Abstract
Cyclic AMP (cAMP)-dependent protein kinase (PKA) and ribosomal S6 kinase 1 (RSK1) share several cellular proteins as substrates. However, to date no other similarities between the two kinases or interactions between them have been reported. Here, we describe novel interactions between subunits of PKA and RSK1 that are dependent upon the activation state of RSK1 and determine its subcellular distribution and biological actions. Inactive RSK1 interacts with the type I regulatory subunit (RI) of PKA. Conversely, active RSK1 interacts with the catalytic subunit of PKA (PKAc). Binding of RSK1 to RI decreases the interactions between RI and PKAc, while the binding of active RSK1 to PKAc increases interactions between PKAc and RI and decreases the ability of cAMP to stimulate PKA. The RSK1/PKA subunit interactions ensure the colocalization of RSK1 with A-kinase PKA anchoring proteins (AKAPs). Disruption of the interactions between PKA and AKAPs decreases the nuclear accumulation of active RSK1 and, thus, increases its cytosolic content. This subcellular redistribution of active RSK1 is manifested by increased phosphorylation of its cytosolic substrates tuberous sclerosis complex 2 and BAD by epidermal growth factor along with decreased cellular apoptosis.
Cyclic AMP (cAMP)-dependent protein kinase (PKA) regulates a wide variety of metabolic and functional processes, including cell proliferation, actin cytoskeleton rearrangements, and gene transcription (41). PKA is a tetramer that consists of two regulatory (R) and two catalytic (PKAc) subunits in its inactive form. Binding of cAMP to the R subunits of PKA results in the dissociation of the PKAc subunits, which can then phosphorylate their substrate proteins. The PKA holoenzyme is localized in the proximity of its target proteins by A-kinase PKA anchoring proteins (AKAPs) that act as scaffolds (2, 52). Moreover, by binding to additional signaling molecules, the AKAPs facilitate the coordination and integration of several signals to regulate biological events (2, 52). The two main forms of the PKA regulatory subunits, RI and RII, have different affinities for the various AKAPs. Although most of the AKAPs appear to have a higher affinity for RII subunits (2), certain AKAPs such as D-AKAP1 and D-AKAP2 preferentially bind the RI subunit (22-24).
In addition to AKAPs, other well-characterized protein interactions are involved in the intracellular targeting and regulation of PKA and its subunits. For instance, RIα associates with cytochrome c oxidase subunit Vb and increases its activity whereas exposure to cAMP inhibits cytochrome c oxidase activity and releases cytochrome c from mitochondria and activates apoptosis (55). PAP7, a protein involved in hormonal regulation of steroid formation, also binds RIα at the outer mitochondrial membrane and helps in cholesterol uptake and transport to the inner mitochondrial membrane (28). Likewise, Rab32, a small GTP binding protein, can act as an AKAP for the RII subunits and target PKA to mitochondria (3). In addition, the amount of RI subunit is increased in certain tumor cells (46) and the RI subunit has also been shown to bind with Grb2, permitting its association with the epidermal growth factor receptor (47). Via its association with the second subunit of replication factor C (RFC40) (18), the RI subunit facilitates the nuclear localization of RFC40 and causes G1 arrest of cells (17). Thus, besides regulating the activity of the catalytic subunit, RI also modulates a number of other signaling pathways in cells.
PKAc also interacts with other signaling pathways through various binding partners. For instance, IκB-α, the inhibitory protein of NFκB, associates with PKAc and inhibits its activity (57). Activation of the NFκB pathway and proteosomal degradation of IκB-α relieves PKAc of the inhibition imposed by IκB-α (57). This mechanism activates PKA in a cAMP-independent manner. Similarly, Rab13, a small GTP binding protein, binds with and inhibits the activity of PKAc during tight junction assembly (26). Recently, it has been shown that PKAc also interacts with and phosphorylates p73α and decreases the transactivation and proapoptotic actions of p73α (19).
Ribosomal S6 kinase 1 (RSK1), a serine/threonine kinase, phosphorylates many of the same substrates as PKA. For example, PKA and RSK1 phosphorylate BAD on Ser112 (20, 29). The phosphorylation of BAD dissociates it from Bcl-2 and inhibits apoptosis (29). Similarly, ETS transcription factor (ER81) is phosphorylated by PKA and RSK1, with a resultant decrease in its DNA binding and transcriptional activity (53). Additionally, PKAc and RSK1 also phosphorylate Ser9 on GSK-3β (56) and Ser431 of LKB1, a causative gene product in Peutz-Jeghers syndrome (39). Moreover, interactions between RSK1 and the β subunit of casein kinase 2 (27) as well as Yersinia factor (YopM) (30) have also been reported. The latter interaction increases RSK1 activity (30). On the other hand, PEA15, which binds RSK2 but not RSK1, inhibits the activity of RSK2 and decreases the nuclear accumulation of RSK2 after stimulation by epidermal growth factor (EGF) (48). Overall, therefore, interactions of RSK1 and its isoform RSK2 with other proteins can alter the activity of these kinases and also regulate their cellular distribution. Notably, however, the interactions of RSK1 and RSK2 with different proteins appear to be specific to the respective isoforms (48).
Herein, we report novel interactions between the regulatory and catalytic subunits of PKA with RSK1. Inactive dephosphorylated RSK1 (dephospho-RSK1) interacts with the RI subunit, and active phospho-RSK1 interacts preferentially with the PKAc subunit of PKA. Moreover, we show that the binding of phospho-RSK1 to PKAc increases the interactions between PKAc and RI subunits and decreases the ability of cAMP to activate the PKA holoenzyme. Conversely, the association of inactive RSK1 with RI decreases the interactions between RI and PKAc. Stimulation of cells with EGF increases phospho-RSK1 levels in the nucleus, and active RSK1 is colocalized with PKAc and RI. The dissociation of the PKA/RSK1 complex from AKAPs decreases the amount of nuclear phospho-RSK1 and increases its cytosolic levels. This is accompanied by increased phosphorylation of the cytosolic RSK1 substrates tuberous sclerosis complex 2 (TSC2) and BAD and decreased cellular apoptosis. Therefore, the association of active RSK1 with PKAc apparently ensures its nuclear localization and, therefore, regulates the biological actions of RSK1.
MATERIALS AND METHODS
Immunoprecipitations.
Mouse lung fibroblasts (B82L) overexpressing the EGF receptor or HeLa cells (250,000 cells/35 mm dish) were treated with or without EGF (100 nM) or H2O2 (0.5 mM) for 10 min at 37°C. Cells were lysed in a buffer containing 50 mM Tris (pH 7.5), 1 mM EGTA, 1 mM EDTA, 1% Triton X-100, 150 mM NaCl, 1 mM sodium orthovanadate, 0.1% β-mercaptoethanol, 50 mM NaF, 5 mM sodium pyrophosphate, 10 μM β-glycerolphosphate, 100 μM phenylmethylsulfonyl fluoride (PMSF), 1 μM microcystin, and 1 μg/ml each pepstatin A, aprotinin, and leupeptin. PKAc, RI, or RSK1 was immunoprecipitated with anti-PKAc or anti-RSK1 (both from Santa Cruz Biotechnology, Inc., Santa Cruz, CA) or anti-RI (Calbiochem, San Diego, CA) antibodies prebound to protein G-agarose. Immune complexes were washed three times with lysis buffer, and proteins were separated on 10% acrylamide gels for immunoblotting.
Kinase activity using purified proteins.
Purified PKAc (1 pmol) and RI (20 pmol) were mixed together in the presence or absence of 3 pmol of purified active RSK1. After 10 min incubation at 4°C, cAMP at the indicated concentrations was added to the reaction mix containing 20 mM HEPES (pH 7.5), 1 mM sodium orthovanadate, 1 mM NaF, 1 mM dithiothreitol (DTT), 25 mM β-glycerophosphate, 5 mM MgSO4, 200 μM Kemptide, 125 μM ATP, and 10 μCi [γ-32P]ATP and incubated for a further 10 min at room temperature. Kemptide phosphorylation was monitored using P81 Whatman filter paper as described previously (4). The assays were performed in the absence or presence of 25 μM of PKA inhibitor, PKI. The PKI-sensitive activity reflected the activity of PKA.
Pull-down of RI subunits with cAMP-agarose.
RI subunits were pulled down as described by Wang et al. (50). Briefly, B82L or HeLa cell lysates (500 μg) were mixed with cAMP-agarose (Sigma Chem. Co., St. Louis, MO) that had been reconstituted with 1.5 mg/ml bovine serum albumin at room temperature for 4 h and washed extensively with lysis buffer. Cell lysates were incubated with cAMP-agarose for 1 h or overnight at 4°C in the presence and absence of 50 mM 8-CPT cAMP (Sigma Chem. Co., St. Louis, MO). The resin was then washed twice with high-salt buffer (10 mM HEPES [pH 7.4], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM NaCl, 0.1% Igepal CA-630, 1 mM DTT, 100 μM PMSF, and 1 μg/ml each pepstatin A, aprotinin, and leupeptin) followed by four washes with no-salt buffer (as described above but without NaCl). Proteins bound to RI were identified by Western analyses using the following antibodies: biotinylated anti-RI (BD Biosciences, Franklin Lakes, NJ) and anti-RSK1 and anti-RPP7 (Cocalico Biologicals Co.). Where indicated, Ht31 and Ht31P peptides (Promega Corp.) or GST-RPP7 and GST-RPP8 (provided by Susan Taylor) were preincubated on ice with lysates (500 μg protein) of B82L cells for 60 min before being mixed with cAMP-agarose.
Pull-down of RI subunits with GST-RPP7.
