Abstract
RAG-1 and RAG-2 initiate V(D)J recombination by cleaving DNA at recombination signal sequences through sequential nicking and transesterification reactions to yield blunt signal ends and coding ends terminating in a DNA hairpin structure. Ubiquitous DNA repair factors then mediate the rejoining of broken DNA. V(D)J recombination adheres to the 12/23 rule, which limits rearrangement to signal sequences bearing different lengths of DNA (12 or 23 base pairs) between the conserved heptamer and nonamer sequences to which the RAG proteins bind. Both RAG proteins have been subjected to extensive mutagenesis, revealing residues required for one or both cleavage steps or involved in the DNA end-joining process. Gain-of-function RAG mutants remain unidentified. Here, we report a novel RAG-1 mutation, E649A, that supports elevated cleavage activity in vitro by preferentially enhancing hairpin formation. DNA binding activity and the catalysis of other DNA strand transfer reactions, such as transposition, are not substantially affected by the RAG-1 mutation. However, 12/23-regulated synapsis does not strongly stimulate the cleavage activity of a RAG complex containing E649A RAG-1, unlike its wild-type counterpart. Interestingly, wild-type and E649A RAG-1 support similar levels of cleavage and recombination of plasmid substrates containing a 12/23 pair of signal sequences in cell culture; however, E649A RAG-1 supports about threefold more cleavage and recombination than wild-type RAG-1 on 12/12 plasmid substrates. These data suggest that the E649A RAG-1 mutation may interfere with the RAG proteins' ability to sense 12/23-regulated synapsis.
V(D)J recombination is the process by which noncontiguous antigen receptor gene coding segments, called variable (V), diversity (D), and joining (J), are assembled during lymphocyte development to produce the variable region exon of a mature antigen receptor gene (3). V(D)J recombination occurs in two distinct phases. In the first phase, two lymphoid cell-specific proteins called RAG-1 and RAG-2 assemble a multiprotein synaptic complex with two different gene segments through interactions with a conserved recombination signal sequence (RSS) that adjoins each gene segment. Each RSS contains a conserved heptamer and nonamer sequence, separated by either 12 or 23 base pairs of intervening DNA of more varied composition (12-RSS and 23-RSS, respectively). Generally, synaptic complexes are assembled with two RSSs whose spacer lengths are different (the 12/23 rule). Subsequently, the RAG proteins catalyze a DNA double-strand break at each RSS (for reviews, see references 10 and 13), yielding a postcleavage complex containing four DNA ends: two blunt, 5′ phosphorylated recombination signal ends and two coding ends terminating in DNA hairpin structures (41, 42, 46). The RAG proteins generate these recombination intermediates by nicking the DNA at the junction between the RSS and the coding sequence and then transferring the resulting 3′-OH to the opposing DNA strand by direct transesterification (32, 57). The second phase of V(D)J recombination involves components of the nonhomologous end-joining repair pathway that function to resect the broken DNA ends. Typically, signal ends are joined to one another heptamer to heptamer to create a signal joint, whereas coding ends are often joined together imprecisely due to variable end processing events that can lead to the gain or loss of nucleotides at the ends prior to joining.
To explore how the RAG proteins mediate RSS synapsis and cleavage and to uncover the roles of the RAG complex in facilitating the joining phase of V(D)J recombination, RAG-1 and RAG-2 have been subjected to extensive mutagenesis. These studies have defined regions of RAG-1 and RAG-2 dispensable for supporting the rearrangement of plasmid V(D)J recombination substrates in cell culture (7, 22, 43, 44); identified various mutations in the RAG proteins that impair RSS binding (9, 11, 49, 52), one or both cleavage steps (12, 20, 24, 28, 39), or the joining phase of V(D)J recombination (21, 39, 47, 54); and refined structural elements required for RAG-1/RAG-2 association or interactions with other proteins (2, 33, 60). RAG mutants that enhance the cleavage or joining phases of V(D)J recombination have not yet been reported, although mutations in RAG-2 that impair its degradation at the G1/S transition of the cell cycle enable the RAG complex to cleave DNA beyond the G0 to G1 phases, where it is normally restricted (25).
Hyperactive transposase mutants have been identified in several “cut-and-paste” transpositional recombination systems, including Tn5 (59, 61, 64), Tn7 (30), P element (4), Himar1 (23), and Sleeping Beauty (62). The ability to isolate hyperactive transposase mutants has led to the suggestion that transposases have not evolved for maximal activity (40). Since V(D)J recombination shares mechanistic similarities with transpositional recombination systems that use a cut-and-paste mechanism (19, 38), it is likely that the RAG complex is also less active than it potentially could be. In support of this idea, we report here the first example of a gain-of-function RAG-1 mutation (E649A) that specifically accelerates the second biochemical step of RAG-mediated cleavage (hairpin formation) in vitro. This rate enhancement is dependent on the type of metal ion cofactor present in the cleavage reaction. Cleavage is significantly elevated in the presence of a Mg2+ ion, thought to be physiologically relevant to V(D)J recombination as it supports RAG-mediated RSS synapsis and cleavage according to the 12/23 rule in vitro (58) but is increased only minimally in the presence of Mn2+, which decouples the 12/23 rule (58). The RAG-1 E649A mutation does not substantially alter the rates of other DNA strand transfer reactions catalyzed by the RAG complex, including transposition, disintegration, and hybrid joint formation. In addition, RAG-RSS complex formation in vitro is also not affected by the RAG-1 mutation. Interestingly, in V(D)J recombination assays performed in cell culture using an extrachromosomal substrate with a complementary pair of RSSs (12/23), wild-type (WT) and E649A RAG-1 support the accumulation of signal ends to comparable levels and mediate signal and coding joint formation similarly. In contrast, the RAG-1 E649A mutant supports higher cleavage and recombination activity than wild-type RAG-1 in the same assay using substrates containing a mispaired (12/12 or 23/23) or an unpaired RSS. Based on the data presented here, we propose that this novel RAG-1 E649A mutation interferes with the ability of the RAG complex to sense 12/23-regulated synapsis, thereby promoting cleavage in violation of the 12/23 rule.
MATERIALS AND METHODS
DNA constructs.
Eukaryotic expression constructs encoding core or full-length RAG-1 and RAG-2, appended at the amino terminus to maltose binding protein (MBP), have been described previously (53). The RAG-1 E649A mutation was generated by recombination PCR mutagenesis (18). The template for PCR was generated by subcloning a KpnI/PstI fragment from pcDNAR1 (encoding residues 384 to 755 of RAG-1) into pBluescript II SK. Template plasmid DNA was linearized with SspI or AflIII and treated with calf intestinal alkaline phosphatase. Template linearized with SspI was subjected to PCR using primer PCS8For (5′-CCCAAGCCCAATTCAGCGCTGTGTTGCAAGCCG-3′) and an antisense primer specific for the ampicillin resistance gene (6). Similarly, PCR was performed with template linearized with AflIII using primer PCS8Rev(5′-CGGCTTGCAACACAGCGCTGAATTGGGCTTGGG-3′) and a sense primer specific for the ampicillin resistance gene. The amplicons were purified using a QIAquick PCR purification kit (QIAGEN), mixed at a 1:1 ratio, and used to transform the Escherichia coli strain DH5a. Recombinant plasmids were screened for the presence of an engineered AfeI restriction site, which was further verified by DNA sequencing. The mutation was introduced into the pcDNA1 expression vectors encoding core or full-length MBP-RAG-1 by cassette replacement using BsrG1. The prokaryotic expression vector, pET11d-HMGB1-T7, encoding full-length HMGB1 tagged at the amino terminus with a polyhistidine sequence, has been previously described (6). The inversional plasmid V(D)J recombination substrate pJH299 has been previously described (16); a version of pJH299 in which the 23-RSS is replaced by a 12-RSS (12/12 pJH299) was generated by annealing two oligonucleotides, 23R12Top (5′-GATCCCACAGTGATACAGCGCTTAACAAAAACCCTCGG-3′) and 23R12Bot (5′-GATCCCGAGGGTTTTTGTTAAGCGCTGTATCACTGTGG-3′), and ligating the duplex to pJH299 digested with BamHI (which removes the 23-RSS). A clone with the 12-RSS in the same orientation as the original 23-RSS was identified by DNA sequencing. A version of pJH299 containing two 23-RSSs (23/23 pJH299) was generously provided by Joanne Hesse (16). To create versions of pJH299 containing only a 12-RSS or a 23-RSS, pJH299 was digested with either BamHI (which removes the 23-RSS) or SalI (which removes the 12-RSS) and religated, forming 12-only pJH299 and 23-only pJH299, respectively.
Protein expression and purification.
MBP-RAG-1 (wild type, catalytically inactive [D600A], or E649A and either core [cMR1] or full-length [FLMR1]) and MBP-RAG-2 (either core [cMR2] or full-length [FLMR2]) were coexpressed in 293 cells and purified as previously described (53). The protein preparations are hereafter termed WT, D600A, or E649A cMR1 (or FLMR1)/cMR2 (or FLMR2). For any given in vitro experiment, the RAG proteins used were expressed and purified in parallel. Full-length HMGB1 was expressed in the E. coli strain BL21(DE3)pLysS and purified by immobilized-metal-affinity chromatography and ion exchange chromatography as previously described (6).
Oligonucleotide cleavage and binding assays.
Radiolabeled intact and nicked 12- and 23-RSS substrates were prepared as described previously (5). A 12-RSS substrate containing a single base-pair substitution at the 5′ end of the heptamer (C17T) has also been described previously (29). The DNA cleavage and binding activities of the RAG preparations were analyzed using in-tube and in-gel cleavage assays or electrophoretic mobility shift assays (EMSA) as indicated in the text, using published procedures (5). To analyze the rates of product accumulation, preparative cleavage reactionswere assembled (50 μl) and incubated at 37°C. Aliquots (5 μl) were removed into 2 volumes of termination solution (95% formamide, 10 mM EDTA, 0.01% bromophenol blue) at the indicated time points, and reaction products were analyzed on sequencing gels as described previously (5). For the EMSAs shown in Fig. 5B, radiolabeled intact 23-RSS was incubated under standard binding conditions with WT or E649A cMR1/cMR2 with or without HMGB1 (150 ng) in the presence of increasing concentrations of cold, nonspecific 50-bp duplex DNA (DAR81/82) or cold 12-RSS as indicated in the text.
