Abstract
The NOTCH1 receptor is cleaved within its extracellular domain by furin during its maturation, yielding two subunits that are held together noncovalently by a juxtamembrane heterodimerization (HD) domain. Normal NOTCH1 signaling is initiated by the binding of ligand to the extracellular subunit, which renders the transmembrane subunit susceptible to two successive cleavages within and C terminal to the heterodimerization domain, catalyzed by metalloproteases and γ-secretase, respectively. Because mutations in the heterodimerization domain of NOTCH1 occur frequently in human T-cell acute lymphoblastic leukemia (T-ALL), we assessed the effect of 16 putative tumor-associated mutations on Notch1 signaling and HD domain stability. We show here that 15 of the 16 mutations activate canonical NOTCH1 signaling. Increases in signaling occur in a ligand-independent fashion, require γ-secretase activity, and correlate with an increased susceptibility to cleavage by metalloproteases. The activating mutations cause soluble NOTCH1 heterodimers to dissociate more readily, either under native conditions (n = 3) or in the presence of urea (n = 11). One mutation, an insertion of 14 residues immediately N terminal to the metalloprotease cleavage site, increases metalloprotease sensitivity more than all others, despite a negligible effect on heterodimer stability by comparison, suggesting that the insertion may expose the S2 site by repositioning it relative to protective NOTCH1 ectodomain residues. Together, these studies show that leukemia-associated HD domain mutations render NOTCH1 sensitive to ligand-independent proteolytic activation through two distinct mechanisms.
The development of multicellular organisms is orchestrated by a limited number of highly conserved signaling pathways. One such pathway involves NOTCH receptors and downstream mediators, which can variously regulate the specification of cell fate, proliferation, self-renewal, survival, and apoptosis in a dose- and context-dependent fashion (3, 47).
Like other members of the NOTCH receptor family, mammalian NOTCH1 is a large multimodular type I transmembrane glycoprotein (Fig. 1A). During maturation, NOTCH1 undergoes proteolytic processing by furin at a site termed S1 that lies ∼70 amino acids external to the transmembrane domain (25), yielding two noncovalently associated extracellular (NEC) and transmembrane (NTM) subunits (6, 25, 37). NEC contains 36 N-terminal epidermal growth factor (EGF)-like repeats that participate in binding to ligands (23, 39, 51) and three iterated LIN-12/NOTCH repeats (LNR), which help to maintain NOTCH receptors in the “off” state prior to ligand binding (13, 24, 40). The association of NEC and NTM is mediated by sequences lying immediately N terminal (HD-N) and C terminal (HD-C) of site S1; together, these sequences constitute the NOTCH subunit association, or “heterodimerization” (HD) domain (40).
FIG. 1.
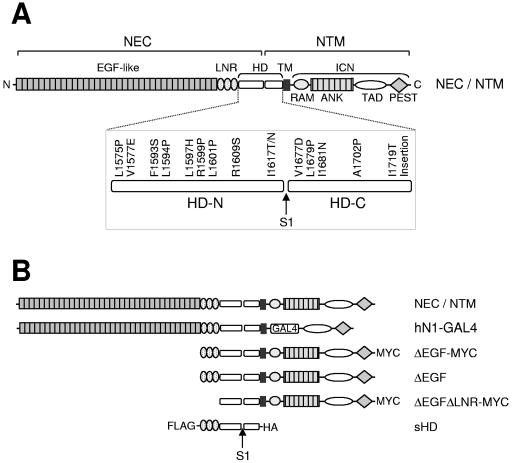
NOTCH1 expression constructs. (A) Schematic representation of the human NOTCH1 receptor. NEC, NOTCH1 extracellular subunit; NTM, NOTCH1 transmembrane subunit; LNR, domain comprising the three LIN12/Notch repeats; HD, heterodimerization domain; HD-N, N-terminal region of the HD domain; HD-C, C-terminal region of the HD domain; TM, transmembrane segment; ICN, intracellular NOTCH1; RAM, RAM domain; ANK, ankyrin repeat domain; TAD, transactivation domain; PEST, PEST domain; S1, furin cleavage site. The 16 leukemia-derived HD domain mutations described in this article are shown. (B) NOTCH1 polypeptides used in these studies.
Binding of ligands to NEC triggers two sequential proteolytic events within the NTM subunit at sites S2 and S3. S2 cleavage occurs just external to the transmembrane domain and is catalyzed by ADAM-type metalloproteases (8, 29). This creates a short-lived intermediate, NTM*, which appears to be recognized through its amino terminus by nicastrin (44), a component of a multiprotein enzyme complex called γ-secretase. NTM* is then cleaved by γ-secretase at several sites within the transmembrane domain (10, 19, 42). The ultimate cleavage (at site S3) releases the intracellular domain of NOTCH1 (ICN1) from the membrane, allowing it to translocate to the nucleus and activate the transcription of target genes through its interaction with the DNA-binding factor CSL [CBF-1/Su(H)/Lag-1] (17, 20, 45, 52) and coactivator proteins of the mastermind family (33, 50).
Although the central events that are necessary for transduction of NOTCH signals are now established, it is still unclear how ligand binding facilitates S2 cleavage, which is an essential prerequisite to S3 cleavage (8, 29). Models involving receptor oligomerization have been proposed, but recent data suggest that both metalloprotease-sensitive and -resistant forms of NOTCH receptors exist primarily as monomers (46). An alternative mechanism supposes that NOTCH1 is rendered sensitive to S2 cleavage by a change in conformation (46), which could occur in response to mechanical forces generated by the endocytic machinery of the ligand-expressing cell (1, 22, 30, 43).
One well-characterized function of NOTCH1 is to direct T-cell development, which relies on NOTCH1 signals at several stages (26, 36). Dysregulated NOTCH1 signaling also figures prominently in T-cell acute lymphoblastic leukemia (T-ALL), an aggressive tumor derived from T-cell progenitors. NOTCH1 was identified initially through its involvement in a rare (7, 9) chromosomal translocation found in human T-ALL (11), and constitutively active forms of NOTCH1 (such as ICN1) are potent inducers of T-ALL in murine models (5, 31). More recent work has established that human T-ALLs often harbor mutations in NOTCH1 (48). The most frequent mutations are single-amino-acid substitutions and small in-frame deletions and insertions in the HD domain, found in both the HD-N and HD-C regions of NEC and NTM, respectively (Fig. 1A).
