Abstract
The k-turn-binding protein 15.5K is unique in that it is essential for the hierarchical assembly of three RNP complexes distinct in both composition and function, namely, the U4/U6 snRNP, the box C/D snoRNP, and the RNP complex assembled on the U3 box B/C motif. 15.5K interacts with the cognate RNAs via an induced fit mechanism, which results in the folding of the surrounding RNA to create a binding site(s) for the RNP-specific proteins. However, it is possible that 15.5K also mediates RNP formation via protein-protein interactions with the complex-specific proteins. To investigate this possibility, we created a series of 15.5K mutations in which the surface properties of the protein had been changed. We assessed their ability to support the formation of the three distinct RNP complexes and found that the formation of each RNP requires a distinct set of regions on the surface of 15.5K. This implies that protein-protein contacts are essential for RNP formation in each complex. Further supporting this idea, direct protein-protein interaction could be observed between hU3-55K and 15.5K. In conclusion, our data suggest that the formation of each RNP involves the direct recognition of specific elements in both 15.5K protein and the specific RNA.
RNA-protein complexes (RNPs) are essential for many critical processes in the cell, including transcription, translation, and RNA processing, and chromatin structure (27). Many RNPs are multiprotein complexes, and therefore, their formation is dependent on both RNA-protein and protein-protein interactions. The assembly of such multiprotein RNPs has been shown to be highly complex and commonly involves a hierarchical assembly pathway (33). This process is often initiated by a primary RNA-binding protein which nucleates the assembly of the complex by creating a platform, often via rearranging the structure of the RNA, for the binding of the remaining proteins (33). One such example of a nucleating factor is 15.5K (NHPX or Snu13p in Saccharomyces cerevisiae), which is associated with both the spliceosomal U4/U6 small nuclear RNP (snRNP) and the box C/D small nucleolar RNPs (snoRNPs) (19, 31). 15.5K belongs to a family of homologous RNA-binding proteins that includes the ribosomal proteins S12, L7a, and L30 as well as the H/ACA snoRNP protein NHP2 (13). A common feature of these proteins is that they all recognize a kink-turn (or k-turn) motif which is comprised of a conserved stem-internal loop-stem structure in which the internal loop commonly contains two G · A base pairs (12, 19, 29, 31). This family of proteins have been shown to bind the k-turn motif using an induced fit interaction with changes in the structures of both the RNA and protein components (19, 28, 34).
The U4/U6 snRNP is an essential component of the pre-mRNA splicing machinery and is comprised of the U4 and U6 snRNAs, Sm and Lsm proteins, 15.5K, hPRP31 (or 61K), and the CYPH/hPRP4/hPRP3 (or 20/60/90K) complex (32). The U4 and U6 snRNAs form a Y-shaped structure through extensive base pairing (Fig. 1). During the formation of the catalytically active spliceosome, the U4-U6 interaction is dissolved to enable U6 base pairing with the U2 snRNA and the pre-mRNA (17, 25, 32). The Sm and Lsm proteins bind the U4 Sm-binding site and the 3′ end of the U6 snRNA, respectively (1, 32). The 15.5K protein binds the k-turn motif in the 5′ stem of the U4 snRNA (Fig. 1), inducing changes in the conformations of both RNA and protein (19, 29). We have previously demonstrated that the binding of 15.5K to the U4 snRNA is essential for the subsequent recruitment of hPRP31 and the CYPH/hPRP4/hPRP3 proteins (20). The full 5′ stem-loop and an adjacent loop region in the U4 snRNA are essential for the binding of hPRP31. In contrast, the binding of the CYPH/hPRP4/hPRP3 proteins also requires stem II of the U4/U6 duplex (Fig. 1) (20).
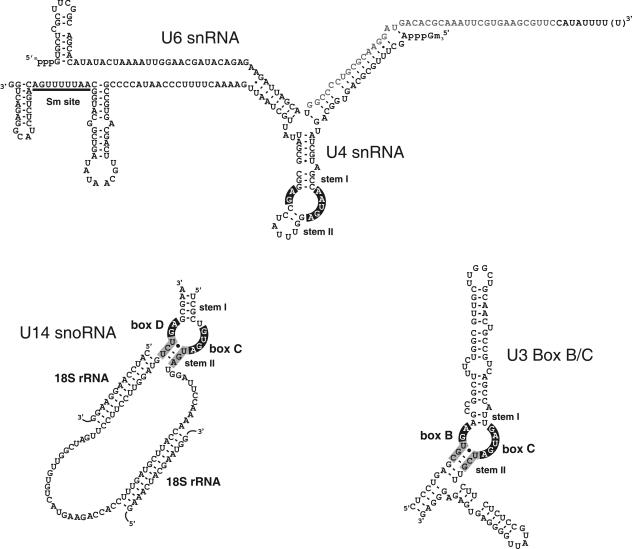
FIG. 1.
Schematic representation of the three RNA motifs bound by 15.5K. The secondary structures of the U4/U6 snRNA (reference 19 and references therein), U14 box C/D snoRNA (30, 31), and U3 snoRNA-specific box B/C motif (6, 31) are shown. The highly conserved internal loop elements recognized by 15.5K are depicted in white type on a black background. The two flanking stem structures (stems I and II) that along with the loop nucleotides are part of the k-turn structure are indicated. The conserved sequence of stem II in the box B/C and box C/D is indicated by black type on a gray background. Base pairing interactions between the U14 snoRNA and the 18S rRNA essential for modification and processing are shown. The Sm-binding site in the U4 snRNA is underlined. In addition, the minimal U6 sequence, which together with the U4 snRNA, is required for the binding of the CYPH/hPRP4/hPRP3 proteins is indicated by gray type (20).
Box C/D snoRNPs are multiprotein complexes essential for the modification and processing of the rRNA (3, 8, 9). The box C/D snoRNAs contain a conserved box C/D motif and often, but not always, a second less homologous copy of this element termed the box C′/D′ motif (8). The box C/D snoRNPs are associated with four core proteins, namely, 15.5K, NOP56, NOP58, and fibrillarin (15, 16, 23, 31, 35). The majority of these complexes direct the site-specific 2′-O methylation of the rRNA by base pairing with the target region. Methylation, which is catalyzed by fibrillarin, occurs 5 nucleotides downstream of either the D or D′ box (Fig. 1) (10, 11). The binding of 15.5K to the k-turn motif present in the box C/D motif is essential for the subsequent association of NOP56, NOP58, and fibrillarin, as well as the binding of the two assembly factors, TIP48 and TIP49 (30). The box C/D motif is a bipartite protein-binding element with internal loop nucleotides are essential for 15.5K binding, while the sequence of stem II is required for the binding of the remaining proteins (26, 30). The U3 box C/D snoRNA, which functions in processing and not modification, contains a U3-specific k-turn motif, termed the box B/C motif, which also binds 15.5K (31). The binding of 15.5K and the small internal loop adjacent to stem I in B/C motif are also essential for the recruitment of the U3-specific hU3-55K (Fig. 1) (6). The box B/C motif, and therefore likely 15.5K and hU3-55K, is essential for the formation of the 80S small-subunit processome and therefore for the processing of the 18S rRNA and the production of the small subunit of the ribosome (7).
