Abstract
In contrast with the early view of metalloproteases as simple extracellular matrix-degrading entities, recent findings show that they are highly specific modulators of different signaling pathways involved, positively or negatively, in tumor development. Thus, before considering a given metalloprotease a therapeutic target, it seems advisable to characterize its function by identifying its repertoire of substrates. Here, we present a proteomic approach to identify ADAM17 substrates by difference gel electrophoresis. We found that the shedding of the extracellular domain of the transferrin receptor and those of two cell-cell adhesion molecules, activated leukocyte cell adhesion molecule (ALCAM) and desmoglein 2 (Dsg-2), is increased in cells overexpressing ADAM17. Genetic evidence shows that while ADAM17 is responsible for the shedding of ALCAM, both ADAM17 and ADAM10 can act on Dsg-2. Activation of the epidermal growth factor receptor leads to the upregulation of the shedding of Dsg-2 and to the concomitant upregulation of ADAM17, but not ADAM10, supporting the ability of overexpressed ADAM17 to shed Dsg-2. These results unveil a role of ADAM10 and ADAM17 in the shedding of cell-cell adhesion molecules. Since loss of cell adhesion is an early event in tumor development, these results suggest that ADAM17 is a useful target in anticancer therapy.
Mammalian extracellular and membrane-anchored metalloproteases are classified in large families; together, the family of the matrix metalloproteases (MMPs) and the related families of ADAMs (a disintegrin and metalloprotease) and ADAM-TS (ADAMs containing thrombospondin motifs) comprise nearly 75 members (21). Metalloproteases have long been considered targets of antitumor therapy because of their frequent upregulation in cancer and their ability to degrade the extracellular matrix, a hypothetical prerequisite for malignant cells to invade surrounding tissue (25). Following this reasoning, several wide-spectrum metalloprotease inhibitors (MPIs) have been clinically tested; unfortunately, no obvious benefit has been observed in treated patients (12).
In contrast with the classical view, accumulating evidence shows that during tumor development, metalloproteases play highly specific roles, not directly related to extracellular matrix degradation. For example, MMP1 seems to promote metastasis through the proteolytic activation of protease-activated receptor 1 (7), a G protein-coupled receptor known to convey signals leading to tumor invasion in a variety of experimental models (11). Further, the proteolytic activity of ADAM17 is required to release ligands that activate the epidermal growth factor receptor (EGFR) (14), a tyrosine kinase receptor which contributes to tumor development by regulating cell proliferation, apoptosis, and migration (32). In contrast to those of MMP1 and ADAM17, the activity of MMP8 seems to play an antitumorigenic role: its absence causes increased tumor susceptibility, probably because of its ability to cleave cytokines that mediate the inflammatory response (4).
In view of these decisive functions, it is not surprising that metalloprotease activity is subjected to tight control. MMPs and ADAMs are synthesized as zymogens containing a prodomain that blocks the catalytic domain. During progression through the secretory pathway, the prodomain is cleaved by furin-like proprotein convertases (9). There are several naturally occurring MPIs, the best characterized of which are known as tissue inhibitors of metalloproteinases, which inhibit all known MMPs and some ADAMs (3). On the other hand, several signaling pathways, including the receptor tyrosine kinase-activated extracellular signal-regulated kinase/mitogen-activated proteins kinase, the protein kinase C, and the G protein-coupled receptor pathways, can upregulate ectodomain shedding (6), a specialized type of limited proteolysis largely mediated by ADAMs that releases the extracellular domains of membrane-anchored proteins (2).
In summary, in contrast with the early view, metalloproteases are currently considered modulators of a variety of signaling pathways affecting, positively or negatively, tumor development. In turn, metalloprotease activity is tightly controlled, by different mechanisms and signaling pathways. Thus, before reevaluating metalloproteases as therapeutic targets, it seems advisable to carefully analyze their function and regulation individually and develop specific inhibitors directed against those contributing to tumor development.
An obvious step towards the characterization of the function of a given metalloprotease is the identification of its repertoire of substrates, also known as the degradome (21). Different proteomic methods have been undertaken to accomplish this goal. Guo et al. identified several substrates of ADAM17 through quantitative comparison by isotope dilution of proteins secreted by control cells and cells treated with activators or inhibitors of ectodomain shedding (18). Similarly, Tam et al. compared proteins in the conditioned media of parental cells and cells overexpressing MT1-MMP. Using isotope-coded affinity tag labeling and multidimensional liquid chromatography inline with tandem mass spectrometry, they found several previously unknown putative substrates of this metalloprotease (29). Difference gel electrophoresis (DIGE) has been recently implemented as an alternative quantitative proteomic technique (30). Its main advantage is that two samples are labeled with different fluorochromes and loaded onto a single two-dimensional (2D) gel, thereby overcoming the lack of reproducibility between different 2D gels but keeping the ability to rapidly compare different samples by using this widespread electrophoretic technique.
