Abstract
Selenoproteins are central controllers of cellular redox homeostasis. Incorporation of selenocysteine (Sec) into selenoproteins employs a unique mechanism to decode the UGA stop codon. The process requires the Sec insertion sequence (SECIS) element, tRNASec, and protein factors including the SECIS binding protein 2 (SBP2). Here, we report the characterization of motifs within SBP2 that regulate its subcellular localization and function. We show that SBP2 shuttles between the nucleus and the cytoplasm via intrinsic, functional nuclear localization signal and nuclear export signal motifs and that its nuclear export is dependent on the CRM1 pathway. Oxidative stress induces nuclear accumulation of SBP2 via oxidation of cysteine residues within a redox-sensitive cysteine-rich domain. These modifications are efficiently reversed in vitro by human thioredoxin and glutaredoxin, suggesting that these antioxidant systems might regulate redox status of SBP2 in vivo. Depletion of SBP2 in cell lines using small interfering RNA results in a decrease in Sec incorporation, providing direct evidence for its requirement for selenoprotein synthesis. Furthermore, Sec incorporation is reduced substantially after treatment of cells with agents that cause oxidative stress, suggesting that nuclear sequestration of SBP2 under such conditions may represent a mechanism to regulate the expression of selenoproteins.
Organisms growing in the presence of oxygen are continuously exposed to oxidative challenge caused by reactive oxygen species (ROS) generated either endogenously through oxygen metabolism or exogenously from environmental stress. Changes in the intracellular redox state lead to protein modifications which in turn affect protein function, interactions, and subcellular localization (43). Upon oxidation, cysteine residues within proteins can be modified to different products, including reversible inter- or intramolecular disulfide bonds (S-S), glutathione-mixed disulfides (S-SG), S-nitrosothiols (S-NO), and sulfenic acids (S-OH), or to the irreversible forms of sulfinic (S-O2H), and sulfonic (S-O3H) acids. Protein disulfides and glutathione-mixed disulfides are, however, thought to be the predominant means of redox modifications within mammalian cells (25, 48). The intracellular redox homeostasis is maintained by the concerted action of antioxidant systems, including thioredoxin (Trx) and thioredoxin reductases (TR), glutaredoxins (Grx), glutathione peroxidases and glutathione reductases (GR), catalase, superoxide dismutase, and the tripeptide glutathione (l-γ-glutamyl-l-cysteinylglycine [GSH]) (26, 35). Members of the antioxidant defense systems include the selenoproteins TR and glutathione peroxidase, which use the redox potential of selenium existent in their catalytic site. In addition, selenoproteins function in thyroid hormone metabolism, transport, and distribution of selenium to remote tissues and have roles as structural proteins (27).
Selenium is cotranslationally incorporated into selenoproteins in the form of the nonstandard amino acid selenocysteine (Sec). Since Sec is encoded by the UGA codon, which most frequently serves as a termination codon, its translation entails a unique decoding process (41). One of the features that dictates Sec incorporation is the Sec insertion sequence (SECIS) element, which in eukaryotes is located in the 3′ untranslated region of selenoprotein mRNAs (5, 6). In addition, several trans-acting factors are involved in the process, including the selenocysteine-specific tRNA, tRNA[Ser]Sec (40, 41); the eukaryotic Sec-specific elongation factor, eEFsec (20, 51); and SECIS binding protein 2 (SBP2) (11). The list of protein factors involved in this mechanism is constantly growing, the most recent members being the ribosomal protein L30 (10), soluble liver antigen, and SECp43 (55).
Despite recent progress in the selenoprotein field, understanding the regulation of selenoprotein synthesis is still in the early phases of discovery. It was recently suggested that Sec incorporation may be inherently inefficient, and a rate-limiting factor has not yet been identified in vivo (45). Current evidence has been somewhat conflicting regarding which of the so-far-identified factors is limiting for selenoprotein synthesis in cell culture models (15). SBP2 is one of the oldest and best-characterized factors in this process, which was shown to bind directly to SECIS elements via its L30-type RNA binding domain. SBP2 was shown to be essential for Sec incorporation in an in vitro model (12, 13); however, its requirement in vivo has not yet been determined. SBP2 is proposed to function by recruiting the eEFsec-tRNASec complex to the ribosome (51) and seems to play a role in dictating Sec incorporation efficiency (44, 56). Interestingly, a recent study found that inherited mutations within SBP2 result in abnormal thyroid hormone metabolism (16), representing the first direct link between SBP2 and human disease.
One aspect of SBP2 regulation that has not been thoroughly addressed has been its subcellular localization in cells. A strong putative nuclear localization signal (NLS) within the SBP2 primary amino acid sequence was reported early on (12), leading to the hypothesis that SBP2 may translocate through the nuclear compartment. However, in later studies, endogenous SBP2 was detected solely in the ribosomal fraction of cells (13, 38) and overexpressed SBP2 was detected within the cytoplasm of cells (45), generating confusion about the localization of SBP2 in cells.
The nuclear or cytoplasmic localization of a protein at any one time depends on the relative rate of nuclear import and export, which may in turn be regulated by the intracellular conditions. This requires the presence of conserved signal sequences such as the NLS and the nuclear export signal (NES) within shuttling proteins, which are recognized by specific receptors and adaptors (18). Transport of proteins into the nucleus is mediated by interactions between members of the importin superfamily of transport proteins and NLSs within cargo proteins (37). The best-characterized pathway for nuclear export of proteins involves the nuclear export receptor CRM1, which binds directly to NESs and enables the translocation of proteins from the nucleus into the cytoplasm (24, 50).
In the present study, we further investigated the localization of SBP2 and report the identification of sequence elements required for nucleocytoplasmic trafficking of SBP2. SBP2 is localized predominantly at the ribosomal sites due to dominant nuclear export signals. SBP2 is depleted from ribosomes and becomes nuclear upon inhibition of CRM1 or after treatment of cells with oxidative stress-causing agents. This nuclear sequestration of SBP2 is associated with a concomitant decrease in selenoprotein synthesis. Consistent with this, we show that SBP2 is a redox-sensitive protein that forms inactivating disulfide bonds and glutathione-mixed disulfides in response to oxidative stress and that the redox status of SBP2 is regulated through the thioredoxin and the glutaredoxin systems. Small interfering RNA (siRNA)-mediated depletion of SBP2 results in decreased selenoprotein synthesis, providing direct evidence for the requirement of SBP2 for incorporation of Sec. Due to lack of efficient selenoprotein synthesis, cells depleted of SBP2 are more sensitive to oxidative stress, suggesting that SBP2 is central to protection against oxidative damage.
MATERIALS AND METHODS
Plasmid construction.
Full-length SBP2 cDNA was produced in two stages. A partial human SBP2 cDNA (AL136881) lacking the 5′ end was obtained as a kind gift of the German Cancer Research Centre (DKFZ), Berlin, Germany. This sequence was subcloned into the EcoRI/SalI sites of the enhanced green fluorescent protein (EGFP)-N1 vector (Clontech). The missing 5′ sequence was generated by PCR using Pfu polymerase from the 293T cell line, and the resulting product was subcloned into the existing EGFP-N1 plasmid using the EcoRI/BclI sites (SBP2-GFP). Deletion constructs NLS-GFP (amino acids [aa] 1 to 583, bp 1 to 1751) and NES-GFP (aa 84 to 854, bp 1752 to 2562) were generated by PCR from the full-length SBP2-GFP plasmid and subcloned into the EGFP-N1 vector using the EcoRI/SalI sites. The NLS mutant (383K→A/K384→A) was generated within SBP2-GFP by site-directed mutagenesis using the QuickChange kit (Stratagene) following the supplier's protocol. For the in vivo NES assay, double-stranded oligonucleotides encoding the amino acid sequence of candidate SBP2 NESs (described in Fig. 2E) were cloned into the pRev(1.4)-GFP plasmid (29) using the BamHI site and screened for orientation by sequencing. For in vitro studies, the carboxy-terminal SBP2 (aa 584 to 854) was cloned into pET24a (Novagen) using the XhoI and NheI restriction sites. For SBP2 siRNA, the sequence AAAATCAAGCTAGAGGTTCACA was cloned into the pSUPER vector (a gift of Reuven Agami, The Netherlands Cancer Institute, Amsterdam) and transfected into cells alongside pSuper-GFP as a control (19).
FIG. 2.
Identification of functional NLS and NES motifs within SBP2. (A) Schematic representation of SBP-GFP fusion constructs generated to contain the putative NLS or NES motifs. (B) Representative microscopy images of localization of the NLS-GFP and NES-GFP fusion proteins in COS7 cells. Cells were treated with LMB (2 ng/ml) where indicated. (C) Representative microscopy images of cells expressing SBP2-GFP with wild-type NLS (382KKKK385) or mutant (mut) NLS (382KAAK385) treated with LMB. (D) Quantitation of number of cells expressing NLS wt or mutant SBP2-GFP with predominantly nuclear fluorescence, as a mean of two independent experiments. (E and F) An in vivo export assay to test the functionality of the five putative candidate SBP2 NES sequences as depicted in panel E. ID, identification. (F) Subcellular localization of functional SBP2 NESs in the Rev1.4-GFP reporter construct within untreated or Act D- or LMB-treated COS7 cells.
Cell culture, treatments, and transfections.
