Abstract
The Notch signaling pathway is an evolutionarily conserved signaling system which has been shown to be essential in cell fate specification and in numerous aspects of embryonic development in all metazoans thus far studied. We recently demonstrated that several components of the Notch signaling pathway, including the four Notch receptors and their five ligands known in mammals, are expressed in mouse oocytes, in mouse preimplantation embryos, or both. This suggested a possible implication of the Notch pathway in the first cell fate specification of the dividing mouse embryo, which results in the formation of the blastocyst. To address this issue directly, we generated zygotes in which both the maternal and the zygotic expression of Rbpsuh, a key element of the core Notch signaling pathway, were abrogated. We find that such zygotes give rise to blastocysts which implant and develop normally. Nevertheless, after gastrulation, these embryos die around midgestation, similarly to Rbpsuh-null mutants. This demonstrates that the RBP-Jκ-dependent pathway, otherwise called the canonical Notch pathway, is dispensable for blastocyst morphogenesis and the establishment of the three germ layers, ectoderm, endoderm, and mesoderm. These results are discussed in the light of recent observations which have challenged this conclusion.
Notch signaling is an evolutionarily conserved system which controls cell fate and has been shown in all metazoans studied so far, in particular in Drosophila melanogaster, Caenorhabditis elegans, and vertebrates, to be mandatory in various developmental processes, including stem cell homeostasis, cell growth, cell differentiation, and survival (1, 18, 28). It is therefore not a surprise that mutations perturbing Notch signaling may result for humans in various pathologies including cancer (9, 12, 24).
Notch signaling takes place through cell-to-cell contact, and the core of the pathway is relatively simple. Indeed, upon binding between the Notch receptor on one cell and a ligand on an adjacent cell, the former, a single-path transmembrane protein, undergoes several proteolytic steps which result in the release of the soluble intracellular domain of Notch (NICD). NICD is then translocated to the nucleus, where it interacts with the transcriptional regulator CSL [CBF1 in humans/Su(H) in Drosophila/LAG1 in C. elegans], also known as RBP-Jκ in mammals. In the absence of a signal-receptor interaction, RBP-Jκ represses transcription through interactions with a corepressor complex containing a histone deacetylase. The binding of NICD to RBP-Jκ in the nucleus displaces the corepressor complex and allows recruitment of histone acetylase, a nuclear protein (Mastermind), and other cofactors, resulting in the transcription of various target genes (2, 20). This core pathway is further regulated at different levels involving many factors (13). Furthermore, interactions with other signaling pathways have been documented (12, 16). Thus, beyond the relative simplicity of its core, the implementation of the Notch signaling pathway is extremely refined, implicating the interplay of many different factors and allowing a very versatile and cellular context-dependent action (13).
We are interested in the first lineage specification of the mouse embryo, which results in the formation of the blastocyst made of two different cell lineages: the trophectoderm, which will give rise to part of the placenta, and the inner cell mass, which is at the origin of all the tissues of the fetus. This process implies cell-cell signaling (17, 25), in which Notch signaling could be implicated. As a first step to address this question, we set out to monitor the expression of various components of the Notch signaling core pathway. We found that the genes coding for the four receptors (Notch1, Notch2, Notch3, and Notch4) and the five ligands (Delta-like 1 [Dll1], Dll3, and Dll4 and Jagged1 and Jagged2) known in mammals (18) as well as the transcription factor RBP-Jκ and the two regulatory proteins Deltex-1 and Deltex-2 were transcribed during oogenesis and/or mouse preimplantation development (4). These observations, which have been subsequently confirmed (34), were consistent with the possibility of Notch signaling being instrumental in blastocyst formation. On the other hand, it is important to note that disruption of these genes by targeted mutagenesis leads either to no obvious phenotype (Notch3 and Notch4) or to embryonic lethality (Notch1, Notch2, Jagged1, Jagged2, Dll1, and Rbpsuh) but not until several days after implantation (35), seemingly dismissing the possibility of Notch signaling being mandatory in preimplantation development. Nevertheless, early embryonic lethality could be rescued for one of the two following reasons. Firstly, there could be functional redundancy due to the expression of a gene(s) compensating for the role of the disrupted gene. However, this hypothesis is unlikely because embryos lacking either Rbpsuh, which codes for RBP-Jκ (21), or both PS1 and PS2 presenilin genes (6) implant and develop beyond gastrulation. The second possibility is that maternal expression of a given component of the Notch pathway could compensate for the zygotic absence of gene function. To address this latter possibility, we decided to disrupt Rbpsuh in such a way that both maternal and zygotic expression would be abrogated. We chose Rbpsuh because it codes for RBP-Jκ, which has been shown to associate with the NICD of all four Notch receptors and consequently to be instrumental in the transcriptional control of target genes mediated by the four receptors (15). RBP-Jκ therefore stands as an essential component of the so-called canonical Notch signaling (21). Thus, monitoring the fate of zygotes lacking both maternal and zygotic RBP-Jκ should allow us to answer the question of the possible participation of canonical Notch signaling in the very early steps of mouse embryonic development.
