Abstract
The Saccharomyces cerevisiae transcription factor Ste12 controls two distinct developmental programs of mating and filamentation. Ste12 activity is regulated by Fus3 and Kss1 mitogen-activated protein kinases through two Ste12 inhibitors, Dig1 and Dig2. Mating genes are regulated by Ste12 through Ste12 binding sites (pheromone response elements [PREs]), whereas filamentation genes are supposedly regulated by the cooperative binding of Ste12 and Tec1 on a PRE adjacent to a Tec1-binding site (TCS), termed filamentous responsive element (FRE). However, most filamentation genes do not contain an FRE; instead, they all have a TCS. By immunoprecipitation, we show that Ste12 forms two distinct complexes, Ste12/Dig1/Dig2 and Tec1/Ste12/Dig1, both in vivo and in vitro. The two complexes are formed by the competitive binding of Tec1 and Dig2 with Ste12, as Tec1 can compete off Dig2 from Ste12 in vitro and in vivo. In the Tec1/Ste12/Dig1 complex, Tec1 binds to the N terminus of Ste12 and to Dig1 indirectly through Ste12. Tec1 has low basal activity, and its transcriptional activation is provided by the associated Ste12, which is under Dig1 inhibition. Filamentation genes are bound by the Tec1/Ste12/Dig1 complex, whereas mating genes are occupied by mostly Ste12/Dig1/Dig2 with some Tec1/Ste12/Dig1. We suggest that Tec1 tethers Ste12 to TCS elements upstream of filamentation genes and defines the filamentation genes as a subset of Ste12-regulated genes.
Key regulators of cell fate determination often control multiple developmental pathways in response to different stimuli. In Saccharomyces cerevisiae, the transcription factor Ste12 is required for both mating and filamentation (11, 13, 14, 19). During the mating of haploid cells, Ste12 induces the expression of pheromone-responsive genes through Ste12 binding sites, or pheromone response elements [PREs; TGAAAC(A/G)], at the promoters of mating genes (41). Ste12 homodimers can bind cooperatively to tandem PREs in vitro, and the PRE is sufficient to induce pheromone-responsive expression of haploid-specific genes in both mating types (18). During filamentous and invasive growth, Ste12 cooperates with Tec1, a TEA/ATTS-family transcription factor (30). Tec1 was first identified as a regulator of the expression of Ty1 transposon insertions. Like other TEA/ATTS family members, it binds to CATTCC or CATTCT (termed TCS, for TEA/ATTS consensus sequence) (25, 26, 29). Enhancer elements containing a PRE adjacent to a TCS are termed filamentation/invasion response element (FREs) (29) or sterile response elements (5). FREs have been found in the promoters of Ty1 and TEC1 and are necessary and sufficient to confer filamentation-associated expression in S. cerevisiae (5, 29). Importantly, recombinant Ste12 and Tec1 bind cooperatively to the FREs of Ty1 and TEC1 in vitro (29). Consistent with the cooperative control of filamentation genes by Ste12 and Tec1, a genome-wide study of Ste12 distribution has localized Ste12 to the promoters of pheromone-induced genes and filamentation genes in vivo, and the binding of Ste12 at the promoters of filamentation genes is Tec1 dependent (47).
Ste12 is regulated by the Fus3 and Kss1 mitogen-activated protein (MAP) kinases (2, 41). Fus3 and Kss1 have overlapping functions in mating (12, 39). Both Fus3 and Kss1 can phosphorylate Dig1 and Dig2, two functionally redundant inhibitors of Ste12 (9, 43). Dig1 binds to the middle region of Ste12 that is responsible for transcriptional induction in response to pheromone, whereas Dig2 binds to the N-terminal region of Ste12 (34). During pheromone induction, the inhibition of Dig1 and Dig2 is relieved, leading to the activation of Ste12 (3, 9, 34, 35, 43). Fus3 and Kss1 play opposing roles on filamentation. Kss1 kinase activity is necessary for filamentation, while Fus3 kinase activity is inhibitory to filamentation (3, 10, 29). During the pheromone response, Fus3, but not Kss1, specifically phosphorylates Tec1 and triggers ubiquitin-mediated Tec1 degradation, preventing the induction of filamentation genes (1, 7, 8).
Although Ste12 is bound to the promoters of filamentation genes (47), we find that promoters of most filamentation genes do not contain FREs, but they all contain TCS. A genetic study by Kohler et al. suggests that Tec1 can regulate the expression of filamentation genes by TCS control when overexpressed (24). Similar to FREs, TCS-driven expression is inhibited by active Fus3 and activated by Kss1 (7, 8). Here, we show that Ste12 forms two distinct complexes, the known Ste12/Dig1/Dig2 complex for mating and also a novel Tec1/Ste12/Dig1 complex that regulates the expression of filamentation genes through TCS. Tec1 by itself does not have significant transcriptional activity, but it tethers Ste12 transcriptional activity to the TCS site to allow the activation of filamentation genes. Our work demonstrates a mechanism for how Ste12 can control transcriptional programs of distinct pathways by its association with different cofactors.
MATERIALS AND METHODS
Yeast strains.
Standard yeast manipulation methods were used. Strains used in this study are listed in Table 1. All the strains constructed in this study are derivatives from 10560-4A in a Σ1278b background unless otherwise specified. Strain HLY2187 was obtained from a cross between strains 10560-6B and L6149. dig1::TRP1, dig2::TRP1, and dig2::KANR were introduced into yeast according to Longtine et al. (28). TRP1 or KANR cassettes were amplified by PCR from plasmid pFA6a-TRP1 or pFA6a-KanMX6 with a pair of primers that included 50 bp upstream and downstream of the DIG1 or DIG2 open reading frame (ORF) and integrated into the genome by homologous recombination. ste12::LEU2 was created by transforming plasmid pSUL16 digested with SacI and SphI (13). The tec1::KANR deletion was introduced by transformation of the PCR product amplified with primers upstream and downstream of the TEC1 ORF from the genomic DNA of a tec1::KAN strain from the S. cerevisiae gene deletion MATa set (Invitrogen).
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| 10560-4A | MATa ura3-52 his3::hisG leu2::hisG trp1::hisG | G. R. Fink |
| 10560-6B | MATα ura3-52 his3::hisG leu2::hisG trp1::hisG | G. R. Fink |
| L6149 | MATatec1::HIS3 ura3-52 his3::hisG leu2::hisG | G. R. Fink |
| HLY3315 | MATadig1::TRP1 ura3-52 his3::hisG leu2::hisG trp1::hisG | This study |
| HLY3316 | MATa dig2::TRP1 ura3-52 his3::hisG leu2::hisG trp1::hisG | This study |
| HLY3317 | MATa dig1::TRP1 dig2::KAN ura-52 his3::hisG leu2::hisG trp1::hisG | This study |
| HLY2187 | MATatec1::HIS3 ura3-52 his3::hisG leu2::hisG trp1::hisG | This study |
| HLY3318 | MATadig1::TRP1 tec1::HIS ura3-52 his3::hisG leu2::hisG trp1::hisG | This study |
| HLY3319 | MATadig2::TRP1 tec1::HIS ura3-52 his3::hisG leu2::hisG trp1::hisG | This study |
| HLY3328 | MATa ura3-52 his3::hisG leu2::hisG trp1::hisG ura3::URA3 lexAops-lacZ | This study |
| HLY3329 | MATadig1::TRP1 ura3-52 his3::hisG leu2::hisG trp1::hisG ura3::URA3 lexAops-lacZ | This study |
| HLY3330 | MATadig2::TRP1 ura3-52 his3::hisG leu2::hisG trp1::hisG ura3::URA3 lexAops-lacZ | This study |
| HLY3331 | MATadig1::TRP1 dig2::KAN ura3-52 his3::hisG leu2::hisG trp1::hisG ura3::URA3 lexAops-lacZ | This study |
| HLY3332 | MATaste12::LEU2 ura3-52 his3::hisG leu2::hisG trp1::hisG ura3::URA3 lexAops-lacZ | This study |
| HLY3333 | MATa ste12::LEU2 dig1::TRP1 ura3-52 his3::hisG leu2::hisG trp1::hisG ura3::URA3 lexAops-lacZ | This study |
| HLY3321 | MATaSTE12-MYC13::HIS3 ura3-52 his3::hisG leu2::hisG trp1::hisG | This study |
| HLY3320 | MATaTEC1-MYC13::HIS3 ura3-52 his3::hisG leu2::hisG trp1::hisG | This study |
| HLY3322 | MATaDIG1-MYC13::HIS3 ura3-52 his3::hisG leu2::hisG trp1::hisG | This study |
| HLY3323 | MATa DIG2-MYC13::HIS3 ura3-52 his3::hisG leu2::hisG trp1::hisG | This study |
| HLY3325 | MATaSTE12-HA3::TRP1 ura3-52 his3::hisG leu2::hisG trp1::hisG | This study |
| HLY3324 | MATaTEC1-HA3::TRP1 ura3-52 his3::hisG leu2::hisG trp1::hisG | This study |
| HLY3326 | MATaDIG1-HA3::TRP1 ura3-52 his3::hisG leu2::hisG trp1::hisG | This study |
| HLY3327 | MATa DIG2-HA3::TRP1 ura3-52 his3::hisG leu2::hisG trp1::hisG | This study |
| HLY3334 | MATaTEC1-HA3::TRP1 STE12-(13MYC)::HIS3 ura3-52 his3::hisG leu2::hisG trp1::hisG | This study |
| HLY3335 | MATaTEC1-HA3::TRP1 DIG1-(13MYC)::HIS3 ura3-52 his3::hisG leu2::hisG trp1::hisG | This study |
| HLY3336 | MATaTEC1-HA3::TRP1 DIG2-(13MYC)::HIS3 ura3-52 his3::hisG leu2::hisG trp1::hisG | This study |
| HLY3337 | MATaDIG2-HA3::TRP1 STE12-(13MYC)::HIS3 ura3-52 his3::hisG leu2::hisG trp1::hisG | This study |
| HLY3338 | MATaDIG2-HA3::TRP1 DIG1-(13MYC)::HIS3 ura3-52 his3::hisG leu2::hisG trp1::hisG | This study |
| HLY3339 | MATaDIG2-HA3::TRP1 TEC1-(13MYC)::HIS3 ura3-52 his3::hisG leu2::hisG trp1::hisG | This study |
| HLY3342 | MATaDIG1- MYC13::HIS3 ste12::LEU2 ura3-52 his3::hisG leu2::hisG trp1::hisG | This study |
| HLY3343 | MATa TEC1- HA3::TRP1 DIG1-(13MYC)::HIS3 ste12::LEU2 ura3-52 his3::hisG leu2::hisG trp1::hisG | This study |
| HLY3341 | MATa STE12-(3HA)::TRP1 DIG2-(13MYC)::HIS3 tec1::KANR ura3-52 his3::hisG leu2::hisG trp1::hisG | This study |
| HLY3350 | MATaste12::LEU2 tec1::KANR ura3-52 his3::hisG leu2::hisG trp1::hisG | This study |
| HLY3405 | MATaTEC1-(3HA)::TRP1 DIG2-(13MYC)::HIS3 dig1::TRP1 ura3-52 his3::hisG leu2::hisG trp1::hisG | This study |
To epitope tag yeast genes, cassettes encoding either a three-hemagglutinin (HA3) tag or a myc13 tag, an ADH1 terminator, and a TRP1 or HIS3 selectable marker were amplified by PCR with target-gene-specific primers from plasmid pF6a-HA3-TRP1 or pF6a-myc13-His3Mx6 and integrated into their respective genomic loci (28). All tagged strains were confirmed by Western blotting and were capable of mating and invasive growth.
