Abstract
Small ubiquitin-related modifier (SUMO) proteins are conjugated to numerous polypeptides in cells, and attachment of SUMO plays important roles in regulating the activity, stability, and subcellular localization of modified proteins. SUMO modification of proteins is a dynamic and reversible process. A family of SUMO-specific proteases catalyzes the deconjugation of SUMO-modified proteins. Members of the Sentrin (also known as SUMO)-specific protease (SENP) family have been characterized with unique subcellular localizations. However, little is known about the functional significance of or the regulatory mechanism derived from the specific localizations of the SENPs. Here we identify a bipartite nuclear localization signal (NLS) and a CRM1-dependent nuclear export signal (NES) in the SUMO protease SENP2. Both the NLS and the NES are located in the nonhomologous domains of SENP2 and are not conserved among other members of the SENP family. Using a series of SENP2 mutants and a heterokaryon assay, we demonstrate that SENP2 shuttles between the nucleus and the cytoplasm and that the shuttling is blocked by mutations in the NES or by treating cells with leptomycin B. We show that SENP2 can be polyubiquitinated in vivo and degraded through proteolysis. Restricting SENP2 in the nucleus by mutations in the NES impairs its polyubiquitination, whereas a cytoplasm-localized SENP2 made by introducing mutations in the NLS can be efficiently polyubiquitinated, suggesting that SENP2 is ubiquitinated in the cytoplasm. Finally, treating cells with MG132 leads to accumulation of polyubiquitinated SENP2, indicating that SENP2 is degraded through the 26S proteolysis pathway. Thus, the function of SENP2 is regulated by both nucleocytoplasmic shuttling and polyubiquitin-mediated degradation.
Reversible posttranslational modification plays an important role in regulating functions of the modified proteins by altering their activity, stability, and localization, as well as their interaction with other molecules. Some modifications consist of small chemical groups, such as phosphate, acetate, lipids, and sugar, that are added to specific amino acids. Another type of modification is the covalent attachment of other polypeptides to the target proteins. The most well known example of such polypeptide modifiers is ubiquitin (Ub), a modifier playing a major role in targeting proteins for proteolytic degradation (13). The group of small ubiquitin-related modifiers (SUMO), like ubiquitin, covalently modify a large number of proteins and play important roles in many cellular processes, including the regulation of gene expression, histone modification, DNA repair, and nucleocytoplasmic shuttling (18). Three members of the SUMO family in vertebrates have been described: SUMO-1 and its close homologs SUMO-2 and SUMO-3. All three can be linked to substrate proteins through a process called sumoylation. The mechanism of sumoylation is very similar to that of ubiquitination, which requires sequential actions of an E1-activating enzyme, an E2-conjugating enzyme, and an E3-ligating enzyme (9, 22). Recently, a fourth member of SUMO (SUMO-4) was discovered, and its expression appears to be restricted to the kidneys (5, 11).
Sumoylation is a dynamic and reversible process. Sentrin (also known as SUMO)-specific proteases (SENPs) participate in both the processing of immature SUMO proteins and the removal of SUMO conjugates from SUMO-modified substrates (23). The first SUMO-specific protease identified was Ulp1, a yeast protein found to be essential for the G2/M cell cycle transition (17). To date, at least seven mammalian orthologs of Ulp1 have been identified (23), and at least four of them have been confirmed to have SUMO protease activity (19). The proteases identified so far possess several interesting features. All contain the conserved catalytic triad in the C-terminal domain consisting of several highly conserved amino acids (cysteine, histidine, and aspartic acid). They possess a large N-terminal domain with minimal or no homology to each other's domain. It has been suggested that the diversified N-terminal domains of SENP proteases determine their substrate specificity by controlling their subcellular localization (2, 12, 24). Mammalian SENPs are found in many different subcellular compartments: the nucleoplasm, nucleolus, nuclear envelope, nuclear bodies, and cytoplasm. It has been shown that SENP1 localizes to the nucleoplasm (10), SENP2 localizes to the nuclear side of the nuclear pore complex (NPC) (24), SENP3 localizes to the nucleolus (20), and SENP6 localizes predominantly to the cytoplasm (16). Although very little is known about the substrate specificity of each of the SENP enzymes, their distinct subcellular localizations have been postulated to play important roles in their function, particularly in substrate selection (19, 23). Nevertheless, how the unique subcellular localization of each SENP is regulated remains unknown.
It has been shown that the N-terminal 63 amino acid residues are required for SENP2 to localize to the NPC (24), indicating that they may contain the nuclear localization signal (NLS) sequence. The same N-terminal area in SENP1, however, is not essential for its nuclear import. Instead, the NLS of SENP1 is found at amino acid residues 171 to 177 (2). The different locations of the NLS in SENP1 and SENP2 underscore the role of the N-terminal domains of SENP enzymes in their diversified cellular functions. Here, we report the identification of a bipartite NLS sequence in the N-terminal domain and a leucine-rich, CRM1-dependent nuclear export signal (NES) sequence in the central domain of SENP2. Consistent with a protein containing both an NLS and an NES, our data demonstrate that SENP2 shuttles between the nucleus and the cytoplasm. Intriguingly, we found that SENP2 can be polyubiquitinated in vivo and that the polyubiquitination can be impaired by restricting the protein's localization to the nucleus. Together, our data indicate that nucleocytoplasmic shuttling is a crucial regulatory mechanism for SENP2 function.
MATERIALS AND METHODS
Plasmids.
The constructs of Flag-tagged SENP2, His-tagged PML and RanGAP1, and hemagglutinin (HA)-tagged SUMO-1 and SUMO-3 were described previously (7). A catalytically inactive SENP2 mutant (SENP2Cat) (R576L, K577M) was prepared by standard cloning and PCR-based mutagenesis. SENP2 cDNA from Flag-tagged constructs was subcloned into pcDNA3 (Invitrogen), which was engineered to contain an in-frame amino-terminal Myc tag to generate Myc-tagged SENP2. SENP2 enzymes with mutations in the NLS and NES (SENP2mNLS and SENP2mNES, respectively) were prepared by standard cloning and PCR-based mutagenesis and verified by DNAsequencing. The oligonucleotides for generating the NES-yellow fluorescent protein (YFP) fusions were synthesized commercially (Invitrogen) [CTAGCATGTACAGATGGCTGGTTAGGATTCTCGGCACCATTTTCCGTTTCGTAC and GAAACGGAAAATGGTGCCGAGAATCCTAACCAGCCATCTGTACATG for SENP2(1-15), CTAGCATGCGCAGAGTCCTGCCTTCCTTTGGTTTTACTTTGAACTCAGTAC and TGAGTTCAAAGTAAAACCAAAGGAAGGCAGGACTCTGCGCATG for SENP2(143-155), CTAGCATGCTGGAGCCTGACCTATCAGAAGAAGTGTCGGCCCGACTCCGCCTGGGCGTAC and GCCCAGGCGGAGTCGGGCCGACACTTCTTCTGATAGGTCAGGCTCCAGCATG for SENP2(317-332), CTAGCATGTACAGATGGCTGGTTAGGGCTGCCGGCACCATTTTCCGTTTCGTAC and GAAACGGAAAATGGTGCCGGCAGCCCTAACCAGCCATCTGTACATG for the SENP2(1-15) mutant, and CTAGCATGCTGGAGCCTGACCTATCAGAAGAAGTGTCGGCCCGAGCTCGCGCCGGCGTAC and GCCGGCGCGAGCTCGGGCCGACACTTCTTCTGATAGGTCAGGCTCCAGCATG for the SENP2(317-332) mutant]. The oligonucleotide pairs were denatured at 95°C for 3 min, annealed by slowly cooling them to room temperature for 30 min, and inserted into the NheI and KpnI sites of the N terminus of the YFP vector to generate in-frame YFP fusion proteins as described previously (21). SENP2 wild-type and SENP2mNES mutant cDNAs from Myc-tagged constructs were subcloned into the pcDNA-EGFP-C4 vector (Invitrogen) to construct green fluorescent protein (GFP) fusion proteins. The cDNA fragments of SENP2 deletion mutants were amplified by PCR using the following primers: AAAAAGATCTGTGCTGCCAGCTTATTTGGATTCCCAT (used as a forward primer for all deletion mutants), CCCGTCTAGATCATAGGGCATTACTGATTTCCTTTTCCATG [used as a reverse primer for the SENP2(66-380) mutant], CCCGTCTAGATCAAGTAATTCGCAATTTGAAAGCACTACTTAGGATT [used as a reverse primer for the SENP2(66-398) mutant], and GCCCTCTAGATCACAGCAACTGCTGATGAAGGATTTC [used as a reverse primer for the SENP2(66-589) mutant]. pcDNA3-Myc-SENP2 wild-type or SENP2mNES mutant plasmids were used for templates and PCR products cloned into the pcDNA3-EGFP-C4 vector. The GFP plasmid construct loading control for transfection efficiency was purchased from BD Bioscience Clontech (pEGFP-N1; catalog no. 6085-1). HA-tagged Ub (HA-Ub) is a gift from Yue Xiong (The University of North Carolina, Chapel Hill, NC).
