Abstract
Within mineralized bone, osteocytes form dendritic processes that travel through canaliculi to make contact with other osteocytes and cells on the bone surface. This three-dimensional syncytium is thought to be necessary to maintain viability, cell-to-cell communication, and mechanosensation. E11/gp38 is the earliest osteocyte-selective protein to be expressed as the osteoblast differentiates into an osteoid cell or osteocyte, first appearing on the forming dendritic processes of these cells. Bone extracts contain large amounts of E11, but immunostaining only shows its presence in early osteocytes compared to more deeply embedded cells, suggesting epitope masking by mineral. Freshly isolated primary osteoblasts are negative for E11 expression but begin to express this protein in culture, and expression increases with time, suggesting differentiation into the osteocyte phenotype. Osteoblast-like cell lines 2T3 and Oct-1 also show increased expression of E11 with differentiation and mineralization. E11 is highly expressed in MLO-Y4 osteocyte-like cells compared to osteoblast cell lines and primary osteoblasts. Differentiated, mineralized 2T3 cells and MLO-Y4 cells subjected to fluid flow shear stress show an increase in mRNA for E11. MLO-Y4 cells show an increase in dendricity and elongation of dendrites in response to shear stress that is blocked by small interfering RNA specific to E11. In vivo, E11 expression is also increased by a mechanical load, not only in osteocytes near the bone surface but also in osteocytes more deeply embedded in bone. Maximal expression is observed not in regions of maximal strain but in a region of potential bone remodeling, suggesting that dendrite elongation may be occurring during this process. These data suggest that osteocytes may be able to extend their cellular processes after embedment in mineralized matrix and have implications for osteocytic modification of their microenvironment.
Even though osteocytes make up more than 90% of all bone cells, little is known concerning their function compared to osteoblasts and osteoclasts. Osteocytes are thought to respond to mechanical strain to send signals of resorption or formation (27). These cells are dispersed throughout the mineralized matrix connected to each other and cells on the bone surface through slender cytoplasmic processes radiating in all directions but generally perpendicular to the bone surface. The cell processes pass through the bone in small tunnels called canaliculi. Osteocytes are thought to function as a network of sensor cells in bone that can mediate the effects of mechanical loading through their extensive communication network. Not only do these cells communicate with each other and with cells on the bone surface, but their dendritic processes are in contact with the bone marrow (22), where they have the potential to stimulate bone resorption (1, 52).
A key regulator of osteoblast and osteoclast activity in bone is mechanical strain. Under normal conditions, bone formation and bone resorption are balanced so that there is no net gain or loss of bone. However, by the process of adaptive remodeling, the skeleton is able to continually adapt to the mechanical loading it receives through daily activity by adding new bone to withstand increased amounts of loading and removing bone in response to unloading or disuse (reviewed in references 4 and 10). Julius Wolff, in 1892, was the first to suggest that bone accommodates or responds to strain. To paraphrase Wolff's law, alteration of internal and external architecture occurs as a consequence of the stressing of bone. This means that, during the process of “adaptive remodeling,” the bone architecture is constantly remodeled so that it is able to resist and withstand the daily habitual strains to which it is subjected, a compromise between safety and functionality and economy of metabolic resources. The cells of bone with the potential for sensing mechanical strain and translating these forces into biochemical signals include bone lining cells, osteoblasts, and osteocytes. Of these, the osteocytes, with their distribution throughout the bone matrix and their high degree of interconnectivity, are thought to be one of the major cell types responsible for sensing mechanical strain and sending out signals that coordinate adaptive remodeling responses in a manner which takes into account not only the intensity of the strain signals but also the distribution of the strain throughout the whole bone (27).
The earliest description of the gene for E11 was in 1990 as an unknown phorbol ester-inducible gene in MC3T3 osteoblast-like cells, called OTS-8 (31). Since that time, this gene or protein has become known by several names, depending on the tissue in which it is expressed. It is expressed in the choroid plexus, in the ciliary epithelium of the eye, in the intestine, in kidney podocytes, in the thyroid, in the esophagus, in type I alveolar lung cells, in the lymphatic endothelium, and in osteocytes in bone. The gene cloned from murine peripheral lymphoid tissue was called Gp38 (11) and, in other species, canine Gp40 or human gp36 (55). When cloned from rat type I epithelial alveolar lung cells, it is known as T1alpha (39) and the protein is known as RTI40 (15). In the mouse kidney, the molecule is known as podoplanin, as it localizes to the foot processes of podocytes (3). Expression of T1alpha or RT140 (E11) occurs on the apical surface of lung epithelial cells, which are the thin, flat, polarized cells that form the air-blood barrier (9, 39). Deletion of this gene results in mice that die at birth because of respiratory failure as their lungs cannot be inflated to normal volumes (38). This is due to a failure of type II alveolar lung cells to differentiate into type I cells. T1alpha (E11) is also expressed on the lymphatic endothelium. Again, expression is polarized on the apical, luminal plasma membrane of intestinal lymphatic endothelial cells. Null mice have defects in lymphatic but not blood vessel pattern formation, with pronounced lymphedema resulting in swelling of the limbs at birth (40).
The E11 protein is extremely hydrophobic. It is a mucin-type glycoprotein with extensive O glycosylation and a high sialic acid content that potentially contributes to its highly negative charge (15). When purified from type I alveolar epithelial lung cells, the protein has a pI of 3 ± 0.5, giving a 1.0 pH unit charge train, suggesting multiple posttranslational modifications. The amino terminus of the molecule is blocked. The protein has putative extracellular and transmembrane domains and a short cytoplasmic tail with putative protein kinase C and cyclic AMP phosphorylation sites. Some information is available regarding the regulation of the gene. Hyperoxia increases gene expression by transcription factor Sp1, while normoxia decreases expression in type I alveolar lung cells (5). The gene is regulated by interleukin-3 in endothelial cells (19).
In osteocytes, the gp38, podoplanin, or T1alpha molecule is known as E11, the name given by Wetterwald and coworkers (49). These investigators made a mouse monoclonal antibody against a rat osteoblast cell line and isolated a clone that recognized only osteocytes in vivo and not other tissue types. Interestingly, this antibody did not recognize this molecule in other tissues known to express it (e.g., lung epithelial cells and kidney podocytes), suggesting that the antibody recognizes a posttranslational modification that is unique to the osteocyte. The E11 antigen was only found on the dendritic processes of osteocytes and not on osteoblasts in vivo with a punctate reaction at the interface between osteocytes and uncalcified osteoid cells (43). This same antibody also reacted with cementocytes (47). Overexpression in an osteoblast-like cell line led to the generation of extended cytoplasmic processes (44). Even though E11 has been shown to be essential for lung and epithelial cell function, very little is known about the function of E11 in mineralized tissues.
In this report, we show that E11 is osteocyte selective compared to osteoblasts, that E11 is increased by mechanical strain in vitro and in vivo, and that E11 is necessary for the elongation of dendritic processes in response to fluid flow shear stress. As dendrite formation is an active, not passive, process, E11 may be critical not only for dendrite formation but also for osteocyte function and viability and therefore essential for normal bone function.
MATERIALS AND METHODS
Primary cells, cell lines, and culture conditions.
MLO-Y4 osteocyte-like cell lines; 2T3, Oct-1, and MC3T3 osteoblast-like cell lines; and MLO-A5 late osteoblast-early osteocyte cells were cultured as described previously (17, 23, 24). Briefly, MLO-Y4 and MLO-A5 cells were plated onto culture dishes coated with rat tail type 1 collagen, whereas the other cell types were plated directly onto the culture dish. All cells were cultured in alpha minimal essential medium supplemented with 5% fetal bovine serum and 5% calf serum for comparison purposes. For long-term cultures of primary osteoblasts and osteoblast-like cell lines, β-glycerol phosphate and ascorbic acid were added and cells were cultured in 10% FBS as described previously (2). 2T3 cells were treated with 40 ng/ml BMP2 as described previously (14).
Isolation of primary cells. (i) Six-week-old mouse long bones.
Sequential digests were performed with long bones of 6-week-old wild-type Blackswiss mice as described previously (52). The mice were sacrificed, femora and humeri were isolated, the attached soft tissues and bone marrow were removed, and the bones were cut into pieces (1 mm by 1 mm), which were then digested in 0.2% collagenase-Hanks balanced salt solution (HBSS) for 20 min six times at 37°C to give six fractions, F1 to F6. The remaining bone pieces were washed twice with phosphate-buffered saline (PBS), incubated with 4 mM EDTA Na2-PBS for 20 min (F7), and further digested with 0.2% collagenase-PBS for 20 min (F8). The cell pellets of F1 to F6 and F7 and F8 were washed by 2× PBS and lysed with radioimmunoprecipitation assay (RIPA) buffer for Western blotting (see below). The remaining bone particles were washed twice in PBS, further minced into very fine bone particles (10 to 50 μm) in PBS, and then boiled with sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis sample buffer (0.5 ml) for 5 min, and the supernatant was loaded onto a 10% SDS-polyacrylamide gel for electrophoresis.
(ii) One-week-old mouse long bones.
Long bones of 7-day-old wild-type Blackswiss mice were stripped of periosteum, cut into pieces approximately 0.3 by 0.3 mm, and digested with 0.2% collagenase-HBSS for 20 min six times. These six fractions were combined, and half of the newly released periosteal cells were used for Western blotting and the other half were used for in vitro culture. After 3 and 7 days of culture, the cells were digested by 0.2% collagenase-HBSS for 20 min. The resultant cell pellets were lysed with RIPA buffer for Western blotting, which was performed as described below.
(iii) Mouse calvaria.