MATERIALS AND METHODS
Mice and embryos.
We used two lines of transgenic mice: a zona pellucida 3-cre (ZP3-Cre) transgenic mouse line (5) and a mouse line containing an exon 6- and 7-floxed Rbpsuh allele (11). These two lines were crossed in order to obtain Rbpsuhf/Δ; ZP3-Cretg/+ females. Embryos were recovered from mating between Rbpsuhf/Δ; ZP3-Cretg/+ females and Rbpsuhf/Δ males. The age of the embryos was determined according to the detection of the vaginal plug (day 1).
Mouse genotyping.
Genotyping was done by PCR analysis of tail tips of mice or placental tissue of embryos. The wild-type, floxed, and deleted Rbpsuh alleles were detected using the following primers: F1, 5′-GTT CTT AAC CTG TTG GTC GGA AAC-3′, and R1, 5′-GCT TGA GGC TTG ATG TTC TGT ATT GC-3′, for the wild-type allele; F1 and R2, 5′-GGG CTG CTA AAG CGC ATG CT-3′, for the floxed allele; F3, 5′-CCT TGG TTT GTT GTT TGG GTT-3′, and R3, 5′-GTG GCT CTC AAC TCC CAA TCG T-3′, for the deleted allele. The Cre transgene was detected using the following primers: Cre1, 5′-GGA CAT GTT CAG GGA TCG CCA GGC G-3′, and Cre2, 5′-GCA TAA CCA GTG AAA CAG CAT TGC TG-3′.
RT-PCR on ovulated oocytes.
Females were superovulated by injection of 5 units of pregnant mare serum gonadotrophin (Calbiochem) followed by injection of 5 units of human chorionic gonadotrophin (Intervet) 48 h later. Sixteen hours after human chorionic gonadotrophin injection, oocytes were collected and the cumulus cells were removed by a hyaluronidase treatment (0.5 mg/ml). Poly(A)+ RNAs were isolated from 50 to 100 oocytes using Dynabeads (mRNA DIRECT kit; DYNAL). Poly(A)+ RNAs were reverse transcribed during 60 min at 42°C using 200 units of SuperScript II (Invitrogen). An equivalent of five oocytes was then used for nested reverse transcription-PCR (RT-PCR). Conditions for RT-PCR were 96°C for 5 min and then 30 cycles of 96°C for 1 min, 60°C for 1 min, and 72°C for 30 s, followed by 10 min at 72°C. A second round of PCR was performed under similar conditions using 1 μl of the first PCR mixture. Two nested primer pairs were used: RBP1, 5′-GGC ACT CCC AAG ATT GAT A-3′, and RBP2, 5′-GGT CCG CCA GCC AGT CCA G-3′, and RBP3, 5′-CAG ACA AGG CCG AGT ACA C-3′, and RBP4, 5′-GTT TCG GCT TCT ACA TCC C-3′. Hypoxanthine phosphoribosyltransferase primers (5′-GTT CTT TGC TGA CCT GCT GGA TTA C-3′ and 5′-GTC AAG GGC ATA TCC AAC AAC AAA C-3′) were used to check the amount and integrity of cDNAs.
RESULTS
Rbpsuh disruption in oocytes.
