Abstract
Although many E2F target genes have been identified recently, very little is known about how any single E2F site controls the expression of an E2F target gene in vivo. To test the requirement for a single E2F site in vivo and to learn how E2F-mediated repression is regulated during development and tumorigenesis, we have constructed a novel series of wild-type and mutant Rb promoter-LacZ transgenic reporter lines that allow us to visualize the activity of a crucial E2F target in vivo, the retinoblastoma tumor suppressor gene (Rb). Two mutant Rb promoter-LacZ constructs were used to evaluate the importance of a single E2F site or a nearby activator (Sp1/Ets) site that is found mutated in low-penetrance retinoblastomas. The activity of the wild-type Rb promoter is dynamic, varying spatially and temporally within the developing nervous system. While loss of the activator site silences the Rb promoter, loss of the E2F site stimulates its activity in the neocortex, retina, and trigeminal ganglion. Surprisingly, E2F-mediated repression of Rb does not act globally or in a static manner but, instead, is a highly dynamic process in vivo. Using neocortical extracts, we detected GA-binding protein α (GABPα, an Ets family member) bound to the activator site and both E2F1 and E2F4 bound to the repressor site of the Rb promoter in vitro. Additionally, we detected binding of both E2F1 and E2F4 to the Rb promoter in vivo using chromatin immunoprecipitation analysis on embryonic day 13.5 brain. Unexpectedly, we detect no evidence for Rb promoter autoregulation in neuroendocrine tumors from Rb+/−; RbP-LacZ mice that undergo loss of heterozygosity at the Rb locus, in contrast to the situation in human retinoblastomas where high RB mRNA levels are found. In summary, this study provides the first demonstration that loss of an E2F site is critical for target gene repression in vivo and underscores the complexity of the Rb and E2F family network in vivo.
Classic E2F target genes include those that regulate cell cycle progression (e.g., CcnE, Cdc6, Cdc25A, Mcm2-7, Orc, CcnA, and Cdc2) or the maintenance of nucleotide pools (e.g., Dhfr, Rnr, Tk, and Ts) (reviewed in references 5, 6, and 56). Typically, these have been identified by mutation of the E2F site [consensus sequence TTT(C/G)(C/G)CGC] in reporter constructs, leading to the deregulated expression of the putative target gene across the cell cycle. A number of E2F target genes have been identified whose products stimulate apoptosis (e.g., p73, Apaf1, Arf, and caspases). Most of the genes encoding E2F family members (E2f1-E2f3a and E2f6-E2f8) are themselves E2F targets, many of which are thought to contribute to a feed-forward amplification loop to generate sufficient E2F activity to stimulate cell cycle progression following pRB phosphorylation (1, 9, 11, 14, 22, 28, 35, 48, 66). More recently, gene expression profiling with inducible E2F expression and chromatin immunoprecipitation (ChIP-on-chip) analysis in cultured cells have greatly expanded the sheer number (estimated to be in the hundreds) and classes (e.g., DNA repair, cell cycle checkpoints, and chromatin dynamics) of E2F target genes substantially (24, 36, 39, 43, 44, 57). However, little is still known about the significance of any single E2F site in the normal regulation of an E2F target gene in vivo.
Interestingly, two Rb family members (Rb and p107) are E2F target genes (20, 65), which suggests that substantial complexity may exist in the transcriptional circuitry connecting the Rb and E2f family members and that E2F may lie upstream and downstream of pRB in a genetic sense. Apart from the well-documented ability of cyclin/cyclin-dependent kinase (CDK)-mediated phosphorylation to regulate pRB function (50), transcription of the human RB gene or mouse Rb gene plays a role in regulating pRB function. Notably, point mutations and deletions in the human RB promoter have been identified in low-penetrance retinoblastomas, emphasizing the importance of the proper levels of RB transcription for tumor suppression (4, 10, 45, 63). Additionally, Rb transcription increases as cells undergo differentiation (e.g., P19 cells with retinoic acid) (41, 52, 62), which is consistent with the role of Rb in promoting differentiation of numerous cell types, particularly the neuronal lineage (18, 32, 37). The presence of elevated levels of mutant RB mRNA in many retinoblastomas has prompted speculation that pRB autoregulates its own promoter, and mutation of the RB gene leads to its increased transcription (15, 20). In light of the recently demonstrated dispensability of G1 cyclins and CDKs during most of development, an exploration of alternative routes to regulating pRB function seems warranted (42, 51). Indeed, transcriptional control of Rb levels during development could provide an alternative mechanism that would bypass the need for G1 cyclin/CDK-mediated phosphorylation in many tissues.
A well-conserved 26-bp cluster of binding sites lying 180 bp upstream of the translational start site accounts for much of the human RB and the mouse Rb promoter activity in vitro (19, 62). Binding sites for Sp1, Ets, ATF, and E2F are present, the first two of which are partially overlapping and are referred to hereafter as Sp1/Ets (see Fig. 1A). A subset of the aforementioned point mutations in low-penetrance retinoblastomas maps into this Sp1/Ets site or into the adjacent ATF site of the RB promoter (45, 63), which is consistent with these being activator sites. In vitro studies have shown that mutation of the E2F site in this cluster activates RB gene expression in cell lines and that overexpression of pRB can repress Rb promoter expression of this putative repressor site (20, 40, 49, 62).
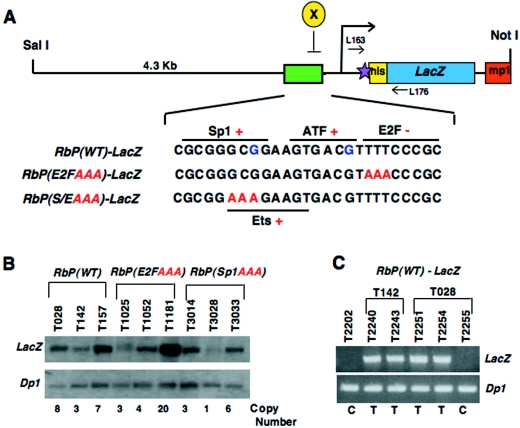
FIG. 1.
Generation of wild-type and mutant Rb promoter reporter lines. To generate the RbP(WT)-LacZ transgene construct, a fragment (4.3 kb) of the wild-type Rb promoter containing a cluster (green box) of transcription factor binding sites (Sp1, Ets sites, ATF site, and E2F site) was subcloned into the pnLacF vector that carries the LacZ reporter gene to which the simian virus 40-T NLS has been fused (38). The pnLacF vector also carries the intron and the 3′ untranslated region with a polyadenylation signal from the mouse protamine (mp1) gene for optimal expression (A). Additionally, to ensure efficient translation, we engineered a Kozak consensus sequence into the 5′ untranslated region surrounding the initiator methionine codon of the Rb gene (violet star). For the mutant RbP-LacZ transgene constructs, AAA substitutions were introduced (sequence positions shown in red) into the Sp1/Ets site or the E2F site. The sequence positions of point mutations found in low-penetrance retinoblastomas are shown in blue. (B) Potential transgenic founders for the wild-type and mutant RbP-LacZ transgenes were identified by Southern analysis using a LacZ probe (top). The transgene copy number was estimated by normalizing the LacZ signal to that of Dp1, an internal genomic control (bottom), using Southern analysis. A subset of these founders was used to establish the RbP(WT)-LacZ lines (T028, T142, and T157), the RbP(E2FAAA)-LacZ lines (T1025, T1052, and T1181), and the RbP(S/EAAA)-LacZ lines (T3014, T3028, and T3033). (C) A genomic PCR assay was designed to detect transgenic progeny from any of the wild-type or mutant RbP-LacZ founder animals using the primers (see panel A) L163 (forward primer lying within the 5′ untranslated region of the Rb gene) and L176 (reverse primer lying within the LacZ transgene). Four transgenic (T) and two control (C) animals are identified using this PCR assay (top). A genomic PCR assay for Dp1 was run in parallel to confirm the presence and quality of the tail DNA used in these reactions (bottom).
To evaluate the importance of a single E2F site in vivo and to understand how E2F-mediated repression of a critical target, such as the Rb tumor suppressor, is regulated during development, we constructed a novel series of wild-type and mutant Rb promoter-LacZ transgenic lines, which allowed us to visualize Rb promoter activity in every tissue throughout development. These novel RbP-LacZ reporter mice express in a tissue- and temporal-specific manner, giving new insight into the role of E2F in vivo and the complex and dynamic balance between transcriptional activation and repression ongoing in the whole animal.
MATERIALS AND METHODS
Construction of RbP-LacZ transgenes.
A phage (φ2) containing the 5′ end of the mouse Rb gene was identified by screening a 129Sv genomic phage library with a 5′ fragment (EcoRI-KpnI fragment of 300 bp) from the mouse Rb cDNA vector (pJ3Ω115.Rox; a gift from R. Bernards). To identify the promoter region of φ2, we generated a PCR probe (481 bp) from the Rb promoter with primers L94 (5′-TAGGCAAGTCTGAAAATTGAAGG-3′) and L95 (5′-GCCTCCTTTCATAATGGTTTCTC-3′) that amplify a promoter region lying 543 bp upstream of the cluster of binding sites of interest. A NotI fragment (4.3 kb) containing this upstream regulatory region of Rb was identified by Southern hybridization with this PCR probe and subcloned into pBSK to yield the pRbP construct. We then subcloned a 500-bp EagI fragment containing the cluster of sites and part of exon 1 of the Rb gene into the pRbP-Eag construct. We then introduced a Kozak consensus sequence (with an embedded NcoI site) in the Rb gene at the initiator methionine codon by site-directed mutagenesis (Quick Change Kit; Stratagene), yielding the vector pRbP-Eag(WT) (where WT is wild type). Triple (AAA) substitutions into the E2F and the Sp1/Ets sites were then introduced in pRbP-Eag(WT) by a new round of site-directed mutagenesis, yielding pRbP-Eag(E2FAAA) and pRbP-Eag(S/EAAA). The EagI fragments containing a perfect Kozak sequence as well as the wild-type and mutant sites were purified and reintroduced into the original pRbP construct (EagI digested), producing the pRbP(WT), pRbP(E2FAAA), and pRbP(S/EAAA) constructs. To facilitate later excision of the RbP-LacZ transgenes, a NotI site was introduced 3′ to the mouse protamine (mP1) terminal exon in the vector pnLacF (38). The pRbP(WT), pRbP(E2FAAA), and pRbP(S/EAAA) constructs were digested with SalI and NcoI to release the 4.3-kb Rb promoter fragments, which were then purified and subcloned into the modified pnLacF, yielding RbP(WT)-LacZ, RbP(E2FAAA)-LacZ, and RbP(S/EAAA)-LacZ transgene constructs.