The methods described by Huang et al. (22) were used. Briefly, bacterial cell lysates containing glutathione S-transferase (GST) or GST-RPP7 were incubated with glutathione resin for 2 h at 4°C in phosphate-buffered saline supplemented with 0.1% (vol/vol) Triton X-100, 1 mM PMSF, 1 mM EDTA, 5 mM benzamidine, and 5 mM β-mercaptoethanol (buffer A). The resin was washed extensively with the same buffer. For pull-down experiments, equimolar amounts (3 pmol) of RI and PKAc were reconstituted in buffer containing 20 mM HEPES (pH 7.4), 20 mM MgSO4, 8 mM MnCl2, 4 mM DTT, 20 mM NaF, and 40 μg/ml each aprotinin and leupeptin for 10 min on ice (final volume, 10 μl). GST-Sepharose or GST-RPP7-Sepharose (10 pmol each) in buffer A was added to the reconstituted proteins (final volume, 150 μl) and incubated for 1 h at 4°C. GST-RPP7 beads were washed once with buffer A to remove unbound RI and/or PKAc followed by the addition of 3 pmol (1×) or 6 pmol (2×) of phospho-RSK1 and further incubated for 1 h at 4°C in a final volume of 150 μl. After three washes with buffer A, proteins in the complex were analyzed by immunoblotting. To dephosphorylate RSK1 with calf intestinal phosphatase (CIP; New England Biolabs, MA), 3 (1×) or 6 pmol (2×) of RSK1 and CIP (12.5 units [1×] or 25 units [2×]) was incubated for 30 min at 37°C in a final volume of 20 μl. The dephospho-RSK was then added to the complex of RI, PKAc, and GST-RPP7.
Immunocytochemistry and cell fractionation.
HeLa and B82L cells were preincubated in the presence and absence of 20 μM each Ht31 or Ht31P followed by treatment with or without 100 nM EGF. After 15 min, cells were fixed with 100% methanol for 10 min at −20°C followed by 1:1 methanol:acetone for 10 min at −20°C. After the cells were permeabilized with 0.3% Triton X-100 in phosphate-buffered saline for 5 min, anti-phospho RSK1 (T573) antibody (Cell Signaling, Danvers, MA) (1:250 dilution), anti-PKAc antibody, and biotinylated anti-RI antibody were added. The secondary goat anti-rabbit antibody conjugated with Alexa Fluor 488, goat anti-mouse antibody conjugated to Alexa Fluor 594, and streptavidin conjugated with Pacific Blue were used to detect phospho-RSK1, PKAc, and RI, respectively. Images were captured using an Olympus fluorescence microscope. Confocal images were obtained using a multiphoton Zeiss LSM 5 Pa laser scanning microscope.
For fractionation, cells treated as described above were lysed and subjected to centrifugation at 14,000 × g for 10 min. Cytosolic fractions were analyzed by immunoblotting with anti-RSK1 antibody, anti-phospho RSK1 antibody (T573), and lactate dehydrogenase antibody (a gift from Rockland Immunologicals Inc., Gilbertsville, PA).
Cellular apoptosis.
HeLa cells (50,000 cells/well) plated in a 24-well dish were serum starved for 24 h and then exposed to 20 μM each Ht31 or Ht31P peptide for 5 min prior to treatment with 100 nM EGF for 15 min. When indicated, MEK inhibitor (PD98059) (50 μM) was added at the time of Ht31 or Ht31P treatment. Cells were then treated with or without 20 ng/ml tumor necrosis factor alpha (TNF-α) plus 25 μg/ml cycloheximide (CHX) (TNF-α/CHX) to induce apoptosis, and DNA fragmentation was measured as described previously (14).
RESULTS
EGF and H2O2 stimulate Kemptide phosphorylation of a PKI-insensitive kinase in PKAc IPs.
EGF can increase cAMP accumulation in certain cell types (reviewed in reference 33). Although EGF does not increase cAMP levels in B82L and HeLa cells (not shown), immunoprecipitates (IPs) of PKAc from these cells treated with EGF demonstrated increased ability to phosphorylate the PKAc substrate Kemptide (see Fig. S1A in the supplemental material). H2O2, which can mediate the downstream signaling by growth factors (16, 43), also increased the ability of the PKAc IPs to stimulate Kemptide phosphorylation two- to threefold (see Fig. S1A in the supplemental material). However, the EGF- or H2O2-mediated increase in Kemptide phosphorylation in these experiments was not inhibited by PKI (see Fig. S1B in the supplemental material). These results suggested that a PKI-insensitive kinase that phosphorylates Kemptide was coimmunoprecipitated with PKAc.
To determine the presence of another kinase in the PKAc IPs, in-gel kinase assays were performed. IPs of PKAc from B82L cells treated with or without H2O2 were separated on casein-containing gels, and kinase activities were monitored. While the PKAc IPs contained PKAc (40 kDa band) that phosphorylated casein, another 90 kDa protein kinase was also co-IP with PKAc (see Fig. S2A in the supplemental material). H2O2 did not alter the activity of PKAc but increased the activity of the 90 kDa kinase that was co-IP (see Fig. S2A in the supplemental material). Moreover, in gels without casein, 32P incorporation into the 90 kDa kinase increased in the H2O2-treated PKAc IPs, indicating that this 90 kDa kinase was autophosphorylated (see Fig. S2B in the supplemental material). Since RSK1 migrates as a 90 kDa protein, can phosphorylate Kemptide (6), and is activated by EGF or H2O2 (1), we reasoned that the 90 kDa kinase in the PKAc IPs may be RSK1. In-gel kinase assays of RSK1 IPs showed that H2O2 autophosphorylated RSK1 and also increased its activity (see Fig. S2A and S2B in the supplemental material). Moreover, the RSK1 IPs contained a 40 kDa kinase whose activity was not stimulated by H2O2 (see Fig. S2A in the supplemental material). Similar results were observed with EGF as an agonist instead of H2O2 and also when HeLa cells were used (data not shown). Notably, the in vitro kinase activities of the PKAc and RSK1 IPs were similar and showed insensitivity to PKI (see Fig. S3A in the supplemental material). These findings suggested that the kinase activity in the PKAc IPs is due to the presence of RSK1. It would be expected that in the PKAc IPs, although RSK1 is present, the PKAc component that phosphorylates Kemptide would be inhibited by PKI. However, the kinase activity of PKAc does not survive the IP procedure. Thus, IPs of even pure PKAc did not phosphorylate Kemptide (see Fig. S3B in the supplemental material). Together, these results strongly suggest that the Kemptide phosphorylation monitored in the PKAc IPs from cells was due to the presence of PKI-insensitive RSK1.
PKAc and RSK1 exist as a complex with RI and D-AKAP1 but RSK1 is not directly associated with D-AKAP1.
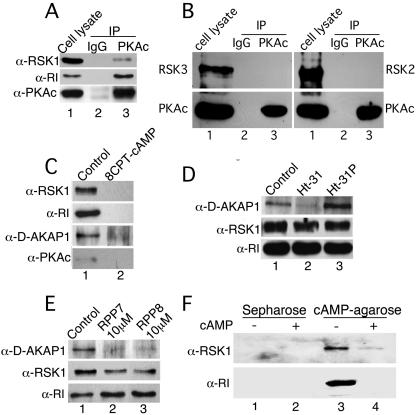
That RSK1 is indeed coimmunoprecipitated with PKAc is shown in Fig. 1A. Essentially, PKAc was IP from HeLa cell lysates and immunoblotted for the presence of PKAc as well as RSK1. Control IPs with nonspecific rabbit immunoglobulin G (IgG) did not show any of these proteins, whereas RSK1 was co-IP with PKAc (Fig. 1A). Although HeLa cells contain RSK2 and RSK3, these proteins did not co-IP with PKAc (Fig. 1B), demonstrating that RSK1 specifically interacts with PKAc in either a direct or an indirect manner. HeLa and B82L cells express the type I regulatory subunit of PKA (RI), and in B82L cells the type II regulatory subunit of PKA (RII) is not detectable (not shown). Therefore, we determined whether RI was also present in the PKAc/RSKI complex. As shown in Fig. 1A, RI was present in the immunocomplex. Because RI binds PKAc, the presence of RSK1 in the PKAc IPs (Fig. 1A) may be due to its association with PKAc or RI or some other proteins such as AKAPs that bind to PKA. Therefore, experiments were designed to identify the binding partners of RSK1.
FIG. 1.
PKAc, RI, D-AKAP1, and RSK1 exist as a complex, and RSK1 associates directly with RI. (A) B82L or HeLa cells were serum starved overnight and subjected to IP with PKAc antibody or nonspecific IgG. RSK1 and RI, which coimmunoprecipitated with PKAc, were detected by immunoblotting. The example shown represents an experiment performed with HeLa cells. (B) Same as panel A except the presence of RSK2 or RSK3 in IPs of PKAc was monitored. Nonspecific rabbit IgG was used for control IPs. (C) RI from B82L cell lysates (500 μg protein) was pulled down with cAMP-agarose in the absence or presence of cAMP (50 mM), and the proteins associated with RI were detected by immunoblotting. (D) Same as panel C except the lysates were incubated in the absence or presence of 100 μM each Ht31 or Ht31P. (E) Same as panel C except RPP7 and RPP8 (10 μM each) replaced Ht31 and Ht31P. Representatives of at least two experiments for each set of conditions are shown. (F) Purified RI (3 pmol) and RSK1 (3 pmol) were incubated together. The RI was then pulled down with cAMP-agarose and, the proteins in the pull-down assay were analyzed by immunoblotting. Control pull-down assays were performed in the presence of 8-CPT-cAMP (50 mM) or Sepharose alone.
Next, we investigated whether RSK1 forms a complex with PKA by interacting with either PKAc or RI or, perhaps, AKAPs. As an example of AKAPs we monitored the presence of D-AKAP1. For this purpose, the RI subunit was pulled down from B82L cell lysates with cAMP-agarose in the absence and presence of free cAMP. As shown in Fig. 1C, the RI pull-down assays contained RSK1 and D-AKAP1. As expected, binding of cAMP-agarose to RI dissociates PKAc; therefore, PKAc was not present in the complex (Fig. 1C). The absence of RSK1, D-AKAP1, and RI in the presence of excess (50 mM) 8-CPT-cAMP shows the specificity of the cAMP-agarose pull-down assay (Fig. 1C). These data suggest that RSK1 may, directly or indirectly, interact with either RI or D-AKAP1.