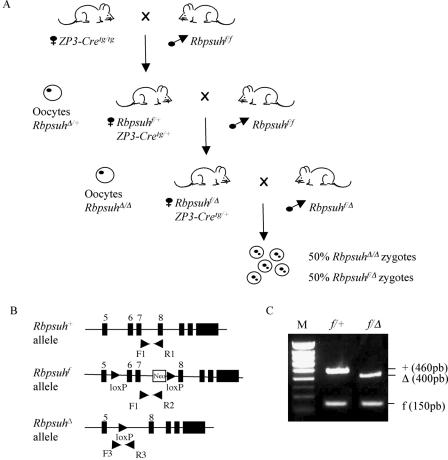
In order to deplete the Rbpsuh maternal store, we adopted a conditional mutagenesis approach. To do this, we used two different lines of transgenic mice: ZP3-Cre transgenic (ZP3-Cretg/tg) mice, which express Cre specifically during early oogenesis (5, 19), and Rbpsuhf/f mice, which carry Rbpsuh floxed alleles in which loxP sites flank exons 6 and 7 coding for the DNA binding domains of RBP-Jκ such that, upon Cre action, these exons are deleted, resulting in a null allele, RbpsuhΔ (31). Thus, as depicted in Fig. 1A, ZP3-Cretg/tg females were crossed with Rbpsuhf/f males, giving rise to Rbpsuhf/+;ZP3-Cretg/+ progeny. Females born from this cross, which produce oocytes with one Rbpsuh-disrupted allele, due to Cre expression in oogenesis (Fig. 1A), were then backcrossed to an Rbpsuhf/f male. The progeny of this cross showed a balanced distribution of Rbpsuhf/Δ and Rbpsuhf/+ animals (25 of the former and 21 of the latter, respectively) (Fig. 1B and 1C), therefore demonstrating consistent expression of Cre in oocytes, with very efficient deletion of the Rbpsuhf allele.
FIG. 1.
Disruption of Rbpsuh in the oocyte. (A) Successive crosses to obtain zygotes deprived of maternal and zygotic Rbpsuh products. (B) Schematic representation of the wild type (+), floxed (f), and deleted (Δ) Rbpsuh alleles. The PCR primers used for genotyping mice are indicated by arrowheads. Black boxes represent exons. (C) PCR of genomic DNA of Rbpsuhf/Δ and Rbpsuhf/+ mice. +, wild-type Rbpsuh allele; f, floxed Rbpsuh allele; Δ, deleted Rbpsuh allele; M, molecular weight markers (Smartladder, small fragment; Eurogentec).
Rbpsuhf/Δ; ZP3-Cretg/+ females produce oocytes deprived of maternal Rbpsuh functional transcripts.
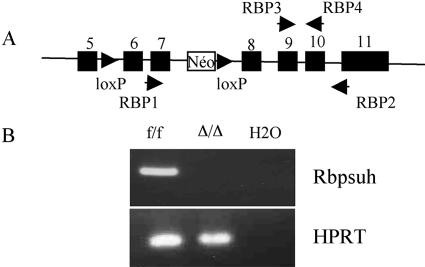
Rbpsuhf/Δ; ZP3-Cretg/+ females obtained in the latter cross were recovered. To demonstrate that these females produce oocytes deprived of a functional Rbpsuh allele, the deletion of coding exons 6 and 7 of Rbpsuh was monitored in ovulated oocytes, using nested RT-PCR. The first step was designed to amplify the 3′ region of the mRNA encompassing exons 7 to 11. RT-PCR should efficiently amplify transcripts produced by wild-type Rbpsuh (Rbpsuh+) and Rbpsuhf alleles but not the nonfunctional transcript produced by the RbpsuhΔ allele, as it lacks exons 6 and 7. As can be seen in Fig. 2, the presence of Rbpsuh transcripts was readily detected in oocytes from Rbpsuhf/f; ZP3-Cre+/+ females, by the presence of a band of the predicted size (181 bp). In contrast, the same primers did not reveal any transcript allowing the amplification of a 181-bp fragment in oocytes from Rbpsuhf/Δ;ZP3-Cretg/+ females. This demonstrated the absence of transcripts containing the region coding for the DNA binding region and therefore the absence of a functional Rbpsuh maternal contribution.
