggest that LPP might affect PEA3 function in tumorigenic breast cells.
LPP regulates the expression of the PEA3 target gene MMP-1.
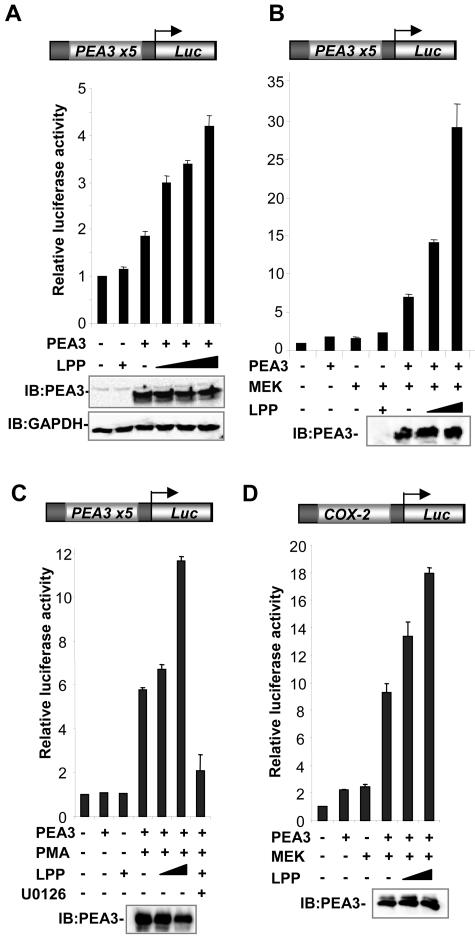
Next, we examined whether LPP is involved in regulating the transcriptional activities of PEA3. First, we tested whether the expression of PEA3 target genes is affected by changes in LPP levels. As a model, we investigated MMP-1 expression as this has been previously implicated as a target for members of the PEA3 subfamily in breast cancer cells (3). MMP expression is, in turn, thought to contribute to the metastatic spreading of tumor cells by degrading the extracellular matrix. First, we overexpressed PEA3 and LPP in 293 cells in the presence of constitutively active MEK to activate the ERK pathway. The transactivation capacity of PEA3 is enhanced in response to the ERK/MAP kinase signaling pathway and is thought to be a functionally important event in PEA3-mediated transcriptional activation of target promoters (28). The expression of PEA3, MEK, or LPP alone had little effect on MMP-1 gene activity (Fig. 3A, lanes 2, 3 and 5). However, coexpression of MEK with PEA3 caused significant upregulation of MMP-1 expression, which was further augmented by cotransfection of LPP (Fig. 3A, lanes 6 and 7). We then tested whether siRNA-mediated downregulation of LPP levels affected the expression of MMP-1 induced by cotransfection of PEA3 and MEK in 293 cells. Three different siRNAs against LPP caused substantial decreases in MMP-1 expression (Fig. 3B, lanes 2 to 4), demonstrating the importance of endogenous LPP in this activation process. Finally, we tested the effect of reductions in endogenous LPP levels on the expression of MMP-1 in the presence of endogenous PEA3 in MDA-MB-231 breast cancer cells. siRNAs against LPP caused reductions in MMP-1 expression in these cells (Fig. 3C). Collectively, these data demonstrate an important role for LPP in activating the expression of the PEA3 target gene MMP-1.
FIG. 3.
LPP is required for PEA3 target gene activity. (A to C) Reverse transcription-PCR analysis of endogenous MMP-1 expression. (A) 293 cells were transfected with the indicated combinations of expression vectors for PEA3 (400 ng), MEK(ΔN-S218E/S222D) (200 ng) and LPP (400 ng). (B) 293 cells were transfected with expression vectors for PEA3 (400 ng), MEK(ΔN-S218E/S222D) (200 ng) and the indicated LPP or control siRNAs. (C) MDA-MB-231 cells were transfected with the indicated LPP or control siRNAs.
LPP potentiates the transactivation activity of PEA3.