Here, we present the results of a proteomic approach to identify ADAM17 substrates by DIGE. Using this approach, we found that the shedding of the transferrin receptor (TfnR) and those of two cell-cell adhesion molecules, activated leukocyte cell adhesion molecule (ALCAM) and desmoglein 2 (Dsg-2), is increased in cells overexpressing ADAM17. The cleavage of ALCAM is severely impaired in cells genetically deficient for ADAM17, confirming a major role of this metalloprotease in the shedding of this cell adhesion molecule. In contrast, the shedding of Dsg-2 was found to be unaffected in these cells, showing that another protease(s) can act on Dsg-2. Genetic evidence shows that ADAM10 can also cleave Dsg-2. Finally, we show that treatment of A431 cells with EGF leads to the upregulation of the shedding of Dsg-2 and to the concomitant upregulation of ADAM17, but not ADAM10, supporting the ability of overexpressed ADAM17 to shed Dsg-2. In summary, these results indicate that DIGE is an effective technique to identify novel substrates of metalloproteases and unveil a role of ADAM10 and ADAM17 in the shedding of cell-cell adhesion molecules. Since loss of cell adhesion is an early event in tumor development, these results suggest that these metalloproteinases are useful targets in anticancer therapy.
MATERIALS AND METHODS
Antibodies and reagents.
The polyclonal antibodies against the intracellular domain of ADAM17 have been described elsewhere (8). The polyclonal antibodies against the extracellular domain of ALCAM and TfnR were from Santa Cruz Biotechnology (Santa Cruz, CA). The monoclonal antibody against the extracellular domain of Dsg-2 was from ABCAM (Cambridge, United Kingdom). Mouse immunoglobulin M antiactin antibody was from Oncogene Research Products. The anti-EGFR antibody C225 was from Merck.
We acknowledge the gifts of anti-mouse PS1 CTF (B32.1), anti-amyloid precursor protein (anti-APP) C terminus (B63.3), and B 42.2 (a polyclonal antibody against the C terminus of ADAM10) by W. Annaert (KU Leuven, Belgium). Polyclonal antibody B32.1 is directed against the murine PS1-CTF (peptide antigen residues 310 to 330, PKNPKYNTQRAERETQDSGSG). B63.3 is a rabbit polyclonal antibody generated using a synthetic peptide mimicking the final 16 amino acids of APP (NGYENPTYKFFEQMQN) coupled to keyhole limpet hemocyanin (Pierce). Polyclonal antibody against EGFR was from Abcam, and monoclonal antibody against phospho-EGFR was from Upstate.
Human Dsg-2 and TfnR cDNAs were kindly provided by K. Green and L. C. Kühn, respectively. BB-94 and recombinant EGF were obtained from British Biotech and R&D Systems, respectively. L685,458 was from Calbiochem. N-Glycosidase F was from Roche Diagnostics (Indianapolis, IN). Additional chemicals were from Sigma-Aldrich (St. Louis, MO).
Cell lines and transfections.
A431 cells were grown in Dulbecco modified Eagle medium (DMEM)-F-12 medium (1:1) supplemented with 10% fetal calf serum and 2 mM l-glutamine. A431/ADAM17 cells were grown as for A431 cells in presence of G-418 (500 μg/ml). Wild-type CHO and TACE-defective mutant cells (M2) were grown in DMEM supplemented with 10% fetal calf serum. Wild-type and ADAM10 knockout mouse embryonic fibroblasts (MEFs) were grown in DMEM supplemented with 5% fetal calf serum on 0.25% gelatin-precoated cell culture plates.
Stable transfection of ADAM17 into A431 cells and transient transfections of CHO/M2 cells were performed as described previously (31). As previously shown (31), transfection of ADAM17 leads to the predominant expression of the pro-ADAM17. We selected a clone with the highest levels of processed ADAM17.
Transient and stable transfections of MEFs were performed using Fugene 6 transfection reagent (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's instructions.