Generation of RbP-LacZ transgenic lines.
To release the transgene inserts (7.9 kb), the RbP(WT)-LacZ, RbP(E2FAAA)-LacZ, and RbP(S/EAAA)-LacZ vectors were digested with SalI and NotI. The insert DNA was purified by electroelution and then dialyzed against TE (10 mM Tris, pH 8.0, 1 mM EDTA) buffer. Microinjection of the transgenes was done several times at New York University's Transgenic Mouse Facility (TgESC; Anna Auerbach) into fertilized eggs on a purebred C57BL/6 genetic background. Early embryos were then implanted into outbred pseudopregnant recipients, which were then imported into Columbia University. All surviving progeny were weaned at 3 weeks of age, ear tagged for identification, and tail clipped to provide DNA for genotyping by Southern analysis and genomic PCR (see below). All transgenic animals were bred to wild-type C57BL/6 animals to establish lines for each RbP-LacZ construct. Transgenic animals that produced LacZ activity in X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining of whole-mount embryos at mid to late gestation were used as founders to establish the following lines for each construct: RbP(WT)-LacZ lines (T028, T142, and T157), RbP(E2FAAA)-LacZ (T1025, T1052, and T1181), and RbP(S/EAAA)-LacZ (T3014, T3028, and T3033). All animals were handled according to protocols approved by the Institutional Animal Care and Use Committee that conform to standard regulatory guidelines.
Southern analysis of the RbP-LacZ transgenics.
Transgenic mice were initially identified by Southern hybridization for the LacZ transgene. Genomic DNA was prepared from tail snips by overnight digestion in tail lysis buffer (100 mM Tris, pH 8.0, 200 mM NaCl, 5 mM EDTA, 0.2% sodium dodecyl sulfate [SDS]) with proteinase K (1 mg/ml), followed by phenol extraction, precipitation with isopropanol, and resuspension overnight in TE buffer at 50°C. Approximately 15 μg of genomic DNA was digested overnight with SacI and KpnI, run on a 0.8% Tris-acetate-EDTA (TAE)-agarose gel for 16 h, and transferred overnight by alkaline transfer to a Hybond-N+ membrane (Amersham Biosciences). The presence of the LacZ transgene was detected with a 32P-labeled LacZ probe (836-bp NcoI-ClaI fragment), prepared with the Redi-Prime II DNA labeling system (Amersham Biosciences). Membranes were hybridized overnight at 60°C in Shackelford buffer supplemented with herring sperm DNA, washed in 2× SSC-1% SDS (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and then 0.1× SSC-0.5% SDS at 60°C, and exposed to autoradiographic film. As an internal loading control, a 32P-labeled Dp1 probe was prepared from a genomic Dp1 fragment (700-bp subcloned SacI-KpnI fragment from V. Criniti) and hybridized to the membrane simultaneously with the LacZ probe. The SacI-KpnI digestion produces a 3.6-kb fragment of the integrated RbP-LacZ transgene and a 0.7-kb fragment of Dp1. Copy number was estimated by comparing hybridization signals from the LacZ probe normalized to the signal from the Dp1 probe using a Storm Phospho-Imager and ImageQuaNT software (Molecular Dynamics).
Genomic PCR genotyping of RbP-LacZ transgenics.
To genotype transgenic progeny, we developed a genomic PCR assay that detects the presence of the LacZ transgene in any of the RbP-LacZ lines. Using forward primer L163 (5′-TCCGGTTTTCCTCGGGGGACGTT-3′) lying 175 bp upstream of the initiator methionine codon in the Rb promoter and reverse primer L176 (5′-TCAGGCTGCCGAACTGTTGGGAA-3′) lying 163 bp into the LacZ reporter gene, we amplified a 380-bp transgene fragment that includes the short region encoding the nuclear localization signal (NLS) according to the following program: melting at 94°C for 5 min, followed by 33 cycles of melting at 94°C for 1 min, annealing at 54°C for 1 min, and extension at 72°C for 1 min, with a final extension for 7 min at 72°C. Due to the extremely high GC content of the Rb promoter, we added betaine to the amplification cocktail to a final concentration of 1.2 M. The PCR products were visualized by running on a 1.9% TAE-agarose gel containing ethidium bromide. To ensure the quality of genomic DNA and the nontransgenic status of animals where no LacZ band was detected, we ran a genomic PCR assay in parallel for the wild-type allele of Dp1 that we developed previously (29) using the L75 and L78 primers (without betaine), which should produce a 200-bp fragment on all samples tested.
Detection of LacZ expression.
For embryonic time points, transgenic males were mated to wild-type C57BL/6 females, and detection of a vaginal plug the next morning was counted as day 0.5. Pregnant females were sacrificed by cervical dislocation, and embryos were fixed as described below following their release from the yolk sac and placenta. For the detection of LacZ expression in whole-mount embryos, embryos were collected from pregnant recipient females, fixed in 10% buffered formalin, rinsed in phosphate-buffered saline (PBS), and then incubated in X-Gal staining solution [20 mM MgCl2, 0.2% NP-40, 50 mM K3Fe(CN)6, 50 mM K4Fe(CN)6, and 1 mg/ml of X-Gal in PBS] overnight at 30°C. The embryos were subsequently washed three times for 5 min in PBS and postfixed for 24 h. For embryonic day 15.5 (E15.5) microdissected brains, embryos were collected, and the brains were dissected and fixed in 10% buffered formalin and stained as above. For detection of LacZ expression in cryosections, E16.5 embryos and dissected brains and eyes from postnatal day 0 (P0) pups as well as 5- and 12-week-old animals were fixed in 2% paraformaldehyde, rinsed in PBS, and equilibrated in 18% sucrose in PBS overnight at 4°C. Tissues were embedded in TissueTek (Ted Pella, Inc.) OCT (22-oxyacalcitriol) and frozen in methylbutane and dry ice, and then frozen sections (10 μm) were prepared on positively charged slides. Cryosections were then incubated overnight in X-Gal staining solution at 30°C, counterstained briefly in Nuclear Fast Red (Vector Laboratories), and dehydrated through a graded series of methanol washes. After a brief dip in xylene, coverslips were mounted with Permount, and the slides were examined using standard light microscopy.
Preparation of nuclear extracts.
Nuclear extracts were prepared as follows from E13.5 and E15.5 microdissected neocortices. Tissue was homogenized in buffer A (10 mM HEPES, pH 7.8, 10 mM NaF, 0.5 mM dithiothreitol [DTT], 0.1 mM EDTA, 0.1 mM EGTA, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 1 μl/ml protease inhibitor cocktail; Sigma). After a 15-min incubation on ice, NP-40 was added to the lysates to a final concentration of 0.5%, incubated for 15 min more on ice, and pelleted at 11,600 rpm for 15 min at 4°C. Pellets were resuspended in 100 μl of buffer B (20 mM HEPES, pH 7.8, 400 mM NaCl, 1 mM DTT, 1 mM EDTA, 1 mM EGTA, 1 mM PMSF, 1 μl/ml protease inhibitor cocktail, 25% glycerol) and incubated on ice for 15 min. Insoluble debris was removed by centrifugation at 13,000 rpm at 4°C, and the supernatants were aliquoted and stored at −80°C.
In vitro binding to the Rb promoter. (i) Competitive gel shift assays.
To evaluate binding to the Rb promoter in vitro, a double-stranded, 38-bp wild-type Rb promoter probe spanning the cluster of binding sites of interest was prepared by first annealing the following complementary primers at 2 μg/μl of each primer: L305 (5′-CGTGAGCGCGGGCGGAAGTGACGTTTTCCCGCGGTTGG-3′) and L306 (5′-CCAACCGCGGGAAAACGTCACTTCCGCCCGCGCTCACG-3′). Annealed primers (20 ng) were labeled with γ-32P-Redivue ATP (20 μCi) using polynucleotide kinase (New England Biolabs) for 30 min at 37°C and purified on a G-50 spin column equilibrated in TE buffer. For the gel shift assay, nuclear extract (1.5 μl) was diluted 12-fold in dilution binding buffer (20 mM HEPES, pH 7.8, 5 mM MgCl2, 0.1 mM EDTA, 0.1 mM EGTA, 0.5 mM NaF, 0.5 mM DTT, 0.5 mM PMSF, 1 μl/ml protease inhibitor cocktail, 3 mg/ml bovine serum albumin) and then incubated with 1 μg of poly(dI-dC) and the 32P-labeled wild-type Rb promoter probe (25,000 cpm is the equivalent of 0.05 ng) in a total volume of 22 μl for 20 min at room temperature. Reactions were mixed with 1/10 volume of glycerol and then loaded onto a 5% Tris-borate-EDTA-polyacrylamide gel, run for 1.5 h at 160 V, dried, and visualized by autoradiography. For competition experiments with this 32P-labeled wild-type Rb promoter probe, we annealed the following primer pairs at 2 μg/μl of each primer, and used 20 ng of annealed primer per competition reaction: for S/EAAA competitor, L293 (5′-GAGCGCGGAAAGAAGTGACGTTTTCCCGCGGT-3′) and L294 (5′-ACCGCGGGAAAACGTCACTTCTTTCCGCGCTC-3′); for ATFAAA competitor, L295 (5′-GAGCGCGGGCGGAAGTGAAAATTTCCCGCGGT-3′) and L296 (5′-ACCGCGGGAAATTTTCACTTCCGCCCGCGCTC-3′); for E2FAAA competitor, L297 (5′-GAGCGCGGGCGGAAGTGACGTAAACCCGCGGT-3′)and L298 (5′-ACCGCGGGTTTACGTCACTTCCGCCCGCGCTC-3′); for Sp1-Mut competitor, L285 (5′-GAGCGATGGCGGAAGTGACGTTTTCCCGCGGT-3′) and L286 (5′-ACCGCGGGAAAACGTCACTTCCGCCATCGCTC-3′); for Ets-Mut competitor, L287 (5′-GAGCGCGGGCGGTCGTGACGTTTTCCCGCGGT-3′) and L288 (5′-ACCGCGGGAAAACGTCACGACCGCCCGCGCTC-3′); for nonspecific competitor, L303 (5′-TATTTTTGTAACGGGAGTCGGGTGAGGACGGG-3′) and L304 (5′-CCCGTCCTCACCCGACTCCCGTTACAAAAATA-3′).