One mechanism of action for LPP in regulating PEA3 target genes would be to act as a coactivator protein, which acts directly on PEA3-regulated promoters. To establish whether LPP functions in this manner to affect the activity of PEA3, a series of reporter gene assays was carried out in which LPP was cotransfected with PEA3. First, we tested whether LPP could affect the activity of PEA3 on a luciferase reporter gene controlled by five ets sites (PEA3-Luc). In the absence of cotransfected PEA3, little enhancement of reporter gene activity by LPP was observed. However, the addition of PEA3 caused an increase in the activity of the reporter gene. This activation was further augmented in a dose-dependent manner upon cotransfection of LPP (Fig. 4A). This increase in activity was not due to changes in PEA3 expression as similar levels were observed in the presence and absence of LPP (Fig. 4A, B, C, and D, bottom panels). As the transactivation capacity of PEA3 is enhanced in response to the ERK/MAP kinase signaling pathway and is thought to be a functionally important event in PEA3-mediated transcriptional activation of target promoters (28), we analyzed the ability of LPP to coactivate PEA3 under conditions in which the ERK pathway was active by cotransfection of constitutively active MEK. PEA3 and MEK alone gave only small increases in reporter gene activity. However, cotransfection of PEA3 and MEK caused a substantial increase in reporter gene activity (sevenfold induction), which was further enhanced by cotransfection of LPP in a dose-dependent manner (30-fold induction) (Fig. 4B). Similarly, when the ERK pathway was activated by PMA stimulation, LPP was able to cause a dose-dependent increase in the activity of the PEA3-Luc reporter in the presence of PEA3 (Fig. 4C). Treatment of cells with the MEK inhibitor U0126 blocked the increase in activity mediated by PMA and LPP, demonstrating that this was ERK pathway-dependent. PMA alone had little effect on reporter gene activity in the absence of transfected PEA3 (data not shown). Finally, we analyzed the ability of LPP to coactivate PEA3 on the promoter of a known target gene, COX-2 (16, 42). The addition of PEA3 or MEK caused a small increase in the activity of the reporter gene. However, cotransfection of PEA3 and MEK caused a substantial increase in reporter gene activity (ninefold induction), which was further enhanced by cotransfection of LPP in a dose-dependent manner (18-fold induction) (Fig. 4D).
FIG. 4.
LPP potentiates the transactivation activity of PEA3. Luciferase reporter gene assays using a PEA3 site-driven (A to C) or a COX-2 promoter-driven luciferase reporter (D) in 293 cells. Data are presented relative to the activity of the reporter alone (taken as 1). Western blots showing the expression levels of GAPDH or PEA3 in the presence and absence of LPP are shown below the graphs. (A) LPP alone (2 μg) or mouse PEA3 (200 ng) and increasing amounts of LPP (0, 0.5, 1, and 2 μg) were cotransfected. (B) Mouse PEA3 (200 ng), constitutively active MEK, and increasing amounts of LPP (0, 1, and 2 μg) were cotransfected. +, addition of 2 μg of LPP. (C) Mouse PEA3 (200 ng) and increasing amounts of LPP (0, 1, and 2 μg) were cotransfected. +, addition of 2 μg of LPP. Cells were either serum starved or stimulated with PMA in the presence or absence of the MEK inhibitor U0126 where indicated. (D) Mouse PEA3 (200 ng), constitutively active MEK, and increasing amounts of LPP (0, 1, and 2 μg) were cotransfected.
Together, these data therefore demonstrate that LPP can act as a coactivator to enhance the activity of PEA3.
LPP is required for PEA3-mediated promoter activation.
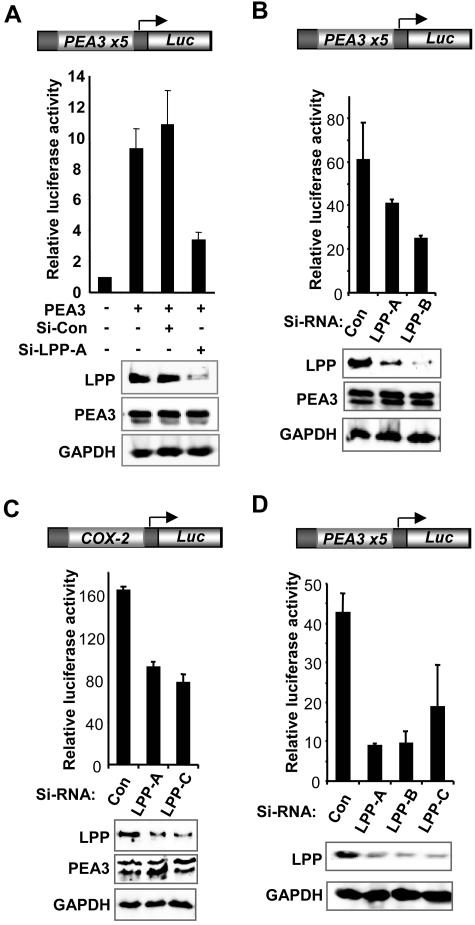
To address whether LPP is required for PEA3-mediated gene expression, we used RNAi to ablate the expression of LPP. LPP is highly expressed in 293 cells, and transfection of RNAi against LPP caused a substantial decrease in the protein levels of LPP but did not affect either cotransfected PEA3 or endogenous GAPDH levels (Fig. 5A, bottom panels). A control RNAi duplex did not affect the levels of any of these proteins. The activity of PEA3 in a reporter gene assay was then examined. In this case, we used high levels of PEA3 to cause substantial activation of the reporter to enable siRNA-mediated reductions in activity to be observed. Transfection of PEA3 alone caused a ninefold increase in the activity of the PEA3-Luc reporter, which was unaffected by cotransfection of a control RNAi duplex (Fig. 5A). However, treatment of the cells with an RNAi duplex against LPP severely reduced reporter gene activation by PEA3. Transfection of siRNA duplexes against LPP had little effect on the activity of the reporter in the absence of cotransfected PEA3 (data not shown). In agreement with these results, the activity of the PEA3-Luc reporter in the presence of PEA3 and ERK pathway activation following PMA treatment was reduced in the presence of siRNA against LPP (Fig. 5B). To establish whether the activity of a PEA3-regulated promoter could be affected by knocking down LPP levels, we examined the COX-2 promoter-reporter construct. As observed with the PEA3 site-driven reporter, siRNAs against LPP caused reductions in the activity of the COX-2 reporter (Fig. 5C). To examine whether LPP plays a role in regulating gene expression in the presence of endogenous levels of PEA3, we examined the expression of the PEA3-Luc reporter in MDA-MB-231 breast cancer cells in the presence of siRNAs against LPP. These cells express high levels of endogenous PEA3 mRNA. Again, reductions in LPP levels caused a decrease in the activity of the PEA3-Luc reporter in these cells (Fig. 5D).
