Abstract
Transcription factors can be sequestered at specific organelles and translocate to the nucleus in response to changes in organellar homeostasis. MondoA is a basic helix-loop-helix leucine zipper transcriptional activator similar to Myc in function. However, unlike Myc, MondoA and its binding partner Mlx localize to the cytoplasm, suggesting tight regulation of their nuclear function. We show here that endogenous MondoA and Mlx associate with mitochondria in primary skeletal muscle cells and erythroblast K562 cells. Interaction between MondoA and the mitochondria is salt and protease sensitive, demonstrating that it associates with the outer mitochondrial membrane by binding a protein partner. Further, endogenous MondoA shuttles between the mitochondria and the nucleus, suggesting that it communicates between these two organelles. When nuclear, MondoA activates transcription of a broad spectrum of metabolic genes, including those for the glycolytic enzymes lactate dehydrogenase A, hexokinase II, and 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3. Regulation of these three targets is mediated by direct interaction with CACGTG sites in their promoters. Consistent with its regulation of glycolytic targets, MondoA is both necessary and sufficient for glycolysis. We propose that MondoA communicates information about the intracellular energy state between the mitochondria and the nucleus, resulting in transcriptional activation of glycolytic target genes.
Retrograde communication between intracellular organelles and the nucleus is a developing signaling paradigm. In several cases, specific transcription factors are associated with organelles and translocate to the nucleus to communicate information about the functional state of the organelle. Subsequent transcriptional activation or repression typically establishes adaptive positive or negative regulatory feedback circuits, resulting in maintenance of cellular homeostasis. For example, in response to increasing inositol levels, the Saccharomyces cerevisiae transcriptional repressor Opi1p translocates from the endoplasmic reticulum (ER) to the nucleus, where it represses genes involved in inositol synthesis (38). Examples from higher eukaryotes include the ER-to-nucleus translocation of sterol regulatory element-binding proteins and ATF6 in response to decreases in intracellular cholesterol levels and unfolded proteins, respectively (9, 57). Nuclear accumulation of sterol regulatory element-binding proteins results in the activation of cholesterol biosynthetic genes, whereas ATF6 regulates genes encoding ER-resident chaperones and folding enzymes.
Our lab recently identified two members of the basic helix-loop-helix leucine zipper (bHLHZip) family of transcription factors, MondoA and Mlx (5, 6). MondoA and Mlx function as a heterodimeric pair, and we have proposed that MondoA-Mlx functions similarly to Myc-Max. For example, like Myc-Max, MondoA-Mlx can activate transcription from CACGTG-dependent reporter genes (5). Furthermore, we have observed synthetic lethality between hypomorphic alleles of the Drosophila orthologs of MondoA and Myc (4). Myc functions in broad aspects of cell physiology including metabolism, growth, and division (22), but whether MondoA contributes to the regulation of each of these diverse processes or only a subset remains to be determined.
In contrast to the primarily nuclear localization of Myc and most other members of the bHLHZip family, MondoA-Mlx heterodimers localize to the cytoplasm in all of the cell types tested (5). However, current evidence argues for their nuclear function. For example, the residues in the basic region required for specific DNA binding are strictly conserved across phyla (our unpublished observation). Consistent with this, MondoA-Mlx heterodimers bind CACGTG E boxes in vitro and activate transcription from CACGTG-dependent synthetic reporter genes in vivo. Furthermore, like many transcription factors, MondoA has a separable and independent transcription activation domain (4, 5). Finally, nuclear accumulation of MondoA-Mlx heterodimers is under tight regulatory control (17), suggesting that the transcriptional targets of MondoA-Mlx heterocomplexes modulate critical aspects of cell physiology. We have proposed that MondoA-Mlx translocates to the nucleus to regulate gene expression in response to an unidentified intra- or extracellular signal(s) (16).
Hints as to the regulatory signal(s) that may control the subcellular localization of MondoA-Mlx come from the work of others examining the regulation of the carbohydrate response element binding protein (ChREBP)/MondoB, which is encoded by Williams-Beuren syndrome critical region gene 14 (WBSCR14) (11, 15). ChREBP is a MondoA paralog originally identified by its inducible CACGTG DNA-binding activity present in liver nuclear extracts prepared from rats fed a high-carbohydrate diet (55). Subsequent experiments showed that a ChREBP-green fluorescent protein (GFP) fusion localized to the cytoplasm of primary hepatocytes but translocated to the nucleus in response to increased glucose levels (30). Despite this high degree of similarity, there appear to be key differences between the functions of these two proteins. For example, subcellular localization of endogenous MondoA is not glucose regulated in either primary human skeletal muscle cells (SkMC) or in C2C12 myoblasts (our unpublished observations) and the phosphorylation sites implicated in the glucose responsiveness of ChREBP are not conserved in MondoA (4, 29, 30).
Here we show that endogenous MondoA and Mlx localize to the outer mitochondrial membrane (OMM) via protein-protein interactions. However, MondoA shuttles between the mitochondria and the nucleus and when nuclear, MondoA activates the transcription of many genes that encode metabolic enzymes. We show that three rate-limiting glycolytic enzymes are direct transcriptional targets of MondoA and furthermore that MondoA is necessary and sufficient for glycolysis. Together, these data suggest that MondoA functions as a sensor of the cellular metabolic state, ultimately communicating between the mitochondria and the nucleus to facilitate changes in the expression of key glycolytic, and likely other, metabolic enzymes.
MATERIALS AND METHODS
Subcloning.
pFLAG:NLSMlx has been described before (5). pcDNA3.1ΔN237MondoA was generated by PCR, and the resulting fragment was cloned into the pcDNA3.1 expression vector (Invitrogen). pBabePuroΔN237NLSMondoA was constructed by fusing an oligonucleotide cassette encoding the nuclear localization signal (NLS) from the simian virus 40 large T antigen in frame to amino acid 237 of MondoA. The resulting chimera was subcloned into the pBabePuro retroviral vector (41). pBabePuroΔN237MondoA(H724P) was constructed with the QuikChange mutagenesis kit (Stratagene). pBabePuroMondoA:TAP was constructed by amplifying the protein A-calmodulin binding protein double-epitope tag (48) from pBS1479 (gift from B. Cairns) and inserting it in frame at the C terminus of the MondoA open reading frame. MondoA-TAP was then subcloned into pBabePuro (41). Amphotropic virus was prepared with the Phoenix packaging cell line (G. Nolan, Stanford University) and used to infect K562 cells. Following selection, single cell clones were isolated by fluorescence-activated cell sorting.
MondoA knockdown.
Two different methods were used for MondoA knockdown. First, small hairpin RNAs (shRNAs) specific for GFP and MondoA, targeting enhanced GFP amino acids 240 to 248 and human MondoA amino acids 667 to 685, were subcloned into a lentiviral vector derivative of pFUGW (39) (gift of R. N. Eisenman) with oligonucleotide cassettes. Lentiviral stocks were made as previously described (39). Following selection in 750 μg/ml G418, cells were prepared for immunofluorescence and Western blotting. Second, shRNAs specific for MondoA and a nonspecific control in the pSM2c retroviral vector were purchased from Open Biosystems. Vesicular stomatitis virus G-pseudotyped virus was made by standard methods and used to infect K562 cells. Following selection in 1 μg/ml puromycin, extracts were prepared for Western blotting and glycolysis measurements. For shRNA sequences, see Fig. S3B in the supplemental material.
Cell culture.