Newborn (<24 h old) mouse calvaria were digested with 0.2% collagenase-HBSS for 20 min six times. Fractions 3 to 6 were combined, and half of the cells were cytocentrifuged onto glass slides and fixed with 4% paraformaldehyde in PBS for 15 min for immunohistochemical staining. The other half of the cells were plated and cultured for 3 days and then subjected to digestion with 0.2% collagenase-HBSS for 20 min before being cytocentrifuged onto glass slides for fixation and immunostaining as described for the freshly isolated cells.
Antibodies to E11.
The 9C11 antibody was generated by injecting MLO-Y4 cells into 6-month-old LOU rats, followed by fusion of the rat spleen with NS-1 mouse myeloma cells to produce hybridomas. The 9C11 clone was identified as producing a monoclonal antibody that was specific for osteocytes and did not react with osteoblasts such as MC3T3 or Oct-1 cells or with other tissues (data not shown). An MLO-Y4 cDNA library in Escherichia coli was made by standard protocols (6) with a pBluescript II XR cDNA library construction kit and pBluescript II KS (+) vector from Stratagene (La Jolla, CA). Screening of the library gave three positive clones, and sequencing of the gene products revealed that this antibody recognized a cDNA sequence similar to the OTS-8 sequence as described previously (31). This information was used to obtain the genomic sequence used in this study. Hamster monoclonal antibody 8.1.1 against mouse thymic type I epithelial cells was a kind gift from Andrew Farr, University of Washington, Seattle.
Western blotting.
Cell pellets were lysed with RIPA buffer containing protease inhibitors (N-ethylmaleimide, leupeptin, and phenylmethylsulfonyl fluoride) and homogenized by being forced through a 23-gauge needle 20 times and freeze-thawed between −80°C and 20°C twice. The protein concentration was determined with a Bio-Rad protein assay kit. Equal amounts of protein were loaded into each well of SDS-polyacrylamide gels (10%), electrophoresed at 200 V for 40 min at room temperature, and transferred to a nitrocellulose membrane at 4°C overnight. Background blocking of the nitrocellulose membrane was performed with 5% bovine serum albumin-1% skim milk-0.05% NaN3-PBS at 4°C overnight. The 9C11 antibody was used undiluted, and monoclonal antibody 8.1.1 was diluted 1:100 in 1% milk-PBS-0.05% NaN3 and incubated with nitrocellulose membrane overnight at 4°C. The second antibody for the 9C11 antibody was horseradish peroxidase-conjugated anti-rat immunoglobulin G (IgG; 1:5,000), and it was developed with a New England Nuclear kit (Dupont NEN Research Products, Boston, MA). The second antibody for the 8.1.1 antibody was peroxidase-conjugated Affinipure goat anti-Syrian hamster IgG (ImmunoResearch Laboratories Inc.) diluted 1:5,000 in 5% milk-PBS without NaN3 and incubated at room temperature for 2 h; a Western Lightning chemiluminescence reagent kit (Perkin-Elmer) was used to detect immunoreactive bands by peroxidase activity.
Northern blot analysis.
Total RNA was isolated from cultured cells with RNA-BEE (Tel-Test, Inc., Friendswood, TX). Two micrograms of mRNA was loaded per lane. Northern blot analysis was performed as described previously (52). The E11 probe was a 519-bp fragment of the coding region of the full-length (2.2-kb) mouse E11 cDNA. Mouse glyceraldehyde-3-phosphate dehydrogenase (1.4-kb fragment) was used as a control.
Immunocytochemistry.
The cells were either cytocentrifuged or fixed onto coverslip slides with 4% paraformaldehyde-PBS before staining. Cells either freshly harvested from bone or from culture were placed in 10% FBS-minimal essential medium to inactivate the collagenase, washed twice in PBS, and cytospun onto slides at 2,000 rpm for 1 min. Endogenous peroxidase was quenched with 3% H2O2-PBS before incubation with 5% normal goat serum-PBS-0.05% NaN3 at 4°C overnight. Samples were incubated in primary antibody 8.1.1 diluted 1:50 in 5% normal goat serum-PBS-0.05% NaN3 at 4°C overnight. The secondary antibody, a peroxidase-conjugated goat anti-Syrian hamster IgG, was used at a 1:200 dilution in 5% normal goat serum-PBS (NaN3 free) at room temperature for 3 h. Peroxidase activity was detected with diaminobenzidine (0.5 mg/ml in 50 mM Tris-HCl containing 0.05% H2O2, pH 7.4). Normal hamster IgG (2 μg/ml) was used as a negative control.
Immunohistochemical staining of tissues.
NIH Swiss mouse brain, lung, kidney, liver, and muscle tissue samples were purchased from Novagen already fixed in 4% paraformaldehyde and embedded in paraffin. The soft-tissue samples were deparaffinized and gradually rehydrated, incubated with 0.1% trypsin-0.1% CaCl2 (pH 7.8) at 37°C for 30 min for antigen retrieval, and endogenous peroxidase quenched with 5% H2O2-PBS. Immunostaining was then performed as described above for cell culture.
In vitro application of mechanical strain.
Fluid flow shear stress was applied as described previously (7, 8), with a Streamer Gold chamber (Flexcell International Corp., Hillsborough, NC). The chamber was connected to a peristaltic pump (Masterflex L/S; Cole-Parmer Instrument Company) controlled by StreamSoft software (Flexcell International Corp., Hillsborough, NC) to control the flow rate. The entire flow system was maintained in a CO2 incubator at 37°C. To determine if E11 can be regulated by mechanical strain, fluid flow shear stress experiments were performed with 4 and 16 dynes/cm2 and the cells were harvested 2 and 24 h after 2 h of shear stress. To determine if E11 is responsible for elongation of dendritic processes, the cells were exposed to 16 dynes/cm2 for 2 h and cultured for 24 h before fixation.
Determination of dendricity.
The cells were fixed in 2% glutaraldehyde, the fixative was washed off with PBS, and the cells were stained with 0.1% crystal violet for 10 min, washed, and dried. Dendricity was determined with the analySIS image analysis software (Soft Imaging Systems Corp.). The mean length of dendrites was determined on a minimum of 125 cells per well.
Generation of functional siRNAs specific for E11.
We generated small interfering RNAs (siRNAs) specific for three regions of E11 in the form of 21-base nucleotides for transient infection. siRNAs A and B were based on the sequence used for human E11 siRNA (40). siRNA C was designed by Ambion (Austin, TX) with the help of the Cenix algorithm. The sequences were as follows: A, +165 ACTGGAGGGCTTAATGAATCT +185; B, +397 AAGATGGCTTGCCAGTAGTCA +417; C, +66 AGGGACTATAGGCGTGAATGA +86.
For siRNA experiments, briefly, MLO-Y4 cells were cultured in 48-well plates with 0.5 ml of growth medium without antibiotics until 30 to 50% confluent at the time of transfection. Diluted siRNA and diluted Lipofectamine 2000 (Invitrogen, Carlsbad, CA) were combined for a total volume of 100 μl per well (50 μl of siRNA plus 1 μl of Lipofectamine 2000 in Opti-MEM medium), mixed gently, and incubated for 20 min at room temperature before being brought to 0.3 ml and added to cells for 24 h of incubation. Maximal blocking of protein expression was observed with a combination of all three nucleotides. We found that a combination of the three siRNAs at 25 nM gave reasonable blocking with no toxicity. Efficiency of blocking was determined by Western blotting of cell lysates as shown below. Reduced E11 protein expression was observed with all three siRNAs at both 250 and 25 nM, but combining siRNAs allowed a decrease in molarity with the same blocking efficiency as using a single siRNA at a higher molarity. When all three were combined, a 70 to 80% reduction of E11 protein was observed. No significant differences were observed in cell number after 24 h of incubation with siRNA or vehicle alone (data not shown).
siRNA experiments performed with MLO-Y4 cells subjected to fluid flow shear stress.
For siRNA experiments performed with MLO-Y4 cells subjected to fluid flow shear stress, the cells were plated at a density or 4 × 105 on collagen-coated slides and preincubated with siRNA and controls for 24 h before the application of fluid flow shear stress. Each experiment is an n = 1; therefore, each experiment was repeated a minimum of three to six times to obtain statistical significance. An siControl RNA-induced silencing complex (RISC)-free siRNA (Dharmacon, Lafayette, CO) was used that has been chemically modified to impair RISC interaction. It is to be used as a negative control to evaluate cellular changes related to siRNA transfection. Diluted siRNA and diluted Oligofectamine (Invitrogen, Carlsbad, CA) were combined for a total volume of 58 μl per well (50 μl of 25 nM siRNA plus 1 μl of Oligofectamine in 7 μl of Opti-MEM), mixed gently, and incubated for 20 min at room temperature before being brought to 0.3 ml with medium for addition to cells for 24 h of incubation. Lipofectamine 2000, Lipofectamine Plus (Invitrogen, Carlsbad, CA), and TransIT-TKO transfection reagent (Mirus, Madison, WI) had an effect on cell morphology and therefore were not used (data not shown). Fluid flow shear stress was applied with the Streamer Gold apparatus at 16 dynes/cm2 for 2 h. After the fluid flow was stopped, cells were cultured for 24 h, fixed, and stained for quantitation of dendritic processes.
Construction of the targeting vector and generation of E11 lacZ knock-in mice.
To generate E11 lacZ knock-in mice, a 0.46-kb fragment (+38 [SmaI] to +496 [SmaI]) of E11 exon 1 and part of intron 1 were replaced with the lacZ and neo (neomycin phosphotransferase gene) cassette. LoxP (recognition sequence for Cre recombinase) sites were added surrounding the neo cassette for future removal of the cassette. To facilitate homologous recombination, a 3.6-kb promoter (−3619 [SalI] to +38 [SmaI]) was inserted upstream of the lacZ-poly(A)-neo cassette as a 5′ arm, and a 2.3-kb intron 1 fragment (+496 [SmaI] to +2832 [HindIII]) was inserted downstream of the lacZ-poly(A)-neo cassette as a 3′ arm. The procedure used for the generation of null mice was described previously (50). E11 heterozygotes were subsequently interbred to generate homozygotes on the C57BL/6 background and on the Blackswiss background. The University of Missouri—Kansas City animal facilities are Association for Assessment and Accreditation of Laboratory Animal Care approved and comply with the Welfare Act to maintain appropriate policies and procedures to ensure the humane care and use of animals.