FIG. 2.
Deletion of floxed Rbpsuh allele in oocytes produced by Rbpsuhf/Δ; ZP3-Cretg/+ females. (A) Primers used to detect Rbpsuh transcripts by nested RT-PCR. (B) cDNA equivalent to that in five eggs was prepared from Rbpsuhf/f (f/f) or from RbpsuhΔ/Δ (Δ/Δ) oocytes obtained from Rbpsuhf/f and RbpsuhfΔ; ZP3-Cretg/+ females, respectively. It was subjected to a nested RT-PCR using the two pairs of primers (RBP1-RBP2 and RBP3-RBP4 indicated in panel A). The experiment was performed twice, using oocytes from different females in each case. As a control for cDNA synthesis and amount, RT-PCR was performed using hypoxanthine phosphoribosyltransferase (HPRT) primers.
RbpsuhΔ/Δ zygotes deprived of maternal transcripts implant and develop beyond gastrulation.
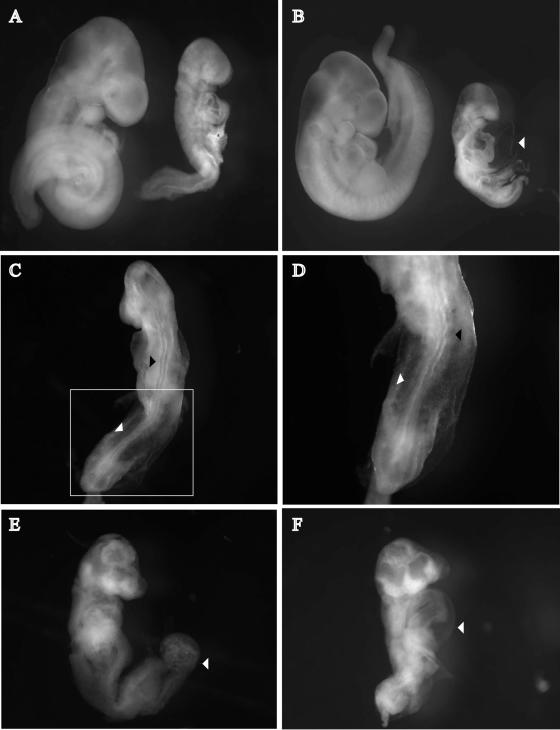
Having obtained females producing oocytes lacking maternal Rbpsuh contribution, we then asked whether zygotes lacking both maternal and zygotic Rbpsuh contribution could survive and develop normally beyond implantation and gastrulation despite the complete absence of RBP-Jκ and therefore the disruption of the RBP-Jκ-dependent canonical Notch signaling. To do this, Rbpsuhf/Δ;ZP3-Cretg/+ females were crossed with RbpsuhΔ/+ males and embryos were recovered at different times during development. An examination of embryos at 4 days postcoitum (dpc) revealed that the blastocysts appeared morphologically normal (data not shown). We then examined embryos at 9 dpc to 11 dpc. All together, 85 embryos were collected, carefully examined, and genotyped. Among them, 45 were homozygous (RbpsuhΔ/Δ) mutant and 40 were heterozygous (RbpsuhΔ/+), further confirming the highly efficient activity of the Cre recombinase in oocytes. At 9 dpc, none of eight mutant embryos examined exhibited obvious morphological anomalies. From 10 dpc, RbpsuhΔ/Δ embryos showed severe growth retardation, and they exhibited a reduction in size compared to RbpsuhΔ/+ embryos with associated defects (Fig. 3A and B). These defects became more pronounced 1 day later. Somites were poorly formed in RbpsuhΔ/Δ embryos, and they were disorganized (Fig. 3C and D). The neural tube of RbpsuhΔ/Δ embryos appeared twisted in shape (Fig. 3C and D), and the anterior neuropore was not closed at 10 dpc whereas closure was evident between 9 and 10 dpc in wild-type embryos (data not shown). Mutant embryos had also a distended pericardium (Fig. 3B and F) and a compact unfused allantois (Fig. 3E). Taken together, these results show that RbpsuhΔ/+ and RbpsuhΔ/Δ zygotes are produced at Mendelian ratios and that the latter, which lack both maternal and zygotic RBP-Jκ, develop normally through the blastocyst stage, implantation, and gastrulation. Later in development, starting at 9.5 dpc, RbpsuhΔ/Δ embryos exhibit gross developmental anomalies, as originally described by Oka et al. for homozygous-null Rbpsuh embryos (21).