Cells were maintained at 37°C in 5% CO2. K562 and C2C12 cells were grown in RPMI medium (Invitrogen) and Dulbecco modified Eagle medium (Invitrogen), respectively, with 10% bovine calf serum (HyClone), penicillin-streptomycin, and glutamine (Invitrogen). SkMC were grown in SKGM medium (Clonetics). BJ cells immortalized with human telomerase reverse transcriptase were grown as previously described (23).
Antibody production.
Two antipeptide rabbit polyclonal antibodies against amino acids 705 to 721 or 902 to 919 of human MondoA were generated and affinity purified (BioSource International, Inc.). Specific immunoreactivity was determined by enzyme-linked immunosorbent assay and Western blotting (data not shown). The Mlx antibody has been described previously (6).
Immunofluorescence and confocal microscopy.
Cells were fixed in 1× phosphate-buffered saline containing 3.7% formaldehyde for 10 min and stained by standard procedures. K562 cells were allowed to adhere to glass coverslips coated with 1 mg/ml poly-l-lysine. Anti-MondoA (amino acids 705 to 721) and anti-cytochrome c (BD Biosciences) antibodies were used at 1:500 and 1:1,000, respectively. MitoTracker Red CMXRos (Molecular Probes) was used at 200 nM. Secondary antibodies (Molecular Probes) were used at 1:500. Stained cells were mounted on slides with Vectashield (Vector Laboratories). Confocal microscopy was performed with an Olympus Fluoview laser scanning microscope.
Western blotting.
Primary antibodies were used at the following dilutions. Anti-MondoA (amino acids 902 to 919) antibody was used at 1:500. Anti-Mlx (5), anti-mSDS3 (19), anti-cytochrome c, antiporin, anti-α-tubulin, anti-Golgin-97 (all four from Molecular Probes), anti-β-actin, anti-mannose-6-phosphate receptor (both from Abcam), and antiaconitase (gift of E. Leibold) antibodies were all used at 1:1,000. Anti-mSin3A antibody was used at 1:2,500 (25). Secondary antibodies (Amersham Biosciences) were used at 1:10,000, and chemiluminescence (Perkin-Elmer) was used for detection.
Isolation of mitochondria.
Crude mitochondrial preparations were obtained by established protocols (7) and resuspended in mitochondrial isolation buffer (10 mM Tris-HCl [pH 7.9], 150 mM KCl, 10 mM MgCl2, 250 mM sucrose) plus protease inhibitors (0.23 μM aprotinin, 2 μM leupeptin, 1.43 μM pepstatin, and 0.5 mM phenylmethylsulfonyl fluoride [PMSF]) for further analysis. Crude mitochondria were resuspended in 0.85 M sucrose and applied to the top of a gradient consisting of 1.0 M, 1.35 M, 1.65 M, and 2.0 M sucrose. Gradients were centrifuged at 80,000 × g for 2 h at 4°C, and purified mitochondria were recovered at the 1.0 M and 1.35 M interface. Purified mitochondria were diluted with 2 volumes of 1 mM EDTA and 10 mM Tris-HCl, pH 7.4, and pelleted by centrifugation at 20,000 × g for 10 min at 4°C.
Protease protection assay.
The protease protection assay was performed as previously described (33). Reaction mixtures contained 50 μg of isolated mitochondria in 50 μl of mitochondrial isolation buffer. Where indicated, samples were treated with 200 μg/ml proteinase K and 1% Triton X-100 on ice for 30 min before stopping the reactions with 2 mM PMSF. For the binding of MondoA-TAP to mitochondria, mitochondria were isolated and treated, where indicated, with 200 μg/ml proteinase K and/or 2 mM PMSF. Reaction mixtures were kept on ice for 30 min before the addition of 2 mM PMSF to stop the reactions. Samples were washed three times and centrifuged, and the pellets were resuspended in 50 μl of cytoplasmic extract from K562 cells expressing MondoA-TAP.
LMB treatment.
Cells were treated with 10 μg/ml leptomycin B (LMB; Sigma) for 4 h and then separated into cytoplasmic and nuclear fractions as previously described (5).
Transcription assays.
Lactate dehydrogenase A (LDH-A) promoter constructs were provided by Chi V. Dang (52). Luciferase reporter assays were performed in triplicate, with the error given as the standard error of the mean as we have reported previously (5).
Chromatin immunoprecipitation (ChIP).
Cells were cross linked and nuclei were prepared as previously described (27). Nuclei were resuspended in sonication buffer (50 mM Tris-HCl [pH 8], 1 mM EDTA, 0.5 mM EGTA) plus protease inhibitors and sonicated on ice to an average length of 500 to 1,000 bp. Chromatin was precleared with a 50% slurry of preblocked protein A/G-Sepharose (Pierce) at 4°C for 4 h and then incubated with 5 μl of each antibody in dilution buffer (20 mM Tris [pH 7.9], 2 mM EDTA, 150 mM NaCl, 1% Triton X-100, 2 mg/ml bovine serum albumin) plus protease inhibitors at 4°C overnight. Fifty microliters of a 50% slurry of protein A/G-Sepharose was added, and immune complexes were recovered after 24 h. Anti-MondoA (amino acids 902 to 919), anti-Myc (N262; Santa Cruz Biotechnology), and anti-Gal4 (RK5C1; Santa Cruz Biotechnology) antibodies were used. Immunoprecipitates were washed five times with wash buffer, cross-links were reversed, and DNA was purified (27). Immunoprecipitated DNA was resuspended in 50 μl of 10 mM Tris (pH 7.9). Real-time PCR was performed with the LightCycler FastStart DNA Master SYBR green I system (Roche) according to the manufacturer's instructions. Average values (± the standard error) from two independent biological replicates were expressed as fold enrichment over a non-CACGTG-containing fragment upstream of the 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase (PFKFB3) promoter. For primer sequences, see Fig. S3B in the supplemental material.
Quantitative PCR.
Vesicular stomatitis virus G-pseudotyped pBabePuroΔN237NLSMondoA and pBabePuroΔN237-NLSMondoA(H724P) retroviruses were used to infect C2C12 cells. Following selection in 1 μg/ml puromycin, total RNA was produced with the RNeasy Mini Kit (QIAGEN). cDNA was produced and purified with the SuperScriptIII reverse transcription kit (Invitrogen) and the QIAquick PCR purification kit (QIAGEN), respectively. Amplification was performed with the Roche LightCycler and LightCycle FastStart DNA Master Mix plus SYBR green (Roche). The comparative Ct method was used for data analysis (37). For primer sequences, see Fig. S3B in the supplemental material.
Measurement of glycolysis.
Glycolysis rates were measured by monitoring the conversion of 5-[3H]glucose to 3H2O as previously described with only minor modifications (53).
Microarray analysis.
RNA was prepared with the RNeasy Mini Kit (QIAGEN) from four biological replicates of C2C12 cells expressing ΔN237NLSMondoA or infected with the pBabePuro vector alone. RNA labeling, hybridization to slides spotted with approximately 25,000 cDNAs, and initial data analysis with the GeneSpring Analysis platform were performed by the Huntsman Cancer Institute Array Core facility. Significance analysis of microarrays (21) was used to identify genes that were regulated more than 1.2-fold with a false-discovery rate of less that 10%.
RESULTS
MondoA and Mlx are resident mitochondrial proteins.
Our previous experiments demonstrated a striking cytoplasmic localization for exogenously expressed MondoA and Mlx (5, 17). To determine the precise cytoplasmic localization of the endogenous proteins, we performed indirect immunofluorescence assays of primary human SkMC. In virtually every cell, endogenous MondoA and Mlx immunoreactivity displayed a reticular pattern in the cytoplasm with only weak background nuclear labeling (Fig. 1A and B and data not shown). For MondoA, blocking the primary antibody with immunizing peptide, but not with a nonrelated peptide, eliminated this staining pattern, demonstrating that the immunoreactivity was specific. MondoA labeling was also reduced to background levels in MondoA knockdown cells, demonstrating the specificity of the MondoA antibody (see Fig. S1 in the supplemental material). For Mlx, no labeling was observed with preimmune serum, demonstrating the specificity of the Mlx labeling pattern (see Fig. S1 in the supplemental material).
