Abstract
Genes involved in the transforming growth factor β (TGF-β) signaling pathway are frequently altered in several types of cancers, and a gastric tumor suppressor RUNX3 appears to be an integral component of this pathway. We reported previously that apoptosis is notably reduced in Runx3−/− gastric epithelial cells. In the present study, we show that a proapoptotic gene Bim was transcriptionally activated by RUNX3 in the gastric cancer cell lines SNU16 and SNU719 treated with TGF-β. The human Bim promoter contains RUNX sites, which are required for its activation. Furthermore, a dominant negative form of RUNX3 comprised of amino acids 1 to 187 increased tumorigenicity of SNU16 by inhibiting Bim expression. In Runx3−/− mouse gastric epithelium, Bim was down-regulated, and apoptosis was reduced to the same extent as that in Bim−/− gastric epithelium. We confirmed comparable expression of TGF-β1 and TGF-β receptors between wild-type and Runx3−/− gastric epithelia and reduction of Bim in TGF-β1−/− stomach. These results demonstrate that RUNX3 is responsible for transcriptional up-regulation of Bim in TGF-β-induced apoptosis.
Transforming growth factor β (TGF-β) regulates a diverse array of biological activities, including cell growth, the cell cycle, early development, differentiation, extracellular matrix formation, hematopoesis, angiogenesis, chemotaxis, and immune functions (34). In many human tumors, many components of the TGF-β pathway are defective. This can occur as the result of mutation or reduced expression of TGF-β receptors or components of the TGF-β signaling pathway (8, 34). Thus, the TGF-β pathway is considered to be a tumor suppressor pathway that negatively regulates cell growth and promotes apoptosis of epithelial cells (41). Acquired resistance toward apoptosis is one of the hallmarks of most and perhaps all types of cancer (18). Thus, understanding how cancer cells either resist or regulate TGF-β-induced apoptosis in carcinogenesis is of central importance to cancer research.
TGF-β-induced apoptosis is a well-documented phenomenon in many cell types in vitro and is known to involve TIEG1 (43), DAXX (36), DAPK (death-associated protein kinase) (25), ARTS (32) SHIP (46), truncated Bad (28), AP1 (51), GADD45b (53), Fas (29), and Bim (49), as well as components of the canonical TGF-β signaling pathway, such as Smad3, Smad4, Smad7, and mitogen-activated protein kinases (MAP kinases), such as Jun N-terminal protein kinase and p38 MAP kinase (39, 41). More recently, interactions between Akt and Smad3 (6) or between Smad7 and β-catenin (10) were shown to be involved in apoptosis induction. The physiological significance of TGF-β-mediated apoptosis in carcinogenesis is unclear since its underlying molecular mechanisms have been described only in in vitro systems, due to a lack of in vivo model systems.
We reported recently that the lack of RUNX3 function is causally related to the genesis and progression of gastric cancer. About 45 to 60% of surgically resected gastric cancer specimens and gastric cancer cell lines do not express RUNX3 due to hemizygous deletion of the gene and hypermethylation of its promoter region (33). A mutation found in a gastric cancer patient, RUNX3(R122C), which results in a single amino acid substitution within the conserved DNA binding domain, completely abolishes the tumor suppressor activity of RUNX3 in a nude mouse assay (33). Primary gastric epithelial cells isolated from Runx3-deficient (Runx3−/−) mice were less sensitive to the growth inhibitory effect and apoptosis-inducing activity of TGF-β. Consistent with these observations, the gastric epithelium of Runx3−/− mice shows hyperplasia. Furthermore, gastric epithelial cell lines (GIF cell lines) isolated from Runx3−/− mice in the p53-null background (Runx3−/− p53−/−) were tumorigenic in nude mice, whereas Runx3+/+ p53−/− cells were not (14, 33).
We have shown previously that RUNX3 forms complexes with Smads that regulate target gene expression; thus, RUNX3 is a downstream target of the TGF-β signaling pathway (19, 24). In TGF-β1-null animals, gastric epithelial hyperplasia is observed (7). Thus, apoptosis and growth of gastric epithelial cells are regulated at least partly by the TGF-β pathway, and molecular interactions between RUNX3 and Smads probably play an important role in gastric carcinogenesis (13, 19). In this study, we identified a proapoptotic gene, Bim, whose transcriptional activation by TGF-β is mediated by RUNX3 in gastric epithelial cells undergoing TGF-β-induced apoptosis.
MATERIALS AND METHODS
Cell lines and mice.
SNU16 and SNU719 (17) cells were maintained in RPMI medium supplemented with 10% fetal bovine serum. GIF cells (Runx3+/+ and Runx3−/−) were maintained in Ham's F12 medium (Sigma) supplemented with 10 or 0.5% horse serum and bovine pituitary extract (100 mg/ml; Gibco-BRL) (14). Cells were transfected with pcDNA3.1/HisC, pcDNA-Flag-RUNX3, pcDNA-Flag-RUNX3(1-187) (a truncated RUNX3 comprised of amino acids 1 to 187), and pEF-BOS-neo-RUNX3-AS using Lipofectamine 2000 and Plus Reagent (Invitrogen) to generate stable transfectants, control SNU16/SNU719 cells, SNU16 cells expressing full-length RUNX3, RUNX3(1-187), or SNU16/SNU719 cells expressing antisense DNA against RUNX3, respectively. pEF-BOS-neo-RUNX3-AS was constructed by subcloning a RUNX3 cDNA fragment (accession number Z35278[1]) into the XbaI site of pEF-BOS-neo (35) in the reverse orientation by blunt-end ligation. Stable transfectants were selected in the presence of 0.5 mg/ml G418 (GIBCO). Independent clones that stably expressed Flag-tagged full-length RUNX3, Flag-tagged RUNX3(1-187) or reduced levels of RUNX3 were identified by Western blot analysis using either an anti-Flag monoclonal antibody (M2; Sigma) or R3-5G4 (23), respectively. SNU16 cells were transfected, respectively, with pSuper-pEF-neo and pSuper-bim73-pEF-neo (4) using the mixture of Lipofectamine 2000 and Plus Reagent to generate stable transfectants, namely, control SNU16 and SNU16 expressing short hairpin RNA (shRNA) targeting Bim. Stable transfectants were selected in the presence of 0.7 mg/ml G418. Reduced levels of Bim were identified by Western blot analysis using an anti-Bim polyclonal antibody (sc-11425; Santa Cruz). All clones were treated with recombinant human TGF-β1 (240-B; R&D Systems).
Runx3- and Bim-deficient mice were described previously (2, 33). FasL(Faslgld) (B6Smn.C3-Tnfsf6gld/J)-deficient and TGF-β1 (Tgfb1tm1Doe) (B6.129S2-Tgfb1tm1Doe/J)-deficient mice were obtained from the Jackson laboratory. To generate TGF-β1 deficient mice in BALB/c background, TGF-β1+/− mice in C57BL/6J background were back-crossed with a wild type of BALB/c mice for more than seven generations. Studies were done in accordance with the guidelines of The Institute of Molecular and Cell Biology, Singapore.
Semiquantitative RT-PCR, quantitative RT-PCR and Western blotting.
RNA was extracted from SNU16 cells, SNU719 cells, GIF cells, and neonatal mouse stomachs using TRIzol (GIBCO) and cDNA was synthesized using Expand Reverse Transcriptase and Primer p[DT]15 (Roche). Semiquantitative reverse transcription-PCR (RT-PCR) for detection of proapoptotic or antiapoptotic gene expression in SNU16 cells and neonatal mouse stomachs was performed using primers shown in Table 1.
TABLE 1.