FIG. 5.
LPP is required for PEA3-mediated promoter activation. Luciferase reporter gene assays were carried out in the presence of the indicated LPP or control siRNAs with the PEA3 site-driven (600 ng; A, B, and D) or Cox-2 promoter-driven (600 ng; C) luciferase reporter constructs. 293 cells were transfected with PEA3 expression constructs (600 ng) in the absence (A and C) and presence (B) of PMA stimulation. (D) MDA-MB-231 cells were transfected with reporter alone. Western blots showing the expression levels of LPP, PEA3, and GAPDH in the presence and absence of the indicated RNAi constructs are shown below the graphs.
Collectively, these loss-of-function approaches complement the overexpression studies and demonstrate that LPP plays an important role in PEA3-mediated transcriptional activation.
LPP binds to PEA3 family subfamily members.
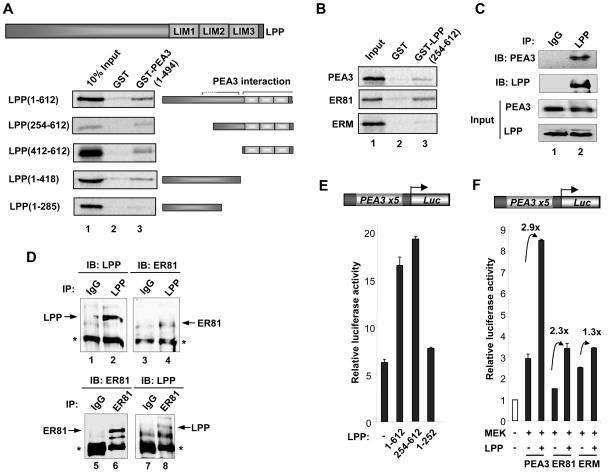
The yeast one-hybrid studies suggest that LPP acts as a coactivator protein by binding to PEA3. GST pull-down assays were therefore carried out to investigate whether direct interactions could be detected in vitro. Interactions between LPP and full-length PEA3 were detected when either PEA3 (Fig. 6A, top panel) or LPP (Fig. 6B, top panel) was immobilized as a GST fusion protein. To identify the binding surface(s) of PEA3 on LPP, we carried out GST pull-down assays using a series of truncated LPP derivatives and a GST fusion to full-length PEA3. The truncated protein LPP(254-612) bound to PEA3 (Fig. 6A), thus confirming the interactions seen in the yeast one-hybrid system (Fig. 1B). A further deletion, LPP(412-612), containing the LIM domains alone, was sufficient for binding to PEA3 (Fig. 6A). However, the N-terminal two-thirds of the protein contained in LPP(1-418) also bound to LPP. This binding was greatly reduced upon deletion of amino acids 286 to 418 in LPP(1-285) (Fig. 6A), indicating that residues comprising a second binding region were located in this deleted region.
FIG. 6.
Mapping the PEA3 binding surface(s) on LPP. (A) GST pull-downs using GST or GST-PEA3 proteins and in vitro translated LPP derivatives. A diagrammatic representation of full-length LPP is shown at the top, and truncated derivatives of this are depicted next to appropriate lanes. (B) GST pull-downs of GST or GST-LPP(254-612) and in vitro translated zebra fish PEA3, human ER81, and human ERM. Ten percent input protein is shown. (C) Coimmunoprecipitation of LPP and PEA3. 293 cells were transfected with a PEA3 expression vector and immunoprecipitations (IP) were carried out with control IgG or LPP antibodies. Immunoprecipitated LPP and PEA3 were detected by immunoblotting (IB). Inputs show equal amounts of LPP and PEA3. (D) Coimmunoprecipitation of endogenous LPP and ER81 from MDA-MB-231 cells. Immunoprecipitations (IPs) were carried using anti-LPP antibody (top panel) or anti-ER81 antibody (bottom panel), and precipitated LPP and ER81 were detected by immunoblotting (IB) with the appropriate antibodies. (E and F) Reporter gene analysis of the PEA3 site-driven luciferase reporter construct in 293 cells. (E) Mouse PEA3 (200 ng) and the indicated Gal4-LPP constructs (2 μg) were cotransfected. (F) Mouse PEA3, human ER81, or human ERM (200 ng) and constitutively active MEK and LPP (2 μg) were cotransfected. The increase in activation (n-fold) by LPP is indicated above each set of bars.