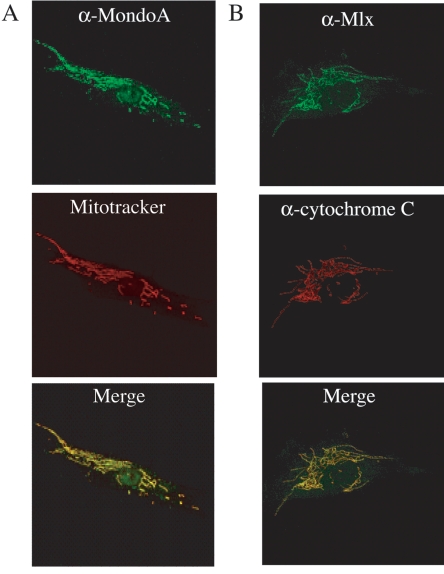
FIG. 1.
MondoA and Mlx colocalize with the mitochondria. (A) Primary human SkMC were labeled with anti-MondoA antibody and MitoTracker, and the two images were merged. (B) Primary human SkMC were labeled with anti-Mlx and anti-cytochrome c antibodies, and the two images were merged. The images in panels A and B were obtained by confocal microscopy.
The reticular, network-like staining pattern of MondoA and Mlx suggested that they localize to the mitochondria. To confirm this, we compared their localization to that of cytochrome c and to the fluorescence of the mitochondrial probe MitoTracker. MondoA immunoreactivity overlapped that of cytochrome c (data not shown) and MitoTracker fluorescence (Fig. 1A). Similarly, Mlx immunoreactivity overlapped that of cytochrome c (Fig. 1B). Therefore, both MondoA and Mlx associate with mitochondria in SkMC. MondoA and Mlx showed a similar mitochondrial localization in a number of lymphoid, myeloid, epithelial, fibroblast, and myoblast cell lines (data not shown), demonstrating that their mitochondrial localization is widespread.
To examine the mitochondrial localization of MondoA and Mlx further, we isolated mitochondria from SkMC and K562 cells by differential centrifugation. Fraction purity was assessed with the mitochondrial protein cytochrome c and the nuclear proteins mSin3A (25) and mSDS3 (19). Consistent with the immunofluorescence data, MondoA was highly enriched in the cytochrome c-containing heavy membrane fraction from both cell lines with negligible amounts present in the cytoplasmic and nuclear fractions (Fig. 2A and B). By contrast, Mlx was detected in both the heavy membrane and nuclear fractions from SkMC (Fig. 2A); a nuclear localization for Mlx is consistent with its interaction with other transcription factors (5, 6, 11).
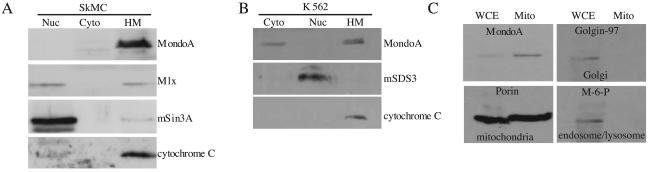
FIG. 2.
MondoA is enriched in the mitochondrial fraction from SkMC and K562 cells. SkMC (A) or K562 cells (B) were separated into nuclear (Nuc), cytoplasmic (Cyto), and heavy membrane (HM) fractions. Levels of MondoA, Mlx, the nuclear markers mSin3A and mSDS3, and the mitochondrial marker cytochrome c were determined by Western blotting. Mitochondria were purified by sucrose density gradient centrifugation (C), and the levels of MondoA, the mitochondrial marker porin, the Golgi marker Golgin-97, and the endosomal/lysosomal marker mannose-6-phosphate receptor (M-6-P) were determined by Western blotting. WCE, whole cell extract; Mito; mitochondrial fraction.
We used sucrose density gradient centrifugation to purify mitochondria from the other membranes present in the heavy membrane fractions. MondoA copurified with the OMM protein porin in a fraction devoid of Golgi (Golgin-97), endosome/lysosome (mannose-6-phosphate receptor), and nuclear (c-Myc) markers (Fig. 2C and data not shown). Together, these data demonstrate that most of the endogenous MondoA is associated with mitochondria.
MondoA associates with an OMM protein.
To determine the submitochondrial localization of MondoA, we performed protease sensitivity experiments. Treatment of intact mitochondria with proteinase K resulted in complete degradation of MondoA and partial degradation of the peripheral OMM protein porin (Fig. 3A). Cytochrome c, which localizes to the inner membrane space, was not degraded, demonstrating that the OMM was intact and the protease only had access to outer membrane-associated proteins. By contrast, permeabilization of mitochondria with Triton X-100 resulted in the complete degradation of all three proteins, demonstrating that they are equally protease sensitive. This experiment demonstrates that MondoA localizes to the OMM rather than to an internal membrane or compartment of the mitochondria.
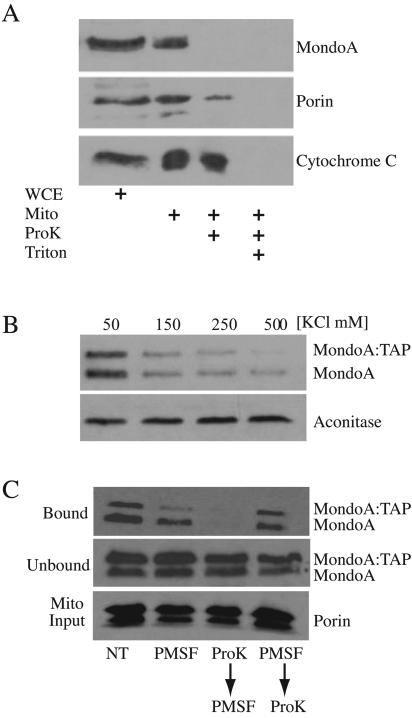
FIG. 3.
MondoA associates with an OMM protein. (A) Mitochondria were prepared from K562 cells and were mock treated or incubated with proteinase K in the presence or absence of Triton X-100 as indicated. Levels of MondoA, the outer membrane marker porin, and the intermembrane space marker cytochrome c were determined by Western blotting. WCE, whole cell extract; Mito; mitochondrial fraction. (B) Isolated mitochondria from K562 cells were incubated with a cytoplasmic lysate containing MondoA-TAP in the presence of increasing KCl concentrations. After binding and washing, the amounts of MondoA, MondoA-TAP, and aconitase that remained associated with mitochondria were determined by Western blotting. (C) Isolated mitochondria were left untreated (NT), treated with PMSF, treated with proteinase K followed by PMSF, or treated with PMSF followed by proteinase K. Treated mitochondria were incubated with a cytoplasmic lysate containing MondoA-TAP. Following binding and washing of the mitochondrial pellets, the amount of MondoA, MondoA-TAP (bound), or porin (Mito Input) that remained associated with mitochondria following each treatment was determined by Western blotting.
To determine whether MondoA is a peripheral or integral membrane-associated protein, we bound cytoplasmic extract containing an epitope-tagged version of MondoA, MondoA-TAP (data not shown), to purified mitochondria in the presence of increasing KCl concentrations. In the presence of 50 mM KCl, MondoA and MondoA-TAP associated with mitochondria; however, the binding of both proteins decreased with increasing KCl concentrations (Fig. 3B). Mitochondrial association of the matrix protein aconitase was unaffected, demonstrating that mitochondrial membranes were not permeabilized by the salt treatment. The salt-sensitive association of MondoA with mitochondria suggests it is a peripheral outer membrane-associated protein.