Primers used for semiquantitative RT-PCR in this study
| Gene | Primer | Sequence |
|---|---|---|
| Bak | Forward | 5′-GAA TTC ATG GCT TCG GGG CAA GGC CCA-3′ |
| Reverse | 5′-GGA TCC TCA TCA TGA TTT GAA GAA TCT TCG T-3′ | |
| Bid | Forward | 5′-CCA TGG ACT GTG AGG TCA AC-3′ |
| Reverse | 5′-GTC CAT CCC ATT TCT GGC TA-3′ | |
| Bax | Forward | 5′-CCT TTT CTA CTT TGT CAG CAA-3′ |
| Reverse | 5′-GAG GCC GTC CCA ACC AC-3′ | |
| Bmf | Forward | 5′-TAG GAG AGA TGG AGC CAT CTC AGT-3′ |
| Reverse | 5′-TTC CTG TTC CAG ACG GTG TTC CTG-3′ | |
| Noxa | Forward | 5′-GAC TGT TCG TGT TCA GCT CGC GTC-3′ |
| Reverse | 5′-GAA GGA GTC CCC TCA TGC AAG TTT-3′ | |
| Bcl-2 | Forward | 5′-GCC CTG TGG ATG ACT GAG TA-3′ |
| Reverse | 5′-ACT TGT GGC TCA GAT AGG CA-3′ | |
| Bcl-xL | Forward | 5′-GCA GGT ATT GGT GAG TCG G-3′ |
| Reverse | 5′-CTG AAG AGT GAG CCC AGC A-3′ | |
| Bcl-w | Forward | 5′-GGA GTT CAC AGC TCT ATA CGG GGA-3′ |
| Reverse | 5′-ACA GCT GCT TAT GTA AGT CCA CCT-3′ | |
| A1 | Forward | 5′-GTT TGA AGA CGG CAT CAT T-3′ |
| Reverse | 5′-ACA AAG CCA TTT TCC CAG-3′ | |
| Mcl-1 | Forward | 5′-CTC TCA TTT CTT TTG GTG CCT-3′ |
| Reverse | 5′-ATT CCT GAT GCC ACC TTC TA-3′ | |
| Bim | Forward | 5′-ATG GCA AAG CAA CCT TCT GA-3′ |
| Reverse | 5′-CGC ATA TCT GCA GGT TCA GCC-3′ | |
| cIAP-1 | Forward | 5′-AGC TGT TGT CAA CTT CAG ATA CCA CT-3′ |
| Reverse | 5′-TGT TTC ACC AGG TCT CTA TTA AAG CC-3′ | |
| cIAP-2 | Forward | 5′-ACT TGA ACA GCT GCT ATC CAC ATC-3′ |
| Reverse | 5′-GTT GCT AGG ATT TTT CTC TGA ACT GTC-3′ | |
| XIAP | Forward | 5′-GGT TCA GTT TCA AGG ACA TT-3′ |
| Reverse | 5′-CAA GGA ACA AAA ACG ATA GC-3′ | |
| TNFR1 | Forward | 5′-TGT GTC TCC TGT AGT AAC TG-3′ |
| Reverse | 5′-ACG AAT TCC TTC CAG CGC AA-3′ | |
| TNF-α | Forward | 5′-CAG AGG GAA GAG TTC CCC CAG-3′ |
| Reverse | 5′-CCT TGG TCT GGT AGG AGA CG-3′ | |
| TRAIL | Forward | 5′-AGA CCT GCG TGC TGA TCG TG-3′ |
| Reverse | 5′-TTA TTT TGC GGC CCA GAG CC-3′ | |
| TRAILR1 | Forward | 5′-CAG AAC GTC CTG GAG CCT GTA AC-3′ |
| Reverse | 5′-ATG TCC ATT GCC TGA TTC TTT GTG-3′ | |
| TRAILR2 | Forward | 5′-GGG AAG AAG ATT CTC CTG AGA TGT G-3′ |
| Reverse | 5′-ACA TTG TCC TCA GCC CCA GGT CG-3′ | |
| DR6 | Forward | 5′-ACA GAA GGC CTC GAA TCT CA-3′ |
| Reverse | 5′-TGC ATT CTC GGT CAG TCA AG-3 | |
| DAPK | Forward | 5′-GAC CGT GTT CAG GCA GGA-3′ |
| Reverse | 5′-TCA GTT GCT TCC TCT TCA GT-3′ | |
| Daxx | Forward | 5′-CAG GAA TTC TGA AAT CCC CAC CAC-3′ |
| Reverse | 5′-TCC TTG GAG ACT GAG AGG CAG TAT-3′ | |
| TIEG1 | Forward | 5′-AAG CAG CCA ACC ATG CTC AA-3′ |
| Reverse | 5′-GTC AGA AGG ACT GTA AGG TG-3′ | |
| GADD45b | Forward | 5′-GGA CCC AGA CAG CGT GGT CCT CTG-3′ |
| Reverse | 5′-GTG ACC AGG AGA CAA TGC AGG TCT-3′ | |
| SHIP | Forward | 5′-GAC ACA GAA AGT GTC GTG TCT C-3′ |
| Reverse | 5′-GGA ACC TCC TTG GCC TCA CAG-3′ | |
| ARTS | Forward | 5′-TGT GGA CAC ACC AGG-3′ |
| Reverse | 5′-TCT TGG CCT GTT CCC TTG AC-3′ | |
| GAPDH | Forward | 5′-ACC ACA GTC CAT GCC ATC AC-3′ |
| Reverse | 5′-TCC ACC ACC CTG TTG CTG TA-3′ | |
| mBad | Forward | 5′-TTC CAG ATC CCA GAG TTT G-3′ |
| Reverse | 5′-GGA GAT CAC TGG GAG GGG GTG G-3′ | |
| mBid | Forward | 5′-ATG GAC TCT GAG G-3′ |
| Reverse | 5′-TTA GTC CAT CTC GTT TCT AAC CAA G-3′ | |
| mBax | Forward | 5′-AGG ATA CAG CTG GAG TCA G-3′ |
| Reverse | 5′-TCT CCT TGT CTA CGC TTT CC-3′ | |
| mBak | Forward | 5′-CCC AGG ACA CAG AGG AGG TC-3′ |
| Reverse | 5′-GCC CAA CAG AAC CAC ACC AAA A-3′ | |
| mBmf | Forward | 5′-CCC TTG GGG AGC AGC CCC CTG-3′ |
| Reverse | 5′-CAA GAC AGT ATC TGT CCT CCC AGA C-3′ | |
| mBcl-2 | Forward | 5′-TAC CGT CGT GAC TTC GCA GAG-3′ |
| Reverse | 5′-GGC AGG CTG AGC AGG GTC TT-3′ | |
| mBcl-xL | Forward | 5′-AGG CAG GCG ATG AGT TTG AAC-3′ |
| Reverse | 5′-GAA CCA CAC CAG CCA CAG TCA-3′ | |
| mBcl-w | Forward | 5′-GTT TCC GCC GCA CCT TCT CT-3′ |
| Reverse | 5′-CCC CGT CAG CAC TGT CCT CA-3′ | |
| mBim | Forward | 5′-TTG CCA TCG TCG CCG TCA C-3′ |
| Reverse | 5′-CAG TTG TAA GAT AAC CAT TTG AGG GTG G-3′ | |
| mFasL | Forward | 5′-TAA AAC CGT TTG CTG GGG C-3′ |
| Reverse | 5′-CTC AGC TCC TTT TTT TCA GGG-3′ | |
| mTNFR1 | Forward | 5′-GAA TTC ATG GGT CTC CCC ACC GTG CC-3′ |
| Reverse | 5′-AAG CTT TCA GGC CAC TTT GAC TGC AAT CT-3′ | |
| mTNFα | Forward | 5′-ATG AGC ACA GAA AGC ATG ATC CGC G-3′ |
| Reverse | 5′-AGA CCT GCC CGG ACT CCG CAA AGT C-3′ | |
| mTRAIL | Forward | 5′-TGA GGA TTT CTG GGA CTC CAC TGA-3′ |
| Reverse | 5′-CTT CAG CTT CCT GAA ATC GG-3′ | |
| mTRAILR2 | Forward | 5′-CAC AAA TAC GGT GTG TCG ATG-3′ |
| Reverse | 5′-CTG GAA CCA GGA GTC CTA TCC-3′ | |
| mDR6 | Forward | 5′-ATG GGG ACC CGG GCA AGC AGC ATCA-3′ |
| Reverse | 5′-TAA CTG TGC CAG AGG AAG GTG GGT T-3′ | |
| mA1 | Forward | 5′-AAT TCC AAC AGC CTC CAG ATA TG-3′ |
| Reverse | 5′-GAA ACA AAA TAT CTG CAA CTC TGG-3′ | |
| mMcl-1 | Forward | 5′-AGA AAT GTG CTG CTG GCT TT-3′ |
| Reverse | 5′-GCA GCT TCA AGT CCA CCT TC-3′ | |
| mDAPK | Forward | 5′-GCA GGA AAA CGT GGA CGA CT-3′ |
| Reverse | 5′-TTG GAT CCT TGA CCA GCA GT-3′ | |
| mTIEG1 | Forward | 5′-GTC TCA GTG CTC CCG TCT GT-3′ |
| Reverse | 5′-CCA CCG CTT CAA AGT CAC TC-3′ | |
| mGADD45b | Forward | 5′-CTT CTG GTC GCA CGG GAA GG-3′ |
| Reverse | 5′-GCT CCA CCG CGG CAG TCA CC-3′ | |
| mSHIP | Forward | 5′-CCT CCA ACC CCT CCC TCC CAA CCA-3′ |
| Reverse | 5′-AAC GCC GGC GGC ATG GCA GTC CTG CCA A-3′ | |
| mcIAP-1 | Forward | 5′-GGA CAT TAG GAG TCT TCC C-3′ |
| Reverse | 5′-GCT GGG AGT GAC AGT GAA C-3′ | |
| mcIAP-2 | Forward | 5′-ATG GTT CAA GAC AGC GCC TTT CTA G-3′ |
| Reverse | 5′-TCC CAT CGC ACG CAA AGC AGG CCA C-3′ | |
| mXIAP | Forward | 5′-GCG GGC GAG GGC GTG GGG CCG GGG C-3′ |
| Reverse | 5′-CTA GGA GGC CCT GGC TGG AAA AGC T-3′ |
Quantitative RT-PCR was performed with cDNA from SNU16 cells using real-time TaqMan technology and an ABI Prism 7000 (Applied Biosystems) instrument to detect expression of all Bim splicing variants (identifier, Hs00708019_s1) and BimL+BimEL (Assays-on-Demand gene expression product; Applied Biosystems; Hs00197982_m1). The relative expression of each was normalized to that of the glyceraldehyde 3-phosphate dehydrogenase gene (GAPDH; Assays-on-Demand gene expression product; Applied Biosystems; Hs99999905_m1) in the same cDNA according to the method described by the manufacturer (Applied Biosystems).
Western blotting for detection of Bim, FasL, and β-actin expression in SNU16 cells and tumors formed by SNU16 cells in nude mice was performed using anti-Bim (Santa Cruz sc-11425 and BD Bioscience 559685 for all three isoforms and EL species, respectively), anti-FasL (sc-834; Santa Cruz), and anti-β-actin (AC-15; Sigma) antibodies, respectively.
