Abstract
The human transcription elongation factor CA150 contains three N-terminal WW domains and six consecutive FF domains. WW and FF domains, versatile modules that mediate protein-protein interactions, are found in nuclear proteins involved in transcription and splicing. CA150 interacts with the splicing factor SF1 and with the phosphorylated C-terminal repeat domain (CTD) of RNA polymerase II (RNAPII) through its WW and FF domains, respectively. WW and FF domains may, therefore, serve to link transcription and splicing components and play a role in coupling transcription and splicing in vivo. In the study presented here, we investigated the subcellular localization and association of CA150 with factors involved in pre-mRNA transcriptional elongation and splicing. Endogenous CA150 colocalized with nuclear speckles, and this was not affected either by inhibition of cellular transcription or by RNAPII CTD phosphorylation. FF domains are essential for the colocalization to speckles, while WW domains are not required for colocalization. We also performed biochemical assays to understand the role of WW and FF domains in mediating the assembly of transcription and splicing components into higher-order complexes. Transcription and splicing components bound to a region in the amino-terminal part of CA150 that contains the three WW domains; however, we identified a region of the C-terminal FF domains that was also critical. Our results suggest that sequences located at both the amino and carboxyl regions of CA150 are required to assemble transcription/splicing complexes, which may be involved in the coupling of those processes.
Expression of protein-encoding genes is a multistep process beginning with transcription by RNA polymerase II (RNAPII). During transcription, the nascent pre-mRNA undergoes several processing steps, including capping, splicing, and polyadenylation. Distinct cellular machines carry out each of the steps in gene expression, but growing evidence indicates that there are coupled interactions between these machineries and it is now believed that most mRNA processing reactions occur cotranscriptionally (23, 51, 54). The C-terminal repeat domain (CTD) of RNAPII has a principal role in coupling eukaryotic transcription and pre-mRNA processing. The CTD of RNAPII is composed of 52 (in mammals) or 26 (in yeast [Saccharomyces cerevisiae]) tandem repeats of the consensus heptapeptide YSPTSPS, and this domain is essential for viability in yeast (11). An RNAPII with an unphosphorylated CTD assembles into preinitiation complexes at the promoters, whereas the transition between initiation and elongation is accompanied by multiple phosphorylation events of the CTD. These events are catalyzed by several protein kinases, including cyclin-dependent kinase 7 (CDK7), which is part of the general transcription factor IIH (TFIIH) (14, 35), and CDK9, which is the kinase component of positive transcription elongation factor b (P-TEFb) (37, 38). Phosphorylation of the CTD on Ser5 correlates with the presence of RNAPII at the promoter, while phosphorylation on Ser2 correlates with RNAPII at the coding regions of genes and thus is considered a mark of elongating polymerases (30). As first proposed by Greenleaf (19), the CTD of RNAPII has been shown to interact with capping, splicing, and polyadenylation factors (8, 9, 25, 39, 40), thus acting as a platform upon which mRNA processing factors bind. It is most likely that the phosphorylation events occurring at the CTD coordinate the recruitment of those processing factors at different stages of mRNA formation.
Several observations link the elongating RNAPII to pre-mRNA splicing. First, hyperphosphorylated RNAPII has been shown to colocalize with splicing factor-rich nuclear speckles (5, 47). Second, truncation of the CTD or overexpression of phospho-CTD peptides results in reduced in vivo splicing (13, 40), and under some circumstances the addition of hyperphosphorylated RNAPII or recombinant CTD can enhance in vitro splicing (24, 71). Third, it has been shown that alterations in the transcription elongation of RNAPII complexes have an effect on alternative mRNA splicing. Thus, transcription complexes with low elongation efficiency promote efficient inclusion of alternatively spliced exons from the fibronectin gene, while complexes with high elongation efficiency have the reverse effect (26). Similarly, pausing a transcription elongation complex in the α-tropomyosin gene or in the fibroblast growth factor receptor 2 gene results in an increase in exon inclusion (56, 57). Recently, functional coupling was reported for an in vitro transcription/splicing system (16). Fourth, hyperphosphorylated CTD binds splicing-related factors (4, 7, 29, 46, 52, 70). The biochemical and functional links between elongation of transcription and splicing support the idea that splicing occurs cotranscriptionally and is mediated by the CTD. This transcriptional coupling to the spliceosomes implies that the elongation and splicing machineries are in close contact and therefore able to interact with each other. However, despite the large macromolecular sizes of those complexes and the known influence of the elongating RNAPII complex on splicing, few transcription elongation factors have been postulated to engage both the elongation and splicing machineries (17).
CA150 (also named TCERG1, for transcription elongation regulator 1; HUGO Gene Nomenclature Committee) was initially purified from HeLa cell nuclear extract as a transcriptional cofactor that regulates human immunodeficiency virus type 1 (HIV-1) Tat gene activation (67). Transient overexpression of CA150 reduces both basal and Tat-activated transcription from the HIV-1 promoter by inhibiting elongation efficiency (66). This repression is promoter specific, as other viral promoters are not affected, and it is also dependent on a specific functional TATA box element (66). The primary sequence of CA150 contains notable motifs and domains, many of which have been also found in other regulators of gene expression, including three WW and six FF domains (1, 3, 67). CA150 interacts directly with the phosphorylated CTD of RNAPII via its FF domains (6), and it also interacts with the splicing factor SF1 through the WW domains (18). Based on those data, it has been proposed that CA150 could tether SF1 to the elongating RNAPII to facilitate mRNA substrate recognition and splicing (17, 18). Very recently, many transcription and splicing components have been found associated with CA150 by use of mass spectrometry (34, 61), thus establishing a new connection between elongation and splicing. Those findings are in agreement with the presence of CA150 in active spliceosomes as demonstrated previously by proteomic approaches (49, 75).
CA150 appears to function in both elongation and splicing stages of mRNA formation. To date, however, very little is known about specific interactions of CA150 with members of either of these cellular machineries. Furthermore, the spatial organization of endogenous CA150 in the cell has not been comprehensively elucidated. In the study presented here, we analyzed the subcellular location of CA150 by laser scanning confocal microscopy and its spatial relationship to nuclear speckles. We found that CA150 colocalized with a number of splicing factors that are clustered in nuclear speckles and was enriched in the peripheral regions of the speckles, and that this distribution was not affected either by inhibition of cellular transcription or by RNAPII CTD phosphorylation. We also show that this colocalization was dependent on the FF domains in CA150. Also, we present biochemical evidence further linking CA150 to the transcription elongation and splicing machineries, thus supporting the hypothesis that CA150 can couple elongation and splicing.
MATERIALS AND METHODS
Plasmids.
The mammalian expression vector pEFBOST7-CA150 contains the cDNA encoding human CA150 and the 11-amino-acid T7 epitope tag at its amino terminus and has been described previously (66). The CA150 deletion expression constructs CA150 (1-1038), CA150 (1-662), CA150 (134-1098), CA150 (428-1098), and CA150 (591-1098), where the numbers in parentheses represent the CA150 amino acids contained in the construct, were cloned by inserting CA150 PCR fragments with BglII ends into the BamHI site of the parental vector pEFBOST7 (45). The CA150 carboxyl-terminal deletion series of CA150 was made in the vector pEF6His6 (Invitrogen), which contains amino-terminal 6-histidine and T7 epitope tags. The constructs made in this way included pEF6HisB CA150 (1-1098), pEF6HisB CA150 (1-1027), pEF6HisB CA150 (1-1004), pEF6HisB CA150 (1-951), pEF6HisB CA150 (1-870), pEF6HisB CA150 (1-787), pEF6HisB CA150 (1-715), and pEF6HisB CA150 (1-663), where the numbers in parentheses represent the CA150 amino acids contained in the construct. The CA150 WW domain mutant constructs (WW1mt, WW2mt, WW3mt, and WW1mt+WW2mt) were created by site-directed mutagenesis and have been described previously (18).
Antibodies.
The CA150 antibody used in this study was generated in rabbits (Pineda Antikörper Service, Germany) by use of a truncated glutathione S-transferase (GST) fusion CA150 protein spanning amino acids 590 to 1098 as described previously (67). For immunofluorescence analysis, immunoglobulins (Igs) were purified from antiserum on protein A-Sepharose columns (Pharmacia) by following standard procedures. CA150 antibodies (0.7 mg/ml) were used at dilutions of 1:2,000 and 1:1,000 for immunofluorescence and immunoblotting, respectively. For the in vitro immunoprecipitations shown below (see Fig. 6 and 7), GST and CA150 antibodies were antigen affinity purified from the same rabbit polyclonal serum according to the procedure of Harlow and Lane (21). Antibodies against the T7 tag were purchased from Novagen and used at dilutions of 1:4,000 and 1:10,000 for immunofluorescence and immunoblotting, respectively. Anti-T7 antibodies shown below (see Fig. 5C) were from Bethyl and were used at a dilution of 1:1,000. Antibody against splicing factor SC35 (Sigma) was used at a dilution of 1:1,000. The mouse monoclonal antibody (Y12) to Smith antigen (Abcam) was used at a dilution of 1:500 for immunofluorescence, and a human anti-Sm serum was used at a dilution of 1:1,000 for immunoblotting. Antibodies against U2AF65 and SF1 were kindly provided by J. Valcárcel (Center for Genomic Regulation [CRG]) and were used at dilutions of 1:500 and 1:2,000, respectively. Antibody to SF2/ASF was kindly provided by J. Ortín (CNB) and used at a dilution of 1:400. Antibody against CDK9 (Santa Cruz Biotechnology) was used at dilutions of 1:100 and 1:250 for immunofluorescence and immunoblotting, respectively. Antibody against cyclin T1 (Santa Cruz Biotechnology) was used at a dilution of 1:250. Rabbit anti-RNAPII serum was kindly provided by M. Dahmus (UC Davis) and used at a dilution of 1:3,000 for immunoblotting. The RAP30, TFIIE-β, and Sp1 antibodies were purchased from Santa Cruz Biotechnology and used at a dilution of 1:100. Antibody to CDK2 (Santa Cruz Biotechnology) was used at a dilution of 1:500. Primary antibodies were detected using appropriate secondary antibodies. For immunofluorescence, we used Alexa 488 (green) and Alexa 564 (red) from Molecular Probes and Cy5 (far red) from Jackson; these were generally used at dilutions of 1:500. For immunoblotting, secondary antibodies to rabbit and mouse were purchased from PerkinElmer Life Sciences, and antibodies to human were obtained from Sigma and were generally used at a dilution of 1:5,000.
FIG. 6.