To determine whether MondoA binds mitochondria via protein-protein interactions or by interacting with phospholipid components of the OMM, we modified the above mixing experiment. Mitochondria from K562 cells were treated with proteinase K for 30 min to degrade OMM proteins, and then the protease was inactivated with PMSF (Fig. 3C). A cytoplasmic extract containing MondoA-TAP was then mixed with the digested mitochondria. Digestion of outer membrane proteins completely eliminated binding of MondoA-TAP (Fig. 3C). Treatment of mitochondria with PMSF, or with PMSF and proteinase K simultaneously, had little effect on MondoA-TAP binding, demonstrating that binding was inhibited by protease digestion of OMM components. MondoA-TAP levels were equal in all of the unbound fractions, demonstrating that it was not degraded by any residual protease activity following PMSF inactivation and was fully available for binding (Fig. 3C). Finally, levels of porin were identical in all of the samples, indicating that equivalent amounts of mitochondria were provided for each condition. Taken together, these experiments demonstrate that MondoA interacts with the OMM via a protein-protein interaction(s).
The association of MondoA and the mitochondria is dynamic.
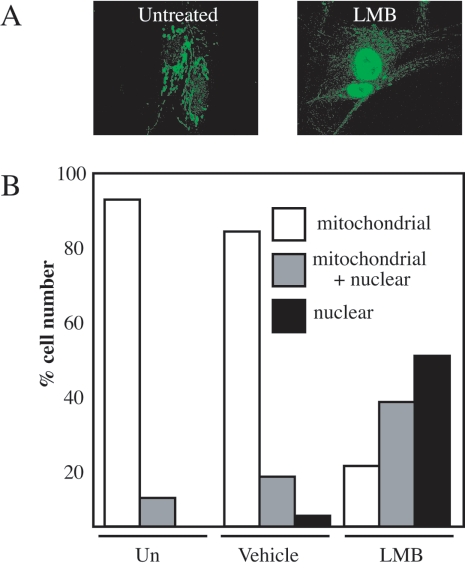
Our previous data demonstrated that ectopically expressed MondoA-Mlx complexes shuttle between the cytoplasm and the nucleus in a manner dependent on the CRM1 nuclear export factor (5). To determine whether endogenous MondoA also shuttles between mitochondria and the nucleus, we treated SkMC for 4 h with the CRM1 inhibitor LMB. MondoA localized to mitochondria in untreated or vehicle-treated cells, with negligible amounts present in the nucleus. By contrast, in the majority of LMB-treated cells, MondoA labeling shifted from the mitochondria to the nucleus (Fig. 4A and B and data not shown). Similar results were obtained with K562 cells (see Fig. S2 in the supplemental material). Therefore, inhibition of CRM1-dependent nuclear export results in the nuclear accumulation of MondoA, demonstrating that MondoA shuttles between the OMM and the nucleus in a dynamic manner, perhaps facilitating communication between these two organelles.
FIG. 4.
The association of MondoA and the mitochondria is dynamic. (A) SkMC were either treated with 10 ng/ml of the nuclear export inhibitor LMB for 4 h or left untreated. Cells were labeled with anti-MondoA antibody, and images were obtained by confocal microscopy. (B) The subcellular localization of endogenous MondoA in SkMC was quantified in a double-blind experiment. Nuclear, predominantly nuclear labeling; mitochondria + nuclear, equivalent mitochondria and nuclear labeling; mitochondria, predominantly mitochondria labeling; Un, untreated.
MondoA regulates transcription of glycolytic enzymes.
To provide information about the nature of the intracellular communication mediated by MondoA, we identified MondoA transcriptional targets. To do this, we expressed a constitutively transcriptionally active version of MondoA, ΔN237NLSMondoA, in the nuclei of C2C12 myoblasts and identified regulated targets by microarray gene profiling. Our previous findings suggest that Mlx is an obligate partner for MondoA (5); therefore, transcriptional effects attributable to MondoA are likely mediated by heterodimerization with endogenous Mlx. We used a commercially available analysis platform, Genesifter (VizXlabs), to identify pathways overrepresented in the list of MondoA-regulated genes. Genes involved in metabolism and cell growth were the two most highly regulated functional categories, representing 53% and 29% of the MondoA targets, respectively; the entire set of MondoA targets regulated more than 1.8-fold is presented in Fig. S3A in the supplemental material. Given the paralogous relationship between MondoA and ChREBP and the established role of c-Myc in glycolysis (31, 44), we focused initially on genes involved in carbohydrate catabolism. Seventy-four array genes are involved in carbohydrate metabolism, and MondoA regulated 39 of these; 34 were upregulated, and 5 were downregulated. The glycolytic enzymes hexokinase II (HKII), PFKFB3, and transketolase were among the most highly regulated MondoA targets, with their expression increasing by 2.8-, 2.0-, and 1.8-fold, respectively.
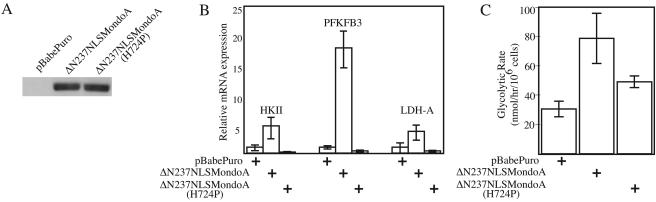
To confirm MondoA's regulation of HKII and PFKFB3, we performed quantitative reverse transcription-PCR. We also examined the expression of LDH-A, which, although weakly regulated on the array, is a well-characterized Myc target (52). To ascertain whether regulation requires specific DNA binding by MondoA, we generated ΔN237NLSMondoA(H724P), which has a mutation in the DNA-binding basic region predicted to abrogate DNA binding by MondoA-Mlx heterodimers (18, 42). ΔN237NLSMondoA and ΔN237NLSMondoA(H724P) were expressed in the nucleus at similar levels in C2C12 myoblast cells by retroviral transduction (Fig. 5A). Relative to C2C12 cells infected with the empty vector, HKII, PFKFB3, and LDH-A levels were increased 4.5-, 17.5-, and 3.5-fold, respectively, by the activity of ΔN237NLSMondoA (Fig. 5B). Similar results were obtained by Northern blotting (data not shown). By contrast, expression of all three targets was reduced in ΔN237NLSMondoA(H724P)-expressing cells, demonstrating a requirement for DNA binding and suggesting that this mutant functioned as a dominant negative regulator at these targets (Fig. 5B). All three genes were regulated to a greater extent than revealed by microarray analysis, suggesting that the array experiment underrepresented the magnitude of the MondoA effect.
FIG. 5.
MondoA regulates the transcription of glycolytic enzymes. (A) Expression of ΔN237NLSMondoA and ΔN237NLSMondoA(H724P) in C2C12 cells was determined by Western blotting. The HKII, PFKFB3, and LDH-A levels (B) and glycolysis rates (C) in C2C12 cells expressing ΔN237NLSMondoA or ΔN237NLSMondoA(H724P) or infected with pBabePuro alone were determined.