Cells were maintained at 37°C in a 5% CO2 incubator and cultured in RPMI 1640 medium supplemented with 3% fetal bovine serum (GIBCO-BRL) and 7% Serum Supreme (Cambrex), 1% l-glutamine, and 100 U/ml penicillin and streptomycin. H2O2 treatments were performed in serum-free medium at a final concentration of 300 μM, unless otherwise indicated. Cells were exposed to 5, 10, or 20 J/m2 type A UV (UVA) irradiation in phosphate-buffered saline (PBS) and allowed to recover in complete medium for 2 h. Leptomycin B (LMB) was kindly supplied by M. Yoshida (Department of Biotechnology, University of Tokyo, Tokyo, Japan) and added to culture medium at a final concentration of 2 ng/ml. Actinomycin D (Act D) (ICN) was dissolved in ethanol and was used at a final concentration of 5 μg/ml. Cycloheximide (Sigma) was added to cells from a stock solution in dimethyl sulfoxide at a final concentration of 15 μg/ml. COS7 cells plated on coverslips in six-well plates 24 h before were transfected with 1 μg/well of plasmid DNA using the FuGene (Roche) reagent according to the manufacturer's instructions.
Fluorescence microscopy.
Cells transfected with GFP-tagged proteins were washed in PBS twice and fixed with 4% paraformaldehyde in PBS for 30 min at room temperature. 4′,6′-Diamidino-2-phenylindole(DAPI) was used to stain the nuclei. Coverslips were mounted with polyvinyl alcohol (Mowiol; Calbiochem) containing 0.6% DABCO (diazobicyclo-octane) and visualized on an inverted fluorescent microscope (IM 1000; Leica).
Generation of anti-SBP2 antibodies.
A highly antigenic region within the SBP2 sequence (aa 279 to 378) was subcloned into the pGEX-4T1 (Pharmacia) expression vector using the EcoRI and XhoI restriction sites. Recombinant protein was purified from Escherichia coli following standard procedures. Two Dutch belted (Oryctolagus cuniculus) rabbits were immunized with 500 μg of the purified glutathione S-transferase (GST) fusion protein suspended in the Sigma MPL+TDM+CWS (monophosphoryl-lipid A plus trehalose dicorynomycolate plus cell wall skeleton) adjuvant system, which was prepared according to the supplier's instructions. Intradermal and intramuscular injections were performed at week 1, followed by intradermal injections at weeks 3, 6, and 9. Blood was collected at weeks 0 (preimmune serum), 5, 10, and 13 (final bleeding by heart puncture), and serum was separated. Polyclonal antibodies against SBP2 were affinity purified from rabbit sera by chromatography through GST-coupled CNBr-activated Sepharose 4B columns (Pharmacia) to remove antibodies against GST, followed by chromatography of the unbound fraction on CNBr-activated Sepharose 4B columns bound with SBP2-fusion protein used for immunization. Antibodies were eluted with Tris-glycine buffer (pH 2.6) and dialyzed against PBS. The final antibody concentration obtained was 1 mg/ml.
Cell lysis, immunoprecipitation, and Western blot analysis.
Cell extracts were prepared by lysis in universal immunoprecipitation (UIP) buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, 2 mM EGTA, 25 mM β-glycerophosphate, 0.2% Triton X-100, and 0.3% NP40) supplemented with 25 mM sodium fluoride, 25 mM sodium orthovanadate, and protease inhibitor cocktail (Sigma). Total cell lysate (40 μg/lane) was resolved by 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to Immobilon-P membranes (Millipore). SBP2 was detected with affinity-purified rabbit polyclonal antibodies at a 1:2,000 dilution in 10% skim milk powder in PBS-Tween 20 (PBST). Anti-RNA polymerase II (Pol II) antibodies (Santa Cruz) were used at a 1:1,000 dilution, antiactin antibodies (Sigma; A-2066) at a 1:500 dilution, and anti-PCNA antibodies (Santa Cruz) at a 1:1,000 dilution. Proteins were detected by chemiluminescence (Perkin-Elmer). Immunoprecipitation was carried out in UIP buffer using 1 μg anti-SBP2 antibodies/mg protein and protein G-Sepharose beads (Amersham) according to the manufacturer's protocol.
Subcellular fractionation.
Subcellular fractions were prepared by a differential centrifugation method (2). Briefly, 5 × 106 293T cells grown in 100-mm2 culture dishes were scraped into phosphate-buffered saline containing 1 mM EDTA and centrifuged at 750 × g for 10 min, washed, and suspended in mitochondrial buffer (MB) (210 mM mannitol, 70 mM sucrose, 1 mM EDTA, 10 mM HEPES-NaOH, pH 7.5) complemented with protease inhibitors. Cells were disrupted by passing through a 26G needle five times followed by centrifugation at 2000 × g for 5 min. This pellet represented the nuclear fraction. The supernatant was subsequently centrifuged at 13,000 × g for 10 min to yield the mitochondrial pellet. The supernatant was further centrifuged at 100,000 × g for 30 min to yield the microsomal pellet containing endoplasmic reticulum and other organelles and centrifuged again at 300,000 × g for 16 h to yield the free cytosolic ribosomes. All pellets were resuspended in equal volumes of UIP lysis buffer containing 2 mM dithiothreitol (DTT), and equal volumes of each fraction, unless otherwise indicated, were loaded on SDS-PAGE for subsequent analysis.
In vivo NES assay.
Analysis of candidate SBP2 NESs was carried out as described previously (29). Briefly, Rev1.4-GFP fusion proteins containing SBP2-NES sequences were expressed in COS7 cells and analyzed by fluorescence microscopy in untreated cells or after 3 h of treatment with either LMB or Act D cotreated with cycloheximide.
Determination of the SBP2 redox state in vivo.
293T cells were treated with oxidants as described above. Iodoacetamide (IAM) at a final concentration of 30 mM was added to cells 20 min prior to harvest to irreversibly alkylate all free thiols in vivo. Cells were washed twice in PBS and lysed in UIP buffer. Residual IAM was removed by passing lysates through a G-25 desalting column (Amersham). Lysates were incubated with 3.5 mM DTT at room temperature for 30 min to reduce any in vivo-formed disulfide bonds and subsequently passed through a G-25 column to remove DTT. Newly reduced thiols were alkylated with biotin-conjugated IAM (BIAM) at 37°C for 30 min. SBP2 was subsequently immunoprecipitated, resolved by SDS-PAGE, and transferred to polyvinylidene difluoride membrane. Membranes were first probed with horseradish peroxidase (HRP)-conjugated streptavidin (Molecular Probes) to detect BIAM-alkylated protein, stripped, and reprobed with anti-SBP2 antibodies for total SBP2.
75Se and [35S]methionine labeling.
HeLa cells were plated onto 10-cm dishes and incubated for 24 h. Where indicated, cells were incubated in the presence of LMB (2 ng/ml) for 16 h or H2O2 as described above. Treatment medium was replaced with fresh complete medium supplemented with 50 nM sodium selenite and 10 μCi 75Se per dish. 75Se was obtained as selenious acid from the Research Reactor Facility (University of Missouri, Columbia). For 35S labeling, medium was changed to methionine-free RPMI medium containing dialyzed fetal calf serum 1 h prior to addition of 100 μCi [35S]methionine (Amersham). Labeling was conducted for 24 h in the presence or absence of LMB. In siRNA experiments, HeLa cells were transfected with siRNA SBP2 and control plasmids and incubated for 72 h to reach maximal SBP2 knockdown. At 72 h posttransfection, cells (∼70% confluent) were treated with H2O2 and labeled with 75Se as described above. Cells were harvested 24 h later and lysed in UIP buffer. Proteins were resolved by SDS-PAGE and analyzed by autoradiography. Densitometry of radioactive signals was performed using the MultiGauge v2.3 software.
Determination of free thiol content and reduction of oxidized six-His-tagged NES (NES-His6) by Trx in vitro.
The number of free thiols in the oxidized and reduced NES-His6 proteins was determined by dithionitrobenzoate (DTNB) assay as described previously (59). Reduction of oxidized NES-His6 by thioredoxin was measured as previously described (33).
Reduction of glutathionylated NES-His6 by glutaredoxin 1 in vitro.
A 78 μM concentration of NES-His6 was reduced with 10 mM DTT at 37°C for 30 min, then DTT was removed by Sephadex G-25 desalting column. The reduced protein was incubated with 100 mM oxidized GSH (GSSG) at 37°C for 2 h and passed through Sephadex G-25 column to remove free GSSG. Reduction by human Grx1 was measured as described previously (36).
Fluorescence emission spectra.
DTT-reduced Trx (0.1 μM) in nitrogen equilibrated Tris-EDTA (TE) buffer was mixed with oxidized NES-His6 (0.1 μM). A thermostatic SPEX-FluoroMax spectrofluorometer was employed to measure the fluorescence as previously described (58). The fluorescence spectra of oxidized Trx were obtained by subtracting the value of oxidized NES-His6 alone from the value of the mixture of reduced Trx with oxidized NES-His6.
Mass spectrometry of glutathionylated NES-His6.
Glutathionylated NES-His6 was prepared as described above and denatured by 8 M guanidinium chloride at 37°C. Denatured protein was diluted 10 times with 10 mM ammonium bicarbonate, pH 7.6, and cleaved by endoproteinase Glu-C (Roche) for 2 h. A Voyager De-PRO matrix-assisted laser desorption ionization (MALDI) mass spectrometer (Applied Biosystems) was used to detect the peptide mass of the complex.
Cell viability assay.