Genotyping and histological evaluation of embryos.
The E11 heterozygotes were bred, the pregnant females (identified by the presence of a vaginal plug) were sacrificed at embryonic day 18.5, and tail genotyping of the embryos was performed by PCR. The primers for E11 are as follows: forward primer, 5′-CTG, GCC, TGA, GGT, CAT, CTT, GT-3′; reverse primer, 5′-TCC, ATC, CCC, ACC, AAC, AAG, TG-3′. The fragment length is 459 bp. The primers for lacZ are as follows: forward primer, same as that for E11; reverse primer, 5′-GGC, AAT, ATC, GCG, GCT, CAG, TTC-3′. The fragment length is 280 bp. The PCR conditions for E11 were 94°C for 5 min; 40 cycles of 94°C for 45 s, 54°C for 45 s, and 72°C for 45 s; and 72°C for 7 min. The PCR conditions for lacZ were 94°C for 5 min; 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s; and 72°C for 7 min.
The hind limbs were fixed with 4% paraformaldehyde-PBS at 4°C for 24 h, decalcified in 1.35 N HCl at room temperature for 24 h, gradually dehydrated in ethanol, and embedded in paraffin for sectioning. Only the center longitudinal section of the femur was used for histological evaluation after hematoxylin and eosin staining. The procedure was performed according to the Manual of Histologic Staining Methods of the Armed Forces Institute of Pathology (27a). After staining of the longitudinal bone sections, the femoral shaft length, midshaft diameter, and maximal cortical thickness were determined with a Nikon Eclipse E800 microscope.
In vivo mechanical loading.
Mechanical loading was applied to 3-month-old wild-type and heterozygotic mice based on the method of Torrance and coworkers (48). Briefly, the animals were anesthetized by intraperitoneal injection of a mixture of ketamine (100 mg/kg) and xylazine (10 mg/kg) in PBS before loading. A compressive load of −3.5 N at 2 Hz was applied for 30 s to the distal end of the right ulna. The left ulna served as an unloaded control. At 4, 24, and 48 h after loading, E11 heterozygotic mice were sacrificed for LacZ (β-galactosidase) staining. At 24, 48, and 96 h after loading, wild-type mice were sacrificed for E11 immunostaining. The bones were sectioned starting at 4.5 mm from the olecranon in seven sections 1 mm apart.
Histochemical and LacZ staining of bone sections.
The epiphysis and diaphysis from each ulna were removed 2 mm from each end, fixed for 1 h in ice-cold 4% paraformaldehyde, washed with PBS for 5 min three times, and incubated in X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) solution (10 ml/ulna) at 37°C in the dark for 24 h as described previously (51). The paraffin-embedded ulnae were sectioned at seven equal distances; the sections (5 μm thick) were counterstained with eosin. The LacZ-positive osteocytes and total osteocytes were counted.
Immunohistochemical staining for E11 protein in bone sections.
The paraffinized bone sections were deparaffinized and gradually rehydrated, incubated with 0.1% trypsin-0.1% CaCl2 (pH 7.8) at 37°C for 30 min for antigen retrieval, and endogenous peroxidase quenched with 5% H2O2-PBS. Immunostaining was performed as described above for nonmineralized tissues. The total number of osteocytes and the total number of stained osteocytes per cross section were determined.
Statistical analysis.
The Student Newman-Keuls test was used for comparison of multiple means. The paired t test was used for comparison of the LacZ- and E11-positive osteocytes in loaded and unloaded control ulnae with the InStat GraphPad software.
Nucleotide sequence accession number.
The sequence determined in this study was submitted to GenBank and assigned accession no. AY115493.
RESULTS
Identification of an osteoblast differentiation marker and osteocyte-selective antigen, E11/gp38.
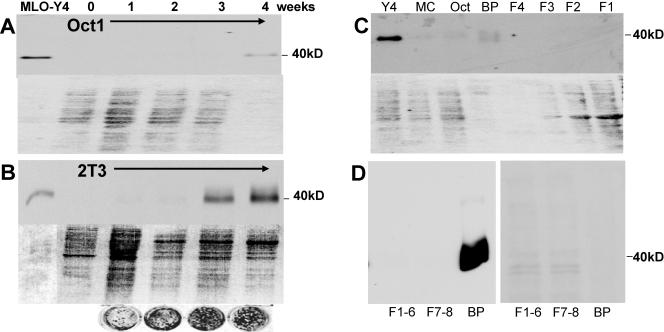
In a search for osteocyte-selective antigens compared to osteoblasts, MLO-Y4 cells were injected into rats in order to generate osteocyte-selective antibodies. One hybridoma, 9C11, produced an antibody that recognized a 40-kDa band that was only observable in MLO-Y4 cells and not other cells or tissues. The MLO-Y4 cells were shown to prominently express this band compared to osteoblasts (Fig. 1). The top of each panel of Fig. 1A to C is a Western blot assay with this antibody, and the bottom is a Ponceau S stain showing relative protein loading. This 40-kDa band became prominent when osteoblast-like cells were cultured under conditions supporting differentiation and mineralization. The band was present by 4 weeks of culture of Oct-1 cells (Fig. 1A) and by 3 weeks of culture of 2T3 cells (Fig. 1B). The circles below the blot are images of von Kossa-stained cultures of 2T3 cells showing that mineralization increases concomitant with 40-kDa band expression.
FIG. 1.
Identification of a 40-kDa protein highly expressed in osteocytes. With monoclonal antibody 9C11, made against MLO-Y4 cells, a 40-kDa band was identified by Western blotting and shown to be highly expressed in MLO-Y4 osteocyte-like cells. The top part of each panel is a Western blot assay, and the bottom is a Ponceau S-stained gel to show relative amounts of loaded protein. Theoretically, as osteoblasts form a mineralized matrix, any cells trapped in that matrix would have the characteristics of osteocytes. Under mineralizing culture conditions, the Oct-1 cells began to express this antigen at 4 weeks (A) and the 2T3 cells began to express this antigen earlier and in large amounts, at 3 and 4 weeks of culture (B). The circles are von Kossa-stained 2T3 cultures showing increased mineralization with time. This suggests that these cells are differentiating into osteocytes in culture. The 9C11 antibody also recognized a band in bone extracts (C). The 40-kDa band is present in MLO-Y4 cells but was not present in osteoblast-like cells such as MC3T3 (MC) and Oct-1 cells (Oct), nor was it visible in extracted cells. These cells were isolated from 6-week-old mouse long bone through serial digestions of collagenase with and without EDTA (F1, F2, F3, and F4). Only the bone particles (BP) containing embedded osteocytes showed the 40-kDa band. Note the relative amounts of protein in the particle fraction compared to protein in the cell fractions. Similar experiments were repeated with the 8.1.1 antibody (D). The Western blot assay is on the left, and the Ponceau stain is on the right. F1 to F6 represent cells on the bone surface that are removed by serial digestions with collagenase. F7 and F8 represent cells on the bone surface that could be removed by EDTA, followed by collagenase. Bone particles (BP) represent the remaining bone that was subjected to boiling in SDS sample buffer. Note that considerably less protein was loaded in the well containing the bone particle extracted protein.
High expression of E11 in extracted bone.
When bone cells were sequentially isolated from 6-week-old mouse calvaria, only the remaining bone particles gave a 40-kDa band (Fig. 1C), suggesting higher expression within the bone matrix. As shown in Fig. 1C, the 40-kDa band was highly expressed in MLO-Y4 cells but not in freshly plated MC3T3 or Oct-1 osteoblast-like cells. The 9C11 antibody was subsequently used to clone the cDNA from an MLO-Y4 library, and the sequence showed similarity to the sequence published by Nose and coworkers as OTS-8 (31). On the basis of this information, the genomic sequence was determined and submitted to GenBank (accession no. AY115493). This hybridoma ceased production, so an antibody to E11/gp38 known as 8.1.1 was obtained from Andrew Farr for further investigation.
To determine if the 8.1.1 antibody recognized the same pattern, a Western blot analysis was performed with isolated fractions of periosteal fibroblasts, osteoblasts, and osteocytes from 6-week-old mouse bone as described previously (52). No expression was observed in the fibroblast-osteoblast (F1 to F6) or the osteoblast (F7 and F8) fractions isolated by collagenase digestion (Fig. 1D). However, extraction of the remaining bone particles with sample buffer showed very high expression of this antigen on the basis of the total protein.
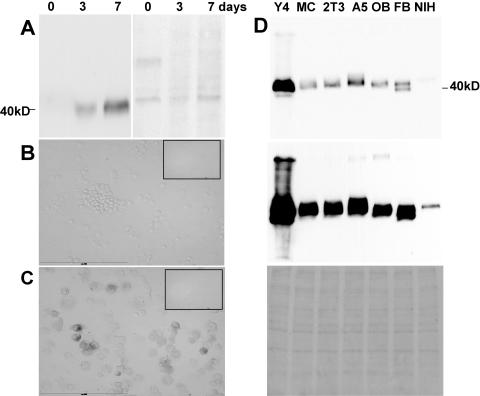
Primary osteoblasts are negative for E11, but protein expression increases with time in culture.
Western blotting showed no reactivity of the 8.1.1 antibody with lysate of freshly isolated periosteal cells from the surface of the long bone (F1 to F6, Fig. 1D); however, increased expression of E11 was observed with time in culture at 3 and 7 days compared to day 0 (Fig. 2A). Similar observations were made with freshly isolated periosteal cells from the calvaria (data not shown). This suggests that these cells are differentiating in culture into osteocyte-like cells.