Reporter assay.
A segment 3.3 kb upstream from the start site of Bim transcription, corresponding to the promoter region of human Bim, was amplified from human genomic DNA by PCR using the primers 5′-CGACCTTTAAGGTACCACACACCTACCTCCACGCACCAA-3′and 5′-AGCGGCGCTGCTCGAGGAGCTCCAACAAACTGCAGACC-3′. The amplified DNA fragment was cloned into pGL3-Basic (E1751; Promega) between the KpnI and XhoI sites. Three RUNX binding sites (sites A, B, and C) and a Smad binding site (site D) were mutated using a QuikChange XL site-directed mutagenesis kit (Stratagene) with primers as follows: 5′-GTGTGGCGGGAAGTGTCCTGGCCCGCCAGCAG-3′and 5′-CTGCTGGCGGGCCAGGACACTTCCCGCCACAC-3′for site A, 5′-CTCACATTCCCAGTGATTTAGAAAAACTGTCCTGCCGAGTGAAAG-3′and 5′-CTTTCACTCGGCAGGACAGTTTTTCTAAATCACTGGGAATGTGAG-3′for site B, 5′-CGTCGGCAAAGCCTGTCCTCCCGAACAAGGGCC-3′and 5′-GGCCCTTGTTCGGGAGGACAGGCTTTGCCGACG-3′for site C, and 5′-GTGCCGCCAAAGGTCTCCTGCTGTTAGCGGTG-3′and 5′-CACCGCTAACAGCAGGAGACCTTTGGCGGCAC-3′for site D.
SNU16 cells were transfected with reporter plasmids and pRL-TK (Promega) using Lipofectamine 2000 and Plus reagent (Invitrogen). A total of 10 ng/ml of TGF-β was added to the culture medium 36 h after transfection. Luciferase activity was measured using a Dual-Luciferase Reporter Assay System (Promega) and normalized to the luciferase activity expressed by the pRL-TK vector.
Xenografts in nude mice.
SNU16 cells (6 × 106 cells of each line) were inoculated subcutaneously into the flank of nude mice subcutaneously. Tumors were dissected 60 days after inoculation. Half of each tumor was used to extract proteins for Western blotting, and the other half was fixed and used for immunohistochemistry and terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL).
TUNEL.
Apoptotic cells were detected with a MEBSTAIN Apoptosis Kit Direct (8445; MBL). Briefly, stomachs of newborn mice and tumors formed by SNU16 cells were fixed with 4% paraformaldehyde, embedded in paraffin, and sectioned at 5 μm. Deparaffinized sections were treated with proteinase K for 30 min at 37°C. SNU16 cells and SNU719 cells were attached on glass slides via Cytospin and fixed with 4% paraformaldehyde. The 3′-OH DNA ends generated by DNA fragmentation were then nick end-labeled with fluorescein isothiocyanate-dUTP, and the nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole). Apoptotic cells were observed under a fluorescent microscope (Olympus), and the ratio of the cell death was determined by making comparisons between the TUNEL signal and the number of nuclei.
Immunocytochemistry.
SNU16 cells deposited onto glass slides by Cytospin and fixed with 4% paraformaldehyde were treated with a serum-free blocking solution (X0909; DAKO) and incubated at 4°C overnight with 1 μg/ml R3-6E9 (23) in diluent (S3022; DAKO). Biotinylated anti-mouse immunoglobulin G (IgG) (BA-9200; VECTOR) and fluorescein avidin D (A-2001; VECTOR) were used for immunofluorescence imaging.
Immunohistochemistry.
For immunohistological analysis, tissues were fixed with 4% paraformaldehyde, embedded in paraffin, and sectioned at 5 μm. Antibodies to Bim (0.4 μg/ml; Santa Cruz sc-11425), Runx3 (R3-1E10, a newly established monoclonal antibody against RUNX3 in our laboratory; 1 μg/ml), TGF-β1 (10 μg/ml; Santa Cruz sc-146), TGF-β receptor type I (RI) (2 μg/ml; Santa Cruz sc-398) and TGF-β RII (4 μg/ml; Santa Cruz sc-400) were used for the immunodetection of antigens on rehydrated sections. An Envision+ system (horseradish peroxidase-diaminobenzidine; DAKO) was used for visualization.
Northern blotting.
Tissues were frozen in liquid nitrogen and treated with TRIzol Reagent (15596; Gibco BRL), and RNAs were extracted. Poly(A)+ RNAs were prepared by using the Oligotex-dT30 Super Kit (48991; Roche). A 1.5-μg aliquot of poly(A)+ RNA was denatured, subjected to electrophoresis on a 1.3% morpholinepropanesulfonic acid-formaldehyde agarose gel, and transferred to a Hybond-XL membrane (RPN2020S; Amersham Pharmacia Biotech). Probes specific for TGFβ1, TGFRI, TGFRII, and G3PDH (internal control) were labeled with a BcaBEST labeling kit (6046; TAKARA). The probes consisted of the following: for TGFβ1, bp 601 1585 of rat TGFβ1 cDNA (38); for G3PDH, human cDNA probe which has been shown to cross-react with the mouse gene (9805-1; Clontech); and TGFRI and TGFRII (12).Hybridization of these probes to their genes was performed with PerfectHyb (HYB-101; TOYOBO).
ChIP.
Chromatin immunoprecipitation (ChIP) was performed using a chromatin immunoprecipitation assay kit (Upstate). Anti-Flag antibody (M2; Sigma) for immunoprecipitation of Flag-tagged RUNX3 protein and mouse normal IgG as a control were added to the extract of SNU16 cells expressing Flag-tagged RUNX3. The following primers were used to amplify by PCR the DNA fragment comprising each RUNX binding site: 5′-GGAGGGTGATCTCGACCTTTAACA-3′and 5′-TTTGCCACCAAAATCCCCGAGAGT-3′for site A, 5′-TGACTGCAACCTCTCCCAACTTCA-3′and 5′-CTCGCGGAAAGAGTGGAGCTTTTT-3′for site B, and 5′-TTAAGCGATGTGGAAGTCGTTCCC-3′and 5′-TCCCTTTCTGTCTCTGCCAAGAGA-3′for site C. Primers for amplification of the GAPDH gene are shown in Table 1.
Electrophoresis mobility shift assay (EMSA).
3′ Biotinylated oligomers were annealed with nonlabeled complementary oligomers after denaturation at 85°C for 5 min. The following pairs of oligonucleotides were used as probes: SiteA, 5′-CGGGAAGTGTGGTGGCCCGC-3′ and 5′-GCGGGCCACCACACTTCCCG-3′; SiteB, 5′-GAAAAACTGTGGTGCCGAGT-3′and 5′-ACTCGGCACCACAGTTTTTC-3′; and SiteC, 5′-CAAAGCCTGTGGTCCCGAAC-3′ and 5′-GTTCGGGACCACAGGCTTTG-3′. The mSiteA, mSiteB, and mSiteC oligonucleotides were identical to SiteA, SiteB, and SiteC except for point mutations in RUNX binding sites (see Fig. 3). The binding reactions were performed in 20 mM HEPES, pH 7.6, 50 mM KCl, 5 mM dithiothreitol, 1 mM EDTA, 4% Ficoll, 0.2 mg/ml bovine serum albumin, 0.1 mg/ml poly(dI-dC), and 50 μg/ml monoclonal antibody as required. Purified FLAG-tagged RUNX3 (1 ng) from COS7 cells expressing the gene exogenously and 10 ng of six-histidine-tagged PEBP2β protein purified from bacterial extracts containing the exogenous protein were added to 10-μl binding reactions. Last, a total of 10 femptomoles of biotinylated probe was added, and the samples were incubated at 4°C for 30 min with or without anti-RUNX3 monoclonal antibody, R3-5G4, or anti-hemagglutinin (HA) monoclonal antibody (12CA5; Roche,) and resolved in a 5% polyacrylamide gel in 0.5× Tris-borate-EDTA buffer.
FIG. 3.
Up-regulation of Bim by RUNX3 is responsible for TGF-β-induced apoptosis of SNU16 cells. (A) Subcellular localization of endogenous RUNX3 in control SNU16 cells (control), SNU16 cells expressing antisense DNA against RUNX3 (RUNX3-AS), and SNU16 cells expressing RUNX3(1-187) [RUNX3(1-187)] was revealed by immunocytochemistry using R3-6E9 over a period of 16 h after treatment with 80 pM TGF-β. Nuclei were visualized by staining with DAPI in controls. (B) Western blots of Bim expression in control, RUNX3(1-187), and RUNX3-AS over a period of 16 h after treatment with 80 pM TGF-β. (C) SNU16 control cells (control) and SNU16 cells expressing shRNA against Bim (shRNA Bim) were cultured with 400 pM TGF-β, and cell numbers were counted using trypan blue at the indicated times. The data shown here represent the mean ± standard deviation (n = 3). (D) Western blot of Bim expression in SNU16 control cells (control) and SNU16 cells expressing shRNA against Bim (shRNA Bim) in the absence (−) and presence (+) (16 h) of 400 pM TGF-β. (E) SNU16 control cells (control) and SNU16 cells expressing shRNA against Bim (shRNA Bim) were cultured with 400 pM TGF-β, and then apoptotic cells were detected by TUNEL and counted. The data shown here represent the mean ± standard deviation (n = 3).