For the detection of E2F activity using the 32P-labeled wild-type Rb promoter probe, we used E13.5 neocortical extracts and sonicated herring sperm DNA (1 μg per reaction) rather than poly(dI-dC) as a nonspecific competitor. To verify that our gel shift activity was E2F, we performed competition reactions with wild-type and mutant AdE2 double-stranded competitors (26 bp) from the adenovirus E2 promoter (7) by annealing the following complementary primers (2 μg/μl of each primer) and using 20 ng of each annealed primer pair per competition reaction: for AdE2 competitor, L275 (5′-ATTTAAGTTTCGCGCCCTTTCTCAA-3′) and L276 (5′-TTGAGAAAGGGCGCGAAACTTAAAT-3′); for AdE2-Mut, AdE2-mut1 (5′-ATTTAAGTTTCGATCCCTTTCTCAA-3′) and AdE2-mut2 (5′-TTGAGAAAGGGATCGAAACTTAAAT-3′).
(ii) Supershift experiments.
To identify which Ets family member bound to the radiolabeled wild-type Rb promoter probe in the gel shift reactions, we preincubated the nuclear extract with the Ets AAA competitor and one of a panel of antibodies against Ets family members at 2 μg per reaction for 5 min at room temperature. The 32P-labeled wild-type Rb promoter probe was then added for an additional 20 min and processed as described above to visualize supershifted complexes. The rabbit polyclonal antibodies used for supershifting Ets family members were anti-Elk1, anti-ERM, anti-GA-binding protein α (anti-GABPα), anti-PEA3 (all from Santa Cruz). Rabbit immunoglobulin G (IgG) was used as a nonspecific antibody control. To identify which E2F family members are present in the neocortical nuclear extract, we preincubated nuclear extract with polyclonal antibodies specific for various E2F and DP family members at 2 μg per reaction for 5 min at room temperature. The polyclonal antibodies used to supershift the E2F family members were anti-E2F1 through anti-E2F4 (Santa Cruz), anti-E2F5 (Neomarkers), and anti-E2F6 (Santa Cruz). Mouse IgG and rabbit IgG were used as nonspecific antibody controls.
Real-time RT-PCR.
Total RNA was isolated from neocortex (E13.5 and E15.5) and limbs (E15.5) using Trizol reagent (Invitrogen), and then cDNA was reverse transcribed from 5 μg of total RNA using Moloney murine leukemia virus reverse transcriptase (Promega). Real-time reverse transcription-PCR (RT-PCR) was performed for actin and individual mouse E2F family members using specific RT-PCR primer pairs that are commercially available (actin, E2f1, E2f3, E2f4, E2f5, and E2f7 [SuperArray]; E2f2 and E2f6 [QIAGEN]), and Power SYBR master mix with Hot Start Taq polymerase (ABI) on an ABI-7300 real-time PCR system. The efficiency of amplification was established for each primer set using 10-fold serial dilutions of cDNA, and then cycle threshold and log input values were calculated for each gene, using the E15.5 cortex values as standards. All E2F log input values were normalized to the actin log input value, and then the ratio of these normalized log input values were expressed for E15.5 cortex/E13.5 cortex and E15.5 cortex/E15.5 limbs.
Western blotting.
Embryos were collected and brains were microdissected and divided in forebrain, midbrain, and hindbrain. For each fraction, total protein was solubilized in 2× Laemmli buffer with repeated rounds of sonication and boiling. An approximately equal amount of total protein (estimated by Coomassie blue staining) was separated by SDS-polyacrylamide gel (10%) electrophoresis and then transferred to Immobilon-P membranes. Western blotting with rabbit polyclonal primary antibodies to GABPα (Santa Cruz), E2F1 (Santa Cruz), E2F4 (Santa Cruz), and actin (Sigma) and a horseradish peroxidase-donkey anti-rabbit IgG (Amersham) secondary antibody was performed to visualize the proteins of interest. The blots were developed with an ECL-Plus kit (Amersham) and exposed to autoradiographic film.
ChIP analysis.
Embryonic brains were microdissected from E13.5 embryos and then trypsinized briefly in 0.25% trypsin, which was then inactivated. Suspensions of primary neurons were fixed with 1% formaldehyde and then pelleted and frozen at −80°C. Cell pellets were resuspended in SDS lysis buffer (Upstate) at 1 × 107 cells per 200 μl and then sonicated for 60 s (three 20-s pulses followed by cooling on ice) using a Branson 250 sonicator (setting 3, 70% output) to shear the chromatin to lengths between 200 and 1,000 bp. Samples were then clarified by centrifugation for 10 min at 13,000 rpm at 4°C and then diluted approximately 10-fold with ChIP dilution buffer (Upstate) with protease inhibitors, such that 2 ml of diluted supernatant is equivalent to 9 × 106 cells. A portion (3%) of this supernatant was removed as the input sample, and then the remainder was precleared by incubating it with salmon sperm DNA-treated protein A-agarose (Upstate) for 30 min at 4°C. Normal rabbit IgG (Zymed) or purified antibody (2 μg) to E2F1 (Santa Cruz), E2F4 (Santa Cruz), or acetylated histone H3 (Upstate) was added to each precleared supernatant (2 ml) and mixed overnight at 4°C. Antibody-bound chromatin was recovered with the addition of salmon sperm DNA and protein A-agarose for 1 h at 4°C; protein A-bound immune complexes were then washed using successive low salt, high salt, and LiCl immune complex wash buffers (Upstate), followed by two washes in TE buffer. Bound chromatin was eluted in 1% SDS-0.1 M NaHC03, adjusted with 20 μl of 5 M NaCl, and chromatin cross-links were reversed by heating at 65°C for 4 h. To isolate DNA, the eluates were treated with proteinase K (5 μg) in 40 mM Tris (pH 6.5)-10 mM EDTA and then phenol-chloroform extracted and ethanol precipitated after the addition of glycogen. DNA was resuspended in 30 μl of water overnight and then used in PCRs to amplify the Rb promoter, intron 3 of the Rb gene, or the Cdc2 promoter. For the Rb promoter, template DNA was mixed with a PCR cocktail containing 1.6 M betaine and the following previously published primers (59): RbChIP1 (5′-GAAAACCGGACGCGCCCGGCAA-3′) and RbChIP2 (5′-CGTTCTCCCAGAGGCCGCGGCT-3′). This was then amplified using a PCR program of denaturation at 94°C for 5 min, followed by 39 cycles of denaturation (92°C for 1 min), annealing (54°C for 1.5 min), and extension (72°C for 1 min). For intron 3 of the Rb gene, template DNA was mixed with a cocktail containing the following primers: L116 (5′-GGGATTTGGGACCAATAATGAAT-3′) and I3L (5′-TGCCCATGTTCGGTCCCTAGCA-3′). This was then amplified using a PCR program of denaturation at 94°C for 5 min, followed by 34 cycles of denaturation (92°C for 1 min), annealing (55°C for 1 min), and extension (72°C for 1 min). For the Cdc2 promoter, template DNA was mixed with a cocktail containing the following primers: Cdc2ChIP3 (5′-GCTCTTGATGTAGTGGTACTGTCAC-3′) and Cdc2ChIP4 (5′-TCCCGGGATCCGCCAATCCGATTGC-3′). This was then amplified using the same PCR program as for intron 3 of the Rb gene. All products were visualized on a 1.9% agarose-TAE gel with ethidium bromide.
RESULTS
Production of wild-type and mutant RbP-LacZ reporter lines.
To evaluate the importance of a single E2F site and understand how the Rb promoter is regulated in vivo, we engineered wild-type and mutant Rb promoter-LacZ transgene constructs (Fig. 1A) to establish reporter lines in which promoter activity could be visualized as β-galactosidase activity (blue staining) in the presence of X-Gal. Such LacZ reporter lines are extremely useful, because they offer greater sensitivity and ease of detection than in situ hybridization. To generate the transgene constructs, we linked a wild-type Rb promoter fragment to a LacZ reporter gene, carrying a simian virus 40-T NLS. Since the wild-type Rb gene lacks a Kozak consensus sequence necessary for efficient translation, we introduced a perfect Kozak sequence at the initiator codon to optimize translation. Additionally, we created two Rb promoter mutations by site-directed mutagenesis to destroy the Sp1/Ets site or the classic E2F site by triple (AAA) substitution (Fig. 1A), mutations that have been used previously in vitro (62). These constructs are referred to hereafter as the RbP(WT)-LacZ, RbP(E2FAAA)-LacZ, and the RbP(S/EAAA)-LacZ transgenes.
Transgenic founder animals were produced from these transgene constructs on a purebred C57BL/6 background. We identified three founder animals (Southern positive and PCR positive) for each of the Rb promoter transgenes that expressed LacZ during mid-gestation. Founders were bred to establish the following transgenic lines: RbP(WT)-LacZ reporters (T028, T142, and T157 lines), RbP(E2FAAA)-LacZ reporters (T1025, T1052, and T1181 lines), and RbP(S/EAAA)-LacZ reporters (T3014, T3028, and T3033 lines). Founders were selected based on their ability to produce progeny that displayed LacZ expression at E12.5, regardless of the expression pattern observed, to avoid bias about where the different transgene constructs should express. We estimated the transgene copy number by comparing the LacZ signal to that of an internal genomic control (Dp1) by Southern blotting (Fig. 1B). Additionally, a genomic PCR assay was designed to follow inheritance of the LacZ transgene in progeny from these founders (Fig. 1C). Rb promoter transgenic lines were generated rather than creating Rb promoter-LacZ knock-in constructs, since loss of a single Rb allele is associated with numerous defects, including neuroendocrine tumor formation upon loss of the remaining wild-type Rb allele. However, any phenotype resulting from the presence of the transgene would have to be present in more than one line from each construct to control for random integration effects.
Dynamic expression of the wild-type Rb promoter in the nervous system.