FIG. 2.
Primary osteoblasts are negative for E11 expression but begin to express this antigen with time in culture. MLO-Y4 osteocyte-like cells express the highest levels of E11. E11 in primary periosteal osteoblasts is not detectable upon isolation but increases with time in culture (A). Cells were isolated from long bones of 1-week-old mice and either lysed after isolation or cultured for 3 to 7 days before processing for Western blot analysis. The left part shows the reaction with the 8.1.1 antibody, and the right part shows Ponceau S staining. Note the lack of expression in the freshly isolated cells but the increased expression with time in culture. Immunocytochemical staining for E11 in freshly isolated cytocentrifuged mouse osteoblasts (B) and osteoblasts cultured for 3 days and then subjected to collagenase treatment and cytocentrifuged (C) was performed. This experiment was performed to show that collagenase treatment of the freshly isolated cells or cultured cells was not removing or having an effect on E11 expression on the cell surface. The insert in the upper right part shows the negative control. Scale bar = 200 μm. E11 expression is much higher in the MLO-Y4 osteocyte-like cell line than in other cell lines, as determined by Western blotting (D). The osteocyte-like cell line MLO-Y4 (Y4), osteoblast-like cell lines MC3T3 (MC) and 2T3, late osteoblast-early osteocyte MLO-A5 (A5) cells, primary osteoblasts (OB), primary fibroblasts (FB), and the fibroblast-like cell line NIH 3T3 (NIH) were cultured for 3 to 4 days before lysis. The upper and middle parts show reaction with the 8.1.1 antibody. The film was exposed for a short period of time to visualize relative expression (top) and for a longer period of time to visualize any additional bands (middle). The lower part shows Ponceau S staining. Note that the highest expression was in the MLO-Y4 osteocyte-like cells compared to less or no expression in the other cell lines. Bands of various sizes were observed, suggesting different extents or forms of posttranslational modification. A 100-kDa band can be seen with the MLO-Y4 cell, MLO-A5 cell, and primary osteoblast lysates after longer exposure.
To ensure that the collagenase used in the isolation and harvesting procedures was not affecting E11 protein expression by potentially removing the protein from the cell surface, primary osteoblasts (fractions 3 to 6) cultured for 3 days were subjected to the same collagenase treatment as freshly isolated cells and then cytocentrifuged for immunostaining. The cells remained positive as shown by E11 antibody staining (Fig. 2C), whereas the freshly isolated, cytocentrifuged cells are negative (Fig. 2B). Similar results were observed with fractions 1 and 2 of periosteal fibroblasts (data not shown), and these results were also validated by Western blotting (data not shown). Therefore, the isolation procedure had little or no effect on E11 expression.
E11 is highly expressed in MLO-Y4 osteocyte-like cells compared to cultured mouse fibroblast-like, osteoblast-like, and osteoid or osteocyte-like cell lines.
Some E11 immunoreactivity in Western blot assays was present in all of the cultured cell lines (Fig. 2D). However, expression was much greater (>10-fold) in MLO-Y4 cells (Y4) compared to osteoblast-like cell lines MC3T3 (MC) and 2T3, late osteoblast-early osteocyte MLO-A5 (A5) cells, primary osteoblasts (OB), primary fibroblasts (FB), and a fibroblast-like cell line, NIH 3T3 (NIH), that had been cultured for 3 to 4 days. Equal amounts of cell lysates were loaded onto SDS-polyacrylamide gels as determined by Ponceau S staining (bottom). As shown by the Western blot assays (top, short exposure; middle, long exposure), considerably more E11 protein is expressed in the osteocyte-like MLO-Y4 cells than in the other cell types. Also, bands of slightly different size were observed, suggesting differences in posttranslational modifications, as described in other tissues (15, 55). A doublet was observed in cultured primary fibroblasts, but very little expression was observed in the fibroblast-like NIH 3T3 cell line. With longer exposure, a 100-kDa band was observed in the MLO-Y4, MLO-A5, and cultured primary osteoblast cell lysates (middle). Immunostaining of the fixed cultured cells showed higher expression of E11 in MLO-Y4 cells compared to the other cell types (data not shown).
E11 expression and distribution in soft tissues compared to bone.
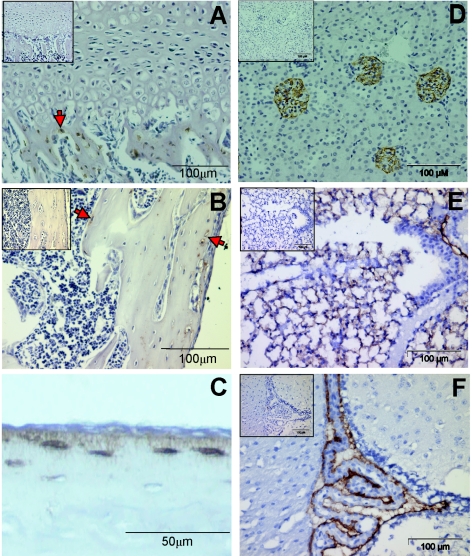
We next validated the specificity of the 8.1.1 antibody originally made against murine thymic epithelial cells (11, 12) and tested its reactivity against bone cells in vivo. In bone, E11 expression was only observed in osteocytes (Fig. 3, arrows). Note that only osteocytes within trabecular bone are positive while the growth plate, marrow, and cartilage are negative (Fig. 3A). In cortical bone, E11 expression is mainly located in the osteocytes near the periosteum and near the endosteum, with decreasing expression in cells deeper within the mineralized matrix (Fig. 3B). Note that the cells on the periosteum and in the bone marrow are negative. E11 is distributed along the osteocyte cell body and along the dendritic processes of the osteocyte (Fig. 3C). This is similar to results obtained by Schulze and coworkers with an antibody made against rat E11 (43).
FIG. 3.
Immunostaining for E11 is only observed in osteocytes, not osteoblasts, cartilage, growth plate, or marrow, in vivo. Localization of E11 in kidney, lung, and brain tissues is shown. Immunohistochemical staining for E11 with the 8.1.1 antibody in a 19-day-old C57BL/6 mouse tibia. Positive staining is brown. The positive osteocytes (arrows) can be seen within the trabecular bone but not in the growth plate (A) and within the cortical bone but not in the marrow (B). Higher magnification of the cortical bone shows that the osteocytic cell body and dendrites are positive, while the cells on the surface are negative (C). Immunohistochemical staining of E11 can be observed in kidney glomeruli because of podocytes (D), in the lung because of type 1 alveolar cells (E), and in the choroid plexus of the brain (F). It is known that E11 is expressed in the podocyte in the kidney, where it is known as podoplanin (3), and in type 1 alveolar lung cells, where it is referred to as T1alpha/RT140 (15, 39). No staining was observed in liver or muscle tissue (data not shown). The upper left insert in panels A, B, D, E, and F shows the negative control with normal hamster IgG in place of the primary antibody.
As various antibodies made against this protein have been shown to be tissue specific (9), we also wanted to verify the specificity of the 8.1.1 antibody. As described previously (11), E11 expression was observed in podocytes in the glomeruli of the kidney (Fig. 3D), in type 1 alveolar lung cells (Fig. 3E), and in the choroid plexus of the brain (Fig. 3F) while liver and muscle tissues were negative (data not shown).
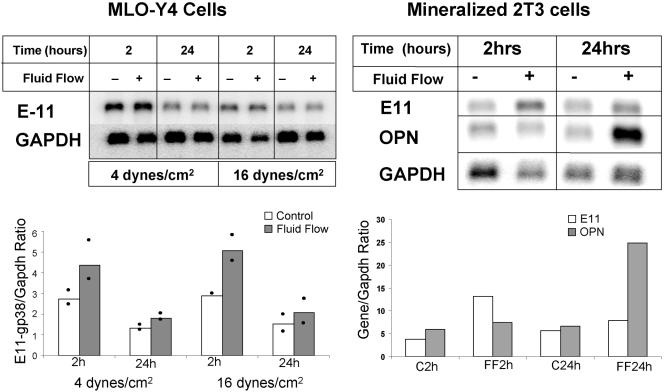
E11 is an early-response gene and is increased in MLO-Y4 cells and mineralizing 2T3 osteoblast-osteocyte cells in response to fluid flow shear stress.
To determine if E11 can be regulated by mechanical strain, fluid flow shear stress experiments were performed with 4 and/or 16 dynes/cm2 as described previously (7, 8). The cells were harvested 2 and 24 h after 2 h of shear stress, and Northern analysis was performed (Fig. 4). A twofold increase in E11 mRNA was observed in MLO-Y4 cells at 2 h after treatment with both 4 and 16 dynes/cm2, but the level returned to the baseline by 24 h. In mineralized 2T3 cells, E11 is also encoded by an early-response gene, showing an elevation only at 2 h after shear stress, whereas osteopontin, which is encoded by a late-response gene, is elevated only at 24 h after shear stress.
FIG. 4.