Due to the high degree of sequence conservation among PEA3 subfamily members, we tested whether LPP could bind to other members of this subfamily. GST pull-down assays revealed that in addition to PEA3, LPP could also bind strongly to ER81, but binding to ERM was barely detectable (Fig. 6B).
To confirm that LPP and PEA3 interact in mammalian cells, coimmunoprecipitation experiments were carried out with endogenous LPP and exogenous PEA3. Anti-LPP antibodies coprecipitated PEA3 (Fig. 6C, lane 2), demonstrating that these two proteins can form a complex. We have been unable to detect high levels of PEA3 in a number of different cell lines (our unpublished data). Thus, to probe interactions between proteins expressed at endogenous levels, we instead tested interactions between LPP and the related protein ER81, which can bind to LPP in vitro (Fig. 6B). Antibodies against LPP were able to precipitate ER81 from MBA-MD-231 cells (Fig. 6D, lane 4), and, reciprocally, anti-ER81 antibodies were able to precipitate LPP (Fig. 6D, lane 8).
Next, we asked whether these in vitro binding data reflected functional in vivo interactions. Reporter gene analysis was used to delineate the regions of LPP important for coactivation of PEA3. We compared the ability of full-length LPP and the N-terminal (residues 1 to 252) and C-terminal (residues 254 to 612) parts to enhance the activity of PEA3-driven transcription. Consistent with the GST pull-down data, only the full-length protein and the C-terminal region of LPP could enhance the activity of PEA3 (Fig. 6E). Next we analyzed whether LPP could coactivate all the members of the PEA3 subfamily. We therefore compared the activity of each family member in the absence and presence of cotransfected LPP. The activity of both PEA3 and ER81 was enhanced by LPP (2.9- and 2.3-fold, respectively) (Fig. 6F). However, the activity of ERM was only moderately increased by LPP (1.3-fold), consistent with the low levels of binding seen in the GST pull-down assays (Fig. 6B).
Collectively, these data therefore demonstrate selectivity in interactions with PEA3 subfamily members and indicate that PEA3 and ER81 functionally interact with LPP through the C-terminal region which includes the LIM domain module.
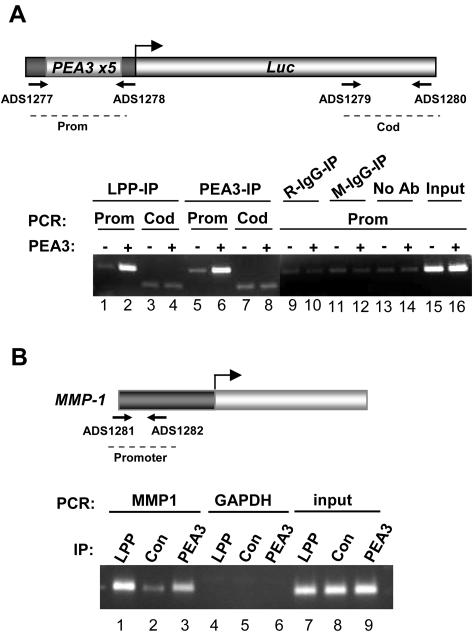
LPP is recruited to PEA3-regulated promoters in vivo.
LPP interacts with PEA3 (Fig. 1 and 6) and regulates its activity (Fig. 3 to 5). To further demonstrate that the two proteins work together and actually co-occupy promoters in the nucleus, we carried out a series of ChIP assays on PEA3-driven promoters. First, we investigated the recruitment of LPP to the PEA3-Luc reporter gene. A basal level of LPP binding to the promoter region is observed, which might be due to recruitment of LPP by endogenous ETS domain transcription factors. However, upon cotransfection of a plasmid encoding PEA3, enhanced PEA3 binding to the promoter region of the reporter could be observed (Fig. 7A, lanes 5 and 6). This increase in PEA3 binding was mirrored by an increase in the recruitment of endogenous LPP to the promoter (Fig. 7A, lanes 1 and 2). In contrast, binding of PEA3 and LPP to the coding region of the luciferase gene was at basal levels, and no enhancement was seen upon PEA3 transfection (Fig. 7A, lanes 3, 4, 7, and 8). In addition, little LPP binding was detected using either control antibodies or protein A beads alone (Fig. 7A, lanes 9 to 14).
FIG. 7.
LPP is recruited to PEA3-regulated promoters in vivo. (A) ChIP assay of endogenous LPP on a PEA3 site-driven luciferase reporter in 293 cells. The presence of transfected PEA3 is indicated. Immunoprecipitations (IP) were carried out with either LPP antibody, PEA3 antibody, rabbit IgG (R), or mouse IgG (M) as controls. Coprecipitating DNA was revealed by PCR with promoter (prom) or coding region (cod)-specific primers as indicated in the schematic. Input DNA was diluted 10-fold before amplification. (B) ChIP assay of endogenous LPP on the MMP-1 promoter in MDA-MB-231 cells. Immunoprecipitations(IPs) were carried out with antibodies against either LPP, PEA3, or IgG as a control (con). Coprecipitating DNA was revealed by PCR with MMP-1 promoter or GAPDH coding region-specific primers.