Elucidating how T-ALL-associated HD domain mutations cause pathophysiologic increases in NOTCH1 function is potentially important in several regards. Such mutations promise to provide further information on how NOTCH receptors are maintained in the “off” state, and some of these insights could prove relevant to understanding how normal receptor activation occurs. Further, because the NOTCH1 signaling pathway is a tractable therapeutic target in T-ALL and possibly other cancers as well, a mechanistic understanding of HD mutations could yield new therapeutic opportunities. We thus undertook the present studies to confirm that T-ALL-associated HD domain mutations cause increased NOTCH1 signaling and to investigate how such increases occur.
MATERIALS AND METHODS
Expression plasmids.
A diagram depicting the various forms of NOTCH1 used in these studies is shown in Fig. 1B. Expression constructs that encode full-length human NOTCH1 (residues M1 to K2555), ΔEGF, a form bearing a deletion that removes the coding region for EGF-like repeats 1 to 36 (residues R23 to I1446), and sHD, a secreted form of NOTCH1 consisting of the leader peptide (M1-P22) fused to the three NOTCH1 LIN-12/NOTCH repeats and the heterodimerization domain (residues E1447 to Q1734), have been described previously (40). These cDNAs were assembled in pcDNA3 (Invitrogen). Mutations were introduced using the QuickChange kit (Stratagene). In some instances, FLAG, hemagglutinin (HA), or c-MYC epitope tag sequences were also introduced at the 5′ or 3′ end of the NOTCH1 cDNAs (Fig. 1B).
Expression of Notch proteins in cultured cells.
U2OS and 293T cells (American Type Culture Collection) were transfected transiently using Lipofectamine 2000 or Lipofectamine Plus reagent (Invitrogen). U2OS and 293T cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), 2 mM l-glutamine (Invitrogen), 100 U of penicillin/ml (Invitrogen), and 100 μg of streptomycin/ml (Invitrogen). Cells were grown at 37°C under 5% CO2.
Transcriptional activation assays.
NOTCH1 expression plasmids were introduced into U2OS cells by transient transfection with Lipofectamine Plus and assessed for their ability to activate a NOTCH-sensitive luciferase reporter gene, as described previously (5). Briefly, cells in 24-well dishes were cotransfected in triplicate with 10 ng of various pcDNA3-NOTCH1 expression constructs, a NOTCH-sensitive firefly luciferase reporter gene (15), and an internal control Renilla luciferase plasmid (Promega). Total introduced DNA was kept constant by adding empty pcDNA3 plasmid. Normalized firefly luciferase activities were measured in whole-cell extracts prepared 44 to 48 h after transfection using the Dual Luciferase kit (Promega) and a specially configured luminometer (Turner Systems). In some experiments, the cells were treated posttransfection with the γ-secretase inhibitor compound E (kind gift of Michael Wolfe) at 1 μM or with carrier alone (0.01% dimethyl sulfoxide).
Ligand stimulation assays.
Chimeric expression constructs were created by ligating a cDNA encoding the DNA-binding domain of Gal4 into a NOTCH1 cDNA cut with the restriction enzymes Bsu36I and NcoI (which removes the coding sequence for most of the RAM domain and all seven ankyrin repeats). NOTCH1-Gal4 cDNAs were assembled in the vector pcDNA5-FRT-TO (invitrogen) and transfected into U2OS TRex cells (kind gift of Jeff Parvin, Department of Pathology, Brigham and Women's Hospital) together with the plasmid pOG44 (Invitrogen), which encodes Flp recombinase. This strain of U2OS cells expresses the Tet repressor and contains a single genomic FRT (Flp recombinase target) site, which permits creation of isogenic recombinants containing a single transgene under the control of tetracycline. After selection with hygromycin, recombinant cells were tested for ligand-dependent and -independent responsiveness as follows. On day 1, cells in 60-mm dishes of U2OS cells were transfected with a mixture containing 1 μg of a GAL4-firefly luciferase reporter and 20 ng of an internal control pRL-TK Renilla luciferase plasmid (Invitrogen). On day 2, the U2OS cells were split onto either OP9 or OP9-DLL1 cells (kind gift of Juan-Carlos Zuniga-Pflücker) in the presence or absence of 1 μg/ml of tetracycline or carrier (70% ethanol) alone. On day 3, dual-luciferase assays were performed on whole-cell lysates. All data points within experiments were obtained in triplicate, and all experiments were repeated three times.
S2 cleavage assays.
The sensitivity of various NOTCH1 polypeptides to cleavage by metalloproteases at site S2 was assessed as described previously (40). Briefly, U2OS cells in 60-mm dishes were transfected transiently with 1 μg of pcDNA3-ΔEGF-MYC plasmids. One day after transfection, the cells were treated with compound E or DAPI (4′,6′-diamidino-2-phenylindole) (1 μM) for 18 h, which acts to stabilize the product of S2 cleavage, NTM*. Cells were then washed once with ice-cold Hank's buffered saline and lysed in buffer A (50 mM Tris, pH 7.5, 100 mM NaCl, 10 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotonin, 10 μg/ml leupeptin, and 1% NP-40) for 15 min on ice. Following centrifugation at 16,000 g for 15 min at 4°C, cleared whole-cell lysates (2 × 106 cells) were incubated for 2 h at 4°C with anti-MYC antibody (9E10) cross-linked to protein G-Sepharose beads (Sigma). After washing twice with buffer A and once with 50 mM Tris, pH 7.5, 100 mM NaCl, the beads were treated with lambda protein phosphatase (New England BioLabs) as recommended by the manufacturer. Following sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), the immunoprecipated polypeptides were stained on Western blots using an antibody against the intracellular domain of NOTCH1 (4).
Heterodimer stability assays.
293T cells in 60-mm dishes were transiently transfected with 1 μg of empty pcDNA3 plasmid or with pcDNA3 plasmids encoding soluble NOTCH1 mini-receptors. To enable detection, N-terminal FLAG and C-terminal HA tags were introduced into each of these polypeptides, which are converted by furin cleavage into heterodimers composed of FLAG-LNR-(HD-N) and (HD-C)-HA (40). Three days after transfection, the conditioned media were cleared by centrifugation at 500 × g and split into two aliquots that were incubated with anti-HA or anti-FLAG beads (Sigma) at 4°C for 2 h. The beads were then washed three times with 1 ml of buffer B (50 mM Tris, pH 7.5, 150 mM NaCl, 0.1% NP-40), and the bound proteins were analyzed by SDS-PAGE followed by Western blotting with antibodies against the epitope tags. To determine the stability of LNR-(HD-N)/(HD-C) heterodimers in the presence of urea, immunoprecipitates prepared from conditioned media as described above were incubated for 30 min at 22°C in 1 ml of buffer B containing 0 to 4 M urea. Following three washes with the incubation buffer, the polypeptides retained on the beads were analyzed by Western blotting.