The 15.5K protein is unique in the fact that it binds three different but related RNA motifs and in doing so nucleates the formation of three functionally distinct RNPs involved in distinct RNA processing pathways in distinct locations in the nucleus. Of the complex-specific proteins, only NOP56, NOP58, and hPRP31 share any similarity (5). Each protein contains a common central region termed the NOP domain, suggesting a common mode of interaction with the 15.5K-RNA complex. However, the lack of sequence identity seen with the remaining proteins suggests that there may be multiple distinct approaches used for the formation of each RNP. The assembly of each specific complex requires unique sequence elements found in the cognate RNA (6, 20, 30). Furthermore, changes in the RNA structure mediated by 15.5K binding are proposed to create the binding site for the complex-specific proteins. The 15.5K protein is evolutionarily highly conserved throughout the length of the polypeptide sequence. However, only 10% of the protein surface is directly involved in RNA binding (29). This suggests that 15.5K may perform roles other than just RNA binding in either the assembly and/or function of one or more of the complexes. In addition, the close proximity of the binding sites for the box C/D proteins and hPRP31 to the 15.5K-binding site makes it likely that protein-protein contacts between these components exist in the box C/D snoRNP and the U4/U6 snRNP, respectively (20). In order to test this possibility, we have used the published crystal structure of 15.5K bound to the U4 5′ stem-loop (29) to target highly conserved amino acids on the surface of the protein for mutagenesis. We have then tested the ability of these mutants to support the formation in the U4/U6 snRNP, the box C/D snoRNP, and the U3 box B/C complex.
MATERIALS AND METHODS
DNA oligonucleotides.
The DNA oligonucleotides used in this study are as follows: 1a, CAAGAAGCTACTGGACCTCGTTCGCAAATCAAAAGAAAGCGGCCAGCTTCGGAAA-GGAGCCAATG; 1b, GTTCTTCGATGACCTGGAGCAAGCGTTTAGTTTTCTTTCGCCGGTCGAAGCCTTTCCTCGGTTAC; 2a, CTGCACCTGCCGCTGCTGTGTCGCAAAATGACTGTGCCCTACGTGTTTGTG; 2b, GACGTGGACGGCGACGACACAGCGTTTTACTGACACGGGATGCACAAACAC; 3a, GTGTTTGTGCGCTCCAAGCAGGATCTGGGGGAAGCCTGTGGGGTCTCCAGG; 3b, CACAAACACGCGAGGTTCGTCCTAGACCCCCTTCGGACACCCCAGAGGTCC; 4a, GAAGGCTCGCAGCTGAAACAGAAAATCAAATCCATTCAGAGCTCCATTGAAAGGCTCTTAG; 4b, CTTCCGAGCGTCGACTTTGTCAAATAGTTTAGGTAAGTCTCGA-GGTAACTTTCCGAGAATC; 5a, TCCAATCCATTCAGCAGTCCATTAGCAGCCTCTTAGTC-TAAACCTGTGGC; and 5b, AGGTTAGGTAAGTCGTCAGGTAATCGTCGGAGAATCAGATTTGGACACCG.
Site-directed mutagenesis and expression and purification of recombinant proteins.
Mutations in pGEX-4T-2-15.5K (19) were generated using the QuikChange mutagenesis system according to the manufacturer's instructions (Stratagene). Primer pairs 1a and 1b, 2a and 2b, 3a and 3b, 4a and 4b, and 5a and 5b were used to generate mutations 1, 2, 3, 4, and 5, respectively. Recombinant wild-type and mutant 15.5K and hPRP31 were overexpressed as described previously (19, 20). Glutathione S-transferase (GST)-tagged proteins were bound to glutathione Sepharose and eluted using 50 mM Tris-HCl (pH 7.9), 100 mM NaCl, 2.5 mM MgCl2, 2 mM dithiothreitol, and 15 mM reduced glutathione. Removal of GST from the GST-15.5K fusion protein was performed on the glutathione Sepharose column. To prepare 15.5K lacking the GST domain, GST-15.K bound to glutathione Sepharose was incubated for 12 h with 10 U of thrombin (Pharmacia) per mg of protein. Thrombin was subsequently removed from the 15.5K preparation using p-aminobenzamidine agarose.
RNA-protein interactions.
Gel mobility shift assays with recombinant 15.5K were performed by the method of Watkins et al. (31). The binding of hPRP31 to the U4 snRNA was assayed by the method of Nottrott et al. (20) by incubating 30 pmol of purified recombinant GSThPRP31 with 32P-labeled in vitro-transcribed U4 snRNA in the presence or absence of 15 pmol of recombinant, purified 15.5K or 15.5K mutant. Complexes were purified using glutathione Sepharose, and U4 snRNA association was analyzed by polyacrylamide gel electrophoresis (PAGE) and autoradiography. CYPH/hPRP4/hPRP3 binding to U4/U6 snRNA was performed by the method of Nottrott et al. (20) by incubating U4/U6 duplex, containing 32P-labeled minimal U6 snRNA (Fig. 1) with 40 pmol of purified CYPH/hPRP4/hPRP3 complex in the presence or absence of 30 pmol of recombinant, purified 15.5K or 15.5K mutant. The complexes were immunoprecipitated using anti-hPRP4 antibodies, and the associated U4/U6 snRNA was analyzed by PAGE and autoradiography. In vitro box C/D snoRNP assembly using nuclear extracts was performed by the method of Watkins et al. (30). Briefly, 800 pmol of SL1 oligonucleotide (19) was added to HeLa nuclear extract to block the endogenous 15.5K. In vitro-transcribed 32P-labeled U14 snRNA, preincubated in the absence or presence of 30 pmol of recombinant 15.5K (or mutant protein), was added to the nuclear extract and incubated for 30 min at 30°C. Protein binding to the U14 snoRNA was assayed by immunoprecipitation using antibodies specific for the core box C/D snoRNP proteins and the two assembly factors TIP48 and TIP49. The copurified RNAs were then analyzed by PAGE and autoradiography. In vitro assembly of the U3 box using nuclear extracts was performed basically as described for box C/D snoRNA assembly, except that 32P-labeled U3 BC RNA was used and the complexes were precipitated using anti-hU3-55K antibodies (6). To assay hU3-55K binding to 15.5K, in vitro-translated 35S-labeled hU3-55K (prepared using TNT reticulocyte lysate system) was incubated with either GST or GST-15.5K (6). Complexes were isolated using glutathione Sepharose, and the copurifying hU3-55K was analyzed by sodium dodecyl sulfate (SDS)-PAGE and autoradiography. All data from pulldown/immunoprecipitation experiments were quantitated using either a phosphorimager or scanned autoradiographs. Where applicable, the signals were adjusted relative to either the input or supernatant for that particular sample.