Next, we determined whether LPP could be detected at the promoter of an endogenous PEA3 family target gene MMP-1 in MDA-MB-231 cells. ChIP analysis revealed that both LPP and PEA3 could be detected above background levels in the promoter region of this gene (Fig. 7B). However, no binding to the GAPDH gene was observed.
Collectively, these data therefore demonstrate that LPP can be recruited to promoters in a PEA3-dependent manner and is present at PEA3-regulated promoters.
DISCUSSION
A large number of mechanisms are employed to transmit signals from the cell surface to the nucleus (reviewed in reference 39). One type of mechanism, typified by the Wnt/β-catenin pathway, involves the shuttling of coactivators from cytoskeletal-plasma membrane junctions to the nucleus (reviewed in reference 13). Here we identify LPP as a novel type of transcriptional coactivator that resides at focal adhesions and shuttles through the nucleus.
The C-terminal part of LPP (amino acids 254 to 612) was initially isolated in a yeast one-hybrid screen for interaction partners for the ETS domain transcription factor PEA3 (Fig. 1B). Subsequent mapping studies identified the LIM domain module (amino acids 411 to 612) and a region within the preceding proline-rich region that were involved in binding to PEA3 (Fig. 6). Interestingly, both of these regions coincided with the two transcription activation domains (TADs) found within LPP (29; data not shown). However, while all three LIM domains of LPP are needed for binding to PEA3, the first LIM domain is sufficient for providing a TAD (data not shown). Similarly, the two C-terminal LIM domains have TAD activity (6). These observations are consistent with the theory that the primary function of LIM domains is to act as protein-protein interaction modules (reviewed in reference 7). In the case of PEA3, at least part of the interaction is mediated by the ETS domain (data not shown). This is consistent with the observation that this region of the protein participates in a wide range of protein-protein interactions in other family members (reviewed in reference 36). However, other regions of PEA3 likely participate in regulating LPP binding, exemplified by the observation that ERM binds poorly to LPP, despite the high homology within the ETS DNA-binding domain. Indeed, the regions flanking the ETS domain modulate DNA binding by PEA3 family proteins and their recruitment by other transcription factors (2, 14). In addition to binding to PEA3, LPP can bind to the related protein ER81 and potentiate its transactivation capacity (Fig. 6B, D, and E). In contrast, less effect is seen on the other PEA3 family member ERM, suggesting that specificity of action exists. Thus, LPP can be regarded as a coactivator for a subset of PEA3 subfamily members rather than PEA3 alone.
A number of observations demonstrate that LPP acts as a coactivator protein to potentiate the transcriptional activation capacity of PEA3. By modulating the levels of LPP in the cell, the activity of PEA3-dependent promoters (Fig. 4 and 5) and activity of target genes (Fig. 3) is altered. LPP can be found at PEA3-regulated promoters, and this recruitment is augmented by exogenous PEA3 (Fig. 7). LPP has previously been shown to contain intrinsic transactivation capacity (29) and to shuttle through the nucleus, suggesting a nuclear role. However, to date, no signals have been identified that trigger the nuclear accumulation of LPP. Nevertheless, LPP can be detected in the nuclear compartment and is found bound to promoters and influences the transcriptional activity of PEA3, despite the apparent transient nature of its shuttling through the nucleus. This suggests a potential role for LPP in continually sensing the events at the cell periphery and communicating these into a nuclear response, although it is possible that, under certain conditions, LPP may be retained in the nucleus when the transcriptional consequences may be more widespread. Indeed, it has previously been shown that although mainly cytoplasmic, around 6% of HeLa cells contain nuclear LPP, and LPP can be detected in the nucleus of keratinocytes (29) and MBA-MD-231 cells (Fig. 2). There is increasing evidence that LIM domain proteins related to LPP have a nuclear role in addition to the well-characterized cytoskeletal role typified by zyxin (reviewed in reference 44). All family members shuttle through the nucleus and are exported in a Crm1-dependent manner, and in the case of zyxin, the E6 oncoprotein has been shown to trigger its nuclear accumulation (8). In addition, several family members have been shown to possess transcriptional activation capacity (reviewed in reference 44). TRIP6 was initially shown to possess coactivator activity for v-Rel (46) and more recently to act as both a coactivator and corepressor protein in the context of AP-1- and NF-κB-mediated gene regulation (20). Normally, TRIP6 is a coactivator but becomes a corepressor in the context of glucocorticoid receptor signaling, where it acts as an adaptor that recruits glucocorticoid receptor to inhibit AP-1 and NF-κB activity. An N-terminally truncated nuclear form of TRIP6 was detected in these studies, suggesting that this is the important coregulatory form of TRIP6 (20). Intriguingly, we can also detect a shorter nuclear form of LPP, which may play an analogous role (data not shown). Our demonstration that LPP is also a bone fide coactivator in mammalian cells suggests that the same may also be true for other family members. However, recent studies on LIMD1 indicate that it acts as a corepressor protein through potentiating the action of retinoblastoma protein (35). Thus, LPP-related proteins can have both corepressive and coactivating properties.