Cell lines and cell culture.
The human osteosarcoma U2OS cell line and human lung tumor H1299 cell line were purchased from the ATCC. Mouse embryonic fibroblast (MEF) cells were isolated in the lab. All cells were routinely maintained in a 37°C incubator with 5% CO2 in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. In all cases, cells were transfected using Effectene transfection reagent (QIAGEN) or Fugene6 (Roche) according to the manufacturer's instructions.
In vivo sumoylation assay.
Cells were lysed in 2% sodium dodecyl sulfate (SDS) lysis buffer (2% [wt/vol] SDS, 50 mM Tris-HCl [pH 6.8], 10% glycerol). Two percent SDS lysis buffer was preheated to 95°C before being added to the cells to protect SUMO-conjugated proteins from desumoylation during protein preparation. Proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose membranes. The blots were blocked in 5% nonfat dried milk in PBST (phosphate-buffered saline [PBS] containing 0.1% [vol/vol] Tween 20) and incubated with primary antibodies and horseradish peroxidase-labeled secondary antibodies in the blocking buffer. The blots were thoroughly rinsed with PBST after each incubation. Antibodies were detected with chemiluminescence detection reagents according to the manufacturer's instructions (Pierce). Flag (M2; Sigma), Myc (9E10.3; NeoMarkers), HA (16B12; COVANCE), His (RGS-His; QIAGEN), actin (Chemicon), Ku80 (NeoMarkers), and GFP (Research Diagnostics) antibodies were purchased. Rabbit anti-Myc antibody was a gift from Yue Xiong (University of North Carolina, Chapel Hill, NC). GFP was used as a loading control for transfection efficiency.
In vivo ubiquitination assay.
H1299 cells were transiently transfected with Myc-tagged SENP2 along with HA-tagged ubiquitin plasmid DNA. GFP plasmid was cotransfected as a control for transfection efficiency. Two days after transfection, cells were treated with 20 μM MG132 for 6 h and then lysed in 1% SDS lysis buffer (1% [wt/vol] SDS and 1% [wt/vol] NP-40 in PBS). The cell lysates were diluted 10 times with 0.1% NP-40-PBS containing 1 mM phenylmethylsulfonyl fluoride (PMSF). The diluted lysates were precleaned with Sepharose CL4B beads (Sigma) for 30 min and then immunoprecipitated with rabbit anti-Myc antibody overnight at 4°C, followed by incubation with protein A/G beads (Pierce) for 2 h at 4°C. The beads were washed four times with cold 0.1% NP-40-PBS containing 1 mM PMSF. The beads were then incubated in 1× SDS loading buffer (100 mM Tris-HCl [pH 6.8], 2% [wt/vol] SDS, 0.2% bromophenol blue, 10% glycerol, 100 mM dithiothreitol) at 100°C for 2 min, followed by SDS-PAGE. After electrophoresis, the proteins were transferred to nitrocellulose membranes. The membranes were blocked in 5% nonfat dry milk in PBST and incubated with primary antibodies and horseradish peroxidase-labeled secondary antibodies in the blocking buffer. Antibodies were detected with chemiluminescence detection reagents according to the manufacturer's instructions (Pierce).
Protein half-life assay.
H1299 cells were transfected with Myc-tagged SENP2 constructs. Twenty-four hours after transfection, cells were pulsed with [35S]methionine for 2 h and then chased for the lengths of time indicated in Fig. 8A and B. Cells were collected and lysed in 0.5% NP-40 lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5% [wt/vol] NP-40, 50 mM NaF, 1 mM NaVO3, 1 mM dithiothreitol, 1 mM PMSF, protease inhibitors). The cell lysates were immunoprecipitated with mouse anti-Myc antibody (9E10) overnight and with protein A/G for 1 h at 4°C. The beads were washed three times with 0.5% NP-40 lysis buffer. The resulting SENP2 immunoprecipitates were separated by SDS-PAGE and visualized by autoradiography. The amount of labeled SENP2 protein at each time point was quantified on a PhosphorImager and normalized relative to the amount of radiolabeled SENP2 present in cells following the 0 h chase.
FIG. 8.
SENP2 is degraded through polyubiquitination-mediated proteolysis. (A) Half-life assay of SENP2. H1299 cells were transfected with plasmid DNA encoding wild-type Myc-tagged SENP2 (WT) or SENP2 mutants. Twenty-four hours after transfection, cells were pulse-labeled with [35S]methionine for 2 h and chased for the indicated lengths of time. Cell lysates were immunoprecipitated with anti-Myc antibody. The resulting SENP2 immunoprecipitates were separated by SDS-PAGE and visualized by autoradiography. (B) Half-life assay of SENP2 as described for panel A except with shorter time intervals, as indicated. The amount of labeled SENP2 protein at each time point was quantified on a PhosphorImager and normalized relative to the amount of radiolabeled SENP2 present in cells following the 0 h chase. Average results of four independent experiments were plotted in panel C. (D) In vivo ubiquitination assay of SENP2. H1299 cells were transfected with the indicated plasmid DNA. The GFP plasmid was also cotransfected as a control for transfection efficiency. The SENP2 constructs were transfected with wild-type Myc-SENP2-SENPmNLS-SENPmNES at a ratio of 1:1.5:0.7 to obtain equal levels of SENP2 protein expression. Two days after transfection, cells were treated with 20 μM MG132 for 6 h. Cells were lysed in 1% SDS lysis buffer and diluted 10 times with 0.1% NP-40-PBS. Diluted lysates were immunoprecipitated (IP) with anti-Myc antibody. The resulting SENP2 immunoprecipitates were separated by SDS-PAGE and immunoblotted (IB) with the indicated antibodies (α). K, in thousands; [Ub]n-SENP2, polyubiquitinated SENP2; expo, exposure. (E) Localization of wild-type SENP2 or its mutants after the treatment with MG132. U2OS cells were transfected with plasmid DNA encoding Myc-tagged SENP2. Twenty-four hours after transfection, cells were treated with 20 μM MG132 for 10 h, fixed, and immunostained with anti-Myc antibody, followed by staining with Cy2-conjugated secondary antibody.
Indirect immunofluorescence.
Cultured cells were seeded in six-well plates and transfected with plasmid DNA. Twenty-four hours after transfection, cells were washed three times with PBS and fixed in PBS containing 4% formaldehyde for 10 min. After fixation, cells were washed twice with PBS, permeabilized in cold PBS containing 0.2% Triton X-100 for 5 min, and incubated with blocking buffer (PBS containing 0.5% bovine serum albumin) for 30 min prior to incubation with the primary antibody in the blocking buffer for 1 h. The cells were then incubated with Cy2-conjugated or rhodamine-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) for 30 min. Cells were washed three times with PBS and finally counterstained with DAPI (4′,6-diamidino-2-phenylindole). Stained cells were covered by antifluorescent mounting medium (DAKO) and examined with an Olympus model IX81 microscope fitted with appropriate fluorescence filters.
Heterokaryon assay.