FIG. 3.
RbpsuhΔ/Δ zygotes lacking both maternal and zygotic Rbpsuh products develop until beyond gastrulation, when they exhibit gross developmental abnormalities. (A and B) RbpsuhΔ/Δ (right) and Rbpsuhf/Δ (left) embryos were isolated from crosses between Rbpsuhf/Δ; ZP3-Cretg/+ females and Rbpsuhf/Δ males at 10 dpc (A) and 11 dpc (B). (C) Defective somitogenesis (white arrowhead) and neurogenesis (black arrowhead) in mutant embryos at 10 dpc. (D) Enlarged view of the boxed part of the embryo in panel C showing abnormal somites (white arrowhead) and neural tube (black arrowhead). (E) Unfused allantois in RbpsuhΔ/Δ embryos. (B and F) Distended pericardium in two different RbpsuhΔ/Δ embryos (white arrowheads).
DISCUSSION
In this study, we addressed the possibility of a role for Notch signaling in early development in the mouse. This was based on our recent observation that several essential actors of the Notch pathway are expressed in oocytes and/or preimplantation embryos from the fertilized oocyte to the blastocyst stage, where the first cell fate specifications of the mouse embryo have taken place (4). To address this question, we generated zygotes lacking both maternal and zygotic expression of Rbpsuh, a gene which has been shown to be an essential component of the core of Notch signaling (21) but also a bottleneck in the Notch cascade, i.e., being implicated in the signaling through all four Notch receptors (15). We showed not only that such zygotes give rise to normal blastocysts but that the latter implant and develop through gastrulation and finally die around day 10 to 11 dpc, exhibiting the same phenotypic anomalies as reported earlier for embryos homozygous for a null mutation in the Rbpsuh gene (21). This strongly suggests that the canonical RBP-Jκ-dependent Notch signaling system is dispensable for the early development of the mouse embryo, including gastrulation and therefore the formation of the three embryonic germ layers. In light of the described expression of the Notch signaling pathway (4, 34), this finding is quite unexpected. It should be noted that a similar conclusion was reached in a very recent study by Shi et al. (29). These authors used a strategy similar to the one used in the present study but targeted Pofut1, a gene which encodes the protein O-fucosyltransferase 1 (O-FucT-1) and has been shown, both in Drosophila (22, 27) and in mouse (30), to act upstream in the canonical Notch signaling pathway and to be an essential component of this pathway. The authors generated embryos lacking both maternal and zygotic Pofut1 gene product and reported that these embryos develop beyond the blastocyst stage until after gastrulation (29). Thus, our study provides further evidence and substantiates the unexpected notion of the dispensability of the canonical RBP-Jκ-dependent Notch pathway for mouse early embryonic development.
Recent studies of Drosophila have given further insight into the molecular mechanisms underlying the function of O-fucosyltransferase 1 in Notch signaling. Firstly, it was shown to be required for the activity of the full-length Notch receptor and essential for the physical interaction of Notch with its ligands Delta and Serrate (23, 27). Secondly, it was demonstrated that, besides its well-established activity of fucosylating serine and threonine in EGF repeats of the Notch extracellular domain (reviewed in reference 10), O-fucosyltransferase 1 has a chaperone activity required for Notch egression from the endoplasmic reticulum and subsequent transport to the membrane. Thus, it would be essential for the proper localization of Notch at the cell surface. To date, these results have not been extended to mammals. However, should it be the case, one would assume that in embryos lacking both maternal and zygotic O-FucT-1, like those generated by Shi et al. (29), both O-fucosylation and chaperone activity of O-FucT-1 could not take place, resulting in defective ligand-receptor binding and therefore the absence of NICD production. Thus, in such embryos, both RBP-Jκ-dependent and RBP-Jκ-independent modes of Notch signaling would be precluded, reinforcing the notion that Notch signaling is dispensable for mammalian early embryonic development.