Analysis of shedding of Dsg-2, ALCAM, and TfnR in A431 and A431/ADAM17 cells.
A431 and A431/ADAM17 cells (5 × 105) were plated in 60-mm dishes. After 24 h, cells were serum starved for 16 h and then treated in the presence or absence of 20 μM BB-94, 50 ng/ml EGF, and/or 200 nM C225 antibody (only for Dsg-2) for 48 h. After that, the cells were lysed in radioimmunoprecipitation assay buffer (20 mM Tris-Cl [pH 7.4], 0.5 mM EDTA, 1% NP-40, 150 mM NaCl, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 10 mM 1,10-phenantroline, 1 mM phenylmethylsulfonyl fluoride). When indicated, 25 μM of BB-94 was added to the lysis buffer. Total protein in the lysates was quantified, and equal amounts of protein from cell lysates were concentrated with wheat germ agglutinin (WGA)-agarose beads (Vector Laboratories, Burlingame, CA) for 2 h at 4°C, which were eluted directly in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis sample buffer for Western blot analysis. In parallel, the conditioned media of the same cells were harvested and concentrated on WGA-agarose as described above. Protein loading for conditioned-medium samples for Western blot analysis was adjusted according to the total protein in cell lysates and checked by Ponceau red staining of the blotted membrane.
Analysis of shedding of Dsg-2 in CHO/M2 cells.
CHO and M2 cells (1.5 × 106) were plated in 100-mm plates and transfected as described previously (9), and then the cells were washed and serum starved for 48 h. After that, WGA-agarose-concentrated cell lysates and conditioned media were analyzed by Western blotting as described above.
Analysis of shedding of ALCAM in CHO/M2 cells.
CHO and M2 cells (4 × 106) were plated in 100-mm plates, and then the cells were washed and serum starved for 48 h. After that, WGA-agarose-concentrated cell lysates and conditioned media were either treated or not with N-glycosidase F and analyzed by Western blotting as described above. N-Glycosidase F treatment (3 units/sample; 16 h at 37°C) was performed on WGA-agarose-concentrated cell lysates or conditioned-medium samples eluted with radioimmunoprecipitation assay buffer at 100°C for 5 min.
Analysis of shedding of Dsg-2 in wild-type and ADAM10 knockout MEFs.
Wild-type and ADAM10 knockout MEFs (5 × 105) were plated in 60-mm dishes, and the cells were then transfected as indicated and serum starved for 48 h. After that, WGA-agarose-concentrated cell lysates and conditioned media were analyzed by Western blotting as described above.
Membrane extractions.
MEFs from ADAM10 knockout mice permanently transfected with Dsg-2 were lysed by sonication. The cell lysates were centrifuged at 1,500 × g for 15 min, and the pellet containing unbroken cells and nuclei was discarded. The resulting supernatants were further centrifuged at 100,000 × g for 1 h, and the pellets (microsomal fraction) were extracted with phosphate-buffered saline (PBS), 1% Triton X-100, 1 M NaCl, or 0.1 M Na2CO3 (pH 12.0) for 30 min on ice. The samples were then centrifuged again at 100,000 × g for 1 h. The resulting supernatants (S) were recovered, and the pellets were washed twice with PBS and extracted with 1% Triton X-100 for 30 min in ice. Finally, the samples were centrifuged at 100,000 × g for 1 h, and both the resulting (P) and the previously obtained (S) supernatants were concentrated with WGA-agarose beads for 1 h at 4°C. The solubilized proteins in both the S and P fractions were analyzed by Western blotting.
Treatment with protease inhibitors.
Cells were treated with 10 nM caspase-3 inhibitor I (Calbiochem, Darmstadt, Germany), 10 μM of MG132 (Calbiochem, Darmstadt, Germany), 125 nM of γ-secretase inhibitor X (Calbiochem, Darmstadt, Germany), or 2 μM of calpain inhibitor I (ALLN) (Sigma, St. Louis, MO) for 4 hours, and the shedding of Dsg-2 was analyzed as described above.
Western blotting.
WGA-agarose-concentrated protein samples from both cell lysate and conditioned medium were loaded onto an 8% SDS-polyacrylamide gel, transferred to nitrocellulose membranes, and blotted with anti-Dsg-2, anti-ALCAM, or anti-TfnR. Concanavalin A-agarose-concentrated protein samples from cell lysates were loaded onto an 8% SDS-polyacrylamide gel, transferred to nitrocellulose membranes, and blotted with anti-ADAM17 or anti-ADAM10 antibody. Detection was performed using the corresponding horseradish peroxidase-conjugated secondary antibody and the SuperSignal chemiluminescence kit from Pierce (Rockford, IL).