Plasmid construction.
The TCS-lacZ plasmid (pHL710) was constructed by inserting the sequence AGAATGTGCATTATCGATTCATTCT into the Xho1 site in the pLG669Z plasmid (17).
To construct plasmids pHL711 (ADH1p-lexA-TEC1 HIS3) and pHL712 (ADH1p-lexA-STE12 HIS3), STE12 and TEC1 ORFs were amplified by PCR and inserted into the yeast vector pEG202 (16) between the EcoRI and XhoI sites.
To construct plasmid pHL731 (ADH1p-TEC1-HA3 TRP1), a 1.5-kb ADH1 promoter region was amplified by PCR and inserted into the yeast expression vector pRS314 (40) between the NotI and EcoRI sites to generate pRS314/ADH1p. Then a Tec1-3HA cassette with an ADH1 terminator sequence was PCR amplified from the epitope-tagged Tec1-HA strain HLY3324 and cloned into the plasmid pRS314/ADH1p between the EcoRI and XhoI sites. To construct pHL732 (ADH1p-TEC1-myc13 TRP1), a Tec1-myc13 cassette with the ADH1 terminator sequence was PCR amplified from the strain HLY3324 and cloned into the plasmid pRS314/ADH1p between the EcoRI and XhoI sites.
For plasmid pHL754 (ADH1p-STE12(1-215)-myc13 HIS3), the Tec1 ORF region of pHL732 was replaced with an STE12(1-215) fragment (STE12 with residues 1 to 215) amplified by PCR. And the ADH1p-Ste12(1-215)-myc13 module was cloned into the plasmid pRS313.
N-terminal and C-terminal Tec1 deletion fusions with HA3 were cloned into pHL731 by replacing the TEC1 coding region of pHL731 with PCR-amplified fragments of TEC1, digested with EcoRI and BamHI. The same TEC1 fragments, flanked with EcoRI and XhoI and followed by a stop codon, were amplified and cloned into the vector pEG202 for transcriptional activity assays. High-fidelity Pfx DNA polymerase (Invitrogen) was used in all PCR amplifications that were used for cloning in this study, and clones used in this study were confirmed by sequencing.
Immunoprecipitation.
Cell cultures (50 ml) were grown to an optical density at 600 nm (OD600) of about 1.0, and cells were washed twice with ice-cold water and then broken with FastPrep (Eppendorf) for 40 s twice in 0.7 ml of high-salt breaking buffer (20 mM Tris-HCl, 300 mM NaCl, 0.1% NP-40, 1m M EDTA) plus protease inhibitors (0.5 mM phenylmethylsulfonyl fluoride [PMSF], 2 mM benzamidine, 1 μM leupeptin, 2 μM pepstatin, 4 μM chymostatin, 2.6 μM aprotinin). After a 10-min spin in a microcentrifuge, the lysates were incubated with 1.5 μg of anti-HA antibody (clone 12C5; Roche) at 4°C for 1.5 h, and subsequently 50 μl of a 1:1 slurry of protein A-Sepharose beads was added and incubated for another 1.5 h at 4°C. Bound proteins were washed five times with breaking buffer and eluted with 1% sodium dodecyl sulfate (SDS) in Tris-EDTA buffer, pH 8.0.
In vitro transcription/translation and in vitro binding.
Plasmids used for in vitro transcription/translation were constructed by PCR amplification of TEC1, STE12, DIG1, and DIG2 with EcoRI and XhoI on each end and cloned in frame into the EcoRI and XhoI sites of the plasmid pCITE4b(+) (Novagen). Tec1-FLAG was constructed by adding a FLAG sequence in frame to the C terminus of Tec1 in a PCR primer for cloning into the pCITE4b(+) vector. mRNA was transcribed from plasmid DNA for 20 min and subsequently translated for 90 min with the STP3 T7 Single Tube Transcription/Translation System (Novagen). For myc-Ste12, mRNA was transcribed from plasmid pGEM4Z-meSTE12 (3) using the STP3 SP6 Transcription Mix (Novagen). In vitro translated proteins were incubated in IPP150 binding buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% Triton X-100, and 1 mM PMSF with rotation at 4°C for 1 h. The proteins were immunoprecipitated with either anti-FLAG M2-agarose (Sigma) or a 1:100 dilution of anti c-myc (A-14) rabbit polyclonal immunoglobulin G (Santa Cruz Biotechnology) with protein A-Sepharose CL-4B (Amersham Biosciences) overnight at 4°C with rotation. The beads were then washed five times with IPP150 binding buffer before being boiled for 5 min in 5× SDS sample buffer. The proteins were resolved on 9% SDS-polyacrylamide gels, fixed, dried, and detected with a phosphorimage cassette.
β-Galactosidase assay.
β-Galactosidase assays were performed as previously described (37) but with the addition of protease inhibitors in the cell-breaking buffer (0.1 M Tris, pH 8.0, 20% glycerol, 0.5 mM PMSF, 2 mM benzamidine, 1 μM leupeptin, 2 μM pepstatin, 4 μM chymostatin, 2.6 μM aprotinin). The following calculation was used: β-galactosidase activity = OD420 × (1.7/0.0045) × 1,000/time × volume × concentration, where time is measured in minutes, volume is in microliters, and concentration is in micrograms/microliter.
Comparative promoter analysis of Tec1 and Ste12 sites.
Ste12- and/or Tec1-regulated genes were obtained from two microarray experiments of dig1 dig2 versus wild type (21), available at http://www.rii.com/publications/2000/cell_hughes.htm. Only genes whose expression levels in dig1 dig2 were at least twofold higher than wild type in both experiments were used in our analysis. Ty genes or genes whose promoters overlap with a Ty element were also excluded. Sequences of promoters 1,000 bp upstream of the selected Ste12-regulated genes in S. cerevisiae were extracted from the yeast genome database (http://www.yeastgenome.org/). Their coding sequences were blasted with the genome sequences of three other Saccharomyces species (http://www.broad.mit.edu/annotation/fungi/compyeasts/) to identify their orthologs and obtain 1,000-bp upstream regions of these orthologs. Promoter sequences from different Saccharomyces species were used to search for Ste12 (TGAAACR) and Tec1 (CATTCY) consensus sequences, and maps of potential sites were created using the regulatory sequence analysis tools website at http://rsat.ulb.ac.be/rsat/ (results are compiled in the figures in the supplemental material).
ChIP assay.
Chromatin immunoprecipitation (ChIP) analysis was performed as previously described (22, 42). The same set of protease inhibitors as described previously for immunoprecipitation was added to the ChIP lysis buffer. Two micrograms of anti-myc antibody (Santa Cruz) was incubated with the cell extract from 50 ml of cells grown to an OD600 of 1.0. PCR primiers used in the ChIP analysis are listed in Table 2.
TABLE 2.
Primers used in ChIP assays
| Primer | Sequence (5′ to 3′) |
|---|---|
| FUS1F | CAACAGAACAATAACGGCAACC |
| FUS1R | ACAAAGCCACTCTTACATTGTC |
| PRM1F | AATGCAAATTTCCGATGATGCC |
| PRM1R | CATCCTAACCAAATATTTCGGG |
| FIG2F | GAACAAATTATTTTGCCTTGTCC |
| FIG2R | AACTGGTCTCTAACAAATCTATAC |
| VPS75F | CTTCGTTATGTGTGCAACTATTC |
| VPS75R | AGCGCCTAATCATTAGGATGC |
| TRA1F | GTGCACGGAAGGCTTCACC |
| TRA1R | TGATTTGGTATTCGATACGTGC |
| AGA1F | CAAGTACGTCCACCCGTTTC |
| AGA1R | CGCTTGTTTTAAGTCATTACTATG |
| KAR4F | GAAGGAAGTTAGTATCGAGCTC |
| KAR4R | GCTGTTCTACTTTCTTCTATGTC |
| TIP1F | CATCAGACGATCTGGTTATGG |
| TIP1R | TCAATTATTTCCATCACAATCTTTG |
| CLN1F | GCATTCCCTTGTTCGCAACAC |
| CLN1R | CATTCTCAATCTTGCCTTCTGC |
| CHS7F | TGTATCGGATGTTCGGATGTC |
| CHS7R | AACACCAGAAGAATGTCAATAAC |
| CWP1F | AGGGATGCTGCAACCACGTC |
| CWP1R | TCGAGCACACAATGTTTGCCG |
RESULTS
Filamentation genes are regulated through TCS elements.