It is currently unclear what role the ERK pathway plays in controlling the activity of the PEA3-LPP complex. However, PEA3 is a direct target of the ERK pathway, and the transactivation capacity of PEA3 is enhanced in response to activation of the ERK/MAP kinase pathway (28). To date, we have been unable to detect a role for the ERK pathway in the shuttling or recruitment of LPP, but it remains possible that the activity of LPP in addition to PEA3 might be directly affected by this pathway.
PEA3 and ER81 have been implicated as important players in breast tumor metastasis (reviewed in references 10 and 22) and HER2/Neu-mediated mammary oncogenesis (37). PEA3 family members are also expressed during normal mammary gland development (reviewed in reference 22). LPP also is expressed in both normal and cancerous breast tissue (Fig. 2). As PEA3 and ER81 functionally interact with LPP, it will be interesting to determine whether LPP also plays a role in these processes. However, one intriguing hypothesis is that LPP might represent a “sensor” that communicates changes in the cytoskeleton or extracellular contacts associated with metastatic tumor formation into PEA3-mediated changes in gene expression profiles. ChIP analysis demonstrates the association of LPP with the MMP-1 promoter in metastatic mammary tumor cells (Fig. 7), which is consistent with a role for LPP in promoting metastasis through MMP-1 upregulation. Future studies will be aimed at studying the potential links of LPP with PEA3-mediated tumorigenesis.
Acknowledgments
We thank Anne Clancy, Margaret Bell, and Linda Shore for excellent technical support. We also thank Michael Sieweke, Yvan de Launoit, Hiroyasu Inoue, and John Hassell for reagents and Shen-Hsi Yang, Stefan Roberts, and Alan Whitmarsh for comments on the manuscript.
R.S. and A.G. were supported by studentships from the MRC and Cancer Research UK, respectively. M.M.R.P. is a Postdoctoral Fellow of the fund for scientific research, Flanders (Belgium) (F.W.O.-Vlaanderen). This work was funded by grants from the Wellcome Trust.
REFERENCES
- 1.Arber, S., D. R. Ladle, J. H. Lin, E. Frank, and T. M. Jessell. 2000. ETS gene Er81 controls the formation of functional connections between group Ia sensory afferents and motor neurons. Cell 101:485-498. [DOI] [PubMed] [Google Scholar]
- 2.Bojovic, B. B., and J. A. Hassell. 2001. The PEA3 Ets transcription factor comprises multiple domains that regulate transactivation and DNA binding. J. Biol. Chem. 276:4509-4521. [DOI] [PubMed] [Google Scholar]
- 3.Bosc, D. G., B. S. Goueli, and R. Janknecht. 2001. HER2/Neu-mediated activation of the ETS transcription factor ER81 and its target gene MMP-1. Oncogene 20:6215-6224. [DOI] [PubMed] [Google Scholar]
- 4.Brown, L. A., A. Amores, T. F. Schilling, T. Jowett, J. L. Baert, Y. de Launoit, and A. D. Sharrocks. 1998. Molecular characterization of the zebrafish PEA3 ETS-domain transcription factor. Oncogene 17:93-104. [DOI] [PubMed] [Google Scholar]
- 5.Chotteau-Lelievre, A., R. Montesano, J. Soriano, P. Soulie, X. Desbiens, and Y. de Launoit. 2003. PEA3 transcription factors are expressed in tissues undergoing branching morphogenesis and promote formation of duct-like structures by mammary epithelial cells in vitro. Dev. Biol. 259:241-257. [DOI] [PubMed] [Google Scholar]
- 6.Crombez, K. R. M. O., E. M. R. Vanoirbeek, W. J. M. Van de Ven, and M. M. R. Petit. 2005. Transactivation functions of the tumor-specific HMGA2/LPP fusion protein are augmented by wild-type HMGA2. Mol. Cancer Res. 3:63-70. [DOI] [PubMed] [Google Scholar]
- 7.Dawid, I. B., J. J. Breen, and R. Toyama. 1998. LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 14:156-162. [DOI] [PubMed] [Google Scholar]
- 8.Degenhardt, Y. Y., and S. Silverstein. 2001. Interaction of zyxin, a focal adhesion protein, with the E6 protein from human papillomavirus type 6 results in its nuclear translocation. J. Virol. 75:11791-11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Launoit, Y., J. L. Baert, A. Chotteau, D. Monte, P. A. Defossez, L. Coutte, H. Pelczar, and F. Leenders. 1997. Structure-function relationships of the PEA3 group of Ets-related transcription factors. Biochem. Mol. Med. 61:127-135. [DOI] [PubMed] [Google Scholar]