HKII, PFKFB3, and LDH-A are key regulators of glycolysis, suggesting that MondoA regulates glucose catabolism. To measure the effect of MondoA on glycolysis directly, we measured the conversion of 5-[3H]glucose to 3H2O in C2C12 cells expressing the two MondoA constructs. Cells expressing ΔN237NLSMondoA converted 5-[3H]glucose to 3H2O at an almost threefold higher rate than cells infected with the empty retroviral vector. By contrast, cells expressing ΔN237NLSMondoA(H724P) showed only a modest increase in 5-[3H]glucose-to-3H2O conversion (Fig. 5B). Therefore, ΔN237NLSMondoA regulates key glycolytic target genes and glycolysis.
MondoA regulates glycolytic targets directly.
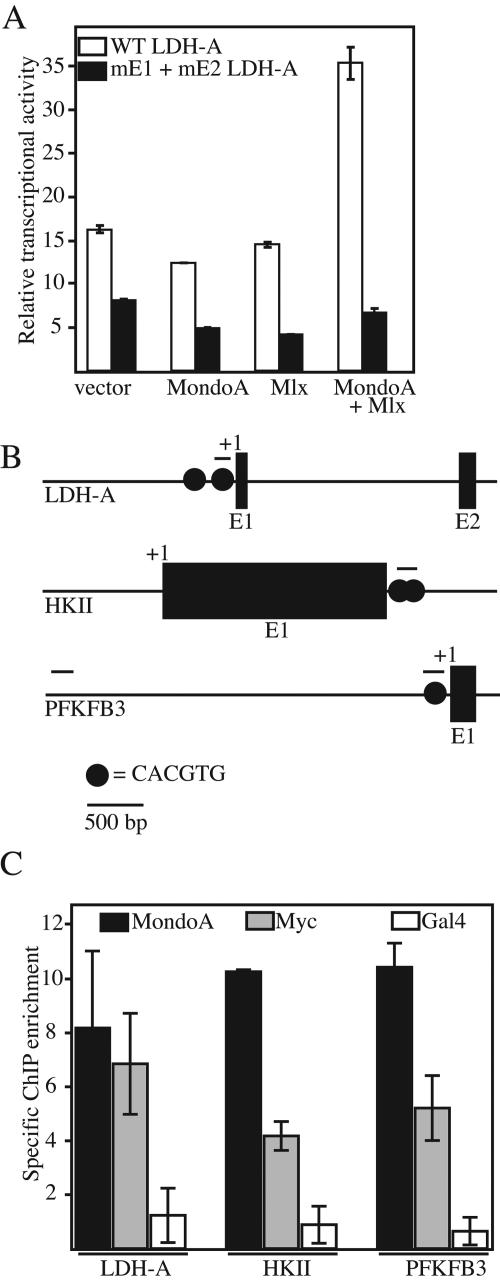
The LDH-A promoter is a well-characterized c-Myc target with activation requiring conserved upstream CACGTG binding sites (31, 52). We used transient expression to determine whether regulation of LDH-A shows a similar dependence on CACGTG sites. Consistent with our previous findings with synthetic promoters (5), expression of ΔN237MondoA or an NLS-tagged version of Mlx alone had little effect on LDH-A luciferase reporter activity. By contrast, coexpression of ΔN237MondoA and NLS-Mlx together resulted in 2.5-fold activation of the reporter (Fig. 6A). An LDH-A promoter construct containing mutations in both upstream CACGTG E boxes (mE1 and mE2) was unresponsive to ΔN237MondoA, NLS-Mlx, or their combination. Therefore, MondoA-Mlx heterodimer activation of the LDH-A promoter is dependent on the two upstream CACGTG elements.
FIG. 6.
MondoA can regulate glycolytic genes directly. (A) The activities of the wild-type (WT) and double CACGTG E-box mutant (mE1 + mE2) LDH-A luciferase reporters were measured in C2C12 cells expressing the indicated combinations of ΔN237MondoA (MondoA) and NLS-Mlx (Mlx) or the corresponding empty expression vectors. (B) Schematic diagrams of the LDH-A, HKII, and PFKFB3 promoters. Exons (E) and transcriptional start sites (+1) and locations of CACGTG E-box elements are indicated. Lines above the promoters indicate the regions amplified by PCR. (C) Real-time PCR assay measuring enrichment of ChIPs from C2C12 cells expressing ΔN237NLSMondoA performed with MondoA, c-Myc, or Gal4p antibodies. Immunoprecipitated DNA was analyzed by real-time PCR with primers specific to the E-box-containing regions of the mouse LDH-A, HKII, and PFKFB3 promoters. Measurements are expressed as fold enrichment over the amount of an upstream region of the PFKFB3 promoter that does not contain a CACGTG site. Each value is the average (±the standard error) from two independent biological replicates.
ChIP was utilized to determine whether MondoA occupies the promoters of the endogenous HKII, PFKFB3, and LDH-A genes. Myc has been shown to regulate HKII directly by binding a region that contains multiple conserved CACGTG E-box elements (31). The PFKFB3 promoter has a putative regulatory E-box that is located approximately 120 bp upstream of the transcriptional start site. This site is conserved between humans and mice (our unpublished observation) and may be occupied by c-Myc and/or MondoA. LDH-A contains the two aforementioned CACGTG E boxes (Fig. 6B, top). We used antibodies specific for MondoA and c-Myc and a negative control antibody specific for the S. cerevisiae transcriptional activator Gal4p to immunoprecipitate cross-linked chromatin prepared from C2C12 cells expressing ΔN237NLSMondoA. Immunoprecipitated DNA was quantified by quantitative PCR with primer pairs flanking the CACGTG E boxes present in each promoter. Specific ChIP enrichment was determined by normalizing to a region approximately 5 kb upstream of the PFKFB3 promoter that lacks CACGTG sites. MondoA and c-Myc occupied each of the promoters, with MondoA showing slightly more binding than c-Myc at each target; no enrichment was observed in the Gal4p ChIP (Fig. 6C). Therefore, this experiment suggests that ΔN237NLSMondoA regulates the glycolytic targets HKII, PFKFB3, and LDH-A directly by interaction with CACGTG-containing regulatory elements.
Endogenous MondoA regulates glycolysis.
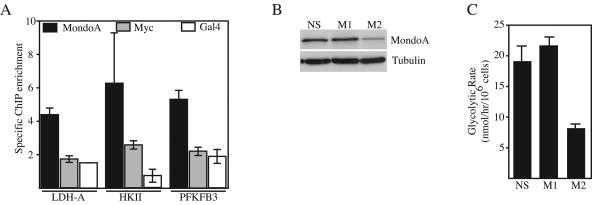
The majority of MondoA and Mlx localizes to mitochondria in most cell types; however, roughly 5 to 10% is present in the nucleus as it shuttles between the OMM and the nuclear compartment (data not shown). To address whether nuclear MondoA is capable of target gene activation, we determined the occupancy of endogenous MondoA at the HKII, PFKFB3, and LDH-A promoters. We used normal diploid human BJ fibroblasts immortalized with human telomerase reverse transcriptase for these experiments, reasoning that these nontransformed cells would closely mimic the physiological regulation of MondoA. Even though the MondoA in BJ cells localized primarily to the mitochondria, with only low levels present in the nucleus (data not shown), we found that it occupied each glycolytic target (Fig. 7A). Therefore, the low level of endogenous nuclear MondoA in BJ cells may be capable of regulating glycolytic genes and glycolysis. By contrast, we only observed significant occupancy by c-Myc at the HKII promoter, suggesting that it does not play a major role in regulating glycolysis in this cell type.
FIG. 7.