Based on the current model that the cooperative binding of Ste12 and Tec1 to FREs regulates the transcription of filamentation genes (29), promoters of filamentation genes should contain FREs (a PRE adjacent to a TCS). However, we noticed that many filamentation genes do not have a sequence that resembles an FRE in their promoter region. To further address this, we extracted 1,000-bp upstream sequences of Ste12-regulated mating and filamentation genes that have been identified by genome-wide transcription analyses (dig1 dig2 versus wild type) (21) and determined the existence of PRE (TGAAACR) and TCS (CATTCY) elements in these promoters. Because both motifs are rather short and can appear randomly at high frequency (PRE, 0.34 sites per 1,000 bp; TCS, 1.04 sites per 1,000 bp), we compared the promoter regions of these genes with the promoters of their orthologs from three other closely related Saccharomyces species (23). The information on PRE and TCS positions in these promoters is compiled in three figures that are available in the supplemental material. Genuine transcription factor binding motifs are likely to be conserved among all four species, while a random occurrence is generally not conserved (23). PREs and TCS that are conserved among Ste12-regulated genes in all four species are compiled in Table 3. All Ste12-regulated genes have either PREs, TCS, or both elements in their promoters. Not surprisingly, all the genes that have only PREs are involved in mating (Table 3; see Fig. S1 in the supplemental material). Only a few mating genes have both PREs and TCS in their promoters, including FUS1, FUS3, and PRM1 (Table 3; see Fig. S3 in the supplemental material). In contrast, most genes that contain only TCS are involved in filamentation (Table 3; see Fig. S2 in the supplemental material). A few filamentation genes also have PREs in their promoters (Table 3; see Fig. S3 in the supplemental material). Therefore, it appears that the major difference between mating and filamentation genes lies in the presence of PREs versus TCS in their promoters.
TABLE 3.
Comparative promoter analysis for conserved Ste12 and Tec1 binding sites in four closely related Saccharomyces speciesa
| Genes with only conserved PREsb
|
Genes with only conserved TCSc
|
Genes with both PREs and TCSd
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | No. of PREs | No. of TCS | Pathway | Expression ratio | Gene | No. of PREs | No. of TCS | Pathway | Expression ratio | Gene | No. of PREs | No. of TCS | Pathway | Expression ratio | |
| FIG1 | 1 | 0 | M | 0.5 | PGU1 | 0 | 2 | F | 44.7 | FUS1 | 3 | 1 | M | 0.7 | |
| SST2 | 1 | 0 | M | 1.0 | YOR296W | 0 | 1 | ? | 5.3 | FUS3 | 2 | 1 | M | NA | |
| PRM1 | 1 | 0 | M | 0.6 | SRL1 | 0 | 1 | F | 4.5 | PRM2 | 1 | 1 | M | 0.5 | |
| PRM3 | 1 | 0 | M | 0.4 | SRL3 | 0 | 2 | ? | 1.9 | PCL2 | 1 | 3 | ? | 0.6 | |
| PRM6 | 1 | 0 | M | 0.8 | CHS7 | 0 | 1 | F | 3.8 | GIC2 | 1 | 1 | M/F | 1.4 | |
| BAR1 | 2 | 0 | M | 1.0 | YNL208W | 0 | 2 | ? | 1.2 | SVS1* | 3 | 2 | ? | 71.9 | |
| FUS2 | 2 | 0 | M | 0.6 | YLR042C | 0 | 2 | F | 14.9 | MSB2 | 2 | 3 | F | 2.8 | |
| CIK1 | 1 | 0 | M | 0.6 | YMR173W | 0 | 1 | ? | 7.2 | YIL117C | 1 | 1 | M/F | 1.2 | |
| PRM4 | 1 | 0 | M | 0.4 | PHD1 | 0 | 1 | F | 2.2 | TEC1* | 4 | 2 | F | 1.1 | |
| AFR1 | 1 | 0 | M | 0.3 | KSS1 | 0 | 1 | F | 8.2 | YDR249C* | 1 | 2 | ? | 1.4 | |
| STE2 | 1 | 0 | M | 1.0 | GSC2 | 0 | 1 | ? | 1.9 | GFA1 | 1 | 1 | ? | 1.4 | |
| AGA1 | 3 | 0 | M | 0.4 | TIP1 | 0 | 1 | ? | 4.3 | ||||||
| KAR4 | 2 | 0 | M | 0.5 | FLO11 | 0 | 2 | F | NA | ||||||
| SCW10 | 2 | 0 | M | 0.5 | CLN1 | 0 | 2 | F | 2.7 | ||||||
| ASG7 | 2 | 0 | M | 0.4 | CWP1 | 0 | 1 | F | NA | ||||||
Microarray data of dig1 dig2 versus wild-type were used to identify Ste12-regulated genes (21). Genes that were upregulated at least twofold in dig1 dig2 versus wild type were included in this study. Ty1 genes were excluded from the list. In addition, FLO11, CLN1, CWP1, and TEC1 were added as they are shown to be regulated by the filamentation pathway and Ste12/Tec1 in other studies (31, 47). The 1,000-bp upstream sequences of each gene from all four Saccharomyces species were extracted (23) and compared to determine potential conserved Ste12 PREs (TGAAACR) and TCS sites (CATTCY). Functional assignments to either the mating (M) or filamentation (F) pathway are based on functional studies and/or transcriptional patterns (27, 31, 36, 45, 47). Ratios of their expression in fus3 mutants treated with 50 nM α-factor versus wild type treated with 50 nM α-factor are from Hughes et al. (21). NA, not available; ?, not determined.
See Fig. S1 in the supplemental material.
See Fig. S2 in the supplemental material.
See Fig. S3 in the supplemental material. Among the genes with both PREs and TCS, TEC1 contains an FRE, which is indicated with an asterisk. In addition, YDR249C and SVS1 also have a potential FRE with 11 bp and 4 bp between the PRE and TCS, respectively. GFA1 promoter contains a PRE and a TCS with 28-bp spacing.
We searched further for potential FREs among the genes that have both a PRE and a TCS in their promoter. In addition to two previously reported genes, TEC1 and Ty1 (5, 29), we found only two additional genes, YDR249C and SVS1, that contain a potential FRE (Table 3; see Fig. S3 in the supplemental material). S. cerevisiae FLO11 contains a single nucleotide mismatch in a potential PRE right upstream of a TCS, and the combination was previously suggested to be a potential FRE. However, the potential PRE is not in the promoters of FLO11 orthologs in the other three species (see Fig. S2 in the supplemental material) and, thus, might not be functional. Because the FLO11 promoter has been shown to be much larger than 1,000 bp (38), we extended our sequence comparison of FLO11 promoters to 3,000 bp. No FREs but only two conserved TCS and a separate PRE were found. Therefore, FLO11 is more likely regulated through TCS than through an FRE, although the upstream PRE may play a role in FLO11 regulation, too. In total, all filamentation genes have TCS in their promoters, but only four of them (Ty1, TEC1, YDR249C, and SVS1) contain an FRE. Our promoter analysis suggests that TCS and not FREs are the prevailing cis elements upstream of filamentation genes.
Our promoter analysis is in agreement with transcription patterns of these genes. Fus3 plays a positive role in the transcriptional activation of Ste12, whereas it specifically phosphorylates Tec1 and triggers ubiquitin-mediated Tec1 degradation during the pheromone response (1, 7, 8). Therefore, transcription of TCS-driven genes in a fus3 mutant is expected to be higher than that in wild type, while PRE-driven transcription in fus3 mutants should not be higher than that in wild type during the pheromone response. Ratios of gene expression in fus3 mutants treated with 50 nM α-factor versus wild type treated with 50 nM α-factor have been determined by Hughes et al. (21) and are listed in Table 3. As expected, all genes with only PREs have a ratio of ≤1.0, and genes with only TCS unanimously have a ratio of >1.0 (Table 3). The ratios also correlate well with the functional assignments for genes with both PREs and TCS (Table 3, right column).
Tec1 is in a complex with Ste12 and Dig1, which is distinct from the complex of Ste12 with Dig1 and Dig2.