Heterokaryon assays were carried out essentially as described previously (25). Human U2OS cells were transfected with plasmid DNA expressing the GFP-SENP2 fusion proteins. After 12 h of incubation with the plasmid, cells were washed twice with prewarmed PBS to remove untransfected DNA. The cells were then trypsinized, mixed with wild-type MEF cells at a 1:1 ratio, and seeded onto a new dish. After another 12 h of incubation, the cells were rinsed once with PBS and covered with a solution of 50% (wt/vol) PEG 8000 (Sigma-Aldrich) for 2 min at 37°C to induce cell fusion. After being washed three times with PBS to remove the PEG, fused cells were continuously cultured in Dulbecco's modified Eagle's medium-10% fetal bovine serum medium for 5 h. Cycloheximide (50 μg/ml; Sigma-Aldrich) was added to the culture 15 min before and immediately after cell fusion to block de novo protein synthesis. Subsequent cell fixation, permeabilization, and immunostaining followed standard immunofluorescence procedures. The cells were stained with a mouse antibody recognizing human but not murine Ku nuclear antigen to distinguish human and murine nuclei. Ku80 antibody was purchased from NeoMarkers.
RESULTS
Identification of a bipartite NLS in the N terminus of SENP2.
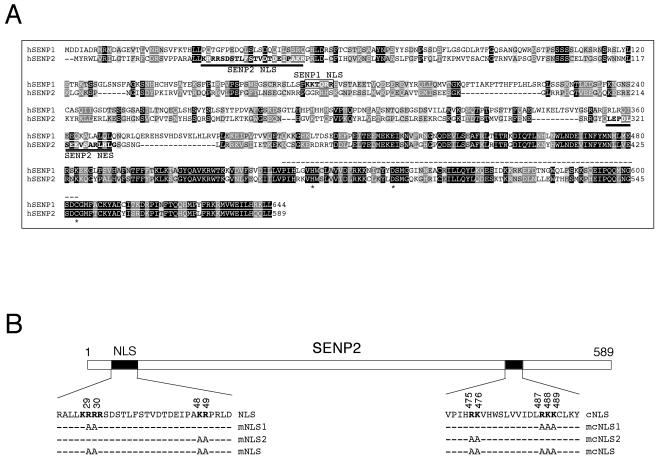
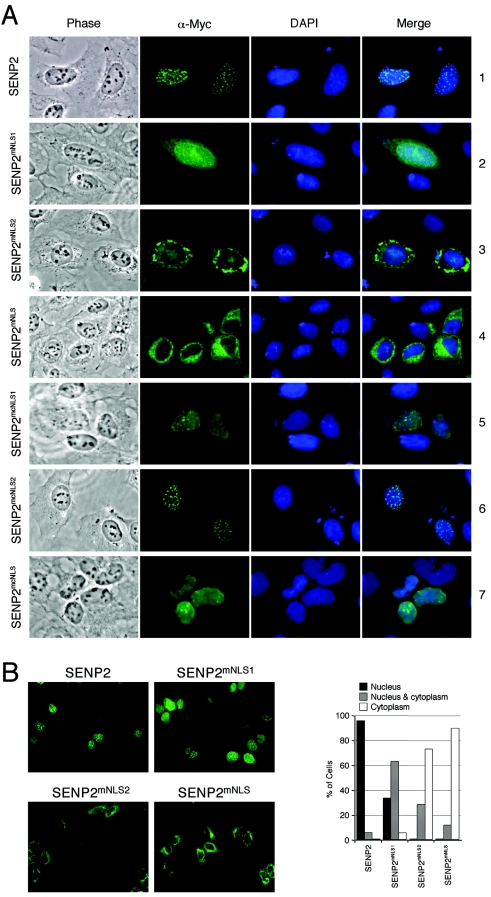
It has been shown recently that SENP1 has an NLS (KKTQRR; see the boxed area in Fig. 1A) that is required for SENP1 to enter the nucleus (2). However, sequence alignment indicates that the SENP1 NLS is not conserved in other SENP proteins, including SENP2 (Fig. 1A and data not shown). It has been previously shown that SENP2 localizes to the inner nuclear membrane side of the NPC and that the N-terminal 63 amino acid residues of SENP2 are required for its nuclear localization (24), indicating that an NLS may exist within this sequence. Inspection of this area identified a stretch of amino acids (KRRR) at residues 28 to 31 that resembles a basic amino acid-rich NLS (Fig. 1B). To determine whether this is the NLS of SENP2, we constructed a SENP2 mutant where the middle two Arg residues were replaced with Ala (KRRR→KAAR; SENP2mNLS1) and tested the localization of the mutant by transient transfection and immunofluorescence staining of U2OS cells. As shown in Fig. 2A, while there was no cytoplasmic staining in the wild-type-SENP2-transfected cells (row 1), cytoplasmic staining was clearly seen in the SENP2mNLS1-transfected cells (row 2), indicating that the KRRR sequence contributes to SENP2 nuclear localization. However, the localization of the SENP2mNLS1 mutant was still predominantly nuclear, indicating that even though the KRRR sequence constitutes an NLS, the mutations only partially affected its function. Further inspection of the SENP2 N-terminal sequence revealed a pair of basic amino acids (KR) (Fig. 1B) 16 residues downstream of the KRRR sequence that, when combined with the KRRR residues (KRRR-X16-KR), would fit the consensus of a classic bipartite NLS (6). To determine whether the KRRR-X16-KR sequence constitutes a bipartite NLS, we generated two SENP2 mutants; SENP2mNLS2 contains a KR→AA mutation, and SENP2mNLS combines the KRRR→KAAR and KR→AA mutations (Fig. 1B). We then examined their localization in transiently transfected U2OS cells by immunofluorescence staining. The SENP2mNLS2 (KR→AA) mutant exhibited considerable cytoplasmic localization (Fig. 2A, row 3), indicating that the mutations significantly affected the function of NLS. When both parts of the putative NLS were mutated (KRRR→KAAR and KR→AA), the mutant (SENP2mNLS) was exclusively localized in the cytoplasm (Fig. 2A, row 4). Hence, we conclude that the KRRR-X16-KR sequence constitutes a bipartite NLS in SENP2. A quantitative summary of the mutational analysis is shown in Fig. 2B. The data presented here are representative of analysis of at least 200 positively transfected cells from six independent experiments. Notably, the SENP2mNLS2 mutant exhibited stronger cytoplasmic staining than did the SENP2mNLS1 mutant, suggesting that the two parts of the bipartite NLS do not contribute equally to the nuclear import activity of SENP2. Alternatively, the two remaining basic amino acids in the SENP2mNLS1 mutant (KAAR) might retain some nuclear import activity. We further tested another NLS-like sequence at residues 475 to 489 of SENP2 that also fits the consensus of a bipartite NLS (Fig. 1B) for its potential nuclear import activity. Mutational analysis indicated that this C-terminally located sequence is not a functional NLS. Instead, for reasons yet unknown, the SENP2 mutant (SENP2mcNLS, where the “c” indicates the C-terminal region) showed diminished speckle formation, and the mutant SENP2 was rather toxic to cells as judged by deformed nuclei and increased cell death (Fig. 2A, rows 5 to 7 and data not shown). Since these mutations fall within the catalytic core region, consisting of a His-Asp-Cys triad (Fig. 1A), the possibility of potential structural and/or enzymatic activity alterations caused by the mutations cannot be ruled out.
FIG. 1.
A sequence alignment of human SENP1 (hSENP1) and SENP2 potentially reveals NLS and NES in SENP2 in unconserved domains. (A) Amino acid sequence alignment of human SENP1 and SENP2 using the CLUSTAL W (version 1.81) multiple-sequence alignment program. The fully conserved residues and the conservation of strong groups are indicated by a black background and a light-gray background, respectively. The NLS and the NES sequences of SENP2 are indicated with underlines. The NLS of SENP1 is indicated with a box. Key amino acid residues involved in the catalytically active site are marked as asterisks. The conserved domain of SENP family proteins is shown with dotted lines above the sequence. (B) Diagram of human SENP2 with positions and amino acid sequences of the two predicted bipartite NLSs. The functionally important basic amino acids of each NLS are shown in boldface. Alanine substitutions to generate SENP2 NLS mutants are indicated. cNLS, C-terminally located putative NLS.