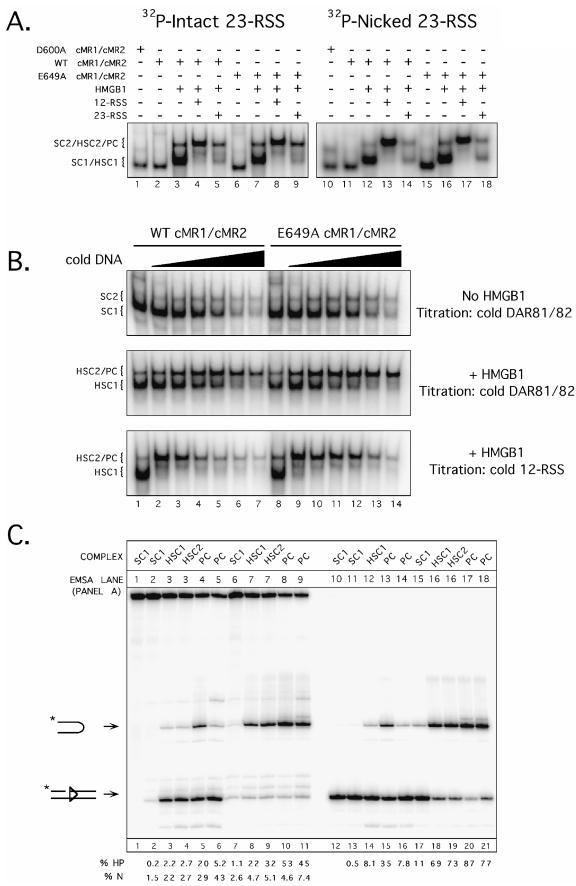
FIG.5.
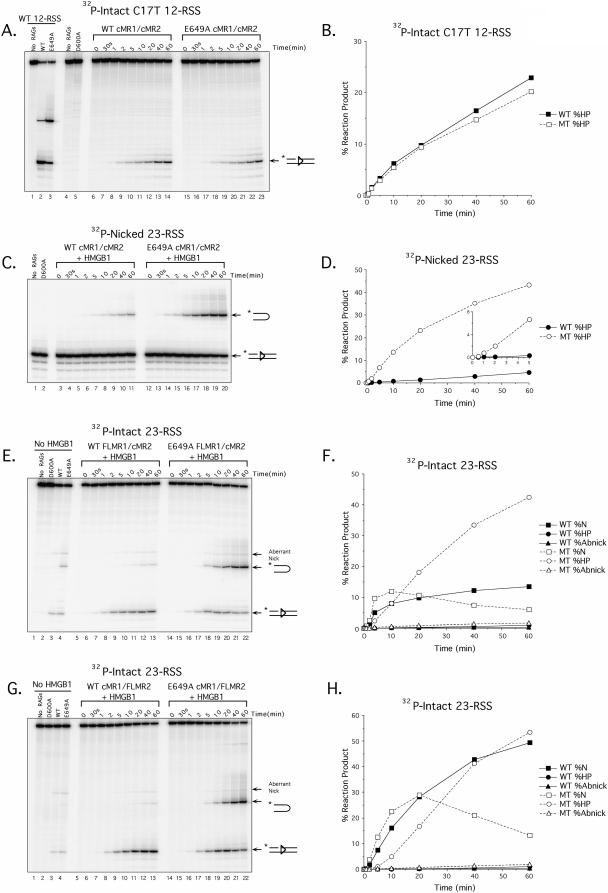
The RAG-1 E649A mutation decreases stimulation of cleavage by 12/23-regulated synapsis in vitro. (A) Radiolabeled intact 23-RSS (lanes 1 to 9) or nicked 23-RSS (lanes 10 to 18) substrates were incubated with D600A, WT, or E649A cMR1/cMR2 in the absence or presence of HMGB1 and/or cold partner DNA (12-RSS or 23-RSS) under binding conditions as indicated above the gel, and protein-DNA complexes were fractionated by EMSA. The positions of SC1, HSC1, SC2, HSC2, and PC species (described in the text) are indicated at left. (B) Radiolabeled intact 23-RSS substrate was incubated with WT or E649A cMR1/cMR2 (∼100 ng) in the absence (top panel) or presence (middle and lower panels) of HMGB1. The binding reactions shown in lanes 2 to 7 and 8 to 14 were further supplemented with increasing amounts (1, 2.5, 5, 10, 25, or 50 pmol) of either unlabeled DAR81/82 (top and middle panels) or unlabeled 12-RSS (lower panel), and the protein-DNA complexes were fractionated by EMSA and are indicated as in panel A. (C) Preparative binding reactions were assembled as shown in panel A, and the cleavage activities of various protein-DNA complexes fractionated by EMSA were compared using an in-gel cleavage assay. Reaction products from protein-DNA complexes of interest shown in the EMSA lanes in panel A (SC1, HSC1, SC2, HSC2, and PC) were recovered, normalized, and fractionated by denaturing gel electrophoresis. Positions of nicked and hairpin products are shown at left. The percentages of nicked (% N) and hairpin (% HP) products shown in each lane are quantified below the gel and account for slight variations in the amounts of DNA actually loaded. The abundance and distribution of the cleavage products observed are representative of independent experiments.
Assays for transposition, disintegration, and hybrid joint formation.
The formation of RAG target capture complexes (TCCs) and strand transfer complexes (STCs) was analyzed using assays described by Matthews et al. (31). Briefly, WT, D600A, or E649A cMR1/cMR2 (∼100 ng) and HMGB1 (∼150 ng) were incubated with 0.02 pmol of both 12- and 23-RSS signal ends for 15 min at room temperature in DNA binding buffer (25 mM morpholinepropanesulfonic acid [MOPS]-KOH [pH 7.0], 1 mM CaCl2, 100 μg/ml bovine serum albumin, 20% dimethyl sulfoxide, 60 mM potassium acetate). Subsequently, the samples were incubated with radiolabeled target DNA assembled from oligonucleotides mm30t and mm30b (37); the duplex containing 32P end-labeled mm30t was prepared as described previously (5), in either 5 mM CaCl2 or MgCl2 for 10 min at 37°C (10 μl final reaction buffer volume). Half of the sample was treated with proteinase K (200 ng/ml final concentration) and sodium dodecyl sulfate (SDS; 1% wt/vol) in the presence of EDTA (10 mM final concentration), and half of the sample was mock treated. Both samples were incubated for an additional 10 min at 37°C and then fractionated on a 4% nondenaturing polyacrylamide gel as described previously (5). Dried gels were analyzed using a Storm 860 phosphorimager. Assays for disintegration and hybrid joint formation were performed as described previously (53).
Detection of signal ends and signal and coding joint formation in cells.
The plasmid V(D)J recombination substrate pJH299 (16), which will undergo inversional rearrangement when cotransfected with mammalian RAG-1 and RAG-2 expression constructs in 293 cells, was used to assay the intermediates and products of V(D)J recombination mediated by wild-type and mutant RAG complexes in cells. Briefly, each 10-cm dish of 293 cells was transfected with 5 μg pJH299 or its derivatives and 2 μg of pcDNA1 encoding MBP-RAG-2 (core or full-length) or 3 μg of pcDNA1 encoding MBP-RAG-1 (wild-type or mutant full-length RAG-1) using 30 μg of polyethylenimine and harvested after 72 h as described previously (5). Plasmid DNA was recovered from cell pellets using a QiaPrep Spin miniprep kit (QIAGEN) and adjusted to 10 ng/μl. Primers 6111F (5′-GCTTGCGGCTCGTATGTTGG-3′) and 6363F (5′-GACCTCAGAACTCGATCTGG-3′) or 6624R (5′-GAACGGTGGTATATCCAGTG-3′) and 6683R (5′-CCAGATGGAGTTCTGAGGTC-3′) were used to amplify coding or signal joints, respectively, using real-time PCR. As a positive control, a fragment of the chloramphenicol acetyltransferase (CAT) gene on pJH299 was amplified with primers CATFOR and CATREV (53). Reaction products assembled for PCR (25 μl) contained recovered plasmid DNA (10 ng) and primer pairs (2.5 pmol each) in 1× SYBR green PCR master mix containing AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA). PCR amplification and data analysis were performed using an ABI Prism 7000 sequence detection system running the Sequence Detection System software (version 1.1; Applied Biosystems). Reaction products were subjected to initial denaturation (95°C, 10 min) and 30 cycles of amplification (95°C for 15 s and 60°C for 60 s). PCR products were fractionated on a 3% agarose gel (1:1 mixture of standard agarose and Low Range Ultra Agarose [Bio-Rad, Hercules, CA]). The target amplicons were TA cloned and sequenced to determine their composition. Signal and coding joints were also sequenced from recombinant plasmids recovered after transformation of E. coli and genetic selection on LB agar containing carbenicillin and chloramphenicol as described previously (15).
Signal ends were detected using ligation-mediated PCR (LM-PCR) according to the method of Roth et al. (42), with minor modifications. Briefly, 40 ng of plasmid DNA recovered after transfection was incubated with 200 pmol of linker DNA (assembled by annealing oligonucleotides DR19 and DR20) in the presence of one unit T4 DNA ligase (20 μl final volume) for 16 h at 15°C. Products from 1/10 of the ligation reaction were amplified by PCR using primers DR20 and 6687R (5′-GGTACATTGAGCAACTGACTG-3′). Initial denaturation was performed at 94°C for 2 min, followed by 26 cycles of amplification (94°C for 15 s, 60°C for 30 s, and 72°C for 30 s) and a final extension (72°C for 10 min). PCR products were fractionated on a 3% agarose gel, TA cloned, and sequenced.
RESULTS
Identification of a gain-of-function RAG-1 mutant that exhibits enhanced cleavage in vitro.
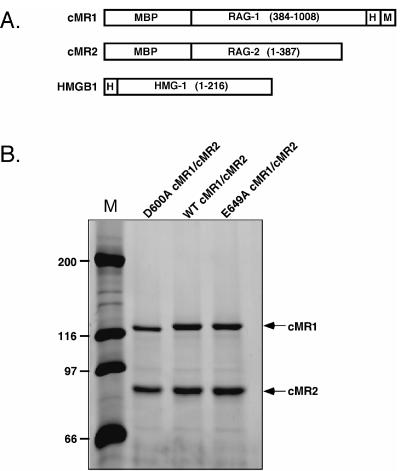
During the course of studies to identify acidic residues in RAG-1 essential for the cleavage activity of the RAG complex, we generated and individually purified an E649A cMR1 mutant that, when incubated with purified cMR2 and a nicked 12-RSS substrate in a standard in vitro cleavage reaction product containing Mg2+, catalyzed hairpin formation more readily than its wild-type counterpart (data not shown). To follow up this initial observation and to obtain more protein of higher specific activity, core forms of RAG-1 (wild type, catalytically inactive [D600A], or E649A) and RAG-2, coexpressed as MBP fusion proteins in 293 cells, were purified by amylose affinity chromatography (the protein preparations are termed WT, D600A, or E649A cMR1/cMR2) (Fig. 1).
FIG. 1.
Proteins used in this study. (A) Schematic diagrams of RAG-1, RAG-2, and HMGB1 fusion proteins are depicted (encoded residues in parentheses) and designated at left. MBP, myc (M), and polyhistidine (H) sequences are also indicated. (B) MBP-RAG fusion proteins were coexpressed in the indicated combinations and purified by amylose affinity chromatography. Protein samples were fractionated by SDS-polyacrylamide gel electrophoresis in parallel with protein standards (M; Bio-Rad) and detected by staining the gel with SYPRO orange. The positions of the RAG-1 and RAG-2 fusion proteins are shown at left. The abundances of WT, D600A, and E649A RAG-1 in FLMR1/cMR2 and cMR1/FLMR2 protein preparations were also similar (data not shown).