Fluid flow shear stress increases mRNA for E11 in MLO-Y4 cells and mineralizing 2T3 cells. Northern blotting of E11 mRNA in MLO-Y4 cells after exposure to fluid flow shear stress at 4 and 16 dynes/cm2 showed a twofold increase at 2 h after shear stress but not at 24 h, indicating that E11 is an early-response gene. The data represent an average of two experiments as shown on the graph. E11 is also an early-response gene in mineralizing 2T3 cells, as shown by an increase at 2 h after exposure to fluid flow shear stress at 16 dynes/cm2, compared to OPN (osteopontin), which is a late-responding gene increased at 24 h, not at 2 h, after shear stress. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
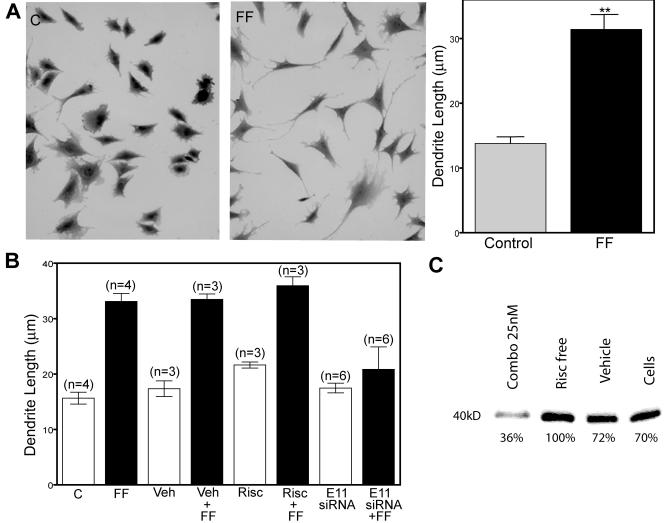
E11 is responsible for the elongation of dendrites in MLO-Y4 cells in response to fluid flow shear stress.
As shown in Fig. 5, the length of dendritic processes of MLO-Y4 cells is increased in response to mechanical strain in vitro. MLO-Y4 cells were subjected to 2 h of fluid flow shear stress at 16 dynes/cm2 and then cultured for 24 h, after which the cells were fixed and stained with crystal violet and the length of the dendrites was quantitated. A dramatic and significant increase in dendrite length was observed (Fig. 5A).
FIG. 5.
Length of dendritic processes of MLO-Y4 cells is increased in response to mechanical strain in vitro. This elongation is blocked by siRNA specific for E11. MLO-Y4 cells stained with crystal violet without fluid flow (part C) and with exposure to 16 dynes/cm2 for 2 h, followed by 24 h of incubation (part FF), show increased length of dendrites (A). The length of the dendrites is significantly increased. The formation of dendritic processes in response to shear stress is blocked by siRNA specific for E11 (B). A combination of three siRNAs specific to E11 when added for 24 h of incubation before 2 h of fluid flow shear stress inhibited the elongation of dendritic processes. No effect was observed with the RISC-free siRNA or the vehicle (Veh). The experiments were repeated three to six times and combined for statistical evaluation. Western blot analysis (C) showed that E11 protein expression was reduced approximately 50 to 70% in these cells. **, significantly different from control (P < 0.005).
To determine if E11 could have a role in dendrite formation in osteocytes, functional siRNAs specific for E11 were generated. Three different siRNAs blocked E11 protein expression at 250 nM, but in order to reduce potential toxicity due to higher molarity, the three siRNAs were combined and used at 25 nM each. Vehicle alone, E11 siRNA, or the control, a RISC-free control siRNA, was added to MLO-Y4 cells for 24 h of incubation before the cells were subjected to fluid flow shear stress for 2 h at 16 dynes/cm2. The cells were fixed and stained 24 h later for quantitation. The formation of dendritic processes in response to shear stress is blocked by siRNA specific for E11 (Fig. 5B). Western blot analysis showed that E11 protein expression was reduced approximately 50 to 70% in cells treated with E11 siRNA compared to cells alone, cells plus vehicle, or cells treated with RISC-free siRNA (Fig. 5C).
Generation of E11-null mice by gene targeting.
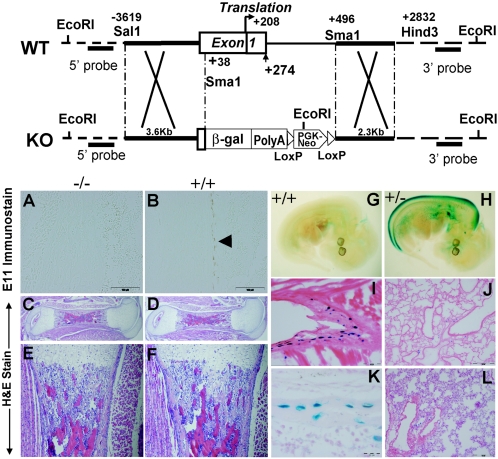
Figure 6 shows the approach we used to generate E11-null mice. As shown by PCR (data not shown) and immunostaining, the null embryos do not express E11 in bone (Fig. 6A and B). These animals die soon after birth. We were successful in generating E11-null mice at about the same time as two publications became available describing the null phenotype (38, 40). These mice most likely die because of a lung defect caused by a lack of functional alveolar type 1 cells. Unfortunately, because of the early perinatal lethality due to E11 gene deletion, it has not been possible to determine the role of E11 in the postnatal skeleton. We were able to use the lacZ knock-in approach to monitor gene expression in osteocytes (see below).
FIG. 6.
Generation and characterization of E11-null mice. A schematic of the mouse E11 wild-type (WT) locus with locations of key restriction enzyme sites and the lacZpA vector with the neo cassette inserted into exon 1 and intron 1 at the SalI and SmaI sites is shown. In the heterozygotes, one of the E11 alleles is replaced with the inserted lacZ cDNA, used to reflect the endogenous E11 expression pattern. Immunohistochemical staining for E11 protein expression was present in the osteocytes in the long bones of wild-type (+/+) embryos (B) but not E11-null (−/−) embryos (A), as shown by the brown staining (arrow). Scale bar = 100 μm. Hematoxylin-and-eosin (H&E)-stained sections of femurs isolated from E11-null and wild-type mouse embryos are compared in panels C, D, E, and F. Histological measurements are shown in Table 1. No significant differences were observed between long bones from E11-null and wild-type embryos. LacZ staining in the E11 heterozygote (+/−) localizes with protein staining in the osteocyte but does not correspond to E11 protein expression in other tissues, such as lung tissue. (G) Background LacZ staining in the 12.5-dpc wild-type embryo. (H) LacZ staining in the 12.5-dpc embryo in the dorsal spinal chord, in the lung bud, and in an area of the ventral spinal chord. Osteocytes in the rib are positive for LacZ (I) (scale bar = 0.2 μm), whereas there is a lack of LacZ staining in the lung (J) from the same newborn (scale bar = 0.5 μm). Lack of LacZ staining in the lung persisted as shown at 3 weeks of age (L) (scale bar = 0.5 μm) but continued in osteocytes as shown in the tails of heterozygote mice (K) (scale bar = 20 μm). KO, knockout; β-gal, β-galactosidase.
Phenotyping of bone in E11-null embryos.
No differences were observed between E11-null and wild-type embryos at any time before birth. Since E11-null neonates die because of respiratory failure soon after birth, 18.5-day postcoitus (dpc) embryos were harvested just before birth to optimize the chance of observing differences. As shown in Table 1 and Fig. 6C, D, E, and F, the general appearance and properties of femurs from E11-null, heterozygote, and wild-type mouse embryos were very similar. No significant differences were observed in the length or diameter of the bone or in cortical thickness. The only significant difference observed was that the null embryos weighed more than the heterozygote and wild-type embryos (Table 1). This may be related to lymphedema resulting from swelling of the limbs (40).
TABLE 1.
Phenotypes of E11-null, heterozygote, and wild-type mouse embryo femurs
| Parameter | E11−/− (n = 5) | E11+/+ (n = 5) | E11+/− (n = 14) |
|---|---|---|---|
| Body wt (g) | 1.45 ± 0.11a | 1.25 ± 0.08 | 1.26 ± 0.04 |
| Femoral shaft length (mm) | 1.58 ± 0.13 | 1.67 ± 0.09 | 1.59 ± 0.06 |
| Midshaft diam (mm) | 0.43 ± 0.02 | 0.43 ± 0.02 | 0.45 ± 0.01 |
| Maximal cortical thickness (mm) | 0.18 ± 0.01 | 0.16 ± 0.01 | 0.17 ± 0.01 |
Each value is the mean ± the standard error of the mean.
lacZ expression in E11+/− heterozygote mice.
In the 12.5-dpc E11+/− heterozygote embryo, lacZ expression was observed in the dorsal spinal cord, in the lung bud, and in the developing choroid plexus (Fig. 6H). In the newborn, high expression of lacZ was observed in osteocytes but, surprisingly, not in tissues shown to express E11/gp38, such as the lung and kidney. LacZ staining is highly visible in osteocytes in the rib of the newborn, as shown in Fig. 6I, but not in the lung at the same age (6J). In the 3-week-old mouse, LacZ staining is present in most, but not all, osteocytes in tail vertebra (6K) but not in lung tissue taken at the same age (Fig. 6L).
Others have observed tissue-specific gene regulation by the E11 promoter. A 1.2-kb portion of the promoter mimics gene expression during development but lacks sequences to enhance expression in perinatal and adult lungs (37). In the present studies, the lack of correlation of E11 gene and protein expression with lacZ expression may be due to the deletion of a portion of intron 1 and/or to the close proximity of the neo promoter. We used our sequence (accession no. AY115493) to generate the construct; therefore, only the fragment between nucleotide 9680 and nucleotide 10137 was deleted (Fig. 7). Our sequence, and therefore our construct, does not have the SmaI site located in the promoter region. A search of the databases revealed that E11 sequences obtained from the 129 mouse strain, such as ours (accession no. AY115493) and others (NT_094216), do not have this potential SmaI cleavage site, whereas sequences obtained from the C57BL/6J mouse strain do. The sequence between nucleotides 9680 and 10137 contains part of the noncoding region of exon 1, all of the coding region for exon 1, and part of intron 1. Therefore, the entire 5′ E11 promoter is present, including the transcription start site. This has been confirmed by sequencing of the construct. One could speculate that a regulatory portion of the gene is present in intron 1 and that when this is deleted, expression in other tissues would be deleted but not expression in osteocytes. Sequence analysis was performed with the Transcription Element Search System. This search revealed several potential transcription factor binding sites, as shown in Fig. 7. It has been shown that the PGKneo cassette can have an effect on neighboring genes (34). As the PGKneo cassette is only separated from the E11 gene by the β-galactosidase gene, the promoter for neo could be having an effect on lacZ expression by decreasing gene expression in other tissues. Therefore, we propose that osteocyte-selective expression and lack of expression in other tissues known to express E11 could be due to either a regulatory element in intron 1 and/or effects of the neo promoter. This observation requires further investigation into the regulation of E11/gp38 gene expression in different tissues by transgenic approaches.