Two sets of observations based on the generation of knockout mice might, however, challenge this contention. In fact, disruption of either Brainiac1 or mouse Notchless (mNle) genes results in a dramatic early phenotype: the embryos homozygous for a null mutation die either before implantation as regards to Brainiac1 (32) or shortly after with a degeneration of the inner cell mass in the case of mNle (3). Importantly, both Brainiac1 and Notchless have been shown in Drosophila to interact genetically with members of the Notch signaling pathway (7, 8, 26). Brainiac was initially characterized as a neurogenic gene, with mutant flies exhibiting neural hyperplasia and epidermal hypoplasia (7), and recently shown to code for a glycosyltranferase, involved in the glycosphingolipid biosynthetic pathway (33). In addition, it has been implicated in the proper differentiation of the epithelial cells surrounding the oocyte (7). Both phenotypes in fly as well as the results of a very recent study of the C. elegans Brainiac gene (bre-5) uncovering its functional implication upstream of the lin12 (Notch) signaling system (14) point to a role for Brainiac in Notch signaling which could be conserved in mammals. It is therefore tempting to speculate that defective preimplantation development of mouse embryos lacking Brainiac might reflect some misfunctioning of the Notch pathway. However, other explanations related to the fact that Brainiac is a glycosyltransferase cannot be excluded whereby Brainiac-dependent glycosphingolipid synthesis might be important in other ways. Other findings suggest a role for Notch in mouse preimplantation development related to the observation that embryos lacking mNle die very shortly after implantation (3). Notchless was initially discovered in Drosophila in a genetic screen for modifiers of Notch activity: loss-of-function dominantly suppressed the wing notching induced by certain Notch alleles (26). It was also shown that depending on the context, Notchless regulates either negatively or positively the Notch pathway. Furthermore, using a biochemical approach, it was demonstrated that Notchless binds to NICD (26). Similarly and importantly, we recently demonstrated that murine Nle protein has conserved its ability to modulate Notch activity (3). Therefore, the phenotypic consequences of disrupting mNle, the mouse ortholog of Notchless, namely, death of embryos shortly after implantation, were consistent with the notion that Notch signaling is implicated in very early development, in apparent contradiction to the delayed phenotypic consequences of Pofut1 or Rbpsuh disruption. One possible explanation to resolve this contradiction would be to assume that mNle acts to repress Notch signaling or maintain it below a certain level and that its abrogation would result in enhanced canonical Notch signaling with detrimental consequences for the development of the embryo.
In conclusion, the phenotypic consequences of the abrogation of zygotic and maternal Rbpsuh contribution (this study) on the one hand and Pofut1 (29) on the other hand strongly suggest that Notch signaling is dispensable in preimplantation mouse development. However and intriguingly, both the expression of various components of the Notch pathway and the phenotypic analysis of embryos mutant for either Brainiac (32) or mNle (3), two genes recognized as modulators of the Notch pathway, are consistent with an opposite conclusion. Presently, it remains unclear how to reconcile these different studies, until the precise molecular mechanisms underlying the phenotypes observed are elucidated.
Acknowledgments
We thank T. Honjo (Kyoto, Japan) for the RBP-Jκf/f mice and B. Knowles (Bar Harbor, Maine) and D. Solter (Freiburg, Germany) for the ZP3-Cretg/tg transgenic line. We thank S. Tajbakhsh for critical reading of the manuscript.
This work was supported by the Centre National de la Recherche Scientifique (CNRS), the Association pour la Recherche contre le Cancer (ARC), the Institut Pasteur GPH07 on stem cells, and the Agence Nationale pour la Recherche (contract number 05-JC05-41835). S.C. received grants from the Pasteur-Negri-Weizmann Council and the ARC. C.S. received grants from CNRS (Bourse de Doctorat pour les Ingénieurs).