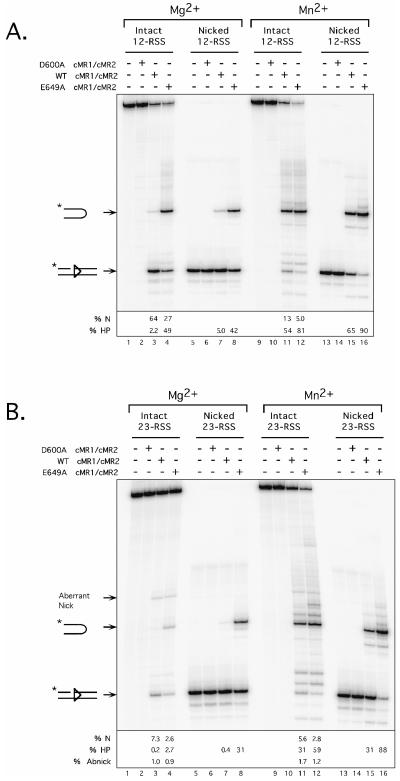
In the first set of experiments, we tested the cleavage activities of WT and E649A cMR1/cMR2 on isolated intact or nicked 12- or 23-RSS substrates (Fig. 2). As a negative control, the cleavage activity of catalytically inactive D600A cMR1/cMR2 was examined in parallel, showing a profile of products identical to samples containing no RAG proteins. As expected from our previous studies (50, 51), incubation of an intact 12-RSS with WT cMR1/cMR2 in the presence of Mg2+ for 1 h at 37°C yields predominantly the nicked product and a small amount of hairpin product. In contrast, E649A cMR1/cMR2 yields about 20-fold more hairpin product under the same conditions and about 2.5-fold less nicked product, likely because the nicked DNA is converted to hairpin products (Fig. 2A, compare lanes 3 and 4). One possible explanation for this result is that RAG-1 E649A mutation accelerates the nicking step. If the production of nicks were rate limiting to the generation of hairpin products, accelerating this step should enhance the abundance of hairpin products observed. To test this scenario, we repeated the experiment using a 12-RSS substrate containing a nick incorporated at the 5′ end of the heptamer (Fig. 2A, lanes 5 to 8). We find that the E649A cMR1/cMR2 converts the nicked substrate to hairpin product about eightfold more efficiently than WT cMR1/cMR2, providing evidence suggesting that the RAG-1 E649A mutation selectively enhances the transesterification step of the cleavage reaction. Previous studies have shown that the RAG proteins catalyze hairpin formation much more readily in the presence of Mn2+ than in the presence of Mg2+, suggesting that Mn2+ relaxes a constraint that ordinarily limits the RAG proteins from mediating this particular strand transfer reaction until a synaptic complex containing both 12- and 23-RSSs is assembled (58). We therefore wondered whether Mn2+ and the RAG-1 E649A mutation share a common mechanism for promoting hairpin formation by the RAG complex. If so, E649A cMR1/cMR2 might be expected to exhibit activity similar to that of WT cMR1/cMR2 in the presence of Mn2+. Consistent with this idea, hairpin formation catalyzed by WT and E649A cMR1/cMR2 in the presence of Mn2+ is more similar than those catalyzed in Mg2+ using both intact and nicked 12-RSS substrates, with the E649A cMR1/cMR2 yielding only slightly more (less than twofold) hairpin product than its wild-type counterpart under Mn2+ conditions (Fig. 2A, compare lane 11 to lane 12 and lane 15 to lane 16). However, since E649A cMR1/cMR2 cleavage activity is enhanced in reactions containing Mn2+ relative to reactions containing Mg2+ (Fig. 2A, compare lane 12 to lane 4 and lane 16 to lane 8), we conclude that Mn2+ and the RAG-1 E649A mutation promote cleavage activity of the RAG complex by distinct, but possibly overlapping, mechanisms. Comparable trends were observed in cleavage assays performed using either an intact or a nicked 23-RSS substrate (Fig. 2B).
FIG. 2.
The RAG-1 E649A mutation enhances RAG complex cleavage activity in vitro. Radiolabeled 12-RSS (A) or 23-RSS (B) substrates that are either intact (lanes 1 to 4 and 9 to 12) or nicked (lanes 5 to 8 and 13 to 16) were incubated for 1 h at 37°C in the absence of RAG proteins or with D600A, WT, or E649A cMR1/cMR2 in reaction buffer containing Mg2+ (lanes 1 to 8) or Mn2+ (lanes 9 to 16) as indicated above the gel. Reaction products were fractionated by denaturing gel electrophoresis and analyzed using a phosphorimager. The percentage of nicked (% N), hairpin (% HP), and aberrantly nicked (% Abnick) products shown in each lane is quantified below the gel. The gel shown is representative of results obtained with several independent protein preparations.
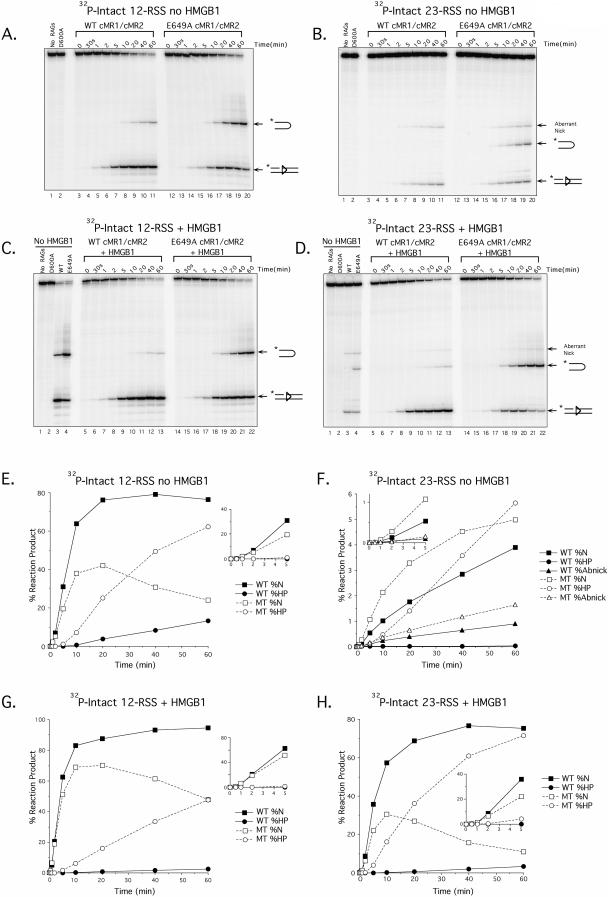
Since differences between WT and E649A cMR1/cMR2 in the ability to nick the RSS are difficult to evaluate because the nicked product is consumed in the reaction, we performed a series of time course experiments to examine the rate at which cleavage products accumulate under various reaction conditions. In the first set of experiments, cleavage product accumulation was analyzed in reactions containing WT or E649A cMR1/cMR2 assembled with intact 12- or 23-RSS substrate in the presence of Mg2+. Under these conditions, WT cMR1/cMR2 introduces nicks in a 12-RSS that are clearly detectable within about 1 min of incubation, accumulating to about 80% of all products after 1 h at 37°C; hairpin products accumulate more slowly, first becoming evident after 10 min of incubation and reaching about 10% of all products after 1 h (Fig. 3A and E). E649A cMR1/cMR2 initially introduces nicks at a rate similar to that of WT cMR1/cMR2, but these products are more rapidly converted to DNA hairpins, which are evident after 5 min of incubation and accumulate to about 60% of all products after 1 h (Fig. 3A, 3E). Because nicks generated on the 12-RSS are rapidly converted to DNA hairpins, we repeated the experiment using a 23-RSS because it is cleaved more slowly than a 12-RSS and because the RAG proteins introduce an aberrant nick in the 23-RSS in the absence of HMGB1 that is not consumed in the reaction (5). In this case, we find that WT cMR1/cMR2 introduces appropriate nicks in a 23-RSS that are detectable within about 2 min of incubation, accumulating to about 3.5% of all reaction products after 1 h at 37°C; hairpin products are almost undetectable (Fig. 3B and F). E649A cMR1/cMR2 introduces appropriate nicks at a slightly faster rate, becoming as much as twofold more abundant than in similar samples containing the WT cMR1/cMR2 before the ratio declines due to conversion of the nicked products to DNA hairpins (Fig. 3B and F). E649A cMR1/cMR2 also exhibits a slightly enhanced rate of aberrant nicking, leading us to conclude that the RAG-1 E649A mutation does indeed modestly accelerate the nicking step on a 23-RSS. For E649A cMR1/cMR2, hairpin products are detectable within about 5 min and accumulate to 6% of all products after 1 h at 37°C, an increase of 150-fold over the result for the comparable sample containing WT cMR1/cMR2.
FIG.3.
The RAG-1 E649A mutation preferentially promotes hairpin formation in vitro. (A to D) Time course assays comparing the cleavage rates between WT and E649A cMR1/cMR2 were performed in reaction buffer containing Mg2+ using either a radiolabeled intact 12-RSS (A and C) or 23-RSS (B and D) substrate in the absence (A and B) or presence (C and D) of HMGB1. Aliquots from preparative in vitro cleavage reactions were removed at the times indicated above the gel and fractionated on a sequencing gel. As negative controls for these experiments, cleavage reactions either lacking RAG proteins or containing D600A, WT, or E649A cMR1/cMR2 without HMGB1, as indicated above the gel, were incubated for 1 h at 37°C. (E to H) From the gels shown in panels A to D, respectively, hairpin (HP), correctly nicked (N), and aberrantly nicked (Abnick) reaction products (positions indicated at right of gels) were quantified using ImageQuant software and plotted as percentages of total reaction products at each time point for WT and E649A cMR1/cMR2 (WT and MT, respectively).
Since HMGB1 is known to stimulate the DNA binding and cleavage activity of the RAG complex (45, 56), we wondered whether the RAG-1 E649A mutation might functionally substitute for HMGB1 in a RAG-RSS complex. If so, one might predict that HMGB1 would fail to stimulate the cleavage activity of E649A cMR1/cMR2. To test this possibility, cleavage reaction products were supplemented with 150 ng of purified recombinant full-length HMGB1 and the time course assay was repeated (Fig. 3C, D, G, and H). As controls for this experiment, WT, D600A, and E649A cMR1/cMR2 were incubated for 1 h at 37°C in the absence of HMGB1 (Fig. 3C and D, lanes 2 to 4). Consistent with previous studies (45, 50, 56, 63), the addition of HMGB1 does not promote 12-RSS cleavage by the RAG complex but does stimulate RAG-mediated 23-RSS cleavage, while concomitantly suppressing aberrant nicking. Interestingly, HMGB1 exerts similar effects when added to cleavage reactions containing E649A cMR1/cMR2 with either a 12- or a 23-RSS substrate. In both cases, reaction products containing E649A cMR1/cMR2 accumulate hairpin products more rapidly than their counterparts containing WT cMR1/cMR2. Taken together, these data suggest that HMGB1 and the RAG-1 E649A mutation function by distinct mechanisms to enhance the cleavage activity of the RAG complex.