RESULTS
T-ALL-associated HD domain mutations generally result in NOTCH1 activation.
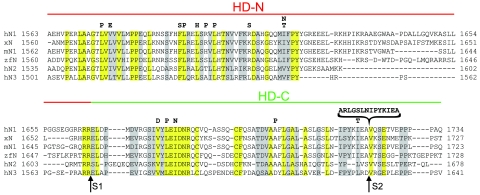
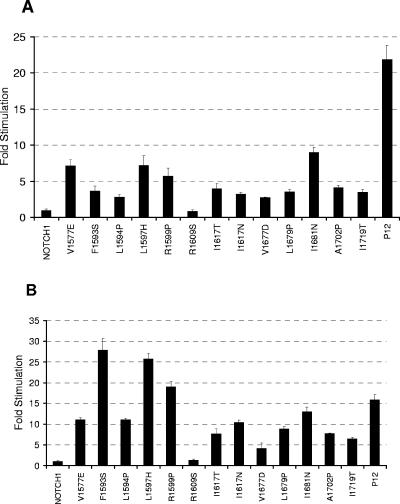
To date, we have identified over 30 different HD domain mutations in primary T-ALL samples or T-ALL cell lines (48; J. C. Aster, unpublished data). From these, we chose 15 representative point mutations and one insertional mutation involving the HD domain of NOTCH1 for further study (Fig. 1A). For the most part, these mutations fall in positions that are highly conserved among vertebrate NOTCH receptors (Fig. 2). When introduced into a full-length NOTCH1 cDNA and expressed transiently in U2OS cells, 15 of these 16 mutations stimulated the activity of a NOTCH-responsive luciferase reporter gene containing multiple iterated CSL-binding sites, as compared to normal NOTCH1 (Fig. 3A). The extent of stimulation varied among the mutations in a reproducible fashion; the insertional mutation (referred to as P12 for the cell of origin, P12-Ichikawa) consistently caused the strongest increase in activity. These results show that the HD domain mutations that occur in T-ALL generally increase NOTCH1 function. Only the R1609S mutation, found in a single primary T-ALL sample, failed to stimulate NOTCH1 signaling. In all other assays, it also behaved like native Notch1, suggesting that it is probably a rare germ line variant, rather than a true gain-of-function mutation.
FIG. 2.
Sequence alignment of the HD domains of various NOTCH receptors. Identical and conserved residues among the sequences are enclosed in yellow and gray boxes, respectively. h, human; x, Xenopus; m, mouse; and zf, zebrafish. Furin (S1) and metalloprotease (S2) cleavage sites are indicated below the sequence. Analyzed leukemia-derived mutations are identified above the sequence.
FIG. 3.
HD domain mutations result in activation of NOTCH1 signaling. HD domain mutations were introduced into (A) full-length or (B) ΔEGF NOTCH1 constructs. Luciferase assays were performed on U2OS cell lysates prepared from cells transfected in triplicate with 10 ng of empty pcDNA3 plasmid or plasmids encoding the indicated forms of (A) full-length or (B) ΔEGF NOTCH1, along with a luciferase reporter plasmid containing iterated CSL-binding sites, and an internal control plasmid expressing Renilla luciferase. Firefly luciferase activity from cell lysates was measured in triplicate, normalized, and expressed relative to the activity in extracts prepared from cells transfected with empty vector, which is arbitrarily set to a value of 1. The mutations L1575P and L1601P, which were previously shown to stimulate signaling in the context of full-length NOTCH1 (48), also stimulated signaling in the context of ΔEGF (data not shown).
Leukemia-associated NOTCH1 HD domain mutations do not require the ligand-binding domain to activate signaling.
Next, we tested whether the ligand-binding domain was required for increased signaling to occur when tumor-associated mutations are present. Thus, we determined the effect of each mutation on the activity of a receptor that lacks the NOTCH1 EGF repeats (ΔEGF), which are necessary for binding of conventional Notch ligands of the Delta-Serrate-Lag2 family (23, 39, 51). Though ΔEGF is quiescent in reporter gene assays, the same 15 mutations that stimulated the activity of full-length NOTCH1 also increased the activity of the ΔEGF form of the receptor (Fig. 3B). With the exception of the P12 mutation, the relative strength of the signaling increase observed with the ΔEGF receptors correlates well with that observed with the full-length receptors. Again, only the R1609S substitution failed to increase signaling above the levels observed with unmutated ΔEGF. Thus, leukemia-associated HD domain mutations activate NOTCH1 independent of stimulation by canonical ligands.
Receptors with leukemia-associated mutations retain the ability to signal upon ligand stimulation.
Because endogenous Notch receptors are expressed on many cultured cell lines, we constructed Notch1-GAL4 chimeras to assess ligand-induced signaling on GAL4 reporters in the absence of background signals from the endogenous receptors. Four different chimeras were constructed: unmutated Notch1-GAL4, Notch1-GAL4 with the relatively weak L1594P mutation, and Notch1-GAL4 with the strong P12 insertion mutation. Each chimeric receptor was then individually introduced into a U20S “flp-in” cell line, creating three isogenic stable U20S cell lines, each expressing one of the chimeric receptors.
Each isogenic flp-in cell line was then assessed for tet-regulated chimera expression and for the ability to activate expression of a GAL4-luciferase reporter gene in the presence of control OP9 cells and OP9 cells stably expressing the NOTCH ligand DLL-1. Consistent with prior transient expression experiments, the L1594P and P12 mutations caused weak and strong activation of the GAL4 reporter gene, respectively, when U2OS cells were cocultivated with OP9 control cells, relative to normal NOTCH1 (see Fig. S1 in the supplemental material). In contrast, all three forms of NOTCH1 produced similar levels of reporter gene activation when cocultivated with OP9-DLL-1 cells (see Fig. S1 in the supplemental material). These data show that NOTCH receptors bearing HD domain mutations also reach the cell surface, respond to ligands, and do not appear to be hypersensitive to ligand stimulation.