The association of regulatory subunits of PKA with AKAPs can be competed off by a 23-amino-acid (aa) peptide (Ht31) (9, 10) that corresponds to a sequence within AKAP-Lbc (15). The replacement of two isoleucines with prolines in this peptide (Ht31P) obliterates its binding to regulatory subunits and thus does not interfere with interactions between PKA and AKAPs (49). Therefore, using Ht31 and Ht31P we determined whether RSK1 was present in the RI pull-down assays due to its interactions with D-AKAP1 or other AKAPs. As expected, cAMP-agarose-bound RI pulled down D-AKAP1 and RSK1 from the complex (Fig. 1D, lane 1). In the presence of Ht31, but not H31P, D-AKAP1 was absent from the RI pull-down assays, demonstrating that Ht31 specifically dissociated the RI subunit from AKAPs (Fig. 1D; cf. lanes 2 and 3). However, even in the absence of D-AKAP1, RSK1 was present in the RI pull-down assay (Fig. 1D, lane 2). These data suggest that the presence of RSK1 in the RI pull-down assays is not due to association of RSK1 with AKAPs such as D-AKAP1; rather, it may be due to the association of RSK1 with RI. This notion was further confirmed by using two short regions of D-AKAP1 and D-AKAP2, namely, RPP7 (aa 284 to 408 of D-AKAP1) and RPP8 (aa 333 to 372 of D-AKAP2) (22, 23). These two regions encompass the RI binding site on D-AKAP1 and D-AKAP2 and compete for the interactions between AKAPs and RI (22, 23). As in the experiments with Ht31, RPP7 and RPP8 dissociated D-AKAP1 from the RI subunit without affecting the amount of RSK1 in the RI pull-down assays (Fig. 1E). Thus, RSK1 does not interact with D-AKAP1 and may be present in the RI pull-down assays due to direct interactions with the RI subunit of PKA.
RI directly interacts with RSK1 but does not modulate the activity of RSK1.
To determine whether RI directly interacts with RSK1 we mixed equimolar amounts of purified RI and RSK1 and pulled down RI with cAMP-agarose in the presence or absence of free excess cAMP. The pure RI (approximately 50% of input) was pulled down along with the pure RSK1 (approximately 60% of input) (Fig. 1F), i.e., stoichiometry revealed that 0.85 pmol RI bound to 1 pmol of RSK1. Neither RSK1 nor RI was present in control pull-down assays with Sepharose alone or with excess free cAMP (Fig. 1F). These results confirm the notion that RI and RSK1 interact directly with each other.
Since binding of RI to PKAc inhibits the activity of PKAc, we investigated the possibility that RI binding to RSK1 may also modulate RSK1 activity in a cAMP-dependent manner. Purified RI and RSK1 or PKAc (positive control) were mixed together, and the kinase activity of the mixtures was then determined in the presence or absence of 8CPT-cAMP. While the PKAc activity was inhibited by RI in the absence of cAMP, in parallel experiments, RI at threefold excess over RSK1 did not alter the RSK1 activity irrespective of whether cAMP was present or not (see Fig. S4 in the supplemental material). Hence, unlike the results seen with PKAc, interactions of RI with RSK1 do not alter RSK1 activity.
Inactive RSK1 preferentially associates with RI, while active RSK1 interacts with PKAc.
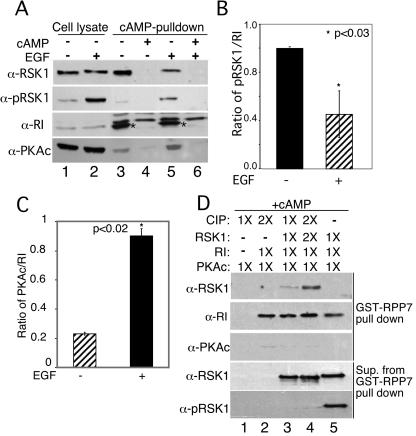
Next, we investigated whether stimulation of cells with EGF altered the interactions between RI and RSK1. Serum-starved HeLa cells were treated with or without EGF (100 nM) for 10 min. RI was pulled down from cell lysates with cAMP-agarose in the presence or absence of free cAMP. As determined with anti-phospho-RSK1 antibody, exposure of cells to EGF resulted in activation of the RSK1 (Fig. 2A, lane 2, second panel from top). The active, phospho-RSK1 migrated slightly slower than its nonphosphorylated counterpart (Fig. 2A, top panel; cf. lanes 1 and 2). In the absence of EGF, RSK1 was detected in the RI pull-down assays performed in the absence of free cAMP (Fig. 2A, lane 3) but not in the presence of excess free cAMP, which prevents binding of RI to cAMP-agarose (Fig. 2A, lanes 4 and 6). Interestingly, when cells were stimulated with EGF, the amount of RSK1 in the cAMP-agarose pull-down assays was markedly diminished (Fig. 2A, top panel; cf. lanes 3 and 5). Quantitative analysis of RI interaction with RSK1 performed by densitometric scanning of the bands from different experiments showed a two- to threefold increase in the interaction between RI and RSK1 in the absence of EGF (Fig. 2B). Surprisingly, however, although the cAMP-agarose pull-down assays of RI would be expected to dissociate PKAc from RI, in the presence of EGF some PKAc was present (Fig. 2A). The reason for this observation is explained later in the context of additional experiments. Overall, however, the data in Fig. 2A and 2B suggest that the affinity of RI for RSK1 decreases when RSK1 is activated by EGF treatment of cells.
FIG. 2.
Inactive RSK1 associates preferentially with RI, while EGF-activated RSK1 interacts with PKAc. Serum-starved HeLa cells were treated with and without EGF (100 nM) for 10 min at 37°C. (A) The RI in the cell lysates was pulled down with cAMP-agarose in the presence or absence of 8-CPT-cAMP (50 mM). Total RSK1, phospho-T573-RSK1, PKAc, and RI in the pull-down assay were analyzed by immunoblotting. Asterisks show the migration of RI. (B) Quantitative analysis of three experiments similar to those whose results are shown in panel A. The ratios of RSK1/RI in the RI pull-down assays were calculated from densitometric scans. *, P < 0.03 (Student's unpaired t test analysis). (C) Quantitative analysis of three experiments similar to those whose results are shown in panel A. The ratios of PKAc/RI in the RI pull-down assays were calculated from densitometric scans. *, P < 0.02 (Student's unpaired t test analysis). (D) Purified RSK1 (3 pmol) was treated with 3 pmol (1×) or 6 pmol (2×) of alkaline CIP before incubation with reconstituted RI, PKAc, and GST-RPP7. cAMP (1 mM) was present in the GST-RPP7 pull-down assays, and the proteins were analyzed by immunoblotting. The supernatants (Sup.) from GST-RPP7 pull-down assays were analyzed for total RSK1 and phospho-RSK1 to confirm dephosphorylation of RSK1.
Additional evidence for preferential interactions between active (phosphorylated) and inactive (dephosphorylated) RSK1 with PKAc and RI is provided by experiments with purified proteins (Fig. 2D). The active phospho-RSK1 (Upstate Biologicals Inc.) was dephosphorylated by treatment with CIP (Fig. 2D; cf. lanes 3, 4, and 5). Both the active and inactive forms of RSK1 were then mixed with equimolar amounts of purified PKA holoenzyme. RI protein was then pulled down using GST-RPP7-linked Sepharose beads in the presence of cAMP to dissociate PKAc from RI and facilitate data interpretation. GST-RPP7 contains the RI binding site in D-AKAP1 (23) and, therefore, pulled down RI (Fig. 2D, lanes 2 to 5). However, only the dephospho-RSK1 was pulled down along with RI (Fig. 2D, top panel; cf. lanes 4 and 5). These data and those in Fig. 2A and 2B clearly show that RI preferentially interacts with the inactive, dephosphorylated RSK1.
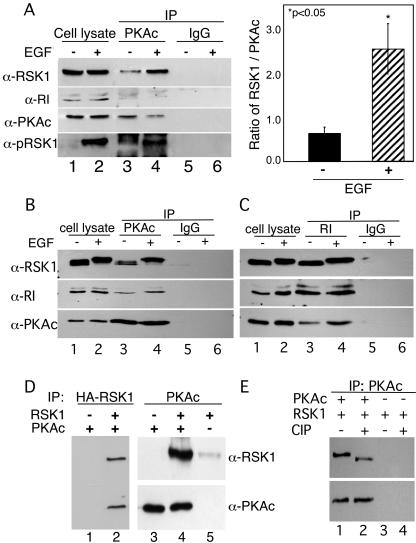
In our initial experiments (see Fig. S1 in the supplemental material), after treatment with EGF, a significant amount of apparent RSK1 activity was present in PKAc IPs. However, as shown in Fig. 2, only the dephosphorylated, inactive form of RSK1 interacts with RI. Therefore, we determined levels of RSK1 in IPs of PKAc from cells treated with or without EGF. The goal here was to determine whether RSK1 was present in PKAc IPs in the absence of RI. Therefore, after treating cells with and without EGF, cAMP was added to the cell lysates to dissociate RI from the PKAc that was subsequently immunoprecipitated. The addition of cAMP to the cell lysates dissociated PKAc from RI, and the latter protein was absent from the PKAc IPs (Fig. 3A, second panel from top, lanes 3 and 4). Most interestingly, in the presence of EGF, the amount of RSK1 in the PKAc IPs was markedly increased (Fig. 3A, top panel; cf. lanes 3 and 4) and this RSK1 was phosphorylated (Fig. 3A, bottom panel; cf. lanes 3 and 4). Quantification of the ratios of RSK1 bound to PKAc from three independent experiments showed that EGF increased by four- to fivefold the binding of RSK1 to PKAc in the presence of EGF (Fig. 3A). These data suggested that while RI preferentially interacts with inactive RSK1, the active, phospho-RSK1 might interact with PKAc. To determine whether active RSK1 in complex with PKAc also pulls down the RI subunit, experiments similar to those whose results are shown in Fig. 3A were performed in the absence of cAMP during the IP procedure. The addition of EGF decreased the migration of RSK1, indicating that this enzyme was phosphorylated (Fig. 3B, top panel). Additionally, consistent with the data in Fig. 3A, the IP of PKAc was accompanied by increased amounts of active RSK1 (Fig. 3B, top panel; cf. lanes 3 and 4). Importantly, the RI subunit was present in the IPs of PKA and the amount of RI was enhanced in the presence of EGF (Fig. 3B, middle panel; cf. lanes 3 and 4). Likewise, in experiments similar to those whose results are shown in Fig. 3B, the IP of RI from cells treated with or without EGF resulted in the co-IP of both RSK1 and PKAc and the presence of EGF increased the amount of PKAc in the complex (Fig. 3C, bottom panel; cf. lanes 3 and 4). All together, the data in Fig. 3A to 3C suggest that EGF-activated phospho-RSK1 interacts with PKAc. The increased amounts of RI in PKAc IPs, along with increased amounts of PKAc in IPs of RI in the presence of EGF, suggest that the interaction of PKAc with RSK1 may increase the association of PKAc with RI. This possibility is further investigated below.