To more clearly illustrate the selective effect of the E649A RAG-1 mutation on hairpin formation, two additional time course experiments were performed. The first experiment examined accumulation of nicked products using a 12-RSS substrate that fails to support hairpin formation due to the presence of a single base-pair substitution at the 5′ end of the heptamer (called C17T) (29). Interestingly, we find that, like WT cMR1/cMR2, E649A cMR1/cMR2 nicks the C17T substrate but cannot convert the nicked products to DNA hairpins. Moreover, E649A cMR1/cMR2 nicks this substrate with slightly delayed kinetics relative to WT cMR1/cMR2, in subtle contrast with results obtained with a consensus 23-RSS. In the second experiment, the time course assay was repeated using a nicked 23-RSS substrate to bypass the need for the RAG proteins to produce this reaction intermediate. Consistent with results obtained with intact substrates, hairpin products accumulate faster in samples containing E649A cMR1/cMR2 than in those containing its wild-type counterpart, both in the absence and in the presence of HMGB1 (Fig. 4C and D and data not shown). Similar trends were observed when these experiments were repeated using purified E649A FLMR1/cMR2 (Fig. 4E and F) or cMR1/FLM2 (Fig. 4G and H), indicating that the presence of noncore portions of RAG-1 or RAG-2, respectively, does not suppress the ability of the E649A RAG-1 mutation to enhance RAG complex cleavage activity in vitro.
FIG. 4.
Effects of composition of RSS substrate and RAG protein on in vitro cleavage activity. Time course assays comparing the cleavage rates between WT and E649A (MT) cMR1/cMR2 (A to D), WT and E649A FLMR1/cMR2 (E and F), or WT and E649A cMR1/FLMR2 (G and H) were performed in reaction buffer containing Mg2+ using either a radiolabeled intact 12-RSS containing a single base-pair substitution (C17T) at the 5′ end of the heptamer (A and B), a nicked 23-RSS substrate (C and D), or an intact 23-RSS substrate (E to H) in the absence (A and B) or presence (C to H) of HMGB1. Experiments were performed and analyzed as shown in Fig. 3.
The E649A RAG-1 mutation does not affect RAG-RSS complex formation but diminishes 12/23-regulated enhancement of RAG-mediated cleavage activity in vitro.
In principle, the enhanced activity of E649A cMR1/cMR2 could be attributed to its ability to support protein-DNA complex formation better than its wild-type counterpart. To examine this possibility, we compared the DNA binding activities of WT and E649A cMR1/cMR2 using EMSA. In these experiments, WT or E649A cMR1/cMR2 was incubated with either a radiolabeled intact or a nicked 23-RSS substrate in binding reactions containing Ca2+ in the absence or presence of HMGB1. After assembling these reaction products, some samples containing HMGB1 were further supplemented with cold partner 12- or 23-RSS to form paired RSS complexes, and then the protein-DNA complexes were fractionated using an EMSA (Fig. 5A). We find that in the absence of HMGB1, both WT and E649A cMR1/cMR2 form two distinct protein-DNA complexes, called SC1 and SC2, similarly when incubated with an isolated RSS substrate, both with intact and with nicked 23-RSS substrates. In a previous study (51), both complexes were shown to contain a RAG-1 dimer, but the RAG-2 stoichiometries differed between SC1 and SC2, with the former containing monomeric RAG-2 and the latter containing two RAG-2 molecules. For both RAG preparations, the SC1 and SC2 complexes are comparably supershifted in the presence of HMGB1, forming HSC1 and HSC2, respectively. Moreover, adding cold 12-RSS partner DNA in both cases comparably promotes the formation of a paired complex (PC) that is slightly supershifted relative to HSC2; this complex is less evident when cold 23-RSS partner is added (Fig. 5A). Similar results were obtained when the EMSA was repeated using Mg2+ instead of Ca2+ in the binding reactions (data not shown). Taken together, these results suggest that the RAG-1 E649A mutation does not dramatically alter the spectrum of protein-DNA complexes that the RAG proteins are capable of assembling on oligonucleotide substrates.
Given these results, we next considered whether the RAG-1 E649A mutation enhances the specificity of the RAG complex, a possibility that the experiments shown in Fig. 5A do not directly address. To test this possibility, we incubated WT or E649A cMR1/cMR2 with a radiolabeled 23-RSS and increasing concentrations of cold, nonspecific 50-bp duplex DNA (DAR81/82) in the absence or presence of HMGB1 (Fig. 5B). We find that the abundance of SC1 and HSC1 (formed in the absence and presence of HMGB1, respectively) decreases similarly for both WT and E649A cMR1/cMR2 as a function of increasing DAR81/82 concentration. Interestingly, in the presence of HMGB1, levels of PC increase as the amounts of DAR81/82 increase to 10 nmol in the binding reactions for both WT and E649A cMR1/cMR2, but whereas the levels of PC decline thereafter in samples containing WT cMR1/cMR2, PC levels remain stable against increasing levels of DAR81/82 with E649A cMR1/cMR2 (Fig. 5B). The same trend is observed when cold 12-RSS partner is titrated into the binding reaction, except that less cold 12-RSS is required to promote PC formation, and PC levels eventually decline in the presence of excess cold 12-RSS (Fig. 5B). These results suggest that the RAG-1 E649A mutation helps stabilize the PC, causing it to resist disassembly in the presence of excess cold DNA.
To exclude any subtle contribution that the RAG-1 E649A mutation provides to enhance the DNA binding activity of the RAG complex, we directly compared the catalytic activities of various preformed protein-DNA complexes assembled with WT or E649A cMR1/cMR2 using an in-gel cleavage assay. In this experiment, WT or E649A cMR1/cMR2 was incubated with radiolabeled intact or nicked 23-RSS substrates in preparative binding reactions assembled as shown in Fig. 5A and fractionated on the same nondenaturing polyacrylamide gel. The gel was then submerged in reaction buffer containing Mg2+ to initiate RSS cleavage, and reaction products were recovered from complexes of interest and analyzed on a sequencing gel (Fig. 5C). Consistent with results obtained from in-tube cleavage experiments, we find that SC1, HSC1, and HSC2 complexes formed with E649A cMR1/cMR2 remain more active than their wild-type counterparts when directly compared in the in-gel cleavage assay. However, the difference in the abundance of hairpin products observed between RAG-RSS complexes assembled with WT and E649A cMR1/cMR2 using the in-gel cleavage assay is more modest than for similarly prepared samples analyzed using the in-tube assay after 1 h at 37°C. Interestingly, however, PCs formed with E649A cMR1/cMR2 assembled in the presence of either cold 12-RSS or 23-RSS partner DNA are similarly active in the in-gel cleavage assay, whereas the PCs formed with WT cMR2/cMR2 assembled with cold 12-RSS partner are about fourfold more active than their counterparts assembled with cold 23-RSS partner. This result suggests that the RAG-1 E649A mutation bypasses the stimulation of RAG-mediated cleavage provided by 12/23-regulated synapsis in vitro.
Other DNA strand transfer reactions are mediated similarly by WT and E649A RAG-1.
The formation of DNA hairpins is physiologically the most important DNA strand transfer reaction that the RAG proteins catalyze. However, they are also known to mediate several alternative DNA strand transfer reactions, including the rejoining of signal ends and coding ends in either the original or the opposing configuration in a reversal of the cleavage reaction (yielding open-shut joints or hybrid joints, respectively) (27, 35), the integration of signal ends into nonhomologous DNA by classical transposition (1, 17), a retrovirus-like disintegration reaction that reverses transpositional strand transfer (34), and an “inverse transposition” reaction, in which a non-RSS DNA strand is transferred into a canonical RSS (48). In principle, one might expect that since the RAG-1 E649A mutation promotes accelerated hairpin formation, this RAG-1 mutant might similarly exhibit enhanced ability to mediate alternative DNA strand transfer reactions. To test the possibility, we evaluated WT and E649A cMR1/cMR2 in assays of RAG-mediated transposition, disintegration, and hybrid joint formation.
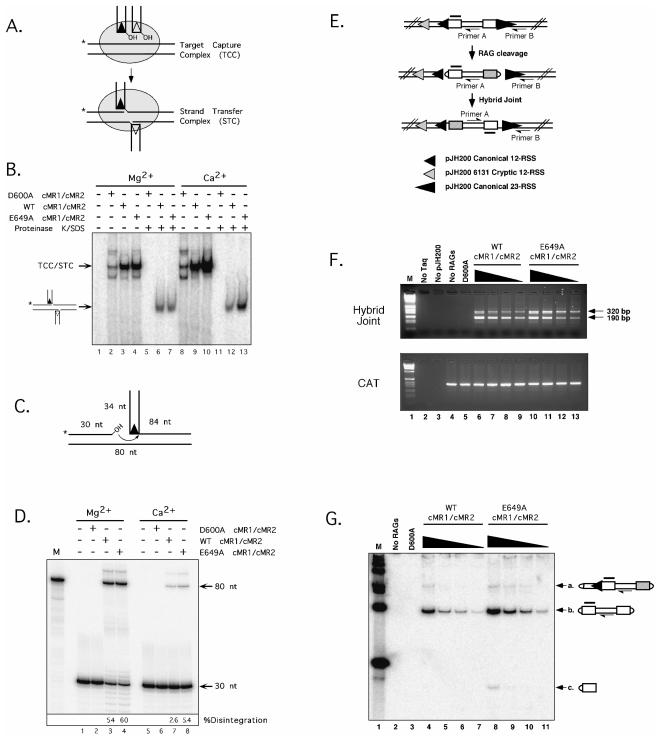
To assay transposition activity, cMR1/cMR2 preparations (D600A, WT, or E649A) were assembled with unlabeled 12- and 23-RSS signal ends in the presence of HMGB1 to form a signal end complex and incubated at 37°C with a 32P-labeled DNA target in reaction buffer containing Mg2+ or Ca2+, and protein-DNA complexes were fractionated on a nondenaturing polyacrylamide gel. A signal end complex that has captured a DNA target is termed a TCC; if the RSS donor becomes covalently linked to the target DNA, the complex is termed an STC (Fig. 6A). The TCC and STC cannot be resolved on a native polyacrylamide gel, but prior treatment with proteinase K and SDS will liberate the transposition product from the STC, enabling it to be visualized on a native gel as a species whose mobility is retarded relative to that of free DNA but faster than that of the TCC/STC. Consistent with previous results (31), catalytically inactive D600A cMR1/cMR2 forms the TCC/STC poorly, as target capture is impaired by mutations in the DDE motif of RAG-1 (Fig. 6B). The residual complexes we observe are attributed to nonspecific binding to target DNA. In the presence of Mg2+, both WT and E649A cMR1/cMR2 form the TCC/STC to comparable levels; in the presence of Ca2+, STC/TCC complex formation is modestly enhanced in samples containing E649A cMR1/cMR2 relative to that in samples containing WT cMR1/cMR2 (Fig. 6B). After treatment with proteinase K and SDS, no transposition product is detected in samples containing D600A cMR1/cMR2 in reaction buffers containing either Mg2+ or Ca2+, a result consistent with data published previously (31). In samples containing WT or E649A cMR1/cMR2 and assembled with Mg2+, the transposition product is detected at similar levels; in Ca2+, this product is about twofold more abundant in the sample containing E649A cMR1/cMR2 than in that containing WT cMR1/cMR2. The difference is likely attributed to the approximately twofold increase in STC/TCC formation observed for E649A cMR1/cMR2 over that for WT cMR1/cMR2 under the Ca2+ conditions (Fig. 6B). These data lead us to conclude that the RAG-1 E649A mutation does not alter the transpositional strand transfer activity of the RAG complex but slightly enhances target site capture activity in the presence of Ca2+.