Leukemia-associated HD domain mutations cause increased S2 and S3 cleavage.
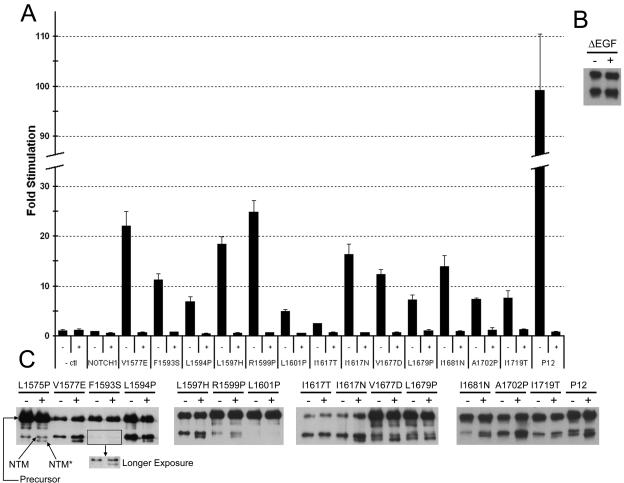
It has been reported that NOTCH receptors are susceptible to activation by noncanonical proteases under some circumstances (16), and we therefore sought to determine if the observed gains in function caused by leukemia-associated HD domain mutations were associated with increased cleavage at sites S2 and S3. In order to determine whether S3 cleavage is required, reporter assays were performed in the presence of two different potent γ-secretase inhibitors, compound E and N-[N-(3,5-difluorophenacetyl)-l-alanyl]-(S)-phenylglycine t-butyl ester (DAPT) (Fig. 4A; see Fig. S2 in the supplemental material). The stimulations caused by all 15 activating mutations were completely abolished by compound E and DAPT, indicating that γ-secretase activity is required uniformly for activity.
FIG. 4.
HD domain mutations cause increased S2 and S3 cleavage. (A) Gamma secretase inhibitor (GSI) treatment abrogates the stimulatory effects of HD domain mutations in full-length NOTCH1. Immediately following transfection of U2OS cells with NOTCH1 expression plasmids encoding the indicated mutations, cells were treated with 1 μM compound E (GSI) or carrier alone. Trancriptional activation assays were performed 44 to 48 h after transfection, as described in the legend to Fig. 3. The increased signaling produced by the L1575P mutation was also inhibited by GSI in other experiments (data not shown). (B) U2OS cells transfected with wild-type ΔEGF-MYC NOTCH polypeptides were incubated in the presence of 1 μM compound E (GSI; +) or carrier (−) for 18 h prior to harvesting. NTM and NTM* polypeptides were then recovered by preparing immunoprecipitates from cell lysates using anti-MYC-coupled beads, resolved by SDS-PAGE under reducing conditions, and detected by Western blotting using an antibody against intracellular NOTCH1. (C) HD domain mutations were introduced into ΔEGF-MYC polypeptides, and the same assay was performed. Precursor, uncleaved NOTCH1 polypeptide; NTM, transmembrane subunit created by furin cleavage at site S1; and NTM*, S2 cleavage product.
NTM*, the product created by metalloprotease cleavage at site S2, is normally short-lived and difficult to detect, but accumulates when γ-secretase activity is inhibited (49). To distinguish polypeptides encoded on the transfected plasmids from endogenous NOTCH1 polypeptides, we introduced a C-terminal c-MYC epitope into ΔEGF expression constructs bearing each of the 16 HD domain mutations. Unmutated ΔEGF is inactive and does not cause the appearance of NTM* (40). As shown in Fig. 4C, NTM* accumulated in the presence of compound E in cells expressing 13 of the 15 mutated ΔEGF-MYC constructs.
The two mutations that did not generate NTM*, L1601P and I1617T, were relatively weak transcriptional activators in reporter gene assays (Fig. 3 and 4A) (48). The L1601P mutation reduced the efficiency of furin cleavage so dramatically that it was not possible to detect the furin-cleaved subunits with this assay. Similar effects on furin cleavage were also seen when L1601P is incorporated into full-length NOTCH1 (not shown). For I1617T, we believe that failure to detect NTM* is simply an issue of assay sensitivity. Although we cannot exclude the possibility that the I1617T mutation activates NOTCH1 in a γ-secretase-dependent, metalloprotease-independent fashion, it seems far more likely that the reporter gene assay is simply more sensitive to small increases in S2-dependent NOTCH1 activation.
Most HD domain mutations reduce heterodimer stability.
Prior studies have shown that polypeptides resembling NTM (often termed ΔE) produce strong gain-of-function phenotypes (9, 12, 31, 38) and that normal ligand-mediated receptor activation can be accompanied by the physical removal of the NEC subunit (21, 30). Thus, one mechanism by which HD domain mutations might induce NOTCH1 activation is by weakening the interaction of NEC and NTM.
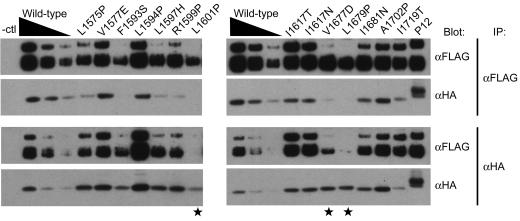
To test whether leukemia-associated mutations reduce the stability of NEC/NTM complexes, we introduced the same set of HD domain mutations into a soluble polypeptide (sHD) consisting of the LNR and HD domains of NOTCH1. When expressed transiently in cultured mammalian cells, unmutated sHD undergoes furin processing and is secreted into conditioned media as an epitope-tagged heterodimer comprising stably associated LNR-(HD-N) and HD-C subunits (40) (Fig. 5), as are a number (n = 12) of the mutated LNR-HD proteins (Fig. 5; see Fig. S3 in the supplemental material). In contrast, three mutations (L1601P, V1677D, and L1679P) induced the dissociation of most or all of the furin-processed sHD into its two subunits.
FIG. 5.
Effect of HD domain mutations on subunit dissociation under native conditions. Conditioned media from 293T cells expressing secreted unmutated and mutated LNR-HD polypeptides (sHDs) were immunoprecipitated (IP) with anti-FLAG (α-FLAG) beads (top two panels) and analyzed following SDS-PAGE by Western blot analysis with the indicated antibodies. To better gauge whether the coprecipitation of the sHD subunits was affected by the mutations, a range of coprecipitated unmutated sHD subunits were run as a control. In addition to the sHD subunits, variable amounts of unprocessed sHD were detected in most of the conditioned media, including the normal control. Mutations inducing subunit dissociation of the HA-tagged HD-C subunit are denoted by stars. Comparable results were observed in parallel experiments in which immunoprecipitates were prepared with anti-HA (α-HA) beads (bottom pair of panels), and coprecipitation of the FLAG-tagged HD-N subunit was assessed by Western blotting.