FIG. 7.
Analysis of the E11 gene sequence (accession no. AY115493) used for these studies. AY115493 is the sequence we submitted and published in GenBank from the 129 mouse strain. NT_094216 (MM4_93853_34, genome) is a Mus musculus strain 129 chromosome 4 genomic contig. NT_039267 (MM4_39307_34) is a M. musculus strain C57BL/6J chromosome 4 genomic contig. AC098724 is M. musculus strain C57BL/6J bacterial artificial chromosome clone RP23-3D14 from chromosome 4. AL611982 is a mouse DNA sequence from clone RP23-348F1 on chromosome 4, also from strain C57BL/6J. Therefore, the C57BL/6J strain contains an SmaI cleavage site at this position in the promoter whereas the 129 mouse strain does not. Sequence analysis with the Transcription Element Search System revealed Ets-2 and Pit-1a binding sites in part of the deleted sequence. Several Sp1 sites and an Ap-1 site are also present in the deleted sequence, nucleotides 9680 to 10137. The nucleotide position numbering is that in the original GenBank sequence file. (R) indicates that the transcription factor recognizes the reverse complement sequence.
E11 response to mechanical loading.
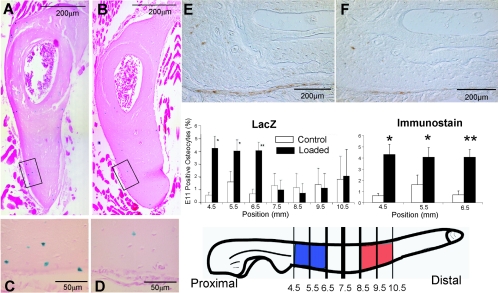
To determine if E11 is regulated by mechanical loading in vivo, loading experiments were next performed with both lacZ E11 heterozygote mice and wild-type mice. To examine gene expression, lacZ-positive cells were quantitated; to determine protein expression, immunostaining of wild-type animals was performed. A well-characterized model of loading that results in new bone formation at sites of maximal strain was used (48). Cross sections were taken at 1-mm intervals starting at 4.5 mm from the olecranon.
Sections were taken from 3-month-old lacZ heterozygote mice sacrificed at 4, 24, and 48 h after loading. Significant increases in expression could be observed at 4 h after loading in proximal sections but not in any section at 24 and 48 h (Fig. 8). Like the in vitro observations (Fig. 4), this finding suggests that E11 is an early-response gene. Surprisingly, not only were osteocytes near the surface of the bone responding to loading with an increase in E11 expression, but so were osteocytes embedded deeper within the bone matrix. Significant differences between loaded and unloaded ulnae were observed at the 4.5-, 5.5-, and 6.5-mm sections (Fig. 8). To validate these findings with lacZ expression, immunostaining was also performed with wild-type animals sacrificed at 24, 48, and 96 h after loading. No significant differences were observed at 48 and 96 h; however, similar to the lacZ sections harvested at 4 h, a significant difference in E11 protein expression in loaded compared to unloaded bone was observed at 24 h at the 4.5-, 5.5-, and 6.5-mm locations (Fig. 8E and F). These data showing protein regulation validate the data on lacZ expression. Maximal expression in response to loading was not observed at sites of maximal strain (8.5 to 10.5 mm from the olecranon, in red) but was observed at sites where potential bone remodeling would occur (4.5 to 6.5 mm from the olecranon, in blue).
FIG. 8.
Mechanical loading of mouse ulnae shows an increase in E11 expression as determined by both lacZ expression and immunostaining for E11 protein. Maximal expression is not observed in areas of maximal strain. E11-lacZ heterozygote mice and wild-type mice at 3 months of age were loaded at 3.5 N for 60 cycles at 2 Hz. The values shown were obtained 4 h after loading for lacZ expression and 24 h after loading for E11 expression. Cross sections of ulnae taken at 6.5 mm from the olecranon show X-Gal staining of osteocytes in a loaded ulna (A) and less staining in a nonloaded control ulna (B). Higher magnifications (C and D) are from the boxed areas of panels A and B, respectively, showing blue staining within osteocytes. The loaded and unloaded sections in panels E and F, respectively, are representative of immunohistochemical staining for E11 protein (brown). The graphs show the percentage of positive osteocytes (y axis) in each section, 4.5, 5.5, 6.5, 7.5, 8.5, 9.5, and 10.5 mm from the olecranon (x axis). A significant increase in E11 expression was observed in the sections 4.5, 5.5, and 6.5 mm from the olecranon. Note that osteocytes within the bone matrix also showed increased expression in response to loading (n = 4 or 5 for lacZ, n = 3 for immunostaining). Data are the mean ± the standard error of the mean. *, P < 0.05; **, P < 0.01. Previously we have noted elevated expression of genes such as those for Dmp1 and MEPE in areas of maximal strain, 8.5 to 10.5 mm from the olecranon (20, 26) (red). However, elevated E11 expression was observed in a region where increased resorption occurs in response to mechanical loading, 4.5 to 6.5 mm from the olecranon (blue). This suggests that increased E11 expression in osteocytes in this area may be associated with remodeling.
DISCUSSION
Here we show that the gene for E11/gp38 is osteocyte selective compared to osteoblasts, that it is regulated by mechanical strain both in vivo and in vitro, and that E11 appears to be responsible for dendrite elongation in response to mechanical strain. These observations provide additional insight into the function of E11/gp38. There has been much speculation regarding the function of E11 in other tissues (28, 54). As the molecule is highly negatively charged and resistant to proteases, it may provide a physical barrier protecting cells from environmental agents (55). The fact that E11 is found in cells that are exposed to an external or internal fluid compartment further supports this hypothesis. However, accumulating evidence, in addition to the data presented here, suggests that a major function of E11/gp38 may be in the formation of dendritic processes. Antibody to podoplanin (E11) causes rapid flattening of podocytes and proteinuria, suggesting that the molecule maintains the shape of the podocyte foot processes (30). Ectopic overexpression of the gene in keratinocytes induces plasma membrane extensions (42) and, in endothelial cells, the formation of long and thin tube-like structures (40) with a major reorganization of the actin cytoskeleton and relocalization of ezrin to cell projections (42). The molecule colocalizes with ezrin, radixin, and moesin (ERM) family proteins (42), molecules that have structural roles and are involved in cell motility (29) and are concentrated in cell surface projections, where they link the actin cytoskeleton to plasma membrane proteins. E11/gp38 was also found to be physically associated with CD44 in tumor vascular endothelial cells (32) and in some cancer cells (46). Together, these data suggest that E11/gp38 associates with CD44 and the ERMs to induce and regulate the formation of dendritic processes. Interestingly, overexpression in keratinocytes leads to the acquisition of a malignant phenotype (41). In the present study, increased E11 expression in vitro and in vivo in response to mechanical strain and blocking of dendrite formation by siRNA specific for E11 suggests that at least one function of E11 in osteocytes is the formation of dendritic processes.
Our present studies suggest that E11 could be playing a role in normal dendrite formation. The early formation of dendrites by embedding osteoid cells or osteocytes is polarized toward the mineralization front and toward blood vessels (35). Osteocyte dendricity changes with static and dynamic bone formation (36). In normal bone, osteocyte connectivity is high and the processes are oriented in the direction of the blood supply (25). In osteoporotic bone, there is a marked decrease in connectivity, as well as disorientation of the dendrites, which increases in severity. In osteoarthritic bone, a decrease in connectivity is observed, but dendrite orientation is intact, whereas in osteomalacic bone, the osteocytes appear viable with high connectivity but the dendrites are distorted and the network is highly disorganized (25). Changes in osteocyte dendricity could have a dramatic effect not only on osteocyte function and viability but also on the mechanical properties of bone. An equilibrium must be obtained between dendrite network complexity to preserve function and viability and the amount that would decrease bone strength. A potential role for E11 in bone disease in unknown.
Dogma exists that the osteocyte is a passive cell that mainly fills space in bone. This raises the question of why E11 would increase in embedded osteocytes in response to strain if the embedded cell cannot make new dendrites. Data are starting to emerge indicating that the osteocyte is a dynamic, not a passive, cell. Okada and colleagues documented an increase in canaliculi in rats between 3 and 12 weeks of age (33). Holmbeck and colleagues (18) have obtained similar results with mice and have shown osteocytogenesis to be an active, invasive process requiring cleavage of collagen and other matrix molecules by MT1-MMP. They propose that MT1-MMP is necessary for the formation of canaliculi as osteocytes in MT1-MMP-null mice have a significantly reduced number and length of dendritic processes. The almost complete lack of dendritic processes in this mouse model did not appear to affect the viability or density of osteocytes, in contrast to studies by Zhao and coworkers (53), where osteocytes in a mouse model of collagenase-resistant type I collagen did show increased apoptosis. Our studies showing that E11 is responsible for osteocyte dendrite formation and previous studies supporting the hypothesis that osteocytes can generate new canaliculi suggest that the osteocyte has the capacity to actively regulate the formation of both dendrites and canaliculi.