FIG. 3.
Active RSK1 interacts with PKAc in the absence of RI. (A) PKAc was IP from lysates of HeLa cells treated with or without EGF (100 nM) for 10 min. 8CPT-cAMP (100 μM) was added to cell lysates and IP buffers to dissociate PKAc from RI. The presence of total and phospho-RSK1 in the IPs was monitored by immunoblotting. Nonspecific rabbit IgG (IgG) was used for control IPs. Results representative of three experiments are shown. The right-hand panel shows the ratios of RSK1 to PKAc in IPs from three similar experiments. *, P < 0.05 (Student's t test). (B and C) Serum-starved HeLa cells were treated with and without EGF (100 nM) for 10 min at 37°C. PKAc (B) or RI (C) was immunoprecipitated from the lysates (500 μg protein). The presence of RI, PKAc, and RSK1 in the IPs was monitored by immunoblotting. Rabbit IgG (B) and mouse IgG (C) were used for control IPs. (D) Purified HA-tagged RSK1 (3 pmol) was mixed with PKAc (3 pmol). Anti-HA and anti-PKAc antibodies were used to IP RSK1 and PKAc, respectively. The presence of PKAc and RSK1 in the IPs was monitored by immunoblotting. (E) Purified RSK1 (3 pmol) treated with and without CIP was mixed with PKAc (3 pmol). Anti-PKAc antibody was used to IP PKAc. The presence of PKAc and RSK1 in the IPs was monitored by immunoblotting.
To determine whether the PKAc and RSK1 interact directly with each other, equimolar amounts of purified PKAc and hemagglutinin (HA)-tagged phospho-RSK1 were mixed and the RSK1 or PKAc was IP. The data in Fig. 3D (lanes 2 and 4) show that pure RSK1 can interact directly with PKAc, as evidenced by the presence of both proteins in the anti-HA and anti-PKAc IPs. Furthermore, when the phospho-RSK1 was dephosphorylated by CIP treatment, its ability to co-IP with PKAc was decreased (Fig. 3E). The effective dephosphorylation of RSK1 by CIP is shown by the higher mobility of RSK1 in the gels (Fig. 3E). Collectively, the data in Fig. 2 and 3 show that active, phospho-RSK1 interacts preferentially with PKAc and that inactive, dephospho-RSK1 associates with RI.
Activated RSK1 increases the interaction between PKAc and RI, while inactive RSK1 decreases interactions between PKAc and RI.
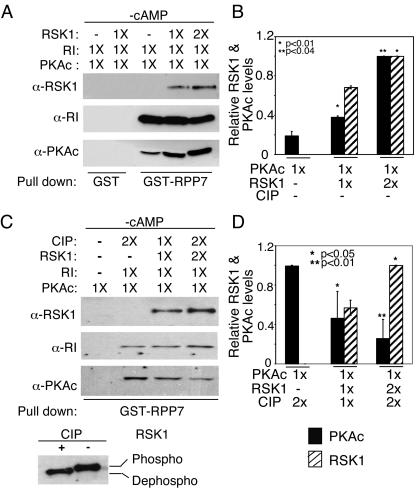
The data in Fig. 3B and 3C suggested that the interactions between PKAc and phospho-RSK1 may alter the association of the RI with PKAc. Therefore, we examined whether the active and inactive forms of RSK1, by associating with the PKAc and RI subunits, respectively, altered the interactions between components of the PKA holoenzyme. The PKA holoenzyme was reconstituted by mixing equimolar amounts of purified PKAc and RI in the presence of GST-RPP7, and the complex was bound to GSH-Sepharose. Then, 3 pmol (1×) or 6 pmol (2×) of active-RSK1 was added to the GST-RPP7-PKA complex. GST-RPP7-bound proteins were pulled down and analyzed by immunoblotting. Figure 4 (panel A, lane 3) shows that RI and PKAc form a complex with GST-RPP7. Active RSK1 was also pulled down with the complex in the proportion in which it had been added (Fig. 4A, top panel, fourth and fifth lanes). Interestingly, the addition of active RSK1 increased the amount of PKAc that was retained in the complex during the washes (Fig. 4A, bottom panel; cf. lanes 3, 4, and 5). Control experiments with GST alone did not pull down any of the proteins (Fig. 4A, lanes 1 and 2). The quantification of data from four similar experiments is shown in Fig. 4B. These findings demonstrate that phospho-RSK1, presumably by associating with PKAc, increases the affinity of interactions between PKAc and RI and therefore prevents the loss of PKAc during subsequent washes of the pull-down procedure. The increased association of PKAc with RI in the presence of active RSK1 also explains the somewhat greater amounts of PKAc and RI in IPs of RI and PKAc, respectively, from lysates of cells treated with EGF (Fig. 3C and 3D).
FIG. 4.
Phospho-RSK1 increases, while dephospho-RSK1 decreases, interactions between PKAc and RI. (A) Equimolar amounts (3 pmol each) of purified PKAc and RI were incubated for 10 min on ice and complexed with GST-RPP7 (10 pmol) as described in Materials and Methods. After the complex was washed, 3 pmol (1×) or 6 pmol (2×) of RSK1 was added. GST-RPP7 was pulled down, and the presence of the various proteins in the complex was determined by immunoblotting. GST-Sepharose served as a control. (B) The results of quantitative analysis of three experiments similar to those whose results are shown in panel A are presented as ratios of RSK1 or PKAc to RI. *, P < 0.01; **, P < 0.04 (Student's t test analysis). (C) CIP-treated RSK1 was mixed with reconstituted RI and PKAc as described for panel A. RI was pulled down with GST-RPP7, and the proteins were analyzed by immunoblotting. The difference in the levels of mobility of phospho- and dephospho-RSK1 before and after CIP treatment are shown in the bottom panel. (D) The results of quantitative analysis of three experiments similar to those whose results are shown in panel C are presented as ratios of RSK1 or PKAc to RI. *, P < 0.05; **, P < 0.01 (Student's t test analysis).
To determine the effect, if any, of the inactive, dephosphorylated RSK1 on PKA holoenzyme, we dephosphorylated active RSK1 by use of CIP (bottom panel in Fig. 4C) in experiments similar to those whose results are shown in Fig. 4A. The data in Fig. 4C show that in contrast to our observations with active, phospho-RSK1, the addition of dephospho-RSK1, in a concentration-dependent manner, decreased the amount of PKAc in the complex (Fig. 4C, bottom panel; cf. the second, third, and fourth lanes). Figure 4D shows the quantification of the amounts of PKAc and dephospho-RSK1 in the GST-RPP7 pull-down assay from three experiments similar to those whose results are shown in Fig. 4C. Thus, by associating with RI, the dephosphorylated, inactive RSK1 apparently decreases the affinity of the interaction between RI and PKAc, resulting in a greater loss of the PKAc during the pull-down assay. The experiments whose results are shown in Fig. 4 were performed in the absence of cAMP. Therefore, the RSK1-mediated changes in association of PKAc with RI are independent of cAMP.
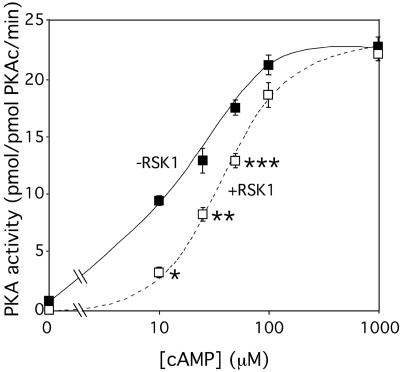
Active RSK1 decreases the ability of cAMP to activate PKA.
Because active RSK1, by interacting with PKAc, increases the association between PKAc and RI, we reasoned that in the presence of active RSK1, the ability of cAMP to activate PKA holoenzyme would be decreased. To test this possibility directly, purified PKAc and RI (1 pmol and 20 pmol, respectively) were mixed together in the presence and absence of 3 pmol of purified active RSK1. The ability of increasing concentrations of cAMP to activate PKA was then monitored. Because PKA and RSK1 both phosphorylate Kemptide, the kinase assays were performed in the presence and absence of PKI, the PKA inhibitor, at a concentration (25 μM) in excess of that required to completely inhibit PKAc activity (see Fig. S1C in the supplemental material). As shown before (see Fig. S3 and S4 in the supplemental material), neither cAMP nor PKI altered RSK1 activity. However, consistent with the finding that active RSK1 increases the association of RI with PKAc, the ability of cAMP to activate PKA was decreased when active RSK1 was present (Fig. 5). Although at maximal concentrations of cAMP (1 mM) the level of activation of the PKA holoenzyme was the same, the presence of RSK1 increased the Ka of cAMP for PKA from ∼17 μM to ∼50 μM. These data (Fig. 5) along with those in Fig. 3 and 4 demonstrate that interactions of active RSK1 with PKAc increase the binding of RI to PKAc and decrease its activity. The regulation of PKA activity by active RSK1 represents a novel mode of regulating PKA activity.
FIG. 5.
Active RSK1 inhibits PKA activity. Purified PKAc (1 pmol) was mixed with 20 pmol of purified RI. The PKA was then assayed for the kinase activity in the presence (+RSK1) and absence (−RSK1) of purified RSK1 (3 pmol) with or without PKI (25 μM). Various concentrations of cAMP were added to the reaction mix, and Kemptide phosphorylation was measured. The PKI-sensitive activity is shown. Data presented represent the means ± standard errors of the means (SEM) of at least three separate experiments. *, P < 0.001; **, P < 0.01; ***, P < 0.03 (Student's t test analyses compared to the corresponding control results).