Of note, L1601P and F1593S, the mutations that greatly reduced furin cleavage in the context of the ΔEGF polypeptide (Fig. 4C), did not preclude furin cleavage in the context of the sHD polypeptide (Fig. 5). This argues further that these polypeptides are fully competent for cleavage if they are exposed to furin. Since furin is localized to the trans-Golgi compartment (7), the observed furin “resistance” could be due to the retention of newly synthesized full-length NOTCH1 polypeptides bearing these mutations in the endoplasmic reticulum (ER). This possibility seems reasonable, especially if the disruptions of folded structure caused by these mutations are propagated more globally to other parts of the full-length protein, facilitating detection and retention by ER-resident quality control mechanisms.
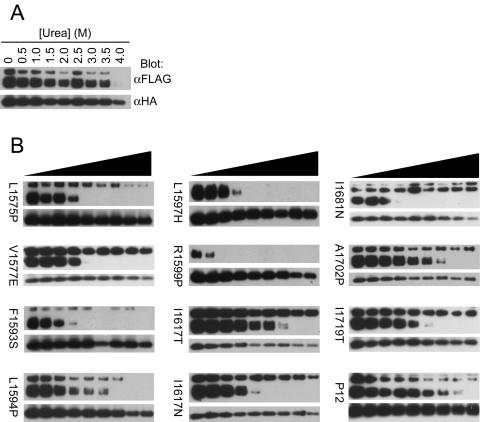
We next evaluated whether the 12 mutations that did not clearly cause overt HD dissociation under native conditions might be associated with more subtle reductions in HD stability. Thus, we compared the stability of the sHDs bearing each of these 12 mutations to that of the native sHD in the presence of increasing concentrations of the denaturant urea. The normal sHD subunits dissociated completely only after treatment with 4 M urea (Fig. 6A). In contrast, 11 of the 12 mutated sHDs that were tested (L1575P, V1577E, F1593S, L1594P, L1597H, R1599P, I1617T, I1617N, I1681N, A1702P, and I1719T) clearly conferred increased sensitivity to urea, as complete subunit dissociation occurred by urea concentrations of 2 to 3.5 M (Fig. 6B).
FIG. 6.
Effect of HD domain mutations on subunit dissociation in the presence of urea. Conditioned media from 293T cells expressing (A) unmutated secreted NOTCH1 heterodimers or (B) mutated sHDs were immunoprecipitated with anti-HA (α-HA)-coupled beads, followed by 30 min of incubation at room temperature in buffer with different concentrations of urea (0 to 4 M). Subunit dissociation was evaluated by SDS-PAGE followed by Western blot analysis with antibodies specific for each of the two subunits. α-FLAG, anti-FLAG.
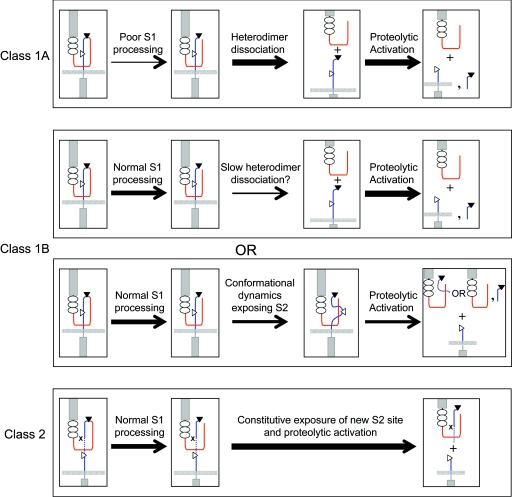
The P12 mutation, which is a 14-residue insertion immediately before the S2 cleavage site, also appeared to confer a slight but reproducible increase in urea sensitivity, as we consistently observed complete dissociation in this assay by 3.5 to 4 M urea (Fig. 6B and see Fig. S4 in the supplemental material), yet this mutation was the strongest activator of NOTCH signaling among the mutations tested in the context of full-length NOTCH1. This insertion stems from a direct duplication (Fig. 2), which is predicted to reposition the endogenous HD-C domain away from the site of S2 cleavage. We postulate that this mutation enhances the access of the S2 site to metalloproteases despite its negligible effect on HD domain stability or folding, and thus represents a distinct class of activating mutation (Fig. 7).
FIG. 7.
Proposed classification of leukemia-associated heterodimerization domain mutations. Activation by class 1 mutations occurs because of heterodimer dissociation or reduced heterodimer stability. Class 1A mutations cause overt heterodimer dissociation, while class 1B mutations cause either slow heterodimer dissociation or an increase in S2 site exposure by inducing local “breathing.” Class 2 mutations have no effect upon the stability of the heterodimer but result in proteolytic activation by positioning an exposed, mutation-associated S2 site away from protective residues that normally prevent metalloprotease access prior to ligand binding. The thickness of the arrow represents the estimated flux of molecules through the indicated step. HD-N, red; HD-C, blue; S1 site, filled triangle; S2 site, open triangle. The dashed line in the bottom panel represents the inserted sequence, and x indicates the position of the original S2 site ablated by the insertion.
DISCUSSION
In prior studies, we showed that three relatively common HD domain mutations found in human T-ALL, L1575P, L1594P, and L1601P each stimulate NOTCH1 signaling in a γ-secretase-dependent fashion (48). Here, we have extended this analysis to an additional set of 13 putative HD domain mutations, which were also identified initially in primary human T-ALL samples or derived cell lines. We find that all but one of these sequence variants behave as true gain-of-function mutations and that they do not require binding to typical ligands of the Delta-Serrate-Lag-2 family to produce NOTCH1 activation. Further, the observed gains in function in reporter assays require S3 cleavage and correlate with an increased sensitivity to cleavage at site S2, indicating that activation likely proceeds through the same steps that take place during normal NOTCH1 activation.