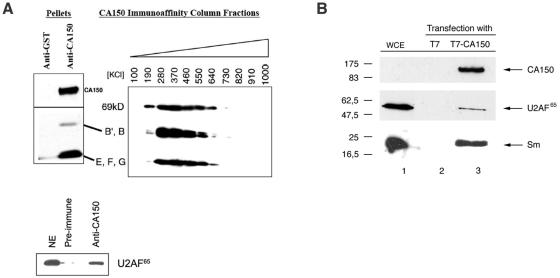
CA150 associates with splicing factors. (A) CA150 interacts with Sm snRNPs and U2AF65 in vitro. The pellet after immunoprecipitation of CA150 with specific antibodies was analyzed by SDS-PAGE and Western blotting using human anti-snRNPs and anti-U2AF65 polyclonal sera. Anti-GST or preimmune serum immunoprecipitations served as negative controls. An anti-CA150 affinity matrix was bound with HeLa cell nuclear extract and washed extensively. The bound protein was eluted with a linear gradient of 100 mM to 1,000 mM KCl, and the composition of a sample of the fractions was determined by Western blotting with human anti-snRNPs polyclonal serum. NE, HeLa nuclear extract; kD, kDa. (B) CA150 interacts with U2AF65 and snRNPs in vivo. 293T cells were transiently transfected with pEFBOST7-CA150 and control plasmids. The expressed CA150 protein contains the 11-amino-acid T7 epitope tag at its amino terminus. Whole-cell extract (WCE) was prepared and subjected to immunoaffinity purification with anti-T7 monoclonal antibodies covalently coupled to cross-linked agarose beads. After being washed, bound proteins were specifically eluted with T7 peptide. A sample of the eluate was subjected to SDS-PAGE and Western blot analysis using antibodies to detect CA150 (anti-T7), U2AF65, and Sm (anti-Y12) proteins. Numbers to the left of the gels indicate molecular masses in kDa.
FIG. 7.
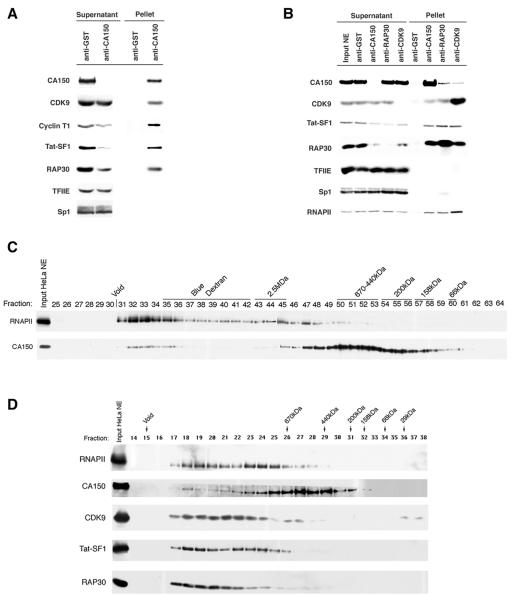
CA150 associates with elongation factors. (A) Immunoprecipitation of CA150 reveals that P-TEFb (CDK9 and cyclin T1), Tat-SF1, and TFIIF (RAP30) associate with CA150 in HeLa nuclear extracts. Immunoprecipitations of CA150 from HeLa nuclear extracts were performed such that all of the CA150 in the extracts was removed from the supernatant and present in the immunoprecipitate pellet. Anti-GST immunoprecipitation served as a negative control. The supernatants after precipitation and the pellets of the immunoprecipitates were analyzed by SDS-PAGE and Western blotting with specific antibodies. The DNA binding factor Sp1 and the general transcription factor TFIIE served as negative controls for abundant nuclear proteins that do not coimmunoprecipitate. (B) CA150, CDK9, and TFIIF were immunoprecipitated from HeLa nuclear extract using anti-CA150, anti-CDK9, and anti-RAP30 under conditions in which each respective protein was depleted from the extracts so as to ascertain the relative proportions of each protein associated with the others. Each precipitate pellet and supernatant were analyzed by Western blotting with specific antibodies against the proteins indicated on the left side of the figure. Anti-Sp1 and anti-TFIIE served as negative controls. CA150, CDK9, Tat-SF1, and RNAPII each coimmunoprecipitated with all three antibodies. Anti-GST immunoprecipitate was used as control for nonspecific interaction with antibodies and beads. (C) Gel filtration chromatography of HeLa nuclear extracts identifies CA150 and elongation factors in high-molecular-mass complexes. Gel filtration of HeLa nuclear extract on a Sephacryl 400HR column reveals two size-differentiated populations of CA150. Ten microliters (15 mg/ml) of HeLa nuclear extract (Input HeLa NE) and 30 μl of the chromatographic fractions were subjected to SDS-PAGE and Western blot analysis. Antisera against RNAPII and CA150 were used to analyze the elution pattern, as indicated on the left of the figure. Molecular mass standards were used to calibrate the columns, and their molecular masses and the fractions of their elution peaks are indicated on the top of the figure above the fraction numbers. The experimentally determined exclusion limit of the columns is also indicated (Void). A minor peak of CA150 elutes with a molecular mass of greater than 1 MDa (fractions 32 to 35). A major peak of CA150 elutes at approximately 600 kDa (fractions 50 to 53). The MDa-sized CA150 peak coelutes with the peak of RNAPII and likely represents a CA150-containing RNAPII complex(es). (D) Superose 6 gel filtration chromatography of HeLa nuclear extract. Specific antibodies were used to detect the proteins indicated on the left side of the figure. Columns were calibrated with molecular mass standards; the fractions containing their elution peaks are indicated on the top of the figure. The experimentally determined exclusion limit of the columns is also indicated (Void). The elongation factors CDK9, TAT-SF1, and TFIIF (RAP30) exist predominantly in large MDa-sized complexes which coelute with RNAPII and the MDa-sized CA150 complex.
FIG. 5.
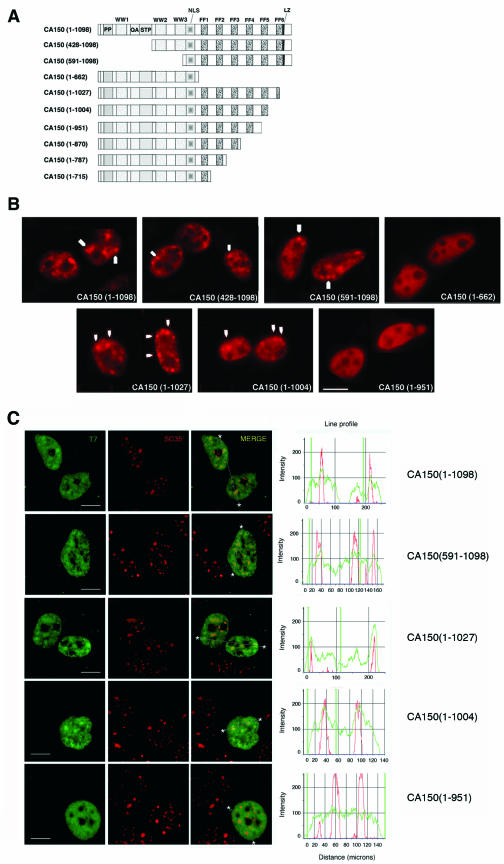
FF5 repeat domain is critical for CA150 localization to nuclear speckles. (A) Diagrammatic representation of the T7-tagged CA150 mutants used. The numbers in parentheses represent the CA150 amino acids contained in the construct. Shown are the proline-rich region (PP); the three WW domains (WW); a glutamine-alanine repeat (QA); a region rich in serine, threonine, and proline (STP); a putative nuclear localization signal (NLS); the six FF domains (FF); and a putative leucine zipper (LZ). (B) Cells were transfected with the indicated plasmids and the expressed CA150 protein was immunolocalized with T7 antibody. Images show a speckled pattern for full-length CA150 (1-1098), CA150 (428-1098), and CA150 (591-1098), but CA150 (1-662) is more diffusely dispersed throughout the nucleoplasm, which demonstrates that CA150 localization to nuclear speckles is dependent on its FF domains. Partial [CA150 (1-1027)] or total [CA150 (1-1004)] deletion of FF6 did not affect the speckle distribution of CA150. A deletion of the FF5 repeat motif [CA150 (1-951)] resulted in a more diffuse pattern of CA150 staining, thereby indicating that the FF5 repeat is important for the association of CA150 with nuclear speckles. (C) Cells were transfected with the indicated plasmids and then processed for immunofluorescence analysis. Cells were dually labeled with antibodies directed against CA150 (green) and SC35 (red). Individual staining and merge images of the cell stained with the indicated antibodies are shown. Line scans showing local intensity distributions of CA150 in green and SC35 in red are shown to the right of the panels. Asterisks in merge panels indicate the positions of the line scans. Note that the overlapping signal at the speckle region in overexpressed CA150 (1-951) is not specific. Scale bars, 10 μm.
Cell culture and transfections.
The human cell line 293T was used in all cell transfection experiments. 293T cells were grown in Dulbecco's modified Eagle medium (Sigma) supplemented with 10% fetal bovine serum (BioWhittaker), l-glutamine at 4 mM (Merck), and penicillin-streptomycin to 100 U and 100 μg per ml, respectively (Sigma). For immunoprecipitation analysis, transfections were performed in 100-mm-diameter plates (Nunc). Each plate was seeded with approximately 2.4 × 106 cells 20 h prior to transfection. Cells were grown to approximately 60 to 70% confluence and transfected with 15 μg of DNA by using Polyfect reagent (QIAGEN) according to the manufacturer's protocols. Transfections were carried out with at least two different preparations of each plasmid DNA purified using kits from QIAGEN, Inc. For immunofluorescence procedures, similar procedures were used, but 293T cells were grown on coverslips to approximately 50% confluence, transfected with 2 μg of DNA by use of calcium phosphate, and processed 18 h after transfection.
Preparation of cell extracts, immunoprecipitations, and Western blotting.
Fifty microliters of HeLa cell nuclear extract prepared using a modified protocol (41) of Dignam et al. (12) was diluted to a 200-μl final volume with IP buffer (20 mM HEPES, pH 7.9, 150 mM KCl, 20% glycerol, 1 mM dithiothreitol, 1% Triton X-100 and 0.5% NP-40, 0.2 mM EDTA). Fifteen micrograms of each antibody was added to the diluted nuclear extract and then incubated with end-over-end rotation for 4 h at 4°C. Immune complexes were collected with 500- to 750-μg magnetic protein A beads (BioMag protein A beads; Perceptive Biosciences) and a magnet. The pellets were washed four or five times with 1 ml of IP buffer by rotation for 5 min at 4°C. The pellets and supernatants were then separated on 10% or 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and analyzed by Western blotting and silver staining. The equivalent of 5 μg IgG of each pellet was loaded per lane, and equal volumes of supernatant were loaded.
Analysis of the in vivo interaction was carried out with whole-cell lysates from 293T cells. Briefly, cells were washed in phosphate-buffered saline (PBS) (pH 7.4), pelleted, and incubated in 500 μl of T7 buffer (20 mM HEPES, pH 7.9, 150 mM NaCl, 5 mM EDTA, 1% NP-40, 1 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride) for 30 min at 4°C. After being spun at 14,000 rpm for 3 min at 4°C, the lysate was used immediately or stored at −80°C for future use. The cell extracts were diluted to a 1-ml final volume with T7 buffer. Fifty microliters of anti-T7 tag monoclonal antibody that was covalently coupled to cross-linked agarose beads (Novagen) was added to the diluted whole-cell extract and then incubated with end-over-end rotation for 4 h at 4°C. After four washes with T7 buffer, proteins bound to the antibody resin were selectively eluted by affinity competition using saturating levels of T7 peptide (1 μg/μl in T7 buffer) in a final volume of 50 μl. Proteins from three consecutive elutions per sample were precipitated by using 1 ml of 0.2% (vol/vol) HCl-acetone overnight at −20°C. The supernatant was carefully discharged and the resulting pellet was briefly dried under vacuum, diluted in a minimal volume of sample buffer, and analyzed by Western blotting.