Cell viability was analyzed using the XTT [2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxyanilide] cell proliferation kit II (Roche). HeLa cells were transfected with siRNA SBP2 or siRNA GFP plasmids. Forty-eight hours after transfections, cells were detached and plated at a density of 4 × 104 cells/well in 96-microwell plates and allowed to grow in the normal growth medium for 24 h. H2O2 and sodium selenite treatments were applied to cells in 96-microwell plates in a serial dilution, and incubation was conducted for another 20 h. The XTT reagent (50 μl/well) was added, and plates were read at 475 nm in a Benchmark Plus microplate reader at 3, 6, and 20 h of XTT incubation. Data points were plotted using the Microsoft Excel program and analyzed for statistical significance using a one-way analysis of variance test.
RESULTS
SBP2 shuttles between the nucleus and the cytoplasm via the CRM1 pathway.
To initiate our studies of SBP2 subcellular localization, we generated a plasmid for expression of GFP-tagged SBP2 in mammalian cells. Localization of the SBP2-GFP fusion protein was determined by fluorescence microscopy after transient expression into various cell types, including COS7, 293T, and HeLa cells. In line with previous observations (45), SBP2-GFP was localized predominantly within the cytoplasm of COS7 cells (Fig. 1A, left panel) and other cells tested (data not shown), despite the presence of a conserved, putative NLS within its amino terminus. Detailed SBP2 amino acid sequence analysis revealed the existence of five putative NESs, reminiscent of the NES identified within the human immunodeficiency virus Rev protein (53), which prompted us to investigate the possibility that SBP2 shuttles between the nucleus and the cytoplasm. Treatment of cells with LMB, a CRM1-specific inhibitor (54), results in nuclear accumulation of proteins that are exported from the nucleus in an NES-dependent manner, via the CRM1 pathway. We first determined the effect of LMB on the subcellular distribution of overexpressed SBP2-GFP, which within 2 h of treatment showed an almost complete nuclear accumulation in COS7 cells (Fig. 1A, right panel) and other cells tested (data not shown). This suggested that SBP2-GFP may contain at least one functional NES, which facilitates its continuous shuttling between the nucleus and the cytoplasm; however, constitutive export by CRM1 confines the steady-state protein localization to the cytoplasm. GFP alone did not respond to the LMB treatment (Fig. 1A, bottom panels). Quantitation of the subcellular distribution of SBP2-GFP (Fig. 1B) showed that LMB treatment significantly increased the proportion of cells with nuclear fluorescence, consequently decreasing the number of cells with cytoplasmic fluorescence.
FIG. 1.
SBP2 shuttles between the nucleus and the cytoplasm via the CRM1 pathway. (A) COS7 cells were transiently transfected with full-length human SBP2 fused to GFP or GFP vector alone, and representative microscopy images are shown with (right panels) or without (left panels) LMB treatment (2 ng/ml). DAPI-stained DNA is shown, and arrows indicate transfected cells. (B) Graphical representation of the percentage of cells showing changes in distribution of SBP2-GFP upon LMB treatment. Fluorescent cells were scored as either nuclear (N), cytoplasmic (C), or nuclear/cytoplasmic (N/C). Values represent the mean of three independent experiments. A minimum of 200 cells were counted per experiment. (C) Western blot (WB) analysis shows specific recognition of overexpressed SBP2-GFP at ∼150 kDa (lane 2) and endogenous SBP2 at ∼120 kDa (lanes 1 and 2) by the anti-SBP2 polyclonal antibodies at a 1:2,000 dilution. (D) Subcellular fractionation of 293T cells. SBP2 was detected as described in panel C. RNA Pol II at a 1:1,000 dilution and actin at a 1:500 dilution were used as markers for the nuclear and cytosolic compartments, respectively. The nuclear and cytosolic fractions were concentrated 5 times (5x conc.), and SBP2 was quantified by comparing it to the original ribosomal (Ribos.) fraction (right panel). The amount of SBP2 as estimated by densitometry is indicated as a percentage below the fraction. (E) Western blot analysis of SBP2 distribution in nuclear and ribosomal fractions from HeLa cells treated with LMB for 20 h. PCNA and actin were used as markers for nuclear and ribosomal loading, respectively. (F) Selenoprotein synthesis and global protein synthesis in HeLa cells in response to LMB treatment were analyzed by 75Se and [35S]methionine labeling, respectively, followed by SDS-PAGE and autoradiography. Densitometry analysis results showing differences (fold) were normalized to the loading of gels, as shown by Coomassie blue (CB) staining.
We next investigated the subcellular distribution of endogenous SBP2 in cells using a polyclonal anti-SBP2 antibody raised in house. Figure 1C shows that the antibodies specifically recognize overexpressed SBP2-GFP and the endogenous 120-kDa SBP2 protein. Since the anti-SBP2 antibodies did not show specificity for use in immunofluorescence microscopy, subcellular fractionation and Western blotting were employed to determine the intracellular distribution of endogenous SBP2. Consistent with previous reports (13, 38), we observed that SBP2 is present in the ribosomal and the endoplasmic reticulum-containing fractions of 293T cells (Fig. 1D). RNA polymerase II and actin were used as markers for the nuclear and cytosolic compartments, respectively, confirming a minimal cross-contamination between the compartments. Since SBP2-GFP was found to be continuously exported from the nucleus by CRM1, we postulated that only a small proportion of endogenous SBP2 may be present within the nuclei of cells at any one time. To address this question, we concentrated the nuclear and cytosolic fractions and quantified the amount of SBP2 by comparing it to the amount in the ribosomal fraction. Interestingly, some SBP2 was indeed present within the nuclear, but not the cytosolic fraction (Fig. 1D, right panel). Using densitometry analysis, we estimate that under the current experimental conditions, in 293T cells ∼60% of the total SBP2 pool was present within the cytosolic ribosomes, ∼30% within the organelles, which may represent the endoplasmic reticulum-attached ribosomes, and ∼10% within the nuclei. These results suggest that the localization of SBP2 is dynamic, not solely confined to ribosomal sites, and that SBP2 does translocate through the nuclear compartment, possibly by using the predicted NLS.
To further analyze the shuttling properties of endogenous SBP2, nuclear extracts were prepared from untreated or LMB-treated HeLa cells. Importantly, Western analysis confirmed the above results, showing that endogenous SBP2 relocates from the ribosomes to the nucleus upon LMB treatment (Fig. 1E, top panels), however, with slower kinetics than overexpressed SBP2-GFP as the accumulation required a 16-h treatment. The nuclear translocation is not due to cell stress or apoptosis in response to unusually long treatment with LMB, since the treatment did not affect the proliferation rate or the viability of cells (data not shown). One possible explanation for this discrepancy is that there is a significant pool of overexpressed SBP2-GFP not bound to ribosomes and therefore free in the cytosol to continuously shuttle between the two compartments. In contrast, the endogenous pool of SBP2 remains stably associated with ribosomes and its nuclear localization may be tightly regulated and only triggered by certain conditions or stimuli.
We next investigated the effect of SBP2 localization on Sec incorporation in cells by 75Se labeling in the presence or absence of LMB. Interestingly, LMB treatment led to a twofold decrease in Sec incorporation, which could be potentially attributed to the depletion of SBP2 from the ribosomes and its accumulation in the nucleus (Fig. 1F, left panels). Since LMB treatment inhibits the CRM1 nuclear export pathway and other general translation factors use this pathway to shuttle in and out of the nucleus (8), we measured the effects of LMB on general protein synthesis by [35S]methionine labeling in the presence or absence of LMB. As shown in Fig. 1F, global protein synthesis was not decreased by the LMB treatment. The translation rate of most proteins remained unchanged, and in particular one band in the 50-kDa range showed an increased synthesis rate in response to the LMB treatment. Together, these data implicate the ribosomal localization of SBP2 in being important for efficient selenoprotein synthesis; however, this does not exclude the possibility that additional Sec incorporation factors may also be affected by the LMB treatment.
Characterization of motifs that regulate subcellular localization of SBP2.
To characterize the identified motifs responsible for SBP2 shuttling activity, we generated GFP-fused deletion constructs containing either the predicted NLS-382KKKK385 (aa 1 to 583), named NLS-GFP, or the predicted NESs (aa 584 to 854), named NES-GFP (Fig. 2A), and analyzed the localization of the truncated proteins within cells. Transiently expressed NLS-GFP localized almost entirely within the nucleus of COS7 cells (Fig. 2B, left panel). In contrast, the NES-GFP fragment was almost exclusively cytoplasmic (Fig. 2B, middle panel). These results indicate that the predicted NLS and NES domains may indeed be functional and regulate SBP2 trafficking between the nucleus and the cytoplasm. To ensure that cytoplasmic NES-GFP arose as a result of NES-mediated nuclear export rather than lack of NLS, we treated NES-GFP-expressing COS7 cells with LMB. Similar to full-length SBP2-GFP, the NES-GFP construct accumulated in the nucleus in response to LMB treatment (Fig. 2B, right panel), suggesting that this region is responsible for the CRM1-dependent nuclear export of SBP2 and that at least one of the putative NESs may be functional. This fragment was able to enter the nucleus despite the lack of sequences that resemble a conventional NLS; therefore, its entry could be mediated by a cryptic NLS that becomes exposed in the truncated protein. Indeed, several lysine and arginine residues which have the potential to form a cryptic NLS are present within this region as part of the RNA binding domain. Another possibility is that the protein enters the nucleus by piggybacking on an NLS-containing binding partner.