Endogenous MondoA regulates glycolysis. (A) Real-time PCR measured enrichment of ChIPs from BJ fibroblasts performed with MondoA, c-Myc, or Gal4p antibodies. Immunoprecipitated DNA was analyzed by real-time PCR with primers specific to the E-box-containing regions of the human LDH-A, HKII, and PFKFB3 promoters. Measurements are expressed as fold enrichment over the amount of an upstream region of the PFKFB3 promoter that does not contain a CACGTG site. Each value is the average (±the standard error) from at least three independent biological replicates. Expression levels of MondoA and tubulin (B) and glycolysis rates (C) in K562 cells expressing the indicated shRNA constructs are also shown. NS, a scrambled shRNA that does not recognize any sequence in the human genome; M1 and M2, shRNA sequences specific for human MondoA.
To test whether endogenous MondoA is necessary for glycolysis, we lowered its expression levels by RNA interference. For this experiment, we used K562 cells, which express MondoA primarily at the mitochondria (Fig. 2; see Fig. S2 in the supplemental material). Cells expressing the M1 shRNA specific for the coding region of MondoA, but not a nonspecific scrambled shRNA, resulted in a reduction of MondoA protein levels and a decrease in the glycolysis rate (Fig. 7B and C). shRNA M1 against the 3′ untranslated region of MondoA was ineffective for MondoA knockdown and did not reduce glycolysis. Therefore, endogenous MondoA levels correlate with the glycolysis rate, suggesting that MondoA is necessary for glycolysis in K562 cells.
DISCUSSION
We show that the bHLHZip transcription factor MondoA localizes to the OMM but shuttles between the OMM and the nucleus. When nuclear, MondoA activates transcription of at least three glycolytic enzymes, LDH-A, HKII, and PFKFB3, via CACGTG E-box elements. Consistent with the upregulation of these targets, MondoA is both necessary and sufficient for glycolysis. Our previous experiments demonstrate that Mlx is an obligate partner for MondoA and is required for its nuclear localization and transcriptional activity (5, 17). We show here that Mlx also associates with mitochondria, presumably complexed with MondoA. On the basis of our findings, we propose that MondoA-Mlx complexes contribute to the regulation of glycolysis in at least two ways. First, the low levels of nuclear MondoA-Mlx complexes are necessary for the basal rates of glycolysis. Second, MondoA-Mlx complexes contribute to the maintenance of energy homeostasis by sensing cellular metabolic status at the OMM and translocating to the nucleus to activate key glycolytic enzymes. The binding of MondoA to glycolytic targets suggests that its regulation of glycolysis is a direct proximal consequence of its function as a transcriptional activator rather than an indirect consequence. We have focused only on three of MondoA's glycolytic targets; however, our microarray analysis suggests a broader role in metabolic and growth control.
To our knowledge, this is the first report of a transcription factor complex that shuttles between mitochondria and the nucleus. Further, the OMM localization of MondoA and Mlx is unique among members of the bHLHZip superfamily and we have been unable to find precedence in the literature of similarly localized transcription factors. There are examples of transcription factors thought to localize primarily to the nucleus that have been subsequently also localized to the mitochondrial matrix; however, these factors appear to regulate transcription of the mitochondrial genome (12, 50, 56). Finally, while signaling pathways downstream of STK11/LKB1 and phosphatidylinositol-3-kinase are clearly implicated in relaying information about the intra- and extracellular bioenergetic and/or nutrient state to the nucleus (1, 47, 51), the transmission of metabolic information to the nucleus by MondoA-Mlx heterodimers may represent an additional mechanism for maintaining cellular energy homeostasis.
Retrograde signaling between mitochondria and the nucleus is not without precedent. For example, the mitochondrial proteins apoptosis-inducing factor and endonuclease G transit from the mitochondria to the nucleus during programmed cell death (28). A subtler example of mitochondria to nuclear signaling is the retrograde response in S. cerevisiae. In this pathway, the transcription factors Rtg1p and Rtg3p localize to the cytoplasm and translocate to the nucleus as an adaptive response to mitochondrial stress (10). Our proposal that MondoA and Mlx communicate information about the functional state of the mitochondria to the nucleus is similar to the yeast retrograde response. Like MondoA and Mlx, Rtg1p and Rtg3p are members of the bHLHZip family. In fact, Rtg3p is the protein most similar to Mlx in the yeast genome; MondoA bears little similarity to Rtg1p outside the bHLHZip domain. Despite these similarities, MondoA-Mlx heterocomplexes remain associated with the mitochondria in respiration-deficient ρ°143B fibrosarcoma cells (32) or in cells treated with a variety of drugs that uncouple electron transport (data not shown). Therefore, despite the parallels between retrograde Rtg1p-Rtg3p signaling and MondoA-Mlx signaling, it seems unlikely that they participate in the mitochondrial stress response.
MondoA contributes to the basal glycolysis rate in K562 cells, but its shuttling between the mitochondria and the nucleus strongly suggests that it communicates between these two organelles. Given the predominant role of mitochondria in intracellular bioenergetics, it seems most likely that MondoA senses aspects of the intracellular energy state and accumulates in the nucleus to activate the transcription of glycolytic, and potentially other, genes as an adaptive response. MondoA's regulation of glycolytic targets suggests that it may translocate to the nucleus when intracellular ATP levels are low or under anaerobic conditions, similar to hypoxia-inducible factor 1α (14). However, lowering intracellular ATP levels, mimicking an increase in intracellular AMP levels (24), or lower oxygen tension had no effect on the mitochondrial localization of MondoA in C2C12 cells (data not shown). Given that glycolysis provides precursors and reducing equivalents for many biosynthetic processes, other pathways remain to be tested. The direct transcriptional targets identified here will allow us to rigorously test different signals and alternative models that may account for the nuclear translocation and activity of MondoA-Mlx heterodimers.
We have characterized a number of similarities between Myc and MondoA and have proposed that MondoA likely functions in a linear or parallel pathway with Myc (4, 5). Our finding that MondoA-Mlx heterodimers can activate transcription of the direct Myc targets LDH-A and HKII provides additional support for this model (31, 52). Similarly, PFKFB3 has not been previously shown to be a Myc target, but we observed a 12-fold upregulation in C2C12 cells overexpressing c-Myc (data not shown). Our ChIP experiments indicate that both Myc and ΔN237NLSMondoA can occupy the promoters of the LDH-A, HKII, and PFKFB3 genes in this population-based assay. In the future, it will be interesting to investigate the determinants of MondoA-Mlx or Myc-Max selectivity at these different promoters and their relative contributions to the regulation of different targets.
In both humans and mice, mondoA mRNA is most highly expressed in skeletal muscle (5), suggesting that one of its main functions is to regulate glucose utilization in this highly metabolic tissue; however, we detected MondoA and Mlx associated with mitochondria in all of the cell lines examined, suggesting a broader function in cell physiology. For example, it is well known that tumors and tumor cell lines are highly glycolytic even under aerobic growth conditions (the Warburg effect) (54). The high glycolysis rate of cancer cells, while inefficient at generating ATP, appears to supply the precursors required for the high levels of nucleotide and lipid biosynthesis needed to support accelerated rates of cell division (3, 13, 20, 26, 43). LDH-A, HKII, and PFKFB3 are often upregulated in cancer cells and tumors, and each has been suggested to contribute to the Warburg effect (2, 13, 14, 35, 45, 52). We demonstrate here that MondoA can upregulate all three of these targets and drives glycolysis. We propose that MondoA may be an important component of the Warburg effect. We are currently examining MondoA levels and localization in different tumor progression models.