For Western blot analysis, proteins were separated on 12.5% SDS-PAGE gels, transferred to a polyvinylidene difluoride (Bio-Rad) or nitrocellulose (Amersham) membrane, and then incubated with the specific antibody. After being washed, the membrane was incubated with a peroxidase-conjugated secondary antibody, and bound antibodies were detected by enhanced chemiluminescence (PerkinElmer Life Sciences).
Gel filtration chromatography.
A Pharmacia fast-performance liquid chromatography system was used for all chromatography. The Superose 6 (Pharmacia) separation range is from 5 kDa to 5 MDa, with column dimensions of 10 mm by 30 cm. Three milligrams of HeLa cell nuclear extract was separated at 4°C with a flow rate of 0.2 ml/min while 0.5-ml fractions were collected. The Sephacryl S400HR column (Pharmacia) has a separation range of 20 kDa to 8MDa, with dimensions of 2 cm2 by 120 cm. Thirty milligrams of HeLa cell nuclear extract was separated at a flow rate of 0.5 ml/min while 2.3-ml fractions were collected. The elution buffer was composed of 20 mM HEPES (pH 7.9), 100 mM KCl, 1 mM dithiothreitol, 20% (vol/vol) glycerol, and 0.2 mM EDTA.
HeLa nuclear extracts depleted of nucleic acids were prepared by a modification of the procedure of Dignam et al. (12). Extraction from nuclei was carried out for 30 min at 30°C with the following modification: buffer C contained 100 mM KCl, instead of 420 mM, in addition to protease inhibitors, 2 mM CaCl2, and 1 unit/μl micrococcal nuclease (USB). After dialysis against buffer D, any remaining nucleic acids were extracted and analyzed by agarose gel electrophoresis. Only small amounts of short oligonucleotides were observed.
Immunofluorescence, image processing, and quantitation.
Immunofluorescence analysis was performed on fixed permeabilized HeLa cells, MDCK cells (NLB2 strain provided by J. Ortín, CNB), 293T cells, and IMR90 fibroblasts (provided by Manuel Collado, Spanish National Cancer Center [CNIO]) as indicated in the legends to the figures. Cells were grown on coverslips in Dulbecco's modified Eagle medium supplemented with 5 to 10% fetal bovine serum for 48 h (approximately 60% confluence). Cells were fixed with 3% paraformaldehyde in PBS buffer (pH 7.4) for 30 min at 4°C, permeabilized in PBS containing 0.5% Triton X-100 for 5 min at 4°C, washed three times with PBS, and blocked in PBS containing 2.5% bovine serum albumin (BSA) for 24 h at 4°C. Cells were incubated with primary antibodies at appropriate dilutions in PBS containing 0.1% BSA and 0.2% saponin for 1 h at room temperature and subsequently washed five times with 0.1% BSA in PBS and incubated with appropriate secondary antibodies under the previously described conditions. Cells were then rinsed three times with 0.1% BSA in PBS and once with PBS. Cell nuclei were identified by staining with TOPRO-3 (Molecular Probes) at a dilution of 1:100 in PBS for 30 min. The coverslip was then mounted onto glass slides using ProLong Antifade (Molecular Probes) mounting medium. For conventional fluorescence microscopy, cells were examined with a Leica DM-XRA microscope. A minimum of 10 fields containing several cells was collected per sample and processed with Qwin imaging software (Leica). For laser scanning confocal microscopy, optical sections (0.3 to 0.5 μm) were collected with a Zeiss Axiovert 200 microscope equipped with an argon ion laser at 488 nm, a krypton-neon laser at 543 nm, and a red diode at 637 nm to excite green, red, and far-red fluorochromes sequentially. Images were taken employing LaserSharp v5.0 software (Bio-Rad) and analyzed using LaserPix v.4 package software (Bio-Rad). A minimum of 10 fields containing several cells was collected per sample. To compare the localizations of the double-labeled images, line scans measuring the local intensity distributions of the two labels were made. In the figures, we show only one of these evaluated line scans. For single-speckle optical sectioning, whole cells were optically dissected with a plane-to-plane step of 0.15 μm. For three-dimensional (3-D) reconstruction, we used ImageJ package software (Bio-Rad). x, y, and z planes were obtained from the same sample with a z-plane step of 0.1 μm.
All images were digitally processed for presentation using Adobe Photoshop v7.0 software.
Preparation of core nuclear matrix and treatment with transcriptional inhibitors.
Extraction of soluble proteins to reveal the residual nuclear core matrix was performed according to the procedures of Wei et al. (68), Belgrader et al. (2), and Fisher et al. (15). Briefly, MDCK cells were grown on coverslips and washed with ice-cold TBS buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM MgCl2, 0.5% BSA) and further washed with glycerol buffer (GB) (20 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 25% glycerol [vol/vol], 0.5 mM EGTA, 0.5 mM phenylmethylsulfonyl fluoride) for 10 min on ice. Washed cells were permeabilized with 0.05% Triton X-100 for 3 min in ice-cold GB. Coverslips were rinsed once with GB, and chromatin was removed by digestion with 80 U/ml of DNase I (Sigma) diluted in GB for 10 min at 30°C. Soluble proteins were extracted by two washes with 0.2 M ammonium sulfate in ice-cold GB for 3 min each. Cells were then processed for immunofluorescence analysis as described before.
Treatment with transcription and kinase inhibitors was performed on cells grown on coverslips by incubation with actinomycin D (Sigma) at 5 μg/ml for 1 h at 37°C, with α-amanitin (Sigma) at 25 μg/ml for 6 h at 37°C, and with 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) at 100 μM for 2 h at 37°C prior to fixation and immunolabeling. Cells were then processed for immunofluorescence analysis.
RESULTS
CA150 localizes to splicing factor-rich nuclear speckles.
We previously reported that CA150 is distributed in the cell nucleus (67). Using confocal laser scanning microscopy, we have further analyzed the spatial distribution of CA150 in the nucleus. HeLa cells were fixed, permeabilized, and immunolabeled with Ig-purified CA150 antibody. Our experiments show that CA150 was excluded from the nucleolar compartment and distributed throughout the nucleoplasm with an increased signal in organized granule-like sites (Fig. 1A). CA150 was located in the interchromatin compartment, where RNA processing is believed to occur, and was excluded from the highly compacted chromatin territories detected with the fluorescent dye TOPRO, which binds to nucleic acids (Fig. 1A).
FIG. 1.
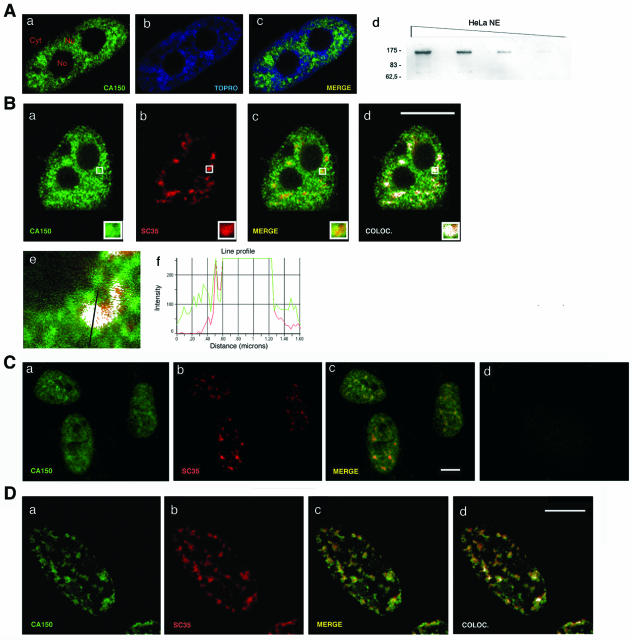
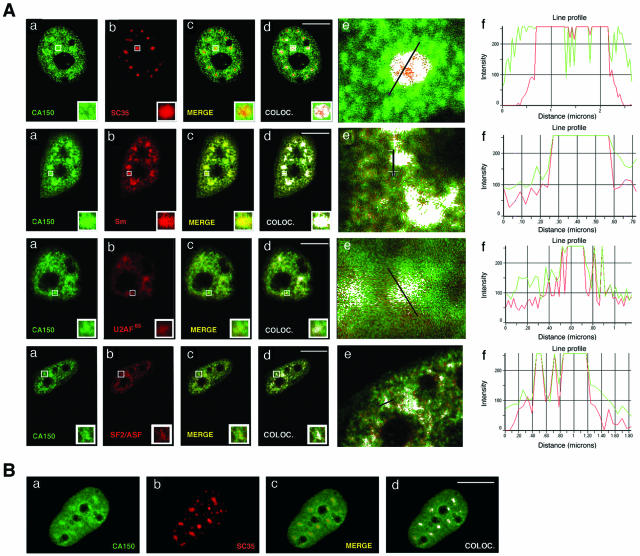
Association of CA150 with splicing factor-rich nuclear speckles. (A) CA150 localizes at numerous foci within the nuclear interchromatin compartment. Endogenous CA150 (green) was detected with a primary Ig-purified polyclonal antibody. TOPRO labeling was used to stain chromosome territory (blue). Locations of nucleoli (No), nucleoplasm (Nu), and cytoplasm (Cyt) are labeled in panel a. (d) NE, nuclear extract. Molecular masses in kDa are indicated to the left. (B) Colocalization (COLOC.) of CA150 with the essential splicing factor SC35. Dual labeling of cells with antibodies directed against CA150 (green) and with the SC35 antibody (red) was performed. Individual staining (a and b), merge (c, yellow), and colocalization (d, white) images of the cell stained with the indicated antibodies are shown. (f) Line scans showing local intensity distributions of CA150 in green and of SC35 in red. A bar in panel e indicates the position of the line scans. Bar in panel d, 10 μm. (C) Colocalization of CA150 with SC35 in IMR90 normal human fibroblasts. Individual staining (a and b) and merge (c, yellow) images of IMR90 cells stained with CA150 (green) and SC35 (red) antibodies are shown. A panel for immunofluorescence analysis performed using preimmune serum as a negative control is also shown (d). (D) A subpopulation of CA150 associates with the insoluble nuclear matrix and localizes to the nuclear speckle region. Dual labeling of cells with antibodies directed against CA150 (green) and SC35 (red) antibodies was performed after extraction of the soluble fraction of nuclear components. The diffuse nucleoplasmic CA150 signal was severely reduced, while the speckle distribution was left intact, as shown in the merge and colocalization images. Cells were also stained with TOPRO to verify removal of DNA (not shown). Bar, 10 μm.