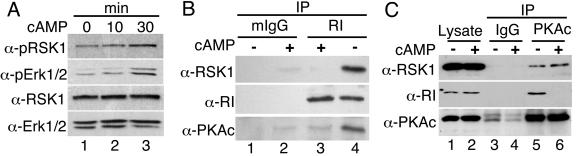
Cyclic AMP activates ERK1/2 and RSK1 and dissociates RSK1 from RI in B82L cells.
In some cell types, cAMP activates extracellular signal-regulated kinases 1 and 2 (ERK1/2) (38, 42). Activation of Erk1/2 results in the phosphorylation and activation of the downstream kinase RSK1. Thus, cAMP can increase the levels of both PKAc and RSK1 activities. Therefore, we investigated whether cAMP treatment of B82L cells resulted in activation of the ERK1/2 pathway and subsequently RSK1 and, if so, whether the interactions between RSK1 and RI were altered. Exposure of cells to 8CPT-cAMP (100 μM) for 30 min increased the amounts of phospho-RSK1 and phospho-ERK1/2 (Fig. 6A). The amounts of total RSK1 and ERK1/2 show equal levels of loading of proteins. In the absence of 8CPT-cAMP, IPs of RI contained RSK1 and PKAc (Fig. 6B, lane 4). In cells treated with 8CPT-cAMP for 30 min, as expected, the PKAc subunit was dissociated from the RI (Fig. 6B, lane 3). Moreover, RSK1 was also not present in the IPs of RI (Fig. 6B; cf. lanes 3 and 4). In similar experiments, after treatment of cells with cAMP for 30 min, IPs of PKAc contained RSK1 but not RI (Fig. 6C, lane 6). Similar results were also observed with HeLa cells (not shown). Thus, as with the experiments with EGF (Fig. 2 and 3), depending upon its activation state the RSK1 associates either with RI or with PKAc. However, unlike the scenario seen with EGF, cAMP-mediated activation of RSK1 is also accompanied by the dissociation of PKAc from RI. Although these findings appear contrary to the notion that active RSK1 increases interactions between RI and PKAc (Fig. 2 to 4), it should be noted that in the experiments with EGF (Fig. 2), cAMP-agarose was added to cell lysates after the activation of RSK1 by EGF whereas for the experiments whose results are shown in Fig. 6, cells were treated with cAMP, which would first activate PKA and then RSK1 at later times. Apparently, therefore, the RI that is already bound to cAMP does not interact with PKAc even in the presence of active RSK1. This would also suggest that the interactions of PKAc and active RSK1 in the presence of EGF that increase association of RI to PKAc may alter the affinity of the RI subunit for cAMP. Indeed, the change in the 50% effective concentration for cAMP-mediated activation of PKA activity (Fig. 5) in the presence of active RSK1 supports this notion.
FIG. 6.
Treatment of B82L cells with 8CPT-cAMP activates Erk1/2 as well as RSK1 and dissociates RSK1 from RI. (A) B82L cells (250,000 cells/35 mm dish) were treated with 8CPT-cAMP (100 μM) for the indicated times. Proteins (30 μg) were immunoblotted for phospho-RSK1, phospho-Erk1/2, total RSK1, and total Erk1/2. The last two served as loading controls. Results representing two similar experiments are shown. (B) B82L cells (1 × 106 cells/100 mm dish) were treated with or without 8CPT-cAMP (100 μM) for 30 min. The RI was then immunoprecipitated from cell lysates (500 μg protein), and the presence of RSK1 in the IPs was monitored by immunoblotting. Mouse IgG2b (mIgG) was used in control IPs. Results representing three experiments are shown. (C) B82L cell lysates (500 μg protein) were treated with or without 8-CPT cAMP (100 μM) for 30 min. PKAc was then IP. RSK1 and RI in the IPs were monitored by immunoblotting. Rabbit IgG (IgG) was used in control IPs. Results representing three experiments is shown.
Functional significance of the binding of RSK1 to PKA.
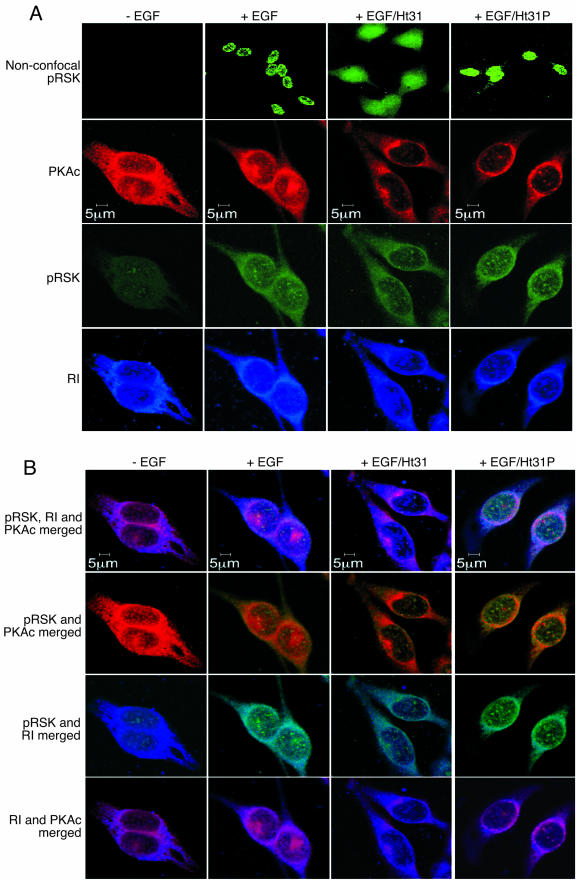
To determine the functional significance of the complex comprising RSK1, PKA, and AKAPs, we first investigated the cellular distribution of a RSK1 and the consequence of dissociating the protein from AKAPs. As shown before by Blenis and coworkers (11, 35), activation of HeLa cells with EGF increased the nuclear amount of active RSK1 and active RSK1 decorated the nucleus in a punctate manner (Fig. 7A, top row). As expected, an identical cellular localization of active RSK1 was observed with cells that had been treated with Ht31P (Fig. 7A). However, when cells were pretreated with Ht31 to dissociate the RSK1/PKA complex from AKAPs, in the presence of EGF the nuclear localization of active RSK1 was decreased and demonstrated a more diffuse, cytoplasmic distribution (Fig. 7A, top row). Similar results were also observed with B82L cells (not shown). These findings were confirmed by the observation that the amount of phosphorylated, active RSK1 in the cytoplasm was increased when the cells were treated with Ht31 compared with the results seen with cells treated with Ht31P or peptide-untreated controls (Fig. 8A, top panel; cf. lanes 2, 3, and 4). In the presence of EGF, active RSK1 was colocalized with PKAc and RI at the periphery and inside the nucleus (Fig. 7B). Although the RI/RSK1 colocalization was more obvious at the periphery of the nucleus, some active RSK1 and RI colocalization was also observed inside the nucleus (Fig. 7B). The abrogation of the interactions of RSK1/PKA complex from AKAPs with Ht31, but not Ht31P, decreased the intranuclear colocalization of active RSK1 with PKAc and RI (Fig. 7B). These data suggest that the tethering of active RSK1 to AKAPs via PKAc is important for its nuclear localization.
FIG. 7.
Active RSK1 and PKAc as well as RI are colocalized in the nucleus, and disruption of the PKA/AKAP decreases nuclear localization of RSK1. Serum-starved HeLa cells were preincubated with nothing or 20 μM each of the cell-permeable, stearated Ht31 or Ht31P peptides for 5 min and then treated with or without EGF (100 nM) for 15 min and fixed. Active phospho-RSK1 was detected using anti-phospho-T573 RSK1 antibody and goat anti-rabbit antibody conjugated to Alexa Fluor 488. PKAc was detected using anti-PKAc antibody (BD Bioscience) and goat anti-mouse antibody conjugated to Alexa Fluor 594. RI was monitored using biotinylated anti-RI antibody (BD Bioscience) and streptavidin-conjugated Pacific Blue. The top row in panel A shows nonconfocal images of the cells, whereas the other rows present confocal images showing the distribution of active RSK1, PKAc and RI. (B) The images in panel A were merged to show colocalization of the three proteins.
FIG. 8.