To further define the role of the amino-terminal consensus NLS (382KKKK385) in nuclear import of SBP2, site-directed mutations of K383 and K384 to alanine were simultaneously introduced in the SBP2-GFP and NLS-GFP constructs, and their effect on SBP2 localization was analyzed in transiently transfected COS7 cells. Inactivation of the NLS in the NLS-GFP construct induced a partial shift of the protein localization to the cytoplasm, the majority of cells showing equal nuclear/cytoplasmic fluorescence (data not shown). The full-length NLS mutant SBP2-GFP was also significantly less efficient in accumulating in the nucleus than wild-type (wt) SBP2-GFP in response to LMB treatment (Fig. 2C). Quantitation of NLS wt or mutant SBP2-GFP in two independent experiments (Fig. 2D) showed that after 2 h of treatment, 75% of cells expressing the wt SBP2-GFP displayed entirely nuclear fluorescence. In contrast, only 30% of NLS mutant SBP2-GFP-expressing cells displayed entirely nuclear fluorescence and the majority of cells showed equal nuclear/cytoplasmic fluorescence. These results establish that the amino-terminal 382KKKK385 sequence is indeed a functional NLS motif that facilitates nuclear translocation of SBP2. The partial nuclear accumulation of the NLS mutant SBP-GFP may occur through partial binding of the importin-α/β complex, the presence of additional NLS sequences, or, as mentioned previously, by piggybacking on an NLS-containing binding partner.
In order to test the candidate SBP2 NES sequences for export activity, we employed a previously described in vivo NES assay (29) which uses a construct containing a nuclear export-deficient HIV Rev protein fused to the GFP reporter protein (Rev1.4-GFP). This fusion protein localizes within the nuclei of cells, and active NESs are identified by their ability to promote export of the protein when inserted between the Rev1.4 and GFP moieties. Candidate SBP2 NES sequences (Fig. 2E) in the Rev1.4-GFP construct were tested for export activity in transiently transfected COS7 cells. As the Rev1.4 protein contains a strong NLS that may not be overcome by a weak NES, cells were cotreated with Act D and cycloheximide, to block nuclear import and protein synthesis, respectively, to ensure that cytoplasmic fluorescence was caused by nuclear export and not by newly synthesized protein. LMB was used in parallel to ensure the specificity of NESs for CRM1-dependent nuclear export. In line with our results from LMB treatment of both full-length and deletion constructs, we identified two SBP2 sequences with nuclear export activity (Fig. 2F) out of the five sequences tested. The sequence representing NES1, 634QMLSKEVDACVTDLLKELVRFQD657, had the ability to partially shift the Rev1.4 protein to the cytoplasm in the absence of Act D treatment, and this effect was greatly enhanced in treated cells (Fig. 2F, middle panels). The NES5 sequence, 756QDQFHKMVELTVAAR770, did so mainly after Act D treatment (Fig. 2F, bottom panels), suggesting that this may represent a weaker, albeit functional, NES. LMB treatment abrogates the cytoplasmic localization of both NESs (Fig. 2F, right panels), confirming their CRM1 specificity. Together, these results show that SBP2 contains functional NES motifs that combined with the NLS enable its trafficking between the nucleus and the cytoplasm via the CRM1 pathway.
Nuclear accumulation of SBP2 coincides with its oxidation in vivo.
To explore the regulation of SBP2 subcellular localization further, we addressed the effects of altered intracellular conditions such as oxidative stress on its localization. Treatment of 293T or HeLa cells with oxidants such as H2O2 or sodium selenite showed no significant changes in SBP2 protein levels for up to 24 h post-stress imposition (data not shown), suggesting that SBP2 is not regulated at the translational level in response to oxidative stress. We next investigated the effect of oxidative stress on overexpressed SBP2-GFP in transiently transfected COS7 or 293T cells. Interestingly, oxidative stress treatments induced the relocation of overexpressed SBP2-GFP from the cytoplasm to the nucleus in both cell types. Figure 3A shows nuclear accumulation of SBP2-GFP in COS7 cells treated with H2O2 (left panel) or UVA (middle panel) and cytoplasmic localization in untreated cells (right panel). Stress caused by osmotic shock did not affect the cytoplasmic localization of SBP2-GFP (data not shown), suggesting that oxidative stress specifically induces nuclear accumulation of SBP2-GFP. To determine if nuclear accumulation of SBP2 is reversible, cells were treated with 100 μM H2O2 for 1 h and SBP2-GFP localization was determined over a 24-h period after release from H2O2. Nuclear localization of SBP2-GFP was maximal at 4 h after treatment, and at later time points (12 and 24 h), cells with mainly cytoplasmic fluorescence were observed, suggesting that nuclear accumulation of SBP2-GFP occurred in a reversible manner (Fig. 3B). Similar effects were observed after UVA treatments at doses of 5, 10, or 20 J/m2. We postulated that nuclear accumulation of SBP2 could be triggered in at least two different ways: by phosphorylation via the stress-induced mitogen-activated protein kinase pathway or by direct oxidation of critical cysteine residues. To test the former model, we treated SBP2-GFP-transfected COS7 cells with either the stress-activated protein kinase (SAPK/p38)-specific inhibitor SB203580 (3) or with the Janus kinase (JNK) specific inhibitor SP600125 (4) prior to H2O2 treatment. These treatments did not prevent nuclear accumulation of SBP2-GFP (data not shown). Thus, phosphorylation, at least if dependent on the p38 or the JNK protein kinases, does not seem to be the cause of nuclear accumulation of SBP2-GFP in response to oxidative stress. In addition, we did not observe any SBP2 mobility changes during reducing electrophoresis following oxidative stress treatments (Fig. 3D, bottom panel), further suggesting that phosphorylation events are not the likely cause of nuclear accumulation of SBP2.
FIG. 3.
Nuclear accumulation of SBP2-GFP in response to oxidative stress correlates with its oxidation in vivo. (A) Representative microscopy images of SBP2-GFP localization in COS7 cells untreated and treated with H2O2 (1 mM) or UVA irradiation (10 J/m2). Nuclei are shown by DAPI stain (blue) (B) Graphical representation of time course analysis of SBP2-GFP subcellular distribution following H2O2 (100 μM) treatment by scoring cells for nuclear (diamond), nuclear/cytoplasmic (triangle), or cytoplasmic (square) fluorescence. (C) Determination of endogenous SBP2 redox status in vivo. 293T cells were treated with 1 mM H2O2, diamide, or sodium selenite for 2 h, and the redox status of SBP2 was analyzed using the double-alkylation method. SBP2 was immunoprecipitated from total cell lysates, and biotinylated protein (i.e., oxidized in vivo) was detected with HRP-conjugated streptavidin (top panel). The membrane was reprobed with anti-SBP2 antibodies as a control for protein loading between samples (bottom panel). WB, Western blotting. (D) 293T cells were treated with sodium selenite to induce oxidative (Ox.) stress, and SBP2 migration was analyzed by Western blotting as described in Fig. 1, under nonreducing and reducing conditions. Sodium selenite concentrations are indicated above each lane. Cross-reacting bands on the same membranes show gel loading and no migration differences.
To address the hypothesis that nuclear accumulation of SBP2 may be caused by direct cysteine oxidation, we investigated the redox state of endogenous SBP2 in untreated cells and in cells exposed to oxidative stress. The two-step alkylation assay (7) was employed using biotin-conjugated IAM (BIAM) as an alkylating agent for detecting potentially oxidized forms of SBP2. As expected, in unstressed cells SBP2 is present in a reduced state, as indicated by the absence of any BIAM-conjugated SBP2 (Fig. 3C, lane 1). Interestingly, all oxidants induced oxidation of SBP2 in vivo, as determined by the presence of BIAM-conjugated SBP2 in protein samples obtained from treated cells (Fig. 3C, lanes 2, 3, and 4). The treatments caused various levels of oxidation of SBP2; the greatest effect was observed in selenite-treated samples. This may reflect the strong oxidizing properties of selenite manifested through the depletion of both the buffering GSH pool and antioxidant enzymes (49). In vivo oxidation of SBP2 occurred at the time point at which SBP2 was first detected in the nucleus after stress treatment, providing further evidence of a redox-regulated nuclear accumulation of SBP2. Moreover, nonreducing SDS-PAGE revealed the formation of oxidation-induced mobility alterations of SBP2, which was noticeable by the appearance of an additional band in samples treated with selenite at a concentration of 100 μM or higher (Fig. 3D), indicating the formation of oxidized forms of the protein in response to oxidant treatment. A cross-reactive band detected on the same blot does not display any similar changes, suggesting that the modifications are specific to SBP2. Importantly, this shift was eliminated by reducing conditions (Fig. 3D, bottom panel), implying that the mobility change is caused by direct oxidation of the protein. The poor detection of SBP2 in samples treated with the highest concentration of selenite treatment (Fig. 3D, top panel, last lane) may be a result of masking of antibody epitope in a highly oxidized protein. Another possibility is that high concentration of selenite causes degradation of SBP2, because in a reducing gel smaller amounts of SBP2, but not cross-reactive proteins, are observed. Collectively, these results suggest that SBP2 is a redox-sensitive protein and direct redox modification occurs within SBP2 in response to oxidative stress.
The carboxy-terminal region of SBP2 mediates its nuclear retention in response to oxidative stress.