RESULTS
Mutation of conserved amino acids on the surface of 15.5K not involved in RNA binding.
The high sequence conservation of 15.5K suggests that this protein may play additional roles in the function and/or assembly of one or more of the 15.5K-containing RNP complexes. Indeed, it is possible that conserved amino acids may be important for protein-protein interactions essential for the formation of the RNP complexes. In order to test this possibility, we identified conserved amino acids on the surface of 15.5K that we could mutate in order to test their function in the assembly of the various RNPs. Conserved amino acids were identified from a sequence alignment in which we have used the homologous, but not functionally related, human L7a ribosomal protein in order to indicate the amino acids that are highly conserved in this family of RNA-binding proteins (Fig. 2A, amino acids on black and gray background). The remaining conserved amino acids are therefore specific to 15.5K (Fig. 2A, amino acids on blue background). Using the crystal structure of 15.5K bound to the 5′ stem-loop of the U4 snRNA (29), we identified and mutated clusters of conserved amino acids present on the surface of the protein (Materials and Methods) (Fig. 2B and C). Importantly, mutations were not made in the conserved amino acids shown to be involved in RNA binding or in close proximity to the RNA-binding pocket (Fig. 2C) (29). In addition, by replacing the conserved residues with amino acids present in the same position of structurally similar proteins, such as the human L7a protein, it was hoped that our changes would not disrupt the overall structure of the protein. However, only 5 of the 21 mutants originally constructed were soluble after overexpression in Escherichia coli (soluble mutants indicated in Fig. 2B; insoluble mutants not shown). The insoluble mutants included both single-amino-acid changes as well as larger mutations, and there was no obvious logic regarding whether the mutants were soluble. We were disappointed with the fact that only one-third of the proteins were soluble. However, it is important to emphasize that the soluble mutants cover the main conserved regions of the protein surface not involved in RNA binding (Fig. 2C). The only conserved region not covered in this series of mutations was the N terminus. Unfortunately, proteins containing either point mutations or larger multiple-amino-acid changes in this region were completely insoluble.
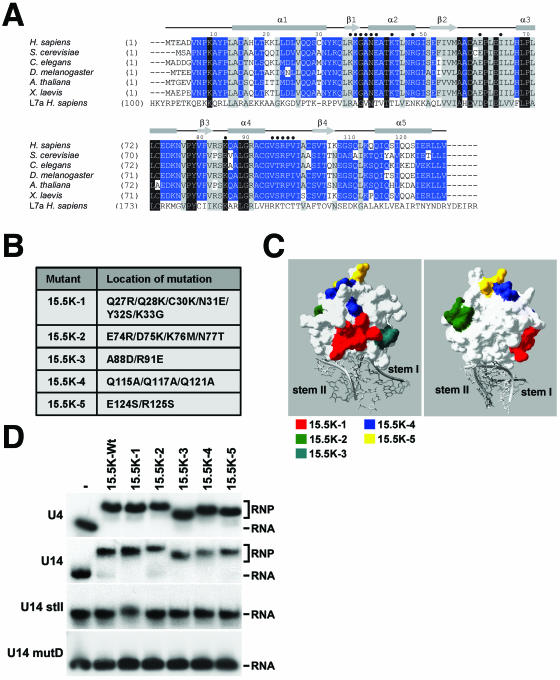
FIG. 2.
Identification and mutation of conserved amino acids on the surface of 15.5K. A) Amino acid alignment of 15.5K from human (Homo sapiens), Saccharomyces cerevisiae (Snu13p), Caenorhabditis elegans (accession no. Q21568), Drosophila melanogaster (accession no. GH03082), Arabidopsis thaliana (accession no. A71421), and Xenopus laevis (accession no. AAH46579) with the human ribosomal protein L7a sequence using the ClustalV program. Conserved residues are shown on a gray background and are grouped as described by Schulz and Schirmer (24). Identical and conserved residues are indicated by white type on black background and by black type on gray background, respectively. Residues specifically conserved in 15.5K are depicted in white type on blue background. Amino acid positions are indicated on the left. The secondary structure of human 15.5K (29) is indicated above the alignment, and amino acids involved in RNA binding are indicated by dark blue circles. B) The five mutations introduced into 15.5K. C) The positions of the residues targeted for mutagenesis on the surface of 15.5K are depicted using the crystal structure of 15.5K bound to the 5′ stem-loop of the U4 snRNA (29). The surface plot of 15.5K is shown in white, and the U4 snRNA is shaded gray. The positions of the amino acids targeted for mutagenesis are indicated in either red (mutant 15.5K-1), green (15.5K-2), turquoise (15.5K-3), blue (15.5K-4), or yellow (15.5K-5). D) Gel mobility shift analysis of the interaction of recombinant 15.5K and 15.5K mutants with U4, U14 and U14 mutants. The mutant proteins outlined in panel B were incubated with 32P-labeled in vitro-transcribed snRNA or snoRNA transcripts, and the resulting RNA-protein complexes were resolved on a 6% native polyacrylamide gel and visualized by autoradiography. The identity of the protein used is indicated above each lane (−, no 15.5K; 15.5K-Wt, wild-type 15.5K). The positions of the protein-RNA complex (RNP) and the free RNA are indicated on the right. The RNA used is indicated on the left.
One important aspect of this approach is that the mutant proteins should not affect RNA binding. Even though we were careful to select regions away from the RNA-binding domain, it was important for the various snRNP and snoRNP assembly assays that the mutants bind the RNA with an affinity and specificity similar to those of the wild-type protein. In order to test this, purified recombinant proteins were incubated with 32P-labeled U4, U3 BC, U14, or mutant U14 RNAs, and the resulting complexes were then resolved on a native polyacrylamide gel. This demonstrated that each of the mutant proteins bound the U4 and U14 RNAs with a specificity similar to that seen for the wild-type protein (Fig. 2D and data not shown). Importantly, titration experiments used to measure the affinity of U4, U14, and U3-BC RNA (31) binding showed that the all of the mutant proteins bind these RNAs with a KD basically the same as that of the wild-type protein (less than twofold variation; data not shown). Interestingly, the migration behavior of the 15.5K-RNA complex in the native gel was slightly different for each mutation. We believe that this is due to changes to the surface properties of the protein affecting the migration behavior in native gels. Importantly, none of the proteins bound the U14 mutD and U14 stII RNAs, in which the conserved GA nucleotides and stem II of the U14 snoRNA box C/D motif have been mutated, respectively (30). Therefore, in creating the mutants, we have changed neither the affinity nor specificity of RNA binding consistent with the fact that the changed amino acids are not part of the RNA-binding domain.