However, we also noted qualitative differences among mutations with respect to their relative strength and their effects on NOTCH1 heterodimer stability. Based on these differences, the mutations can be tentatively placed into two broad classes (which are summarized in Fig. 7): (i) mutations that destabilize NOTCH1 heterodimers, which can be further subdivided into mutations that enhance subunit dissociation under native conditions (class 1A; n = 3) or in the presence of a denaturant (class 1B; n = 11); and (ii), insertions adjacent to the S2 cleavage site that act by enhancing access of metalloproteases to site S2 without greatly affecting heterodimer stability (class 2; n = 1).
The simplest way for class 1A mutants to act is to cause the spontaneous dissociation of NEC from NTM, thereby exposing the S2 site on NTM to ligand-independent cleavage by ADAM-type metalloproteases. However, the activity of some class IA mutations may be tempered by diminished furin processing (Fig. 4C), which is an essential prerequisite for subunit dissociation. Because furin is localized to the trans-Golgi compartment (7), the diminished furin processing could stem from the retention of newly synthesized mutated NOTCH1 polypeptides in the endoplasmic reticulum. This mechanism supposes that there is a sensor for HD domain folding; given the importance of this domain in maintaining NOTCH receptors in the “off state,” the existence of such a quality control mechanism seems plausible. An alternative possibility would be for the mutations to modify the structure of the HD domain such that furin does not recognize the cleavage site. We have recently determined that furin cleavage occurs in an unfolded loop (S. C. Blacklow, unpublished data), making the second explanation seem less likely, but further work will be necessary to delineate between these possibilities.
The 11 class 1B mutations also destabilize the NOTCH1 heterodimer, but this difference is only revealed in the presence of a denaturant. Although such mutations could act by promoting the slow spontaneous dissociation of NEC, loosening of contacts between NEC and NTM might also allow S2 cleavage by increasing the dynamic “breathing” or exposure of the HD domain, which could enhance the access of metalloproteases to the S2 site. One way of distinguishing between these possibilities is to determine whether cells expressing NOTCH1 receptors with class IB mutations shed (i) NEC monomers or (ii) complexes containing NEC and the portion of HD-C that is released by S2 cleavage. We are currently attempting to develop antibody reagents that can be used to address this issue. Alternatively, it would be of interest to determine whether class 1B mutations depend strictly on furin cleavage for their activity; structure-based deletions and/or mutations that render NOTCH1 furin resistant without perturbing the overall structure of the HD domain will enable such studies.
The one mutation assigned to class 2 is a 14-residue insertion immediately N terminal to the S2 cleavage site that leads to strong gains in signaling activity despite causing a minimal change in NOTCH1 heterodimer stability. It seems likely that the inserted sequence (which leaves HD-C essentially intact) circumvents an endogenous restraint on activation by repositioning the S2 site away from protective residues in the HD domain (and possibly the LNR domain), rendering the S2 site accessible to metalloproteases. A similar mutation found in a primary T-ALL sample inserts 13 amino acids at the same position (48), suggesting that receptor activation through this mechanism requires the insertion of a spacer of a particular size. One interesting possibility is that a similar displacement of the LNR/HD negative regulatory unit might result from a mechanical force that is transmitted along the long axis of the receptor in response to ligand binding and endocytosis. If so, such forces could promote S2 cleavage prior to physical removal of the NEC subunit, a possibility that is in line with several studies from the fly (1, 18).
It is noteworthy that all of the gain-of-function HD domain mutations analyzed to date activate signaling in a ligand-independent fashion. Unlike solid tumors, leukemia cells infiltrate and grow in diverse tissues and fluids, such as blood and malignant effusions, as single dyscohesive cells that are likely to rely on cell autonomous growth signals. Cell autonomous activation of NOTCH signals by HD domain mutations could contribute to the induction or progression of T-ALL in several ways. NOTCH1 signals in marrow lymphoid progenitors are normally too weak to induce T-cell development. NOTCH1 HD domain mutations in multipotent bone marrow progenitors are predicted to redirect these cells towards T-cell fate (34), thus expanding an ectopic pool of cells that are at risk for acquisition of additional genetic events and eventual outgrowth as T-ALL. In support of this scenario, patients with T-ALL do not always have thymic involvement at the time of diagnosis (suggesting that these tumors sometimes originate within the bone marrow), and the thymus is expendable for induction of T-ALL by constitutively active forms of NOTCH1 in mouse models (34). Alternatively, the thymus is rich in stromal cells expressing NOTCH ligands (2, 4, 14, 28); thus, the acquisition of cell autonomous gain-of-function NOTCH1 mutations might be important in the spread of T-ALL of thymic origin to extrathymic sites.
Of practical importance, our work shows that a broad spectrum of leukemia-associated HD domain mutations retain dependence on γ-secretase to produce gains in function. This provides a further rationale for γ-secretase inhibitor therapy in T-ALL, which is highly associated with mutations in NOTCH1. Specifically, HD domain mutations are found in 40 to 50% of T-ALLs (48), which appear to rely on NOTCH1 signals for growth and survival. Recently, the Notch signaling pathway has been linked to other human cancers, including tumors of the breast (32), brain (35), prostate (41), and pancreas (27), as well as tumor-induced angiogenesis (53). Thus, detailed functional understanding of the mechanisms by which leukemia-associated HD mutations act may also provide insights relevant to other more common forms of human cancer.
Supplementary Material
Acknowledgments
We thank Didem Vardar and Wendy Gordon for helpful discussions.