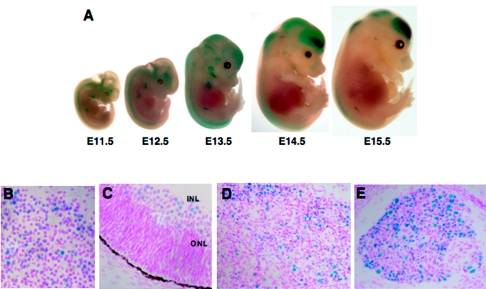
To characterize the activity of the wild-type Rb promoter during development, we examined RbP(WT)-LacZ embryos from E11.5 through E15.5 using whole-mount X-Gal staining. LacZ expression is evident in the developing forebrain and spinal cord at all time points tested (Fig. 2A). The intensity and the position of the positive regions within the nervous system change with gestational age. Expression of the LacZ transgene within the developing nervous system occurs in all three of the RbP(WT)-LacZ reporter lines. Furthermore, in embryonic cryosections at E16.5, we detected LacZ expression in the neurons of the central nervous system (CNS) (e.g., cortex and the retina) and the peripheral nervous system (PNS) (e.g., trigeminal ganglion and dorsal root ganglia) (Fig. 2B to E). Importantly, the RbP(WT)-LacZ transgene does not direct pan-neuronal expression; instead, high-level expression occurs in a subset of neurons in the developing forebrain (neocortex), retina, trigeminal ganglion, and dorsal root ganglia. Thus, the Rb promoter is responsible for a highly dynamic pattern of LacZ expression in the CNS and PNS of embryos in a temporally and spatially specific manner. A neuronal pattern of Rb expression is consistent with the requirement of Rb for the development of the neuronal lineage (12, 18, 37, 64) and with the in situ hybridization experiments from our laboratory (data not shown) and from others (27). During the course of this work, a neuronal pattern of Rb expression was observed by other investigators using wild-type promoter transgenics (26). Importantly, our RbP(WT)-LacZ transgene drives a neuronal-specific pattern of LacZ expression to which our mutant RbP(E2FAAA)- and RbP(S/EAAA)-LacZ transgenes could be compared to test the importance of these cis acting elements in vivo.
FIG. 2.
The wild-type Rb promoter is expressed dynamically in the nervous system. The RbP(WT)-LacZ reporter lines all express in the CNS and PNS as visualized with X-Gal staining of either whole-mount embryos (A) or embryonic cryosections (B to E). For example, the strongest-expressing wild-type Rb promoter reporter line, T157, is presented with prominent staining in the neocortex and developing spine from E11.5 to E15.5. Hemisected, E16.5 embryos of this same wild-type RbP(WT)-LacZ reporter line show strong, but restricted, LacZ expression in the neocortex (B) and the inner neuroblastic layer (INL) of the developing retina (ONL, outer neuroblastic layer) (C). A subset of neurons in the trigeminal ganglia (D) and the dorsal ganglia (E) express the RbP(WT)-LacZ transgene at E16.5.
Deregulation of the Rb promoter in the nervous system.
To determine whether mutation of the E2F site or the Sp1/Ets site deregulated the Rb promoter in vivo, we analyzed the mutant RbP(E2FAAA)- and RbP(S/EAAA)-LacZ reporter embryos from E11.5 to E16.5 using whole-mount embryo X-Gal staining. While all three RbP(S/EAAA)-LacZ lines showed only low levels of LacZ expression, all three RbP(E2FAAA)-LacZ lines exhibited strong LacZ expression relative to that seen in all of the RbP(WT)-LacZ lines in the nervous system, particularly the developing forebrain or neocortex (Fig. 3). The lateral edges of the neocortex from RbP(WT)-LacZ embryos display moderate LacZ expression, while the midline of the neocortex from RbP(E2FAAA)-LacZ embryos exhibits robust LacZ expression in whole-mount embryos at E13.5 to E15.5 (Fig. 3A, E13.5) and in microdissected brains (Fig. 3B, E15.5). Only weak LacZ expression is apparent anywhere in the neocortex of RbP(S/EAAA)-LacZ embryos, even upon X-Gal staining of embryonic cryosections; yet all three RbP(S/EAAA)-LacZ lines express in the CNS or PNS at E16.5 and in adulthood, albeit at extremely low levels that are visible in embryonic cryosections (data not shown).
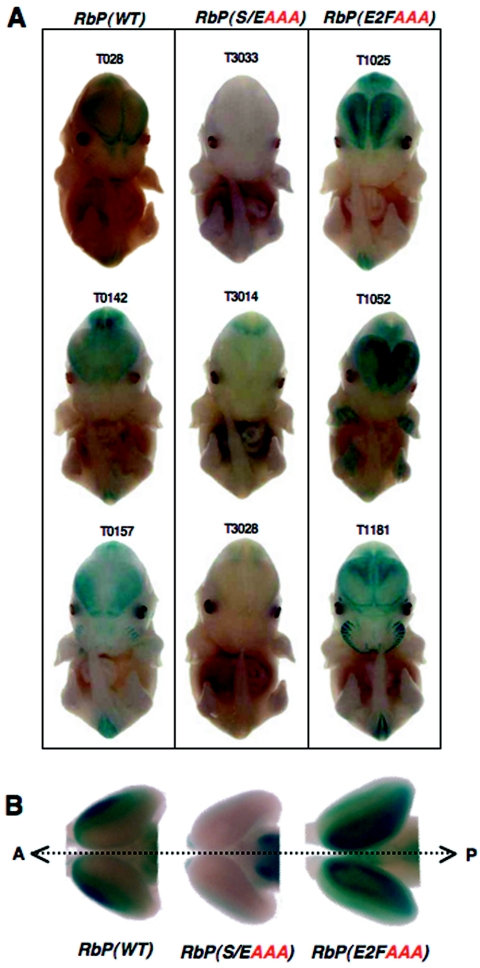
FIG. 3.
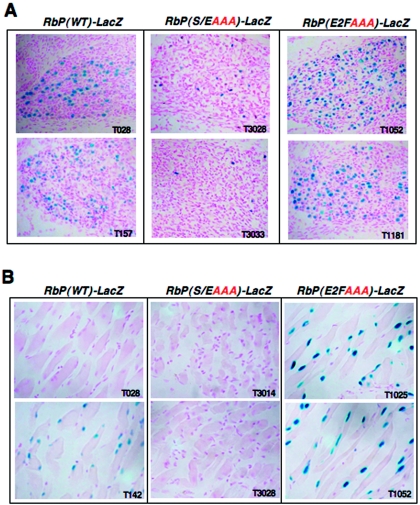
Deregulation of the Rb promoter with loss of the E2F site in the cortex. Expression of the RbP(WT)-LacZ transgene occurs at the lateral edges of the neocortex, as seen using whole mount X-Gal staining of embryos at E13.5 (A, left column) or of microdissected neocortices at E15.5 (B, left sample). All of the RbP(E2FAAA)-LacZ lines exhibit strong derepression of the transgene in the neocortex at the midline (A, right column, and B, right sample). In contrast, all of the RbP(S/EAAA)-LacZ lines display little X-Gal staining in the neocortex of whole-mount embryos (A, middle column) or of microdissected neocortices (B, middle sample). The dashed arrow in panel B indicates the midline (A, anterior; P, posterior). Thus, loss of the E2F site results in strong activation of the Rb promoter, while loss of the Sp1/Ets site severely impairs reporter activity in the neocortex.
The differential expression of LacZ within the neocortex cannot be explained simply by a change in the transgene copy number in the wild-type and mutant RbP-LacZ lines. For example, the deregulation of LacZ expression in the cortex occurs in two of the RbP(E2FAAA)-LacZ lines (T1025 and T1052) that have low transgene copy numbers (three and four copies, respectively) but show strong elevation of LacZ activity relative to that found in the cortex of all three RbP(WT)-LacZ lines (T028, T142, and T157) that have moderate transgene copy numbers (eight, three, and seven copies, respectively). Thus, loss of the Sp1/Ets site results in a loss of activation of the Rb promoter, while loss of the E2F site results in a loss of repression of the Rb promoter within the neocortex.
A strikingly similar change in LacZ expression can be seen in the adult retina (12 weeks) from the wild-type and mutant Rb-LacZ lines (Fig. 4A). Two of the RbP(WT)-LacZ reporter lines (T028 and T157) have discrete expression patterns in the the retinal neuroepithelium, particularly in the ganglionic cell layer (Fig. 4A, G). This is in agreement with the retinal expression pattern seen for endogenous Rb (54). All of the RbP(S/EAAA)-LacZ reporter lines show weak or absent retinal staining. In contrast, the RbP(E2FAAA)-LacZ reporter lines (T1025 and T1052) display strong, widespread expression in all three layers of the adult retina (ganglionic cell layer, inner nuclear layer, and the outer nuclear layer containing the rod and cone photoreceptors). All but a small population of neurons present in the inner nuclear layer express LacZ in these RbP(E2FAAA)-LacZ lines. Since this inner nuclear layer is composed of three neuronal cell types (bipolar, amacrine, and horizontal neurons) and Muller glial cells (17), it appears likely that the Rb promoter is deregulated by loss of the E2F site in almost all retinal cell types. Expression is seen in photoreceptors in only one RbP(WT)-LacZ line (T142) but at a much lower level than that seen in the RbP(E2FAAA)-LacZ lines (T1025 and T1052) (data not shown). Thus, loss of the Sp1/Ets site results in lower activation of the Rb promoter, while loss of the E2F site strongly derepresses the Rb promoter in the adult retina.
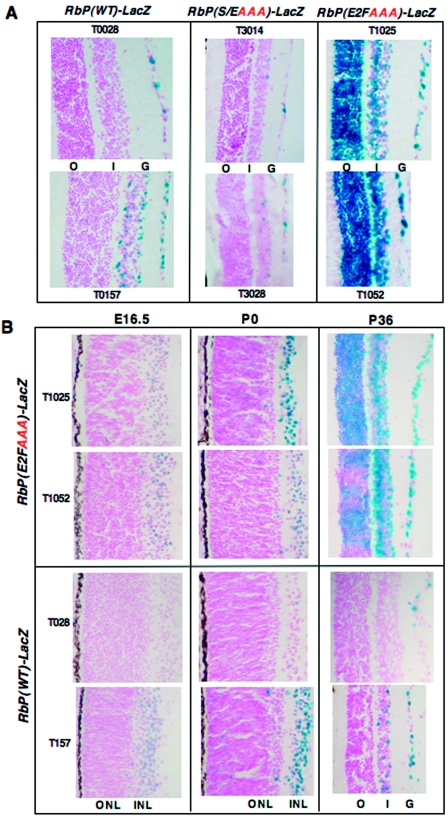
FIG. 4.
Deregulation of the Rb promoter with loss of the E2F site in the retina. Expression of the RbP(WT)-LacZ transgene occurs in the outer layers of the adult retina (A, left column), including the ganglionic cell layer (G) and/or the inner nuclear layer (I). Loss of the E2F site results in a strong derepression of LacZ expression throughout the adult retina (A, right column), especially the outer nuclear layer that contains the rod and cone photoreceptors (O). Loss of the Sp1/Ets site dampens LacZ expression, particularly in the ganglionic layer (A, middle column). Derepression of the LacZ transgene by loss of the E2F site is not static but developmentally regulated in the RbP(E2FAAA)-LacZ lines (T1025 and T1052) (B). In RbP(WT)-and RbP(E2FAAA)-LacZ lines, LacZ expression is restricted to the inner neuroblastic layer (INL) at E16.5 and in the ganglionic cell layer at postnatal day zero (P0). However, mosaic LacZ expression with loss of the E2F site is evident in the outer photoreceptor layer by 5 weeks of age (P36) (top half of right column), eventually reaching robust levels in adulthood at 12 weeks of age (A, right column). RbP(WT)-LacZ lines (T028 and T157) do not express in the outer photoreceptor layer at P36 (lower half of right column) or at 12 weeks of age (A, left column).