The increased accumulation at numerous foci of CA150 is reminiscent of the nuclear speckled pattern observed for splicing components (32). To determine whether CA150 colocalizes with the speckles, we carried out double-labeled immunofluorescence experiments using CA150-specific antibodies and antibodies against splicing factors in HeLa cells. A merge of the individual images shows the overlap between CA150 and the other marker in yellow. Colocalization of signals is shown in white. A merge of signals is defined as the overlap of two or more of the emission signals due to their close proximity. Colocalization of signals is defined by the presence of two or more different signals in the same image pixel with an intensity profile above that of a given fluorescence background (see Materials and Methods). A quantitative assessment of fluorophore colocalization in confocal optical sections was obtained using the information obtained from scatter plots and selected regions of interest. We compared the signals obtained with anti-CA150 with those of anti-SC35, which recognizes the phosphorylated form of SC35, an essential splicing factor that commonly serves to define nuclear speckles. CA150 was concentrated at the splicing factor-rich nuclear speckles (Fig. 1B, panels a to d). The same result was obtained with antibodies against other splicing factors (anti-Sm, anti-U2AF65, and anti-SF2/ASF) that are enriched within nuclear speckles (data not shown; also see Fig. 3). To confirm those data, we performed semiquantitative analysis of the spatial relationship between the relative spatial distributions of CA150 and the splicing factor as a way of determining the degree of colocalization (Fig. 1B, panels e and f; also data not shown). These results suggest that there are at least two populations of CA150 in the nucleus: a population that shows diffuse nucleoplasmic distribution and a population that shows a speckle pattern of localization. The distribution of CA150 in the nucleus described before as well as the colocalization of CA150 with SC35 was also seen in the nontransformed IMR90 cell line (Fig. 1C).
FIG. 3.
Colocalization of CA150 with nuclear speckles is not perturbed following inhibition of transcription. (A) HeLa cells were treated with 5 μg/ml of actinomycin D for 1 h at 37°C and then processed for immunofluorescence analysis. Cells were stained for CA150, SC35, and Sm snRNPs, U2AF65, and SF2/ASF. Staining with anti-CA150 was detected with Alexa 488-conjugated anti-rabbit Ig (green), and SC35, Sm, U2AF65, and SF2/ASF were detected with Alexa 594-conjugated anti-mouse Ig (red). Individual staining (a and b), merge (c), and colocalization (COLOC.) (d) images of the cell stained with the indicated antibodies are shown. Panels f show line scans showing local intensity distributions of CA150 in green and of the splicing factor in red. Bars in panels e indicate the positions of the line scans. Note that endogenous CA150 is redistributed to enlarged and rounded foci that match the shape and size of nuclear speckles. (B) HeLa cells were treated with 25 μg/ml of α-amanitin for 6 h at 37°C and then processed for immunofluorescence analysis. Cells were stained for CA150 and SC35 as described above. Individual staining (a and b), merge (c), and colocalization (d) images of the cell stained with the indicated antibodies are shown. Bar, 10 μm.
To further analyze the CA150 spatial distribution, we extracted the soluble fraction under mild conditions of nonionic detergent and removed most of the chromatin fraction by in situ digestion with DNase I and ammonium sulfate washes (see Materials and Methods). This treatment did not modify the speckle pattern in the nuclei, as shown by the anti-SC35 staining (Fig. 1D, panel b). The CA150 distribution, however, was clearly affected. The diffuse nucleoplasmic signal was greatly reduced, while the speckle distribution was left intact (Fig. 1D, panel a), as demonstrated by the extensive colocalization of SC35 and CA150 staining (Fig. 1D, panel d). These data together with the previously described immunofluorescence studies confirm the presence of two populations of CA150: a nucleoplasmic pool of CA150 and a less-soluble CA150 population that colocalizes with nuclear speckles.
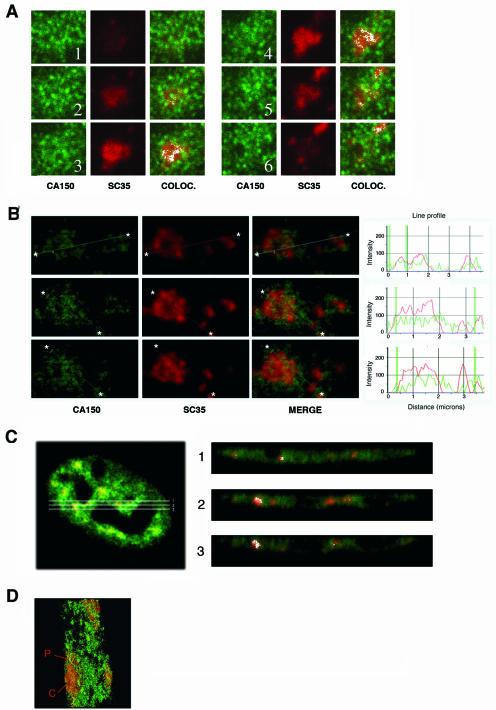
A widely held view is that nuclear speckles function as storage/assembly/modification compartments that can supply splicing factors to surrounding active transcription sites (32). Splicing factors are proposed to be recruited from nuclear speckles to other nucleoplasmic regions for coordinated transcription/splicing. A significant number of active RNAPII-dependent transcription sites have been found located in the periphery of nuclear speckles (68), and hyperphosphorylated RNAPII has been localized to nuclear speckles (5, 47). To further analyze the spatial distribution of CA150, we examined individual nuclear speckles by confocal microscopy optical sectioning (0.15-μm sections) and found that the speckles had peripheral and internally located CA150, which is excluded from the core region of the speckles (Fig. 2A). This experiment was repeated to analyze CA150 localization at multiple speckles by performing semiquantitative analysis of the spatial relationship between the relative spatial distributions of CA150 and SC35. The panels in Fig. 2B show individual stains for CA150 and SC35 and the merge images for this experiment. The analysis of those images showed that the CA150 peaks partially overlap, but do not coincide with, the SC35 peaks (Fig. 2B), supporting the notion that CA150 is enriched at the speckle periphery.
FIG. 2.
The speckle-associated CA150 subpopulation localizes mainly to the periphery and outside the core region of speckles. (A) Optical section series (z-plane step, 0.15 μm) through individual nuclear speckles. Staining with anti-CA150 was detected with Alexa 488-conjugated anti-rabbit Ig (green), and SC35 was detected with Alexa 594-conjugated anti-mouse Ig (red). Colocalization (COLOC.) is also shown (white). (B) The same experiment described for panel A was repeated in a large area with multiple speckles. Seriated optical sections were analyzed at low signal intensity. Individual staining and merge images of a cell stained with antibodies to CA150 (green) and SC35 (red) are shown. Line scans showing local intensity distributions of CA150 in green and SC35 in red are shown to the right of the panels. Asterisks in panels indicate the positions of the line scans. (C) z planes from the peripheral and inner areas of nuclear speckles. Single z-plane cross-section images from the peripheral (1 and 3) and inner (2) areas (right panel) of a HeLa cell labeled with antibodies directed against CA150 (green) and SC35 (red) antibodies (left panel) are shown. The merge image of the cell stained with the indicated antibodies and the colocalization images of the z planes are shown. (D) Three-dimensional visualization of speckle regions of cells dually labeled with antibodies against CA150 (green) and SC35 (red). Optical sections (0.15 μm) were reconstructed to form a three-dimensional image. A section of a three-dimensional reconstruction of a group of speckles from merged focal planes is shown. The image was pseudocolored using Adobe Photoshop so that SC35 is shown in red and CA150 is shown in green. Speckle core (C) and peripheral (P) regions are labeled in the image.
To lend additional support to the observation that CA150 is enriched at the speckle periphery, we analyzed the speckles through serial z-axis optical sections (Fig. 2C and data not shown). For Fig. 2C, we generated three single z-plane cross-sections across the periphery and central portion of a group of speckles from a cell double labeled for CA150 and SC35. The overlay images of those sections are shown (Fig. 2C, right panel). Analysis of those images shows that CA150 was distributed at the periphery of the speckles, which could be seen better in sections corresponding to the central region of speckles (Fig. 2C, right panels 2 and 3). Finally, we analyzed the 3-D organization of CA150 in the speckles by reconstructing optical sections to form a 3-D image, thereby allowing a closer examination of the speckle. A 3-D reconstruction of a double-labeled HeLa cell was created from a stack of confocal merged serial planes (z-plane step; 0.15 μm). A section of the 3-D image displaying a speckle is shown in Fig. 2D. This approach allowed us to discern the speckle core from the periphery of the speckle and confirmed that CA150 was preferentially peripheral and internally located in the speckle region (Fig. 2D). In addition, we note that this location was maintained after removing the soluble fraction of nuclei as described above (data not shown).
The association of CA150 with nuclear speckles is not dependent on active transcription.
There is significant evidence that actively transcribing genes are associated with splicing factor-rich nuclear speckles (60, 68). Taking into account our results described above, we sought to determine the effect of transcriptional activity on the distribution of the speckle-associated CA150. HeLa cells were treated with actinomycin D, which blocks RNAPII transcription by binding to DNA. After 5 h, we examined the distribution of endogenous CA150, SC35, Sm, U2AF65, and SF2/ASF. As shown in Fig. 3, the actinomycin D treatment reduced the number of speckles, which were larger and more rounded (compare panels b, from the SC35 staining, in Fig. 1B and 3A). This result is consistent with previous studies that show an alteration of the speckle morphology when cells are treated with transcriptional inhibitors (64). Actinomycin D treatment redistributed CA150 to the enlarged and rounded granule-like sites, which coincided with splicing factor labeling (Fig. 3A). Under those conditions, the CA150 structures appeared larger than the SC35 structures and colocalized best with structures seen with anti-Sm staining. These results suggest that the colocalization of CA150 with speckles was not dependent on active transcription; however, we noted a redistribution of CA150 from nucleoplasmic regions towards the speckles.
We further examined the distribution of CA150 in α-amanitin-treated cells. HeLa cells were treated with α-amanitin at 25 μg/ml, which strongly inhibits RNAPII-dependent transcription. After 6 h, we performed immunofluorescence localization of CA150 and SC35. As shown in Fig. 3B, α-amanitin treatment had effects similar to those of actinomycin D treatment on speckle morphology. Under those conditions, endogenous CA150 colocalized with SC35 in the speckle region (Fig. 3B), which confirms the previous RNAPII inhibition results. In addition, peripheral localization of CA150 persists upon transcription inhibition (data not shown).
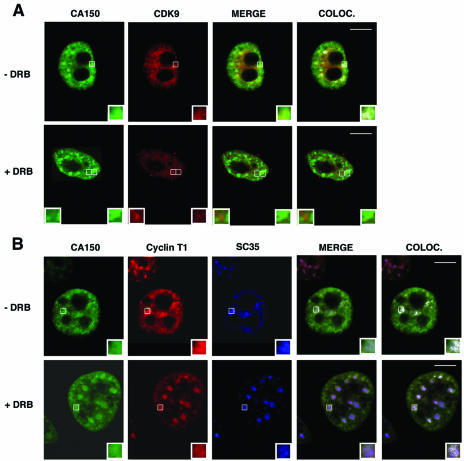
RNAPII-directed transcription can also be blocked by low levels of DRB. DRB is a nucleoside analog that inhibits RNAPII CTD phosphorylation and the production of mRNAs in vivo by targeting the kinase activity of P-TEFb (36, 76). We sought to determine the effect of DRB on the nuclear localization of CA150. In untreated cells, CDK9 and cyclin T1 subunits of P-TEFb localize to splicing factor-rich nuclear speckle regions (Fig. 4) (22). In DRB-treated cells, CDK9 was diffuse in the nucleoplasm and no longer concentrated in speckles (Fig. 4A) (22). Under these conditions, CA150 was localized in a speckled pattern of staining (Fig. 4A), which coincided with the more enlarged SC35 stain (Fig. 4B). As seen with the actinomycin D treatment, the nucleoplasmic pool of CA150 was depleted, while the speckle-associated CA150 was clearly augmented. Cyclin T1 was also associated with speckles (Fig. 4B) as previously reported (22). Those results demonstrate that colocalization of CA150 with nuclear speckles is not dependent on CTD phosphorylation by CDK9.