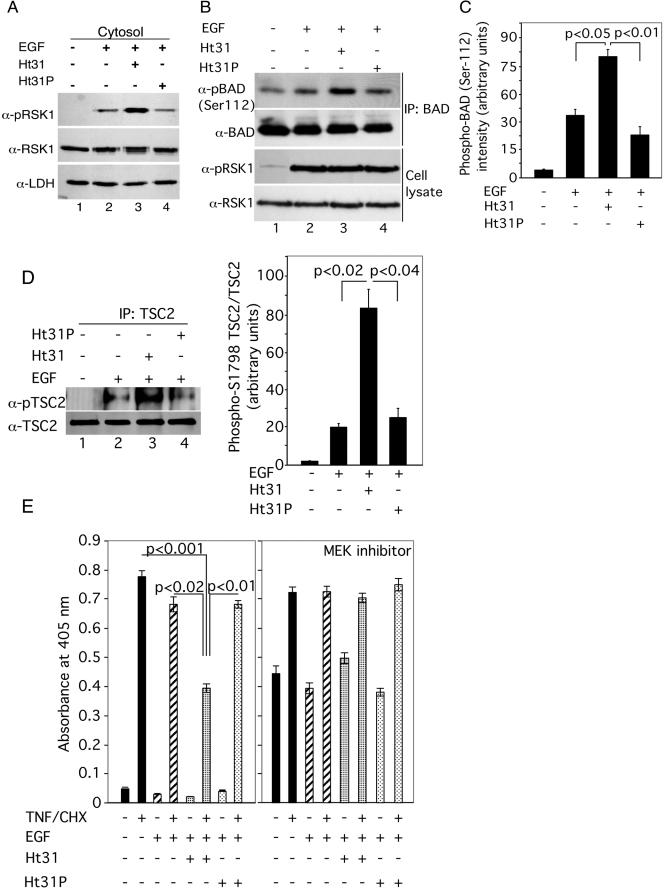
Dissociation of AKAPs from regulatory subunits of PKA alters the subcellular distribution of active RSK1, increases EGF-elicited phosphorylation of BAD and TSC2, and decreases cellular apoptosis. (A) HeLa cells were incubated exactly as described for Fig. 7 except that at the end of the incubations with or without EGF, the cells were lysed and cytosolic fractions were isolated. A representative Western blot with anti-pRSK1 antibody is shown. The same blot was reprobed with anti-lactate dehydrogenase (LDH) antibody as a loading control. (B) Serum-starved HeLa cells were treated with Ht31 or Ht31P for 10 min prior to treatment with EGF (100 nM) for 1 h. BAD was immunoprecipitated from cell lysates, and the amount of phospho-S112 BAD as well as of total BAD was monitored. Nonspecific rabbit IgG was used for control IPs. Lower panels: Western analyses of total cell lysates (20 μg protein) for the amounts of pRSK1 and RSK1. (C) Quantitative analyses of the ratio of phospho-S112 BAD/BAD in IPs from three experiments similar to those whose results are shown in panel B. Data represent the means ± SEM (n = 3). (D) Serum-starved HeLa cells were treated with Ht31 or Ht31P 10 min prior to treatment with EGF (100 nM) for 30 min. TSC2 was immunoprecipitated from cell lysates, and the amount of phospho-TSC2 at Ser1798 was monitored using anti-phospho-Akt substrate antibody (36). The blot was stripped and probed for total TSC2. Nonspecific rabbit IgG was used for control IPs. The right-hand panel shows quantitative analyses of the ratios of phospho-TSC2/TSC2 in the TSC2 IPs from three experiments. (E) Serum-starved HeLa cells (50,000 cells/well) were preincubated in the presence and absence of 20 μM concentrations each of the stearated Ht31 or Ht31P and, when present, 50 μM of MEK inhibitor PD98059 for 15 min followed by treatment with or without 100 nM EGF for 15 min. TNF-α (20 ng/ml) and cycloheximide (25 μg/ml) (TNF-α/CHX) were then added for 3 h, and DNA fragmentation was monitored. Data represent means ± SEM of the optical density at 405 nm per 50,000 cells (n = 6). Significant differences were assessed by Student's unpaired t test.
RSK1 phosphorylates the proapoptotic protein BAD and dissociates it from Bcl-2 (44). Likewise, RSK1 also phosphorylates TSC2 (36, 37). To determine whether changes in cellular distribution of active RSK1 caused by its dissociation from AKAPs (Fig. 7 and 8A) alter the phosphorylation of its cytosolic substrates such as BAD and TSC2, HeLa cells were treated with EGF in the presence and absence of Ht31 and Ht31P. BAD was immunoprecipitated from cell lysates, and its phosphorylation on Ser112, the RSK1 site (20, 29), was analyzed by immunoblotting. Cell lysates were also analyzed to determine the phospho-RSK1 and total RSK1 levels. The phosphorylation of RSK1 by EGF was not altered by Ht31 or Ht31P (Fig. 8B, lanes 1 to 3). However, addition of Ht31 increased EGF-elicited phosphorylation of BAD on Ser112 compared to the results seen with control untreated cells or cells exposed to Ht31P (Fig. 8B [cf. lanes 1, 2, and 3] and Fig. 8C). Similarly, EGF-elicited phosphorylation of TSC2 on Ser1798, the RSK1 site (detected by the anti-phospho-Akt substrate antibody) (36), was also increased by Ht31 but not by Ht31P (Fig. 8D). This is consistent with the observation that Ht31 increases the amount of active RSK1 in the cytoplasm (Fig. 7 and 8A) and, therefore, augments phosphorylation of its cytosolic substrates BAD and TSC2. Because increased phosphorylation of BAD results in increased protection against apoptosis (20), we investigated whether the ability of EGF to protect against cell apoptosis induced by TNF-α/CHX was enhanced by Ht31. DNA fragmentation in HeLa cells exposed for 3 h to TNF-α/CHX in the presence or absence of EGF and Ht31 or Ht31P was monitored as described before (14). As shown in Fig. 8E, TNF-α/CHX treatment resulted in a significant increase in DNA fragmentation. The antiapoptotic activity of EGF was significantly enhanced by Ht31 but not by Ht31P. To determine the contribution of RSK1 via the ERK kinase cascade, experiments similar to those whose results are shown in Fig. 8E were performed with the MEK inhibitor PD098059. As shown in Fig. 8E, the MEK inhibitor by itself caused some apoptosis that was further enhanced by TNF/CHX. However, EGF in the presence or absence of Ht31 did show any antiapoptotic effects. These data show that the Ht31-mediated increase in the antiapoptotic actions of EGF without the MEK inhibitor in Fig. 8E were due to redistribution of RSK1, the downstream kinase of the ERKs.
DISCUSSION
In this report, we describe four major and novel findings. First, we have shown that inactive RSK1 interacts directly with the RI subunit while active RSK1 directly interacts with PKAc. This is the first demonstration of an activation-dependent association of RSK1 with different subunits of the PKA holoenzyme. Moreover, by switching the partners that the inactive and active RSK1 associate with, irrespective of its activation state, RSK1 ensures its indirect association with PKA holoenzyme and, therefore, AKAPs. Because B82L cells express only the RIα subunit, in this report we have focused on the interactions of RSK1 with this subunit. However, given the similarity in the structures of the different regulatory subunit isoforms, it is possible that RSK1 may also interact with RII subunits.
A second important finding of our studies is that the associations between inactive RSK1 and RI and between active RSK1 and PKAc modulate the interactions between the subunits of the PKA holoenzyme. Thus, inactive RSK1, by interacting with RI, decreases the associations between PKAc and RI, whereas active RSK1, which associates with PKAc, increases the interactions between PKAc and RI. The increase in interactions between subunits of PKA resulting from the presence of active RSK1 also decreases the ability of cAMP to activate PKA (Fig. 5). These findings demonstrate one of the functional significances of the interactions we have unraveled and identify a novel mode of regulating PKA activity.
Although the regions on PKA subunits that RSK1 interacts with are not known, from the structure of the type I (RI-containing) PKA holoenzyme (25) it is apparent that upon association with PKAc, the RI subunit undergoes major conformation changes; also, the ordering of the linker region on RI is important in the RI/PKAc interactions (25). Because inactive RSK1, by interacting with RI, decreases the association of RI with PKAc, it is tempting to suggest either that RSK1 binds to the linker region of RI or that the binding of RSK1 to some other region of RI inhibits the ordering of the linker region on RI and, therefore, association with PKAc. On the other hand, active RSK1, by interacting with PKAc, may somehow alter the conformation of the large lobe where the major interactions with RI occur to facilitate the formation of the holoenzyme (25). Future studies which identify the interacting regions on RSK1 and PKA subunits will provide mechanistic insights into how RSK1 regulates the interactions between PKA subunits.
A third major functional role of the interactions between RSK1 and PKA subunits elucidated by our experiments is that the indirect interactions (via PKA subunits) of RSK1 with AKAPs are essential for the nuclear localization of active RSK1. Hence, when the interactions of RI with AKAPs are disrupted by Ht31, active RSK1 is excluded from the nucleus and accumulates in the cytoplasm (Fig. 7 and 8A). The resultant increase in cytosolic active RSK1 levels is accompanied by increased phosphorylation of its cytosolic substrates TSC-2 and BAD and an increased ability of EGF to protect against cellular apoptosis. These findings underscore the importance of the RSK1/PKA subunit interactions in regulating the cellular distribution and biological functions of RSK1.
Because AKAPs localize PKA in the proximity of its substrates (2, 52) the dissociation of PKA from AKAPs has been shown to decrease BAD phosphorylation (20). However, the dissociation of RSK1 from AKAPs increases its cytosolic content and phosphorylation of BAD (Fig. 8A and B). These findings show that the dissociation of proteins from scaffolds such as AKAPs, by increasing their concentrations in other compartments, can augment some signals. On the other hand, the exclusion of active RSK1 from the nucleus by dissociating the RSK1/PKA complex from AKAPs may decrease the phosphorylation of the nuclear substrates of RSK1 such as c-Fos, Mit1, Bub1, and histone 3. RSK1 also phosphorylates CREB on Ser133 (54). However this site on CREB is also phosphorylated by several other kinases, including Ca2+-calmodulin-dependent protein kinases, mitogen- and stress-activated protein kinase 1, and mitogen kinase-activated protein kinase-activated protein 2 (13, 34, 45). Because EGF, via increases in cytosolic free-Ca2+ levels, activates Ca2+-calmodulin-dependent protein kinases and downstream of mitogen-activated protein kinases can activate mitogen- and stress-activated protein kinase 1 and mitogen kinase-activated protein kinase-activated protein 2 (8, 13, 32, 40), we were unable to conclusively decipher changes in CREB phosphorylation on S133 when intracellular distribution of RSK1 was altered by the presence of Ht31 (not shown).
Since active RSK1 is colocalized in the nucleus and perinuclear region with PKAc and RI, an intriguing possibility that arises is that the nuclear AKAP7γ that binds RI (7) or some other, as-yet-unidentified, nuclear RI-specific AKAP may play a central role in the nuclear localization of RSK1. In this context, a recent study has shown that muscle-selective AKAP (mAKAP) binds and localizes PDK1, ERK, and RSK3 to the nuclear periphery and regulates the activation of RSK3 (31). However, it is unlikely that mAKAP is involved in the interactions we report since mAKAP is mainly expressed in muscle tissue. Secondly, RSK3, directly or indirectly, binds mAKAP (31) whereas in our experiments, neither RSK2 and nor RSK3 was present in IPs of PKAc (Fig. 1B). Nevertheless, in tissues such as myocardium that are rich in mAKAP and RSK1, it is possible that the mAKAP may also associate with RSK1 via PKA subunits. The mAKAP-mediated colocalization of RSK1, PKAc, ERK, and PDK1 at the periphery of the nucleus may be important in bringing these kinases in proximity of each other, especially because PDK1 plays an integral role in the activation of these two kinases (5, 12). In this regard, unlike the colocalization of ERK, PDK1, and RSK3 in the complex with mAKAP (31), we do not presently know whether ERK or PDK1 is also present in the complex of PKA subunits and RSK1 investigated here. Moreover, although in the studies of Michel et al. (31) activation of RSK3 by PDK1 released the enzyme from the mAKAP complex, we do not know whether ERK or PDK1 alters the interactions between PKA subunits and RSK1 studied here. Future experiments will address these possibilities.