Interestingly, derepression of the Rb promoter with loss of the E2F site is dynamic (Fig. 4B). At E16.5, only the inner neuroblastic layer (destined to become the ganglionic and inner nuclear layers) of the developing retina expresses LacZ in all of the RbP(E2FAAA)-LacZ and RbP(WT)-LacZ lines, and little more is evident by birth (P0), when at least four of the seven cell types are present in the retina (16). By 5 weeks of age (Fig. 4B, P36), retinal histogenesis is complete (all seven cell types are present), and the retina is postmitotic; yet mosaic expression of the LacZ transgene in the photoreceptor layer is obvious in two of the RbP(E2FAAA)-LacZ lines (T1025 and T1052) but not the RbP(WT)-LacZ lines (T028 and T157). By 12 weeks of age, the postmitotic retina displays robust LacZ expression in all retinal layers in these RbP(E2FAAA)-LacZ lines (Fig. 4A).
Similar to the situation in the neocortex and adult retina, two of the RbP(E2FAAA)-LacZ lines (T1052 and T1181) display elevated LacZ expression in the trigeminal ganglion (the fifth cranial nerve) at E16.5 relative to that seen in the RbP(WT)-LacZ lines (T028 and T157) (Fig. 5A). Not only is the number of LacZ-expressing neurons greater with loss of the E2F site, but the intensity of the individual LacZ positive neurons is also obviously stronger in the RbP(E2FAAA)-LacZ lines, as judged by the examination of X-Gal-stained, serial embryo cryosections (10 to 25 per embryo). None of the RbP(S/EAAA)-LacZ reporter lines exhibits frequent or robust LacZ expression in the trigeminal ganglion. These changes are also seen generally in other regions of the PNS, including the dorsal root ganglia at E16.5; however, levels of LacZ expression in the dorsal root ganglion cannot always be predicted by levels of LacZ observed in the trigeminal ganglion for all lines (data not shown).
FIG. 5.
Deregulation of the Rb promoter with loss of the E2F site in the PNS and muscle. At E16.5, expression of the RbP(WT)-LacZ transgene occurs in a subset of neurons of the trigeminal ganglion that belongs to the peripheral nervous system (PNS) (A, left column). Loss of the E2F site results in a larger subset of neurons expressing higher levels of LacZ activity in the trigeminal ganglion (A, right column). In contrast, loss of the Sp1/Ets site strongly suppresses the level and the number of neurons expressing LacZ in the trigeminal ganglion (A, middle column). In muscle surrounding the adult eye, RbP(WT)-LacZ transgene expression is either low or moderate (B, left column). Loss of the E2F site greatly derepresses LacZ expression (B, right column), while loss of the Sp1/Ets site results in no detectable LacZ expression (B, middle column). Thus, deregulation of the Rb promoter occurs inside and outside of the CNS at a limited number of sites.
Lastly, one of the RbP(E2FAAA)-LacZ lines (T1181, with 20 transgene copies) expresses in the peripheral neurons surrounding the whiskers (Fig. 3A and cryosection staining data not shown), a site where we along with others see low levels of staining of the RbP(WT)-LacZ lines (26); however, we attribute this difference to the presence of more integrated transgenes in the T1181 line rather than the loss of Rb promoter repression.
Deregulation of the Rb promoter outside of the nervous system.
Importantly, mutation of the E2F site in the Rb promoter does not lead to global derepression of the LacZ transgene, which is still predominantly expressed in the nervous system (Fig. 3A). Although not global, derepression of the Rb promoter with loss of the E2F site is apparent outside of the nervous system in adult muscle surrounding the eye (Fig. 5B). It is less clear whether mutation of the Sp1/Ets site in the Rb promoter compromises Rb promoter activation, since two of the RbP(WT)-LacZ reporters (T028 and T157) and all of the RbP(S/EAAA)-LacZ reporter lines do not express LacZ in the adult muscle surrounding the eye. The diaphragm and heart muscle display increased levels of LacZ in two of the RbP(E2FAAA)-LacZ reporter lines (T1025 and T1052; data not shown). Since developing muscle (e.g., somites and heart) does not express LacZ in any of our reporter lines, it is likely that, again, derepression of the Rb promoter is dynamic as described above for the retina. The absence of LacZ expression in developing muscle, for instance, could be due to the absence of a muscle-specific activator or coactivator (e.g., HCF-1) important for Rb expression in muscle (13). Alternatively, fixation of the muscle may be suboptimal, although we observe ample expression of the transgene in the CNS and PNS of these same embryos. Two of the RbP(E2FAAA)-LacZ mutant lines (T1052 and T1025) also express LacZ in the developing digits of whole-mount stained embryos (data not shown); however, cryosectioning revealed that the cells expressing LacZ in these two mutant lines do not appear to be the same. In summary, limited deregulation of the Rb promoter with loss of the E2F site occurs outside of the CNS and PNS in muscle.
Identifying putative activators and repressors of the Rb promoter in vitro and in vivo.
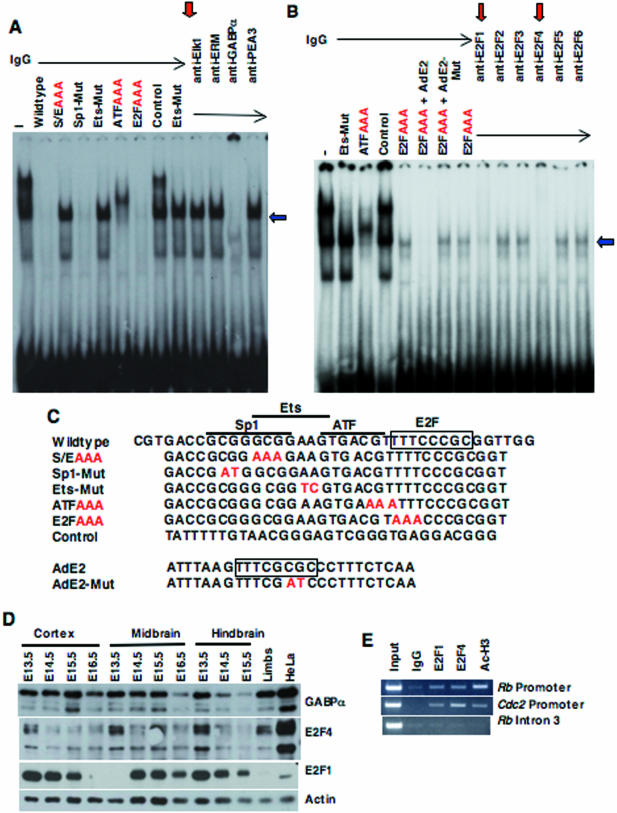
To identify activators and repressors of the Rb promoter present in the developing neocortex, we used gel shift analysis with neocortical nuclear extracts and a wild-type Rb promoter probe (38 bp) that spans the cluster of binding sites of interest. We can detect specific binding to the Sp1/Ets site or the ATF site as judged by competition analysis using wild-type and mutant double-stranded (DS) oligonucleotides containing triple (AAA) substitutions into the Sp1/Ets, ATF, or E2F sites of interest (Fig. 6A, left lanes). By using mutant DS oligonucleotides bearing substitutions in only the Sp1 or Ets site, we can show that it is the Ets site that is bound by a putative activator in the neocortical extracts at E15.5. Furthermore, we have used antibodies specific for different Ets family members to identify by supershift analysis the putative activator bound to the Ets site in the neocortical extracts as GABPα (Fig. 6A, right lanes). Thus, GABPα is likely to be the putative activator whose binding to the RbP(S/EAAA)-LacZ transgene is prevented in vivo. Previously, GABPα (also known as E4TF1 or RBF1) binding activity has been found on the Rb promoter in cell lines (46, 53), while Fli-1, another Ets family member, has been identified in an erythroblastic cell line (55). More recently, GABPα has been shown to activate Rb expression during myogenesis in vitro (13). However, GABPα is well expressed in most tissues (Fig. 6D), and, thus, it is unlikely to be responsible alone for tissue-specific activation of the Rb promoter.
FIG. 6.
Identification of putative regulators of the Rb Promoter. Gel shift analysis with neocortical nuclear extracts at E15.5 and an Rb promoter probe (wild-type, 38 bp) containing the Sp1, Ets, ATF, and E2F sites identified activator binding to the Rb promoter (A). Competition with wild-type or mutant DS oligonucleotides that inactivate these sites (individually or in combination) demonstrated efficient occupancy of the Ets and ATF sites (A, left lanes). Antibodies to various Ets family members were used for supershifts in the presence of the Ets mutant competitor, which identified GABPα as the activator bound to the critical Ets site on the Rb promoter (A, right lanes). (B) Gel shift analysis with the wild-type Rb promoter probe and neocortical extracts at E13.5 identified modest E2F activity that could be competed with the wild-type but not mutant adenovirus E2 DS oligonucleotides containing a classical E2F site. Antibodies specific for various E2F family members were used for supershifts in the presence of the E2F mutant competitor, which identified E2F1 and E2F4 binding to the Rb promoter (right lanes). (C) The sequence of all oligonucleotides used in panels A and B is displayed with mutated positions indicated in red. The E2F sites are indicated with a box, and the Sp1, Ets, and ATF sites are marked with bars above their positions in the wild-type Rb promoter probe. (D) Strong expression of GABPα, E2F1, and E2F4 is seen throughout the developing brain using Western blotting and actin as a loading control. (E) ChIP analysis demonstrates that E2F1 and E2F4, as well as acetylated histone H3 are bound to the Rb and Cdc2 promoters, but not to intron 3 of the Rb gene in the E13.5 embryonic brain.
Additionally, we can detect specific binding to the E2F site in the Rb promoter using E13.5 neocortical extracts and a different competitor for nonspecific DNA binding [sonicated herring sperm DNA rather than poly(dI-dC)] (Fig. 6B, middle lanes). By first competing other GABPα and ATF complexes from this 38-bp Rb promoter probe using the Rb promoter E2FAAA competitor, we were able to visualize modest E2F gel shift activity that was specifically competed by a wild-type, but not a mutant, adenovirus E2 competitor (7). Furthermore, we were able to supershift this E2F complex in the presence of the E2FAAA competitor using E2F-specific antibodies to E2F1 or E2F4 but not with antibodies to E2F2, E2F3, E2F5, or E2F6 (Fig. 6B, right lanes).