In the present study, no bone phenotype was observed in late embryonic E11-null mice. Mice null for dentin matrix protein 1 (Dmp1), matrix extracellular phosphoglycoprotein (MEPE), or OF45, Sost, and other proteins that are highly expressed in osteocytes do not show a phenotype until days to weeks or even months after birth (13, 16, 45). One potential explanation for this is that osteocytes in the embryo may not require extensive dendrite connections because the bone cortices and trabeculae are relatively thin and poorly mineralized and the cells are near the bone surface. Thus, nutrients may be able to readily diffuse to the osteocytes without requiring an extensive canalicular system. Also, in utero, although subjected to some mechanical loading via muscle insertions, the skeleton is not subjected to significant loading from weight-bearing activity. It may therefore be that the responses of load-related bone remodeling are less significant in the developing embryo, where the overriding signals are for bone growth and development. Molecules that play a role in the response of osteocytes to mechanical strain may not reveal their importance for normal skeletal physiology until postnatally or in the adult animal. Thus, E11, like other osteocyte-selective molecules, may play a more important role in the adult skeleton, specifically, in responses to mechanical strain. To overcome the perinatal lethality of E11 gene deletion via its effects in the lung, a conditional-knockout approach is required. Our present approach is to eliminate E11 expression in osteocytes, with E11-floxed mice crossed with transgenic mice expressing Cre recombinase driven by the osteocalcin promoter (OC-Cre) that is expressed very late in osteoblast differentiation, just before E11. An alternative approach is to use the 8-kb Dmp1 promoter, which has been shown to be osteocyte selective (21). These transgenic approaches are the focus of ongoing studies.
Acknowledgments
We thank Andrew Farr at the University of Washington, Seattle, for the 8.1.1 antibody and acknowledge the technical assistance of Dayong Guo, University of Missouri at Kansas City, and Yiqiang Zhang, University of Texas Health Science Center at San Antonio.
This study was supported by National Institutes of Health grant PO1 AR46798 to S.E.H., J.Q.F., and L.F.B.
REFERENCES
- 1.Baylink, D., J. Sipe, J. Wergedal, and O. J. Whittemore. 1973. Vitamin D-enhanced osteocytic and osteoclastic bone resorption. Am. J. Physiol. 224:1345-1357. [DOI] [PubMed] [Google Scholar]
- 2.Bonewald, L. F., S. E. Harris, J. Rosser, M. R. Dallas, S. L. Dallas, N. P. Camacho, B. Boyan, and A. Boskey. 2003. von Kossa staining alone is not sufficient to confirm that mineralization in vitro represents bone formation. Calcif. Tissue Int. 72:537-547. [DOI] [PubMed] [Google Scholar]
- 3.Boucherot, A., R. Schreiber, H. Pavenstadt, and K. Kunzelmann. 2002. Cloning and expression of the mouse glomerular podoplanin homologue gp38P. Nephrol. Dial. Transplant. 17:978-984. [DOI] [PubMed] [Google Scholar]
- 4.Burr, D. B., A. G. Robling, and C. H. Turner. 2002. Effects of biomechanical stress on bones in animals. Bone 30:781-786. [DOI] [PubMed] [Google Scholar]
- 5.Cao, Y. X., M. I. Ramirez, and M. C. Williams. 2003. Enhanced binding of Sp1/Sp3 transcription factors mediates the hyperoxia-induced increased expression of the lung type I cell gene T1alpha. J. Cell. Biochem. 89:887-901. [DOI] [PubMed] [Google Scholar]
- 6.Carninci, P. H., and Y. Hayashizaki. 1999. High efficiency full-length cDNA cloning. Methods Enzymol. 303:19-44. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, B., Y. Kato, S. Zhao, J. Luo, E. Sprague, L. F. Bonewald, and J. X. Jiang. 2001. PGE2 is essential for gap junction-mediated intercellular communication between osteocyte-like MLO-Y4 cells in response to mechanical strain. Endocrinology 142:3464-3473. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, B., S. Zhao, J. Luo, E. Sprague, L. F. Bonewald, and J. X. Jiang. 2001. Expression of functional gap junctions and regulation by fluid flow in osteocyte-like MLO-Y4 cells. J. Bone Miner. Res. 16:249-259. [DOI] [PubMed] [Google Scholar]
- 9.Dobbs, L. G., M. C. Williams, and R. Gonzalez. 1988. Monoclonal antibodies specific to apical surfaces of rat alveolar type I cells bind to surfaces of cultured, but not freshly isolated, type II cells. Biochim. Biophys. Acta 970:146-156. [DOI] [PubMed] [Google Scholar]
- 10.Ehrlich, P. J., B. S. Noble, H. L. Jessop, H. Y. Stevens, J. R. Mosley, and L. E. Lanyon. 2002. The effect of in vivo mechanical loading on estrogen receptor alpha expression in rat ulnar osteocytes. J. Bone Miner. Res. 17:1646-1655. [DOI] [PubMed] [Google Scholar]
- 11.Farr, A., A. Nelson, and S. Hosier. 1992. Characterization of an antigenic determinant preferentially expressed by type I epithelial cells in the murine thymus. J. Histochem. Cytochem. 40:651-664. [DOI] [PubMed] [Google Scholar]
- 12.Farr, A. G., M. L. Berry, A. Kim, A. J. Nelson, M. P. Welch, and A. Aruffo. 1992. Characterization and cloning of a novel glycoprotein expressed by stromal cells in T-dependent areas of peripheral lymphoid tissues. J. Exp. Med. 176:1477-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reference deleted.
- 14.Ghosh-Choudhury, N., J. J. Windle, B. A. Koop, M. A. Harris, D. L. Guerrero, J. M. Wozney, G. R. Mundy, and S. E. Harris. 1996. Immortalized murine osteoblasts derived from BMP 2-T-antigen expressing transgenic mice. Endocrinology 137:331-339. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez, R. F., and L. G. Dobbs. 1998. Purification and analysis of RTI40, a type I alveolar epithelial cell apical membrane protein. Biochim. Biophys. Acta 1429:208-216. [DOI] [PubMed] [Google Scholar]
- 16.Gowen, L. C., D. N. Petersen, A. L. Mansolf, H. Qi, J. L. Stock, G. T. Tkalcevic, H. A. Simmons, D. T. Crawford, K. L. Chidsey-Frink, H. Z. Ke, J. D. McNeish, and T. A. Brown. 2003. Targeted disruption of the osteoblast/osteocyte factor 45 gene (OF45) results in increased bone formation and bone mass. J. Biol. Chem. 278:1998-2007. [DOI] [PubMed] [Google Scholar]
- 17.Harris, S. E., L. F. Bonewald, M. A. Harris, M. Sabatini, S. Dallas, J. Q. Feng, N. Ghosh-Choudhury, J. Wozney, and G. R. Mundy. 1994. Effects of transforming growth factor beta on bone nodule formation and expression of bone morphogenetic protein 2, osteocalcin, osteopontin, alkaline phosphatase, and type I collagen mRNA in long-term cultures of fetal rat calvarial osteoblasts. J. Bone Miner. Res. 9:855-863. [DOI] [PubMed] [Google Scholar]
- 18.Holmbeck, K., P. Bianco, I. Pidoux, S. Inoue, R. C. Billinghurst, W. Wu, K. Chrysovergis, S. Yamada, H. Birkedal-Hansen, and A. R. Poole. 2005. The metalloproteinase MT1-MMP is required for normal development and maintenance of osteocyte processes in bone. J. Cell Sci. 118:147-156. [DOI] [PubMed] [Google Scholar]
- 19.Hong, Y. K., and M. Detmar. 2003. Prox1, master regulator of the lymphatic vasculature phenotype. Cell Tissue Res. 314:85-92. [DOI] [PubMed] [Google Scholar]
- 20.Hsieh, Y. F., A. G. Robling, W. T. Ambrosius, D. B. Burr, and C. H. Turner. 2001. Mechanical loading of diaphyseal bone in vivo: the strain threshold for an osteogenic response varies with location. J. Bone Miner. Res. 16:2291-2297. [DOI] [PubMed] [Google Scholar]
- 21.Kalajzic, I., A. Braut, D. Guo, X. Jiang, M. S. Kronenberg, M. Mina, M. A. Harris, S. E. Harris, and D. W. Rowe. 2004. Dentin matrix protein 1 expression during osteoblastic differentiation, generation of an osteocyte GFP-transgene. Bone 35:74-82. [DOI] [PubMed] [Google Scholar]
- 22.Kamioka, H., T. Honjo, and T. Takano-Yamamoto. 2001. A three-dimensional distribution of osteocyte processes revealed by the combination of confocal laser scanning microscopy and differential interference contrast microscopy. Bone 28:145-149. [DOI] [PubMed] [Google Scholar]
- 23.Kato, Y., A. Boskey, L. Spevak, M. Dallas, M. Hori, and L. F. Bonewald. 2001. Establishment of an osteoid preosteocyte-like cell MLO-A5 that spontaneously mineralizes in culture. J. Bone Miner. Res. 16:1622-1633. [DOI] [PubMed] [Google Scholar]
- 24.Kato, Y., J. J. Windle, B. A. Koop, G. R. Mundy, and L. F. Bonewald. 1997. Establishment of an osteocyte-like cell line, MLO-Y4. J. Bone Miner. Res. 12:2014-2023. [DOI] [PubMed] [Google Scholar]
- 25.Knothe Tate, M. L., J. R. Adamson, A. E. Tami, and T. W. Bauer. 2004. The osteocyte. Int. J. Biochem. Cell. Biol. 36:1-8. [DOI] [PubMed] [Google Scholar]
- 26.Kotha, S. P., Y. F. Hsieh, R. M. Strigel, R. Muller, and M. J. Silva. 2004. Experimental and finite element analysis of the rat ulnar loading model—correlations between strain and bone formation following fatigue loading. J. Biomech. 37:541-548. [DOI] [PubMed] [Google Scholar]
- 27.Lanyon, L. E. 1993. Osteocytes, strain detection, bone modeling and remodeling. Calcif. Tissue Int. 53:S102-S107. [DOI] [PubMed] [Google Scholar]
- 27a.Luna, L. G. 1968. Manual of histologic staining methods of the Armed Forces Institute of Pathology. McGraw-Hill, New York, N.Y.