The PKA subunits and RSK1 also interact with other proteins (see the introduction). Therefore, it is possible that these other interactions may also influence the ability of RSK1 to associate with PKA subunits in the cells and permit the spatial localization of RSK1 at its cellular targets. For instance, the interactions between RI and cytochrome c oxidase subunit Vc or PAP7 (28, 55) and between RI and RSK1 may localize RSK1 to the mitochondria to permit phosphorylation of BAD (23). In this respect, D-AKAP1, which specifically binds RI, also contains a mitochondrial localization sequence and could also permit the colocalization of RSK1 to the mitochondria. Future experimentation will elucidate whether the other proteins that interact with PKA subunits (see the introduction) also modulate the interactions between RSK1 and PKA subunits or regulate the spatiotemporal localization of RSK1 and augment the repertoire of signaling events.
A fourth important point that our studies brought forth is that in certain cell types in which cAMP, by increasing ERK1/2 activity, activates RSK1, both PKAc and active RSK1 reside as a complex. Because these kinases share common phosphorylation sites on certain substrates such as CREB, BAD, and Nur77, the phosphorylation of these substrates by agonists that increase intracellular cAMP content may be erroneously ascribed solely to PKAc. This could account for the ambiguity in the literature concerning the identity of the kinase that actually phosphorylates some of the common substrates of PKA and RSK1 (see examples with respect to Nur77 [21, 51] and Ser155 [29, 44, 58] phosphorylation of BAD).
In conclusion, herein, we report novel interactions of inactive and active RSK1 with RI and PKAc subunits of PKA, respectively. The functional significance of these interactions is demonstrated by the following. First, by modulating the interactions between PKAc and RI, the inactive and active RSK1 may alter the ability of cAMP to activate PKA. Indeed, in the presence of active RSK1, the Ka of cAMP for PKA activation is increased. This represents a novel mechanism for regulating PKA activity. Second, the indirect (via PKA subunit) interactions of active RSK1 with AKAPs are essential for its nuclear localization and phosphorylation of its substrates. Thus, when its interactions with AKAPs are disrupted, active RSK1 is excluded from the nucleus and its cytosolic content is augmented, with a resultant increase in phosphorylation of its cytosolic substrates such as BAD that enhance the antiapoptotic actions of EGF. Finally, in certain cell types in which cAMP activates RSK1, because RSK1 and PKAc are in the same complex, the identity of the kinase that phosphorylates their common substrates may be erroneously ascribed to PKAc.
Supplementary Material
Acknowledgments
We thank the following individuals for their help: Susan S. Taylor, University of California San Diego for the purified RIα, PKAc, constructs for GST-RPP7, GST-RPP8, and D-AKAP 1 antibody; John Shabb, University of North Dakota, Grand Forks, ND, for purified RI subunit; Rockland Immunochemicals Inc., for PA anti-lactate dehydrogenase conjugated to horseradish; and Don Bers and Sanda Despa (Loyola University, Chicago) for their help with confocal imaging. We are also indebted to Susan S. Taylor, Mary Hunzicker-Dunn (Northwestern University, Chicago), and Ravi Iyengar, Mt. Sinai Medical School, NY, for reading the original version of our manuscript and providing constructive criticisms.
This research was supported by grants GM 073181 and HL 59679 from the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abe, J., M. Okuda, Q. Huang, M. Yoshizumi, and B. C. Berk. 2000. Reactive oxygen species activate p90 ribosomal S6 kinase via Fyn and Ras. J. Biol. Chem. 275:1739-1748. [DOI] [PubMed] [Google Scholar]
- 2.Alto, N. M., and J. D. Scott. 2004. The role of A-kinase anchoring proteins in cAMP-mediated signal transduction pathways. Cell Biochem. Biophys. 40:201-208. [DOI] [PubMed] [Google Scholar]
- 3.Alto, N. M., J. Soderling, and J. D. Scott. 2002. Rab32 is an A-kinase anchoring protein and participates in mitochondrial dynamics. J. Cell Biol. 158:659-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbier, A. J., H. M. Poppleton, Y. Yigzaw, J. B. Mullenix, G. J. Wiepz, P. J. Bertics, and T. B. Patel. 1999. Transmodulation of epidermal growth factor receptor function by cyclic AMP-dependent protein kinase. J. Biol. Chem. 274:14067-14073. [DOI] [PubMed] [Google Scholar]
- 5.Biondi, R. M., A. Kieloch, R. A. Currie, M. Deak, and D. R. Alessi. 2001. The PIF-binding pocket in PDK1 is essential for activation of S6K and SGK, but not PKB. EMBO J. 20:4380-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjorbaek, C., Y. Zhao, and D. E. Moller. 1995. Divergent functional roles for p90rsk kinase domains. J. Biol. Chem. 270:18848-18852. [DOI] [PubMed] [Google Scholar]
- 7.Brown, R. L., S. L. August, C. J. Williams, and S. B. Moss. 2003. AKAP7gamma is a nuclear RI-binding AKAP. Biochem. Biophys. Res. Commun. 306:394-401. [DOI] [PubMed] [Google Scholar]
- 8.Cano, E., Y. N. Doza, R. Ben-Levy, P. Cohen, and L. C. Mahadevan. 1996. Identification of anisomycin-activated kinases p45 and p55 in murine cells as MAPKAP kinase-2. Oncogene 12:805-812. [PubMed] [Google Scholar]
- 9.Carr, D. W., Z. E. Hausken, I. D. Fraser, R. E. Stofko-Hahn, and J. D. Scott. 1992. Association of the type II cAMP-dependent protein kinase with a human thyroid RII-anchoring protein. Cloning and characterization of the RII-binding domain. J. Biol. Chem. 267:13376-13382. [PubMed] [Google Scholar]
- 10.Carr, D. W., R. E. Stofko-Hahn, I. D. Fraser, S. M. Bishop, T. S. Acott, R. G. Brennan, and J. D. Scott. 1991. Interaction of the regulatory subunit (RII) of cAMP-dependent protein kinase with RII-anchoring proteins occurs through an amphipathic helix binding motif. J. Biol. Chem. 266:14188-14192. [PubMed] [Google Scholar]
- 11.Chen, R. H., C. Sarnecki, and J. Blenis. 1992. Nuclear localization and regulation of erk- and rsk-encoded protein kinases. Mol. Cell. Biol. 12:915-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng, X., Y. Ma, M. Moore, B. A. Hemmings, and S. S. Taylor. 1998. Phosphorylation and activation of cAMP-dependent protein kinase by phosphoinositide-dependent protein kinase. Proc. Natl. Acad. Sci. USA 95:9849-9854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deak, M., A. D. Clifton, L. M. Lucocq, and D. R. Alessi. 1998. Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 17:4426-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng, W., H. Poppleton, S. Yasuda, N. Makarova, Y. Shinozuka, D. A. Wang, L. R. Johnson, T. B. Patel, and G. Tigyi. 2004. Optimal lysophosphatidic acid-induced DNA synthesis and cell migration but not survival require intact autophosphorylation sites of the epidermal growth factor receptor. J. Biol. Chem. 279:47871-47880. [DOI] [PubMed] [Google Scholar]
- 15.Diviani, D., J. Soderling, and J. D. Scott. 2001. AKAP-Lbc anchors protein kinase A and nucleates Gα12-selective Rho-mediated stress fiber formation. J. Biol. Chem. 276:44247-44257. [DOI] [PubMed] [Google Scholar]
- 16.Goldkorn, T., N. Balaban, K. Matsukuma, V. Chea, R. Gould, J. Last, C. Chan, and C. Chavez. 1998. EGF-receptor phosphorylation and signaling are targeted by H2O2 redox stress. Am. J. Respir. Cell Mol. Biol. 19:786-798. [DOI] [PubMed] [Google Scholar]
- 17.Gupte, R. S., P. Pozarowski, J. Grabarek, F. Traganos, Z. Darzynkiewicz, and M. Y. Lee. 2005. RIα influences cellular proliferation in cancer cells by transporting RFC40 into the nucleus. Cancer Biol. Ther. 4:429-437. [DOI] [PubMed] [Google Scholar]
- 18.Gupte, R. S., Y. Weng, L. Liu, and M. Y. Lee. 2005. The second subunit of the replication factor C complex (RFC40) and the regulatory subunit (RIα) of protein kinase A form a protein complex promoting cell survival. Cell Cycle 4:323-329. [PubMed] [Google Scholar]
- 19.Hanamoto, T., T. Ozaki, K. Furuya, M. Hosoda, S. Hayashi, M. Nakanishi, H. Yamamoto, H. Kikuchi, S. Todo, and A. Nakagawara. 2005. Identification of protein kinase A catalytic subunit beta as a novel binding partner of p73 and regulation of p73 function. J. Biol. Chem. 280:16665-16675. [DOI] [PubMed] [Google Scholar]
- 20.Harada, H., B. Becknell, M. Wilm, M. Mann, L. J. Huang, S. S. Taylor, J. D. Scott, and S. J. Korsmeyer. 1999. Phosphorylation and inactivation of BAD by mitochondria-anchored protein kinase A. Mol. Cell 3:413-422. [DOI] [PubMed] [Google Scholar]
- 21.Hirata, Y., K. Kiuchi, H. C. Chen, J. Milbrandt, and G. Guroff. 1993. The phosphorylation and DNA binding of the DNA-binding domain of the orphan nuclear receptor NGFI-B. J. Biol. Chem. 268:24808-24812. [PubMed] [Google Scholar]