Finally, by virtue of its association with the OMM, we propose that MondoA functions as an intracellular metabolic sensor; however, we cannot rule out additional functions for MondoA at the mitochondria. Several proteins have been described with both nuclear and mitochondrial functions, providing precedent for such a model. For example, in anterograde signaling responses, p53, Nur77, and histone H1.2 translocate from the nucleus to mitochondria as cell death effectors in response to apoptosis induction (34, 36, 40). As MondoA is tethered to the OMM via a protein-protein interaction, it is most likely that it regulates the activity of one of the multiprotein complexes embedded in the OMM (8, 46, 49). The development of a dominant nuclear version of MondoA and identification of effective RNA interference sequences specific for MondoA will allow us to determine the breadth of the MondoA transcriptional response and determine whether it has additional activities in regulating mitochondrial function.
Supplementary Material
Acknowledgments
We thank Brad Cairns, Elizabeth Leibold, Bob Eisenman, Steve Lessnick, and Chi Dang for reagents; Eleanor Sundwall, Amber Lewis, and Chris Peterson for technical assistance; David Virshup, Steve Lessnick, and Elizabeth Leibold for reviewing the manuscript; and members of the Ayer lab for helpful discussions.
This work was supported by NIH grant GM55668 and funds from the Huntsman Cancer Foundation. DNA sequencing and oligonucleotide synthesis were supported by Cancer Center support grant 2P30 CA42014.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Altomare, D. A., and J. R. Testa. 2005. Perturbations of the AKT signaling pathway in human cancer. Oncogene 24:7455-7464. [DOI] [PubMed] [Google Scholar]
- 2.Atsumi, T., J. Chesney, C. Metz, L. Leng, S. Donnelly, Z. Makita, R. Mitchell, and R. Bucala. 2002. High expression of inducible 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (iPFK-2; PFKFB3) in human cancers. Cancer Res. 62:5881-5887. [PubMed] [Google Scholar]
- 3.Baron, A., T. Migita, D. Tang, and M. Loda. 2004. Fatty acid synthase: a metabolic oncogene in prostate cancer? J. Cell. Biochem. 91:47-53. [DOI] [PubMed] [Google Scholar]
- 4.Billin, A. N., and D. E. Ayer. 2006. The Mlx network: evidence for a parallel Max-like transcriptional network that regulates energy metabolism, p. 255-278. In R. N. Eisenman (ed.), The Myc/Max/Mad transcription factor network, vol. 302. Springer, Heidelberg, Germany. [DOI] [PubMed] [Google Scholar]
- 5.Billin, A. N., A. L. Eilers, K. L. Coulter, J. S. Logan, and D. E. Ayer. 2000. MondoA, a novel basic helix-loop-helix-leucine zipper transcriptional activator that constitutes a positive branch of a Max-like network. Mol. Cell. Biol. 20:8845-8854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Billin, A. N., A. L. Eilers, C. Queva, and D. E. Ayer. 1999. Mlx, a novel Max-like BHLHZip protein that interacts with the Max network of transcription factors. J. Biol. Chem. 274:36344-36350. [DOI] [PubMed] [Google Scholar]
- 7.Bonifacino, J. S., M. Dasso, J. Lippincott-Schwartz, J. B. Harford, and K. M. Yamada. 2000. Isolation of mitochondria from tissues and cells by differential centrifugation. John A. Wiley & Sons, Inc., New York, N.Y.
- 8.Brdiczka, D., G. Beutner, A. Ruck, M. Dolder, and T. Wallimann. 1998. The molecular structure of mitochondrial contact sites. Their role in regulation of energy metabolism and permeability transition. Biofactors 8:235-242. [DOI] [PubMed] [Google Scholar]
- 9.Brown, M. S., and J. L. Goldstein. 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89:331-340. [DOI] [PubMed] [Google Scholar]
- 10.Butow, R. A., and N. G. Avadhani. 2004. Mitochondrial signaling: the retrograde response. Mol. Cell 14:1-15. [DOI] [PubMed] [Google Scholar]
- 11.Cairo, S., G. Merla, F. Urbinati, A. Ballabio, and A. Reymond. 2001. WBSCR14, a gene mapping to the Williams-Beuren syndrome deleted region, is a new member of the Mlx transcription factor network. Hum. Mol. Genet. 10:617-627. [DOI] [PubMed] [Google Scholar]
- 12.Chen, J. Q., M. Eshete, W. L. Alworth, and J. D. Yager. 2004. Binding of MCF-7 cell mitochondrial proteins and recombinant human estrogen receptors alpha and beta to human mitochondrial DNA estrogen response elements. J. Cell. Biochem. 93:358-373. [DOI] [PubMed] [Google Scholar]
- 13.Chesney, J., R. Mitchell, F. Benigni, M. Bacher, L. Spiegel, Y. Al-Abed, J. H. Han, C. Metz, and R. Bucala. 1999. An inducible gene product for 6-phosphofructo-2-kinase with an AU-rich instability element: role in tumor cell glycolysis and the Warburg effect. Proc. Natl. Acad. Sci. USA 96:3047-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dang, C. V., and G. L. Semenza. 1999. Oncogenic alterations of metabolism. Trends Biochem. Sci. 24:68-72. [DOI] [PubMed] [Google Scholar]
- 15.de Luis, O., M. C. Valero, and L. A. Jurado. 2000. WBSCR14, a putative transcription factor gene deleted in Williams-Beuren syndrome: complete characterisation of the human gene and the mouse ortholog. Eur. J. Hum. Genet. 8:215-222. [DOI] [PubMed] [Google Scholar]
- 16.Eilers, A. L., A. N. Billin, J. Liu, and D. E. Ayer. 1999. A 13-amino acid amphipathic alpha-helix is required for the functional interaction between the transcriptional repressor Mad1 and mSin3A. J. Biol. Chem. 274:32750-32756. [DOI] [PubMed] [Google Scholar]
- 17.Eilers, A. L., E. Sundwall, M. Lin, A. A. Sullivan, and D. E. Ayer. 2002. A novel heterodimerization domain, CRM1, and 14-3-3 control subcellular localization of the MondoA-Mlx heterocomplex. Mol. Cell. Biol. 22:8514-8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferre, D. A. A. R., G. C. Prendergast, E. B. Ziff, and S. K. Burley. 1993. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature 363:38-45. [DOI] [PubMed] [Google Scholar]
- 19.Fleischer, T. C., U. J. Yun, and D. E. Ayer. 2003. Identification and characterization of three new components of the mSin3A corepressor complex. Mol. Cell. Biol. 23:3456-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gatenby, R. A., and R. J. Gillies. 2004. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 4:891-899. [DOI] [PubMed] [Google Scholar]
- 21.Gottardo, R., J. A. Pannucci, C. R. Kuske, and T. Brettin. 2003. Statistical analysis of microarray data: a Bayesian approach. Biostatistics 4:597-620. [DOI] [PubMed] [Google Scholar]
- 22.Grandori, C., S. M. Cowley, L. P. James, and R. N. Eisenman. 2000. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16:653-699. [DOI] [PubMed] [Google Scholar]
- 23.Hahn, W. C., C. M. Counter, A. S. Lundberg, R. L. Beijersbergen, M. W. Brooks, and R. A. Weinberg. 1999. Creation of human tumour cells with defined genetic elements. Nature 400:464-468. [DOI] [PubMed] [Google Scholar]
- 24.Hardie, D. G., J. W. Scott, D. A. Pan, and E. R. Hudson. 2003. Management of cellular energy by the AMP-activated protein kinase system. FEBS Lett. 546:113-120. [DOI] [PubMed] [Google Scholar]