RESULTS
SNU16 and SNU719 cells show RUNX3-dependent TGF-β-induced apoptosis.
Almost all gastric cancer-derived cell lines are resistant to TGF-β, and the TGF-β-induced signal transduction pathway is frequently impaired in many cases of gastric cancer. The cell line SNU16 is exceptional among gastric cancer-derived cell lines in that the cells respond to TGF-β and express endogenous nonmutated Smad2, Smad3, Smad4, TGF-β RII (17), and RUNX3 (33). Therefore, SNU16 is useful for studying the role of RUNX3 in the TGF-β signaling pathway.
In earlier studies, we found that RUNX3 is localized primarily in the cytoplasm in SNU16 cells. TGF-β in SNU16 cells induces nuclear translocation of RUNX3, which can then function as a transcription factor. On the other hand, RUNX3(1-187), the C-terminally truncated form of RUNX3 lacking the transactivation domain (16, 19), is retained in the cytoplasm regardless of the presence or absence of TGF-β in these cells (23). In the presence of RUNX3(1-187), endogenous RUNX3 is also retained in the cytoplasm (23). Therefore, RUNX3(1-187) functions as a dominant negative form of RUNX3 in this system, although the mechanism is presently unclear.
When SNU16 cells were exposed to TGF-β, cell number started to fall around 16 h posttreatment (Fig. 1A). A general caspase inhibitor, z-VAD, inhibited cell loss (Fig. 1D), and the population of TUNEL-positive cells (Fig. 1A and B) or annexin V-positive cells (data not shown) increased, thus showing that the reduction in cell number was largely due to apoptosis. The timing of apoptosis induction coincided well with that of the nuclear translocation of RUNX3 by TGF-β (Fig. 1A and B; see also Fig. 3A). On the other hand, either stable clones of SNU16 cells expressing exogenous RUNX3(1-187) or those expressing antisense DNA against RUNX3 (RUNX3-AS) showed significant resistance to TGF-β, resulting in a reduction in the TUNEL-positive population (Fig. 1A and B). Thus, these data suggest that apoptosis induced by TGF-β treatment in SNU16 cells may be mediated by RUNX3. Another gastric cancer cell line, SNU719, was found to express endogenous RUNX3. TGF-β also induced apoptosis in SNU719 probably mediated by RUNX3 (Fig. 1C).
FIG. 1.
SNU16 cells expressing RUNX3(1-187) polypeptide or antisense DNA against RUNX3 and SNU719 cells expressing antisense DNA against RUNX3 are resistant to apoptosis-inducing action of TGF-β. (A) SNU16 control clone and SNU16 clones expressing RUNX3(1-187) were cultured with 400 pM TGF-β, and cell numbers were counted using trypan blue at the indicated times. The data shown here represent the means ± standard deviations (n = 3). A Western blot shows exogenous expression of RUNX3(1-187) in each clone (top). Apoptotic cells were detected by TUNEL and counted. The data shown here represent the means ± standard deviations (n = 3) (bottom). (B) SNU16 control clones and SNU16 clones expressing antisense DNA against RUNX3 were cultured with 400 pM TGF-β, and the cell numbers were counted using trypan blue at the indicated times. The data shown here represent the means ± standard deviations (n = 3). A Western blot shows endogenous expression of RUNX3 in each clone (top). Apoptotic cells were detected by TUNEL and counted. The data shown here represent the means ± standard deviations (n = 3). (C) SNU719 control clones and SNU719 clones expressing antisense DNA against RUNX3 were cultured with 400 pM TGF-β, and cell numbers were counted using trypan blue at the indicated times. The data shown here represent the means ± standard deviations (n = 3). A Western blot shows endogenous expression of RUNX3 in each clone (top). Apoptotic cells were detected by TUNEL and counted. The data shown here represent the means ± standard deviations (n = 3). (D) SNU16 control clones were cultured with 400 pM TGF-β with or without the caspase inhibitor z-VAD, and viable cell numbers were counted using trypan blue at the indicated times.
RUNX3-dependent induction of a proapoptotic gene, Bim, during TGF-β-induced apoptosis.
Next, we investigated which apoptosis-related genes are regulated by RUNX3 during TGF-β-induced apoptosis. We examined the effect of TGF-β treatment in the following three populations: (i) control SNU16 cells (control), (ii) those stably expressing exogenous RUNX3(1-187) [RUNX3(1-187)], and (iii) those expressing exogenous Flag-tagged full-length RUNX3 (RUNX3). The cells were treated with TGF-β, and expression profiles of proapoptotic and antiapoptotic genes were compared by semiquantitative RT-PCR as shown in Fig. 2A. The expression patterns of each gene were compared in the three populations 48 h after the addition of TGF-β. We selected Bcl-2 family genes, IAP family genes, and death receptor genes and their ligands as well as TIEG1, GADD45b, DAPK, DAXX, ARTS, and SHIP which are thought to be involved in TGF-β-mediated apoptosis (see above). Among them, Bim was found to be the only one whose expression was induced by TGF-β treatment in the control cells and further induced in cells expressing exogenous full-length RUNX3, but the expression was repressed in cells expressing RUNX3(1-187) (Fig. 2A). TGF-β induced Bcl-w and A1 expression in SNU16 cells, but the pattern was not affected in the presence of RUNX3(1-187), suggesting that these two genes are not involved in the RUNX3-dependent TGF-β-induced apoptosis in SNU16 cells. Bim is a member of the Bcl-2 homology domain 3-only protein family whose members induce apoptosis by neutralizing the activity of antiapoptotic members of the Bcl-2 family (21). Three bands of Bim RNA represent three major splice variants, BimEL, BimL, and BimS (3). The expression levels of Bmf, the tumor necrosis factor α gene, ARTS, and SHIP were below the detectable level throughout the experiments (Fig. 2A), but Fas was detected consistently (data not shown) in SNU16 cells. Likewise, the RUNX3-dependent induction of Bim by TGF-β was observed in SNU719 cells, where two splice variants, BimEL and BimL were clearly detected by RT-PCR (Fig. 2B). Up-regulation of Bim by TGF-β was confirmed by Western blotting using the Bim antibody recognizing only EL species (Fig. 2C) and that recognizing all three isoforms (Fig. 3B and D), as well as by quantitative RT-PCR using two different primer sets to detect all splicing variants of either Bim or BimL+BimEL (Fig. 2D). The timing of Bim induction by TGF-β in the control cells coincided well with that of RUNX3 nuclear translocation (Fig. 3A and B). In SNU16 cells in which Bim expression was knocked down by an shRNA, TGF-β-induced apoptosis was greatly reduced, indicating that Bim is responsible for the TGF-β-induced apoptosis in SNU16 cells (Fig. 3C to E).
FIG. 2.
Induction of Bim by TGF-β in SNU16 cells depends on RUNX3 function. (A) Expression profile of proapoptotic and antiapoptotic genes in SNU16 control clones (control), SNU16 clones expressing RUNX3(1-187) [RUNX3(1-187)], and SNU16 clones expressing exogenous RUNX3 in addition to endogenous RUNX3 (RUNX3) revealed by RT-PCR. (B) Expression of Bim in SNU719 control clones (control) and SNU719 clones expressing antisense DNA against RUNX3 (RUNX3-AS) revealed by RT-PCR. (C) Expression of Bim in the absence or presence of TGF-β in control, RUNX3(1-187), and RUNX3 revealed by Western blotting. (D) Comparison of Bim mRNA induction by TGF-β between control and RUNX3(1-187) by quantitative RT-PCR using two primer sets for detection of all splicing variants of Bim or BimL+BimEL.
Using RUNX3-negative gastric cancer cell lines, MNK28 and AGS (33), we observed that TGF-β treatment of the cells did not increase apoptosis (data not shown). However, when inducible RUNX3 in AGS cells is activated, Bim expression was induced (52). Similarly, Wei et al. recently described that exogenous expression of RUNX3 in AGS cells induces apoptosis (47).
These results suggest that Bim is a positive target of RUNX3 and that up-regulated Bim induces apoptosis attenuating by the antiapoptotic Bcl-2 family in SNU16 cells.
Transcriptional stimulation of Bim requires RUNX proteins and Smads.
In a previous study, we found that RUNX proteins form complexes with Smads to stimulate transcription of target genes in a cooperative manner (19, 54). To examine whether RUNX3 directly regulates Bim expression, 3.3 kb of the human Bim promoter region located upstream of the transcription start site was cloned and mutated. This region contains three putative RUNX binding sites (sites A to C) and one Smad binding site (site D) (Fig. 4A). We mutated these sites one by one and evaluated the effects on promoter activity in luciferase assays. From 8 to 24 h after addition of TGF-β, the promoter activity increased in controls but not in RUNX3(1-187) or RUNX3-AS (Fig. 4B). The luciferase activities from each mutated construct suggested that RUNX sites A and B and a Smad site, D, are responsible for the activation of the Bim promoter (Fig. 4C). We found that murine Bim promoter (3) also has Runx and Smad sites (corresponding to the sites B and D in Fig. 4A, respectively) within the conserved region of the Bim promoter, although the Runx site in murine sequence is 62 nucleotides away from the position in the human sequence.
FIG. 4.