FIG. 4.
Colocalization (COLOC.) of CA150 with nuclear speckles is not perturbed upon inhibition of CTD phosphorylation. HeLa cells were treated with DRB at 100 μM for 2 h at 37°C and then processed for immunofluorescence analysis. (A) Cells were dually labeled with antibodies directed against CA150 (green) and CDK9 (red). Individual staining, merge, and colocalization images of the cell stained with the indicated antibodies are shown. In the absence of DRB, CA150 and CDK9 colocalize into speckles. In DRB-treated cells, CA150 localizes in a speckled pattern of staining, whereas CDK9 appears to be more diffusely distributed and less present at speckles. (B) Cells were triple stained for CA150 (green), cyclin T1 (red), and SC35 (blue). Individual, merge, and colocalization images are shown. As shown, CA150 and cyclin T1 are redistributed to enlarged and rounded nuclear speckles in DRB-treated cells. Bar, 10 μm.
FF domains are essential for the association of CA150 with nuclear speckles.
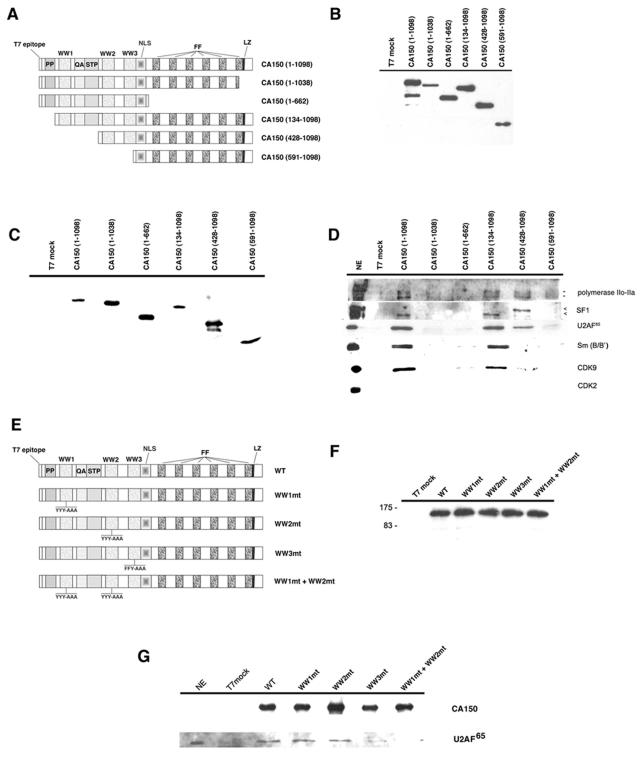
To characterize the regions in CA150 responsible for association with nuclear speckles, we transiently transfected 293T cells with a series of epitope-tagged CA150 deletion mutants. The constructs are diagramed in Fig. 5A. All mutants retain the putative nuclear localization signal found in the middle of the protein (67). We examined the localization of the different CA150 constructs by immunofluorescence staining, and the results for some of those mutants are shown in Fig. 5. The localization of full-length T7-tagged CA150 was similar to that of the endogenous CA150. CA150 mutants with amino-terminal truncations also produced a speckled pattern of staining within the nucleus which was similar to that observed for endogenous CA150 [i.e., CA150 (428-1098) and CA150 (591-1098) in Fig. 5B]. A different labeling pattern was observed for the CA150 mutants with deletions in the carboxyl-terminal half. A CA150 deletion that completely eliminated the FF domains was localized in a diffuse pattern throughout the nucleus without any evident accumulation on the speckles [CA150 (1-662); Fig. 5B]. These results indicate the importance of FF domains for targeting CA150 to nuclear speckles and are consistent with previous work that suggests that FF domains are specifically involved in targeting CA150 to nuclear speckles (61).
To further characterize the FF domains, we generated a carboxyl-terminal deletion series of the FF repeat motifs of CA150 (Fig. 5A) and examined their nuclear localizations. Partial [CA150 (1-1027)] or total [CA150 (1-1004)] deletion of FF6 did not affect the speckle distribution of CA150. A deletion of the FF5 repeat motif [CA150 (1-951)] or larger deletions [CA150 (1-870), CA150 (1-787), and CA150 (1-715)], however, did affect the speckle distribution of CA150 (Fig. 5B and data not shown). These results indicate that a deletion of the FF5 and FF6 repeat domains disrupted the localization to nuclear speckles and imply that FF5 is important for this association.
To demonstrate that CA150 proteins indeed were locating to speckles upon overexpression of DNA constructs, we repeated the experiment to show colabeling of SC-35. The results with some of the mutant constructs are shown in Fig. 5C. As shown before, a deletion of FF6 did not affect speckle distribution of CA150, but a deletion of the FF5 motif resulted in perturbation of CA150 localization to speckles (Fig. 5C). It is noteworthy that transfected nuclei exhibit a more diffuse localization of overexpressed CA150 proteins, which resulted in a partially overlapping signal at the speckle region [Fig. 5C, merge panel for CA150 (1-951)]. This overlapping signal was not specific, as demonstrated by semiquantitative analysis of colocalization signals (Fig. 5C and data not shown).
CA150 interacts with splicing factors.
The immunofluorescence staining of CA150 revealed that this protein colocalizes with the speckles, consistent with the possibility that CA150 and splicing factors do interact in the cell. To investigate this hypothesis, we immunoprecipitated CA150 from HeLa cell nuclear extracts by using antigen affinity-purified anti-CA150 polyclonal antibodies. Anti-GST antibodies were also purified from the same antiserum and used as a negative control for nonspecific interactions. Supporting our hypothesis, we observed that CA150 could associate strongly with Sm snRNPs and U2AF65 proteins in HeLa nuclear extracts (Fig. 6A). Other splicing components, such as SF1, were also detected in the immunoprecipitates (data not shown and reference 18). To address the relative stability of the interaction of CA150 with Sm snRNPs, we passed HeLa nuclear extract over a column containing anti-CA150 coupled to protein A-Sepharose. The bound proteins were subsequently eluted with a linear gradient of KCl, and the fractions were assayed by Western blotting. Sm snRNPs eluted from the column with a peak salt sensitivity of approximately 300 mM KCl (Fig. 6A).
To test whether CA150 could interact with splicing factors in cell lysates, we examined 293T cells transiently transfected with CA150 tagged with an amino-terminal T7 epitope. Upon immunoprecipitation with anti-T7 antibodies and elution with an excess of T7 peptide, we observed coprecipitation of CA150 with endogenous Sm and U2AF65 splicing factors (Fig. 6B). We also found SF1 coprecipitating with CA150 (see Fig. 8) as previously reported (18). These data suggest that CA150 can associate physically with splicing factors.
FIG. 8.
The amino and carboxyl moieties of CA150 are required to interact with splicing and elongation factors in cell lysates. (A). Schematic of the T7-tagged CA150 deletion mutants used in this study. The numbers in parentheses represent the CA150 amino acids contained in the construct. Domains and motifs in CA150 are described in the legend to Fig. 5. (B) Protein expressions were similar among the truncated CA150 constructs. 293T cells were transfected with CA150 mutants or with empty vector (T7 mock), and a sample of the cell extracts was analyzed by SDS-PAGE and immunoblotting using T7-specific antibodies. (C) Affinity purifications by competitive elution with the T7 peptide were similar among the truncated CA150 constructs. Whole-cell extracts were subjected to immunoprecipitation with anti-T7 antibody linked to agarose beads, and proteins were then eluted with T7 peptide. A sample was subjected to SDS-PAGE and Western blotting with T7-specific antibody. (D) Association of CA150 with RNAPII, SF1, U2AF65, Sm snRNPs, and CDK9 depends on specific amino acid sequences located at the amino and carboxyl parts of the protein. Proteins from the affinity purification scheme were subjected to SDS-PAGE and Western blotting with the specific antibodies indicated on the right side of the panel. The T7-tagged CA150 deletion mutants used are shown at the top of the figure. NE, HeLa nuclear extract. (E) Diagrams of CA150 mutants containing mutations in the WW domains. Abbreviations are the same as those used in panel A. YYY-AAA indicates the mutations of tyrosine to alanine in the central aromatic residues of WW1 and WW2. FFY-AAA indicates mutations of phenylalanine and tyrosine to alanine in the central aromatic residues of WW3. Single (WW1mt, WW2mt, and WW3mt) and double (WW1mt+WW2mt) mutants are indicated at the right side of the panel. (F) A representative anti-T7 immunoblot of cell extracts indicating equal levels of expression for each CA150 protein. Molecular masses in kDa are indicated to the left. (G) WW domains mediate the interaction between CA150 and U2AF65. Cell extracts were prepared and subjected to immunoprecipitation with anti-T7 antibody and selective elution with T7 peptide as described in Materials and Methods. Proteins were resolved by using SDS-PAGE and Western blotting with anti-T7 and anti-U2AF65 antibodies. Positions for CA150 and U2AF65 are indicated at the right side of the figure. WT, wild type.
CA150 interacts with transcription elongation factors.
As mentioned above, P-TEFb localizes to nuclear speckles. From our data, it is evident that CA150 also localizes to nuclear speckles. Moreover, CA150 binds the hyperphosphorylated CTD (6), which is a target of CDK9 phosphorylation, and can also affect transcription elongation (66). Those data prompted us to investigate whether CA150 might have a physical connection to other transcription elongation factors. Western blot analysis of the anti-CA150 immunoprecipitates and supernatants from HeLa nuclear extracts revealed the presence of both CDK9 and cyclin T1 components of P-TEFb associated with CA150 (Fig. 7A). CDK9 and cyclin T1 levels in the supernatants were slightly diminished, indicating that a small fraction of the total P-TEFb is associated with CA150 in HeLa nuclear extracts (Fig. 7A). Tat-specific splicing factor 1 (Tat-SF1) coprecipitated with CA150 and was significantly depleted by anti-CA150 immunoprecipitation (Fig. 7A). Tat-SF1, which is the mammalian ortholog of the yeast splicing factor CUS2, has been shown to play roles in the assembly of the spliceosome (33, 69, 72) and implicated in transcription elongation and in the mechanism of trans activation by HIV-1 Tat (36, 67, 72, 74, 76). Moreover, P-TEFb was also found to interact with and phosphorylate Tat-SF1 (73). Very recently, a direct interaction between CA150 and Tat-SF1 was reported, thus corroborating our data (61). The RAP30 subunit of the general transcription factor TFIIF also coimmunoprecipitated with CA150 (Fig. 7A). A significant portion of RAP30 associated with CA150, as evidenced by the levels of RAP30 remaining in the supernatant fraction. It is also noteworthy that TFIIF, like CA150, Tat-SF1, and P-TEFb, has been implicated in the mechanism by which HIV-1 Tat activates transcription elongation (28).