To measure the relative levels of E2F family members (E2f1 to E2f7), we generated cDNA pools from developing cortex (E13.5 and E15.5) and limbs (E15.5) and used real time RT-PCR with primer pairs specific for individual mouse E2F family members or for actin to amplify each gene of interest. After normalizing to actin, we expressed the relative ratios of each E2F family member in the E15.5 cortex/E13.5 cortex and the E15.5 cortex/E15.5 limbs (Table 1). Clearly, the developing cortex expresses all seven E2F family members; however, only E2F1 and E2F4 interact with the E2F site in the Rb promoter in vitro (Fig. 6B). Using Western blotting, E2F1 and E2F4 are present throughout the developing brain (Fig. 6D). Since E2F4 can be found in complex with all three Rb family members, the binding of E2F4 to the Rb promoter in neocortical extracts does not necessarily indicate which Rb family member is responsible for repression of the Rb promoter. However, the binding of E2F1 to the Rb promoter in neocortical extracts would appear to implicate pRB as the family member responsible for repression of the Rb promoter.
TABLE 1.
Relative expression of E2F family members in the developing cortex
| E2F | Relative ratio of expression:
|
|
|---|---|---|
| E15.5 cortex/E13.5 cortex | E15.5 cortex/limbs | |
| E2F1 | 0.525 | 0.260 |
| E2F2 | 0.310 | 0.216 |
| E2F3 | 2.602 | 0.815 |
| E2F4 | 1.083 | 0.539 |
| E2F5 | 1.029 | 0.571 |
| E2F6 | 0.579 | 0.630 |
| E2F7 | 0.519 | 0.330 |
To determine whether E2F1 or E2F4 binds to the Rb promoter in vivo, we performed ChIP analysis using E13.5 microdissected brains (Fig. 6E). First, we validated our ChIP procedure by demonstrating that acetylated histone H3, a marker of active genes, is bound to the Cdc2 promoter in vivo. Next, we showed that E2F1 and E2F4 are both bound to the Cdc2 promoter in vivo, as shown by others previously in cultured cells and in adult mouse tissues (44, 59). Finally, we showed that both E2F1 and E2F4 as well as acetylated histone H3 are bound to the Rb promoter in vivo (using primers that flank the cluster of binding sites containing the Ets, ATF, and E2F sites) but not to intron 3 of the Rb gene, a region lying ∼28 kb downstream of the Rb promoter (Fig. 6E). Thus, both E2F1 and E2F4 bind to the Rb promoter in vitro and in vivo.
In summary, this cluster of binding sites in the Rb promoter binds widely expressed factors, GABPα, E2F4, and E2F1 (bound to an Rb family member), interactions alone which are unlikely to dictate such narrow domains of LacZ expression in the cortex. A reasonable notion is that these factors are necessary but not sufficient to specify Rb promoter activity in the nervous system and are likely to require another factor, in combination with which the pattern of Rb promoter activity is specified. Whether the additional factor is an ATF family member remains to be seen, but it is less likely given the overlapping positions of the Ets and ATF sites in the Rb promoter. However, use of a truncated Rb promoter 200-bp fragment that includes the Sp1/Ets, ATF, and E2F sites is sufficient to drive nervous system expression (26).
Absence of Rb promoter autoregulation.
To determine whether Rb represses its own promoter, we crossed the RbP(WT)-LacZ lines with Rb+/− mice to generate Rb+/−; RbP(WT)-LacZ animals that were aged to allow the development of neuroendocrine tumors. Analysis of LacZ expression is hindered in Rb−/−; RbP(WT)-LacZ embryos (data not shown) due to the primary requirement for Rb in the placenta (61) and by the technical difficulty of visualizing LacZ activity in cryosections of E13.5 embryos. Instead, neuroendocrine tumors in Rb+/− mice, particularly tumors originating in the intermediate lobe of the pituitary, display loss of heterozygosity at the Rb locus (21, 25, 31). Thus, we expected to detect increased LacZ expression within the Rb-deficient tumors of Rb+/−; RbP(WT)-LacZ mice. Recently, we have reported that the C57BL/6 background enhances the development of thyroid C-cell tumors and tumors originating in the anterior lobe of the pituitary (33). Given the C57BL/6 background of our RbP-LacZ transgenic lines and the mixed 129Sv × C57BL/6 genetic background of our Rb+/− mice, we anticipated that our Rb+/−; RbP(WT)-LacZ animals would actually develop all three tumor types in which we could test for Rb promoter deregulation. This was the case, but, surprisingly, we did not detect increased LacZ expression in any of the neuroendocrine tumor types that developed in our Rb+/−; RbP(WT)-LacZ animals (n = 13 thyroid C-cell tumors; n = 3 tumors in the anterior lobe of the pituitary; n = 3 tumors in the intermediate lobe of the pituitary), strongly suggesting that Rb does not autoregulate its promoter in these tumor types (see Fig. S1 in the supplemental material). Normal neuroendocrine tissue expressed little if any LacZ activity in the Rb+/− or Rb+/+ background. Given that autoregulation of the RB promoter was first proposed from studies in human retinoblastomas (15, 20), it is possible that the absence of Rb promoter autoregulation in our transgenic lines is due to differences between humans and the mice or to differences between retinoblastomas and neuroendocrine tumors. Our detection of E2F1 binding to the Rb promoter in vitro (Fig. 6B, neocortical extracts) and in vivo (Fig. 6E, embryonic brain) suggests that differences in tissue-specific repression of the Rb promoter may be the most likely answer.
DISCUSSION
Given the high number of E2F target genes and the frequent deregulation of E2F activity in human tumors, it is important to consider how critical any single E2F site might be. We sought to test the in vivo significance of a single E2F site in a particularly crucial E2F target gene, that encoding the pRB tumor suppressor. Clearly, mutation of this E2F site in the Rb promoter deregulates expression in vivo, leading to elevated activity in regions of the CNS and PNS, as well as in specific muscle sites. Although derepression of the Rb promoter has been reported previously in tissue culture, our novel transgenic lines demonstrate how loss of the E2F site impacts Rb expression in all tissues throughout development and even in tumorigenesis; this has led us to several surprising conclusions.
First, loss of the E2F site does not lead to the global derepression of the Rb promoter in all tissues. Expression is not seen in fetal liver that contains the developing hematopoietic system nor is it seen in the developing gut, for instance. Second, loss of the E2F site does not give static derepression of the Rb promoter in sites at which we observed deregulation (e.g., the neocortex, retina, and muscle). In fact, in the retinal neuroepithelium, derepression is dynamic, beginning in patches within the photoreceptor layer 5 weeks after birth, well after proliferation has ceased in the eye (P10), and eventually becoming uniformly distributed throughout the photoreceptor layer by 12 weeks of age. These results cannot simply be explained by random transgene integration or the number of transgenes integrated in the wild-type or mutant RbP-LacZ lines. Instead, these results suggest that E2F-mediated repression is not actually needed unless some activator that is presumably in limiting concentrations becomes expressed. Such an activator is unlikely to be GABPα, which is expressed ubiquitously, but may be another activator or coactivator that depends or cooperates with GABPα for strong activation of the Rb promoter. Downstream of the E2F repressor site lies another Sp1 binding site that is found mutated in a separate cohort of low-penetrance retinoblastomas (10); however, this site is not conserved in the mouse Rb promoter. Alternatively, repression through the E2F site may be apparent only after another repressor is removed. During myogenesis in vitro, for instance, GABPα recruits the HC-1 coactivator to increase Rb expression, overcoming repression by another repressor, YY1 (13). There is an additional E2F site that lies ∼400 bp upstream of the Ets site of interest, which may mediate additional repression of the mouse Rb promoter; however, this site is not conserved in the human RB promoter. In summary, activation and repression enact a delicate balance in vivo to perform a temporally and spatially coordinated dance.
Third, loss of the E2F site does not lead to less activation of the Rb promoter in vivo, suggesting that this E2F site is not a switch allowing both E2F-mediated activation and pRB family mediated repression, as has been modeled previously for more complex, bifunctional E2F sites in the CcnE1 or Dhfr promoters. Rather, this E2F site appears to be a purely repressive module. The generation of an RbP(E2FAAA) mutant knock-in allele into the endogenous Rb locus would rigorously test the functional consequence of derepressing Rb expression through this critical E2F site during development.
Fourth, Rb transcription does not appear to be autoregulated through pRB binding to this critical E2F site in neuroendocrine tumors developing in Rb+/−; RbP(WT)-LacZ reporter lines, although previous reports had suggested that pRB binds and represses its own promoter in human retinoblastomas (15, 20). Neuroendocrine tumors in Rb+/−; RbP(WT)-LacZ mice develop after loss of heterozygosity at the Rb locus, yet such tumors do not display increased LacZ expression. It is important to note that our RbP(WT)-LacZ lines do not express uniformly in normal neuroendocrine tissue, which may be linked to the inability to visualize deregulation of this transgene in neuroendocrine tumors in the Rb+/−; RbP(WT)-LacZ lines. Loss of Rb may indirectly affect Rb transcription by changing the levels of activators or repressors. For example, levels of p107 mRNA are known to increase with loss of Rb (23, 47); this involves loss of direct binding of pRB to the p107 promoter (60), but this would lead to less Rb transcription if p107 were to be part of a repressor complex for the Rb promoter.
We have identified both E2F1 and E2F4 bound to the Rb promoter in vitro and in vivo, suggesting that multiple pRB family members may cooperate to repress the Rb promoter in the nervous system. While p107/E2F4 and p130/E2F4 complexes bind the human RBL2 (p107) promoter, they are not found on the human RB promoter in cycling cells in vitro using ChIP-on-chip analysis (2). Interestingly, both E2F4 and E2F1 are bound to the human RB promoter in quiescent fibroblasts, suggesting that multiple pRB family member complexes may interact with the E2F site in the RB promoter (44, 58). In contrast to the situation in neuroendocrine tumors, pRB may bind E2F1 and E2F4 to repress the Rb promoter in embryonic tissues such as the developing brain. Upon loss of Rb and E2F4, E2F1 can form complexes with p107, suggesting that substantial flexibility exists within the Rb and E2f families to compensate for the loss of various family members (30). The use of a conditional Rb allele in combination with our RbP(WT)-LacZ lines would allow the requirement for Rb in repression of the Rb promoter to be tested during development. Additionally, the p107 or p130 deficiency can be combined with the RbP(WT)-LacZ lines to test the requirement for p107 or p130 in repression of the Rb promoter in vivo.