Dependency of human bim promoter activation by TGF-β on RUNX and Smad sites. (A) Human bim promoter upstream of the start site of transcription containing three RUNX sites and a Smad site. These sites were mutated as indicated by underlining for the experiment shown in panel C. (B) The promoter activity in control SNU16 cells (control), SNU16 cells expressing RUNX3(1-187) [RUNX3(1-187)], and SNU16 cells expressing antisense DNA against RUNX3 (RUNX3-AS) was measured at the indicated times up to 24 h after addition of 400 pM TGF-β. The data shown here represent the means ± standard deviation (n = 3). (C) The promoter activity of wild type and each mutant as indicated in control SNU16 cells was measured in the absence or presence of 400 pM TGF-β. The measurement was done 24 h after the TGF-β treatment. The data shown here represent the means ± standard deviations (n = 3). (D) ChIP assay showing direct binding of RUNX3 to RUNX sites (sites A to C) of the human bim promoter in SNU16 cells induced by TGF-β. PCR was performed for the indicated number of cycles (×32, ×35, and ×37) to detect expression in a semiquantitative fashion. Twenty percent of total chromatin DNA as templates, the GAPDH gene, and normal mouse IgG were used as controls for ChIP. ChIP was performed using three independent cell extracts, and essentially the same results were obtained in all cases. (E) Direct binding of RUNX3 on RUNX sites (sites A to C) of the human bim promoter revealed by EMSA. Shifted bands were observed on sites A to C but not on mutated sites A to C (mSiteA to mSiteC), and a band was supershifted by anti-RUNX3 antibody, R3-5G4 (αRUNX3), but not by anti-HA antibody (αHA).
Binding of RUNX3 to these putative binding sites was confirmed by the ChIP assay and EMSA. As shown in Fig. 4D, RUNX3 bound to each of the three sites, A, B, and C, in vivo in a TGF-β-dependent manner. Sites B and C were found to be the preferred sites, although the significance of this observation, if any, is currently unclear. On the other hand, in the in vitro EMSAs, site C seemed to be the preferred site, although RUNX3 also bound well to sites A and B (Fig. 4E). The appearance of a supershifted band after the addition of a RUNX3-specific monoclonal antibody to the EMSAs confirmed that RUNX3 protein was present in the DNA-protein complexes (Fig. 4E). Since naked DNA was used in the EMSA, whereas chromatin DNA was used in the ChIP assay, the results obtained from these two assays would not necessarily be identical, depending on the chromatin structure of each site.
Inhibition of the nuclear translocation of RUNX3 increases the tumorigenicity of SNU16 cells.
The exogenous expression of RUNX3 in MKN28, a gastric cancer-derived cell line lacking endogenous RUNX3, resulted in reduced tumorigenicity compared to the parental cell line in nude mice (33). However, the mechanism of this tumor-suppressive effect was unclear. Since SNU16 cells are also tumorigenic in nude mice, we compared the tumorigenic potential of SNU16 cells (control) with SNU16 cells expressing RUNX3(1-187), in which RUNX3-dependent TGF-β-mediated apoptosis is suppressed. Tumors formed by SNU16 cells expressing antisense DNA against RUNX3 (data not shown) or by RUNX3(1-187) were significantly larger than those formed by control cells (Fig. 5A). In tumors formed by SNU16 cells expressing RUNX3(1-187), Bim was expressed at lower levels than in the control (Fig. 5A). Using immunohistochemistry, Bim protein expression in the tumors formed by SNU16 cells expressing RUNX3(1-187) was found to be reduced compared to that in control cells (Fig. 5B). Consistent with these observations, apoptosis was greatly reduced in RUNX3(1-187) tumor cells (Fig. 5B). Furthermore, SNU16 cells expressing shRNA against Bim increased the tumorigenicity significantly (Fig. 5C).
FIG. 5.
Enhancement of tumorigenicity of SNU16 cells by repressing the expression of Bim. (A) Expression of exogenous RUNX3(1-187), Bim, and FasL as revealed by Western blotting in tumors formed in nude mice by control SNU16 clones (control) or SNU16 clones expressing RUNX3(1-187) [RUNX3(1-187)] (top). Note that expression of FasL was not changed between the control and RUNX3(1-187). The weight of each tumor is shown (right graph) and a plot of all tumor weights (left graph). Averages are indicated by bars (P < 0.01). Two independently cloned control or RUNX3(1-187) clones were used. One clone was injected into the flank of one side of 5 mice and the other clone was injected into the other 5 mice. (B) Immunodetection of Bim and detection of apoptotic cells by TUNEL in tumors formed by control or RUNX3(1-187) clones. The enlargements are shown in the insets. Scale bars are equal to 150 μm or 20 μm (inset). (C) The weight of each tumor formed by SNU16 control cells (control) and by SNU16 cells expressing shRNA against Bim (shBim) in 5 nude mice is shown. Each pair was from the same mouse (P < 0.05 by paired t test).
Taken together, these results show that Bim is up-regulated by RUNX3 to induce apoptosis, most likely in response to TGF-β in vivo, and blocking Bim induction increased the tumorigenicity of SNU16 cells.
Reduced expression of Bim and apoptosis in Runx3−/− gastric epithelial cells.
The observations described above were made using a human gastric cancer-derived cell line, SNU16. To verify that Bim is a positive target of RUNX3 in TGF-β-induced apoptosis in vivo, we examined expression of endogenous Bim protein in neonate stomach tissues from wild-type (WT) and Runx3−/− mice. We reported earlier that WT, but not Runx3−/− stomach epithelial cells of neonate mice could undergo apoptosis that was suggested to be TGF-β dependent (33). Consistent with our previous observations, Bim was expressed in the stomach epithelium of WT but not in that of Runx3−/− mice, and it was the only gene whose expression was uniformly absent in all three Runx3−/− mice (Fig. 6A). We also examined the expression of 23 other apoptosis-related genes. None of them showed specific changes of expression in Runx3−/− mice (Fig. 6A).
FIG. 6.
Expression of apoptosis-related genes in gastric epithelial cells of WT and Runx3−/− neonate mice. (A) Detection of mRNA by semiquantitative RT-PCR. One WT and three independent Runx3−/− stomachs were used. (B) Immunodetection of Bim protein. Counter-staining was done with hematoxylin. (C) Apoptotic cells in WT, Bim−/−, Runx3−/−, or FasL−/− stomachs as revealed by TUNEL. Note that apoptotic cells were detected in the FasL−/− stomach to the same extent as in WT. Scale bar, 200 μm. (D) Induction of Bim by TGF-β in Runx3+/+ but not in Runx3−/− GIF cells in the medium containing 0.5% (low) serum as revealed by RT-PCR (top). Sensitivity to TGF-β of Runx3+/+ and Runx3−/− GIF cells in the medium containing 0.5% (low) serum (bottom). The growth of cells in the presence of TGF-β (10 ng/ml) for 48 h was normalized to growth in its absence (P < 0.05).
Consistent with the results described in Fig. 6A, immunohistochemical analysis revealed that the expression of Bim was undetectable in Runx3−/− gastric mucosa and resembled that seen in Bim−/− mice (Fig. 6B). Concurrent with this observation, a significant level of apoptosis was observed in WT neonate gastric epithelial cells facing the lumen but not in Bim−/− cells. This result suggests that, out of numerous proapoptotic and antiapoptotic genes, the apoptosis observed in this tissue under the conditions used is primarily mediated by Bim. We previously observed that this apoptosis was also greatly reduced in Runx3−/− stomach cells (33) (Fig. 6C). For further confirmation, we examined Bim induction during TGF-β-induced apoptosis in mouse gastric epithelial (GIF) cell lines isolated from Runx3+/+ or Runx3−/− mice in the p53-null background (14, 33). In the presence of 0.5% (low) serum, TGF-β induced Bim expression with concomitant induction of apoptosis in Runx3+/+ but not in Runx3−/− GIF cells (Fig. 6D). Thus, the results suggest that Runx3-mediated apoptosis as observed here is primarily executed through the Bim pathway.
Immunohistochemical analysis using the monoclonal antibody against Runx3, R3-1E10, detected endogenous Runx3 in almost all cells from the neonate stomach, including mesenchymal and epithelial cells (Fig. 7A). The expression levels of TGF-β signaling pathway components, including the TGF-β1 ligand and TGF-β RI/II, were indistinguishable between WT and Runx3−/− cells. These proteins were detected only in the cells facing the lumen, which also showed apoptosis in WT (Fig. 7B and C). These in vivo results show that lack of Runx3 does not alter the expression of TGF-β signaling pathway components but greatly affects Bim expression. Furthermore, a high level of apoptosis occurs only in cells that express Runx3, TGF-β1, and TGF-β RI/II, suggesting that Bim is a downstream target of Runx3 which, in turn, functions downstream of TGF-β to induce apoptosis. Finally, we examined Bim expression in TGF-β1−/− stomach. TGF-β1−/− mice of BALB/c but not C57BL/6J strain can survive at the Mendelian ratio till embryonic day 18.5 when TGF-β-induced apoptosis was observed in the gastric epithelial cells (33). Bim expression was greatly reduced in TGF-β1−/− gastric epithelial cells, suggesting that Bim is a downstream target of TGF-β1 in vivo (Fig. 7D).
FIG. 7.