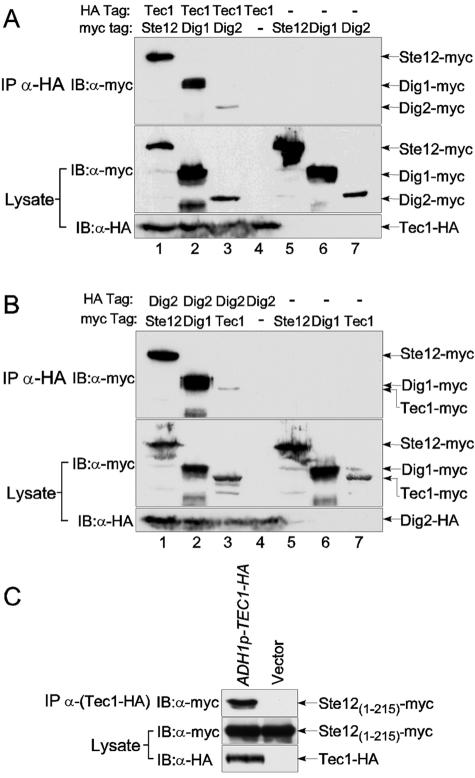
Since Ste12 can regulate filamentation genes without binding to PREs, there must be an alternative way for Ste12 to associate with the promoters. One likely model for Ste12 to control TCS-driven transcription is by direct association of Ste12 with Tec1. Ste12 is suspected to interact with Tec1 because Tec1 and Ste12 can bind cooperatively to FREs both in vitro and in vivo (5, 29), although it has not been shown whether the two proteins can interact in the absence of their binding to adjacent cis elements on DNA. The only report of Ste12 association with Tec1 independent of cis elements was from a systematic identification of protein complexes by mass spectrometry in cells that overexpressed TEC1-FLAG from the GAL1 promoter (20). To further investigate whether Tec1 interacts with Ste12 and Ste12-associated proteins Dig1 and Dig2, we tagged these proteins at their C termini with either HA3 or myc13 on chromosomes. The tagged proteins were expressed from their own promoters and were functional (data not shown). Fractionation experiments with epitope-tagged Ste12 showed that over 90% of Ste12 was in the pellet fraction with DNA after an ultracentrifugation when 150 mM NaCl was used for protein extraction, whereas Ste12 was effectively extracted when 300 mM NaCl was used (data not shown). Therefore, 300 mM NaCl was used in all of our in vivo immunoprecipitation experiments in this study. Immunoprecipitation of Tec1-HA with an anti-HA antibody was able to bring down similar amounts of Dig1-myc and Ste12-myc (Fig. 1A), despite the fact that Dig1 was more abundant than Ste12 in the whole-cell lysate. The interaction was specific, as immunoprecipitation of the cell extracts of untagged strains did not pull down any myc-tagged proteins. Reciprocal immunoprecipitation of Dig1 or Ste12 also detected Tec1 (data not shown). In contrast to Dig1 and Ste12, immunoprecipitation of Tec1-HA did not bring down a substantial amount of Dig2 (Fig. 1A, lane 3). Repeating the Tec1 immunoprecipitation showed that Tec1 consistently pulled down Dig1 and Ste12, but the Dig2 signal in the Tec1 immunoprecipitation (IP) was sometimes weak and sometimes undetectable. As Dig1 is more abundant than Dig2 in whole-cell lysate, one may argue that the lack of detectable Dig2 in the Tec1 IP could be caused by the difference in protein abundance. But this is unlikely because immunoprecipitation of Dig2-HA specifically brought down Ste12-myc, as previously reported (43), as well as Dig1-myc (Fig. 1B), but the amount of Tec1 in the Dig2 IP was barely detectable, despite the fact that similar levels of Ste12 and Tec1 were in the cell lysate (Fig. 1). Therefore, Tec1 and Dig2 are likely in two different complexes with Ste12 and Dig1. The Ste12/Dig1/Dig2 complex is the known Ste12 complex for the mating program (34), while the newly identified Tec1/Ste12/Dig1 complex is likely for the filamentation program.
FIG. 1.
Tec1 forms a complex with Ste12 and Dig1 but not with Dig2. (A) Immunoprecipitation of Tec1-HA. Protein lysates were subjected to immunoprecipitation with an anti-HA antibody, and the precipitation products were resolved by SDS-polyacrylamide gel electrophoresis and probed with an anti-myc antibody. As controls, cell lysates were subjected to Western blotting with anti-myc and anti-HA antibodies. The following yeast strains were used: lane 1, HLY3334 (TEC1-HA STE12-myc); lane 2, HLY3335 (TEC1-HA DIG1-myc); lane 3, HLY3336 (TEC1-HA DIG2-myc); lane 4, HLY3320 (TEC1-myc); lane 5, HLY3321 (STE12-myc); lane 6, HLY3322 (DIG1-myc); and lane 7, HLY3323 (DIG2-myc). The lower band in lane 2 is a breakdown product of Dig1-myc. (B) Immunoprecipitation of Dig2-HA, as in panel A. The yeast strains in each lane are as follows: lane 1, HLY3337 (DIG2-HA STE12-myc); lane 2, HLY3338 (DIG2-HA DIG1-myc); lane 3, HLY3339 (DIG2-HA TEC1-myc); lane 4, HLY3327 (DIG2-HA); lane 5, HLY3321 (STE12-myc); lane 6, HLY3322 (DIG1-myc); and lane 7, HLY3324 (TEC1-HA). The lower bands in lanes 2 and 6 are a breakdown product of Dig1-myc. (C) Tec1 binds to the N-terminal region of Ste12. The yeast strain HLY3350 (ste12 tec1) carrying plasmid pHL754 (ADH1p-Ste12(1-215)-myc) was transformed with either pHL731 (ADH1p-TEC1-HA) or a vector (pRS313), and the transformed strains were used in immunoprecipitation with an anti-HA antibody. The precipitated proteins were analyzed by Western blotting with an anti-myc antibody. IB, immunoblot.
TCS-lacZ expression and LexA-Tec1 transcriptional activity are inhibited by Dig1 but not Dig2.
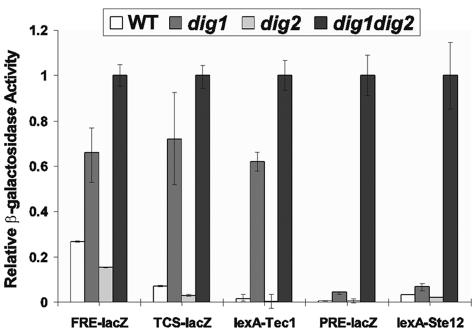
Dig1 and Dig2 are two functionally redundant inhibitors of Ste12, and PRE-lacZ expression or Ste12 activity is high in dig1 dig2 double mutants but not in either dig1 or dig2 single mutants (9, 43). FRE-lacZ is also highly expressed in a dig1 dig2 double mutant (4), but its expression in dig1 or dig2 single mutants has not been reported. If only Dig1, but not Dig2, is present in the complex with Tec1 and Ste12, TCS-driven expression is expected to be high in a dig1 strain. To investigate whether Dig1 and Dig2 play different roles in the regulation of TCS-driven transcription, we assayed TCS-lacZ (8) expression in dig1, dig2, and dig1 dig2 mutants. For comparison, we also assayed the expression of PRE(FUS1)-lacZ (44) and FRE(Ty1)-lacZ (5, 29, 32) reporters in the dig1, dig2, and dig1 dig2 mutants. Expression of TCS-lacZ and FRE(Ty1)-lacZ, but not PRE(FUS1)-lacZ, was significantly elevated in dig1 mutants (Fig. 2). In contrast, TCS-lacZ and FRE(Ty1)-lacZ expression was not increased in dig2 mutants. Deletion of both DIG1 and DIG2 increased the expression of all three reporters, as expected for TCS-lacZ and as previously reported for FRE(Ty1)-lacZ and PRE(FUS1)-lacZ (4, 34). Therefore, a dig1 single mutant is able to release the inhibition on FRE- or TCS-driven expression. This is consistent with a genome-wide transcription analysis which shows that Dig1 is the primary negative regulator for the expression of filamentation genes (6).
FIG. 2.
Differential regulation of Ste12 and Tec1 transcriptional activities by Dig1 and Dig2. Relative β-galactosidase activities of PRE(FUS1)-lacZ, FRE(Ty1)-lacZ, and TCS-lacZ (pHL710) in wild-type (10560-4A), dig1 (HLY3315), dig2 (HLY3316), and dig1 dig2 (HLY3317) strains and relative β-galactosidase activities of LexA-Tec1 (pHL711) and LexA-Ste12 (pHL712) in lexAops-lacZ-integrated wild-type (HLY3328), dig1 (HLY3329), dig2 (HLY3330), and dig1 dig2 (HLY3331) strains are shown. The activity for each strain was an average from three independent transformants, and the relative activity was calculated by dividing activity by that of the dig1 dig2 strain for each reporter.
To determine whether the high TCS-lacZ expression in the dig1 strain reflects the inhibitory effect of Dig1 on Tec1 transcriptional activity, we constructed a fusion of Tec1 to the DNA-binding domain of bacterial LexA, and the lexA-TEC1 expression was under the control of the ADH1 promoter so that the lexA-TEC1 expression was not influenced by Ste12 activity. Tec1 transcriptional activity was assayed in strains carrying a lacZ reporter under the regulation of lexA operators. Similar to the TCS reporter, LexA-Tec1 activity was up by ∼45-fold in the dig1 mutant (Fig. 1). In contrast to LexA-Tec1, LexA-Ste12 activity was high only in the dig1 dig2 double mutant and not in the dig1 single mutant, as previously reported (34). Therefore, Dig1 is the major inhibitor of Tec1 activity. Since PRE-lacZ expression or LexA-Ste12 activity was similar in dig1 and dig2 single mutants, the observed difference in TCS-lacZ expression or LexA-Tec1 activity between dig1 and dig2 strains is unlikely due to the difference in Dig1 and Dig2 protein abundance but could be explained by the protein composition in the Tec1/Ste12/Dig1 and Ste12/Dig1/Dig2 complexes.
Tec1 binds to the same region of Ste12 as Dig2.
Since the two Ste12 complexes differ in Tec1 and Dig2, a possible mechanism that could give two distinct Ste12 complexes is one whereby Tec1 and Dig2 bind to the same region on Ste12 in a mutually exclusive way. Dig2 is known to bind to the N-terminal DNA binding region of Ste12 (34). We found that Tec1 also associated with the N-terminal region of Ste12 by immunoprecipitation (Fig. 1C). A myc-tagged N-terminal fragment of Ste12(1-215) in an ste12 mutant was used for IP to avoid the potential interaction of the Ste12 N terminus with full-length Ste12. Because Ste12(1-215) is not sufficient for the expression of Ste12-regulated genes, HA-tagged Tec1 was expressed from the ADH1 promoter. The N-terminal DNA binding region (residues 1 to 215) of Ste12 is sufficient for interaction with Tec1; other regions of Ste12 were not required for the interaction with Tec1, as immunoprecipitation of Ste12 with deletions between residues 253 to 355, 387 to 512, and 512 to 669 could still pull down Tec1 (data not shown).
Dig1 interaction with Tec1 requires Ste12.
Because Dig1 can bind Ste12 and inhibit Ste12 activity (9, 43) and because Ste12 is a component of the Tec1 immunocomplex, it is possible that the interaction of Dig1 with Tec1 is not direct but is mediated through Ste12. To test this possibility, we examined whether Dig1 and Tec1 still interact in the absence of Ste12. Because TEC1 expression is Ste12 dependent, we placed TEC1-HA under the control of the ADH1 promoter. Dig1-myc was detected in the Tec1-HA immunoprecipitation in wild type but not in an ste12 mutant (Fig. 3A). Therefore, Ste12 is required for Tec1 interaction with Dig1.