To further define the mechanism involved in redox regulation of SBP2, we initially applied an in silico method (23) to predict redox-sensitive cysteine residues within SBP2 using the web-based server at http://manaslu.aecom.yu.edu/cysredox.html. Four cysteines (C633, C644, C691, and C698) with putative redox properties were predicted using this method; they were located within the carboxy-terminal region of SBP2. Further analysis of SBP2 amino acid sequence revealed that SBP2 contains 12 cysteines in total and that 7 of these, including the ones predicted in silico, are present in a cluster within the C-terminal region of SBP2. Interestingly, this cluster overlaps with the RNA binding domain and the identified NESs and may constitute a redox-sensitive cysteine-rich domain (CRD) with potential to regulate SBP2 localization and/or function. This domain is reminiscent of the CRD identified within the Yap-1 transcription factor, which was shown to be directly involved in regulating Yap-1 transactivation activity in response to oxidative stress (39). The location of the CRD and other cysteine residues within the SBP2 amino acid sequence is schematically presented in Fig. 4A.
FIG. 4.
Carboxy-terminal region of SBP2 contains a CRD that can modulate SBP2 subcellular localization. (A) Cysteine residues and their distribution within SBP2 are schematically indicated. Cysteines within the CRD are shown in boldface. The amino acid sequence of the C-terminal SBP2 region (NES-GFP) shows the location of the RNA binding domain (italic), the NESs (underlined), and the cysteines within the CRD. (B) Representative microscopy images of COS7 cells expressing the NES-GFP, with or without H2O2 (100 μM) or IAM (1 mM) treatment. DAPI stain shows the nuclei, and arrows indicate transfected cells.
To explore the role of CRD in regulation of SBP2, its contribution to the nuclear sequestration of SBP2 in response to oxidative stress was investigated using the NES-GFP construct, which encompasses the CRD as well as the RNA binding domain and the identified NESs. As predicted, we found that oxidative stress imposed by H2O2 (Fig. 4B, middle panel), diamide, and sodium selenite (data not shown) caused nuclear accumulation of the otherwise cytoplasmic NES-GFP (Fig. 4B, left panel), suggesting that the CRD may be mediating nuclear retention of SBP2 by masking the NES through disulfide bond formation. Consistently, treatment with the irreversible thiol alkylator IAM induced nuclear retention of NES-GFP (Fig. 4B, right panel), confirming that nuclear export requires the presence of free cysteine residues. Similar results using IAM were observed for the full-length SBP2-GFP (data not shown). Together, these results suggest that cysteine residues within the CRD are directly involved in regulation of SBP2 nuclear export. The location of C644 within NES 1 identified further supports this model.
Oxidized SBP2 is reduced by the thioredoxin system in vitro.
To assess direct redox-dependent modifications of SBP2, we conducted a detailed in vitro biochemical analysis of the His-tagged, C-terminal region of SBP2 (NES-His6 aa 584 to 854). Nonreducing SDS-PAGE and thiol-specific alkylation with the fluorescent probe 5-iodoacetofluorescein (5-IAF) revealed the existence of oxidized, faster-migrating NES-His6 monomer (Fig. 5A, left panel). Likewise, an oxidized dimer migrating at double the molecular mass and oxidized protein aggregates were detected. Moreover, the absence of fluorescent signal under nonreducing conditions confirms the existence of NES-His6 in an oxidized state. Reduction with DTT or the physiological reductant GSH resulted in complete alkylation of the protein, as indicated by the strong fluorescent labeling and the electrophoretic mobility retardation (Fig. 5A, right panel). Dimerization of NES-His6 was further confirmed by electrospray ionization tandem mass spectrometry (ESI MS/MS) (data not shown), which demonstrated that in the presence of DTT, most of the protein was detected in the monomeric form (31,693 Da). However, in the absence of reductant, the dimeric form (63,385 Da) prevailed. Dimerization of SBP2 has previously been reported using recombinant proteins (1, 42); however, since then, a more recent study indicated that dimerization of SBP2 was an artifact caused by the tag used to purify the protein (38). The data presented here clearly demonstrate that the C-terminal region of SBP2 is able to form dimers in the absence of reductant and suggest that SBP2 has the potential to dimerize under conditions of oxidative stress. Whether this occurs in vivo and whether it has any implications for SBP2 Sec incorporation function would be interesting topics of future investigations.
FIG. 5.
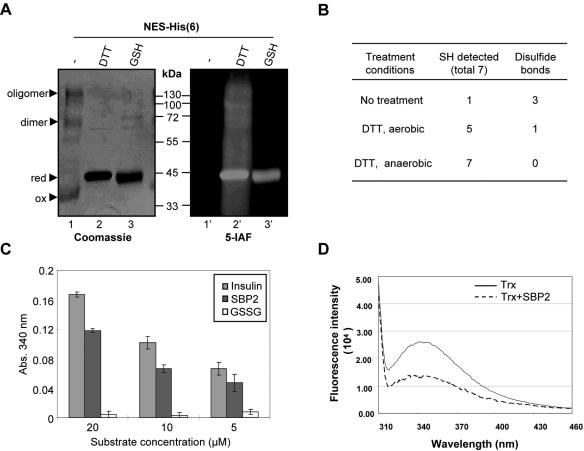
Redox-sensitive cysteine residues within carboxy-terminal SBP2 act as a substrate of the thioredoxin system. (A) Analysis of the redox state of His6-tagged C-terminal region of SBP2 (NES-His6) by alkylation with the fluorescent thiol alkylator 5-IAF during nonreducing (lanes 1 and 1′) or reducing (lanes 2 and 2′ and 3 and 3′) SDS-PAGE. The redox states of the protein as reduced (red) or oxidized (ox) monomers, dimers, and oligomers are indicated by arrows. Loading is shown by Coomassie blue staining, and 5-IAF incorporation is shown as detected with UV light (right panel). (B) Fifty microliters of NES-His6 (63 μM) pretreated as indicated was denatured in 450 μl 8 M guanidinium hydrochloride (Gn-HCl) containing DTNB. The thiol content was analyzed by measuring the absorbance at 412 nm and calculated using the extinction coefficient of thionitrobenzoate (TNB) at 412 nm. (C) Graphical representation of a comparative thioredoxin activity assayed using insulin, NES-His6, or GSSG as the substrate. Kinetics were measured in a master mix containing 50 mM Tris-HCl, pH 7.5, 2 mM EDTA, 0.1 mg/ml bovine serum albumin, 50 nM TR, 200 μM NADPH, and 5 μM Trx. (D) Changes in fluorescence emission spectra of E. coli Trx reduced (solid line) and oxidized by NES-His6 (dashed line).
Quantitative determination of thiols and disulfide bonds in SBP2 using the DTNB method (33) revealed the presence of one cysteine in its thiol form, without prior reduction. DTT reduction followed by immediate processing in aerobic solutions leads to the detection of five free thiols, and incubation in aerobic solutions for as brief as 1 to 2 min leads to a rapid reoxidation to the fully oxidized form. Detection of seven thiols was only measurable following preparation under anaerobic conditions, indicating that the NES-His6 protein is extremely sensitive to oxidation.
We next sought to determine whether the oxidation of cysteines within the CRD of SBP2 may have an in vivo relevance in terms of their reversibility. We tested whether NES-His6 could act as a substrate of the major cellular protein-disulfide reductase system comprising thioredoxin (Trx), thioredoxin reductase (TR), and NADPH (34). Since Trx and TR are both disulfide-reducing enzymes, we determined that oxidized NES-His6 is not a direct substrate of TR (data not shown). In contrast, oxidized NES-His6 is efficiently reduced by Trx, as shown in a comparative NADPH oxidation assay with insulin, an efficient Trx substrate, at various concentrations of substrates. The results of three independent kinetic measurement experiments are presented in Fig. 5C. Direct redox reactions between NES-His6 disulfides and Trx dithiols were also confirmed by apparent differences in the fluorescence spectra of reduced E. coli Trx upon incubation with oxidized NES-His6 (Fig. 5D). These results not only identify SBP2 as a novel substrate of the Trx system but also imply that SBP2 redox status in vivo may be modulated by the Trx system.
SBP2 forms glutaredoxin-reversible glutathione-mixed disulfides.
In addition to the Trx system, GSH and Grx represent another essential cellular redox-controlling system (21). This system, comprising NADPH, GSH, GR and Grx, acts in concert with the Trx system in maintaining the intracellular redox balance. Although they have overlapping functions, one distinct feature between the two systems is the higher specificity of Grx for glutathionylated protein disulfides, also referred to as GSH-mixed disulfides (48). Protein S glutathionylation is believed to be the predominant reversible oxidative modification during oxidative stress, but also acts as a regulatory mechanism during normal metabolism (25). To address whether SBP2 can be modified by glutathionylation, DTT-reduced NES-His6 was incubated with oxidized GSH (GSSG) and the products were analyzed by nonreducing SDS-PAGE and Western blotting with anti-GSH antibodies. Interestingly, approximately half the total NES-His6 protein formed GSH-mixed disulfides, evident by slower electrophoresis migration similar to that of the IAM-alkylated protein (Fig. 6A) and recognition by the anti-GSH antibodies, suggesting that glutathionylation may represent an additional means of regulation of SBP2. To address the significance of glutathionylated SBP2, we investigated its reversibility using the Grx system, which has a high specificity in reducing glutathionylated protein substrates. Enzyme kinetic measurements showed that GSSG-oxidized NES-His6 was approximately fivefold more efficiently reduced by human Grx1 than oxidized NES-His6 (Fig. 6B). Consistent with these data, MS/MS analysis by nano-ESI-quadrupole-time of flight (TOF) mass spectrometry identified the existence of a single or double GSH-linked NES-His6 protein (data not shown). Moreover, MALDI-TOF analysis of endo-Glu-C-digested GSSG-oxidized NES-His6 identified two glutathionylated sites (Fig. 6C), at positions C691 or C698 and C803. The close proximity between C691 and C698 did not allow the determination of the exact GSH-bound cysteine residue. Collectively, these results demonstrate that SBP2 forms reversible GSH-mixed disulfides in vitro and identify SBP2 as a novel substrate of human Grx1, suggesting that glutathionylation of SBP2 may have a significant regulatory role in vivo.