Immunoprecipitation experiments with anti-RAP30 demonstrated that CA150, Tat-SF1, and CDK9 are associated with TFIIF (Fig. 7B), confirming our observations with CA150 immunoprecipitates. Moreover, the anti-CDK9 pelleted CA150, Tat-SF1, and RAP30 (Fig. 7B). A relatively small fraction of CA150 coimmunoprecipitated with anti-RAP30 or anti-CDK9 (Fig. 7B). Therefore, it is likely that only a subset of CA150 is associated with complexes containing P-TEFb and TFIIF. RNAPII was found in the immunoprecipitates with anti-CA150, anti-RAP30, and anti-CDK9 (Fig. 7B), which is consistent with our previous data that CA150 binds to the large subunit of RNAPII (18). Other transcription factors, such as TFIIB, TFIIE, TFIID, Sp1, DSIF, and NELF, did not associate with CA150 (Fig. 7B and data not shown). Taken together, these data suggest that CA150 associates with an RNAPII complex containing transcription elongation factors P-TEFb, Tat-SF1, and TFIIF.
To assess the protein complexes formed by CA150 and associated factors, we employed gel filtration chromatography of HeLa nuclear extracts. First, a high-capacity Sephacryl S400HR column was used to fractionate nuclear extract. CA150 and RNAPII were detected in fractions by Western blotting. CA150 was detected in two distinct size populations (Fig. 7C). The majority of CA150 eluted with a peak molecular mass of approximately 600 kDa. A second population of CA150 was found in very-high-molecular-mass fractions in the MDa range (greater than 1 MDa). RNAPII was found predominantly in large MDa-sized fractions, with a major peak overlapping the MDa-sized CA150 peak (Fig. 7C). We also note that hyperphosphorylated RNAPII was observed in the MDa-sized RNAPII peak that coeluted with the MDa-sized form of CA150 (fractions 31 to 35), as indicated by the shift in mobility of the largest subunit of RNAPII in the Western blot (Fig. 7C). The coelution and coimmunoprecipitation of CA150 with hyperphosphorylated RNAPII support the notion that the CA150 association with the MDa-sized population of RNAPII may be dictated by phosphorylation.
To increase the resolution of this approach, we used a Superose 6 gel filtration column to separate HeLa nuclear extract. The fractions were probed by Western assay with antibodies against CA150, RNAPII, P-TEFb, Tat-SF1, and TFIIF. Again, we observed a minor MDa-sized peak (Fig. 7D, fraction 18) and a major 600-kDa peak (Fig. 7D, fraction 27) of CA150. We also found that RAP30 and Tat-SF1 were almost exclusively in large MDa-sized complexes, consistent with their association with RNAPII complexes (Fig. 7D). Finally, the large majority of CDK9 eluted as MDa-sized complexes, while two minor peaks were detected at approximately 600 to 700 kDa and less than 29 kDa. These data lend additional support to the observation that CA150 can exist in a large MDa-sized complex containing RNAPII, RAP30, Tat-SF1, and P-TEFb. We note that the patterns of elution of the different proteins indicate that multiple high-molecular-mass complexes containing these factors exist in the extract, possibly with distinct protein compositions. This proposition is supported by the immunoprecipitation data presented above, which demonstrated that a subset of P-TEFb, Tat-SF1, TFIIF, and RNAPII associates with CA150 and that these factors can exist in other complexes.
To determine if nucleic acids present in the nuclear extract might influence the apparent molecular mass of these protein complexes, we made a HeLa cell nuclear extract depleted of nucleic acids. Protein was extracted from purified HeLa cell nuclei in the presence of a high concentration of micrococcal nuclease. This treatment degraded nucleic acids to small oligonucleotides or free nucleotides (data not shown). The elution pattern of CA150 and RNAPII was unaffected in the nucleic acid-free nuclear extract (data not shown). The immunoprecipitation Western data presented above were recapitulated in this modified nuclear extract (data not shown). Therefore, the coimmunoprecipitation of RNAPII, P-TEFb, Tat-SF1, and TFIIF with CA150 is not the result of these factors being tethered together by nucleic acids.
These results demonstrate that at least two forms of CA150 exist: a large RNAPII complex containing CA150 and associated elongation factors and a second, more abundant CA150 complex whose composition is currently unknown.
The amino- and carboxyl-terminal halves of CA150 are required for interaction with splicing/elongation factors.
To characterize the regions in CA150 responsible for the interaction of this protein with splicing and elongation factors, we constructed a series of mutants in the amino- and carboxyl-terminal parts of CA150 tagged with the T7 epitope. We performed immunoprecipitation experiments with human 293T cells that were transfected with either full-length CA150 or the mutated constructs. First, we carried out the analysis with a collection of truncated CA150 proteins (Fig. 8A). Expressions in 293T cells (Fig. 8B) and affinity purifications by competitive elution with the T7 peptide (Fig. 8C) were similar among the truncated CA150 constructs. We next examined the interaction of CA150 with the splicing factors SF1, U2AF65, and Sm, as well as with RNAPII and the elongation factor CDK9 (Fig. 8D). Deletion analysis revealed that the amino-terminal region is necessary for the interaction of CA150 with splicing and elongation factors. Deletion of the first 133 amino acids, which eliminates the polyproline-rich region of CA150 [CA150 (134-1098); Fig. 8D], did not affect interactions. Deletion of the first 427 amino acids [CA150 (428-1098); Fig. 8D], which eliminates the first WW domain and the QA/STP region, abrogated the interaction of CA150 with Sm and CDK9, but it did not affect the interaction with RNAPII and affected the interaction with U2AF65 only slightly. Interestingly, we observed a change in the interaction profile of this truncated protein with the splicing factor SF1. Alternatively spliced variants of human SF1 are expressed in a cell-type-specific manner, and at least two different protein products of around 67 and 75 kDa are expressed in HeLa cells (20, 55). Overexpressed CA150 interacted mainly with the smaller SF1 isoform in 293T cells (Fig. 8D). However, the truncated CA150 (428-1098) protein interacted with the 75-kDa isoform (Fig. 8D). At present, we do not know the relevance of this intriguing finding. Further deletion of the second WW eliminated the interaction with SF1 (data not shown), which agrees with previously published data showing that the WW1 or WW2 domain of CA150 was required for binding to SF1 (18). Finally, deletion of the first 590 amino acids [CA150 (591-1098) in Fig. 8A], which eliminates the three WW domains, disrupted the interaction of CA150 with all the RNA processing factors tested (Fig. 8D). RNAPII binding, however, was only slightly affected, which agrees with previous data showing that the principal phospho-CTD-interacting portion of CA150 is the FF domain-containing segment (6). These data demonstrate that transcription and splicing components bound to CA150 through a region in the amino-terminal part of the protein that contains the three WW domains.
Further deletion analysis showed that a truncated CA150 protein in which the FF domains have been eliminated failed to show any interaction [CA150 (1-662); Fig. 8D]. Interestingly, we found that a truncation of the last 59 amino acids of the protein abolished the interaction with splicing/elongation factors [CA150 (1-1038); Fig. 8D]. This deletion eliminates part of the last FF domain and a putative leucine zipper which partially overlaps with the former. Those results suggest an essential role of amino acids located at the 3′ end of the FF domains in the binding of CA150 with transcription/splicing components, including that with the RNAPII.
To assess the contribution of the WW domains of CA150 in the interaction with splicing/elongation factors, we performed immunoprecipitation experiments with 293T cells that were transfected with mutated CA150 proteins. We used CA150 constructs that contained mutations in the central aromatic residues of the WW domains, which are critical for ligand binding (Fig. 8E). All the WW mutant constructs expressed equal levels of protein, as demonstrated by Western blot analysis (Fig. 8F). From those experiments, we determined that the WW3 domain of CA150 is specifically required for interaction with U2AF65, since the WW3mt protein was not capable of binding to the splicing factor (Fig. 8G). We also show that WW1mt and WW2mt bound to U2AF65 as well as the wild type did, thus suggesting that those individual WW domains are not required to recognize U2AF65. A double-mutant CA150 protein containing mutations in both WW1 and WW2 (WW1mt+WW2mt) resulted in loss of U2AF65 binding. Our data suggest that U2AF65 binds CA150 in vivo mainly through the WW3 domain but also suggest that the WW1 and WW2 domains contribute to its association. We conclude that multiple protein interaction domains in the amino and carboxyl regions of CA150 mediate assembly of transcription/splicing components into macromolecular complexes.
DISCUSSION
A growing body of evidence supports the notion that transcriptional elongation and pre-mRNA splicing are linked within the cell temporally, spatially, and functionally (51, 55). Less information is available about the physical interactions between these complex machines. In the studies reported here, we have investigated those connections by studying the transcriptional cofactor CA150 and its relationship to the elongation and splicing machineries. We have found that CA150 is present in the splicing factor-rich nuclear speckle region and that CA150 interacts with both transcription elongation and splicing factors. Our results are suggestive of a role for CA150 in coupling the elongating RNAPII complexes to the spliceosomes through its binding to the phosphorylated CTD of RNAPII.
Why is CA150 in speckles?
CA150 is distributed in dot-like structures throughout the nucleoplasm. This distribution is somewhat different from the more diffuse nucleoplasmic staining usually seen for splicing factors such as ASF/SF2 (Fig. 3). We have found, however, that CA150 has a distribution pattern similar to that of Sm proteins (Fig. 3). Speckles are subnuclear structures that are enriched in pre-mRNA splicing factors and are located in the interchromatin regions of the nucleoplasm of mammalian cells (32, 63). The speckles correspond to previously characterized interchromatin granule clusters (IGCs), which are seen by electron microscopy. The IGCs/speckles contain a subset of splicing and transcription factors which includes the large subunit of RNAPII (5, 47, 62); however, transcription and pre-mRNA splicing take place outside the IGC/speckle compartment. Thus, previous studies using live imaging of cells expressing a fluorescence-tagged splicing factor have shown that splicing factors are recruited from speckles to active sites of transcription (53). Interestingly, the hyperphosphorylated form of RNAPII has been implicated in the targeting of splicing factors to transcription sites in vivo (43). Splicing factors accumulate in speckles when transcription or pre-mRNA splicing is inhibited (50, 64), and posttranscriptional modifications, such as phosphorylation, seem to be required for efficient recruitment of splicing factors at the sites of transcription (44). Those lines of evidence support the notion that speckles function as dynamic storage/assembly/modification compartments of transcription and splicing factors (32, 62, 63). This is further supported by electron microscopy pulse-labeling experiments that showed that newly synthesized mRNA occurs outside the speckles in fibrillar structures known as perichromatin fibrils (10).
Our results on the spatial distribution of CA150 in speckles and its association with splicing factors, as well as data showing that CA150 is a component of the spliceosome (49, 75), suggest that CA150 could be recruited to active transcription sites from the speckle region. In doing so, CA150 could recruit elongation/splicing factors from the speckles to transcription sites. Indeed, many of the factors that coimmunoprecipitate with CA150, including transcription elongation factor P-TEFb and splicing factors SF1 and U2AF65, are also present in the speckles. Speckles are dynamic, with rapid flux of some components having been documented (32). Future experiments will address the question of whether CA150 also shuttles between speckles and transcription sites.
We have analyzed in detail individual nuclear speckles by confocal microscopic optical sectioning and three-dimensional visualization and found the majority of speckle-associated CA150 distributed along the periphery of the speckles. This finding may agree with recent proteomic analysis of the speckle core that has revealed the absence of CA150 (42, 58). The distribution of CA150 overlaps that seen for some of the transcription sites, where the majority of speckle-associated transcription is found along the periphery or juxtaposed to the speckles (68). This association seems to be independent of the presence of DNA (the CA150 localization pattern did not change upon removal of most of the chromatin [Fig. 1]) or active transcription (Fig. 3), suggesting that the speckle localization is not the result of the association of CA150 with actively transcribed genes at the perichromatin fibrils.