Besides the CNS and PNS, Rb mRNA is evident in the fetal liver (26), where little if any LacZ expression is detected in our RbP(WT)-LacZ lines. Since we can detect pRB immunoreactivity in fetal liver using immunohistochemistry and Western blotting and can detect full-length Rb mRNA using RT-PCR from fetal liver (data not shown), it is possible that the 4.3-kb genomic fragment used to construct the Rb promoter transgenic constructs is lacking a region that dictates high-level expression outside of the nervous system. Other possible explanations include alternative promoter usage or increased translation or stability of Rb mRNA in the muscle and fetal liver. Also, an alternative form of pRB, ΔRB-p70, has been described recently in the myeloid lineage, a major component of the fetal liver, that is thought to result from usage of alternative, downstream AUG codons found in the Rb mRNA transcript containing at least exon 2 to exon 27 (34). The reason for this discrepancy is yet to be resolved.
Approximately 50% of human tumors deregulate the RB pathway by overexpressing G1 cyclin/CDK activity or by inactivating INK4A/INK4B and or degrading CIP/KIP family members that normally restrain G1 cyclin/CDK activity (50). This means that in half of human tumors, no RB mutations have been detected, but endogenous levels of pRB are simply overwhelmed by G1 cyclin/CDK-mediated phosphorylation of pRB. One of our long-term interests is to deregulate the RB promoter in this subset of human tumors that retain a wild-type RB allele. Dialing up levels of pRB may help restore pRB-mediated tumor suppression by simply increasing the substrate (pRB) to make the cyclin/CDK complexes again limiting. Indeed, overexpression of pRB in transgenic mice using the human RB promoter leads to dwarfism, yet it also leads to protection from neuroendocrine tumorigenesis (3, 8). Given the roles of pRB in promoting cell cycle arrest and differentiation, it is possible that only a transient rise in pRB expression will induce a more slowly growing and/or more differentiated tumor cell, leading to a less aggressive tumor with a better clinical prognosis.
Finally, the dispensability of the G1 cyclins (D or E type) and G1 CDKs (Cdk4/6 or Cdk2) throughout most or all tissues during mouse development has challenged the field to propose alternative mechanisms for pRB-mediated growth control (42, 51). This study proposes one such alternative, in that the levels of pRB are not static during development, though they change very little during the oft-used fibroblast cell culture models. Thus, it is quite possible that G1 cyclin/CDKs are dispensable due to the capacity of the temporal and spatial fluctuation of pRB levels during development to control growth. One mechanism for fluctuating pRB levels is clearly transcriptional control, as we have demonstrated, perhaps through the normal regulation of the activators and repressors of the Rb promoter. However, equally plausible mechanisms for fluctuating pRB levels during development include use of an alternate Rb promoter, differential stability of pRB protein or Rb mRNA, and differential translational control of Rb mRNA. These seemingly basic possibilities should be revisited, given the obviously nonessential nature of the G1 cyclins and CDKs in most developing tissues.
Supplementary Material
Acknowledgments
This work was supported by grants from the Pew Scholars Program in the Biomedical Sciences, the Human Frontiers Science Program, the Charlotte Geyer Foundation, and the National Institutes of Health (R01-CA079646).
We thank R. Palmiter for supplying the pnlacF vector, and we acknowledge A. Auerbach and A. Joyner of the New York University Transgenic Facility for the injection of the transgene constructs and L. Yang of the Histopathology Core Facility of the Herbert Irving Cancer Center at Columbia University for the preparation of serial cryosections. We acknowledge S. Leung, A. Fitzmaurice, P. Nguyen, A. Beltran, and R. Bruckner for their assistance. We also thank M. Classon, C. Prives, D. Wolgemuth, D. Kalderon, R. Prywes, and M. Pagano for helpful discussions. M.A. thanks her family and J. Martin-Serrano for their support. L.Y. thanks Isabella and Michele and Marie Yamasaki for their continual support and encouragement.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adams, M. R., R. Sears, F. Nuckolls, G. Leone, and J. R. Nevins. 2000. Complex transcriptional regulatory mechanisms control expression of the E2F3 locus. Mol. Cell. Biol. 20:3633-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balciunaite, E., A. Spektor, N. H. Lents, H. Cam, H. Te Riele, A. Scime, M. A. Rudnicki, R. Young, and B. D. Dynlacht. 2005. Pocket protein complexes are recruited to distinct targets in quiescent and proliferating cells. Mol. Cell. Biol. 25:8166-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bignon, Y. J., Y. Chen, C. Y. Chang, D. J. Riley, J. J. Windle, P. L. Mellon, and W. H. Lee. 1993. Expression of a retinoblastoma transgene results in dwarf mice. Genes Dev. 7:1654-1662. [DOI] [PubMed] [Google Scholar]
- 4.Bookstein, R., P. Rio, S. A. Madreperla, F. Hong, C. Allred, W. E. Grizzle, and W. H. Lee. 1990. Promoter deletion and loss of retinoblastoma gene expression in human prostate carcinoma. Proc. Natl. Acad. Sci. USA 87:7762-7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bracken, A. P., M. Ciro, A. Cocito, and K. Helin. 2004. E2F target genes: unraveling the biology. Trends Biochem. Sci. 29:409-417. [DOI] [PubMed] [Google Scholar]
- 6.Cam, H., and B. D. Dynlacht. 2003. Emerging roles for E2F: beyond the G1/S transition and DNA replication. Cancer Cell 3:311-316. [DOI] [PubMed] [Google Scholar]
- 7.Cao, L., B. Faha, M. Dembski, L. H. Tsai, E. Harlow, and N. Dyson. 1992. Independent binding of the retinoblastoma protein and p107 to the transcription factor E2F. Nature 355:176-179. [DOI] [PubMed] [Google Scholar]
- 8.Chang, C. Y., D. J. Riley, E. Y. Lee, and W. H. Lee. 1993. Quantitative effects of the retinoblastoma gene on mouse development and tissue-specific tumorigenesis. Cell Growth Differ. 4:1057-1064. [PubMed] [Google Scholar]
- 9.Christensen, J., P. Cloos, U. Toftegaard, D. Klinkenberg, A. P. Bracken, E. Trinh, M. Heeran, L. Di Stefano, and K. Helin. 2005. Characterization of E2F8, a novel E2F-like cell-cycle regulated repressor of E2F-activated transcription. Nucleic Acids Res. 33:5458-5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cowell, J. K., B. Bia, and A. Akoulitchev. 1996. A novel mutation in the promoter region in a family with a mild form of retinoblastoma indicates the location of a new regulatory domain for the RB1 gene. Oncogene 12:431-436. [PubMed] [Google Scholar]
- 11.de Bruin, A., B. Maiti, L. Jakoi, C. Timmers, R. Buerki, and G. Leone. 2003. Identification and characterization of E2F7, a novel mammalian E2F family member capable of blocking cellular proliferation. J. Biol. Chem. 278:42041-42049. [DOI] [PubMed] [Google Scholar]
- 12.de Bruin, A., L. Wu, H. I. Saavedra, P. Wilson, Y. Yang, T. J. Rosol, M. Weinstein, M. L. Robinson, and G. Leone. 2003. Rb function in extraembryonic lineages suppresses apoptosis in the CNS of Rb-deficient mice. Proc. Natl. Acad. Sci. USA 100:6546-6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delehouzee, S., T. Yoshikawa, C. Sawa, J. Sawada, T. Ito, M. Omori, T. Wada, Y. Yamaguchi, Y. Kabe, and H. Handa. 2005. GABP, HCF-1 and YY1 are involved in Rb gene expression during myogenesis. Genes Cells 10:717-731. [DOI] [PubMed] [Google Scholar]
- 14.Di Stefano, L., M. R. Jensen, and K. Helin. 2003. E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. EMBO J. 22:6289-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunn, J. M., R. A. Phillips, X. Zhu, A. Becker, and B. L. Gallie. 1989. Mutations in the RB1 gene and their effects on transcription. Mol. Cell. Biol. 9:4596-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dyer, M. A., and R. Bremner. 2005. The search for the retinoblastoma cell of origin. Nat. Rev. Cancer 5:91-101. [DOI] [PubMed] [Google Scholar]
- 17.Dyer, M. A., and C. L. Cepko. 2001. Regulating proliferation during retinal development. Nat. Rev. Neurosci. 2:333-342. [DOI] [PubMed] [Google Scholar]
- 18.Ferguson, K. L., J. L. Vanderluit, J. M. Hebert, W. C. McIntosh, E. Tibbo, J. G. MacLaurin, D. S. Park, V. A. Wallace, M. Vooijs, S. K. McConnell, and R. S. Slack. 2002. Telencephalon-specific Rb knockouts reveal enhanced neurogenesis, survival and abnormal cortical development. EMBO J. 21:3337-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gill, R. M., P. A. Hamel, J. Zhe, E. Zacksenhaus, B. L. Gallie, and R. A. Phillips. 1994. Characterization of the human RB1 promoter and of elements involved in transcriptional regulation. Cell Growth Differ. 5:467-474. [PubMed] [Google Scholar]
- 20.Hamel, P. A., R. M. Gill, R. A. Phillips, and B. L. Gallie. 1992. Transcriptional repression of the E2-containing promoters EIIaE, c-myc, and RB1 by the product of the RB1 gene. Mol. Cell. Biol. 12:3431-3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison, D. J., M. L. Hooper, J. F. Armstrong, and A. R. Clarke. 1995. Effects of heterozygosity for the Rb-1t19neo allele in the mouse. Oncogene 10:1615-1620. [PubMed] [Google Scholar]
- 22.Hsiao, K. M., S. L. McMahon, and P. J. Farnham. 1994. Multiple DNA elements are required for the growth regulation of the mouse E2F1 promoter. Genes Dev. 8:1526-1537. [DOI] [PubMed] [Google Scholar]
- 23.Hurford, R. K., Jr., D. Cobrinik, M. H. Lee, and N. Dyson. 1997. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 11:1447-1463. [DOI] [PubMed] [Google Scholar]