Expression of Runx3, TGF-β1, and TGF-β RI/II in gastric epithelial cells of neonate mice and lack of Bim expression in TGF-β1−/− embryonic day 18.5 stomach. (A) Expression of Runx3 in WT and Runx3−/− stomachs revealed by immunostaining with R3-1E10. Immunostaining with normal mouse IgG as a primary antibody in a WT stomach is shown as a negative control. (B) Expression of TGF-β1, TGFβRI, and TGFβRII in WT and Runx3−/− stomachs as revealed by Northern blotting. (C) Expression of TGF-β1 and TGF-β RI/II in WT and Runx3−/− stomachs. Immunostaining with normal mouse IgG as a primary antibody is shown as a negative control (control). (D) Expression of Bim in WT and TGF-β1−/− stomachs. Immunostaining with normal mouse IgG as a primary antibody in a WT stomach is shown as a negative control (IgG). Counter-staining was done with hematoxylin. Scale bar, 200 μm.
Bim−/− mice are born normally, but it was reported that the number of lymphocytes, macrophages, and granulocytes in Bim−/− mice is larger than that in normal mice and that these cells show resistance to apoptosis by cytokine withdrawal. Additionally, aged Bim−/− mice on a mixed genetic background display autoimmune glomerulonephritis (2). Our observation that Bim−/− mice have reduced gastric epithelial apoptosis is new. Therefore, these mice may provide an attractive in vivo model to study the involvement of TGF-β-induced apoptosis in gastric carcinogenesis, particularly with respect to the involvement of Runx3 as a tumor suppressor (see Discussion).
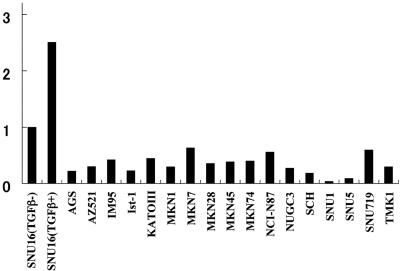
Consistent with this notion, all gastric cancer cell lines we examined showed lower levels of Bim expression than that of SNU16 (considered to be a basal level of expression), which is the only cell line responsive to TGF-β (Fig. 8). We described above that RUNX3(1-187) and antisense RUNX3 inhibited TGF-β-induced expression of Bim but not the basal level of the expression (Fig. 2C). Our recent report may increase our understanding of the reasons for the different basal levels. We reported recently that in more than 80% of gastric cancer cases, RUNX3 is inactivated by promoter methylation and protein mislocalization (23). In fact, we have preliminary evidence that most of the cell lines shown in Fig. 8 do not have functional RUNX3. On the other hand, in cells expressing RUNX3(1-187) and antisense RUNX3, RUNX3 is only partially inactivated. The difference in the residual level of RUNX3 probably explains why, in the cancer cells shown in Fig. 8, the level of Bim is lower than the basal level, whereas in transient experiments, the basal level is not affected by RUNX3(1-187) and antisense RUNX3. We are currently investigating how frequently Bim levels are attenuated in gastric cancer.
FIG. 8.
Expression of Bim mRNA in gastric cancer cell lines revealed by quantitative RT-PCR using a primer set for detection of all Bim splicing variants (see Materials and Methods). All relative expression was normalized to that of non-TGF-β-treated SNU16 cells (TGFβ-). SNU16 (TGFβ+) refers to expression in SNU16 cells treated with TGF-β for 24 h as shown in Fig. 2C.
Taken together, our results indicate that the Bim pathway is primarily responsible for the TGF-β-induced apoptosis mediated by RUNX3 in vivo and in vitro. Therefore, at least a part of the tumor suppressor activity associated with RUNX3 in gastric epithelial cells is due to its ability to regulate apoptosis through a proapoptotic gene, Bim.
DISCUSSION
In the present study, we present evidence suggesting that a gastric tumor suppressor, RUNX3, mediates apoptosis induced by TGF-β by up-regulating transcription of Bim in a gastric cancer-derived cell line, SNU16. This observation was validated by in vivo studies using Runx3-null, Bim-null, and TGF-β1-null mice, suggesting that the phenomenon is not unique to SNU16 cells but is more generally applicable to the gastric epithelium. Validation of the results carried out by using gastric cancer cell lines was put forward by the following evidence. We reported earlier that primary gastric epithelial cells isolated from a Runx3+/+ mouse responded to TGF-β stimulation to induce apoptosis in vitro as revealed by TUNEL. This apoptosis was greatly reduced in Runx3−/− gastric epithelial cells (33), suggesting that Runx3-mediated apoptosis is induced by TGF-β in vivo. This is the closest to in vivo experiments that can be carried out technically. Here in this report, we showed the reduction of Bim expression in TGF-β1−/− gastric epithelial cells compared with TGF-β1+/+ cells in vivo. The data suggest that the proapoptotic gene involved in TGF-β-induced, Runx3-mediated apoptosis of gastric cells is Bim. Additionally, in GIF, a cell line isolated from gastric epithelial cells of mouse embryo of p53−/− background (14, 33), TGF-β was shown to induce Bim in Runx3+/+ but only at very low level in Runx3−/− GIF cells in this study. Furthermore, cell death induced by TGF-β was greatly reduced in Runx3−/− cells compared to that of WT cells. These results altogether strongly support the conclusion obtained by gastric cancer cell lines that there is a linear cascade of TGF-β, Runx3, Bim, and apoptosis in gastric epithelial cells.
In the tumors formed in nude mice by SNU16 cells harboring RUNX3(1-187), expression of Bim is greatly reduced compared with those induced by control SNU16 cells (Fig. 5). This reduction does not seem to be due to inhibition of basal level of Bim expression by RUNX3(1-187) but, rather, due to inhibition of Bim induction by TGF-β in vivo. Nuclear translocation of RUNX3 in SNU16 cells was shown to be induced by TGF-β, and the tumor cells harboring RUNX3(1-187) largely had endogenous RUNX3 in the cytoplasm, whereas the control SNU16 cells growing as tumors displayed a significant percentage of RUNX3 in the nucleus (23), suggesting that nuclear translocation triggered by TGF-β in vivo must have induced Bim and that this induction is blocked by RUNX3(1-187). However, some other mechanisms cannot be ruled out.
Transcriptional control of Bim appears to be complex, since different groups have reported different modes of regulation. Growth factor withdrawal causes increased Bim expression in various neuronal (37, 48) and hematopoietic cell types (9, 40), and in neurons, this expression requires either Jun N-terminal protein kinase activation (20, 37, 48) or phosphatidylinositol 3′-kinase inhibition (15). On the other hand, interleukin-3 stimulation represses Bim expression through activation of the MAP kinase pathway and/or the phosphatidylinositol 3′-kinase pathway in hematopoietic cell lines (40). Furthermore, a forkhead transcription factor, FKHR-L1 (FOXO3a), up-regulates Bim in hematopoietic cells (9), neurons (15), and paclitaxel-treated breast cancer cells (42). More recently, Smad3 was reported to induce Bim in TGF-β2-mediated apoptosis in WEHI 231 cells, a B lymphocyte cell line (49). However, these observations were made only by in vitro experiments using either chemical inhibitors or overexpression of signaling molecules. For a better understanding of the role of these pathways in the control of Bim expression, more rigorous studies using in vivo systems are needed. The data presented in this communication were primarily obtained by using cell line SNU16 but were supported by in vivo studies using mice lacking Runx3, Bim, and TGF-β1, suggesting that our observations are valid.
Recently, the involvement of Bim as a tumor suppressor in Myc-induced mouse B cell leukemia, mantle cell lymphoma, and epithelial solid tumors was reported (11, 44, 45). Experiments using Bim−/− mice have shown that Bim is essential for apoptosis of T lymphocytes, B lymphocytes, myeloid cells, and neurons (2, 37, 48). In this study, we have shown a lack of apoptosis in the gastric epithelia in neonate Bim−/− mice; however, apoptosis in adult Bim−/− stomach remains to be investigated.
The observation that the inhibitory effect of RUNX3(1-187) on TGF-β-induced apoptosis was larger than that of antisense DNA against RUNX3 suggests the possibility that RUNX1 and RUNX2 also regulate apoptosis in SNU16 cells (Fig. 1A and C). It would be interesting to see whether RUNX1 and RUNX2 are involved in regulation of Bim in T lymphocytes, B lymphocytes, and myeloid cells, in which they are expressed, as well as in gastric epithelial cells. We reported previously that Runx3−/− dorsal root ganglion neurons do not extend their axons to peripheral muscle, suggesting that neurons fail to receive survival factor, neurotrophin 3. However, data showed the presence of neuronal soma, raising the question as to how neurons survived without neurotrophin 3 (22). Although the underlying mechanism remains unknown, Krieglstein et al. (31) reported that neurotrophin-depleted apoptosis is controlled by TGF-β (31). In dorsal root ganglion neurons whose death is modulated by Bim (37, 48), Runx3 may also regulate Bim expression.
It is known that tumor suppressors have roles in the regulation of apoptosis either as proapoptotic proteins or as up- or down-regulators of proapoptotic or antiapoptotic genes. For example, tumor suppressors, such as p53, p19ARF, ATM, Chk2, Rb, and PTEN, are known to regulate expression and/or function of proapoptotic or antiapoptotic genes and others, such as Bax, Bak, Apaf-1, CD-95/Fas, TRAIL-R1/R2, and caspase-8, are themselves proapoptotic proteins (26). Similarly, the TGF-β signaling pathway, which is known as a tumor suppressor pathway, induces apoptosis. However, the involvement of TGF-β-induced apoptosis in carcinogenesis is poorly understood. In this study, we showed that RUNX3, a downstream target of the TGF-β tumor suppressor pathway, mediates apoptosis through up-regulation of Bim, the Bcl-2 interacting mediator of cell death. In addition, we recently found that RUNX3 up-regulates p21WAF1/Cip1, an important factor in CDK inhibition and cell cycle control, and that it does this in collaboration with Smads downstream of TGF-β in gastric cancer (5). In this respect, it is noteworthy that RUNX3 has a critical role in apoptosis induction as well as in the regulation of cell growth arrest. These observations strengthen the significance of RUNX3 as a tumor suppressor in gastric carcinogenesis.