How does CA150 associate with speckles?
A question that arises from this study is what targets CA150 to nuclear speckle regions. We (this study) and others (61) have demonstrated the importance of the FF domains of CA150 in its localization at the nuclear speckle region. We further characterized the FF repeat motifs in detail and found that a CA150 protein lacking the FF5 and FF6 repeat motifs is not able to colocalize to speckles. Several interpretations are possible: FF5 is essential for the subnuclear localization of CA150, FF6 and FF5 are both important but neither is essential, or a minimum number of FF repeats are required for the localization to nuclear speckles.
The FF5 domain of CA150 bound to hyperphosphorylated RNAPII CTD most strongly in vitro (6). However, the interaction between CA150 and the CTD is not necessarily responsible for targeting CA150 to nuclear speckles. Our coimmunoprecipitation data show that truncation of just FF6 [CA150 (1-1038)] abrogates RNAPII association (Fig. 8D); however, this mutant protein localizes to speckles (data not shown). Another factor(s) may bind to the FF5 domain and be responsible for targeting CA150 to the splicing factor-rich speckle region, perhaps by competing with the CTD binding. In fact, Tat-SF1 and other splicing factors interact with CA150 through the FF repeats (62). With our data, we cannot conclude that RNAPII binding is responsible for the speckle localization of CA150.
CA150 connects splicing and transcription machineries through multiple protein interaction domains.
There are data that show a physical interaction between the transcription and splicing machineries (27, 31), and CA150 itself has been associated with transcription and splicing components (6, 18, 34, 61). In this report, we found that CA150 is able to form macromolecular complexes with factors involved in elongation and splicing, which is consistent with a potential role for CA150 in linking both processes. Our biochemical data show the importance of the FF6 domain and/or the surrounding regions for the interaction between CA150 and transcription/splicing factors in vivo, including the binding to the CTD. Those results suggest that while the FF5 domain can bind autonomously to the phospho-CTD (6), other sequences must also be capable of binding to the phospho-CTD or collaborate with FF5 to bind to the phospho-CTD. In agreement with this, we have found that internal deletion of FF5 had no effect on in vitro phospho-CTD binding in the context of the carboxyl-terminal half of CA150 (A. C. Goldstrohm, S. M. Carty, M. A. Garcia-Blanco, and A. L. Greenleaf, unpublished results). Thus, FF domains other than FF5 must also recognize the phospho-CTD.
Our data also suggest that the carboxyl-terminal region of CA150, while sufficient for localization to speckles, is not sufficient for the assembly of CA150 into large macromolecular complexes. Truncation of the amino-terminal part of CA150 eliminates the interaction of CA150 with elongation/splicing components. It is possible that amino acids within this region interact with the CTD of RNAPII in vivo, thus modulating the assembly of a multiprotein complex. In fact, a WW domain-containing portion of CA150 binds to the phospho-CTD in vitro, albeit less strongly than FF domains (6). The yeast nuclear protein Prp40, which is structurally related to CA150, can also bind the phospho-CTD at multiple locations, including sites in both the WW and the FF domain regions (46). Other proteins have also been shown to bind the CTD or phospho-CTD at least in part through interactions mediated by WW domains (46, 48).
Our results obtained with the WW mutant constructs (Fig. 8E) support a model in which CA150 mediates a network of protein interactions between transcription/splicing components. WW1 and WW2 bind SF1 (18), SF1 binds to U2AF65 (59), and U2AF65 bound WW3 (Fig. 8E), suggesting that there are multiple interactions through WW3-U2AF65 and WW1/WW2-SF1 to efficiently bind those proteins. However, none of those factors bound to a truncated CA150 protein in which the FF domains had been eliminated [CA150 (1-662); Fig. 8D]. Therefore, CA150 may nucleate the assembly of transcription/splicing factors with the collaboration of amino acid residues located at the amino and carboxyl protein interaction domains. This would be consistent with previous functional data demonstrating that transcription regulation by CA150 requires both amino- and carboxyl-terminal domains to affect elongation (18).
The in vitro (18) and in vivo (this study) interactions of CA150 with SF1 and the reported association of the WW2 of CA150 with the 17S U2 snRNP (34) are suggestive of a role for CA150 in regulating this early step in spliceosome formation. SF1 specifically recognizes the branch point region and facilitates recruitment of U2 snRNP at the 3′ splice site by a mechanism not well understood. We have detected a specific change in the interaction of CA150 with two previously reported SF1 isoforms (20, 55) that is dependent on WW1 and QA/STP regions in CA150. Further investigation is required to know the SF1 isoform that associates preferentially with CA150, which in turn may provide insights about the role of this factor in splicing. It is noteworthy that the assembly of splicing factors at the 3′ splice site of the intron can be affected by the RNAPII elongation rate or processivity. Thus, the RNAPII elongation rate controls the inclusion/exclusion of alternative spliced exons by recognizing different 3′ splice sites in the pre-mRNA sequence (31), emphasizing a very close link between transcription and control of alternative splicing. Recent data have shown that the WW2 of CA150 activated exon inclusion with a β-tropomyosin minigene, albeit with low efficiency, thus supporting a putative role for CA150 in regulating this early step of spliceosome formation (34).
CA150 associates with Tat cofactors.
The association of CA150 with P-TEFb may explain the presence of CA150 in Tat affinity chromatographic fractions. CA150 was originally identified by virtue of its ability to bind to a wild-type Tat affinity column but not to a mutant Tat column (67). CA150 eluted from the wild-type Tat column in a fraction that contained an activity required for transcription activation by Tat (67). However, CA150 did not bind directly to Tat; therefore, its association with the Tat affinity matrix must have been indirect, i.e., mediated by other proteins. We and others have also shown that a complex containing P-TEFb, Tat-SF1, and other factors bound to Tat affinity columns (33, 65, 72). The immunoprecipitation data presented here demonstrate that CA150 associates with P-TEFb subunits and with Tat-SF1. Therefore, the most likely explanation for the presence of CA150 in the Tat affinity column fraction containing the Tat coactivators is that CA150 was associated with the P-TEFb, which bound to Tat via the cyclin T1 subunit. In relation to this, we previously showed that immunodepletion of CA150 decreased Tat activation of RNAPII elongation efficiency in vitro, suggesting that CA150 may have a positive role in this process (67). Here, we have shown that depletion of CA150 from nuclear extract was accompanied by a substantial decrease in the amount of Tat-SF1 as well as by a significant decrease in TFIIF and a diminution of P-TEFb subunits. These results suggest that removal of the CA150-associated Tat coactivators, and not removal of CA150 per se, resulted in decreased Tat activation. Of course, these findings do not exclude the possibility that a complex containing CA150, Tat-SF1, and P-TEFb is involved in HIV-1 Tat activation.
Acknowledgments
This research was supported by grants from the Spanish Ministry of Education and Science (SAF2002-02641 and BFU2005-02806) to C.S. and NIH (RO1GM071037) to M.A.G.-B. M.S.-Á. is supported by a fellowship from the Spanish Ministry of Education and Science (FPU program).
The technical assistance of Sylvia Gutiérrez-Erlandsson in the confocal microscopy studies is gratefully acknowledged. We thank Andrés Aguilera, Amelia Nieto, Juan Ortín, and Juan Valcárcel for critical reading of the manuscript. We also thank Cristina Hernández-Munain for useful discussion during the course of this work.
REFERENCES
- 1.Bedford, M. T., and P. Leder. 1999. The FF domain: a novel motif that often accompanies WW domains. Trends Biochem. Sci. 24:264-265. [DOI] [PubMed] [Google Scholar]
- 2.Belgrader, P., A. J. Siegel, and R. Berezney. 1991. A comprehensive study on the isolation and characterization of the HeLa S3 nuclear matrix. J. Cell Sci. 98:281-291. [DOI] [PubMed] [Google Scholar]
- 3.Bohne, J., S. E. Cole, C. Suñe, B. R. Lindman, V. D. Ko, T. F. Vogt, and M. A. Garcia-Blanco. 2000. Expression analysis and mapping of the mouse and human transcriptional regulator CA150. Mamm. Genome 11:390-393. [DOI] [PubMed] [Google Scholar]
- 4.Bourquin, J. P., I. Stagljar, P. Meier, P. Moosmann, J. Silke, T. Baechi, O. Georgiev, and W. Schaffner. 1997. A serine/arginine-rich nuclear matrix cyclophilin interacts with the C-terminal domain of RNA polymerase II. Nucleic Acids Res. 25:2055-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bregman, D. B., L. Du, S. van der Zee, and S. L. Warren. 1995. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J. Cell Biol. 129:287-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carty, S. M., A. C. Goldstrohm, C. Suñe, M. A. Garcia-Blanco, and A. L. Greenleaf. 2000. Protein-interaction modules that organize nuclear function: FF domains of CA150 bind the phosphoCTD of RNA polymerase II. Proc. Natl. Acad. Sci. USA 97:9015-9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carty, S. M., and A. L. Greenleaf. 2002. Hyperphosphorylated C-terminal repeat domain-associating proteins in the nuclear proteome link transcription to DNA/chromatin modification and DNA processing. Mol. Cell. Proteomics 1:598-610. [DOI] [PubMed] [Google Scholar]
- 8.Cho, E. J., C. R. Rodriguez, T. Tagaki, and S. Buratowski. 1998. Allosteric interactions between capping enzyme subunits and the RNA polymerase II carboxy-terminal domain. Genes Dev. 15:3482-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho, E. J., T. Takagi, C. R. Moore, and S. Buratowski. 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 15:3319-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cmarko, D. 1999. Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection. Mol. Biol. Cell 10:211-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corden, J. L. 1990. Tails of RNA polymerase II. Trends Biochem. Sci. 15:383-387. [DOI] [PubMed] [Google Scholar]
- 12.Dignam, J. D., R. N. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du, L., and S. L. Warren. 1997. A functional interaction between the carboxy-terminal domain of RNA polymerase II and pre-mRNA splicing. J. Cell Biol. 136:5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feaver, W. J., O. Gileadi, Y. Li, and R. D. Kornberg. 1991. CTD kinase associated with yeast RNA polymerase II initiation factor b. Cell 67:1223-1230. [DOI] [PubMed] [Google Scholar]
- 15.Fisher, P. A., L. Lin, M. McConnell, A. Greenleaf, J. M. Lee, and D. E. Smith. 1989. Heat shock-induced appearance of RNA polymerase II in karyoskeletal protein-enriched (nuclear “matrix”) fractions correlates with transcriptional shutdown in Drosophila melanogaster. J. Biol. Chem. 264:3464-3469. [PubMed] [Google Scholar]
- 16.Ghosh, S., and M. A. Garcia-Blanco. 2000. Coupled in vitro synthesis and splicing of RNA polymerase II transcripts. RNA 9:1325-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldstrohm, A. C., A. L. Greenleaf, and M. A. Garcia-Blanco. 2001. Co-transcriptional splicing of pre-messenger RNAs: consideration for the mechanism of alternative splicing. Gene 277:31-47. [DOI] [PubMed] [Google Scholar]