FIG. 2.
Identification of a bipartite NLS in SENP2. (A) Localization of wild-type SENP2 and SENP2 NLS mutants. U2OS cells were transfected with the indicated Myc-tagged SENP2 plasmids. Twenty-four hours after transfection, cells were fixed and immunostained with an anti-Myc antibody (α-Myc; 9E10.3), followed by staining with Cy2-conjugated secondary antibody. Immunofluorescent, phase-contrast, and DAPI-stained images are shown. (B) Quantification of SENP2 mutants in nuclear and cytoplasmic fractions. Low-magnification pictures of immunostained SENP2 are shown. The graphs show the percentage of cells in each subcellular fraction as determined by counting at least 200 cells in each sample.
Identification of a CRM1-dependent NES in SENP2.
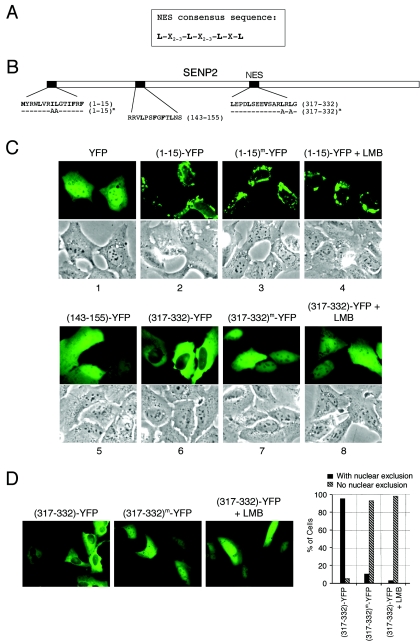
We further examined whether SENP2 contains an NES. Proteins undertaking nuclear export usually have a consensus sequence comprising several hydrophobic amino acid residues (Leu, Ile, Phe, etc.) separated by defined spaces (Fig. 3A) (4). Visual examination of the SENP2 sequence found three potential NES sequences that fit the consensus (Fig. 3B). To determine whether any of these sequences comprise a functional NES, we employed a previously described technique (25) by attaching short peptides representing each of the three putative NES domains to the N terminus of YFP and examining the localization of the fusion proteins in transiently transfected cells. In the absence of a functional NLS and NES, a “naked” YFP is passively diffused throughout the nucleus and the cytoplasm, with mild accumulation in the nucleus (Fig. 3C, panel 1). When each of the NES-like peptides was fused with YFP, the fusion of residues 143 to 155 with YFP [(143-155)-YFP] did not alter YFP's diffused localization pattern, indicating that it is not a functional NES in the context of YFP (Fig. 3C, panel 5). The (1-15)-YFP and the (317-332)-YFP fusions, on the other hand, displayed nuclear exclusion, suggesting that they may be functional NES domains in the context of YFP (Fig. 3C, panels 2 and 6). To further confirm whether these NES sequences are genuine, leucine-rich, CRM1-dependent NES domains, we introduced mutations into putative essential leucine residues or treated cells with the CRM1 inhibitor leptomycin B (LMB) and examined fusion protein localization in live cells. Mutations of the leucine residues or treatment of the cells with LMB did not affect the nuclear exclusion of (1-15)-YFP (Fig. 3C, panels 3 and 4) but completely abrogated the nuclear exclusion of (317-332)-YFP (Fig. 3C, panels 7 and 8), demonstrating that amino acid residues 317 to 332 comprise an autonomous CRM1-dependent NES but that the sequence within residues 1 to 15 does not. The cause of nuclear exclusion of the (1-15)-YFP fusion protein remains unclear. A quantitative summary of the mutational analysis completed by randomly scoring more than 200 cells positively transfected with the NES-YFP fusion is shown in Fig. 3D. (The data shown here represent one of three independent experiments). Taken together, these results demonstrate that SENP2 contains an autonomous NES, at least in the context of a YFP fusion, in the nonconserved central domain of the protein (residues 317 to 332) that functions through the CRM1-dependent nuclear export pathway.
FIG. 3.
Identification of a CRM1-dependent NES in SENP2. (A) The consensus sequence for the CRM1-dependent nuclear export signal is shown; the conserved hydrophobic amino acid can be leucine (L), isoleucine (I), methionine (M), or phenylalanine (F). (B) Diagram of SENP2 with the positions and amino acid sequences of three NES-like sites indicated. Alanine substitutions for NES functional essential hydrophobic residues tested in the study are indicated. Superscript “m” indicates a mutant protein. (C) The SENP2 NES is a CRM1-dependent NES. Short peptides of the putative SENP2 NES were fused with a YFP vector, and localization of the fusion proteins was observed in transfected U2OS cells. Transfected cells were also treated with 10 nM LMB for 7 h to determine possible CRM1 dependency. Pictures were taken with living cells 24 h after transfection. Corresponding phase-contrast pictures are shown. (D) Quantification of cells. Low-magnification pictures of transfected cells are shown. The graph shows the percentages of cells with nuclear exclusion as determined by counting at least 200 cells.
SENP2 is a shuttling protein.
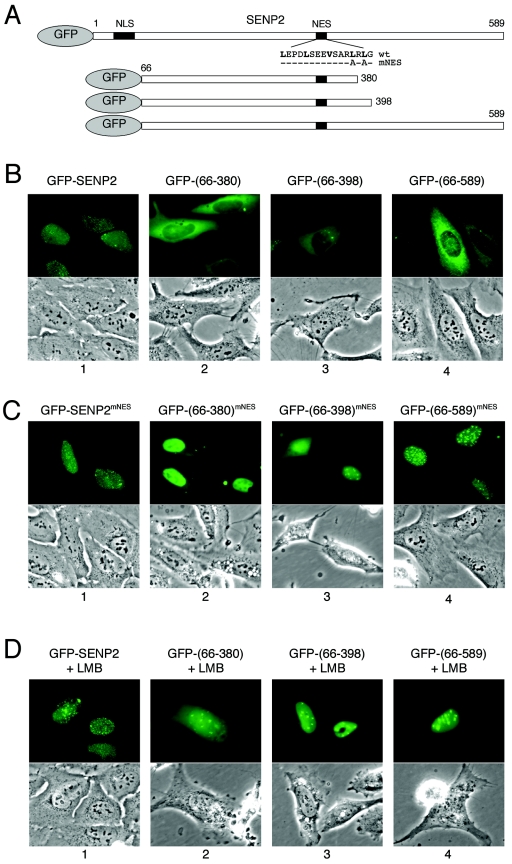
Having found both an NLS and an NES in SENP2, we hypothesized that SENP2 might be a shuttling protein undergoing nuclear import and export via the NLS and the NES. To determine whether SENP2 indeed shuttles between the nucleus and the cytoplasm, we carried out two experiments that are commonly used to demonstrate protein shuttling. First, we determined whether the NES identified at residues 317 to 332 acts as a true nuclear export signal sequence in the context of SENP2 protein. To this end, we generated green fluorescent protein (GFP) fusion constructs expressing either full-length SENP2 or SENP2 deletions lacking the NLS but retaining the NES (Fig. 4A). When transiently expressed in U2OS cells, the full-length GFP-SENP2 fusion protein showed typical nuclear punctate localization, indicating that attachment of a GFP moiety has not affected SENP2's normal subcellular localization (Fig. 4B, panel 1). On the other hand, GFP fused with each of the three SENP2 deletion mutants lacking the NLS displayed nuclear exclusion (Fig. 4B, panels 2 to 4). The nuclear exclusion pattern indicated that the fusion protein was unable to enter the nucleus due to a lack of NLS; alternatively, the relative strength of the NES activity in the absence of the NLS may have enabled more-efficient export of the fusion protein. To distinguish between the two possibilities, we introduced mutations to the essential leucine residues in the SENP2 NES and examined the localization of the mutant proteins (GFP-SENP2mNES). As shown in Fig. 4C, while the localization of full-length SENP2-GFP was not affected by the NES mutation as expected (panel 1), localization of the three NLS deletion mutants was altered by the NES mutation and all exhibited substantial nuclear accumulation (Fig. 4C, panels 2 to 4). This indicates that the NES indeed functions in the context of the SENP2 protein, at least when the NLS is deleted. To demonstrate that the function of NES is CRM1 dependent, we treated the transfected cells with LMB to specifically inhibit the CRM1-dependent nuclear export activity of the NES (8). LMB treatment effectively reversed the nuclear exclusion pattern of the fusion protein (Fig. 4D), demonstrating that the function of the NES, and thus the nuclear export of SENP2, is CRM1 dependent. We noticed that the SENP2 fragments with C-terminal deletions were less capable of forming speckles, similar to what occurs with the SENP2mcNLS mutant, which has a mutated putative C-terminal NLS (Fig. 2A, row 7). The mechanism by which the mutations disrupt SENP2 speckles remains to be determined.