REFERENCES
- 1.Artavanis-Tsakonas, S., M. D. Rand, and R. J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science 284:770-776. [DOI] [PubMed] [Google Scholar]
- 2.Baron, M. 2003. An overview of the Notch signalling pathway. Semin. Cell Dev. Biol. 14:113-119. [DOI] [PubMed] [Google Scholar]
- 3.Cormier, S., S. Le Bras, C. Souilhol, S. Vandormael-Pournin, B. Durand, C. Babinet, P. Baldacci, and M. Cohen-Tannoudji. 2006. The murine ortholog of Notchless, a direct regulator of the Notch pathway in Drosophila melanogaster, is essential for survival of inner cell mass cells. Mol. Cell. Biol. 26:3541-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cormier, S., S. Vandormael-Pournin, C. Babinet, and M. Cohen-Tannoudji. 2004. Developmental expression of the Notch signaling pathway genes during mouse preimplantation development. Gene Expr. Patterns 4:713-717. [DOI] [PubMed] [Google Scholar]
- 5.de Vries, W. N., L. T. Binns, K. S. Fancher, J. Dean, R. Moore, R. Kemler, and B. B. Knowles. 2000. Expression of Cre recombinase in mouse oocytes: a means to study maternal effect genes. Genesis 26:110-112. [PubMed] [Google Scholar]
- 6.Donoviel, D. B., A. K. Hadjantonakis, M. Ikeda, H. Zheng, P. S. Hyslop, and A. Bernstein. 1999. Mice lacking both presenilin genes exhibit early embryonic patterning defects. Genes Dev. 13:2801-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goode, S., M. Melnick, T. B. Chou, and N. Perrimon. 1996. The neurogenic genes egghead and brainiac define a novel signaling pathway essential for epithelial morphogenesis during Drosophila oogenesis. Development 122:3863-3879. [DOI] [PubMed] [Google Scholar]
- 8.Goode, S., D. Wright, and A. P. Mahowald. 1992. The neurogenic locus brainiac cooperates with the Drosophila EGF receptor to establish the ovarian follicle and to determine its dorsal-ventral polarity. Development 116:177-192. [DOI] [PubMed] [Google Scholar]
- 9.Gridley, T. 2003. Notch signaling and inherited disease syndromes. Hum. Mol. Genet. 12(special no. 1):R9-R13. [DOI] [PubMed] [Google Scholar]
- 10.Haines, N., and K. D. Irvine. 2003. Glycosylation regulates Notch signalling. Nat. Rev. Mol. Cell Biol. 4:786-797. [DOI] [PubMed] [Google Scholar]
- 11.Han, H., K. Tanigaki, N. Yamamoto, K. Kuroda, M. Yoshimoto, T. Nakahata, K. Ikuta, and T. Honjo. 2002. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int. Immunol. 14:637-645. [DOI] [PubMed] [Google Scholar]
- 12.Hansson, E. M., U. Lendahl, and G. Chapman. 2004. Notch signaling in development and disease. Semin. Cancer Biol. 14:320-328. [DOI] [PubMed] [Google Scholar]
- 13.Kadesch, T. 2004. Notch signaling: the demise of elegant simplicity. Curr. Opin. Genet. Dev. 14:506-512. [DOI] [PubMed] [Google Scholar]
- 14.Katic, I., L. G. Vallier, and I. Greenwald. 2005. New positive regulators of lin-12 activity in Caenorhabditis elegans include the BRE-5/Brainiac glycosphingolipid biosynthesis enzyme. Genetics 171:1605-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato, H., T. Sakai, K. Tamura, S. Minoguchi, Y. Shirayoshi, Y. Hamada, Y. Tsujimoto, and T. Honjo. 1996. Functional conservation of mouse Notch receptor family members. FEBS Lett. 395:221-224. [DOI] [PubMed] [Google Scholar]
- 16.Kluppel, M., and J. L. Wrana. 2005. Turning it up a Notch: cross-talk between TGF beta and Notch signaling. Bioessays 27:115-118. [DOI] [PubMed] [Google Scholar]
- 17.Kunath, T., D. Strumpf, and J. Rossant. 2004. Early trophoblast determination and stem cell maintenance in the mouse—a review. Placenta 25(Suppl. A):S32-S38. [DOI] [PubMed] [Google Scholar]