- 24.Ishida, S., E. Huang, H. Zuzan, R. Spang, G. Leone, M. West, and J. R. Nevins. 2001. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 21:4684-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacks, T., A. Fazeli, E. M. Schmitt, R. T. Bronson, M. A. Goodell, and R. A. Weinberg. 1992. Effects of an Rb mutation in the mouse. Nature 359:295-300. [DOI] [PubMed] [Google Scholar]
- 26.Jiang, Z., Z. Guo, F. A. Saad, J. Ellis, and E. Zacksenhaus. 2001. Retinoblastoma gene promoter directs transgene expression exclusively to the nervous system. J. Biol. Chem. 276:593-600. [DOI] [PubMed] [Google Scholar]
- 27.Jiang, Z., E. Zacksenhaus, B. L. Gallie, and R. A. Phillips. 1997. The retinoblastoma gene family is differentially expressed during embryogenesis. Oncogene 14:1789-1797. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, D. G., K. Ohtani, and J. R. Nevins. 1994. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 8:1514-1525. [DOI] [PubMed] [Google Scholar]
- 29.Kohn, M. J., R. T. Bronson, E. Harlow, N. J. Dyson, and L. Yamasaki. 2003. Dp1 is required for extra-embryonic development. Development 130:1295-1305. [DOI] [PubMed] [Google Scholar]
- 30.Lee, E. Y., H. Cam, U. Ziebold, J. B. Rayman, J. A. Lees, and B. D. Dynlacht. 2002. E2F4 loss suppresses tumorigenesis in Rb mutant mice. Cancer Cell 2:463-472. [DOI] [PubMed] [Google Scholar]
- 31.Lee, E. Y., C. Y. Chang, N. Hu, Y. C. Wang, C. C. Lai, K. Herrup, W. H. Lee, and A. Bradley. 1992. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature 359:288-294. [DOI] [PubMed] [Google Scholar]
- 32.Lee, E. Y., N. Hu, S. S. Yuan, L. A. Cox, A. Bradley, W. H. Lee, and K. Herrup. 1994. Dual roles of the retinoblastoma protein in cell cycle regulation and neuron differentiation. Genes Dev. 8:2008-2021. [DOI] [PubMed] [Google Scholar]
- 33.Leung, S. W., E. H. Wloga, A. F. Castro, T. Nguyen, R. T. Bronson, and L. Yamasaki. 2004. A dynamic switch in Rb+/− mediated neuroendocrine tumorigenesis. Oncogene 23:3296-3307. [DOI] [PubMed] [Google Scholar]
- 34.Liu, H. P., A. M. Thompson, and K. F. Macleod. 2005. A novel form of pRb expressed during normal myelopoiesis and in tumour-associated macrophages. Cell Prolif. 38:13-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lyons, T. E., M. Salih, and B. S. Tuana. 2006. Activating E2Fs mediate the transcriptional regulation of the human E2F6 repressor. Am. J. Physiol. Cell Physiol. 290:C189-C199. [DOI] [PubMed] [Google Scholar]
- 36.Ma, Y., R. Croxton, R. L. Moorer, Jr., and W. D. Cress. 2002. Identification of novel E2F1-regulated genes by microarray. Arch. Biochem. Biophys. 399:212-224. [DOI] [PubMed] [Google Scholar]
- 37.MacPherson, D., J. Sage, D. Crowley, A. Trumpp, R. T. Bronson, and T. Jacks. 2003. Conditional mutation of Rb causes cell cycle defects without apoptosis in the central nervous system. Mol. Cell. Biol. 23:1044-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mercer, E. H., G. W. Hoyle, R. P. Kapur, R. L. Brinster, and R. D. Palmiter. 1991. The dopamine beta-hydroxylase gene promoter directs expression of E. coli lacZ to sympathetic and other neurons in adult transgenic mice. Neuron 7:703-716. [DOI] [PubMed] [Google Scholar]
- 39.Muller, H., A. P. Bracken, R. Vernell, M. C. Moroni, F. Christians, E. Grassilli, E. Prosperini, E. Vigo, J. D. Oliner, and K. Helin. 2001. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 15:267-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohtani-Fujita, N., T. Fujita, R. Takahashi, P. D. Robbins, T. P. Dryja, and T. Sakai. 1994. A silencer element in the retinoblastoma tumor-suppressor gene. Oncogene 9:1703-1711. [PubMed] [Google Scholar]
- 41.Okuyama, Y., Y. Sowa, T. Fujita, T. Mizuno, H. Nomura, T. Nikaido, T. Endo, and T. Sakai. 1996. ATF site of human RB gene promoter is a responsive element of myogenic differentiation. FEBS Lett. 397:219-224. [DOI] [PubMed] [Google Scholar]
- 42.Pagano, M., and P. K. Jackson. 2004. Wagging the dogma; tissue-specific cell cycle control in the mouse embryo. Cell 118:535-538. [DOI] [PubMed] [Google Scholar]
- 43.Polager, S., Y. Kalma, E. Berkovich, and D. Ginsberg. 2002. E2Fs up-regulate expression of genes involved in DNA replication, DNA repair and mitosis. Oncogene 21:437-446. [DOI] [PubMed] [Google Scholar]
- 44.Ren, B., H. Cam, Y. Takahashi, T. Volkert, J. Terragni, R. A. Young, and B. D. Dynlacht. 2002. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 16:245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakai, T., N. Ohtani, T. L. McGee, P. D. Robbins, and T. P. Dryja. 1991. Oncogenic germline mutations in Sp1 and ATF sites in the human retinoblastoma gene. Nature 353:83-86. [DOI] [PubMed] [Google Scholar]
- 46.Savoysky, E., T. Mizuno, Y. Sowa, H. Watanabe, J. Sawada, H. Nomura, Y. Ohsugi, H. Handa, and T. Sakai. 1994. The retinoblastoma binding factor 1 (RBF-1) site in RB gene promoter binds preferentially E4TF1, a member of the Ets transcription factors family. Oncogene 9:1839-1846. [PubMed] [Google Scholar]
- 47.Schneider, J. W., W. Gu, L. Zhu, V. Mahdavi, and B. Nadal-Ginard. 1994. Reversal of terminal differentiation mediated by p107 in Rb−/− muscle cells. Science 264:1467-1471. [DOI] [PubMed] [Google Scholar]
- 48.Sears, R., K. Ohtani, and J. R. Nevins. 1997. Identification of positively and negatively acting elements regulating expression of the E2F2 gene in response to cell growth signals. Mol. Cell. Biol. 17:5227-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shan, B., C. Y. Chang, D. Jones, and W. H. Lee. 1994. The transcription factor E2F-1 mediates the autoregulation of RB gene expression. Mol. Cell. Biol. 14:299-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sherr, C. J., and F. McCormick. 2002. The RB and p53 pathways in cancer. Cancer Cell 2:103-112. [DOI] [PubMed] [Google Scholar]
- 51.Sherr, C. J., and J. M. Roberts. 2004. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 18:2699-2711. [DOI] [PubMed] [Google Scholar]
- 52.Slack, R. S., P. A. Hamel, T. S. Bladon, R. M. Gill, and M. W. McBurney. 1993. Regulated expression of the retinoblastoma gene in differentiating embryonal carcinoma cells. Oncogene 8:1585-1591. [PubMed] [Google Scholar]
- 53.Sowa, Y., Y. Shiio, T. Fujita, T. Matsumoto, Y. Okuyama, D. Kato, J. Inoue, J. Sawada, M. Goto, H. Watanabe, H. Handa, and T. Sakai. 1997. Retinoblastoma binding factor 1 site in the core promoter region of the human RB gene is activated by hGABP/E4TF1. Cancer Res. 57:3145-3148. [PubMed] [Google Scholar]
- 54.Spencer, C., S. Pajovic, H. Devlin, Q. D. Dinh, T. W. Corson, and B. L. Gallie. 2005. Distinct patterns of expression of the RB gene family in mouse and human retina. Gene Expr. Patterns 5:687-694. [DOI] [PubMed] [Google Scholar]
- 55.Tamir, A., J. Howard, R. R. Higgins, Y. J. Li, L. Berger, E. Zacksenhaus, M. Reis, and Y. Ben-David. 1999. Fli-1, an Ets-related transcription factor, regulates erythropoietin-induced erythroid proliferation and differentiation: evidence for direct transcriptional repression of the Rb gene during differentiation. Mol. Cell. Biol. 19:4452-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trimarchi, J. M., and J. A. Lees. 2002. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell. Biol. 3:11-20. [DOI] [PubMed] [Google Scholar]
- 57.Weinmann, A. S., P. S. Yan, M. J. Oberley, T. H. Huang, and P. J. Farnham. 2002. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev. 16:235-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wells, J., and P. J. Farnham. 2002. Characterizing transcription factor binding sites using formaldehyde crosslinking and immunoprecipitation. Methods 26:48-56. [DOI] [PubMed] [Google Scholar]
- 59.Wells, J., C. R. Graveel, S. M. Bartley, S. J. Madore, and P. J. Farnham. 2002. The identification of E2F1-specific target genes. Proc. Natl. Acad. Sci. USA 99:3890-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams, J. P., T. Stewart, B. Li, R. Mulloy, D. Dimova, and M. Classon. 2006. The retinoblastoma protein is required for Ras-induced oncogenic transformation. Mol. Cell. Biol. 26:1170-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu, L., A. de Bruin, H. I. Saavedra, M. Starovic, A. Trimboli, Y. Yang, J. Opavska, P. Wilson, J. C. Thompson, M. C. Ostrowski, T. J. Rosol, L. A. Woollett, M. Weinstein, J. C. Cross, M. L. Robinson, and G. Leone. 2003. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature 421:942-947. [DOI] [PubMed] [Google Scholar]
- 62.Zacksenhaus, E., R. M. Gill, R. A. Phillips, and B. L. Gallie. 1993. Molecular cloning and characterization of the mouse RB1 promoter. Oncogene 8:2343-2351. [PubMed] [Google Scholar]
- 63.Zajaczek, S., A. Jakubowska, G. Kurzawski, Z. Krzystolik, and J. Lubinski. 1998. Age at diagnosis to discriminate those patients for whom constitutional DNA sequencing is appropriate in sporadic unilateral retinoblastoma. Eur. J. Cancer 34:1919-1921. [DOI] [PubMed] [Google Scholar]
- 64.Zhang, J., J. Gray, L. Wu, G. Leone, S. Rowan, C. L. Cepko, X. Zhu, C. M. Craft, and M. A. Dyer. 2004. Rb regulates proliferation and rod photoreceptor development in the mouse retina. Nat. Genet. 36:351-360. [DOI] [PubMed] [Google Scholar]
- 65.Zhu, L., E. Xie, and L. S. Chang. 1995. Differential roles of two tandem E2F sites in repression of the human p107 promoter by retinoblastoma and p107 proteins. Mol. Cell. Biol. 15:3552-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu, W., P. H. Giangrande, and J. R. Nevins. 2005. Temporal control of cell cycle gene expression mediated by E2F transcription factors. Cell Cycle 4:633-636. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.