FIG. 3.
Ste12 interaction with Tec1 is essential for the transcriptional activation of Tec1. (A) The Dig1 and Tec1 interaction is Ste12 dependent. Yeast strains HLY3335 (Tec1-HA Dig1-myc) and HLY3343 (Tec1-HA Dig1-myc ste12) with TEC1 under its endogenous promoter and strains HLY3322 (Dig1-myc) and HLY3342 (Dig1-myc ste12) bearing the plasmid pHL731 (ADH1p-Tec1-HA) were used for immunoprecipitation with an anti-HA antibody. The lysates and IP products were analyzed on Western blots with an anti-myc antibody. (B) Dig1 inhibition of LexA-Tec1 activity is Ste12 dependent. β-Galactosidase assays of LexA-Tec1 (ADH1p-lexA-TEC1) in strains HLY3328 (wild type; lexAops-lacZ), HLY3329 (dig1 lexAops-lacZ), HLY3332 (ste12 lexAops-lacZ), and HLY3333 (dig1 ste12 lexAops-lacZ). (C) Ste12 interacts with a Tec1 C-terminal region. Yeast strain HLY3321 (STE12-myc) carrying plasmids expressing various fragments of Tec1-HA or pRS314 (vector) were grown to mid-log phase and subjected to immunoprecipitation with an anti-HA antibody. N- and C-terminal residues of each remaining Tec1 fragment are indicated. (D) β-Galactosidase assays of the LexA-Tec1 fragments in the yeast strains HLY3328 (wild type; lexAops-lacZ) and HLY3329 (dig1 lexAops-lacZ). IB, immunoblot.
Tec1 transcriptional activity is dependent on its association with Ste12.
Not only was the interaction of Dig1 and Tec1 dependent on Ste12, but deleting STE12 in a dig1 mutant also blocked the elevated LexA-Tec1 transcriptional activity that was otherwise observed in a dig1 strain (Fig. 3B, dig1 versus dig1 ste12). It is possible that Tec1-associated Ste12 is directly responsible for the induction of Tec1 transcriptional activity in the dig1 strain. In this case, the region of Tec1 that interacts with Ste12 should be required for Tec1 transcriptional activation. To test this possibility, we generated deletions of Tec1 from either the N or the C terminus. Deletions within the first 300 residues did not affect its association with Ste12, whereas a deletion of up to 400 residues was unable to bind Ste12 (Fig. 3C). A deletion from residue 401 to the C terminus was able to bind to Ste12, but deleting to residue 301 abolished Ste12 binding (Fig. 3C). These data suggest that the region between residues 301 to 400 of Tec1 is required for Tec1 to interact with Ste12. Although required, the region was not sufficient for binding with Ste12 (data not shown). The Tec1 deletions were then fused in frame to the DNA binding domain of LexA, and the LexA-Tec1 fusions were analyzed for Tec1 transcriptional activity in the dig1 strain. We found that C-terminal deletions of Tec1 abolished the elevated LexA-Tec1 transcriptional activity in the dig1 strain (Fig. 3D). In contrast, the region from the N terminus to residue 300 was not required for the high LexA-Tec1 activity in the dig1 mutant (Fig. 3D, Tec1 regions 101 to 486, 201 to 486, and 301 to 486). These data suggest that the Ste12 binding region (residues 301 to 400) is required for LexA-Tec1 transcriptional activity. Therefore, Tec1 transcriptional activity is dependent on its association with Ste12, which is under the negative regulation of Dig1. Although the C terminus of Tec1 (from 401 to 486) was not essential for Ste12 binding, it was still required for lexA-Tec1 transcriptional induction in the dig1 strain (Fig. 3D). This suggests that, besides Ste12, there might be additional regulations on the C terminus of Tec1.
Stoichiometry of Ste12 interaction with Tec1, Dig1, and Dig2 in vitro.
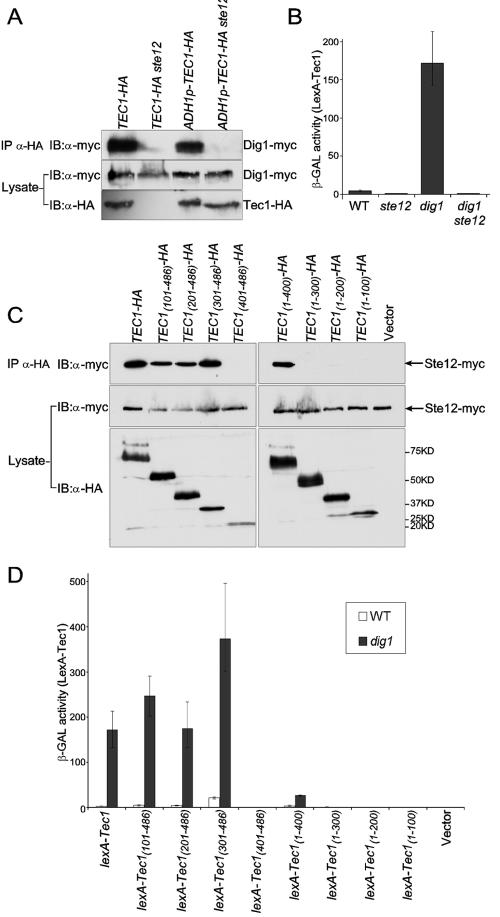
To further characterize the interaction between Tec1 and Ste12, we generated Tec1-FLAG and Ste12 by in vitro transcription/translation and examined whether Tec1-FLAG could interact with Ste12 in vitro. As shown in Fig. 4A, lane 4, IP with an anti-FLAG antibody brought down similar levels of 35S-labeled Tec1-FLAG and Ste12. Reciprocally, IP of 35S-labeled myc-Ste12 also brought down 35S-Tec1 at the molar ratio of about 1 Ste12 to 1.3 Tec1 (Fig. 4D, lane 8).
FIG. 4.
Formation of distinct Ste12 complexes in vitro. (A) Tec1 binds to Ste12 with an equal molar ratio. One microliter of in vitro translated and 35S-labeled Tec1-FLAG or Ste12 was loaded as input. A total of 15 μl of each was used for immunoprecipitation with anti-FLAG M2-agarose (Sigma). (B) Tec1 does not bind Dig1 or Dig2 directly. Dig1 association with Tec1-Ste12 is mediated through Ste12. Either 1 μl or 15 μl of in vitro translated product was used as input or in IP for each protein as described in panel A. (C) Tec1-Ste12 complex does not bind Dig2. Limited Dig2 is tethered to the complex through Dig1. Both 1 μl and 15 μl of in vitro translated product were used as input or in IP for each protein as described in panel A. (D) myc-Ste12 association with Tec1, Dig1, and Dig2. Either 2 μl or 30 μl of in vitro translated myc-Ste12 was loaded as input or used in IP with c-myc rabbit polyclonal immunoglobulin G(Santa Cruz Biotechnology) and protein A-Sepharose CL-4B (Amersham Biosciences). For other proteins, 1 μl or 15 μl of in vitro translated product was used as input or in IP. The percent adjusted intensity/number of methionines relative to Ste12 has been calculated for lanes 8 to 11.
We also used in vitro translated proteins to confirm that Ste12 is required to mediate the interaction between Tec1 and Dig1. IP of Tec1-FLAG did not bring down Dig1 in the absence of Ste12 (Fig. 4 B, lane 8); the weak Dig1 band in lane 8 was nonspecific, as a similar level of Dig1 was also seen in the IP with beads without Tec1-FLAG in lane 5. However, a significant amount of Dig1 was precipitated with Tec1-FLAG in the presence of Ste12 (Fig. 4B, lane 10). Therefore, Tec1 does not interact with Dig1 directly; rather, Ste12 bridges Tec1 and Dig1 in the complex.
In contrast to Dig1, Dig2 was not detected in the Tec1-FLAG immunoprecipitation even in the presence of Ste12 (Fig. 4B, lane 11). Interestingly, when both Ste12 and Dig1 were present, Tec1-FLAG could bring down a small, but detectable amount of Dig2 (Fig. 4B, lane 12 as indicated by the arrow). Because 35S-Ste12 IP also produced a faint band at the same position as Dig2 (Fig. 4B, lanes 7, 10, and 11), we repeated the Tec1-FLAG IP experiment with unlabeled Ste12 (Fig. 4C). IP of Tec1-FLAG in the presence of Ste12 could bring down Dig1 (lane 7) but not Dig2 (lane 5). In the presence of both Ste12 and Dig1, Tec1-FLAG could bring down a small amount of Dig2 (Fig. 4C, lane 6). Therefore, a small amount of Dig2 is tethered to the Tec1/Ste12/Dig1 complex through its association with Dig1.
We also investigated the ability of Ste12 to interact with Dig1 and Dig2 by using a myc-tagged Ste12 for IP (3). As shown by Bardwell et al., a small amount of Dig1 bound to myc-Ste12 (Fig. 4D, lane 9). The molar ratio of this binding is about 1 Ste12 to 0.3 Dig1. The amount of Dig2 in the myc-Ste12 IP was also very small, at about 1 Ste12 to 0.2 Dig2 (Fig. 4D, lane 10). Surprisingly, in the presence of both Dig1 and Dig2, the amount of Dig1 and Dig2 associated with myc-Ste12 reached a molar ratio of 1.4 Dig1 and 1.5 Dig2 to 1 Ste12 (Fig. 4D, lane 11). Therefore, there is synergy between Dig1 and Dig2 in binding to myc-Ste12 in vitro. Identical results, both in terms of low levels of binding with Dig1 or Dig2 and the synergy between them, were observed with an Ste12-myc in which we fused myc13 to the C terminus of Ste12 (data not shown).
Ste12-associated Dig2 can be competed off by Tec1.