FIG. 6.
Intermolecular DNA strand transfer reactions are not significantly enhanced by the RAG-1 E649A mutation. (A) Diagram of TCC/STC formation. The RAG proteins and HMGB1 assemble a protein-DNA complex (shaded oval) with 12- and 23-signal ends (the donor DNA), capture a radiolabeled DNA target (the TCC; the position of 32P is indicated by an asterisk), and integrate the signal ends into target DNA (the STC). (B) Donor and target DNA were incubated in reaction buffer containing Mg2+ (lanes 1 to 7) or Ca2+ (lanes 8 to 13) with D600A (inactive), WT, or E649A cMR1/cMR2 in the presence of HMGB1 as indicated. Untreated samples (lanes 2 to 4 and 8 to 10) and samples further incubated with proteinase K and SDS (lanes 5 to 7 and 11 to 13) were fractionated on a native 4% polyacrylamide gel. The positions of the mixed TCC/STC species and the transposition products released by proteinase K/SDS treatment are shown at left. (C) Schematic diagram of the disintegration substrate and reaction outcome. (D) The disintegration substrate was incubated in reaction buffer containing Mg2+ (lanes 1 to 4) or Ca2+ (lanes 5 to 8) with D600A, WT, or E649A cMR1/cMR2 as indicated. Reaction products were fractionated on a sequencing gel and analyzed using a phosphorimager. The radiolabeled 80-nucleotide bottom strand oligonucleotide serves a sizing marker (M) for the position of the expected disintegration product. The percentage of disintegration product shown in each lane is quantified below the gel. (E) Diagram of the PCR assay used to detect hybrid joints formed on pJH200. The positions of PCR primers A and B (half arrowhead) and the Southern hybridization probe (shaded overline) are shown. The relative positions and orientations of the canonical 12- and 23-RSS are indicated by small and large filled triangles; the “6131” cryptic 12-RSS (26) is indicated by a shaded triangle. (F) Supercoiled pJH200 was incubated with D600A (100 ng), WT, or E649A cMR1/cMR2 (100, 50, 25, and 12 ng) under conditions that permit coupled cleavage. PCR was performed on a portion of the reaction products using primers designed to detect hybrid joints (top panel) or CAT (bottom panel). Samples were run in parallel with molecular sizing markers (M; 1-kb ladder; Invitrogen); the positions of the ∼190- and 320-bp amplicons are denoted by arrows at right. (G) The portion of the cleavage reaction products not used for PCR detection of hybrid joints shown in panel F was fractionated on a 7% nondenaturing polyacrylamide gel and analyzed by Southern hybridization using the probe shown in panel E. The composition of the three major cleavage products is shown at right.
We next considered whether the RAG-1 E649A mutation enhances a disintegration reaction that models the reversal of transpositional strand transfer (34). For these experiments, a substrate containing a 12-RSS integrated into a radiolabeled target DNA was incubated with the RAG proteins in the presence of either Mg2+ or Ca2+. In this reaction, RAG-mediated disintegration regenerates the “top” strand of the labeled target, yielding an 80-nucleotide product (Fig. 6C). In the presence of Mg2+, both WT and E649A cMR1/cMR2 support the disintegration reaction to similar levels; in the presence of Ca2+, the E649A cMR1/cMR2 yields about twofold more disintegration product than WT cMR1/cMR2 (Fig. 6D), most likely attributed to slightly enhanced substrate binding under these conditions (data not shown). Thus, we conclude that the RAG-1 E649A mutation does not alter the disintegration activity of the RAG complex.
Since the RAG-1 E649A mutation facilitates hairpin formation, we wondered whether this mutation would also affect the rate at which this reaction is reversed. As discussed above, the outcome of reversing this reaction depends upon which signal end attacks the hairpin. In the experiments described here, we chose to examine cleavage and rejoining in a plasmid V(D)J recombination substrate called pJH200, which normally yields signal joints after V(D)J rearrangement but can yield hybrid joints detectable by PCR if the RAG proteins mediate the inversion of the intervening sequence in vitro (the strategy is outlined in Fig. 6E). We previously demonstrated that PCR amplifies three major products from pJH200 after incubation with the RAG proteins (53): an ∼190-bp “canonical hybrid joint” product corresponding to the rejoining of the 23-RSS to the coding sequence originally abutting the 12-RSS, an ∼320-bp “cryptic hybrid joint” product that results from the rejoining of the 23-RSS with the coding sequence abutting a cryptic 12-RSS called 6121 (usually incorporating a single P nucleotide), and an ∼400-bp product reflecting a wider spectrum of integration products upstream of the cryptic 12-RSS whose underlying molecular mechanisms remain uncertain. Using this PCR assay with a semiquantitative approach, we find that E649A cMR1/cMR2 supports formation of the “ca nonical hybrid joint” at levels comparable to those supported by its wild-type counterpart, but the “cryptic hybrid joint” is formed slightly more readily (approximately twofold) by E649A cMR1/cMR2 than by WT cMR1/cMR2 (Fig. 6F). This difference may be attributed in part to the approximately twofold increase in substrate cleavage levels observed in reaction products containing E649A cMR1/cMR2 over those in reaction products containing WT cMR1/cMR2 as assessed by Southern hybridization (Fig. 6G) but does not entirely explain why the distribution of amplicons observed after PCR is skewed toward the “cryptic hybrid joint” product for E649A cMR1/cMR2. Thus, these results raise the possibility that the RAG-1 E649A mutation eases a constraint that normally limits hybrid joint formation with a noncanonical RSS.
The E649A RAG-1 mutation does not enhance canonical V(D)J recombination but impairs adherence to the 12/23 rule in vivo.
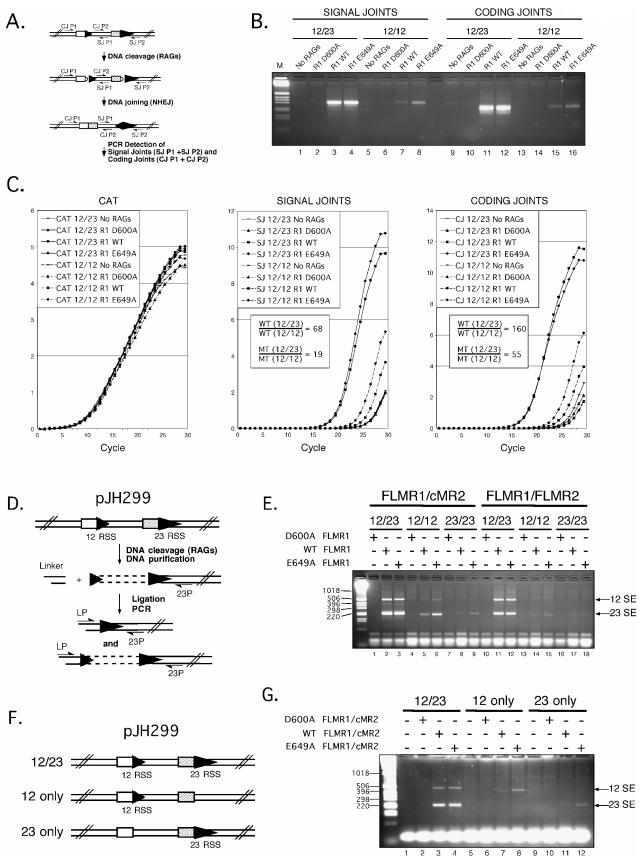
Since E649A cMR1 (and FLMR1)/cMR2 exhibits a significant rate enhancement in hairpin formation in vitro, we naturally wondered whether this mutant would support higher levels of V(D)J recombination in vivo. To test this possibility, we used a well-established cell culture-based assay of V(D)J recombination which utilizes a plasmid substrate, called pJH299 (16), that undergoes RAG-mediated inversional rearrangement when the substrate is cotransfected with RAG-1 and RAG-2 expression constructs in 293 cells. At 72 h after transfection, DNA harvested from the cells was subjected to PCR to detect signal joint and coding joint formation using the strategy outlined in Fig. 7A. In control reactions containing DNA obtained from cells transfected with pJH299 and either the empty mammalian expression vector pcDNA1 or versions of pcDNA1 encoding core MBP-RAG-2 and an MBP-tagged version of full-length D600A RAG-1, specific amplification of signal joints or coding joints is not detected, although a very low level of nonspecific amplification is observed (Fig. 7B). In contrast, signal joints and coding joints are readily amplified in PCR samples containing DNA from cells transfected with pJH299, the core MBP-RAG-2 expression construct, and pcDNA1 encoding an MBP-tagged version of full-length RAG-1 (WT or E649A). Interestingly, the levels of reaction products observed after PCR are quite similar in both cases (Fig. 7B, compare lanes 3 and 4 and lanes 11 and 12). This outcome is consistent with an earlier report showing that a comparable E649Q RAG-1 mutant did not exhibit a hyper-recombination phenotype (24). As expected from previous studies (16), when rearrangement is assayed using a form of pJH299 containing a pair of 12-RSSs (12/12), we find that full-length WT and E649A RAG-1 support lower levels of signal joint and coding joint formation detectable by PCR than when similar experiments are performed using 12/23 pJH299. However, in contrast to results obtained with 12/23 pJH299, E649A RAG-1 consistently supports slightly more rearrangement than WT RAG-1 on a 12/12 version of pJH299 (Fig. 7B, compare lanes 7 and 8 and lanes 15 and 16). To more carefully evaluate these differences, we employed real-time PCR to analyze the rate of amplicon accumulation by the comparative threshold approach (14), using amplification of a fragment from the CAT gene in pJH299 as a calibrator (Fig. 7C). These data confirm that while the 12/23 pJH299 signal joint and coding joint amplicons accumulate at comparable rates from WT and E649A RAG-1 samples, 12/12 signal joint and coding joint amplicons accumulate about threefold faster from E649A RAG-1 samples than from their wild-type counterparts (Table 1).These differences cannot be attributed to variations in the amounts of template in the reaction products, as PCR amplification of CAT proceeds similarly in all samples tested (Fig. 7C). Whether E649A RAG-1 exhibits elevated recombination activity on a 23/23 version of pJH299 could not be ascertained unambiguously, as levels of signal and coding joint amplicons obtained with WT or E649A RAG-1 using this substrate were very close to background levels obtained with inactive D600A RAG-1 (Table 1). In the presence of full-length MBP-RAG-2, WT and E649A RAG-1 also exhibited similar recombination activities using 12/23 pJH299, although the abundances of signal and coding joint amplicons detected were consistently two- to threefold lower, but 12/12 and 23/23 rearrangements were virtually undetectable above background levels (Table 1).