FIG. 4.
Function of NES in SENP2 export. (A) Schematic diagram of human SENP2 and its deletion mutants. Positions of the NLS and NES and of the N-terminal fusion of GFP are indicated. (B and C) Localization of GFP-SENP2 fusion proteins in transiently transfected U2OS cells. Plasmid DNA encoding each of the indicated GFP-SENP2 fusion proteins was transfected into U2OS cells, and the GFP signal was examined with a fluorescence microscope 24 h after transfection into live cells. Corresponding phase-contrast pictures are also shown. (D) The NES-mediated nuclear export of SENP2 is blocked by LMB. Plasmids expressing various GFP-SENP2 fusion proteins were transfected into U2OS cells. Twenty-four hours after transfection, the cells were treated with 10 nM LMB for 7 h and microscope pictures were taken with living cells.
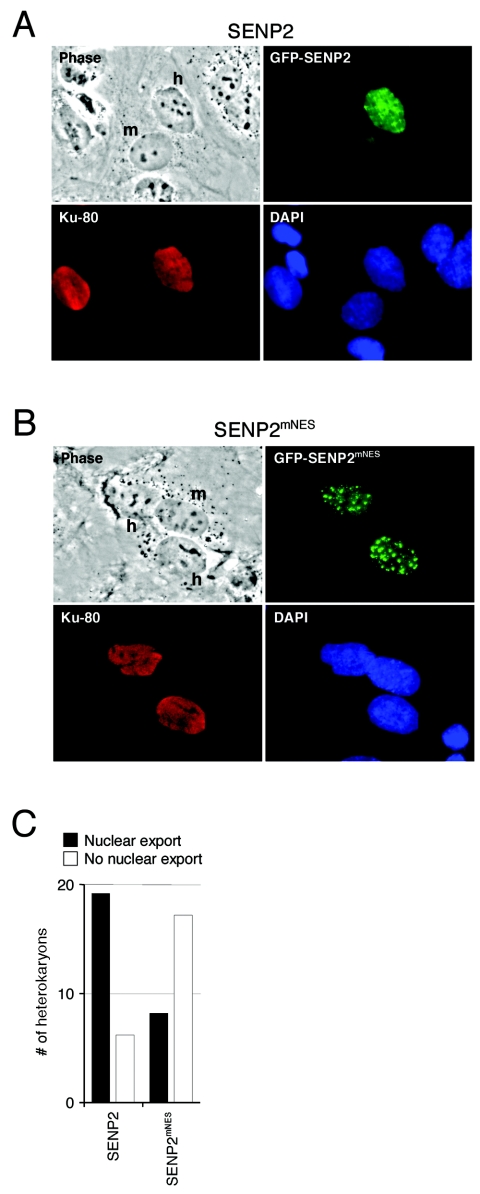
Next, we carried out an interspecies heterokaryon assay to directly show the nucleocytoplasmic shuttling of SENP2. Human U2OS cells were transiently transfected with either wild-type GFP-SENP2 or an NES mutant GFP-SENP2mNES fusion construct. One day after transfection, the cells were fused with MEF cells by PEG in the presence of cycloheximide (50 μg/ml) to block de novo protein synthesis. Accumulation of the GFP-tagged SENP2 protein in the mouse nucleus of the heterokaryon indicates that the protein has been exported out of the human nucleus into the shared cytoplasm and imported into the mouse nucleus. Using this technique, we found that a significant amount of GFP-SENP2 relocalized to the mouse nucleus after 5 h of incubation in the majority of the heterokaryons randomly examined (Fig. 5A and C). GFP-SENP2mNES, where the NES is mutated, remained in the human nucleus in the heterokaryons (Fig. 5B and C). Thus, together with the results from Fig. 1 to 5, we conclude that SENP2 shuttles between the nucleus and the cytoplasm through an NLS identified at residues 29 to 49 and an NES at residues 317 to 332.
FIG. 5.
Interspecies heterokaryon assay shows that SENP2 is a shuttling protein. U2OS cells were transfected with plasmids expressing wild-type GFP-SENP2 (A) or the NES mutant GFP-SENP2mNES (B) fusion proteins. Twenty-four hours after transfection, the cells were washed and fused to wild-type MEF cells, and de novo protein synthesis was blocked by cycloheximide. Five hours after fusion, the cells were fixed and stained with an antibody to human Ku antigen, followed by staining with a rhodamine-conjugated secondary antibody. Representative heterokaryons were photographed. Nuclei of human (h) and mouse (m) origin in each heterokaryon are indicated. (C) Quantification of cells exported from the nucleus. Twenty-five randomly chosen heterokaryons from four independent heterokaryon assays described for panels A and B were examined, and the results were plotted.
SUMO-modified PML, but not RanGAP1, can be efficiently deconjugated by SENP2.
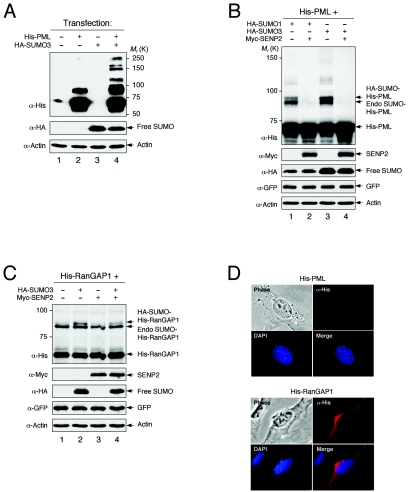
SENP2 desumoylates a large number of cellular proteins modified by either SUMO-1 or SUMO-3 (our unpublished results), indicating the involvement of SENP2 in various aspects of cellular function. In a study of substrate specificity of SENP2, we examined the activity of SENP2 in desumoylating PML and RanGAP1, two of the most abundantly sumoylated proteins in cells. An added advantage of using PML and RanGAP1 as SENP2 substrates in the study is that the endogenous sumoylation activity is sufficient to modify both proteins when they are exogenously expressed. We carried out an in vivo sumoylation assay with H1299 cells transiently transfected with His-tagged PML or RanGAP1 along with SUMO-1, SUMO-3, and SENP2 in various combinations. Cells were lysed with hot SDS lysis buffer to prevent SUMO-conjugated proteins from desumoylation during sample preparation, and the SUMO-modified PML and RanGAP1 were detected by a Western blotting assay using an anti-His antibody. Using this assay, His-PML was efficiently modified by endogenous sumoylation activity and detected by an anti-His antibody. SUMO-modified PML was detected as several discrete bands presumably resulting from the monosumoylation of three SUMO sites on PML (15) (Fig. 6A, lanes 2 and 4). When SENP2 was coexpressed, the SUMO-1- and SUMO-3-modified PML was completely deconjugated (Fig. 6B, lanes 2 and 4), indicating that PML is either a direct substrate of SENP2 in vivo or indirectly affected by SENP2 activity. In contrast to PML and consistent with a previous report (12), the sumoylated RanGAP1 was mostly resistant to SENP2 desumoylation activity (Fig. 6C). Thus, in cell culture, the SUMO-modified PML, but not the SUMO-modified RanGAP1, is efficiently deconjugated by SENP2. It was postulated that the distinct subcellular localization of SENP enzymes plays an important role in their function, particularly in substrate selection (19, 23). It is possible that SENP2, a nuclear protein, efficiently deconjugates only nucleus-localized PML and not cytoplasm-localized RanGAP1, as shown with the distinct subcellular localizations in cells (Fig. 6D). However, our experiments (see below) suggest that the resistance of RanGAP1 to desumoylation by SENP2 was not just due to its discrete localization from SENP2.