Because both Tec1 and Dig2 bind to the N-terminal region of Ste12 (Fig. 1C), we reasoned that they might bind to Ste12 in a competitive and, therefore, mutually exclusive manner to generate two distinct Ste12 complexes. To determine whether Tec1 can compete with Dig2 for binding to Ste12, an increasing amount of Tec1 was added to IP reaction mixtures that contained an equal amount of myc-Ste12 and 35S-labeled Dig1 and Dig2. Decreasing levels of Dig2 and Dig1 were found to be associated with myc-Ste12 in the presence of an increasing amount of Tec1 (Fig. 5A). This suggests that excess Tec1 could compete off Dig2 from Ste12. Interestingly, a significant amount of Dig1 also fell off with Dig2 from the Ste12 complex in the presence of a large excess of Tec1 (Fig. 5A, lane 8 and 9), consistent with the observed synergistic interaction of Dig1 and Dig2 with Ste12. When we carried out the Tec1 competition experiment with myc-Ste12 and Dig1, the Ste12-bound Dig1 level did not change with an increasing amount of Tec1 (data not shown). When an increasing amount of Tec1 was added to myc-Ste12 and Dig2, Ste12-bound Dig2 decreased; however, the initial amount of Ste12-bound Dig2 was very small (data not shown).
FIG. 5.
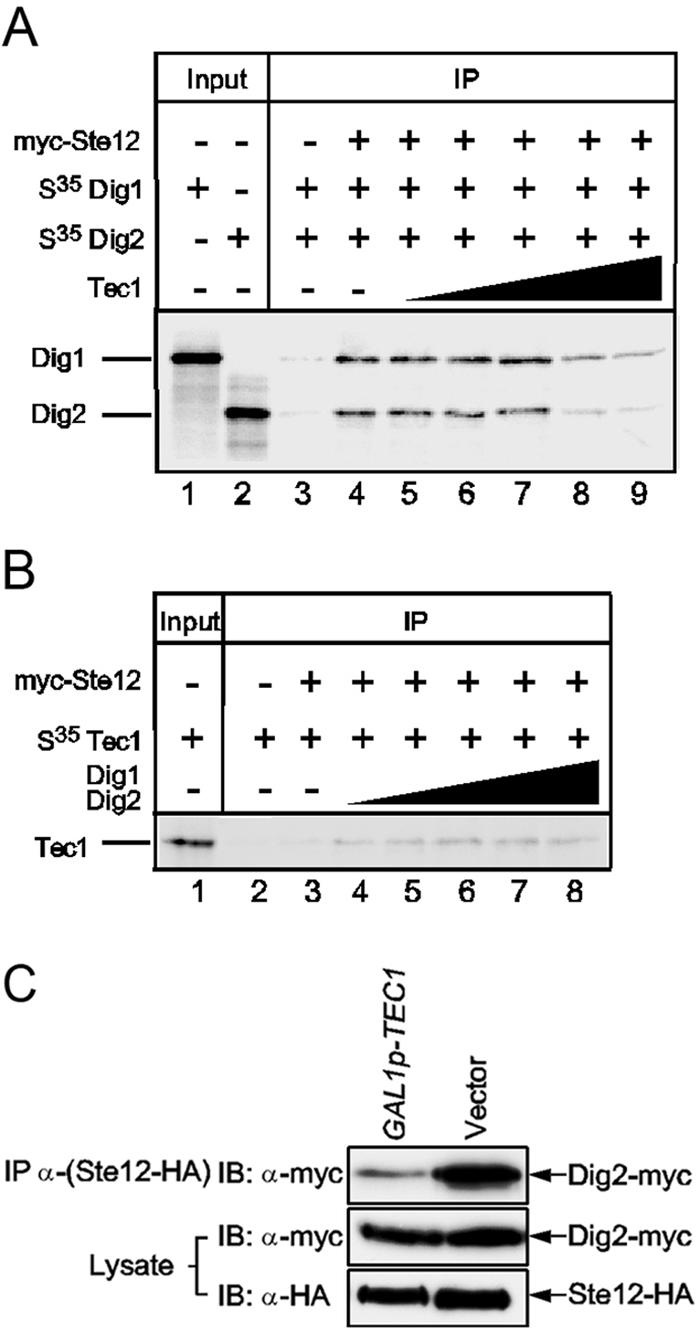
Tec1 competes with Dig2 for Ste12 binding both in vitro and in vivo. (A) Tec1 can compete off Dig2 from Ste12. Twenty microliters of myc-Ste12 and 10 μl of in vitro translated and 35S-labeled Dig1 and Dig2 were used in each immunoprecipitation as described in the legend of Fig. 4D, and 1 μl of Dig1 or Dig2 was loaded as input. Unlabeled in vitro translated Tec1 was added to each IP in increasing amounts: 0 μl, 5 μl, 10 μl, 20 μl, 50 μl, and 100 μl. (B) Dig2 cannot compete off Tec1 from Ste12. Twenty microliters of myc-Ste12 and 10 μl of 35S-labeled Tec1 were used in each IP as described in the legend of Fig. 4D, and 1 μl of Tec1 was loaded as input. Increasing amounts (0 μl, 5 μl, 10 μl, 20 μl, 50 μl, and 100 μl) of both Dig2 and Dig1 were added to each IP for competition. (C) Tec1 competes with Dig2 for Ste12 binding in vivo. Strain HLY3340 (STE12-HA DIG2-myc tec1) carrying either a GAL-TEC1 (27c-2A) (33) or a vector was grown in YEPD until log phase; cells were washed several times with water and resuspended into YEP +2% raffinose to grow overnight. Galactose (2%) was added, and cells were grown for an additional 4.5 h before harvest for IP with an anti-HA antibody. The lysate and IP eluate were blotted with either anti-HA (anti-Ste12) or anti-myc (anti-Dig2) antibodies. IB, immunoblot.
We also examined whether Dig2 and Dig1 proteins could compete off Tec1 from Ste12. Interestingly, Tec1 could not be competed off from Ste12 with excess Dig2 and Dig1 proteins (Fig. 5B). The competition data suggest that Tec1 and Dig2 share an overlapping region of Ste12 for binding because Tec1 could replace Dig2 from Ste12, but their affinities are not identical. Tec1 may have a higher Ste12 binding affinity than that of Dig2, and, therefore, it cannot be replaced by Dig2 in binding to Ste12.
We further tested the ability of Tec1 to compete with Dig2 for Ste12 binding in vivo by an Ste12 IP in cells with and without overexpression of TEC1. When overexpressed from the GAL1 promoter, Tec1 significantly reduced the amount of Dig2 that was associated with Ste12 in yeast cells (Fig. 5C). Thus, our data show that Tec1 competes with Dig2 in binding with Ste12 both in vivo and in vitro.
The Tec1/Ste12/Dig1 complex binds to TCS of filamentation genes and PREs of mating genes.
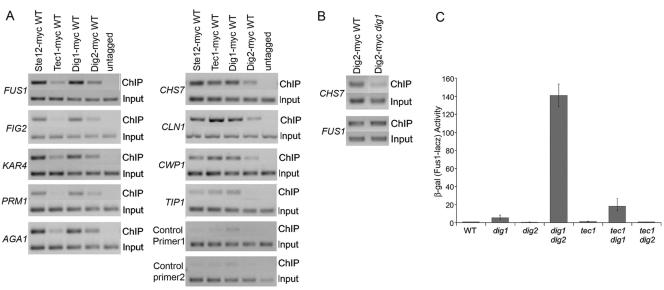
If filamentation genes are regulated by the Tec1/Ste12/Dig1 complex via the TCS and mating genes are regulated by the Ste12/Dig1/Dig2 complex via the PRE, we would expect to find Dig2 at the promoters of mating genes and Tec1 at those of filamentation genes. Ste12 and Dig1 should be present at the promoters of both groups. A study of the genome-wide location of Ste12, Dig1, and Tec1 has shown that Ste12 and Dig1 are present at both mating and filamentation genes, and Tec1 is present at filamentation genes as well as at some of the mating genes (47). The localization of Dig2 in these genes is not known. Therefore, we compared the distribution of Tec1 and Dig2 at the promoters of mating and filamentation genes using ChIP analysis. Ste12 and Dig1 were included as controls. STE12-myc, TEC1-myc, DIG1-myc, and DIG2-myc strains were grown in YEPD (yeast extract, peptone, and dextrose) medium and harvested for ChIP analysis. The immunoprecipitated DNA was analyzed by PCR using primer pairs that are located about 100 bp upstream and downstream of either the TCS elements of filamentation genes or PRE elements of mating genes. Because we wanted to determine the differences between Ste12 complexes at PREs and at TCS, promoters containing both PRE and TCS sites were excluded from the analysis. In addition, only promoters that were efficiently bound by Ste12 and Tec1 in the whole-genome ChIP experiment were used (47). Ste12, Tec1, and Dig1 were present in nearly equal amounts at the TCS of filamentation genes, whereas Dig2 was detected at a lower level (Fig. 6A). Since our in vitro binding experiments have shown that Tec1, Ste12, and Dig1 were present at a near equal ratio in the Tec1/Ste12 complex and that limited Dig2 was bound to the complex through the association with Dig1, the relative amounts of the four proteins detected at the filamentation genes are consistent with the localization of the Tec1/Ste12/Dig1 complex to the filamentation genes.
FIG. 6.
Distribution and function of the Tec1/Ste12/Dig1 complex at TCS sites of filamentation genes and PREs of mating genes (A) Yeast strains HLY3320 (TEC1-myc), HLY3321 (STE12-myc), HLY3322 (DIG1-myc), HLY3323 (DIG2-myc), and 10560-4A (wild type) were grown to mid-log phase in YEPD for ChIP with an anti-myc antibody. The input and ChIP products were amplified by PCR with promoter-specific primers (sequences are listed in Table 2) and resolved on an agarose gel. Promoters of VPS75 and TRA1 were used as negative controls. (B) Yeast strains HLY3323 (DIG2-myc) and HLY3405 (DIG2-myc dig1) were grown to log phase and subjected to ChIP analysis with an anti-myc antibody. (C) β-Galactosidase assays of PRE(FUS1)-lacZ (29) in yeast strains 10560-4A (wild type), HLY3315 (dig1), HLY3316 (dig2), HLY3317 (dig1 dig2), HLY2187 (tec1), HLY3318 (dig1 tec1), and HLY3319 (dig2 tec1). WT, wild type.