- 18.Lai, E. C. 2004. Notch signaling: control of cell communication and cell fate. Development 131:965-973. [DOI] [PubMed] [Google Scholar]
- 19.Lan, Z. J., X. Xu, and A. J. Cooney. 2004. Differential oocyte-specific expression of Cre recombinase activity in GDF-9-iCre, Zp3cre, and Msx2Cre transgenic mice. Biol. Reprod. 71:1469-1474. [DOI] [PubMed] [Google Scholar]
- 20.Mumm, J. S., and R. Kopan. 2000. Notch signaling: from the outside in. Dev. Biol. 228:151-165. [DOI] [PubMed] [Google Scholar]
- 21.Oka, C., T. Nakano, A. Wakeham, J. L. de la Pompa, C. Mori, T. Sakai, S. Okazaki, M. Kawaichi, K. Shiota, T. W. Mak, and T. Honjo. 1995. Disruption of the mouse RBP-J kappa gene results in early embryonic death. Development 121:3291-3301. [DOI] [PubMed] [Google Scholar]
- 22.Okajima, T., and K. D. Irvine. 2002. Regulation of notch signaling by o-linked fucose. Cell 111:893-904. [DOI] [PubMed] [Google Scholar]
- 23.Okajima, T., A. Xu, and K. D. Irvine. 2003. Modulation of notch-ligand binding by protein O-fucosyltransferase 1 and fringe. J. Biol. Chem. 278:42340-42345. [DOI] [PubMed] [Google Scholar]
- 24.Radtke, F., and K. Raj. 2003. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat. Rev. Cancer 3:756-767. [DOI] [PubMed] [Google Scholar]
- 25.Rossant, J. 2004. Lineage development and polar asymmetries in the peri-implantation mouse blastocyst. Semin. Cell Dev. Biol. 15:573-581. [DOI] [PubMed] [Google Scholar]
- 26.Royet, J., T. Bouwmeester, and S. M. Cohen. 1998. Notchless encodes a novel WD40-repeat-containing protein that modulates Notch signaling activity. EMBO J. 17:7351-7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sasamura, T., N. Sasaki, F. Miyashita, S. Nakao, H. O. Ishikawa, M. Ito, M. Kitagawa, K. Harigaya, E. Spana, D. Bilder, N. Perrimon, and K. Matsuno. 2003. neurotic, a novel maternal neurogenic gene, encodes an O-fucosyltransferase that is essential for Notch-Delta interactions. Development 130:4785-4795. [DOI] [PubMed] [Google Scholar]
- 28.Schweisguth, F. 2004. Notch signaling activity. Curr. Biol. 14:R129-R138. [PubMed] [Google Scholar]
- 29.Shi, S., M. Stahl, L. Lu, and P. Stanley. 2005. Canonical Notch signaling is dispensable for early cell fate specifications in mammals. Mol. Cell. Biol. 25:9503-9508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi, S., and P. Stanley. 2003. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc. Natl. Acad. Sci. USA 100:5234-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanigaki, K., H. Han, N. Yamamoto, K. Tashiro, M. Ikegawa, K. Kuroda, A. Suzuki, T. Nakano, and T. Honjo. 2002. Notch-RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat. Immunol. 3:443-450. [DOI] [PubMed] [Google Scholar]
- 32.Vollrath, B., K. J. Fitzgerald, and P. Leder. 2001. A murine homologue of the Drosophila brainiac gene shows homology to glycosyltransferases and is required for preimplantation development of the mouse. Mol. Cell. Biol. 21:5688-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wandall, H. H., S. Pizette, J. W. Pedersen, H. Eichert, S. B. Levery, U. Mandel, S. M. Cohen, and H. Clausen. 2005. Egghead and brainiac are essential for glycosphingolipid biosynthesis in vivo. J. Biol. Chem. 280:4858-4863. [DOI] [PubMed] [Google Scholar]
- 34.Wang, Q. T., K. Piotrowska, M. A. Ciemerych, L. Milenkovic, M. P. Scott, R. W. Davis, and M. Zernicka-Goetz. 2004. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev. Cell 6:133-144. [DOI] [PubMed] [Google Scholar]
- 35.Yoon, K., and N. Gaiano. 2005. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat. Neurosci. 8:709-715. [DOI] [PubMed] [Google Scholar]