FIG. 6.
SUMO-modified PML, but not RanGAP1, is efficiently deconjugated by SENP2. (A to C) H1299 cells were transfected with the indicated plasmids, and cells were lysed 2 days after transfection with hot SDS (2%) lysis buffer. The cell lysate was resolved by SDS-PAGE and blotted with the indicated antibodies (α). GFP was used as a loading control for transfection efficiency. K, in thousands. (A and B) Unmodified and sumoylated His-tagged PML were detected with anti-His antibody. Unconjugated SUMO (free SUMO) was detected with anti-HA antibody. (C) Unmodified RanGAP1 and sumoylated His-tagged RanGAP1 were detected with an anti-His antibody. Unconjugated SUMO (free SUMO) was detected with anti-HA antibody. (D) Localization of PML and RanGAP1. U2OS cells were transfected with His-PML or His-RanGAP1. Twenty-four hours after transfection, cells were fixed and immunostained with an anti-His antibody, followed by staining with Cy2-conjugated or rhodamine-conjugated secondary antibody. Immunofluorescent, phase-contrast, and DAPI-stained images are shown.
Disruption of the NLS, but not the NES, impairs the protease function of SENP2.
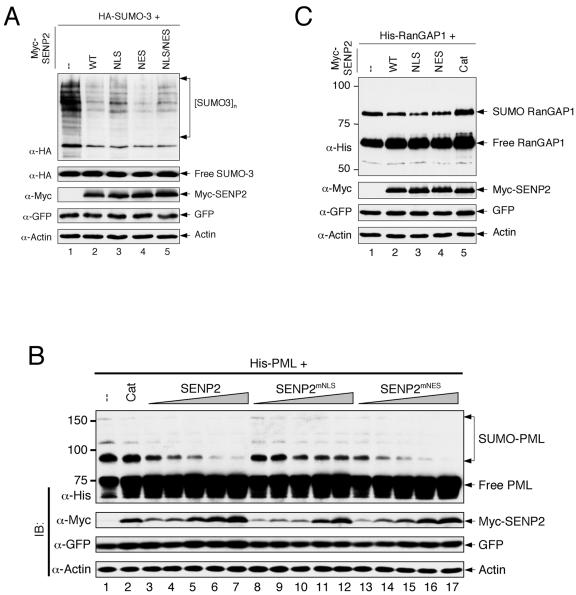
It is possible that the discrete cellular localizations of SENP2 (in the nucleus) and RanGAP1 (in the cytoplasm) contributed to RanGAP1's resistance to SENP2 desumoylation. With the identification of the NLS and construction of a cytoplasm-localized SENP2mNLS mutant, we wanted to determine whether the nuclear localization of SENP2 plays a role in its protease function. To this end, we constructed a Myc-tagged, wild-type SENP2 (Myc-SENP2), an NLS mutant (Myc-SENP2mNLS), an NES mutant (Myc-SENP2mNES), and an NLS/NES double mutant (Myc-SENP2mNLS/mNES) and tested the function of the SENP2 constructs by an in vivo sumoylation assay. We first determined whether nuclear import and/or export of SENP2 affects its desumoylation activity toward total SUMO-modified cellular proteins. The various SENP2 proteins, along with HA-SUMO-3, were expressed in H1299 cells. Total sumoylated cellular proteins were detected with an anti-HA antibody in a Western blotting assay. The SENP2mNES mutant appeared to have desumoylation activity similar to that of wild-type SENP2 (Fig. 7A, compare lane 4 with lane 2). The SENP2mNLS mutant, on the other hand, exhibited substantially reduced activity compared to those of wild-type and NES mutant SENP2 (Fig. 7A, lane 3), indicating that the nuclear import of SENP2 is important for its protease function. Because the SENP2mNLS mutant localizes to the cytoplasm (Fig. 2), this result also indicated that many of the SENP2 substrates might reside in the nucleus. To substantiate this possibility, a SENP2 mutant containing mutations in both the NLS and NES (SENP2mNLS/mNES) was generated, resulting in a mutant with a more-diffuse subcellular localization than that of either SENP2mNLS or SENP2mNES alone (data not shown). The desumoylation activity exhibited by SENP2mNLS/mNES was somewhat in between that of the SENP2mNLS and the SENP2mNES mutants (Fig. 7A, lane 5). The impairment of the desumoylation function by the NLS mutation, however, appeared to be limited. We believe that this is most likely due to a combination of incomplete disruption of the NLS by the point mutations and a leakage of the mutant protein caused by overexpression. A small amount of the SENP2mNLS mutant protein entering the nucleus could have sufficient protease activity to deconjugate a large amount of SUMO-modified proteins.
FIG. 7.
Disruption of the NLS, but not NES, impairs the protease function of SENP2. (A) H1299 cells were cotransfected with plasmids encoding HA-SUMO-3 and various Myc-SENP2 enzymes as indicated. The SENP2 constructs were transfected with wild-type SENP2 [WT]-SENPmNLS-SENPmNES-SENPmNLS/mNES at a ratio of 1:1.5:0.7:1.1 to obtain equal levels of SENP2 protein expression. Cells were lysed 2 days after transfection with a 2% hot SDS lysis buffer. Cell lysates were separated by SDS-PAGE, and the proteins were detected by immunoblotting with the indicated antibodies (α). GFP was used as a loading control for transfection efficiency. High-molecular-weight protein species detected with an anti-HA antibody indicate sumoylated proteins. Unconjugated SUMO-3 (free SUMO-3) is shown. (B) Desumoylation of PML by SENP2. H1299 cells were cotransfected with plasmids encoding His-PML and various Myc-SENP2 enzymes as indicated. The SENP2 constructs were transfected with wild-type Myc-SENP2-SENPmNLS-SENPmNES at a ratio of 1:1.2:0.7 to obtain similar levels of SENP2 protein expression. Cells were lysed 2 days after transfection with a 2% hot SDS lysis buffer. Cell lysates were subjected to SDS-PAGE and immunoblotted (IB) with the indicated antibodies. Unmodified and sumoylated His-PML was detected with anti-His antibody. Cat, a catalytically inactive SENP2 mutant. (C) Desumoylation of RanGAP1 by SENP2. H1299 cells were cotransfected with plasmids encoding His-RanGAP1 and the wild-type or mutant Myc-SENP2. The SENP2 constructs were transfected with wild-type Myc-SENP2-SENPmNLS-SENPmNES-SENP2Cat [Cat]) at a ratio of 1:1.5:0.7:1.5 to obtain equal levels of SENP2 protein expression. Cells were lysed 2 days after transfection with a 2% hot SDS lysis buffer, and the cell lysates were assayed as described for panel B.
To further examine functional alterations of the SENP2 NLS and NES mutants, we compared their activities in desumoylating PML and RanGAP1, a nuclear protein and a cytoplasmic protein, respectively. His-PML and His-RanGAP1 were each coexpressed with SENP2 or SENP2 mutants in H1299 cells, and SUMO-modified His-PML and His-RanGAP1 were detected by an anti-His antibody. A catalytically inactive SENP2Cat mutant was used as a negative control. As shown in Fig. 7B and C, ectopically expressed His-PML and His-RanGAP1 were efficiently modified by endogenous sumoylation activity in the absence of SENP2 (lane 1 in Fig. 7B and C).
With coexpression of increasing amounts of SENP2, the SUMO-modified PML was reduced in a dose-dependent manner, indicating that PML is a cellular substrate of SENP2 (Fig. 7B). Importantly, among the three SENP2 proteins (SENP2, SENP2mNLS, and SENP2mNES), SENP2mNLS exhibited the lowest desumoylation activity toward PML, even though the protein expressed at comparable or higher levels (Fig. 7B, compare lanes 11 to 12 with lanes 5 to 6 and 15 to 16). On the other hand, SENP2mNES appeared to be the most active in desumoylating PML. Given that PML is a nuclear protein (Fig. 6D), these results indicate that colocalization with PML in the nucleus is important for SENP2 to desumoylate PML. In contrast to PML, SUMO-modified RanGAP1 was resistant to desumoylation by any of the three catalytically active SENP2 enzymes. However, we did notice that the SENP2mNLS mutant, which is the least effective in desumoylating PML, appeared to be more effective in desumoylating RanGAP1 than were wild-type SENP2 and SENP2mNES (compare lane 3 with lanes 2 and 4 in Fig. 7C). Together, these results suggest that the colocalization of SENP2 with its substrates in the same subcellular compartment facilitates SUMO deconjugation.