To test whether the presence of Dig2 at the promoters of the filamentation gene promoters was due to its association with Dig1 in the Tec1/Ste12/Dig1 complex, instead of with Ste12, we compared Dig2-myc localization at the promoters of filamentation genes in a wild-type strain and a dig1 mutant by ChIP (Fig. 6B). The deletion of DIG1 greatly decreased the amount of Dig2 bound to the promoter of CHS7 (Fig. 6B) as well as CWP1 (data not shown), indicating that Dig2 associates with Dig1 to TCS. The observed decrease in Dig2 at the CHS7 promoter in the dig1 mutant was not due to a potential Dig1-dependent interaction of Dig2 with Ste12, as the same amount of Dig2 was detected at the PREs of FUS1 in wild-type and dig1 mutant cells (Fig. 6B).
PCR of selected mating genes from the above ChIPs shows that Tec1 was present, but at a much lower level than Ste12, Dig1, and Dig2, at the promoters of mating genes (Fig. 6A). Because Tec1 is only present in the Tec1/Ste12/Dig1 complex, the detection of the small amount of Tec1 at the mating genes suggested that there was some Tec1/Ste12/Dig1 complex at the promoters of mating genes via Ste12 binding to PREs. We suggest that both types of Ste12 complexes are present at the promoters of mating genes. The Ste12/Dig1/Dig2 complex is the major form, while the Tec1/Ste12/Dig1 complex is the minor form.
The biological significance for the presence of the Tec1/Ste12/Dig1 complex at the PREs of mating genes is not clear. But we did find that the PRE(FUS1)-lacZ expression level was slightly elevated in tec1 and was even higher in a tec1 dig1 double mutant (Fig. 6C). The synergistic effect between tec1 and dig1 was specific, as PRE(FUS1)-lacZ expression was not increased in a tec1 dig2 mutant, suggesting that Tec1 might function together with Dig1 in inhibiting Ste12 activity at the promoters of mating genes.
DISCUSSION
Formation of two distinct Ste12 complexes by competitive binding of Tec1 and Dig2 to Ste12.
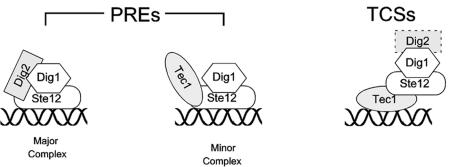
In this study, we show that Tec1 and Ste12, along with Dig1, form an Ste12 complex that is different from the known Ste12/Dig1/Dig2 complex that regulates mating genes (Fig. 7). In vivo IP of Tec1 or Dig2 could pull down Ste12 and Dig1 but not much of each other. Using in vitro translated proteins, we show that Tec1 can bind directly to Ste12 at a nearly 1:1 molar ratio but cannot bind to Dig1 or Dig2. Dig1 is associated with the Tec1-Ste12 complex through Ste12, probably with the middle region of Ste12 as previously defined for the Ste12/Dig1/Dig2 complex (34). Limited Dig2 is tethered to the Tec1 complex through its interaction with Dig1. This explains why residual Dig2 is found in the Tec1 complex in vivo in both IP and ChIP experiments.
FIG. 7.
Proposed regulation of mating and filamentation genes by Ste12/Dig1/Dig2 and Tec1/Ste12/Dig1 complexes in S. cerevisiae. Ste12 forms two distinct transcriptional complexes: Ste12/Dig1/Dig2 at the PREs of mating genes and Tec1/Ste12/Dig1 at the TCS of filamentation genes. A small amount of Tec1/Ste12/Dig1 is also present at the PREs of mating genes. The expression of filamentation genes is regulated by Tec1-associated Ste12 on TCS, which is inhibited by Dig1. A smaller amount of Dig2 is associated with the Tec1 complex through Dig1; thus, Dig2 does not regulate Ste12 activity.
Ste12 can form two distinct complexes because Tec1 and Dig2 bind to Ste12 in a competitive manner so that Ste12 interacts with either Tec1 or Dig2 but not both. Like Dig2 (34), Tec1 binds to the N-terminal DNA binding region of Ste12. Excess Tec1 can replace Dig2 from binding to Ste12 both in vitro and in vivo. This provides an underlying mechanism for the existence of two distinct Ste12 complexes. Interestingly, excess Dig2 cannot replace Tec1 from the associated Ste12, indicating that Tec1 may have a higher affinity for Ste12 than Dig2. The different Ste12 binding affinities between Dig2 and Tec1 may imply that the amount of Tec1 in a cell determines the ratio of the two Ste12 complexes. The number of Tec1, Dig2, Dig1, and Ste12 molecules inside a MATa cell, not exposed to alpha pheromone, is estimated to be about 530, 1310, 1460, and 1920 molecules/cell, respectively (15). Therefore, Tec1 seems to be the limiting protein among the three Ste12 binding proteins. Considering that Tec1 transcription and protein stability are tightly regulated by the mating and filamentation MAP kinase pathways and that Tec1 has a higher affinity for Ste12 than Dig2, the amount of Tec1 may determine the ratio of the two Ste12 complexes in a cell, thus facilitating either mating or filamentation.
The Tec1/Ste12/Dig1 complex regulates TCS-driven transcription.
We show that TCS-driven transcription is regulated by the Tec1/Ste12/Dig1 complex. Tec1 by itself has minimal transcriptional activity. Its activity is determined by the associated Ste12 because Tec1 transcriptional activity is low in ste12 mutants, and STE12 deletion abolishes all elevated Tec1 activity in a dig1 mutant. This result is different from the reported Ste12-independent transcriptional activity of Tec1 in TCS control (24). The Ste12-independent activity of Tec1 appears to be significant only when Tec1 is highly overproduced (24), whereas we showed Ste12-dependent Tec1 transcriptional activity and TCS transcription under normal circumstances and in a dig1 strain. The Ste12-independent activation of Tec1 could be mediated through the C terminus (residues 401 to 486) of Tec1, as we found the region is not essential for Ste12 interaction but is required for Tec1 transcriptional activity. In the Tec1/Ste12/Dig1 complex, the main function of Tec1 is to associate with Ste12 through its C-terminal region to bring Ste12 and its transcriptional activity to TCS sites. Removal of the Tec1 interaction domain with Ste12 abolishes its transcriptional activity. The same C-terminal domain of Tec1 is termed the TCS-control region by Kohler et al. (24). We suggest that Ste12 acts as the transcriptional activator in the Tec1/Ste12/Dig1 complex and, thus, places the TCS control under the filamentation MAP kinase pathway. Active Kss1 removes Dig1 inhibition on Ste12 and allows transcription from a TCS. In fact, Fus3 and Kss1 have an equal and exchangeable role in activating transcription from TCS, as revealed by a stable Tec1 mutant in kss1 and fus3 (8), which is also consistent with the finding that both Fus3 and Kss1 are able to phosphorylate Dig1 (9, 43).
A limited amount of Dig2 is present in the Tec1/Ste12/Dig1 complex via interaction with Dig1, based on our in vitro IP (Fig. 4D) and ChIP analysis of Dig2 in dig1 cells (Fig. 6B). The lack of direct Dig2 interaction on Ste12 in the Tec1/Ste12/Dig1 complex may explain the difference in basal transcription levels of mating and filamentation genes, as filamentation genes are moderately expressed in vegetative growing cells, whereas mating genes are more stringently regulated with very low levels of basal expression. It also explains why Dig1 is the major inhibitor for the expression of filamentation genes, while Dig2 has minimal effects (6).
Transcriptional regulation of most filamentation genes is through TCS.
The transcription of filamentation genes is thought to be regulated by cooperative interaction of Ste12 and Tec1 via the FRE cis elements (29). However, most filamentation genes do not have FREs in their promoters (Table 3). Many filamentation genes do not even have a PRE; rather, they all have TCS. Since a single TCS is sufficient to provide the expression pattern of filamentation genes (24), most filamentation genes are likely regulated through TCS by the Tec1/Ste12/Dig1 complex. In support of this, we show by ChIP assay that Tec1, Ste12, and Dig1 are present at the promoters of filamentation genes, while Dig2 is detected at a lower level, and the binding is Dig1 dependent. Our model of the Tec1/Ste12/Dig1 complex binding to TCS elements gives an alternative and more general explanation for why the binding of Ste12 to filamentation genes is Tec1 dependent (47).
Ste12 controls diverse transcriptional programs by association with different cofactors that are differentially regulated at the transcription and protein stability levels.
The regulation of Ste12-mediated transcriptional programs provides a prime example for us to understand how a transcription factor selectively activates distinct developmental programs in response to different stimuli. In the case of Ste12, it controls different transcriptional programs by selective partnership with different cofactors. We show that Tec1 brings Ste12 to TCS elements to activate filamentation genes. This mechanism is similar to the regulation of α-specific genes, where Ste12 is brought by α1 to the promoters of α-specific genes (46). Ste12 activation by the pheromone-responsive MAP kinase pathway is responsible for the induction of a-specific, α-specific, and haploid-specific genes. Active Ste12, therefore, is capable of activating all genes under its control through interaction with its cofactors. Thus, a key to the signaling specificity is selective regulation of the associated cofactors. Degradation of Tec1 specifically disassociates Ste12 from the promoters of filamentation genes and turns off the filamentation transcriptional program (1, 7, 8). Degradation of Tec1 may also allow maximal induction of mating genes (7). Therefore, signaling specificity for Ste12 in regulating multiple developmental pathways is achieved by its ability to associate with other transcription factors and by selective regulation of the associated factors.
Conclusion.
Ste12 forms two distinctive complexes, Ste12/Dig1/Dig2 and Tec1/Ste12/Dig1, by competitive binding of Dig2 and Tec1 to the N terminus of Ste12. Most filamentation genes are regulated by the Tec1/Ste12/Dig1 complex via Tec1 binding sites.