FIG. 7.
The RAG-1 E649A mutation relaxes enforcement of the 12/23 rule in vivo. (A) PCR assay of coding joint (CJ) and signal joint (SJ) formation. V(D)J rearrangement of the plasmid substrate pJH299 results in inversion of the DNA sequence between the RSSs, yielding signal and coding joints detectable with primer pairs SJP1/SJP2 and CJP1/CJP2, respectively. (B and C) Plasmid DNA isolated from 293 cells 72 h after transfection with RAG expression constructs and 12/23 or 12/12 versions of pJH299 was subjected to real-time PCR to detect signal and coding joint formation using the SYBR green dye. As a control, a fragment of the CAT gene in pJH299 was also amplified. (B) Amplicons representing signal joints and coding joints were visualized on an agarose gel by staining with ethidium bromide. (C) Amplification curves charting fluorescence emission at each reaction cycle were generated for each PCR to detect CAT, signal joint formation, or coding joint formation from samples shown in panel B containing 12/23 pJH299 (solid line) or 12/12 pJH299 (dashed line). The difference (n-fold) in the relative abundances of signal or coding joints detected by real-time PCR in samples containing 12/23 versus 12/12 pJH299 recovered after transfection with WT or E649A (MT) RAG-1 is shown in the inset. (D) Ligation-mediated PCR strategy to detect signal ends. Cleavage at either RSS in vivo yields a DNA break that can be detected by ligating linker DNA to the blunt signal end and amplifying the ligated end using a linker primer (LP) and a primer specific to the sequence 3′ of the 23-RSS (23P). (E) Plasmid DNA recovered from 293 cells cotransfected with various pcDNA1 expression constructs encoding FLMR1 (D600A, WT, or E649A) and either cMR2 or FLMR2 and various forms of pJH299 (12/23, 12/12, or 23/23; the combinations are as indicated above the gel) was subjected to LM-PCR using the LP/23P primer pair shown in panel D, and the amplicons were visualized on an agarose gel by staining with ethidium bromide. The positions of PCR products corresponding to linker ligation to signal ends produced at the 12-RSS (12-SE) and 23-RSS (23-SE) are indicated at right. (F) Diagram of forms of pJH299 containing a 12/23 pair of RSSs or a single 12- or 23-RSS (12 only and 23 only, respectively). (G) LM-PCR products obtained from plasmid DNA recovered from cells cotransfected with the pcDNA1 expression constructs shown in panel E and the forms of pJH299 shown in panel F. Note that removal of the 23-RSS shortens the length of the PCR product that detects the cleaved single 12-RSS.
TABLE 1.
Comparisons of signal and coding joint formation determined by real-time PCR
| Protein preparation, pJH299 substrate, and joint analyzeda | Relative amplicon level obtained with indicated RAG-1/2 combinationd
|
|
|---|---|---|
| FLMR1/cMR2 | FLMR1/FLMR2 | |
| E649A RAG-1 12/23 vs WT RAG-1 12/23 | ||
| SJ | 1.13 ± 0.42b | 0.82 ± 0.19 |
| CJ | 1.03 ± 0.30c | 0.93 ± 0.24 |
| E649A RAG-1 12/12 vs WT RAG-1 12/12 | ||
| SJ | 2.90 ± 0.52b | 1.94 ± 1.17 |
| CJ | 2.69 ± 0.46c | 1.75 ± 0.41 |
| E649A RAG-1 23/23 vs WT RAG-1 23/23 | ||
| SJ | 1.81 ± 0.58 | 0.84 ± 0.13 |
| CJ | 1.57 ± 0.40 | 1.54 ± 0.19 |
| WT RAG-1 12/23 vs D600A RAG-1 12/23 | ||
| SJ | 319 ± 21 | 145 ± 76 |
| CJ | 501 ± 278 | 232 ± 193 |
| WT RAG-1 12/12 vs D600A RAG-1 12/12 | ||
| SJ | 5.21 ± 2.83 | 1.73 ± 0.77 |
| CJ | 5.55 ± 3.42 | 1.59 ± 0.77 |
| WT RAG-1 23/23 vs D600A RAG-1 23/23 | ||
| SJ | 1.55 ± 0.71 | 1.78 ± 0.59 |
| CJ | 2.06 ± 0.71 | 1.56 ± 0.57 |
SJ, signal joint; CJ, coding joint.
P of <0.003 comparing FLMR1/cMR2 SJ between 12/23 and 12/12 using the paired t test (n = 5).
P of <0.004 comparing FLMR1/cMR2 CJ between 12/23 and 12/12 using the paired t test (n = 5).
Relative amplican levels for WT FLMR1/cMR2 12/23 versus WT FLMR1/FLMR2 12/23 are as follows: SJ, 3.71 ± 1.69; CJ, 2.38 ± 0.33.
In principle, the failure to observe greater recombination of 12/23 pJH299 with E649A RAG-1 than with WT RAG-1 in vivo may be because the activity of the nonhomologous end-joining factors is rate limiting for V(D)J recombination in this assay. Hence, greater levels of RAG-mediated cleavage may not necessarily result in higher levels of DNA end joining. To test this possibility, we used LM-PCR to determine the abundance of signal end intermediates in the same DNA preparations used for evaluating signal and coding joint formation (schematically depicted in Fig. 7D). We find that two major amplicons are detected after LM-PCR from DNA isolated from cells transfected with pJH299 and WT or E649A RAG-1 expression constructs, which are not evident in samples derived from cells transfected with a construct encoding inactive D600A RAG-1 (Fig. 7E). Sequencing of the TA-cloned products reveals that the smaller of the two amplicons is the expected product derived from linker ligation to the 23-RSS. The larger product is derived from linker ligation to a cleaved 12-RSS in which the 23-RSS partner is intact or nicked (PCR cannot distinguish between these possibilities). Since the abundance and distribution of the two PCR products are comparable between WT and E649A RAG-1 samples, we conclude that the enhancement in cleavage activity apparent with E649A RAG-1 in vitro is not evident in vivo when assayed using recombination substrates bearing a 12/23-RSS pair. However, when the assay is performed using the 12/12 or 23/23 form of pJH299, E649A RAG-1 is found to support about two- to threefold higher levels of signal end formation detectable by LM-PCR than WT RAG-1, which is consistent with assays of signal and coding joint formation, but the levels of these products remain lower than those observed using 12/23 pJH299 (Fig. 7E). Interestingly, in the presence of core RAG-2, E649A RAG-1 also supports two- to threefold more cleavage of pJH299 containing either a single 12- or 23-RSS (Fig. 7F, 7G), suggesting that cleavage of 12/12 and 23/23 substrates in this case may be independent of synapsis. E649A RAG-1 also supports levels of signal end formation on 12/23 pJH299 comparable to those supported by WT RAG-1 in the presence of full-length RAG-2, but the abundance of the amplicons (especially the 23SE) is consistently lower than when core RAG-2 is present (Fig. 7E). Moreover, signal ends generated from 12/12 and 23/23 forms of pJH299 in the presence of full-length RAG-2 are essentially undetectable above background levels. Taken together, these data suggest that the RAG-1 E649A mutation relaxes adherence to the 12/23 rule, thereby promoting greater cleavage and recombination of substrates containing a mispaired or an unpaired RSS.
Finally, we wondered whether E649A RAG-1 might promote excessive processing of signal and/or coding ends in vivo if it exhibits poor postcleavage complex stability. To examine this possibility, we compared the sequences of signal joints and coding joints in rearranged pJH299 plasmids (both 12/23 and 12/12) recovered from E. coli transformants selected on LB agar plates containing carbenicillin and chloramphenicol. No striking differences in the abundance or distribution of insertions or deletions in signal joints or coding joints were apparent in 12/23 pJH299 plasmids recovered from cells transiently expressing WT or E649A RAG-1 in the presence of core RAG-2 (Table 2). Unexpectedly, we failed to recover any 12/12 pJH299 recombinants that had undergone inversional recombination, even though sequence analysis of signal joints and coding joints detected by PCR clearly indicated the presence of such molecules. Instead, all 12/12 pJH299 recombinants recovered after antibiotic selection from both WT and E649A RAG-1 samples had undergone a deletion event in which the heptamer of the 12-RSS that replaced the 23-RSS was joined precisely to the 3′ end of the heptamer of the other 12-RSS (Fig. 8). Visual inspection of the sequence reveals that the heptamer at this site might service a nonamer-like sequence in the opposite orientation, creating a cryptic 23-RSS in a deletional configuration with the 12-RSS (Fig. 8). This outcome would not be detected by our PCR strategy, as the internal primer sites are deleted after recombination, but must be at least 10-fold more frequent than inversional recombination, since no inversional recombinants were recovered from more than 15 colonies identified and characterized after genetic screening.
TABLE 2.
Sequence analysis of 12/23 pJH299 coding joints obtained from recombination assays of WT or E649A FLMR1/cMR2
| Clone | Result for 23-RSS coding enda | P nucleotide(s) | Result for 12-RSS coding endb |
|---|---|---|---|
| WT 12/23 | |||
| DA7.2 | −0 | −3 | |
| PS3.15 | −0 | −3 | |
| PS3.2 | −0 | −4 | |
| PS3.3 | −0 | −6 | |
| DA7.4 | −0 | −10 | |
| PS3.10 | −1 | −3 | |
| PS3.4 | −1 | −4 | |
| PS3.1 | −1 | −8 | |
| PS3.5 | −3 | C | −0 |
| DA7.5 | −6 | −5 | |
| PS3.13 | −6 | −5 | |
| PS3.6 | HJ | −8 | |
| PS3.9 | HJ | −3 | |
| E649A 12/23 | |||
| PS4.7 | −0 | GGA | −1 |
| DA10.2 | −0 | −3 | |
| PS4.1 | −0 | −7 | |
| PS4.12 | −0 | −7 | |
| PS.4.9 | −0 | −8 | |
| PS4.11 | −1 | −10 | |
| PS4.10 | −3 | C | −0 |
| DA10.3 | −3 | −5 | |
| DA10.1 | −4 | −1 | |
| PS4.13 | −4 | −1 | |
| PS4.3 | HJ | −2 | |
| PS4.14 | HJ | −3 |
Coding end sequence TCGATGAGAGGATCC.
Coding end sequence GTCGACCTGCAGCCC.
HJ, hybrid joint formed by deletion to 23-RSS.
FIG. 8.