Conserved amino acids on the surface of 15.5K are required for hPRP31 association with the U4 snRNA.
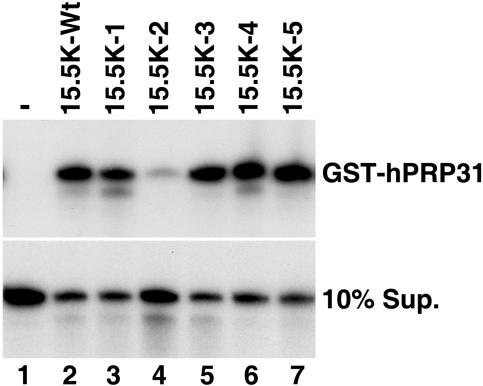
Next, we were interested in determining whether any of these mutations would block the formation of 15.5K-dependent RNP complexes. For the first approach, we analyzed the ability of these mutants to support hPRP31 binding to the U4 snRNA. Recombinant 15.5K protein (either wild type or mutant) was incubated with GST-hPRP31 and 32P-labeled U4 snRNA. Importantly, the amount of 15.5K used was shown to be in excess of that required by both the wild-type and mutant proteins to fully bind the RNA in the experiment. After incubation, GST-hPRP31 was isolated using glutathione Sepharose, and the bound RNAs were analyzed by polyacrylamide gel electrophoresis. Consistent with our earlier work (20), 15.5K is required for the coprecipitation of the U4 snRNA with GST-hPRP31 (Fig. 3, cf. lanes 1 and 2). Quantitation revealed that mutants 15.5K-1, 15.5K-3, 15.5K-4, and 15.5K-5 also supported hPRP31 association with the U4 snRNA with similar efficiencies, with only 15.5K-1 showing a slight reduction (85% of wild-type levels) compared to that of the wild-type protein (Fig. 3, cf. lane 2 with lanes 3, 5, 6, and 7). In contrast, a fourfold reduction of U4 snRNA association with hPRP31 was observed with 15.5K-2 (Fig. 3, lane 4). Importantly, gel shift analysis of the binding reaction mixtures showed that all mutant proteins efficiently bound the U4 snRNA and with the exception of 15.5K-2, recruited hPRP31, confirming our result and demonstrating that the lack of 15.K-2 recruitment of hPRP31 is not due to a lack of RNA binding under these conditions and that each of the proteins was stable under our reaction conditions (data not shown). Therefore, the surface amino acids mutated in 15.5K-2 (α-helix 3 and the adjacent loop α3-β3) are required for hPRP31 binding. The fact that this region of the protein is important for the recruitment of hPRP31 in this simple three-component system supports our hypothesis that 15.5K mediates the assembly of RNP complexes, in part, through protein-protein contacts.
FIG. 3.
Binding of hPRP31 to the U4 snRNA requires residues in α-helix 3 of 15.5K. GST-hPRP31 was incubated with 32P-labeled U4 snRNA in the presence (lanes 2, 3, 4, 5, 6 and 7) or absence (lane 1) of either wild-type or mutant 15.5K. GST-hPRP31 was isolated using glutathione resin, and bound RNA was extracted and ethanol precipitated. The bound (GST-hPRP31) and unbound RNA (10% supernatant [10% Sup.]) was analyzed on a denaturing 8% polyacrylamide-7 M urea gel and visualized by autoradiography. The identity of the 15.5K protein used is indicated above each lane (15.5K-Wt, wild-type 15.5K).
Multiple mutations on the surface of 15.5K disrupt CYPH/hPRP4/hPRP3 complex binding to the U4/U6 snRNA.
Having demonstrated that changes in the surface amino acids can affect hPRP31 binding to the U4 snRNA, we next analyzed whether the mutations would support binding of the CYPH/hPRP4/hPRP3 heteromeric complex to the U4/U6 snRNA utilizing the reconstitution assay developed by Nottrott et al. (20). Briefly, recombinant 15.5K protein was incubated with radiolabeled U4/U6 snRNA duplex, in which the minimal fragment of U6 snRNA required for CYPH/hPRP4/hPRP3 binding was radiolabeled (Fig. 1), and biochemically purified CYPH/hPRP4/hPRP3 protein complex. The assembled complexes were then immunoprecipitated using anti-hPRP4 antibodies, and the coprecipitated RNAs were analyzed by polyacrylamide gel electrophoresis.
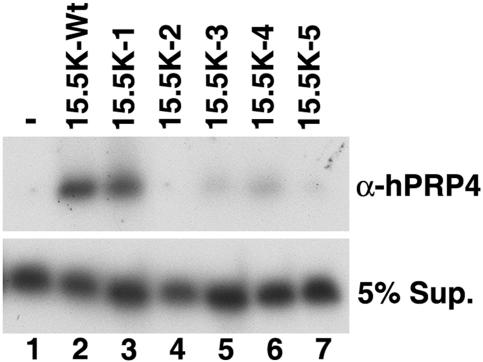
The U4/U6 snRNA duplex was coprecipitated by anti-hPRP4 antibodies when incubated with CYPH/hPRP4/hPRP3 and 15.5K but not in the presence of CYPH/hPRP4/hPRP3 alone (Fig. 4, lanes 1 and 2), consistent with our previous findings. Analysis of the mutants in this assay revealed that 15.5K-1 supported the U4/U6 snRNA-CYPH/hPRP4/hPRP3 interaction to 80% of the level seen with the wild-type protein (Fig. 3, lane 4). In contrast, a reduction in the formation of complex of greater than fivefold, relative to that of the wild-type protein, was observed using 15.5K-3 and 15.5K-4 (Fig. 4, lanes 4 and 7), and the interaction was effectively abolished in the presence of 15.5K-2 and 15.5K-5 (lanes 5 and 8). Therefore, four of the five regions, including α-helix 3 and the adjacent loop α3-β3, α-helix 4, and α-helix 5, are required for the interaction between U4/U6 snRNA and the CYPH/hPRP4/hPRP3 complex. This suggests that significant contacts are made between one or more of the proteins in the CYPH/hPRP4/hPRP3 complex and 15.5K that are important for U4/U6 snRNP formation.