Supplementary Material
Acknowledgments
We thank Gerald Fink, Hiten Madhani, Hans Mosch, and Lee Bardwell for strains and plasmids and Peter Kaiser for helpful discussions.
This work is supported by National Institutes of Health Grant GM55155. S. Chou was a predoctoral fellow of a training grant from the University of California Systemwide Biotechnology Research and Education program.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bao, M. Z., M. A. Schwartz, G. T. Cantin, J. R. Yates III, and H. D. Madhani. 2004. Pheromone-dependent destruction of the Tec1 transcription factor is required for MAP kinase signaling specificity in yeast. Cell 119:991-1000. [DOI] [PubMed] [Google Scholar]
- 2.Bardwell, L. 2005. A walk-through of the yeast mating pheromone response pathway. Peptides 26:339-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardwell, L., J. G. Cook, D. Voora, D. M. Baggott, A. R. Martinez, and J. Thorner. 1998. Repression of yeast Ste12 transcription factor by direct binding of unphosphorylated Kss1 MAPK and its regulation by the Ste7 MEK. Genes Dev. 12:2887-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bardwell, L., J. G. Cook, J. X. Zhu-Shimoni, D. Voora, and J. Thorner. 1998. Differential regulation of transcription: repression by unactivated mitogen-activated protein kinase Kss1 requires the Dig1 and Dig2 proteins. Proc. Natl. Acad. Sci. USA 95:15400-15405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baur, M., R. K. Esch, and B. Errede. 1997. Cooperative binding interactions required for function of the Ty1 sterile responsive element. Mol. Cell. Biol. 17:4330-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breitkreutz, A., L. Boucher, B. J. Breitkreutz, M. Sultan, I. Jurisica, and M. Tyers. 2003. Phenotypic and transcriptional plasticity directed by a yeast mitogen-activated protein kinase network. Genetics 165:997-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruckner, S., T. Kohler, G. H. Braus, B. Heise, M. Bolte, and H. U. Mosch. 2004. Differential regulation of Tec1 by Fus3 and Kss1 confers signaling specificity in yeast development. Curr. Genet. 46:331-342. [DOI] [PubMed] [Google Scholar]
- 8.Chou, S., L. Huang, and H. Liu. 2004. Fus3-regulated Tec1 degradation through SCFCdc4 determines MAPK signaling specificity during mating in yeast. Cell 119:981-990. [DOI] [PubMed] [Google Scholar]
- 9.Cook, J. G., L. Bardwell, S. J. Kron, and J. Thorner. 1996. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes Dev. 10:2831-2848. [DOI] [PubMed] [Google Scholar]
- 10.Cook, J. G., L. Bardwell, and J. Thorner. 1997. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature 390:85-88. [DOI] [PubMed] [Google Scholar]
- 11.Dolan, J. W., C. Kirkman, and S. Fields. 1989. The yeast STE12 protein binds to the DNA sequence mediating pheromone induction. Proc. Natl. Acad. Sci. USA 86:5703-5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elion, E. A., J. A. Brill, and G. R. Fink. 1991. Functional redundancy in the yeast cell cycle: FUS3 and KSS1 have both overlapping and unique functions. Cold Spring Harb. Symp. Quant. Biol. 56:41-49. [DOI] [PubMed] [Google Scholar]
- 13.Fields, S., and I. Herskowitz. 1987. Regulation by the yeast mating-type locus of STE12, a gene required for cell-type-specific expression. Mol. Cell. Biol. 7:3818-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fields, S., and I. Herskowitz. 1985. The yeast STE12 product is required for expression of two sets of cell-type specific genes. Cell 42:923-930. [DOI] [PubMed] [Google Scholar]
- 15.Ghaemmaghami, S., W. K. Huh, K. Bower, R. W. Howson, A. Belle, N. Dephoure, E. K. O'Shea, and J. S. Weissman. 2003. Global analysis of protein expression in yeast. Nature 425:737-741. [DOI] [PubMed] [Google Scholar]
- 16.Golemis, E. A., and R. Brent. 1992. Fused protein domains inhibit DNA binding by LexA. Mol. Cell. Biol. 12:3006-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guarente, L., and M. Ptashne. 1981. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 78:2199-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagen, D. C., G. McCaffrey, and G. F. Sprague, Jr. 1991. Pheromone response elements are necessary and sufficient for basal and pheromone-induced transcription of the FUS1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 11:2952-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herskowitz, I. 1995. MAP kinase pathways in yeast: for mating and more. Cell 80:187-197. [DOI] [PubMed] [Google Scholar]
- 20.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 21.Hughes, T. R., M. J. Marton, A. R. Jones, C. J. Roberts, R. Stoughton, C. D. Armour, H. A. Bennett, E. Coffey, H. Dai, Y. D. He, M. J. Kidd, A. M. King, M. R. Meyer, D. Slade, P. Y. Lum, S. B. Stepaniants, D. D. Shoemaker, D. Gachotte, K. Chakraburtty, J. Simon, M. Bard, and S. H. Friend. 2000. Functional discovery via a compendium of expression profiles. Cell 102:109-126. [DOI] [PubMed] [Google Scholar]
- 22.Kaiser, P., K. Flick, C. Wittenberg, and S. I. Reed. 2000. Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCF(Met30)-mediated inactivation of the transcription factor Met4. Cell 102:303-314. [DOI] [PubMed] [Google Scholar]
- 23.Kellis, M., N. Patterson, M. Endrizzi, B. Birren, and E. S. Lander. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423:241-254. [DOI] [PubMed] [Google Scholar]
- 24.Kohler, T., S. Wesche, N. Taheri, G. H. Braus, and H. U. Mosch. 2002. Dual role of the Saccharomyces cerevisiae TEA/ATTS family transcription factor Tec1p in regulation of gene expression and cellular development. Eukaryot. Cell 1:673-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laloux, I., E. Dubois, M. Dewerchin, and E. Jacobs. 1990. TEC1, a gene involved in the activation of Ty1 and Ty1-mediated gene expression in Saccharomyces cerevisiae: cloning and molecular analysis. Mol. Cell. Biol. 10:3541-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laloux, I., E. Jacobs, and E. Dubois. 1994. Involvement of SRE element of Ty1 transposon in TEC1-dependent transcriptional activation. Nucleic Acids Res. 22:999-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lo, W. S., and A. M. Dranginis. 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 9:161-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 29.Madhani, H. D., and G. R. Fink. 1997. Combinatorial control required for the specificity of yeast MAPK signaling. Science 275:1314-1317. [DOI] [PubMed] [Google Scholar]
- 30.Madhani, H. D., and G. R. Fink. 1998. The riddle of MAP kinase signaling specificity. Trends Genet. 14:151-155. [DOI] [PubMed] [Google Scholar]
- 31.Madhani, H. D., T. Galitski, E. S. Lander, and G. R. Fink. 1999. Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc. Natl. Acad. Sci. USA 96:12530-12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mosch, H. U., and G. R. Fink. 1997. Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. Genetics 145:671-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mosch, H. U., E. Kubler, S. Krappmann, G. R. Fink, and G. H. Braus. 1999. Crosstalk between the Ras2p-controlled mitogen-activated protein kinase and cAMP pathways during invasive growth of Saccharomyces cerevisiae. Mol. Biol. Cell 10:1325-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olson, K. A., C. Nelson, G. Tai, W. Hung, C. Yong, C. Astell, and I. Sadowski. 2000. Two regulators of Ste12p inhibit pheromone-responsive transcription by separate mechanisms. Mol. Cell. Biol. 20:4199-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pi, H., C. T. Chien, and S. Fields. 1997. Transcriptional activation upon pheromone stimulation mediated by a small domain of Saccharomyces cerevisiae Ste12p. Mol. Cell. Biol. 17:6410-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts, C. J., B. Nelson, M. J. Marton, R. Stoughton, M. R. Meyer, H. A. Bennett, Y. D. He, H. Dai, W. L. Walker, T. R. Hughes, M. Tyers, C. Boone, and S. H. Friend. 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287:873-880. [DOI] [PubMed] [Google Scholar]
- 37.Rose, M., and D. Botstein. 1983. Construction and use of gene fusions to lacZ (beta-galactosidase) that are expressed in yeast. Methods Enzymol. 101:167-180. [DOI] [PubMed] [Google Scholar]
- 38.Rupp, S., E. Summers, H. J. Lo, H. Madhani, and G. Fink. 1999. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 18:1257-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sabbagh, W., Jr., L. J. Flatauer, A. J. Bardwell, and L. Bardwell. 2001. Specificity of MAP kinase signaling in yeast differentiation involves transient versus sustained MAPK activation. Mol. Cell 8:683-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sprague, G. F., and J. Thorner. 1992. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae, p. 657-744. In J. R. Broach, J. R. Pringle, and E. Jones (ed.), The molecular biology of the yeast Saccharomyces, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Tanaka, T., D. Knapp, and K. Nasmyth. 1997. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell 90:649-660. [DOI] [PubMed] [Google Scholar]
- 43.Tedford, K., S. Kim, D. Sa, K. Stevens, and M. Tyers. 1997. Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Curr. Biol. 7:228-238. [DOI] [PubMed] [Google Scholar]
- 44.Trueheart, J., J. D. Boeke, and G. R. Fink. 1987. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol. Cell. Biol. 7:2316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White, J. M., and M. D. Rose. 2001. Yeast mating: getting close to membrane merger. Curr. Biol. 11:R16-R20. [DOI] [PubMed] [Google Scholar]
- 46.Yuan, Y. O., I. L. Stroke, and S. Fields. 1993. Coupling of cell identity to signal response in yeast: interaction between the alpha 1 and STE12 proteins. Genes Dev. 7:1584-1597. [DOI] [PubMed] [Google Scholar]
- 47.Zeitlinger, J., I. Simon, C. T. Harbison, N. M. Hannett, T. L. Volkert, G. R. Fink, and R. A. Young. 2003. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell 113:395-404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.