FIG. 6.
Distinct SBP2 carboxy-terminal cysteines form glutathione-mixed disulfides that are reduced by human glutaredoxin 1 (hGrx1). (A) Reduced NES-His6 was incubated with GSSG or IAM. Complexes were resolved by nonreducing SDS-PAGE. Western blot (WB) analysis using anti-His antibodies (top panel) shows the different redox forms of NES-His6: oxidized (ox) or reduced (red) monomer and oxidized dimer. Reprobing with anti-GSH antibodies (bottom panel) confirmed glutathionylated protein (NES-His6-S-SG). (B) Reduction of glutathionylated NES-His6-S-SG versus oxidized NES-His6-S-S by hGrx1. Kinetics were measured in a solution containing 50 mM Tris-HCl, pH 7.5, 2 mM EDTA, 0.1 mg/ml bovine serum albumin, 6 μg/ml glutathione reductase, 200 μM NADPH, 10 μM NES-His6 or glutathionylated NES-His6, and the indicated concentration of hGrx1. (C) MALDI-TOF analysis of endoproteinase-Glu-C-digested GSSG-oxidized NES-His6. Peaks representing peptides encompassing glutathionylated cysteine residues C691 or C698 and C803, their amino acid sequences, and molecular masses are outlined in the bottom panels.
SBP2 is required for Sec incorporation in vivo and regulates selenoprotein synthesis during oxidative stress.
Several lines of evidence from in vitro and cell culture models have indicated SBP2 as the central regulator of selenoprotein synthesis. However, conclusive evidence in vivo has so far not been provided. Recently, using siRNA technology, two novel factors, Secp43 and the soluble liver antigen proteins, were shown to be required for selenoprotein biosynthesis (55) in cell lines, and the same technology was used in the present study to characterize the cellular phenotype of SBP2-depleted cells.
First, we undertook a comparative analysis of cellular survival following H2O2 and selenite exposure in SBP2-depleted (transfected with SBP2 siRNA) and control cells (transfected with GFP siRNA). A number of target siRNA sequences within SBP2 were tried, and only a partial knockdown of SBP2 (∼50%) was achieved in cells (Fig. 7A), suggesting either negative selection against SBP2 depletion or SBP2 mRNA is protected against the siRNA effect. Partial depletion of SBP2 in HeLa cells had no effect on cellular viability when propagated under normal conditions. On the other hand, H2O2 and sodium selenite stimulated a dose-dependent decrease in cell survival and SBP2-depleted cells exhibited significantly greater sensitivity to H2O2 and selenite exposure than control cells, as evidenced by reduced survival at most dose points (Fig. 7A). In order to investigate the possible contribution of selenoprotein synthesis to enhanced sensitivity of SBP2-depleted cells to oxidative stress, we compared the effect of H2O2 on Sec incorporation in SBP2-depleted and control cells by 75Se labeling. We found a modest decrease in Sec incorporation in untreated, SBP2-depleted HeLa cells (Fig. 7B, lanes 1 and 3) with an ∼1.5-fold decrease in the low-molecular-weight selenoproteins and an ∼1.3-fold decrease in TR, correlating with the partial effect of siRNA on SBP2 levels. To address the effects of oxidative stress, we initially monitored Sec incorporation in H2O2-treated HeLa cells by 75Se labeling, and, unexpectedly, we observed a general decrease in selenoprotein synthesis during a time course experiment in stressed cells at several time points tested (data not shown), suggesting a compromised translation process. Importantly, H2O2 treatment combined with siRNA-mediated depletion of SBP2 showed a significant decrease in Sec incorporation (Fig. 7B, lanes 2 and 4), with an ∼3.5-fold decrease in 75Se labeling of the low-molecular-weight selenoproteins and an ∼1.8-fold decrease in TR (55 kDa) labeling. These results provide for the first time direct evidence for the requirement of SBP2 for Sec incorporation in vivo. The increased sensitivity toward oxidative injury in SBP2-depleted cells may therefore arise as a direct consequence of the decreased selenoprotein synthesis capacity in these cells, suggesting a vital role of SBP2 in the protection against oxidative stress.
FIG. 7.
SBP2 is required for incorporation of Sec into selenoproteins in vivo. (A and B) HeLa cells were transfected with SBP2 siRNA (siSBP2) or GFP siRNA (siGFP) as a control and incubated for 72 h. (A) Cell viability assay of SBP2-depleted or control cells in response to oxidative stress caused by H2O2 or sodium selenite. Western blot analysis of SBP2 protein levels was performed as described in the legend to Fig. 1 (bottom panels). Actin was used as a loading control. (B) Autoradiography of SBP2-depleted or control cells labeled with 75Se with or without prior treatment with H2O2. Coomassie staining shows the gel loading. Shown are the densitometry signal intensity for TR at 55 kDa (top panel and graph) and the low-molecular-weight (MW) selenoproteins (middle panel and graph).
DISCUSSION
In spite of the progress that has been made in delineating the mechanism of Sec insertion in mammalian systems, a number of central questions remain to be answered: in particular, the molecular events that regulate this process in its entirety and the individual roles played by its components. In this study, we have characterized the regulation of SBP2 subcellular localization and function and showed for the first time that SBP2 is required for Sec incorporation in vivo. The findings presented included identification of functional NLS and CRM1-dependent NES motifs facilitating a dynamic subcellular localization of SBP2, as well as redox regulation of SBP2. Several models for Sec incorporation have been put forward in the literature. The most recent and well-validated model proposed that SBP2 is stably bound to the ribosomes, except during the Sec delivery to the ribosomal A site, when its release facilitates the delivery of the eEFSec-tRNASec complex and incorporation of Sec (9, 38). Our data agree with this model and in addition support earlier proposed models by demonstrating that SBP2 subcellular localization is not solely confined to ribosomal sites; in fact, SBP2 shuttles between the nucleus and the cytoplasm in a motif-regulated manner. We propose that SBP2 translocates through the nuclear compartment either immediately after its synthesis or during its release from the ribosome during the process of Sec incorporation. Within the nucleus, the dominant NES is exposed, which in turn triggers its rapid export to the cytoplasm dependent on the presence of functional CRM1 (Fig. 8A). A question that remains unanswered, however, is the role of nuclear SBP2. It was previously suggested that SBP2 may enter the nucleus to associate with selenoprotein-encoding mRNAs and facilitate their transport to the cytoplasm for translation. mRNAs are generally exported from the nucleus as ribonucleoprotein complexes, including RNA binding proteins and transport factors (57). Because translation of selenoproteins requires unique factors, the transport of their cognate mRNAs from the nucleus may involve interactions with some of these factors prior to their arrival at the ribosomal sites. We showed that SBP2 contains NLS and NES motifs to perform such functions; consequently, its shuttling properties may be involved in transport of mRNAs and be directly linked to its Sec incorporation function. Interestingly, the NES motifs are located close to the RNA binding domain, raising the possibility that their availability for CRM1 binding may be related to conformational changes caused by the binding of SBP2 to SECIS elements. In that respect, inhibition of transcription using Act D for up to 24 h did not lead to nuclear retention of overexpressed SBP2-GFP (data not shown), suggesting that binding of mRNAs may not be a prerequisite for nuclear export of SBP2. Since SBP2 is also known to associate with rRNA (13, 38), we cannot exclude a possible role of this association in the nuclear export of SBP2. Alternatively, SBP2 nuclear localization may serve additional functions, possibly unrelated to Sec incorporation, which should be an interesting topic for future studies. Our data, together with previous reports (13, 38), provide strong evidence that SBP2 is predominantly associated with the ribosomes and its depletion from ribosomes significantly decreased selenoprotein synthesis, while global protein synthesis was unaffected by similar conditions, suggesting that a ribosome-associated pool of SBP2 directs the synthesis of selenoproteins.
FIG. 8.
A proposed model for the regulation of SBP2 subcellular localization and function. (A) Under normal conditions, the majority of the SBP2 pool is localized within the cytoplasm, at ribosomal sites, where it functions as part of the Sec incorporation complex. SBP2 contains functional NLS that mediates its nuclear translocation to potentially bind SECIS-containing mRNAs. This may trigger exposure of the NES, which would lead to rapid nuclear export of SBP2 via the CRM1 pathway and restore the ribosomal steady-state levels of SBP2. (B) During oxidative stress, redox-sensitive cysteine residues within SBP2 become oxidized to disulfide linkages (S-S) and/or form glutathione-mixed disulfides (S-SG), leading to a conformational change, disrupting its ribosomal localization and triggering its nuclear translocation. The lack of sufficient reducing equivalents maintains SBP2 in an oxidized state which masks the NES, inhibiting its export and leading to its nuclear retention and inhibition of selenoprotein synthesis. (C) During the cell recovery phase, Trx and Grx relocate to the nucleus and reduce SBP2, leading to exposure of the NES and promoting CRM1-dependent nuclear export. In the cytoplasm, reassembly of SBP2-containing translationally active ribosomal complexes takes place and synthesis of selenoproteins can commence.