This work was supported by NIH grants CA92433 and CA119130 (to S.C.B.) and CA82308 (to J.C.A.).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ahimou, F., L. P. Mok, B. Bardot, and C. Wesley. 2004. The adhesion force of Notch with Delta and the rate of Notch signaling. J. Cell Biol. 167:1217-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, G., J. Pongracz, S. Parnell, and E. J. Jenkinson. 2001. Notch ligand-bearing thymic epithelial cells initiate and sustain Notch signaling in thymocytes independently of T cell receptor signaling. Eur. J. Immunol. 31:3349-3354. [DOI] [PubMed] [Google Scholar]
- 3.Artavanis-Tsakonas, S., M. D. Rand, and R. J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science 284:770-776. [DOI] [PubMed] [Google Scholar]
- 4.Aster, J. C., E. S. Robertson, R. P. Hasserjian, J. R. Turner, E. Kieff, and J. Sklar. 1997. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jkappa or nuclear localization sequences retain the ability to associate with RBP-Jkappa and activate transcription. J. Biol. Chem. 272:11336-11343. [DOI] [PubMed] [Google Scholar]
- 5.Aster, J. C., L. Xu, F. G. Karnell, V. Patriub, J. C. Pui, and W. S. Pear. 2000. Essential roles for ankyrin repeat and transactivation domains in induction of T-cell leukemia by Notch1. Mol. Cell. Biol. 20:7505-7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaumueller, C. M., H. Qi, P. Zagouras, and S. Artavanis-Tsakonas. 1997. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell 90:281-291. [DOI] [PubMed] [Google Scholar]
- 7.Bresnahan, P. A., R. Leduc, L. Thomas, J. Thorner, H. L. Gibson, A. J. Brake, P. J. Barr, and G. Thomas. 1990. Human fur gene encodes a yeast KEX2-like endoprotease that cleaves pro-beta-NGF in vivo. J. Cell Biol. 111:2851-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brou, C., F. Logeat, N. Gupta, C. Bessia, O. LeBail, J. R. Doedens, A. Cumano, P. Roux, R. A. Black, and A. Israel. 2000. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol. Cell 5:207-216. [DOI] [PubMed] [Google Scholar]
- 9.Coffman, C. R., P. Skoglund, W. A. Harris, and C. R. Kintner. 1993. Expression of an extracellular deletion of Xotch diverts cell fate in Xenopus embryos. Cell 73:659-671. [DOI] [PubMed] [Google Scholar]
- 10.De Strooper, B., W. Annaert, P. Cupers, P. Saftig, K. Craessaerts, J. S. Mumm, E. H. Schroeter, V. Schrijvers, M. S. Wolfe, W. J. Ray, A. Goate, and R. Kopan. 1999. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature 398:518-522. [DOI] [PubMed] [Google Scholar]
- 11.Ellisen, L. W., J. Bird, D. C. West, A. L. Soreng, T. C. Reynolds, S. D. Smith, and J. Sklar. 1991. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 66:649-661. [DOI] [PubMed] [Google Scholar]
- 12.Fortini, M. E., I. Rebay, L. A. Caron, and S. Artavanis-Tsakonas. 1993. An activated Notch receptor blocks cell-fate commitment in the developing Drosophila eye. Nature 365:555-557. [DOI] [PubMed] [Google Scholar]
- 13.Greenwald, I., and G. Seydoux. 1990. Analysis of gain-of-function mutations of the lin-12 gene of Caenorhabditis elegans. Nature 346:197-199. [DOI] [PubMed] [Google Scholar]
- 14.Harman, B. C., E. J. Jenkinson, and G. Anderson. 2003. Microenvironmental regulation of Notch signalling in T cell development. Semin. Immunol. 15:91-97. [DOI] [PubMed] [Google Scholar]
- 15.Hsieh, J. J., T. Henkel, P. Salmon, E. Robey, M. G. Peterson, and S. D. Hayward. 1996. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol. Cell. Biol. 16:952-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huppert, S. S., M. X. Ilagan, B. De Strooper, and R. Kopan. 2005. Analysis of Notch function in presomitic mesoderm suggests a gamma-secretase-independent role for presenilins in somite differentiation. Dev. Cell. 8:677-688. [DOI] [PubMed] [Google Scholar]
- 17.Jarriault, S., C. Brou, F. Logeat, E. H. Schroeter, R. Kopan, and A. Israel. 1995. Signalling downstream of activated mammalian Notch. Nature 377:355-358. [DOI] [PubMed] [Google Scholar]
- 18.Kidd, S., and T. Lieber. 2002. Furin cleavage is not a requirement for Drosophila Notch function. Mech. Dev. 115:41-51. [DOI] [PubMed] [Google Scholar]
- 19.Kimberly, W. T., W. P. Esler, W. Ye, B. L. Ostaszewski, J. Gao, T. Diehl, D. J. Selkoe, and M. S. Wolfe. 2003. Notch and the amyloid precursor protein are cleaved by similar gamma-secretase(s). Biochemistry 42:137-144. [DOI] [PubMed] [Google Scholar]
- 20.Kopan, R., E. H. Schroeter, H. Weintraub, and J. S. Nye. 1996. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc. Natl. Acad. Sci. USA 93:1683-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kramer, H. 2000. RIPping notch apart: a new role for endocytosis in signal transduction? Sci. STKE 2000:PE1. [Online.] [DOI] [PubMed]
- 22.Lai, E. C., G. A. Deblandre, C. Kintner, and G. M. Rubin. 2001. Drosophila neuralized is a ubiquitin ligase that promotes the internalization and degradation of delta. Dev. Cell 1:783-794. [DOI] [PubMed] [Google Scholar]
- 23.Lawrence, N., T. Klein, K. Brennan, and A. Martinez Arias. 2000. Structural requirements for notch signalling with delta and serrate during the development and patterning of the wing disc of Drosophila. Development 127:3185-3195. [DOI] [PubMed] [Google Scholar]
- 24.Lieber, T., S. Kidd, E. Alcamo, V. Corbin, and M. W. Young. 1993. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 7:1949-1965. [DOI] [PubMed] [Google Scholar]
- 25.Logeat, F., C. Bessia, C. Brou, O. LeBail, S. Jarriault, N. G. Seidah, and A. Israel. 1998. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl. Acad. Sci. USA 95:8108-8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maillard, I., T. Fang, and W. S. Pear. 2005. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu. Rev. Immunol. 23:945-974. [DOI] [PubMed] [Google Scholar]
- 27.Miyamoto, Y., A. Maitra, B. Ghosh, U. Zechner, P. Argani, C. A. Iacobuzio-Donahue, V. Sriuranpong, T. Iso, I. M. Meszoely, M. S. Wolfe, R. H. Hruban, D. W. Ball, R. M. Schmid, and S. D. Leach. 2003. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell 3:565-576. [DOI] [PubMed] [Google Scholar]