TGF-β-induced apoptosis has been observed in prostate epithelial cells and hepatic cells (39). Interestingly, the involvement of RUNX3 as a tumor suppressor in prostate cancer and hepatocellular carcinoma was reported (27, 30, 50). If RUNX3 regulates Bim expression in TGF-β-induced apoptosis in these cancers, as shown for gastric epithelial cells in this study, then the signaling cascade described in this paper, namely, that involving TGF-β, Smads, RUNX3, and Bim, would be an important carcinogenic process of endoderm-derived tissues.
Recently, we showed that RUNX3 is inactivated in more than 80% of gastric cancers through gene silencing and protein mislocalization (23). If inactivation of RUNX3 occurs at the early stages of carcinogenesis, the extremely high frequency of RUNX inactivation could be used as a molecular marker for the early diagnosis of gastric cancer. Since Bim is a positive target of RUNX3 in gastric epithelial cells, Bim might also be a good marker for this purpose.
Acknowledgments
We thank S. Yonehara for critical reading of the manuscript and for helpful discussion. We also thank Y. K. Loh, E. Tai, and the staff in the animal holding unit in IMCB for their technical help.
This work was supported by A*STAR (Agency for Science, Technology and Research), Singapore and Human Frontier Science Program, RGP0375/2001-M202.
REFERENCES
- 1.Bae, S. C., E. Takahashi, Y. W. Zhang, E. Ogawa, K. Shigesada, Y. Namba, M. Satake, and Y. Ito. 1995. Cloning, mapping and expression of PEBP2αC, a third gene encoding the mammalian Runt domain. Gene 159:245-248. [DOI] [PubMed] [Google Scholar]
- 2.Bouillet, P., D. Metcalf, D. C. Huang, D. M. Tarlinton, T. W. Kay, F. Köntgen, J. M. Adams, and A. Strasser. 1999. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 286:1735-1738. [DOI] [PubMed] [Google Scholar]
- 3.Bouillet, P., L. C. Zhang, D. C. S. Huang, G. C. Webb, C. D. K. Bottema, P. Shore, H. J. Eyre, G. R. Sutherland, and J. M. Adams. 2001. Gene structure, alternative splicing, and chromosomal localization of pro-apoptotic Bcl-2 relative Bim. Mamm. Genome 12:163-168. [DOI] [PubMed] [Google Scholar]
- 4.Bouillet, P., M. Robati, M. Bath, and A. Strasser. 2005. Polycystic kidney disease prevented by transgenic RNA interference. Cell Death Differ. 12:831-833. [DOI] [PubMed] [Google Scholar]
- 5.Chi, X. Z., J. O. Yang, K. Y. Lee, K. Ito, C. Sakakura, Q. L. Li, H. R. Kim, E. J. Cha, A. Kaneda, T. Ushijima, W. J. Kim, Y. Ito, and S. C. Bae. 2005. RUNX3 suppresses gastric epithelial cell growth by inducing p21WAP1/Cip1 expression in cooperation with TGF-β-activated SMAD. Mol. Cell. Biol. 25:8097-8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conery, A. R., Y. Cao, E. A. Thompson, C. M. Townsend, T. C. Ko, and K. Luo. 2004. Akt interacts directly with Smad3 to regulate the sensitivity to TGF-β-induced apoptosis. Nat. Cell Biol. 6:366-372. [DOI] [PubMed] [Google Scholar]
- 7.Crawford, S. E., V. Stellmach, J. E. Murphy-Ullrich, S. M. Ribeiro, J. Lawler, R. O. Hynes, G. P. Boivin, and N. Bouck. 1998. Thrombospondin-1 is a major activator of TGF-β1 in vivo. Cell 93:1159-1170. [DOI] [PubMed] [Google Scholar]
- 8.Derynck, R., R. J. Akhurst, and A. Balmain. 2001. TGF-β signaling in tumor suppression and cancer progression. Nat. Genet. 29:117-129. [DOI] [PubMed] [Google Scholar]
- 9.Dijkers, P. F., R. H. Medema, J. W. Lammers, L. Koenderman, and P. J. Coffer. 2000. Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr. Biol. 10:1201-1204. [DOI] [PubMed] [Google Scholar]
- 10.Edlund, S., S. Y. Lee, S. Grimsby, S. Zhang, P. Aspenström, C. H. Heldin, and M. Landström. 2005. Interaction between Smad7 and β-catenin: importance for transforming growth factor β-induced apoptosis. Mol. Cell. Biol. 25:1475-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egle, A., A. W. Harris, P. Bouillet, and S. Cory. 2004. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc. Natl. Acad. Sci. USA 101:6164-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franzén, P., P. tenDijke, H. Ichijo, H. Yamashita, P. Schulz, C. H. Heldin, and K. Miyazono. 1993. Cloning of a TGFβ type I receptor that forms a heteromeric complex with the TGFβ type II receptor. Cell 75:681-692. [DOI] [PubMed] [Google Scholar]
- 13.Fukamachi, H., and K. Ito. 2004. Growth regulation of gastric epithelial cells by Runx3. Oncogene 23:4330-4335. [DOI] [PubMed] [Google Scholar]
- 14.Fukamachi, H., K. Ito, and Y. Ito. 2004. Runx3−/− gastric epithelial cells differentiate into intestinal type cells. Biochem. Biophys. Res. Commun. 321:58-64. [DOI] [PubMed] [Google Scholar]
- 15.Gilley, J., P. J. Coffer, and J. Ham. 2003. FOXO transcription factors directly activate Bim gene expression and promote apoptosis in sympathetic neurons. J. Cell Biol. 162:613-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo, W. H., L. Q. Weng, K. Ito, L. F. Chen, H. Nakanishi, M. Tatematsu, and Y. Ito. 2002. Inhibition of growth of mouse gastric cancer cells by Runx3, a novel tumor suppressor. Oncogene 21:8351-8355. [DOI] [PubMed] [Google Scholar]
- 17.Han, S. U., H. T. Kim, D. H. Seong, Y. S. Kim, Y. S. Park, Y. J. Bang, H. K. Yang, and S. J. Kim. 2004. Loss of the Smad3 expression increases susceptibility to tumorigenicity in human gastric cancer. Oncogene 23:1333-1341. [DOI] [PubMed] [Google Scholar]
- 18.Hanahan, D., and R. A. Weinberg. 2000. The hallmarks of cancer. Cell 100:57-70. [DOI] [PubMed] [Google Scholar]
- 19.Hanai, J., L. F. Chen, T. Kanno, N. Ohtani-Fujita, W. Y. Kim, W. H. Guo, T. Imamura, Y. Ishidou, M. Fukuchi, M. J. Shi, J. Stavnezer, M. Kawabata, K. Miyazono, and Y. Ito. 1999. Interaction and functional cooperation of PEBP2/CBF with Smads. Synergistic induction of the immunoglobulin germline Cα promoter. J. Biol. Chem. 274:31577-31582. [DOI] [PubMed] [Google Scholar]
- 20.Harris, C. A., and M. J. Eugane. 2001. BH3-only Bcl-2 family members are coordinately regulated by the JNK pathway and require Bax to induce apoptosis in neurons. J. Biol. Chem. 276:37754-37760. [DOI] [PubMed] [Google Scholar]
- 21.Huang, D. C., and A. Strasser. 2000. BH3-only proteins—essential initiators of apoptotic cell death. Cell 103:839-842. [DOI] [PubMed] [Google Scholar]
- 22.Inoue, K., S. Ozaki, T. Shiga, K. Ito, T. Masuda, N. Okado, T. Iseda, S. Kawaguchi, M. Ogawa, S. C. Bae, N. Yamashita, S. Itohara, N. Kudo, and Y. Ito. 2002. Runx3 controls the axonal projection of proprioceptive dorsal root ganglion neurons. Nat. Neurosci. 5:946-954. [DOI] [PubMed] [Google Scholar]
- 23.Ito, K., Q. Liu, M. Salto-Tellez, T. Yano, K. Tada, H. Ida, C. Huang, N. Shah, M. Inoue, A. Rajnakova, K. C. Hiong, B. K. Peh, H. C. Han, T. Ito, M. Teh, K. G. Yeoh, and Y. Ito. 2005. RUNX3, a novel tumor suppressor, is highly inactivated in gastric cancer by protein mislocalization. Cancer Res 65:7743-7750. [DOI] [PubMed] [Google Scholar]
- 24.Ito, Y., and K. Miyazono. 2003. RUNX transcription factors as key targets of TGF-β superfamily signaling. Curr. Opin. Genet. Dev. 13:43-47. [DOI] [PubMed] [Google Scholar]
- 25.Jang, C. W., C. H. Chen, C. C. Chen, J. Y. Chen, Y. H. Su, and R. H. Chen. 2002. TGF-β induces apoptosis through Smad-mediated expression of DAP-kinase. Nat. Cell Biol. 4:51-58. [DOI] [PubMed] [Google Scholar]
- 26.Johnstone, R. W., A. A. Ruefli, and S. W. Lowe. 2002. Apoptosis: a link between cancer genetics and chemotherapy. Cell 108:153-164. [DOI] [PubMed] [Google Scholar]