Furthermore, we have also identified a novel link between the redox status of SBP2 and its intracellular localization. As several selenoproteins are directly involved in the antioxidant defense mechanism and detoxification of reactive oxygen species, an increase in selenoprotein expression in response to oxidative stress was expected. Paradoxically, however, a clear reduction in selenoprotein synthesis after oxidative stress was observed, indicating that translation ceased during oxidizing conditions within the cytoplasm. Analysis of data from a previous study (46) revealed no changes in mRNA transcript levels of 12 selenoprotein genes under similar stress conditions, suggesting that the transcriptional response is not affected. Therefore, it is likely that the reduced protein levels observed herein are due to inhibition at the level of translation. In accordance with these observations, a growing body of evidence suggests that global protein translation is reduced in response to most types of cellular stresses, allowing the cells to conserve resources and to initiate a reconfiguration of gene expression to effectively manage stress conditions (32, 52). These events are mainly regulated through inhibitory phosphorylation of the global initiation factor 2α (eIF-2α) (17, 22), and simultaneously, a switch to the cap-independent, internal ribosomal entry site (IRES)-mediated translation occurs to allow production of a select set of proteins required for cell survival, proliferation, or death, depending on the severity of the stress (22, 47). Transient stress conditions (such as used in this study) were shown to favor the translation of prosurvival IRES elements such as those present in the mRNA of the antiapoptotic molecule XIAP, whereas severe stress supports translation of prodeath IRES elements such as the proapoptotic molecule Apaf-1 (47). To date, no selenoprotein mRNAs have been shown to contain IRES elements within their mRNAs. It is thus possible that their decreased translation may reflect the absence of such elements. Moreover, since Sec is an extremely redox-sensitive amino acid, it seems feasible for its translation to be restrained by oxidizing conditions. It would be interesting to investigate how tRNA[Ser]Sec biosynthesis is affected by oxidative stress, as tRNA[Ser]Sec availability is believed to be a main regulatory point in selenoprotein synthesis (28). Since the requirement of SBP2 for selenoprotein synthesis in vivo was demonstrated, we propose that translational inhibition during oxidative stress is mediated through direct oxidation of SBP2 and by its sequestration from the ribosomes into the nuclear compartment. Its nuclear retention may be mediated by oxidized cysteines and masking of the NES in a manner that interferes with CRM1 transport and possibly also mRNA binding (Fig. 8B). We hence propose that SBP2 Sec incorporation function is directed by the ribosome-associated pool of SBP2, which is regulated by its redox state. Accordingly, several translation factors form inactivating disulfide linkages within cells exposed to oxidative stress (14), and in yeast, H2O2 stress causes a rapid, but reversible inhibition of protein synthesis mediated by glutathionylation of translation factors (48). Since SBP2 is able to form GSH-mixed disulfides in vitro, it is tempting to speculate that glutathionylation, by introducing a steric hindrance, may have an inhibitory effect on SBP2 Sec incorporation function. The reversibility of oxidative modifications in vitro by Trx and Grx led us to speculate that nuclear export of SBP2 and its relocation to the ribosomes during the cell recovery from oxidative stress are dependent on the reducing capacity of these two systems (Fig. 8C), which also accumulate in the nucleus in response to stress (30, 31).
In conclusion, this study provides a novel level of regulation of selenoprotein synthesis through redox regulation of SBP2 and will most certainly serve as a platform for future, more elaborate studies to understand the different aspects of SBP2 redox regulation, such as ribosomal localization and RNA binding, as well as the regulation by glutathionylation. As is apparent from the data presented herein, regulation of SBP2 localization and function is more complicated than previously thought and most likely requires multiple events as well as several pathways involving Trx and Grx, all working in concert.
Acknowledgments
We thank Beric Henderson for providing the REVm1.4-GFP plasmid and assisting in identification of candidate SBP2 NES sequences and David Jans for critical review of the manuscript.
REFERENCES
- 1.Allmang, C., P. Carbon, and A. Krol. 2002. The SBP2 and 15.5 kD/Snu13p proteins share the same RNA binding domain: identification of SBP2 amino acids important to SECIS RNA binding. RNA 8:1308-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antonsson, B., S. Montessuit, B. Sanchez, and J. C. Martinou. 2001. Bax is present as a high molecular weight oligomer/complex in the mitochondrial membrane of apoptotic cells. J. Biol. Chem. 276:11615-11623. [DOI] [PubMed] [Google Scholar]
- 3.Badger, A. M., J. N. Bradbeer, B. Votta, J. C. Lee, J. L. Adams, and D. E. Griswold. 1996. Pharmacological profile of SB 203580, a selective inhibitor of cytokine suppressive binding protein/p38 kinase, in animal models of arthritis, bone resorption, endotoxin shock and immune function. J. Pharmacol. Exp. Ther. 279:1453-1461. [PubMed] [Google Scholar]
- 4.Bennett, B. L., D. T. Sasaki, B. W. Murray, E. C. O'Leary, S. T. Sakata, W. Xu, J. C. Leisten, A. Motiwala, S. Pierce, Y. Satoh, S. S. Bhagwat, A. M. Manning, and D. W. Anderson. 2001. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc. Natl. Acad. Sci. USA 98:13681-13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berry, M. J., L. Banu, Y. Y. Chen, S. J. Mandel, J. D. Kieffer, J. W. Harney, and P. R. Larsen. 1991. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature 353:273-276. [DOI] [PubMed] [Google Scholar]
- 6.Berry, M. J., L. Banu, J. W. Harney, and P. R. Larsen. 1993. Functional characterization of the eukaryotic SECIS elements which direct selenocysteine insertion at UGA codons. EMBO J. 12:3315-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bersani, N. A., J. R. Merwin, N. I. Lopez, G. D. Pearson, and G. F. Merrill. 2002. Protein electrophoretic mobility shift assay to monitor redox state of thioredoxin in cells. Methods Enzymol. 347:317-326. [DOI] [PubMed] [Google Scholar]
- 8.Bohnsack, M. T., K. Regener, B. Schwappach, R. Saffrich, E. Paraskeva, E. Hartmann, and D. Gorlich. 2002. Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J. 21:6205-6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caban, K., and P. R. Copeland. 2006. Size matters: a view of selenocysteine incorporation from the ribosome. Cell Mol. Life Sci. 63:73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chavatte, L., B. A. Brown, and D. M. Driscoll. 2005. Ribosomal protein L30 is a component of the UGA-selenocysteine recoding machinery in eukaryotes. Nat. Struct. Mol. Biol. 12:408-416. [DOI] [PubMed] [Google Scholar]
- 11.Copeland, P. R., and D. M. Driscoll. 1999. Purification, redox sensitivity, and RNA binding properties of SECIS-binding protein 2, a protein involved in selenoprotein biosynthesis. J. Biol. Chem. 274:25447-25454. [DOI] [PubMed] [Google Scholar]
- 12.Copeland, P. R., J. E. Fletcher, B. A. Carlson, D. L. Hatfield, and D. M. Driscoll. 2000. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 19:306-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Copeland, P. R., V. A. Stepanik, and D. M. Driscoll. 2001. Insight into mammalian selenocysteine insertion: domain structure and ribosome binding properties of Sec insertion sequence binding protein 2. Mol. Cell. Biol. 21:1491-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cumming, R. C., N. L. Andon, P. A. Haynes, M. Park, W. H. Fischer, and D. Schubert. 2004. Protein disulfide bond formation in the cytoplasm during oxidative stress. J. Biol. Chem. 279:21749-21758. [DOI] [PubMed] [Google Scholar]
- 15.Driscoll, D. M., and P. R. Copeland. 2003. Mechanism and regulation of selenoprotein synthesis. Annu. Rev. Nutr. 23:17-40. [DOI] [PubMed] [Google Scholar]
- 16.Dumitrescu, A. M., X. H. Liao, M. S. Abdullah, J. Lado-Abeal, F. A. Majed, L. C. Moeller, G. Boran, L. Schomburg, R. E. Weiss, and S. Refetoff. 2005. Mutations in SECISBP2 result in abnormal thyroid hormone metabolism. Nat. Genet. 37:1247-1252. [DOI] [PubMed] [Google Scholar]
- 17.Dunand-Sauthier, I., C. A. Walker, J. Narasimhan, A. K. Pearce, R. C. Wek, and T. C. Humphrey. 2005. Stress-activated protein kinase pathway functions to support protein synthesis and translational adaptation in response to environmental stress in fission yeast. Eukaryot. Cell 4:1785-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabbro, M., and B. R. Henderson. 2003. Regulation of tumor suppressors by nuclear-cytoplasmic shuttling. Exp. Cell Res. 282:59-69. [DOI] [PubMed] [Google Scholar]