Regulation of SENP2 by polyubiquitination-mediated degradation.
Although we have demonstrated SENP2 shuttling between the nucleus and cytoplasm, the predominance of wild-type SENP2 nuclear localization, combined with the results of our mutational analysis, suggests that the protease activity of SENP2 occurs in the nucleus. Based on the characterization of other shuttling proteins, such as cyclin B, E2F1, p53, and MDM2, whose shuttling activities are linked to protein degradation, we sought to determine whether SENP2 protein stability could be affected by its shuttling. To this end, we examined SENP2 protein stability by a half-life assay. SENP2 and SENP2 mutants were transiently expressed in H1299 cells, pulse-labeled with [35S]methionine for 2 h, and chased every 6 h for up to 24 h. The assay showed that the SENP2mNLS mutant has a much shorter half-life than wild-type SENP2 and the SENP2mNES mutant (Fig. 8A). Reducing the time interval in the half-life assay revealed that the SENP2mNLS mutant has a half-life of approximately 3 h (Fig. 8B and C). The shorter half-life of the cytoplasm-localized SENP2mNLS mutant than those of wild-type SENP2 and the nucleus-localized SENP2mNES mutant suggests that the degradation of SENP2 occurs in the cytoplasm. It also argues that the decreased stability of the SENP2mNLS mutant could contribute to the observed overall lower activity of this mutant in deconjugating total cellular SUMO-modified proteins (Fig. 7A). The shorter half-life of the SENP2mNLS mutant was also consistent with the observation that a larger amount of the SENP2mNLS mutant DNA than of SENP2 or SENP2mNES DNA is required to achieve equal levels of protein expression following transient transfection. Ectopically expressed proteins tend to have a longer half-life than those that are endogenously expressed; therefore, under physiological conditions, SENP2's half-life could be shorter than those that we observed in this assay. The lack of antibodies able to immunoprecipitate endogenous SENP2 limited our ability to determine its half-life under physiological conditions.
The short half-life of SENP2 suggests that it might undergo ubiquitin-mediated proteolysis. To determine whether SENP2 is ubiquitinated and degraded through proteolysis, we carried out an in vivo ubiquitination assay by transfecting H1299 cells with plasmid DNA expressing HA-Ub and Myc-tagged SENP2. Two identical sets of samples were prepared: one untreated control and one treated with the 26S proteasome inhibitor MG132 to allow ubiquitinated proteins to accumulate. After cells were lysed in hot SDS lysis solution (to protect ubiquitinated protein species from deubiquitination), Myc-tagged SENP2 was pulled down with an anti-Myc antibody and separated on an SDS-PAGE gel, and HA-Ub-conjugated SENP2 was detected with an anti-HA antibody. As shown in Fig. 8D, polyubiquitinated SENP2 was barely seen without MG132 treatment (lanes 3 to 5). In samples treated with MG132, however, high-molecular-weight polyubiquitinated SENP2 was readily detected, demonstrating that SENP2 can be efficiently polyubiquitinated in vivo (lanes 8 to 10). Immunofluorescent staining indicated that MG132 treatment did not affect the localization of SENP2 or the SENP2 mutants (Fig. 8E). Notably, we found that among the three SENP2 proteins, SENP2mNLS was ubiquitinated to the greatest extent, suggesting that the polyubiquitination of SENP2 occurs more efficiently in the cytoplasm. Alternatively, this effect could be due to a higher deubiquitination activity in the nucleus, which would be consistent with an observed lower level of polyubiquitination in the SENP2mNES mutant than in wild-type SENP2 (Fig. 8D, compare lanes 8 and 10), yet both proteins have similar half-lives, and our results indicate that ubiquitination and proteasomal degradation might not be entirely linked in this case. The higher level of ubiquitination of SENP2mNLS correlates with its shorter half-life, indicating that SENP2 is degraded through an ubiquitin-mediated proteolysis pathway. Given that the ubiquitination assay was performed without coexpressing an E3 ubiquitin ligase, the observed polyubiquitination of SENP2 must be achieved by an endogenous ubiquitin ligase. Thus, it appears that the in vivo ubiquitination activity of SENP2 is rather high. Taken together, our data demonstrate that SENP2 can be polyubiquitinated in vivo and degraded by 26S proteasome-mediated proteolysis. These data also indicate that the polyubiquitination of SENP2 takes place preferentially in the cytoplasm.
DISCUSSION
In analyzing the mechanisms controlling SENP2 function, we examined the nucleocytoplasmic shuttling of SENP2. Several lines of evidence have emerged from this study: (i) SENP2 has a bipartite NLS and a CRM1-dependent NES, both of which are located in nonconserved domains in SENP2 and are not found in other members of the SENP family; (ii) SENP2 shuttles between the nucleus and the cytoplasm via its NLS and NES; (iii) colocalization of SENP2 with substrates in the same subcellular compartment facilitates SUMO deconjugation; and (iv) SENP2 is polyubiquitinated in vivo and degraded by the 26S proteasome-dependent pathway.
An NES-mediated nuclear export of SENP2 may serve two purposes. The first possible role may be to add another layer of regulation for SENP2 activity and substrate selectivity. In theory, SENP2 could have substrates in both the nucleus and the cytoplasm. When and how much of these substrates must be desumoylated could be influenced by cellular functions, such as cell cycle progression. By controlling the function of the nucleocytoplasmic shuttling of SENP2, cells may be able to selectively desumoylate specific substrates in the nucleus and/or in the cytoplasm, thereby achieving a greater flexibility in regulating SENP2 activity. The second purpose concerns the role of nucleocytoplasmic shuttling in controlling SENP2 protein turnover. Our data show that SENP2 is efficiently polyubiquitinated in vivo and undergoes polyubiquitin-mediated proteasomal degradation and that the polyubiquitination of SENP2 appears to occur preferentially in the cytoplasm. Based on our data, we envision a model for SENP2 nucleocytoplasmic shuttling whereby SENP2 enters the nucleus via NLS-mediated nuclear import and functions as a nucleus-specific SUMO protease. When SENP2's activity is no longer required or needs to be reduced, the protein is exported out of the nucleus via NES-dependent nuclear export. As a result, polyubiquitination occurs in the cytoplasm and SENP2 is degraded through the 26S proteolysis pathway. This mechanism could effectively separate the locations of SENP2's function (in the nucleus) and destruction (in the cytoplasm), so that premature degradation of SENP2 can be prevented when its activity is needed in the nucleus.
We have noticed that even though the impairment of nuclear localization of SENP2 by NLS mutation is considerably thorough, the impairment of the desumoylation function of the mutant appears to be limited. Two possibilities may contribute to this discrepancy. First, it is possible that the SUMO-modified protein substrates travel between the nucleus and the cytoplasm, allowing a cytoplasm-restricted SENP2mNLS to deconjugate nuclear substrates in the cytoplasm without entering the nucleus. The second alternative may involve only partial inhibition of SENP2mNLS nuclear localization due to either incomplete inactivation of the NLS or protein expression levels far in excess of physiological levels. In this case, even a minor fraction of ectopically expressed SENP2 entering the nucleus could have sufficient protease activity to process a large quantity of substrate. An earlier report has shown that elimination of the first 70 amino acids of SENP2, the region containing the NLS, affected SENP2's association with the nuclear pore and increased the capacity of SENP2 to deconjugate SUMO-1-conjugated proteins in cells. Hang and Dasso suggested that the association of SENP2 to the nuclear pore may have a negative impact on the function of SENP2 (12). Our results, using conjugated point mutant SENP2 and SUMO-3 proteins, indicate that the cytoplasmically confined SENP2 mutant was less active in desumoylating SUMO-3-conjugated cellular proteins (Fig. 7A). Whether the discrepancy reflects functional differences between an N-terminally truncated SENP2 and a full-length SENP2 or differential activities of the SENP2 mutants toward SUMO-1- and SUMO-3-conjugated cellular proteins remains to be determined.