- 28.Mohtashami, M., and J. C. Zuniga-Pflucker. 2006. Cutting edge: three-dimensional architecture of the thymus is required to maintain delta-like expression necessary for inducing T cell development. J. Immunol. 176:730-734. [DOI] [PubMed] [Google Scholar]
- 29.Mumm, J. S., E. H. Schroeter, M. T. Saxena, A. Griesemer, X. Tian, D. J. Pan, W. J. Ray, and R. Kopan. 2000. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol. Cell 5:197-206. [DOI] [PubMed] [Google Scholar]
- 30.Parks, A. L., K. M. Klueg, J. R. Stout, and M. A. Muskavitch. 2000. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development 127:1373-1385. [DOI] [PubMed] [Google Scholar]
- 31.Pear, W. S., J. C. Aster, M. L. Scott, R. P. Hasserjian, B. Soffer, J. Sklar, and D. Baltimore. 1996. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J. Exp. Med. 183:2283-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pece, S., M. Serresi, E. Santolini, M. Capra, E. Hulleman, V. Galimberti, S. Zurrida, P. Maisonneuve, G. Viale, and P. P. Di Fiore. 2004. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J. Cell Biol. 167:215-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petcherski, A. G., and J. Kimble. 2000. LAG-3 is a putative transcriptional activator in the C. elegans Notch pathway. Nature 405:364-368. [DOI] [PubMed] [Google Scholar]
- 34.Pui, J. C., D. Allman, L. Xu, S. DeRocco, F. G. Karnell, S. Bakkour, J. Y. Lee, T. Kadesch, R. R. Hardy, J. C. Aster, and W. S. Pear. 1999. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity 11:299-308. [DOI] [PubMed] [Google Scholar]
- 35.Purow, B. W., R. M. Haque, M. W. Noel, Q. Su, M. J. Burdick, J. Lee, T. Sundaresan, S. Pastorino, J. K. Park, I. Mikolaenko, D. Maric, C. G. Eberhart, and H. A. Fine. 2005. Expression of Notch-1 and its ligands, Delta-like-1 and Jagged-1, is critical for glioma cell survival and proliferation. Cancer Res. 65:2353-2363. [DOI] [PubMed] [Google Scholar]
- 36.Radtke, F., A. Wilson, G. Stark, M. Bauer, J. van Meerwijk, H. R. MacDonald, and M. Aguet. 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 10:547-558. [DOI] [PubMed] [Google Scholar]
- 37.Rand, M. D., L. M. Grimm, S. Artavanis-Tsakonas, V. Patriub, S. C. Blacklow, J. Sklar, and J. C. Aster. 2000. Calcium depletion dissociates and activates heterodimeric Notch receptors. Mol. Cell. Biol. 20:1825-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rebay, I., R. G. Fehon, and S. Artavanis-Tsakonas. 1993. Specific truncations of Drosophila Notch define dominant activated and dominant negative forms of the receptor. Cell 74:319-329. [DOI] [PubMed] [Google Scholar]
- 39.Rebay, I., R. J. Fleming, R. G. Fehon, L. Cherbas, P. Cherbas, and S. Artavanis-Tsakonas. 1991. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell 67:687-699. [DOI] [PubMed] [Google Scholar]
- 40.Sanchez-Irizarry, C., A. C. Carpenter, A. P. Weng, W. S. Pear, J. C. Aster, and S. C. Blacklow. 2004. Notch subunit heterodimerization and prevention of ligand-independent proteolytic activation depend, respectively, on a novel domain and the LNR repeats. Mol. Cell. Biol. 24:9265-9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santagata, S., F. Demichelis, A. Riva, S. Varambally, M. D. Hofer, J. L. Kutok, R. Kim, J. Tang, J. E. Montie, A. M. Chinnaiyan, M. A. Rubin, and J. C. Aster. 2004. JAGGED1 expression is associated with prostate cancer metastasis and recurrence. Cancer Res. 64:6854-6857. [DOI] [PubMed] [Google Scholar]
- 42.Schroeter, E. H., J. A. Kisslinger, and R. Kopan. 1998. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393:382-386. [DOI] [PubMed] [Google Scholar]
- 43.Seugnet, L., P. Simpson, and M. Haenlin. 1997. Requirement for dynamin during Notch signaling in Drosophila neurogenesis. Dev. Biol. 192:585-598. [DOI] [PubMed] [Google Scholar]
- 44.Shah, S., S. F. Lee, K. Tabuchi, Y. H. Hao, C. Yu, Q. Laplant, H. Ball, C. E. Dann III, T. Sudhof, and G. Yu. 2005. Nicastrin functions as a gamma-secretase-substrate receptor. Cell 122:435-447. [DOI] [PubMed] [Google Scholar]
- 45.Struhl, G., and I. Greenwald. 1999. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature 398:522-525. [DOI] [PubMed] [Google Scholar]
- 46.Vooijs, M., E. H. Schroeter, Y. Pan, M. Blandford, and R. Kopan. 2004. Ectodomain shedding and intramembrane cleavage of mammalian Notch proteins is not regulated through oligomerization. J. Biol. Chem. 279:50864-50873. [DOI] [PubMed] [Google Scholar]
- 47.Weng, A. P., and J. C. Aster. 2004. Multiple niches for Notch in cancer: context is everything. Curr. Opin. Genet. Dev. 14:48-54. [DOI] [PubMed] [Google Scholar]
- 48.Weng, A. P., A. A. Ferrando, W. Lee, J. P. T. Morris, L. B. Silverman, C. Sanchez-Irizarry, S. C. Blacklow, A. T. Look, and J. C. Aster. 2004. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306:269-271. [DOI] [PubMed] [Google Scholar]
- 49.Weng, A. P., Y. Nam, M. S. Wolfe, W. S. Pear, J. D. Griffin, S. C. Blacklow, and J. C. Aster. 2003. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of Notch signaling. Mol. Cell. Biol. 23:655-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu, L., J. C. Aster, S. C. Blacklow, R. Lake, S. Artavanis-Tsakonas, and J. D. Griffin. 2000. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet. 26:484-489. [DOI] [PubMed] [Google Scholar]
- 51.Xu, A., L. Lei, and K. D. Irvine. 2005. Regions of Drosophila notch that contribute to ligand binding and the modulatory influence of fringe. J. Biol. Chem. 280:30158-30165. (First published 30 June 2005; doi: 10.1074/jbc.M505569200.) [DOI] [PubMed] [Google Scholar]
- 52.Ye, Y., N. Lukinova, and M. E. Fortini. 1999. Neurogenic phenotypes and altered Notch processing in Drosophila Presenilin mutants. Nature 398:525-529. [DOI] [PubMed] [Google Scholar]
- 53.Zeng, Q., S. Li, D. B. Chepeha, T. J. Giordano, J. Li, H. Zhang, P. J. Polverini, J. Nor, J. Kitajewski, and C. Y. Wang. 2005. Crosstalk between tumor and endothelial cells promotes tumor angiogenesis by MAPK activation of Notch signaling. Cancer Cell 8:13-23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.