- 19.Fabbro, M., B. B. Zhou, M. Takahashi, B. Sarcevic, P. Lal, M. E. Graham, B. G. Gabrielli, P. J. Robinson, E. A. Nigg, Y. Ono, and K. K. Khanna. 2005. Cdk1/Erk2- and plk1-dependent phosphorylation of a centrosome protein, cep55, is required for its recruitment to midbody and cytokinesis. Dev. Cell 9:477-488. [DOI] [PubMed] [Google Scholar]
- 20.Fagegaltier, D., N. Hubert, K. Yamada, T. Mizutani, P. Carbon, and A. Krol. 2000. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 19:4796-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandes, A. P., and A. Holmgren. 2004. Glutaredoxins: glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxid. Redox Signal. 6:63-74. [DOI] [PubMed] [Google Scholar]
- 22.Fernandez, J., I. Yaman, R. Mishra, W. C. Merrick, M. D. Snider, W. H. Lamers, and M. Hatzoglou. 2001. Internal ribosome entry site-mediated translation of a mammalian mRNA is regulated by amino acid availability. J. Biol. Chem. 276:12285-12291. [DOI] [PubMed] [Google Scholar]
- 23.Fiser, A., and I. Simon. 2000. Predicting the oxidation state of cysteines by multiple sequence alignment. Bioinformatics 16:251-256. [DOI] [PubMed] [Google Scholar]
- 24.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 25.Fratelli, M., L. O. Goodwin, U. A. Orom, S. Lombardi, R. Tonelli, M. Mengozzi, and P. Ghezzi. 2005. Gene expression profiling reveals a signaling role of glutathione in redox regulation. Proc. Natl. Acad. Sci. USA 102:13998-14003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghezzi, P. 2005. Regulation of protein function by glutathionylation. Free Radic. Res. 39:573-580. [DOI] [PubMed] [Google Scholar]
- 27.Gromer, S., J. K. Eubel, B. L. Lee, and J. Jacob. 2005. Human selenoproteins at a glance. Cell Mol. Life Sci. 62:2414-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hatfield, D. L., and V. N. Gladyshev. 2002. How selenium has altered our understanding of the genetic code. Mol. Cell. Biol. 22:3565-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henderson, B. R., and A. Eleftheriou. 2000. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp. Cell Res. 256:213-224. [DOI] [PubMed] [Google Scholar]
- 30.Hirota, K., M. Matsui, S. Iwata, A. Nishiyama, K. Mori, and J. Yodoi. 1997. AP-1 transcriptional activity is regulated by a direct association between thioredoxin and Ref-1. Proc. Natl. Acad. Sci. USA 94:3633-3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirota, K., M. Matsui, M. Murata, Y. Takashima, F. S. Cheng, T. Itoh, K. Fukuda, and J. Yodoi. 2000. Nucleoredoxin, glutaredoxin, and thioredoxin differentially regulate NF-kappaB, AP-1, and CREB activation in HEK293 cells. Biochem. Biophys. Res. Commun. 274:177-182. [DOI] [PubMed] [Google Scholar]
- 32.Holcik, M., and N. Sonenberg. 2005. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 6:318-327. [DOI] [PubMed] [Google Scholar]
- 33.Holmgren, A. 1984. Enzymatic reduction-oxidation of protein disulfides by thioredoxin. Methods Enzymol. 107:295-300. [DOI] [PubMed] [Google Scholar]
- 34.Holmgren, A. 1985. Thioredoxin. Annu. Rev. Biochem. 54:237-271. [DOI] [PubMed] [Google Scholar]
- 35.Holmgren, A., C. Johansson, C. Berndt, M. E. Lonn, C. Hudemann, and C. H. Lillig. 2005. Thiol redox control via thioredoxin and glutaredoxin systems. Biochem. Soc. Trans. 33:1375-1377. [DOI] [PubMed] [Google Scholar]
- 36.Johansson, C., C. H. Lillig, and A. Holmgren. 2004. Human mitochondrial glutaredoxin reduces S-glutathionylated proteins with high affinity accepting electrons from either glutathione or thioredoxin reductase. J. Biol. Chem. 279:7537-7543. [DOI] [PubMed] [Google Scholar]
- 37.Johnson, H. M., P. S. Subramaniam, S. Olsnes, and D. A. Jans. 2004. Trafficking and signaling pathways of nuclear localizing protein ligands and their receptors. Bioessays 26:993-1004. [DOI] [PubMed] [Google Scholar]
- 38.Kinzy, S. A., K. Caban, and P. R. Copeland. 2005. Characterization of the SECIS binding protein 2 complex required for the co-translational insertion of selenocysteine in mammals. Nucleic Acids Res. 33:5172-5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuge, S., N. Jones, and A. Nomoto. 1997. Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 16:1710-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, B. J., M. Rajagopalan, Y. S. Kim, K. H. You, K. B. Jacobson, and D. Hatfield. 1990. Selenocysteine tRNA[Ser]Sec gene is ubiquitous within the animal kingdom. Mol. Cell. Biol. 10:1940-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee, B. J., P. J. Worland, J. N. Davis, T. C. Stadtman, and D. L. Hatfield. 1989. Identification of a selenocysteyl-tRNA(Ser) in mammalian cells that recognizes the nonsense codon, UGA. J. Biol. Chem. 264:9724-9727. [PubMed] [Google Scholar]
- 42.Lescure, A., C. Allmang, K. Yamada, P. Carbon, and A. Krol. 2002. cDNA cloning, expression pattern and RNA binding analysis of human selenocysteine insertion sequence (SECIS) binding protein 2. Gene 291:279-285. [DOI] [PubMed] [Google Scholar]
- 43.Linke, K., and U. Jakob. 2003. Not every disulfide lasts forever: disulfide bond formation as a redox switch. Antioxid. Redox Signal. 5:425-434. [DOI] [PubMed] [Google Scholar]
- 44.Low, S. C., E. Grundner-Culemann, J. W. Harney, and M. J. Berry. 2000. SECIS-SBP2 interactions dictate selenocysteine incorporation efficiency and selenoprotein hierarchy. EMBO J. 19:6882-6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mehta, A., C. M. Rebsch, S. A. Kinzy, J. E. Fletcher, and P. R. Copeland. 2004. Efficiency of mammalian selenocysteine incorporation. J. Biol. Chem. 279:37852-37859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murray, J. I., M. L. Whitfield, N. D. Trinklein, R. M. Myers, P. O. Brown, and D. Botstein. 2004. Diverse and specific gene expression responses to stresses in cultured human cells. Mol. Biol. Cell 15:2361-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nevins, T. A., Z. M. Harder, R. G. Korneluk, and M. Holcik. 2003. Distinct regulation of internal ribosome entry site-mediated translation following cellular stress is mediated by apoptotic fragments of eIF4G translation initiation factor family members eIF4GI and p97/DAP5/NAT1. J. Biol. Chem. 278:3572-3579. [DOI] [PubMed] [Google Scholar]
- 48.Shelton, M. D., P. B. Chock, and J. J. Mieyal. 2005. Glutaredoxin: role in reversible protein s-glutathionylation and regulation of redox signal transduction and protein translocation. Antioxid. Redox Signal. 7:348-366. [DOI] [PubMed] [Google Scholar]
- 49.Shen, H. M., C. F. Yang, and C. N. Ong. 1999. Sodium selenite-induced oxidative stress and apoptosis in human hepatoma HepG2 cells. Int. J. Cancer 81:820-828. [DOI] [PubMed] [Google Scholar]
- 50.Stade, K., C. S. Ford, C. Guthrie, and K. Weis. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90:1041-1050. [DOI] [PubMed] [Google Scholar]
- 51.Tujebajeva, R. M., P. R. Copeland, X. M. Xu, B. A. Carlson, J. W. Harney, D. M. Driscoll, D. L. Hatfield, and M. J. Berry. 2000. Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep. 1:158-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wek, R. C., H. Y. Jiang, and T. G. Anthony. 2006. Coping with stress: eIF2 kinases and translational control. Biochem. Soc Trans. 34:7-11. [DOI] [PubMed] [Google Scholar]
- 53.Wen, W., J. L. Meinkoth, R. Y. Tsien, and S. S. Taylor. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82:463-473. [DOI] [PubMed] [Google Scholar]
- 54.Wolff, B., J. J. Sanglier, and Y. Wang. 1997. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 4:139-147. [DOI] [PubMed] [Google Scholar]
- 55.Xu, X. M., H. Mix, B. A. Carlson, P. J. Grabowski, V. N. Gladyshev, M. J. Berry, and D. L. Hatfield. 2005. Evidence for direct roles of two additional factors, SECp43 and soluble liver antigen, in the selenoprotein synthesis machinery. J. Biol. Chem. 280:41568-41575. [DOI] [PubMed] [Google Scholar]
- 56.Zavacki, A. M., J. B. Mansell, M. Chung, B. Klimovitsky, J. W. Harney, and M. J. Berry. 2003. Coupled tRNA(Sec)-dependent assembly of the selenocysteine decoding apparatus. Mol. Cell 11:773-781. [DOI] [PubMed] [Google Scholar]
- 57.Zenklusen, D., and F. Stutz. 2001. Nuclear export of mRNA. FEBS Lett. 498:150-156. [DOI] [PubMed] [Google Scholar]
- 58.Zhao, R., H. Masayasu, and A. Holmgren. 2002. Ebselen: a substrate for human thioredoxin reductase strongly stimulating its hydroperoxide reductase activity and a superfast thioredoxin oxidant. Proc. Natl. Acad. Sci. USA 99:8579-8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhong, L., E. S. Arner, and A. Holmgren. 2000. Structure and mechanism of mammalian thioredoxin reductase: the active site is a redox-active selenolthiol/selenenylsulfide formed from the conserved cysteine-selenocysteine sequence. Proc. Natl. Acad. Sci. USA 97:5854-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]