- 10.de Launoit, Y., A. Chotteau-Lelievre, C. Beaudoin, L. Coutte, S. Netzer, C. Brenner, I. Huvent, and J. L. Baert. 2000. The PEA3 group of ETS-related transcription factors. Role in breast cancer metastasis. Adv. Exp. Med. Biol. 480:107-116. [DOI] [PubMed] [Google Scholar]
- 11.Firnberg, N., and A. Neubuser. 2002. FGF signaling regulates expression of Tbx2, Erm, Pea3, and Pax3 in the early nasal region. Dev. Biol. 247:237-250. [DOI] [PubMed] [Google Scholar]
- 12.Galanis, A., S.-H. Yang, and A. D. Sharrocks. 2001. Selective targeting of MAPKs to the ETS domain transcription factor SAP-1. J. Biol. Chem. 276:965-973. [DOI] [PubMed] [Google Scholar]
- 13.Giles, R. H., J. H. van Es, and H. Clevers. 2003. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim. Biophys. Acta 1653:1-24. [DOI] [PubMed] [Google Scholar]
- 14.Greenall, A., N. Willingham, E. Cheung, D. Boam, and A. D. Sharrocks. 2001. Intermolecular and intramolecular protein-protein interactions regulate DNA binding by the ETS-domain transcription factor PEA3. J. Biol. Chem. 276:16207-16215. [DOI] [PubMed] [Google Scholar]
- 15.Guan, K. L., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192:262-267. [DOI] [PubMed] [Google Scholar]
- 16.Howe, L. R., H. C. Crawford, K. Subbaramaiah, J. A. Hassell, A. J. Dannenberg, and A. M. Brown. 2001. PEA3 is up-regulated in response to Wnt1 and activates the expression of cyclooxygenase-2. J. Biol. Chem. 276:20108-20115. [DOI] [PubMed] [Google Scholar]
- 17.Inoue, H., C. Yokoyama, S. Hara, Y. Tone, and T. Tanabe. 1995. Transcriptional regulation of human prostaglandin-endoperoxide synthase-2 gene by lipopolysaccharide and phorbol ester in vascular endothelial cells. Involvement of both nuclear factor for interleukin-6 expression site and cAMP response element. J. Biol. Chem. 270:24965-24971. [DOI] [PubMed] [Google Scholar]
- 18.Janknecht, R., D. Monte, J. L. Baert, and Y. de Launoit. 1996. The ETS-related transcription factor ERM is a nuclear target of signaling cascades involving MAPK and PKA. Oncogene 13:1745-1754. [PubMed] [Google Scholar]
- 19.Janknecht, R. 1996. Analysis of the ERK-stimulated ETS transcription factor ER81. Mol. Cell. Biol. 16:1550-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kassel, O., S. Schneider, C. Heilbock, M. Litfin, M. Gottlicher, and P. Herrlich. 2004. A nuclear isoform of the focal adhesion LIM-domain protein Trip6 integrates activating and repressing signals at AP-1- and NF-kappaB-regulated promoters. Genes Dev. 18:2518-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasza, A., A. O'Donnell, K. Gascoigne, L. A. Zeef, A. Hayes, and A. D. Sharrocks. 2005. The ETS domain transcription factor Elk-1 regulates the expression of its partner protein, SRF. J. Biol. Chem. 280:1149-1155. [DOI] [PubMed] [Google Scholar]
- 22.Kurpios, N. A., N. A. Sabolic, T. G. Shepherd, G. M. Fidalgo, and J. A. Hassell. 2003. Function of PEA3 Ets transcription factors in mammary gland development and oncogenesis. J. Mammary Gland Biol. Neoplasia 8:177-190. [DOI] [PubMed] [Google Scholar]
- 23.Li, B., L. Zhuang, M. Reinhard, and B. Trueb. 2003. The lipoma preferred partner LPP interacts with alpha-actinin. J. Cell Sci. 116:1359-1366. [DOI] [PubMed] [Google Scholar]
- 24.Livet, J., M. Sigrist, S. Stroebel, V. De Paolo, S. R. Price, C. E. Henderson, T. M. Jessell, and S. Arber. 2002. ETS gene Pea3 controls the central position and terminal arborization of specific motor neuron pools. Neuron 35:877-892. [DOI] [PubMed] [Google Scholar]
- 25.Monte, D., L. Coutte, J. L. Baert, I. Angeli, D. Stehelin, and Y. de Launoit. 1995. Molecular characterization of the ets-related human transcription factor ER81. Oncogene 11:771-779. [PubMed] [Google Scholar]
- 26.Munchberg, S. R., E. A. Ober, and H. Steinbeisser. 1999. Expression of the Ets transcription factors erm and pea3 in early zebrafish development. Mech. Dev. 88:233-236. [DOI] [PubMed] [Google Scholar]
- 27.Munchberg, S. R., and H. Steinbeisser. 1999. The Xenopus Ets transcription factor XER81 is a target of the FGF signaling pathway. Mech. Dev. 80:53-65. [DOI] [PubMed] [Google Scholar]
- 28.O'Hagan, R. C., R. G. Tozer, M. Symons, F. McCormick, and J. A. Hassell. 1996. The activity of the Ets transcription factor PEA3 is regulated by two distinct MAPK cascades. Oncogene 13:1323-1333. [PubMed] [Google Scholar]