FIG. 4.
The association of CYPH/hPRP4/hPRP3 to the U4/U6 snRNA requires multiple regions on the surface of 15.5K. Purified CYPH/hPRP4/hPRP3 complex was incubated with U4/U6 snRNA duplex (U4 snRNA base paired with 32P-labeled U6 nucleotides 58 to 87 [Fig. 1]) in the presence (lanes 2 to 7) or absence (lane 1) of wild-type or mutant 15.5K. The CYPH/hPRP4/hPRP3 complex was then immunoprecipitated using anti-hPRP4 antibody (α-hPRP4) and the coprecipitating RNA and the RNA present in the supernatant (5% Sup.) were analyzed on a denaturing 8% polyacrylamide-7 M urea gel and visualized by autoradiography. The identity of the 15.5K protein used is indicated above each lane (−, no 15.5K; 15.5K-Wt, wild-type 15.5K).
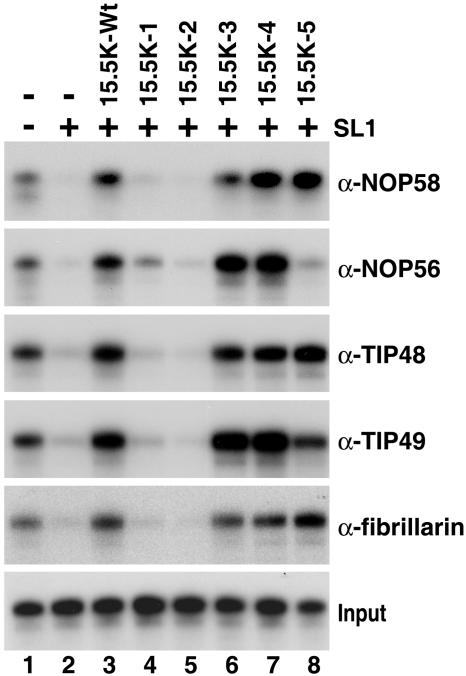
Box C/D snoRNP assembly requires multiple regions on the surface of 15.5K.
We next turned our attention to the assembly of the box C/D snoRNP. For this we used a system that we had previously established to analyze U14 snoRNP assembly (30). Briefly, in vitro-transcribed, 32P-labeled U14 snoRNA was incubated with HeLa nuclear extract, and snoRNP assembly was determined by immunoprecipitation using antibodies specific to the core box C/D proteins and the assembly factors TIP48 and TIP49. Using this system, we were able to selectively block the binding of the endogenous 15.5K in the extract using an excess of the 5′ stem-loop of the U4 snRNA (SL1) (30). Box C/D snoRNP assembly in the blocked extract was restored by the addition of recombinant 15.5K. Therefore, we used this approach to analyze the ability of the mutant proteins to support box C/D snoRNP assembly. Protein binding was subsequently analyzed using antifibrillarin, anti-NOP56, anti-NOP58, anti-TIP48, and anti-TIP49 antibodies, and coprecipitated RNAs were analyzed by polyacrylamide gel electrophoresis.
Consistent with our previous observations, the addition of U4 SL1 RNA inhibited box C/D snoRNP assembly, while the binding of all proteins was restored by the addition of recombinant 15.5K (Fig. 5, lanes 1, 2, and 3) (30). Similar levels of NOP56, NOP58, fibrillarin, TIP48, and TIP49 association with the U14 snoRNA were also observed when the wild-type protein was replaced by either mutant 15.5K-3 or 15.5K-4 (Fig. 5, lanes 6 and 7). In most cases, the level of precipitation was slightly higher than that seen for the wild-type protein but slightly reduced for fibrillarin and NOP58 (90% of wild-type levels) when mutant 15.5K-3 was used. In contrast, the binding of all proteins was either reduced by greater than 20-fold or effectively abolished when mutants 15.5K-1 and 15.5K-2 were used in this assay (lanes 4 and 5). Interestingly, 15.5K-5 supported the binding of just a subset of the box C/D-associated proteins (lane 8). In the presence of 15.5K-5, NOP58, TIP48, and fibrillarin association was equivalent to that seen with the wild-type protein, whereas the binding of TIP49 and NOP56 was either reduced (80% of wild-type levels) or effectively abolished (∼2% of wild-type levels), respectively. These results imply that multiple regions of the 15.5K surface are important for box C/D snoRNP assembly, including amino acids in α-helix 1 (15.5K-1), α-helix 3, and the adjacent loop between α3-b3 for complete box C/D snoRNP formation and α-helix 5 for the specific association of NOP56 and TIP49.
FIG. 5.
Multiple regions on the surface of 15.5K are required for box C/D snoRNP assembly. HeLa nuclear extract was preincubated with either 800 pmol of U4-SL1 (SL1) RNA oligonucleotide (+) or buffer (−). Radiolabeled U14 snoRNA was subsequently added in the presence (lanes 2 to 8) or absence (−) (lanes 1 and 2) of wild-type or mutant 15.5K. The binding of individual snoRNP proteins was then assayed by immunoprecipitation (see Materials and Methods). Bound RNAs were recovered and then separated on an 8% polyacrylamide-7 M urea gel. Antibodies used for immunoprecipitation are indicated on the right. Ten percent of the RNA after incubation in nuclear extract was used as input in the bottom gel. The 15.5K protein used is indicated above each lane (15.5K-Wt, wild-type 15.5K).
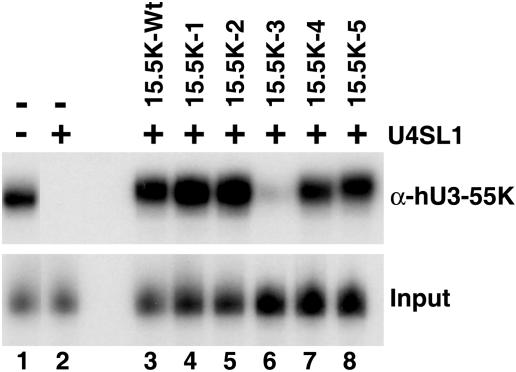
α-Helix 4 of 15.5K is required for hU3-55K binding to the U3 snoRNA.