- 18.Goldstrohm, A. C., T. R. Albrecht, C. Suñe, M. T. Bedford, and M. A. Garcia-Blanco. 2001. The transcription elongation factor CA150 interacts with RNA polymerase II and the pre-mRNA splicing factor SF1. Mol. Cell. Biol. 21:7617-7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenleaf, A. L. 1993. Positive patches and negative noodles: linking RNA processing to transcription? Trends Biochem. Sci. 18:117-119. [DOI] [PubMed] [Google Scholar]
- 20.Guth, S., and J. Valcarcel. 2000. Kinetic role for mammalian SF1/BBP in spliceosome assembly and function after polypyrimidine tract recognition by U2AF. J. Biol. Chem. 275:38059-38066. [DOI] [PubMed] [Google Scholar]
- 21.Harlow, E., and D. Lane. 1999. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Herrmann, C. H., and M. A. Mancini. 2001. The CDK9 and cyclin T subunits of TAK/P-TEFb localize to splicing factor-rich nuclear speckle regions. J. Cell Sci. 114:1491-1503. [DOI] [PubMed] [Google Scholar]
- 23.Hirose, Y., and J. L. Manley. 2000. RNA polymerase II and the integration of nuclear events. Genes Dev. 14:1415-1429. [PubMed] [Google Scholar]
- 24.Hirose, Y., R. Tacke, and J. L. Manley. 1999. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev. 13:1234-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho, C. K., and S. Shuman. 1999. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol. Cell 3:405-411. [DOI] [PubMed] [Google Scholar]
- 26.Kadener, S., P. Cramer, G. Nogues, D. Cazalla, M. Mata, J. P. Fededa, S. E. Werbajh, A. Srebrow, and A. R. Kornblihtt. 2001. Antagonistic effects of T-Ag and VP16 reveal a role for RNA pol II elongation on alternative splicing. EMBO J. 20:5759-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kameoka, S., P. Duque, and M. M. Konarska. 2004. p54(nrb) associates with the 5′ splice site within large transcription/splicing complexes. EMBO J. 23:1782-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kato, H., H. Sumimoto, P. Pognonec, C. H. Chen, C. A. Rosen, and R. G. Roeder. 1992. HIV-1 Tat acts as a processivity factor in vitro in conjunction with cellular elongation factors. Genes Dev. 6:655-666. [DOI] [PubMed] [Google Scholar]
- 29.Kim, E., L. Du, D. B. Bregman, and S. L. Warren. 1997. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J. Cell Biol. 136:19-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kornblihtt, A. R., M. de la Mata, J. P. Fededa, M. J. Muñoz, and G. Nogues. 2004. Multiple links between transcription and splicing. RNA 10:1489-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamond, A. I., and D. L. Spector. 1993. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4:605-612. [DOI] [PubMed] [Google Scholar]
- 33.Li, X. Y., and M. R. Green. 1998. The HIV-1 Tat cellular coactivator Tat-SF1 is a general transcription elongation factor. Genes Dev. 12:2992-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin, K. T., R. M. Lu, and W. Y. Tarn. 2004. The WW domain-containing proteins interact with the early spliceosome and participate in pre-mRNA splicing in vivo. Mol. Cell. Biol. 24:9176-9185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu, H., L. Zawel, L. Fisher, J. M. Egly, and D. Reinberg. 1992. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature 358:641-645. [DOI] [PubMed] [Google Scholar]
- 36.Mancebo, H. G. L., J. Flygare, J. Tomassini, P. Luu, Y. Zhu, C. Blau, D. Hazuda, D. Price, and O. Flores. 1997. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 11:2633-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marshall, N. F., and D. H. Price. 1995. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 270:12335-12338. [DOI] [PubMed] [Google Scholar]
- 38.Marshall, N. F., J. Peng, Z. Xie, and D. H. Price. 1996. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 271:27176-27183. [DOI] [PubMed] [Google Scholar]
- 39.McCracken, S., N. Fong, K. Yankulov, S. Ballantyne, G. Pan, J. Greenblatt, S. D. Patterson, M. Wickens, and D. L. Bentley. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385:357-361. [DOI] [PubMed] [Google Scholar]
- 40.McCracken, S., N. Fong, E. Rosonina, K. Yankulov, G. Brothers, D. Siderovski, A. Hessel, S. Foster, S. Shuman, and D. L. Bentley. 1997. 5′-capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 15:3306-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller, C. R., S. F. Jamison, and M. A. Garcia-Blanco. 1997. HeLa nuclear extract: a modified protocol. Academic Press, New York, N.Y.
- 42.Mintz, P. J., S. D. Patterson, A. F. Neuwald, C. S. Spahr, and D. L. Spector. 1999. Purification and biochemical characterization of interchromatin granule clusters. EMBO J. 18:4308-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Misteli, T., and D. L. Spector. 1999. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol. Cell 3:697-705. [DOI] [PubMed] [Google Scholar]
- 44.Misteli, T., J. F. Caceres, J. D. Clement, A. R. Krainer, M. F. Wilkinson, and D. L. Spector. 1998. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J. Cell Biol. 143:297-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizushima, S., and S. Nagata. 1990. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 18:5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morris, D. P., and A. L. Greenleaf. 2000. The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 275:39935-39943. [DOI] [PubMed] [Google Scholar]
- 47.Mortillaro, M. J., B. J. Blencowe, X. Wei, H. Nakayasu, L. Du, S. L. Warren, P. A. Sharp, and R. Berezney. 1996. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc. Natl. Acad. Sci. USA 93:8253-8257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Myers, J. K., D. P. Morris, A. L. Greenleaf, and T. G. Oas. 2001. Phosphorylation of RNA polymerase II CTD fragments results in tight binding to the WW domain from the yeast prolyl isomerase Ess1. Biochemistry 40:8479-8486. [DOI] [PubMed] [Google Scholar]
- 49.Neubauer, G., A. King, J. Rappsilber, C. Calvio, M. Watson, P. Ajuh, J. Sleeman, A. Lamond, and M. Mann. 1998. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat. Genet. 20:46-50. [DOI] [PubMed] [Google Scholar]
- 50.O'Keefe, R. T., A. Mayeda, C. L. Sadowski, A. R. Krainer, and D. L. Spector. 1994. Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J. Cell Biol. 124:249-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Orphanides, G., and D. Reinberg. 2002. A unified theory of gene expression. Cell 108:439-451. [DOI] [PubMed] [Google Scholar]
- 52.Patturajan, M., X. Wei, R. Berezney, and J. L. Corden. 1998. A nuclear matrix protein interacts with the phosphorylated C-terminal domain of RNA polymerase II. Mol. Cell. Biol. 18:2406-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404:604-609. [DOI] [PubMed] [Google Scholar]
- 54.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 55.Rain, J. C., Z. Rafi, Z. Rhani, P. Legrain, and A. Kramer. 1998. Conservation of functional domains involved in RNA binding and protein-protein interactions in human and Saccharomyces cerevisiae pre-mRNA splicing factor SF1. RNA 4:551-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roberts, G. C., C. Gooding, H. Y. Mak, N. J. Proudfoot, and C. W. Smith. 1998. Co-transcriptional commitment to alternative splice site selection. Nucleic Acids Res. 26:5568-5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robson-Dixon, N. D., and M. A. Garcia-Blanco. 2004. MAZ elements alter transcription elongation and silencing of the fibroblast growth factor receptor 2 exon IIIb. J. Biol. Chem. 279:29075-29084. [DOI] [PubMed] [Google Scholar]
- 58.Saitoh, N., C. S. Spahr, S. D. Patterson, P. Bubulya, A. F. Neuwald, and D. L. Spector. 2004. Proteomic analysis of interchromatin granule clusters. Mol. Biol. Cell 15:3876-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Selenko, P., G. Gregorovic, R. Sprangers, G. Stier, Z. Rhani, A. Kramer, and M. Sattler. 2003. Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP. Mol. Cell 4:965-976. [DOI] [PubMed] [Google Scholar]
- 60.Smith, K. P., P. T. Moen, K. L. Wydner, J. R. Coleman, and J. B. Lawrence. 1999. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J. Cell Biol. 144:617-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith, M. J., S. Kulkarni, and T. Pawson. 2004. FF domains of CA150 bind transcription and splicing factors through multiple weak interactions. Mol. Cell. Biol. 24:9274-9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spector, D. L. 1993. Macromolecular domains within the cell nucleus. Annu. Rev. Cell Biol. 9:265-315. [DOI] [PubMed] [Google Scholar]
- 63.Spector, D. L. 1993. Nuclear domains. J. Cell Sci. 114:2891-2893. [DOI] [PubMed] [Google Scholar]
- 64.Spector, D. L., W. H. Schrier, and H. Busch. 1983. Immunoelectron microscopic localization of snRNPs. Biol. Cell 49:1-10. [DOI] [PubMed] [Google Scholar]
- 65.Suñe, C., A. C. Goldstrohm, J. Peng, D. H. Price, and M. A. Garcia-Blanco. 2000. An in vitro transcription system that recapitulates equine infectious anemia virus Tat-mediated inhibition of human immunodeficiency virus type 1 Tat activity demonstrates a role for positive transcription elongation factor b and associated proteins in the mechanism of Tat activation. Virology 274:356-366. [DOI] [PubMed] [Google Scholar]
- 66.Suñé, C., and M. A. Garcia-Blanco. 1999. Transcriptional cofactor CA150 regulates RNA polymerase II elongation in a TATA-box-dependent manner. Mol. Cell. Biol. 19:4719-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suñé, C., R. Hayashi, Y. Liu, W. S. Lane, R. A. Young, and M. A. Garcia-Blanco. 1997. CA150, a nuclear protein associated with the RNA polymerase II holoenzyme, is involved in Tat-activated human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 17:6029-6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wei, X., S. Somanathan, J. Samarabandu, and R. Berezney. 1999. Three-dimensional visualization of transcription sites and their association with splicing factor-rich nuclear speckles. J. Cell Biol. 146:543-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yan, D., R. Perriman, H. Igel, K. J. Howe, M. Neville, and M. Ares, Jr. 1998. CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif. Mol. Cell. Biol. 16:818-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuryev, A., M. Patturajan, Y. Litingtung, R. V. Joshi, C. Gentile, M. Gebara, and J. L. Corden. 1996. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc. Natl. Acad. Sci. USA 93:6975-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zeng, C., and S. M. Berget. 2000. Participation of the C-terminal domain of RNA polymerase II in exon definition during pre-mRNA splicing. Mol. Cell. Biol. 20:8290-8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou, Q., and P. A. Sharp. 1996. Tat-SF1: cofactor for stimulation of transcription elongation by HIV-1 Tat. Science 274:605-610. [DOI] [PubMed] [Google Scholar]
- 73.Zhou, Q., D. Chen, E. Pierstorff, and K. Luo. 1998. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 17:3681-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou, Q., and P. A. Sharp. 1995. Novel mechanism and factor for regulation by HIV-1 Tat. EMBO J. 14:321-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou, Z., L. J. Licklider, S. P. Gygi, and R. Reed. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419:182-185. [DOI] [PubMed] [Google Scholar]
- 76.Zhu, Y., T. Pe'ery, J. Peng, Y. Ramanathan, N. Marshall, T. Marshall, B. Amendt, M. Mathews, and D. Price. 1997. Transcription elongation factor P-TEFb is required for HIV-1 Tat transactivation in vitro. Genes Dev. 11:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]