- 25.Hassig, C. A., T. C. Fleischer, A. N. Billin, S. L. Schreiber, and D. E. Ayer. 1997. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell 89:341-347. [DOI] [PubMed] [Google Scholar]
- 26.Hatzivassiliou, G., F. Zhao, D. E. Bauer, C. Andreadis, A. N. Shaw, D. Dhanak, S. R. Hingorani, D. A. Tuveson, and C. B. Thompson. 2005. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell 8:311-321. [DOI] [PubMed] [Google Scholar]
- 27.Hollenhorst, P. C., D. A. Jones, and B. J. Graves. 2004. Expression profiles frame the promoter specificity dilemma of the ETS family of transcription factors. Nucleic Acids Res. 32:5693-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang, X., and X. Wang. 2004. Cytochrome c-mediated apoptosis. Annu. Rev. Biochem. 73:87-106. [DOI] [PubMed] [Google Scholar]
- 29.Kawaguchi, T., K. Osatomi, H. Yamashita, T. Kabashima, and K. Uyeda. 2002. Mechanism for fatty acid “sparing” effect on glucose-induced transcription: regulation of carbohydrate-responsive element-binding protein by AMP-activated protein kinase. J. Biol. Chem. 277:3829-3835. [DOI] [PubMed] [Google Scholar]
- 30.Kawaguchi, T., M. Takenoshita, T. Kabashima, and K. Uyeda. 2001. Glucose and cAMP regulate the L-type pyruvate kinase gene by phosphorylation/dephosphorylation of the carbohydrate response element binding protein. Proc. Natl. Acad. Sci. USA 98:13710-13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, J. W., K. I. Zeller, Y. Wang, A. G. Jegga, B. J. Aronow, K. A. O'Donnell, and C. V. Dang. 2004. Evaluation of myc E-box phylogenetic footprints in glycolytic genes by chromatin immunoprecipitation assays. Mol. Cell. Biol. 24:5923-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.King, M. P., and G. Attardi. 1996. Isolation of human cell lines lacking mitochondrial DNA. Methods Enzymol. 264:304-313. [DOI] [PubMed] [Google Scholar]
- 33.Kondo-Okamoto, N., J. M. Shaw, and K. Okamoto. 2003. Mmm1p spans both the outer and inner mitochondrial membranes and contains distinct domains for targeting and foci formation. J. Biol. Chem. 278:48997-49005. [DOI] [PubMed] [Google Scholar]
- 34.Konishi, A., S. Shimizu, J. Hirota, T. Takao, Y. Fan, Y. Matsuoka, L. Zhang, Y. Yoneda, Y. Fujii, A. I. Skoultchi, and Y. Tsujimoto. 2003. Involvement of histone H1.2 in apoptosis induced by DNA double-strand breaks. Cell 114:673-688. [DOI] [PubMed] [Google Scholar]
- 35.Lee, M. G., and P. L. Pedersen. 2003. Glucose metabolism in cancer: importance of transcription factor-DNA interactions within a short segment of the proximal region of the type II hexokinase promoter. J. Biol. Chem. 278:41047-41058. [DOI] [PubMed] [Google Scholar]
- 36.Lin, B., S. K. Kolluri, F. Lin, W. Liu, Y. H. Han, X. Cao, M. I. Dawson, J. C. Reed, and X. K. Zhang. 2004. Conversion of Bcl-2 from protector to killer by interaction with nuclear orphan receptor Nur77/TR3. Cell 116:527-540. [DOI] [PubMed] [Google Scholar]
- 37.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 38.Loewen, C. J., M. L. Gaspar, S. A. Jesch, C. Delon, N. T. Ktistakis, S. A. Henry, and T. P. Levine. 2004. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science 304:1644-1647. [DOI] [PubMed] [Google Scholar]
- 39.Lois, C., E. J. Hong, S. Pease, E. J. Brown, and D. Baltimore. 2002. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295:868-872. [DOI] [PubMed] [Google Scholar]
- 40.Mihara, M., S. Erster, A. Zaika, O. Petrenko, T. Chittenden, P. Pancoska, and U. M. Moll. 2003. p53 has a direct apoptogenic role at the mitochondria. Mol. Cell 11:577-590. [DOI] [PubMed] [Google Scholar]
- 41.Morgenstern, J. P., and H. Land. 1990. A series of mammalian expression vectors and characterisation of their expression of a reporter gene in stably and transiently transfected cells. Nucleic Acids Res. 18:1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nair, S. K., and S. K. Burley. 2003. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell 112:193-205. [DOI] [PubMed] [Google Scholar]
- 43.Ookhtens, M., R. Kannan, I. Lyon, and N. Baker. 1984. Liver and adipose tissue contributions to newly formed fatty acids in an ascites tumor. Am. J. Physiol. 247:R146-R153. [DOI] [PubMed] [Google Scholar]
- 44.Osthus, R. C., H. Shim, S. Kim, Q. Li, R. Reddy, M. Mukherjee, Y. Xu, D. Wonsey, L. A. Lee, and C. V. Dang. 2000. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J. Biol. Chem. 275:21797-21800. [DOI] [PubMed] [Google Scholar]
- 45.Pastorino, J. G., and J. B. Hoek. 2003. Hexokinase II: the integration of energy metabolism and control of apoptosis. Curr. Med. Chem. 10:1535-1551. [DOI] [PubMed] [Google Scholar]
- 46.Pfanner, N., N. Wiedemann, C. Meisinger, and T. Lithgow. 2004. Assembling the mitochondrial outer membrane. Nat. Struct. Mol. Biol. 11:1044-1048. [DOI] [PubMed] [Google Scholar]
- 47.Plas, D. R., and C. B. Thompson. 2005. Akt-dependent transformation: there is more to growth than just surviving. Oncogene 24:7435-7442. [DOI] [PubMed] [Google Scholar]
- 48.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 49.Rehling, P., K. Brandner, and N. Pfanner. 2004. Mitochondrial import and the twin-pore translocase. Nat. Rev. Mol. Cell. Biol. 5:519-530. [DOI] [PubMed] [Google Scholar]
- 50.Scheller, K., P. Seibel, and C. E. Sekeris. 2003. Glucocorticoid and thyroid hormone receptors in mitochondria of animal cells. Int. Rev. Cytol. 222:1-61. [DOI] [PubMed] [Google Scholar]
- 51.Shamji, A. F., P. Nghiem, and S. L. Schreiber. 2003. Integration of growth factor and nutrient signaling: implications for cancer biology. Mol. Cell 12:271-280. [DOI] [PubMed] [Google Scholar]
- 52.Shim, H., C. Dolde, B. C. Lewis, C. S. Wu, G. Dang, R. A. Jungmann, R. Dalla-Favera, and C. V. Dang. 1997. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc. Natl. Acad. Sci. USA 94:6658-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vander Heiden, M. G., D. R. Plas, J. C. Rathmell, C. J. Fox, M. H. Harris, and C. B. Thompson. 2001. Growth factors can influence cell growth and survival through effects on glucose metabolism. Mol. Cell. Biol. 21:5899-5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Warburg, O. 1956. On the origin of cancer cells. Science 123:309-314. [DOI] [PubMed] [Google Scholar]
- 55.Yamashita, H., M. Takenoshita, M. Sakurai, R. K. Bruick, W. J. Henzel, W. Shillinglaw, D. Arnot, and K. Uyeda. 2001. A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proc. Natl. Acad. Sci. USA 98:9116-9121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang, S. H., R. Liu, E. J. Perez, Y. Wen, S. M. Stevens, Jr., T. Valencia, A. M. Brun-Zinkernagel, L. Prokai, Y. Will, J. Dykens, P. Koulen, and J. W. Simpkins. 2004. Mitochondrial localization of estrogen receptor beta. Proc. Natl. Acad. Sci. USA 101:4130-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, K., and R. J. Kaufman. 2004. Signaling the unfolded protein response from the endoplasmic reticulum. J. Biol. Chem. 279:25935-25938. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.