Having analyzed the effects of the mutants on U4 snRNP, U4/U6 snRNP, and box C/D snoRNP formation, we next addressed the assembly of the U3-specific box B/C RNP complex. Using the U4 5′ stem-loop as a competitor, we have shown that the binding of hU3-55K to the U3 B/C motif in nuclear extract is dependent on 15.5K (6). Therefore, we used this approach to analyze the ability of the 15.5K mutants to support hU3-55K binding. Briefly, 32P-labeled U3 box B/C RNA was incubated with nuclear extract, which had been pretreated with the U4 5′ stem-loop RNA SL1, and then protein binding was determined by immunoprecipitation using anti-hU3-55K antibodies. Coprecipitated RNAs were then analyzed by polyacrylamide gel electrophoresis. Consistent with our earlier observations, the addition of SL1 RNA inhibited the coprecipitation of the U3 box B/C snoRNA with anti-hU3-55K antibodies (6). This interaction was restored by the addition of recombinant 15.5K. Analysis of the ability of the mutant proteins to restore protein binding revealed that 15.5K-1, 15.5K-2, 15.5K-4, and 15.5K-5 all effectively restored the association of the hU3-55K with the U3 box B/C RNA to 90 to 100% of wild-type levels (Fig. 6, lanes 4, 5, 7, and 8, respectively). In contrast, 15.5K-3 resulted in a reduction in the association of the hU3-55K protein of greater than 10-fold (lane 6). These results therefore show that the conserved region of α-helix 4, found on the protein surface, is important for interaction of hU3-55K protein with the U3 box B/C RNP complex.
FIG. 6.
hU3-55K binding to the U3 box B/C motif requires a distinct region on the surface of 15.5K. HeLa nuclear extract was preincubated with either 800 pmol of U4-SL1 RNA oligonucleotide (+) or buffer (−). Radiolabeled U3 box B/C motif RNA was subsequently added in the presence (lanes 3 to 8) or absence (lanes 1 and 2) of recombinant 15.5K or mutant 15.5K. The binding of hU3-55K was then assayed by immunoprecipitation (see Materials and Methods). Bound RNAs were recovered and then separated on an 8% polyacrylamide-7 M urea gel. Five percent of the RNA after incubation in nuclear extract was used as input in the bottom gel. The 15.5K protein used is indicated above each lane (15.5K-Wt, wild-type 15.5K). α-hU3-55K, anti-hU3-55K antibody.
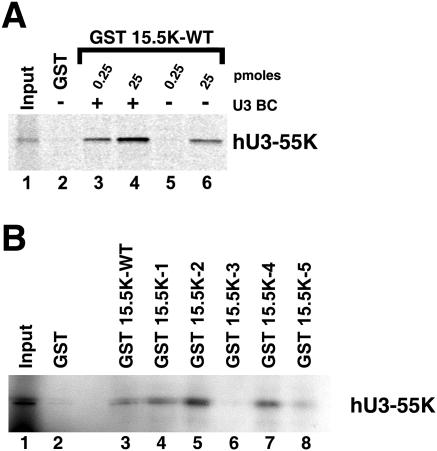
Direct interaction between hU3-55K and 15.5K.
We had previously developed an in vitro assembly system, using recombinant 15.5K and in vitro-translated hU3-55K, to analyze the assembly of the box B/C RNP complex (6). Therefore, we used this system to further investigate the relative importance of the RNA and 15.5K protein in the recruitment of hU3-55K. GST-tagged 15.5K was incubated with in vitro-translated 35S-labeled hU3-55K in the presence or absence of box B/C RNA. GST-15.5K was isolated using glutathione Sepharose, and hU3-55K association was determined by SDS-PAGE, followed by autoradiography. As previously described, using 0.25 pmol GST-15.5K, copurification of the hU3-55K was dependent on the presence of the box B/C RNA (6) (Fig. 7A, lanes 3 and 5). This amount of protein was also previously shown to still be capable of binding the U3 BC RNA in the absence of hU3-55K, indicating that the protein is stable in the absence of the U3-specific protein. However, we found that increasing the amount of GST-15.5K (25 pmol) resulted in a significant coprecipitation of 15.5K in the absence of the B/C RNA (Fig. 7A, lane 6). This interaction was not susceptible to RNase A treatment of the translation product, suggesting that this is not due to endogenous RNA present in the reticulocyte lysate (data not shown). Interestingly, under these conditions the binding of hU3-55K was stimulated more than threefold by the presence of the B/C RNA (Fig. 7A, lanes 4 and 6), suggesting that the full interaction is mediated by both the RNA and 15.5K. Importantly, the use of 25 pmol of GST alone failed to pull down hU3-55K, showing that this interaction is specific (lane 2).
FIG. 7.
hU3-55K directly interacts with 15.5K. A) 35S-labeled hU3-55K was incubated with either GST-15.5K (0.25 or 25 pmol) in the absence (−) or presence (+) of U3 B/C motif RNA (U3BC) (lanes 3 to 6) or 25 pmol of GST (lane 2). Bound, radiolabeled proteins were purified using glutathione resin, resolved on a 12% polyacrylamide-SDS gel, and revealed by autoradiography. The amount of recombinant 15.5K used is indicated above each lane. Ten percent of the input material was used in the Input lane. GST 15.5K-WT, GST fused to wild-type 15.5K. B) 35S-labeled hU3-55K was incubated with 25 pmol of either wild-type (WT) or mutant GST-15.5K (lanes 3 to 8) or GST (lane 2). Bound, radiolabeled proteins were purified using glutathione resin and resolved on a 12% polyacrylamide-SDS gel. Ten percent of the input material was used in the Input lane. The identity of the GST protein used is indicated above each lane.
If the interaction observed with the in vitro-translated product were the same as the proposed interaction in nuclear extract essential for hU3-55K recruitment, then this would predict that 15.5K-3 would not bind this protein. Therefore, we analyzed the ability of the 15.5K mutations to interact with in vitro-translated hU3-55K using 25 pmol of the GST-tagged protein. We found that GST-15.5K-1, -2, and -4 associated with hU3-55K either to an equivalent level or more efficiently (approximately twofold higher for GST-15.5K-2) than the wild-type protein did (Fig. 7B, cf. lane 2 and lanes 4, 5, and 7). GST-15.5K bound hU3-55K to about 60% of the levels seen for the wild-type protein (Fig. 7B, lane 8). This suggests that while this mutant does not function as efficiently as the wild-type protein, it also does not significantly reduce the association of hU3-55K. However, GST-15.5K-3 showed a 20-fold reduction in hU3-55K copurification levels (lane 6). These data therefore provide strong evidence that a protein-protein interaction with 15.5K is important for hU3-55K binding to the U3 snoRNA.