To date, all the SENP species identified in mammalian cells have divergent N-terminal domains, believed to be involved in determining the substrate specificity of each SENP protease (14, 23). Individual SENP species have been found in different subcellular compartments. For example, SENP1 localizes to the nuclear foci and the nuclear rim (3), SENP2 localizes to the nuclear side of the nuclear pore complex (12, 24), SENP3 localizes to the nucleolus (20), and SENP6 is found primarily in the cytoplasm (16). It is conceivable that the diverse subcellular localizations of the proteases play important roles in determining their substrate specificities. Nevertheless, how a specific subcellular localization is linked to the function of each SENP protease remains poorly understood.
How sumoylation, and particularly desumoylation, might affect the biological processes of cells is unclear. Sumoylation has been implicated in many cellular functions, including regulation of transcription, formation of PML nuclear bodies, chromosome organization, DNA repair, and nuclear transport (14, 22). A common problem in examining the functional consequences of sumoylation/desumoylation is that, with rare exceptions, the proportion of most SUMO-modified proteins, compared with that of those lacking SUMO modifications, is always extremely low. Therefore, it is not clear in most cases if any specific biological function can be attributed to SUMO modifications or whether the fraction of sumoylated protein bears any physiological significance. There may be multiple potential explanations concerning the nature of low sumoylated-protein levels, but at the very least we will consider two of them. First, much like polyubiquitinated proteins, SUMO-modified proteins may escape detection by becoming unstable and short-lived during sample preparations. It is of interest to note that sumoylation and ubiquitination may be intimately linked (9), considering the fact that many sumoylated proteins are also ubiquitinated and degraded by proteolysis. Whether sumoylation competes with ubiquitination on the same lysine residues or is a prerequisite for ubiquitination and subsequent degradation is unknown. A second potential factor for the low level of sumoylation could be high desumoylation activity in cells. We postulate that protein sumoylation might occur constitutively in cells and that the control steps for SUMO modification might be at the steps of desumoylation, but not sumoylation. If so, SUMO proteases could represent a group of new targets for therapeutic intervention in certain human diseases, including cancer, for which studies have suggested that altered sumoylation may contribute to both the onset and progression of the disease (1).
Acknowledgments
We thank Koji Itahana for his contribution and helpful discussions to the study, as well as Chad Deisenroth and Hilary Clegg for critical readings of the manuscript.
Y.Z. is a recipient of a Career Award in Biomedical Science from the Burroughs Wellcome Fund and a Howard Temin Award from the National Cancer Institute. This study was supported by NIH grants CA100302 and CA87580 (to Y.Z.).
REFERENCES
- 1.Alarcon-Vargas, D., and Z. Ronai. 2002. SUMO in cancer—wrestlers wanted. Cancer Biol. Ther. 1:237-242. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, D., and P. O'Hare. 2004. Characterization of the localization and proteolytic activity of the SUMO-specific protease, SENP1. J. Biol. Chem. 279:692-703. [DOI] [PubMed] [Google Scholar]
- 3.Bailey, D., and P. O'Hare. 2002. Herpes simplex virus 1 ICP0 co-localizes with a SUMO-specific protease. J. Gen. Virol. 83:2951-2964. [DOI] [PubMed] [Google Scholar]
- 4.Bogerd, H. P., R. A. Fridell, R. E. Benson, J. Hua, and B. R. Cullen. 1996. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol. Cell. Biol. 16:4207-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohren, K. M., V. Nadkarni, J. H. Song, K. H. Gabbay, and D. Owerbach. 2004. A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J. Biol. Chem. 279:27233-27238. [DOI] [PubMed] [Google Scholar]
- 6.Boulikas, T. 1993. Nuclear localization signals (NLS). Crit. Rev. Eukaryot. Gene Expr. 3:193-227. [PubMed] [Google Scholar]
- 7.Cheng, J., D. Wang, Z. Wang, and E. T. Yeh. 2004. SENP1 enhances androgen receptor-dependent transcription through desumoylation of histone deacetylase 1. Mol. Cell. Biol. 24:6021-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 9.Gill, G. 2004. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 18:2046-2059. [DOI] [PubMed] [Google Scholar]
- 10.Gong, L., S. Millas, G. G. Maul, and E. T. Yeh. 2000. Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J. Biol. Chem. 275:3355-3359. [DOI] [PubMed] [Google Scholar]
- 11.Guo, D., M. Li, Y. Zhang, P. Yang, S. Eckenrode, D. Hopkins, W. Zheng, S. Purohit, R. H. Podolsky, A. Muir, J. Wang, Z. Dong, T. Brusko, M. Atkinson, P. Pozzilli, A. Zeidler, L. J. Raffel, C. O. Jacob, Y. Park, M. Serrano-Rios, M. T. Larrad, Z. Zhang, H. J. Garchon, J. F. Bach, J. I. Rotter, J. X. She, and C. Y. Wang. 2004. A functional variant of SUMO4, a new I kappa B alpha modifier, is associated with type 1 diabetes. Nat. Genet. 36:837-841. [DOI] [PubMed] [Google Scholar]
- 12.Hang, J., and M. Dasso. 2002. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J. Biol. Chem. 277:19961-19966. [DOI] [PubMed] [Google Scholar]
- 13.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, E. S. 2004. Protein modification by SUMO. Annu. Rev. Biochem. 73:355-382. [DOI] [PubMed] [Google Scholar]
- 15.Kamitani, T., K. Kito, H. P. Nguyen, H. Wada, T. Fukuda-Kamitani, and E. T. Yeh. 1998. Identification of three major sentrinization sites in PML. J. Biol. Chem. 273:26675-26682. [DOI] [PubMed] [Google Scholar]
- 16.Kim, K. I., S. H. Baek, Y. J. Jeon, S. Nishimori, T. Suzuki, S. Uchida, N. Shimbara, H. Saitoh, K. Tanaka, and C. H. Chung. 2000. A new SUMO-1-specific protease, SUSP1, that is highly expressed in reproductive organs. J. Biol. Chem. 275:14102-14106. [DOI] [PubMed] [Google Scholar]
- 17.Li, S. J., and M. Hochstrasser. 1999. A new protease required for cell-cycle progression in yeast. Nature 398:246-251. [DOI] [PubMed] [Google Scholar]
- 18.Melchior, F. 2000. SUMO—nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16:591-626. [DOI] [PubMed] [Google Scholar]
- 19.Melchior, F., M. Schergaut, and A. Pichler. 2003. SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem. Sci. 28:612-618. [DOI] [PubMed] [Google Scholar]
- 20.Nishida, T., H. Tanaka, and H. Yasuda. 2000. A novel mammalian Smt3-specific isopeptidase 1 (SMT3IP1) localized in the nucleolus at interphase. Eur. J. Biochem. 267:6423-6427. [DOI] [PubMed] [Google Scholar]
- 21.O'Keefe, K., H. Li, and Y. Zhang. 2003. Nucleocytoplasmic shuttling of p53 is essential for MDM2-mediated cytoplasmic degradation but not ubiquitination. Mol. Cell. Biol. 23:6396-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seeler, J. S., and A. Dejean. 2003. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell. Biol. 4:690-699. [DOI] [PubMed] [Google Scholar]
- 23.Yeh, E. T., L. Gong, and T. Kamitani. 2000. Ubiquitin-like proteins: new wines in new bottles. Gene 248:1-14. [DOI] [PubMed] [Google Scholar]
- 24.Zhang, H., H. Saitoh, and M. J. Matunis. 2002. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol. Cell. Biol. 22:6498-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, Y., and Y. Xiong. 2001. A p53 amino terminal nuclear export signal inhibited by DNA damage-induced phosphorylation. Science 292:1910. [DOI] [PubMed] [Google Scholar]