- 29.Petit, M. M., J. Fradelizi, R. M. Golsteyn, T. A. Ayoubi, B. Menichi, D. Louvard, W. J. Van de Ven, and E. Friederich. 2000. LPP, an actin cytoskeleton protein related to zyxin, harbors a nuclear export signal and transcriptional activation capacity. Mol. Biol. Cell 11:117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petit, M. M., S. M. Meulemans, and W. J. Van de Ven. 2003. The focal adhesion and nuclear targeting capacity of the LIM-containing lipoma-preferred partner (LPP) protein. J. Biol. Chem. 278:2157-2168. [DOI] [PubMed] [Google Scholar]
- 31.Petit, M. M., S. M. Meulemans, P. Alen, T. A. Ayoubi, E. Jansen, and W. J. Van de Ven. 2005. The tumor suppressor Scrib interacts with the zyxin-related protein LPP, which shuttles between cell adhesion sites and the nucleus. BMC Cell Biol. 6:1. [Online.] doi: 10.1186/1471-2121-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petit, M. M. R., R. Mols, E. F. Schoenmakers, N. Mandahl, and W. J. Van de Ven. 1996. LPP, the preferred fusion partner gene of HMGIC in lipomas, is a novel member of the LIM protein gene family. Genomics 36:118-129. [DOI] [PubMed] [Google Scholar]
- 33.Raible, F., and M. Brand. 2001. Tight transcriptional control of the ETS domain factors Erm and Pea3 by Fgf signaling during early zebrafish development. Mech. Dev. 107:105-117. [DOI] [PubMed] [Google Scholar]
- 34.Roehl, H., and C. Nusslein-Volhard. 2001. Zebrafish pea3 and erm are general targets of FGF8 signaling. Curr. Biol. 11:503-507. [DOI] [PubMed] [Google Scholar]
- 35.Sharp, T. V., F. Munoz, D. Bourboulia, N. Presneau, E. Darai, H. W. Wang, M. Cannon, D. N. Butcher, A. G. Nicholson, G. Klein, S. Imreh, and C. Boshoff. 2004. LIM domains-containing protein 1 (LIMD1), a tumor suppressor encoded at chromosome 3p21.3, binds pRB and represses E2F-driven transcription. Proc. Natl. Acad. Sci. USA 101:16531-16536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharrocks, A. D. 2001. The ETS-domain transcription factor family. Nat. Rev. Mol. Cell. Biol. 2:827-837. [DOI] [PubMed] [Google Scholar]
- 37.Shepherd, T. G., L. Kockeritz, M. R. Szrajber, W. J. Muller, and J. A. Hassell. 2001. The pea3 subfamily ets genes are required for HER2/Neu-mediated mammary oncogenesis. Curr. Biol. 11:1739-1748. [DOI] [PubMed] [Google Scholar]
- 38.Shore, P., and A. D. Sharrocks. 1994. The transcription factors Elk-1 and serum response factor interact by direct protein-protein contacts mediated by a short region of Elk-1. Mol. Cell. Biol. 14:3283-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shore, P., and A. D. Sharrocks. 2000. Regulation of transcription by extracellular signals, p. 113-135. In J. Locker, Transcription factors. Bios Scientific Publishers, Ltd., Oxford, United Kingdom.
- 40.Sieweke, M. H., H. Tekotte, J. Frampton, and T. Graf. 1996. MafB is an interaction partner and repressor of Ets-1 that inhibits erythroid differentiation. Cell 85:49-60. [DOI] [PubMed] [Google Scholar]
- 41.Spencer, V. A., J. M. Sun, L. Li, and J. R. Davie. 2003. Chromatin immunoprecipitation: a tool for studying histone acetylation and transcription factor binding. Methods 31:67-75. [DOI] [PubMed] [Google Scholar]
- 42.Subbaramaiah, K., L. Norton, W. Gerald, and A. J. Dannenberg. 2002. Cyclooxygenase-2 is overexpressed in HER-2/neu-positive breast cancer: evidence for involvement of AP-1 and PEA3. J. Biol. Chem. 277:18649-18657. [DOI] [PubMed] [Google Scholar]
- 43.Wang, Y., and T. D. Gilmore. 2001. LIM domain protein Trip6 has a conserved nuclear export signal, nuclear targeting sequences, and multiple transactivation domains. Biochim. Biophys. Acta 1538:260-272. [DOI] [PubMed] [Google Scholar]
- 44.Wang, Y., and T. D. Gilmore. 2003. Zyxin and paxillin proteins: focal adhesion plaque LIM domain proteins go nuclear. Biochim. Biophys. Acta 1593:115-120. [DOI] [PubMed] [Google Scholar]
- 45.Yang, S.-H., P. R. Yates, A. J. Whitmarsh, R. J. Davis, and A. D. Sharrocks. 1998. The Elk-1 ETS-domain transcription factor contains a mitogen-activated protein kinase targeting motif. Mol. Cell. Biol. 18:710-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao, M. K., Y. Wang, K. Murphy, J. Yi, M. C. Beckerle, and T. D. Gilmore. 1999. LIM domain-containing protein Trip6 can act as a coactivator for the v-Rel transcription factor. Gene Expr. 8:207-217. [PMC free article] [PubMed] [Google Scholar]