DISCUSSION
In this study, we set out to investigate the role protein-protein interactions play in the 15.5-K-mediated assembly of four distinct RNP complexes. Using the crystal structure of 15.5K bound to the 5′ stem-loop of U4 snRNA (29), we have identified and mutated regions of conserved amino acids on the surface of the protein. Analysis of the ability of these mutants to support RNP formation revealed that different regions on the surface of the protein are required for the assembly of each complex. This implies that, in each of the RNPs, substantial and essential contacts are made between this protein and one or more of the complex-specific proteins. Importantly, these mutations were not in the areas of the protein involved in RNA binding, and none of the mutations altered either the affinity or specificity of protein binding to the U4 snRNA, U14 box C/D snoRNA, and U3 BC RNA. In addition, the changes have not altered either the stability or the gross structural organization of the protein, as each of the mutations supported the formation of at least one RNP complex. Indeed, in many cases the binding reaction mixtures using the mutant 15.5K proteins, especially those presented in Fig. 3 and 4, were analyzed by native gel electrophoresis in order to monitor that the proteins were stable and retained their RNA-binding ability under our assay conditions (data not shown). This therefore implies that the effects we observed are the result of specific changes and not due to nonspecific disruption of the overall structure of the protein.
We had previously noted that distinct structures and/or sequences in the RNA are required for the specific assembly of each complex (6, 20, 30). We have now found that distinct regions of the surface of 15.5K are necessary for the assembly of each complex. This suggests that, in each case, the formation of the RNP requires the complex recognition of both protein and RNA. The identification of the RNA-independent hU3-55K-15.5K interaction supports our notion that protein-protein interactions within each of the complexes are important. Importantly, a similar approach was used to detect direct interactions between 15.5K and hPRP31, hPRP3, hPRP4, CYPH, NOP56, NOP58, and fibrillarin in the absence of RNA. However, no specific interaction could be detected between 15.5K and these proteins (data not shown). This therefore implies that the interaction between 15.5K and these proteins was not sufficiently stable to allow detection and that a stable interaction requires the presence of both 15.5K and RNA. The differential recognition of RNA and 15.5K is not surprising, considering that the proteins involved in each complex are very different and use distinct domains/structures to mediate complex formation. The only exception to this is the NOP domain proteins, namely, NOP56, NOP58, and hPRP31 (5). Interestingly, NOP58 and NOP56 recruitment to U14 snoRNA and hPRP31 binding to the U4 snRNA require the highly conserved amino acids present in α-helix 3. However, NOP56 and NOP58 binding in HeLa nuclear extract also required other regions of 15.5K, and we cannot at this point separate the requirements for binding of the individual snoRNP components. Further analysis of the role of the conserved surface amino acids in protein recruitment has been severely hampered by solubility problems observed with the vast majority of the mutations constructed during this study. Indeed, 21 mutant constructs were generated during this study; of the 21 mutant constructs, only the five mutants used in this study were soluble (see above). Therefore, while it is interesting to speculate that NOP58 or NOP56 and hPRP31 contact the same region of 15.5K, we cannot at present make any conclusions about the nature of the protein-protein interactions in box C/D snoRNPs.
It is clear from our data that much of the surface of 15.5K is essential for the formation of the U4/U6 snRNP, implying that 15.5K is surrounded by proteins in the U4/U6.U5 tri-snRNP. This is consistent with our earlier observation for both HeLa and yeast extracts that 15.5K was not accessible for immunoprecipitation in the tri-snRNP. Interestingly, 15.5K-2 inhibits the binding of both hPRP31 and the CYPH/hPRP4/hPRP3 complex. The binding of these two activities has been shown not to be mutually exclusive (20), and since all of these proteins are present in the U4/U6.U5 tri-snRNP, it is likely that this mutation could disrupt two adjacent protein-binding sites on the surface of 15.5K.
The 15.5K mutations that block snoRNP assembly either totally abolish the binding of all proteins or inhibit the recruitment of NOP56 and TIP49. This therefore implies that two or more activities bind the 15.5K box C/D snoRNA complex. Indeed, the data could suggest that the binding of NOP58, TIP48, and fibrillarin is essential for the subsequent recruitment of NOP56 and TIP49. Furthermore, NOP56 and NOP58 proteins and TIP48 and TIP49 proteins are two pairs of homologous proteins (18), respectively. Our data show that recruitment of TIP49 and NOP56 is linked. This therefore suggests that TIP48 might be involved in the assembly of NOP58. NOP56 and NOP58 are proposed to specifically contact the box C′/D′ and C/D motifs, respectively, and interact with one another through a conserved coiled-coil domain (2, 4). Therefore, on the basis of this model, NOP56 recruitment is linked to an interaction with NOP58 bound at the C/D motif. However, our finding that α-helix 5 of 15.5K is important for the specific recruitment of NOP56 and efficient binding of TIP49 suggests that NOP56 directly contacts 15.5K bound to the box C/D motif and that its recruitment is not solely due to contact with NOP58. Further work is required to define the various activities involved in snoRNP assembly.
Our data imply that 15.5K is involved in multiple interactions in the four distinct RNP complexes. The evolutionary pressure required to maintain these interactions therefore explains the high degree of conservation of 15.5K in eukaryotes. In archaea, a homologous protein, namely, L7ae, is present in the ribosome, box C/D RNPs, and H/ACA RNPs (14, 21, 22). Therefore, this protein performs the functions of the eukaryotic L7a, 15.5K, and NHP2, which implies that the three eukaryotic proteins are derived from a common ancestor. Interestingly, the amino acid composition of α-helix 3 of the eukaryotic 15.5K, which is required for snoRNP formation, is similar to that of the archaeal L7ae (data not shown), suggesting that this interaction is evolutionarily conserved.
Acknowledgments
We thank T. Conrad, P. Kempkes, and H. Kohansal for their technical support. We are also grateful to Klaus Hartmuth for critically reading the manuscript.
This work was supported by grants from the DFG (LU294/12-1), Ernst-Jung-Stiftung, and Fonds der Chemischen Industrie to R.L.