- 27.Kang, G. H., S. Lee, H. J. Lee, and K. S. Hwang. 2004. Aberrant CpG island hypermethylation of multiple genes in prostate cancer and prostatic intraepithelial neoplasia. J. Pathol. 202:233-240. [DOI] [PubMed] [Google Scholar]
- 28.Kim, B. C., M. Mamura, K. S. Choi, B. Calabretta, and S. J. Kim. 2002. Transforming growth factor β1 induces apoptosis through cleavage of BAD in a Smad3-dependent mechanism in FaO hepatoma cells. Mol. Cell. Biol. 22:1369-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, S. G., H. S. Jong, T. Y. Kim, J. W. Lee, N. K. Kim, S. H. Hong, and Y. J. Bang. 2004a. Transforming growth factor β1 induces apoptosis through Fas ligand-independent activation of the Fas death pathway in human gastric SNU-620 carcinoma cells. Mol. Biol. Cell 15:420-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, T. Y., H. J. Lee, K. S. Hwang, M. Lee, J. W. Kim, Y. J. Bang, and G. H. Kang. 2004b. Methylation of RUNX3 in various types of human cancers and premalignant stages of gastric carcinoma. Lab. Investig. 84:479-484. [DOI] [PubMed] [Google Scholar]
- 31.Krieglstein, K., S. Richter, L. Farkas, N. Schuster, N. Dünker, R. W. Oppenheim, and K. Unsicker. 2000. Reduction of endogenous transforming growth factors β prevents ontogenetic neuron death. Nat. Neurosci. 3:1085-1090. [DOI] [PubMed] [Google Scholar]
- 32.Larisch, S., Y. Yi, R. Lotan, H. Kerner, S. Eimerl, W. TonyParks, Y. Gottfried, S. Birkey Reffey, M. P. de Caestecker, D. Danielpour, N. Book-Melamed, R. Timberg, C. S. Duckett, R. J. Lechleider, H. Steller, J. Orly, S. J. Kim, and A. B. Roberts. 2000. A novel mitochondrial septin-like protein, ARTS, mediates apoptosis dependent on its P-loop motif. Nat Cell Biol. 2:915-921. [DOI] [PubMed] [Google Scholar]
- 33.Li, Q. L., K. Ito, C. Sakakura, H. Fukamachi, K. Inoue, X. Z. Chi, K. Y. Lee, S. Nomura, C. W. Lee, S. B. Han, H. M. Kim, W. J. Kim, H. Yamamoto, N. Yamashita, T. Yano, T. Ikeda, S. Itohara, J. Inazawa, T. Abe, A. Hagiwara, H. Yamagishi, A. Ooe, A. Kaneda, T. Sugimura, T. Ushijima, S. C. Bae, and Y. Ito. 2002. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell 109:113-124. [DOI] [PubMed] [Google Scholar]
- 34.Massagué, J., S. W. Blain, and R. S. Lo. 2000. TGF-β signaling in growth control, cancer, and heritable disorders. Cell 103:295-309. [DOI] [PubMed] [Google Scholar]
- 35.Mizushima, S., and S. Nagata. 1990. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 18:5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perlman, R., W. P. Schiemann, M. W. Brooks, H. F. Lodish, and R. A. Weinberg. 2001. TGF-β-induced apoptosis is mediated by the adapter protein Daxx that facilitates JNK activation. Nat. Cell Biol. 3:708-714. [DOI] [PubMed] [Google Scholar]
- 37.Putcha, G. V., K. L. Moulder, J. P. Golden, P. Bouillet, J. A. Adams, A. Strasser, and E. M. Johnson. 2001. Induction of BIM, a proapoptotic BH3-only BCL-2 family member, is critical for neuronal apoptosis. Neuron 29:615-628. [DOI] [PubMed] [Google Scholar]
- 38.Qian, S. W., P. Kondaiah, A. B. Roberts, and M. B. Sporn. 1990. cDNA cloning by PCR of rat transforming growth factor β-1. Nucleic Acids Res. 18:3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schuster, N., and K. Krieglstein. 2002. Mechanisms of TGF-β-mediated apoptosis. Cell Tissue Res. 307:1-14. [DOI] [PubMed] [Google Scholar]
- 40.Shinjyo, T., R. Kuribara, T. Inukai, H. Hosoi, T. Kinoshita, A. Miyajima, P. J. Houghton, A. T. Look, K. Ozawa, and T. Inaba. 2001. Downregulation of Bim, a proapoptotic relative of Bcl-2, is a pivotal step in cytokine-initiated survival signaling in murine hematopoietic progenitors. Mol. Cell. Biol. 21:854-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siegel, P. M., and J. Massagué. 2003. Cytostatic and apoptotic actions of TGF-β in homeostasis and cancer. Nat. Rev. Cancer 3:807-821. [DOI] [PubMed] [Google Scholar]
- 42.Sunters, A., S. Fernandez de Mattos, M. Stahl, J. J. Brosens, G. Zoumpoulidou, C. A. Saunders, P. J. Coffer, R. H. Medema, R. C. Coombes, and E. W. Lam. 2003. FoxO3a transcriptional regulation of Bim controls apoptosis in paclitaxel-treated breast cancer cell lines. J. Biol. Chem. 278:49795-49805. [DOI] [PubMed] [Google Scholar]
- 43.Tachibana, I., M. Imoto, P. N. Adjei, G. J. Gores, M. Subramaniam, T. C. Spelsberg, and R. Urrutia. 1997. Overexpression of the TGFβ-regulated zinc finger encoding gene, TIEG, induces apoptosis in pancreatic epithelial cells. J. Clin. Investig. 99:2365-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tagawa, H., S. Karnan, R. Suzuki, K. Matsuo, X. Zhang, A. Ota, Y. Morishima, S. Nakamura, and M. Seto. 2004. Genome-wide array-based CGH for mantle cell lymphoma: identification of homozygous deletions of the proapoptotic gene BIM. Oncogene 24:1348-1358. [DOI] [PubMed] [Google Scholar]
- 45.Tan, T. T., K. Degenhardt, D. A. Nelson, B. Beaudoin, W. Nieves-Neira, P. Bouillet, A. Villunger, J. M. Adams, and E. White. 2005. Key roles of BIM-driven apoptosis in epithelial tumors and rational chemotherapy. Cancer Cell 7:227-238. [DOI] [PubMed] [Google Scholar]
- 46.Valderrama-Carvajal, H., E. Cocolakis, A. Lacerte, E. H. Lee, G. Krystal, S. Ali, and J. J. Lebrun. 2002. Activin/TGF-β induce apoptosis through Smad-dependent expression of the lipid phosphatase SHIP. Nat. Cell Biol. 4:963-969. [DOI] [PubMed] [Google Scholar]
- 47.Wei, D., W. Gong, S. C. Oh, Q. Li, W. D. Kim, L. Wang, X. Le, J. Yao, T. T. Wu, S. Huang, and K. Xie. 2005. Loss of RUNX3 expression significantly affects the clinical outcome of gastric cancer patients and its restoration causes drastic suppression of tumor growth and metastasis. Cancer Res. 65:4809-4816. [DOI] [PubMed] [Google Scholar]
- 48.Whitfield, J., S. J. Neame, L. Paquet, O. Bernard, and J. Ham. 2001. Dominant-negative c-Jun promotes neuronal survival by reducing BIM expression and inhibiting mitochondrial cytochrome c release. Neuron 29:629-643. [DOI] [PubMed] [Google Scholar]
- 49.Wildey, G. M., S. Patil, and P. H. Howe. 2003. Smad3 potentiates transforming growth factor β (TGFβ)-induced apoptosis and expression of the BH3-only protein Bim in WEHI 231 B lymphocytes. J. Biol. Chem. 278:18069-18077. [DOI] [PubMed] [Google Scholar]
- 50.Xiao, W. H., and W. W. Liu. 2004. Hemizygous deletion and hypermethylation of RUNX3 gene in hepatocellular carcinoma. World J. Gastroenterol. 10:376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamamura, Y., X. Hua, S. Bergelson, and H. F. Lodish. 2000. Critical role of Smads and AP-1 complex in transforming growth factor-β-dependent apoptosis. J. Biol. Chem. 275:36295-36302. [DOI] [PubMed] [Google Scholar]
- 52.Yamamura, Y., W. L. Lee, K. I. Inoue, H. Ida, H. Jiang, P. K. Vogt, and Y. Ito. 2006. RUNX3 cooperates with FoxO3a to induce apoptosis in gastric cancer cells. J. Biol. Chem. 281:5267-5276. [DOI] [PubMed] [Google Scholar]
- 53.Yoo, J., M. Ghiassi, L. Jirmanova, A. G. Balliet, B. Hoffman, D. A. FornaceLiebermann, E. P. Bottinger, and A. B. Roberts. 2003. Transforming growth factor-β-induced apoptosis is mediated by Smad-dependent expression of GADD45b through p38 activation. J. Biol. Chem. 278:43001-43007. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, Y. W., N. Yasui, K. Ito, G. Huang, M. Fujii, J. Hanai, H. Nogami, T. Ochi, K. Miyazono, and Y. Ito. 2000. A RUNX2/PEBP2αA/CBFA1 mutation displaying impaired transactivation and Smad interaction in cleidocranial dysplasia. Proc. Natl. Acad. Sci. USA 97:10549-10554. [DOI] [PMC free article] [PubMed] [Google Scholar]