- 28.Ma, T., B. Yang, M. A. Matthay, and A. S. Verkman. 1998. Evidence against a role of mouse, rat, and two cloned human t1alpha isoforms as a water channel or a regulator of aquaporin-type water channels. Am. J. Respir. Cell Mol. Biol. 19:143-149. [DOI] [PubMed] [Google Scholar]
- 29.Mangeat, P., C. Roy, and M. Martin. 1999. ERM proteins in cell adhesion and membrane dynamics. Trends Cell Biol. 9:187-192. [DOI] [PubMed] [Google Scholar]
- 30.Matsui, K., S. Breiteneder-Geleff, and D. Kerjaschki. 1998. Epitope-specific antibodies to the 43-kD glomerular membrane protein podoplanin cause proteinuria and rapid flattening of podocytes. J. Am. Soc. Nephrol. 9:2013-2026. [DOI] [PubMed] [Google Scholar]
- 31.Nose, K., H. Saito, and T. Kuroki. 1990. Isolation of a gene sequence induced later by tumor-promoting 12-O-tetradecanoylphorbol-13-acetate in mouse osteoblastic cells (MC3T3-E1) and expressed constitutively in ras-transformed cells. Cell Growth Differ. 1:511-518. [PubMed] [Google Scholar]
- 32.Ohizumi, I., N. Harada, K. Taniguchi, Y. Tsutsumi, S. Nakagawa, S. Kaiho, and T. Mayumi. 2000. Association of CD44 with OTS-8 in tumor vascular endothelial cells. Biochim. Biophys. Acta 1497:197-203. [DOI] [PubMed] [Google Scholar]
- 33.Okada, S., S. Yoshida, S. H. Ashrafi, and D. E. Schraufnagel. 2002. The canalicular structure of compact bone in the rat at different ages. Microsc. Microanal. 8:104-115. [DOI] [PubMed] [Google Scholar]
- 34.Olson, E. N., H. H. Arnold, P. W. Rigby, and B. J. Wold. 1996. Know your neighbors: three phenotypes in null mutants of the myogenic bHLH gene MRF4. Cell 85:1-4. [DOI] [PubMed] [Google Scholar]
- 35.Palumbo, C. 1986. A three-dimensional ultrastructural study of osteoid-osteocytes in the tibia of chick embryos. Cell Tissue Res. 246:125-131. [DOI] [PubMed] [Google Scholar]
- 36.Palumbo, C., M. Ferretti, and G. Marotti. 2004. Osteocyte dendrogenesis in static and dynamic bone formation: an ultrastructural study. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 278:474-480. [DOI] [PubMed] [Google Scholar]
- 37.Ramirez, M. I., Y. X. Cao, and M. C. Williams. 1999. 1.3 kilobases of the lung type I cell T1alpha gene promoter mimics endogenous gene expression patterns during development but lacks sequences to enhance expression in perinatal and adult lung. Dev. Dyn. 215:319-331. [DOI] [PubMed] [Google Scholar]
- 38.Ramirez, M. I., G. Millien, A. Hinds, Y. Cao, D. C. Seldin, and M. C. Williams. 2003. T1alpha, a lung type I cell differentiation gene, is required for normal lung cell proliferation and alveolus formation at birth. Dev. Biol. 256:61-72. [DOI] [PubMed] [Google Scholar]
- 39.Rishi, A. K., M. Joyce-Brady, J. Fisher, L. G. Dobbs, J. Floros, J. VanderSpek, J. S. Brody, and M. C. Williams. 1995. Cloning, characterization, and development expression of a rat lung alveolar type I cell gene in embryonic endodermal and neural derivatives. Dev. Biol. 167:294-306. [DOI] [PubMed] [Google Scholar]
- 40.Schacht, V., M. I. Ramirez, Y. K. Hong, S. Hirakawa, D. Feng, N. Harvey, M. Williams, A. M. Dvorak, H. F. Dvorak, G. Oliver, and M. Detmar. 2003. T1alpha/podoplanin deficiency disrupts normal lymphatic vasculature formation and causes lymphedema. EMBO J. 22:3546-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scholl, F. G., C. Gamallo, and M. Quintanilla. 2000. Ectopic expression of PA2.26 antigen in epidermal keratinocytes leads to destabilization of adherens junctions and malignant progression. Lab. Investig. 80:1749-1759. [DOI] [PubMed] [Google Scholar]
- 42.Scholl, F. G., C. Gamallo, S. Vilaró, and M. Quintanilla. 1999. Identification of PA2.26 antigen as a novel cell-surface mucin-type glycoprotein that induces plasma membrane extensions and increased motility in keratinocytes. J. Cell Sci. 112(Pt. 24):4601-4613. [DOI] [PubMed] [Google Scholar]
- 43.Schulze, E., M. Witt, M. Kasper, C. W. Lowik, and R. H. Funk. 1999. Immunohistochemical investigations on the differentiation marker protein E11 in rat calvaria, calvaria cell culture and the osteoblastic cell line ROS 17/2.8. Histochem. Cell Biol. 111:61-69. [DOI] [PubMed] [Google Scholar]
- 44.Sprague, L., A. Wetterwald, U. Heinzman, and M. J. Atkinson. 1996. Phenotypic changes following over-expression of sense or antisense E11 cDNA in ROS 17/2.8 cells. J. Bone Miner. Res. 11:S132. [Google Scholar]
- 45.Sun, N., Y. Gao, J. Pretoriu, S. Morony, P. J. Kostenuik, S. Simonet, D. L. Lacey, I. Sarosi, C. Kurahara, and C. Patszty. 2003. High bone mineral density in SOST knock-out mice demonstrates functional conservation of osteocyte mediated bone homeostasis in mouse and human. J. Bone. Miner. Res. 18(Suppl. 2):S7. [Google Scholar]
- 46.Taniguchi, K., N. Harada, I. Ohizumi, M. Kinoshita, Y. Tsutsumi, S. Nakagawa, S. Kaiho, and T. Mayumi. 2000. Molecular cloning and characterization of antigens expressed on rat tumor vascular endothelial cells. Int. J. Cancer 86:799-805. [DOI] [PubMed] [Google Scholar]
- 47.Tenorio, D., A. Cruchley, and F. J. Hughes. 1993. Immunocytochemical investigation of the rat cementoblast phenotype. J. Periodontal Res. 28:411-419. [PubMed] [Google Scholar]
- 48.Torrance, A. G., J. R. Mosley, R. F. Suswillo, and L. E. Lanyon. 1994. Noninvasive loading of the rat ulna in vivo induces a strain-related modeling response uncomplicated by trauma or periostal pressure. Calcif. Tissue Int. 54:241-247. [DOI] [PubMed] [Google Scholar]
- 49.Wetterwald, A., W. Hoffstetter, M. G. Cecchini, B. Lanske, C. Wagner, H. Fleisch, and M. Atkinson. 1996. Characterization and cloning of the E11 antigen, a marker expressed by rat osteoblasts and osteocytes. Bone 18:125-132. [DOI] [PubMed] [Google Scholar]
- 50.Ye, L., Y. Mishina, D. Chen, H. Huang, S. L. Dallas, M. Dallas, T. W. Kunieda, T. Tsutsui, A. Boskey, L. F. Bonewald, and J. Q. Feng. 2004. Dentin matrix protein 1 (Dmp1) deficient mice display severe defects in cartilage formation responsible for a chondrodysplasia-like phenotype. J. Biol. Chem. 280:6197-6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50a.Ye, L., Y. Mishina, D. Chen, H. Huang, S. L. Dallas, M. R. Dallas, P. Sivakumar, T. Kunieda, T. W. Tsutsui, A. Boskey, L. F. Bonewald, and J. Q. Feng. 2005. Dmp1-deficient mice display severe defects in cartilage formation responsible for a chondrodysplasia-like phenotype. J. Biol. Chem. 280:6197-6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, J., X. Tan, C. H. Contag, Y. Lu, D. Guo, S. E. Harris, and J. Q. Feng. 2002. Dissection of promoter control modules that direct Bmp4 expression in the epithelium-derived components of hair follicles. Biochem. Biophys. Res. Commun. 293:1412-1419. [DOI] [PubMed] [Google Scholar]
- 52.Zhao, S., Y. K. Zhang, S. Harris, S. S. Ahuja, and L. F. Bonewald. 2002. MLO-Y4 osteocyte-like cells support osteoclast formation and activation. J. Bone Miner. Res. 17:2068-2079. [DOI] [PubMed] [Google Scholar]
- 53.Zhao, W., M. H. Byrne, Y. Wang, and S. M. Krane. 2000. Osteocyte and osteoblast apoptosis and excessive bone deposition accompany failure of collagenase cleavage of collagen. J. Clin. Investig. 106:941-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zimmer, G., H. D. Klenk, and G. Herrler. 1995. Identification of a 40-kDa cell surface sialoglycoprotein with the characteristics of a major influenza C virus receptor in a Madin-Darby canine kidney cell line. J. Biol. Chem. 270:17815-17822. [DOI] [PubMed] [Google Scholar]
- 55.Zimmer, G., F. Oeffner, V. Von Messling, T. Tschernig, H. J. Groness, H. D. Klenk, and G. Herrler. 1999. Cloning and characterization of gp36, a human mucin-type glycoprotein preferentially expressed in vascular endothelium. Biochem. J. 341(Pt 2):277-284. [PMC free article] [PubMed] [Google Scholar]