REFERENCES
- 1.Achsel, T., H. Brahms, B. Kastner, A. Bachi, M. Wilm, and R. Lührmann. 1999. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 18:5789-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aittaleb, M., R. Rashid, Q. Chen, J. R. Palmer, C. J. Daniels, and H. Li. 2003. Structure and function of archaeal box C/D sRNP core proteins. Nat. Struct. Biol. 10:256-263. [DOI] [PubMed] [Google Scholar]
- 3.Bachellerie, J. P., J. Cavaille, and A. Huttenhofer. 2002. The expanding snoRNA world. Biochimie 84:775-790. [DOI] [PubMed] [Google Scholar]
- 4.Cahill, N. M., K. Friend, W. Speckmann, Z. H. Li, R. M. Terns, M. P. Terns, and J. A. Steitz. 2002. Site-specific cross-linking analyses reveal an asymmetric protein distribution for a box C/D snoRNP. EMBO J. 21:3816-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gautier, T., T. Berges, D. Tollervey, and E. Hurt. 1997. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol. 17:7088-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Granneman, S., G. J. Pruijn, W. Horstman, W. J. van Venrooij, R. Lührmann, and N. J. Watkins. 2002. The hU3-55K protein requires 15.5K binding to the box B/C motif as well as flanking RNA elements for its association with the U3 small nucleolar RNA in vitro. J. Biol. Chem. 277:48490-48500. [DOI] [PubMed] [Google Scholar]
- 7.Granneman, S., J. Vogelzangs, R. Lührmann, W. J. van Venrooij, G. J. Pruijn, and N. J. Watkins. 2004. Role of pre-rRNA base pairing and 80S complex formation in subnucleolar localization of the U3 snoRNP. Mol. Cell. Biol. 24:8600-8610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henras, A. K., C. Dez, and Y. Henry. 2004. RNA structure and function in C/D and H/ACA s(no)RNPs. Curr. Opin. Struct. Biol. 14:335-343. [DOI] [PubMed] [Google Scholar]
- 9.Kiss, T. 2002. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109:145-148. [DOI] [PubMed] [Google Scholar]
- 10.Kiss-Laszlo, Z., Y. Henry, J. P. Bachellerie, M. Caizergues-Ferrer, and T. Kiss. 1996. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85:1077-1088. [DOI] [PubMed] [Google Scholar]
- 11.Kiss-Laszlo, Z., Y. Henry, and T. Kiss. 1998. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J. 17:797-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein, D. J., T. M. Schmeing, P. B. Moore, and T. A. Steitz. 2001. The kink-turn: a new RNA secondary structure motif. EMBO J. 20:4214-4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koonin, E. V., P. Bork, and C. Sander. 1994. A novel RNA-binding motif in omnipotent suppressors of translation termination, ribosomal proteins and a ribosome modification enzyme? Nucleic Acids Res. 22:2166-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhn, J. F., E. J. Tran, and E. S. Maxwell. 2002. Archaeal ribosomal protein L7 is a functional homolog of the eukaryotic 15.5kD/Snu13p snoRNP core protein. Nucleic Acids Res. 30:931-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lafontaine, D. L., and D. Tollervey. 1999. Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA 5:455-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lafontaine, D. L., and D. Tollervey. 2000. Synthesis and assembly of the box C+D small nucleolar RNPs. Mol. Cell. Biol. 20:2650-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madhani, H. D., and C. Guthrie. 1994. Dynamic RNA-RNA interactions in the spliceosome. Annu. Rev. Genet. 28:1-26. [DOI] [PubMed] [Google Scholar]
- 18.Newman, D. R., J. F. Kuhn, G. M. Shanab, and E. S. Maxwell. 2000. Box C/D snoRNA-associated proteins: two pairs of evolutionarily ancient proteins and possible links to replication and transcription. RNA 6:861-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nottrott, S., K. Hartmuth, P. Fabrizio, H. Urlaub, I. Vidovic, R. Ficner, and R. Lührmann. 1999. Functional interaction of a novel 15.5kD [U4/U6.U5] tri-snRNP protein with the 5′ stem-loop of U4 snRNA. EMBO J. 18:6119-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nottrott, S., H. Urlaub, and R. Lührmann. 2002. Hierarchical, clustered protein interactions with U4/U6 snRNA: a biochemical role for U4/U6 proteins. EMBO J. 21:5527-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Omer, A. D., S. Ziesche, H. Ebhardt, and P. P. Dennis. 2002. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc. Natl. Acad. Sci. USA 99:5289-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rozhdestvensky, T. S., T. H. Tang, I. V. Tchirkova, J. Brosius, J. P. Bachellerie, and A. Huttenhofer. 2003. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: a shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res. 31:869-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schimmang, T., D. Tollervey, H. Kern, R. Frank, and E. C. Hurt. 1989. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 8:4015-4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schulz, G. E., and R. H. Schirmer. 1979. Amino acids, p. 1-16. In C. R. Cantor (ed.), Principles of protein structure. Springer-Verlag, New York, N.Y.
- 25.Staley, J. P., and C. Guthrie. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92:315-326. [DOI] [PubMed] [Google Scholar]
- 26.Szewczak, L. B., S. J. DeGregorio, S. A. Strobel, and J. A. Steitz. 2002. Exclusive interaction of the 15.5 kD protein with the terminal box C/D motif of a methylation guide snoRNP. Chem. Biol. 9:1095-1107. [DOI] [PubMed] [Google Scholar]
- 27.Szymanski, M., M. Z. Barciszewska, M. Zywicki, and J. Barciszewski. 2003. Noncoding RNA transcripts. J. Appl. Genet. 44:1-19. [PubMed] [Google Scholar]
- 28.Turner, B., S. E. Melcher, T. J. Wilson, D. G. Norman, and D. M. Lilley. 2005. Induced fit of RNA on binding the L7Ae protein to the kink-turn motif. RNA 11:1192-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vidovic, I., S. Nottrott, K. Hartmuth, R. Lührmann, and R. Ficner. 2000. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol. Cell 6:1331-1342. [DOI] [PubMed] [Google Scholar]
- 30.Watkins, N. J., A. Dickmanns, and R. Lührmann. 2002. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol. Cell. Biol. 22:8342-8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watkins, N. J., V. Segault, B. Charpentier, S. Nottrott, P. Fabrizio, A. Bachi, M. Wilm, M. Rosbash, C. Branlant, and R. Lührmann. 2000. A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103:457-466. [DOI] [PubMed] [Google Scholar]
- 32.Will, C. L., and R. Lührmann. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13:290-301. [DOI] [PubMed] [Google Scholar]
- 33.Williamson, J. R. 2000. Induced fit in RNA-protein recognition. Nat. Struct. Biol. 7:834-837. [DOI] [PubMed] [Google Scholar]
- 34.Wozniak, A. K., S. Nottrott, E. Kuhn-Holsken, G. F. Schroder, H. Grubmuller, R. Luhrmann, C. A. Seidel, and F. Oesterhelt. 2005. Detecting protein-induced folding of the U4 snRNA kink-turn by single-molecule multiparameter FRET measurements. RNA 11:1545-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu, P., J. S. Brockenbrough, A. C. Metcalfe, S. Chen, and J. P. Aris. 1998. Nop5p is a small nucleolar ribonucleoprotein component required for pre-18 S rRNA processing in yeast. J. Biol. Chem. 273:16453-16463. [DOI] [PMC free article] [PubMed] [Google Scholar]