Structure of pJH299 recombinants obtained after genetic selection. Diagrams of 12/23 and 12/12 versions of pJH299 are shown at left and right, respectively. Heptamer and nonamer elements are boxed, and the spacer lengths are indicated. V(D)J recombination of 12/23 pJH299 results primarily in the inversion of the DNA sequence between the RSSs. For 12/12 pJH299, all recombinants recovered after genetic selection showed precise joining of the replaced 23-RSS heptamer to the 3′ end of the original 12-RSS heptamer. We speculate that a cryptic nonamer may service the original 12-RSS heptamer in the inverse orientation, forming a cryptic 23-RSS in a deletional configuration. The putative cryptic nonamer lies 22 bp from the heptamer and is shown aligned with a canonical nonamer sequence. Sequence identities between canonical and cryptic heptamer and nonamer sequences are underlined.
DISCUSSION
RAG-1 and RAG-2 have been subjected to extensive mutagenesis by many laboratories (2, 7, 9, 11, 12, 20-22, 24, 28, 33, 39, 43, 44, 47, 49, 52, 54, 60). Through these efforts, several classes of mutations have been identified, including mutations that impair the DNA binding activity of the RAG complex, abolish one or both biochemical steps of RAG-mediated DNA cleavage (nicking and transesterification), disrupt the joining phase of V(D)J recombination, or alter RAG-1-RAG-2 association or interactions between the RAG complex and other proteins. To date, no mutant forms of RAG-1 or RAG-2 that enhance the cleavage or joining phases of V(D)J recombination have been reported in the literature. Since V(D)J recombination shares mechanistic similarities with transposition, for which gain-of-function mutants have been reported in several systems (4, 23, 30, 36, 61, 64), it is likely that the RAG complex in its current form is less active than it potentially could be. In support of this possibility, we report here that mutation of a single residue in RAG-1, E649A, specifically enhances the ability of the RAG complex to catalyze hairpin formation in vitro and facilitate greater cleavage of plasmid V(D)J recombination substrates containing an unpaired RSS or a mispaired (12/12 or 23/23) RSS in vivo, in violation of the 12/23 rule. The E649A RAG-1 mutation is located within the central domain of RAG-1 (8), which encompasses two residues critical for the catalytic activity of the recombinase, D600 and D708 (12, 21, 24). This colocalization suggests that the central domain plays an active role not only in recognition and catalysis at the site of DNA cleavage but also in perceiving 12/23-regulated synapsis.
E649A RAG-1 as a separation-of-function mutant for RAG-mediated DNA strand transfer reactions.
The core RAG complex is capable of mediating several DNA strand transfer reactions, including the hairpin formation step of the V(D)J cleavage reaction, open-shut and hybrid joint formation, transpositional strand transfer, and a retroviral-like disintegration reaction. All of these reactions involve the attack of a 3′-OH on a phosphodiester bond. Despite this mechanistic similarity, the observation that the E649A cMR1/cMR2 complex selectively enhances the hairpin formation step of V(D)J cleavage in vitro, while not substantially altering other DNA strand transfer reactions mediated by the RAG complex, clearly illustrates that these reactions are biochemically distinct.
The feature that separates hairpin formation from other DNA strand transfer reactions mediated by the RAG complex is that hairpin formation is an intramolecular strand transfer reaction, whereas the other reactions can be considered intermolecular strand transfer reactions. We speculate that the RAG-1 E649A mutation may relax a constraint that normally limits the efficiency of hairpin formation. One constraint placed on this reaction is the steric requirements involved in positioning the 3′ terminal nucleotide in an orientation that allows for in-line attack of the 3′-OH on the opposing phosphodiester strand. In principle, the RAG-1 E649A mutation might ease the ability of the RAG complex to induce the conformational changes necessary in the DNA to facilitate this reaction. However, if this were true in vivo, one might expect that E649A RAG-1 would support enhanced cleavage and recombination of the pJH299 plasmid V(D)J recombination substrate in vivo, which is not observed. While this outcome could be attributed simply to a failure of the RAG-1 E649A mutation's effect in vitro to be recapitulated in vivo, an outcome with precedents in studies of other transposases (55), we consider an alternative possibility in which the RAG-1 E649A mutation disrupts the sensing of 12/23-regulated synapsis by the RAG complex, which is normally required to promote cleavage at both RSSs. In this scenario, the RAG-1 E649A mutation “tricks” the RAG complex into falsely perceiving synapsis, causing it to cleave a bound RSS as though it were integrated into a paired complex. Three lines of evidence support this hypothesis. First, in the presence of Mn2+, which is known to uncouple the 12/23 rule, the cleavage activities of WT and E649A cMR1/cMR2 using isolated RSS substrates are quite similar. Second, unlike that of WT cMR1/cMR2, the cleavage activity of E649A cMR1/cMR2 is not significantly stimulated by paired-complex formation as assessed by in-gel cleavage assays. Third, the cleavage and recombination activities supported by E649A RAG-1 on V(D)J recombination substrates containing an unpaired or a mispaired RSS in vivo are enhanced relative to those supported by WT RAG-1 (at least in the presence of core RAG-2). However, we recognize that the 12/23 rule is still partially enforced by E649A RAG-1 in vivo, since the levels of cleavage and recombination of 12/12 pJH299 supported by E649A RAG-1 still remain below those for 12/23 pJH299. How can this result be explained mechanistically? Since the ability of the RAG complex to support 12/23-regulated synapsis is not dramatically perturbed by the RAG-1 E649A mutation based on mobility shift assays, we speculate that in the presence of RAG-2, E649A RAG-1 mediates synapsis of 12/23 pJH299 similarly to the WT RAG-1 and cleaves it with comparable efficiency. However, when confronted with pJH299 containing an unpaired or mispaired RSS, we suggest that E649A RAG-1 and RAG-2 may assemble paired complexes inefficiently, but the assembled complex may able to cleave the RSS(s) independently of synapsis more efficiently than WT RAG-1.
Comparisons with other gain-of-function mutants in transpositional recombination systems.
V(D)J recombination exhibits many similarities to “cut-and-paste” transpositionalrecombination systems. In many of these systems, gain-of-function transposase mutants have been identified and characterized, revealing a spectrum of biochemical mechanisms leading to transposase hyperactivity. In some cases, such as for the Sleeping Beauty (62) and Tn5 transposases (36, 64), mutations that enhance the DNA binding activity of the transposase, thereby improving the efficiency of donor DNA cleavage, have been identified. In other cases, such as for the Tn5 transposase (61), the P-element transposase (4), and TnsA and TnsB involved in Tn7 transposition (30), mutations that bypass negative regulatory mechanisms without altering the intrinsic DNA binding activity of the transposase have been identified. A third type of mutation is one that enhances transposase activity by promoting interactions between the subunits that comprise the functional transposase. This type of mutation is exemplified by the LP372 mutant Tn5 transposase (59), whose hyperactivity has been attributed to the increasing of the dimerization potential of the transposase. The RAG-1 E649A mutation exhibits most similarities to the latter two classes of mutations, as the mutant RAG complex appears to bypass the 12/23 rule that regulates the DNA cleavage activity of the recombinase. The 12/23 rule is likely enforced by key protein-protein and/or protein-DNA interactions that help prevent inappropriate synapsis. Dissecting what interactions are altered by the RAG-1 E649A mutation will provide greater insight into how the RAG complex mediates 12/23-regulated synapsis and coupled cleavage.
Acknowledgments
We thank Joanne Hesse for providing 12/23 and 23/23 versions of pJH299 and Prafulla Raval for technical assistance, as well as anonymous referees for helpful comments and suggestions.
This work was initiated with support from an American Cancer Society Research Scholar Grant (RSG-01-020-01-CCE, to P.C.S.) and completed with support from the NIH (R01 AI055599, to P.C.S.). This investigation was conducted in a facility constructed with support from the Research Facilities Improvement Program of the NIH National Center for Research Resources (C06 RR17417-01).