- 22.Huang, L. J., K. Durick, J. A. Weiner, J. Chun, and S. S. Taylor. 1997. D-AKAP2, a novel protein kinase A anchoring protein with a putative RGS domain. Proc. Natl. Acad. Sci. USA 94:11184-11189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, L. J., K. Durick, J. A. Weiner, J. Chun, and S. S. Taylor. 1997. Identification of a novel protein kinase A anchoring protein that binds both type I and type II regulatory subunits. J. Biol. Chem. 272:8057-8064. [DOI] [PubMed] [Google Scholar]
- 24.Huang, L. J., L. Wang, Y. Ma, K. Durick, G. Perkins, T. J. Deerinck, M. H. Ellisman, and S. S. Taylor. 1999. NH2-terminal targeting motifs direct dual specificity A-kinase-anchoring protein 1 (D-AKAP1) to either mitochondria or endoplasmic reticulum. J. Cell Biol. 145:951-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, C., N. H. Xuong, and S. S. Taylor. 2005. Crystal structure of a complex between the catalytic and regulatory (RIα) subunits of PKA. Science 307:690-696. [DOI] [PubMed] [Google Scholar]
- 26.Kohler, K., D. Louvard, and A. Zahraoui. 2004. Rab13 regulates PKA signaling during tight junction assembly. J. Cell Biol. 165:175-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kusk, M., R. Ahmed, B. Thomsen, C. Bendixen, O. G. Issinger, and B. Boldyreff. 1999. Interactions of protein kinase CK2β subunit within the holoenzyme and with other proteins. Mol. Cell. Biochem. 191:51-58. [PubMed] [Google Scholar]
- 28.Li, H., B. Degenhardt, D. Tobin, Z. X. Yao, K. Tasken, and V. Papadopoulos. 2001. Identification, localization, and function in steroidogenesis of PAP7: a peripheral-type benzodiazepine receptor- and PKA (RIα)-associated protein. Mol. Endocrinol. 15:2211-2228. [DOI] [PubMed] [Google Scholar]
- 29.Lizcano, J. M., N. Morrice, and P. Cohen. 2000. Regulation of BAD by cAMP-dependent protein kinase is mediated via phosphorylation of a novel site, Ser155. Biochem. J. 349:547-557. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.McDonald, C., P. O. Vacratsis, J. B. Bliska, and J. E. Dixon. 2003. The yersinia virulence factor YopM forms a novel protein complex with two cellular kinases. J. Biol. Chem. 278:18514-18523. [DOI] [PubMed] [Google Scholar]
- 31.Michel, J. J., I. K. Townley, K. L. Dodge-Kafka, F. Zhang, M. S. Kapiloff, and J. D. Scott. 2005. Spatial restriction of PDK1 activation cascades by anchoring to mAKAPα. Mol. Cell 20:661-672. [DOI] [PubMed] [Google Scholar]
- 32.Nishibe, S., M. I. Wahl, S. M. Hernandez-Sotomayor, N. K. Tonks, S. G. Rhee, and G. Carpenter. 1990. Increase of the catalytic activity of phospholipase C-gamma 1 by tyrosine phosphorylation. Science 250:1253-1256. [DOI] [PubMed] [Google Scholar]
- 33.Patel, T. B. 2004. Single transmembrane spanning heterotrimeric G protein-coupled receptors and their signaling cascades. Pharmacol. Rev. 56:371-385. [DOI] [PubMed] [Google Scholar]
- 34.Ribar, T. J., R. M. Rodriguiz, L. Khiroug, W. C. Wetsel, G. J. Augustine, and A. R. Means. 2000. Cerebellar defects in Ca2+/calmodulin kinase IV-deficient mice. J. Neurosci. 20:RC107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richards, S. A., V. C. Dreisbach, L. O. Murphy, and J. Blenis. 2001. Characterization of regulatory events associated with membrane targeting of p90 ribosomal S6 kinase 1. Mol. Cell. Biol. 21:7470-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rolfe, M., L. E. McLeod, P. F. Pratt, and C. G. Proud. 2005. Activation of protein synthesis in cardiomyocytes by the hypertrophic agent phenylephrine requires the activation of ERK and involves phosphorylation of tuberous sclerosis complex 2 (TSC2). Biochem. J. 388:973-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roux, P. P., B. A. Ballif, R. Anjum, S. P. Gygi, and J. Blenis. 2004. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc. Natl. Acad. Sci. USA 101:13489-13494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rudolph, J. A., J. L. Poccia, and M. B. Cohen. 2004. Cyclic AMP activation of the extracellular signal-regulated kinases 1 and 2: implications for intestinal cell survival through the transient inhibition of apoptosis. J. Biol. Chem. 279:14828-14834. [DOI] [PubMed] [Google Scholar]
- 39.Sapkota, G. P., A. Kieloch, J. M. Lizcano, S. Lain, J. S. Arthur, M. R. Williams, N. Morrice, M. Deak, and D. R. Alessi. 2001. Phosphorylation of the protein kinase mutated in Peutz-Jeghers cancer syndrome, LKB1/STK11, at Ser431 by p90RSK and cAMP-dependent protein kinase, but not its farnesylation at Cys433, is essential for LKB1 to suppress cell growth. J. Biol. Chem. 276:19469-19482. [DOI] [PubMed] [Google Scholar]
- 40.Schultz, H., K. Engel, and M. Gaestel. 1997. PMA-induced activation of the p42/44ERK- and p38RK-MAP kinase cascades in HL-60 cells is PKC dependent but not essential for differentiation to the macrophage-like phenotype. J. Cell Physiol. 173:310-318. [DOI] [PubMed] [Google Scholar]
- 41.Skalhegg, B. S., and K. Tasken. 1997. Specificity in the cAMP/PKA signaling pathway. Differential expression, regulation, and subcellular localization of subunits of PKA. Front. Biosci. 2:d331-d42. [DOI] [PubMed] [Google Scholar]
- 42.Stork, P. J., and J. M. Schmitt. 2002. Crosstalk between cAMP and MAP kinase signaling in the regulation of cell proliferation. Trends Cell Biol. 12:258-266. [DOI] [PubMed] [Google Scholar]
- 43.Sundaresan, M., Z. X. Yu, V. J. Ferrans, K. Irani, and T. Finkel. 1995. Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science 270:296-299. [DOI] [PubMed] [Google Scholar]
- 44.Tan, Y., M. R. Demeter, H. Ruan, and M. J. Comb. 2000. BAD Ser-155 phosphorylation regulates BAD/Bcl-XL interaction and cell survival. J. Biol. Chem. 275:25865-25869. [DOI] [PubMed] [Google Scholar]
- 45.Tan, Y., J. Rouse, A. Zhang, S. Cariati, P. Cohen, and M. J. Comb. 1996. FGF and stress regulate CREB and ATF-1 via a pathway involving p38 MAP kinase and MAPKAP kinase-2. EMBO J. 15:4629-4642. [PMC free article] [PubMed] [Google Scholar]
- 46.Tortora, G., and F. Ciardiello. 2002. Protein kinase A type I: a target for cancer therapy. Clin. Cancer Res. 8:303-304. [PubMed] [Google Scholar]
- 47.Tortora, G., V. Damiano, C. Bianco, G. Baldassarre, A. R. Bianco, L. Lanfrancone, P. G. Pelicci, and F. Ciardiello. 1997. The RIα subunit of protein kinase A (PKA) binds to Grb2 and allows PKA interaction with the activated EGF-receptor. Oncogene 14:923-928. [DOI] [PubMed] [Google Scholar]
- 48.Vaidyanathan, H., and J. W. Ramos. 2003. RSK2 activity is regulated by its interaction with PEA-15. J. Biol. Chem. 278:32367-32372. [DOI] [PubMed] [Google Scholar]
- 49.Vijayaraghavan, S., K. D. Trautman, S. A. Goueli, and D. W. Carr. 1997. A tyrosine-phosphorylated 55-kilodalton motility-associated bovine sperm protein is regulated by cyclic adenosine 3′,5′-monophosphates and calcium. Biol. Reprod. 56:1450-1457. [DOI] [PubMed] [Google Scholar]
- 50.Wang, L., R. K. Sunahara, A. Krumins, G. Perkins, M. L. Crochiere, M. Mackey, S. Bell, M. H. Ellisman, and S. S. Taylor. 2001. Cloning and mitochondrial localization of full-length D-AKAP2, a protein kinase A anchoring protein. Proc. Natl. Acad. Sci. USA 98:3220-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wingate, A. D., D. G. Campbell, M. Peggie, and J. S. Arthur. 2006. Nur77 is phosphorylated in cells by RSK in response to mitogenic stimulation. Biochem. J. 393:715-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong, W., and J. D. Scott. 2004. AKAP signalling complexes: focal points in space and time. Nat. Rev. Mol. Cell Biol. 5:959-970. [DOI] [PubMed] [Google Scholar]
- 53.Wu, J., and R. Janknecht. 2002. Regulation of the ETS transcription factor ER81 by the 90-kDa ribosomal S6 kinase 1 and protein kinase A. J. Biol. Chem. 277:42669-42679. [DOI] [PubMed] [Google Scholar]
- 54.Xing, J., J. M. Kornhauser, Z. Xia, E. A. Thiele, and M. E. Greenberg. 1998. Nerve growth factor activates extracellular signal-regulated kinase and p38 mitogen-activated protein kinase pathways to stimulate CREB serine 133 phosphorylation. Mol. Cell. Biol. 18:1946-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang, W. L., L. Iacono, W. M. Tang, and K. V. Chin. 1998. Novel function of the regulatory subunit of protein kinase A: regulation of cytochrome c oxidase activity and cytochrome c release. Biochemistry 37:14175-14180. [DOI] [PubMed] [Google Scholar]
- 56.Yusta, B., J. Estall, and D. J. Drucker. 2002. Glucagon-like peptide-2 receptor activation engages bad and glycogen synthase kinase-3 in a protein kinase A-dependent manner and prevents apoptosis following inhibition of phosphatidylinositol 3-kinase. J. Biol. Chem. 277:24896-24906. [DOI] [PubMed] [Google Scholar]
- 57.Zhong, H., H. SuYang, H. Erdjument-Bromage, P. Tempst, and S. Ghosh. 1997. The transcriptional activity of NF-κB is regulated by the IkappaB-associated PKAc subunit through a cyclic AMP-independent mechanism. Cell 89:413-424. [DOI] [PubMed] [Google Scholar]
- 58.Zhou, X. M., Y. Liu, G. Payne, R. J. Lutz, and T. Chittenden. 2000. Growth factors inactivate the cell death promoter BAD by phosphorylation of its BH3 domain on Ser155. J. Biol. Chem. 275:25046-25051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.