REFERENCES
- 1.Agrawal, A., Q. M. Eastman, and D. G. Schatz. 1998. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394:744-751. [DOI] [PubMed] [Google Scholar]
- 2.Aidinis, V., D. C. Dias, C. A. Gomez, D. Bhattacharyya, E. Spanopoulou, and S. Santagata. 2000. Definition of minimal domains of interaction within the recombination-activating genes 1 and 2 recombinase complex. J. Immunol. 164:5826-5832. [DOI] [PubMed] [Google Scholar]
- 3.Bassing, C. H., W. Swat, and F. W. Alt. 2002. The mechanism and regulation of chromosomal V(D)J recombination. Cell 109(Suppl.):S45-S55. [DOI] [PubMed] [Google Scholar]
- 4.Beall, E. L., M. B. Mahoney, and D. C. Rio. 2002. Identification and analysis of a hyperactive mutant form of Drosophila P-element transposase. Genetics 162:217-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergeron, S., D. K. Anderson, and P. C. Swanson. The RAG and HMGB1 proteins: purification and biochemical analysis of recombination signal complexes. Methods Enzymol., in press. [DOI] [PubMed]
- 6.Bergeron, S., T. Madathiparambil, and P. C. Swanson. 2005. Both high mobility group (HMG)-boxes and the acidic tail of HMGB1 regulate recombination-activating gene (RAG)-mediated recombination signal synapsis and cleavage in vitro. J. Biol. Chem. 280:31314-31324. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuomo, C. A., and M. A. Oettinger. 1994. Analysis of regions of RAG-2 important for V(D)J recombination. Nucleic Acids Res. 22:1810-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De, P., and K. K. Rodgers. 2004. Putting the pieces together: identification and characterization of structural domains in the V(D)J recombination protein RAG1. Immunol. Rev. 200:70-82. [DOI] [PubMed] [Google Scholar]
- 9.Difilippantonio, M. J., C. J. McMahan, Q. M. Eastman, E. Spanopoulou, and D. G. Schatz. 1996. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell 87:253-262. [DOI] [PubMed] [Google Scholar]
- 10.Fugmann, S. D., A. I. Lee, P. E. Shockett, I. J. Villey, and D. G. Schatz. 2000. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol. 18:495-527. [DOI] [PubMed] [Google Scholar]
- 11.Fugmann, S. D., and D. G. Schatz. 2001. Identification of basic residues in RAG2 critical for DNA binding by the RAG1-RAG2 complex. Mol. Cell 8:899-910. [DOI] [PubMed] [Google Scholar]
- 12.Fugmann, S. D., I. J. Villey, L. M. Ptaszek, and D. G. Schatz. 2000. Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol. Cell 5:97-107. [DOI] [PubMed] [Google Scholar]
- 13.Gellert, M. 2002. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem. 71:101-132. [DOI] [PubMed] [Google Scholar]
- 14.Giulietti, A., L. Overbergh, D. Valckx, B. Decallonne, R. Bouillon, and C. Mathieu. 2001. An overview of real-time quantitative PCR: applications to quantify cytokine gene expression. Methods 25:386-401. [DOI] [PubMed] [Google Scholar]
- 15.Hesse, J. E., M. R. Lieber, M. Gellert, and K. Mizuuchi. 1987. Extrachromosomal DNA substrates in pre-B cells undergo inversion or deletion at immunoglobulin V-(D)-J joining signals. Cell 49:775-783. [DOI] [PubMed] [Google Scholar]
- 16.Hesse, J. E., M. R. Lieber, K. Mizuuchi, and M. Gellert. 1989. V(D)J recombination: a functional definition of the joining signals. Genes Dev. 3:1053-1061. [DOI] [PubMed] [Google Scholar]
- 17.Hiom, K., M. Melek, and M. Gellert. 1998. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell 94:463-470. [DOI] [PubMed] [Google Scholar]
- 18.Jones, D. H., and S. C. Winistorfer. 1992. Recombinant circle PCR and recombination PCR for site-specific mutagenesis without PCR product purification. BioTechniques 12:528-530, 532, 534-535. [PubMed] [Google Scholar]
- 19.Jones, J. M., and M. Gellert. 2004. The taming of a transposon: V(D)J recombination and the immune system. Immunol. Rev. 200:233-248. [DOI] [PubMed] [Google Scholar]
- 20.Kale, S. B., M. A. Landree, and D. B. Roth. 2001. Conditional RAG-1 mutants block the hairpin formation step of V(D)J recombination. Mol. Cell. Biol. 21:459-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, D. R., Y. Dai, C. L. Mundy, W. Yang, and M. A. Oettinger. 1999. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 13:3070-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirch, S. A., P. Sudarsanam, and M. A. Oettinger. 1996. Regions of RAG1 protein critical for V(D)J recombination. Eur. J. Immunol. 26:886-891. [DOI] [PubMed] [Google Scholar]
- 23.Lampe, D. J., B. J. Akerley, E. J. Rubin, J. J. Mekalanos, and H. M. Robertson. 1999. Hyperactive transposase mutants of the Himar1 mariner transposon. Proc. Natl. Acad. Sci. USA 96:11428-11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landree, M. A., J. A. Wibbenmeyer, and D. B. Roth. 1999. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 13:3059-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, J., and S. Desiderio. 1999. Cyclin A/CDK2 regulates V(D)J recombination by coordinating RAG-2 accumulation and DNA repair. Immunity 11:771-781. [DOI] [PubMed] [Google Scholar]
- 26.Lewis, S. M., E. Agard, S. Suh, and L. Czyzyk. 1997. Cryptic signals and the fidelity of V(D)J joining. Mol. Cell. Biol. 17:3125-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis, S. M., and J. E. Hesse. 1991. Cutting and closing without recombination in V(D)J joining. EMBO J. 10:3631-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, W., F. C. Chang, and S. Desiderio. 2001. Rag-1 mutations associated with B-cell-negative SCID dissociate the nicking and transesterification steps of V(D)J recombination. Mol. Cell. Biol. 21:3935-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, W., P. Swanson, and S. Desiderio. 1997. RAG-1 and RAG-2-dependent assembly of functional complexes with V(D)J recombination substrates in solution. Mol. Cell. Biol. 17:6932-6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu, F., and N. L. Craig. 2000. Isolation and characterization of Tn7 transposase gain-of-function mutants: a model for transposase activation. EMBO J. 19:3446-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matthews, A. G., S. K. Elkin, and M. A. Oettinger. 2004. Ordered DNA release and target capture in RAG transposition. EMBO J. 23:1198-1206. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McBlane, J. F., D. C. van Gent, D. A. Ramsden, C. Romeo, C. A. Cuomo, M. Gellert, and M. A. Oettinger. 1995. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell 83:387-395. [DOI] [PubMed] [Google Scholar]
- 33.McMahan, C. J., M. J. Sadofsky, and D. G. Schatz. 1997. Definition of a large region of RAG1 that is important for coimmunoprecipitation of RAG2. J. Immunol. 158:2202-2210. [PubMed] [Google Scholar]
- 34.Melek, M., and M. Gellert. 2000. RAG1/2-mediated resolution of transposition intermediates: two pathways and possible consequences. Cell 101:625-633. [DOI] [PubMed] [Google Scholar]
- 35.Morzycka-Wroblewska, E., F. E. Lee, and S. V. Desiderio. 1988. Unusual immunoglobulin gene rearrangement leads to replacement of recombinational signal sequences. Science 242:261-263. [DOI] [PubMed] [Google Scholar]
- 36.Naumann, T. A., and W. S. Reznikoff. 2002. Tn5 transposase with an altered specificity for transposon ends. J. Bacteriol. 184:233-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neiditch, M. B., G. S. Lee, M. A. Landree, and D. B. Roth. 2001. RAG transposase can capture and commit to target DNA before or after donor cleavage. Mol. Cell. Biol. 21:4302-4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oettinger, M. A. 2004. Molecular biology: hairpins at split ends in DNA. Nature 432:960-961. [DOI] [PubMed] [Google Scholar]
- 39.Qiu, J., S. B. Kale, H. Y. Schultz, and D. B. Roth. 2001. Separation-of-function mutants reveal critical roles for RAG2 in both the cleavage and joining steps of V(D)J recombination. Mol. Cell 7:77-87. [DOI] [PubMed] [Google Scholar]
- 40.Reznikoff, W. S., A. Bhasin, D. R. Davies, I. Y. Goryshin, L. A. Mahnke, T. Naumann, I. Rayment, M. Steiniger-White, and S. S. Twining. 1999. Tn5: A molecular window on transposition. Biochem. Biophys. Res. Commun. 266:729-734. [DOI] [PubMed] [Google Scholar]
- 41.Roth, D. B., J. P. Menetski, P. B. Nakajima, M. J. Bosma, and M. Gellert. 1992. V(D)J recombination: broken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell 70:983-991. [DOI] [PubMed] [Google Scholar]
- 42.Roth, D. B., C. Zhu, and M. Gellert. 1993. Characterization of broken DNA molecules associated with V(D)J recombination. Proc. Natl. Acad. Sci. USA 90:10788-10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sadofsky, M. J., J. E. Hesse, and M. Gellert. 1994. Definition of a core region of RAG-2 that is functional in V(D)J recombination. Nucleic Acids Res. 22:1805-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sadofsky, M. J., J. E. Hesse, J. F. McBlane, and M. Gellert. 1993. Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res. 21:5644-5650. (Erratum, 22: 550, 1994.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sawchuk, D. J., F. Weis-Garcia, S. Malik, E. Besmer, M. Bustin, M. C. Nussenzweig, and P. Cortes. 1997. V(D)J recombination: modulation of RAG1 and RAG2 cleavage activity on 12/23 substrates by whole cell extract and DNA-bending proteins. J. Exp. Med. 185:2025-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schlissel, M., A. Constantinescu, T. Morrow, M. Baxter, and A. Peng. 1993. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev. 7:2520-2532. [DOI] [PubMed] [Google Scholar]
- 47.Schultz, H. Y., M. A. Landree, J. Qiu, S. B. Kale, and D. B. Roth. 2001. Joining-deficient RAG1 mutants block V(D)J recombination in vivo and hairpin opening in vitro. Mol. Cell 7:65-75. [DOI] [PubMed] [Google Scholar]
- 48.Shih, I. H., M. Melek, N. D. Jayaratne, and M. Gellert. 2002. Inverse transposition by the RAG1 and RAG2 proteins: role reversal of donor and target DNA. EMBO J. 21:6625-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spanopoulou, E., F. Zaitseva, F. H. Wang, S. Santagata, D. Baltimore, and G. Panayotou. 1996. The homeodomain region of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell 87:263-276. [DOI] [PubMed] [Google Scholar]
- 50.Swanson, P. C. 2002. Fine structure and activity of discrete RAG-HMG complexes on V(D)J recombination signals. Mol. Cell. Biol. 22:1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swanson, P. C. 2002. A RAG-1/RAG-2 tetramer supports 12/23-regulated synapsis, cleavage, and transposition of V(D)J recombination signals. Mol. Cell. Biol. 22:7790-7801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swanson, P. C., and S. Desiderio. 1998. V(D)J recombination signal recognition: distinct, overlapping DNA-protein contacts in complexes containing RAG1 with and without RAG2. Immunity 9:115-125. [DOI] [PubMed] [Google Scholar]
- 53.Swanson, P. C., D. Volkmer, and L. Wang. 2004. Full-length RAG-2, and not full-length RAG-1, specifically suppresses RAG-mediated transposition but not hybrid joint formation or disintegration. J. Biol. Chem. 279:4034-4044. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 54.Tsai, C. L., A. H. Drejer, and D. G. Schatz. 2002. Evidence of a critical architectural function for the RAG proteins in end processing, protection, and joining in V(D)J recombination. Genes Dev. 16:1934-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Twining, S. S., I. Y. Goryshin, A. Bhasin, and W. S. Reznikoff. 2001. Functional characterization of arginine 30, lysine 40, and arginine 62 in Tn5 transposase. J. Biol. Chem. 276:23135-23143. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 56.van Gent, D. C., K. Hiom, T. T. Paull, and M. Gellert. 1997. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 16:2665-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Gent, D. C., K. Mizuuchi, and M. Gellert. 1996. Similarities between initiation of V(D)J recombination and retroviral integration. Science 271:1592-1594. [DOI] [PubMed] [Google Scholar]
- 58.van Gent, D. C., D. A. Ramsden, and M. Gellert. 1996. The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell 85:107-113. [DOI] [PubMed] [Google Scholar]
- 59.Weinreich, M. D., A. Gasch, and W. S. Reznikoff. 1994. Evidence that the cis preference of the Tn5 transposase is caused by nonproductive multimerization. Genes Dev. 8:2363-2374. [DOI] [PubMed] [Google Scholar]
- 60.West, K. L., N. C. Singha, P. De Ioannes, L. Lacomis, H. Erdjument-Bromage, P. Tempst, and P. Cortes. 2005. A direct interaction between the RAG2 C terminus and the core histones is required for efficient V(D)J recombination. Immunity 23:203-212. [DOI] [PubMed] [Google Scholar]
- 61.Wiegand, T. W., and W. S. Reznikoff. 1992. Characterization of two hypertransposing Tn5 mutants. J. Bacteriol. 174:1229-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yant, S. R., J. Park, Y. Huang, J. G. Mikkelsen, and M. A. Kay. 2004. Mutational analysis of the N-terminal DNA-binding domain of Sleeping Beauty transposase: critical residues for DNA binding and hyperactivity in mammalian cells. Mol. Cell. Biol. 24:9239-9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoshida, T., A. Tsuboi, K. Ishiguro, F. Nagawa, and H. Sakano. 2000. The DNA-bending protein, HMG1, is required for correct cleavage of 23 bp recombination signal sequences by recombination activating gene proteins in vitro. Int. Immunol. 12:721-729. [DOI] [PubMed] [Google Scholar]
- 64.Zhou, M., and W. S. Reznikoff. 1997. Tn5 transposase mutants that alter DNA binding specificity. J. Mol. Biol. 271:362-373. [DOI] [PubMed] [Google Scholar]