DIGE.
Cells (2 × 108) were seeded, and after 24 h they were extensively washed with serum-free medium. Cells were then treated with or without 20 μM BB-94 for 48 h, and the conditioned media (500 ml each) were harvested and incubated overnight at 4°C with WGA-agarose beads (1 μl of stirred suspension per ml of conditioned medium). Glycoproteins were eluted by stirring for 2 h at 4°C with 0.5 ml of 0.5 M N-acetylglucosamine in 10 mM HEPES (pH 7.5)-0.15 M NaCl buffer. The glycoprotein solutions were then concentrated fivefold by ultrafiltration (Amicon Ultra-4, 5-kDa cutoff; Millipore, Carrigtwohill, Ireland), followed by a modified acetone-trichloroacetic acid precipitation (2D-CleanUp kit; GE Healthcare). The protein pellets were resuspended in the DIGE labeling buffer {8 M urea, 4% [wt/vol] 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS], 30 mM Tris [pH 8.0]}, and the protein concentration was determined using the Bio-Rad RCDC protein assay as described by the manufacturer (Bio-Rad, United Kingdom). The protein concentration in the samples was then adjusted to 2 mg/ml by addition of DIGE labeling buffer. Fifty micrograms of each sample was labeled by addition of 400 pmol of either Cy3 or Cy5 cyanine dye (GE Healthcare)in 1 μl of anhydrous dimethylformamide. After 30 min of incubation on ice in the dark, the reaction was quenched by addition of 10 mM lysine followed by a further 10 min of incubation. After labeling, the samples corresponding to untreated cells (Cy3) and treated cells (Cy5) were mixed and diluted twofold with isoelectric focusing sample buffer (8 M urea, 4% [wt/vol] CHAPS, 2% dithiothreitol, 2% Pharmalytes [pH 3 to 10]).
2D electrophoresis was performed using GE Healthcare reagents and equipment. First-dimension isoelectric focusing was performed on immobilized pH gradient strips (24 cm; pH 3 to 10 or 4 to 7) using an Ettan IPGphor system. Samples were applied via cup loading near the basic ends of the strips, which were previously rehydrated overnight in 450 μl of rehydration buffer (8 M urea, 4% [wt/]vol CHAPS, 1% Pharmalytes [pH 3 to 10], 100 mM DeStreak). After focusing for a total of 67 kV-h, the strips were equilibrated first for 15 min in 6 ml of reducing solution (6 M urea, 100 mM Tris-HCl [pH 8], 30% [vol/vol] glycerol, 2% [wt/vol] SDS, 5 mg/ml DTT) and then for a further 15 min in 6 ml of alkylating solution (6 M urea, 100mM Tris-HCl [pH 8], 30% [vol/vol] glycerol, 2% [wt/vol] SDS, 22.5 mg/ml iodoacetamide) on a rocking platform. Second-dimension SDS-polyacrylamide gel electrophoresis was run by overlaying the strips on 12.5% isocratic Laemmli gels (24 by 20 cm), cast in low-fluorescence glass plates, on an Ettan DALT VI system. Gels were run at 20°C and at a constant power of 2.5 W per gel for 30 min, followed by 17 W per gel until the bromophenol blue tracking front had run off the bottoms of the gels (about 5 h).
Fluorescence images of the gels were acquired on a Typhoon 9400 scanner (GE Healthcare). Cy3 and Cy5 images were scanned at 532 nm excitation/580 nm emission and 633 nm excitation/670 nm emission, respectively, at a 100-μm resolution. Image analysis was performed using DeCyder V. 5.0 software (GE Healthcare).
Protein identification by mass spectrometry.
Protein spots of interest were excised from the gel by using an automated Spot Picker (GE Healthcare). In-gel trypsin digestion was performed essentially as described by Shevchenko et al. (27), using autolysis-stabilized trypsin (Promega, Madison, WI). Tryptic digests were purified using ZipTip microtiter plates (Millipore, Carrigtwohill, Ireland).
Matrix-assisted laser desorption ionization mass spectrometric analysis of tryptic peptides was performed on an Ultraflex TOF-TOF instrument (Bruker, Germany). Samples were prepared using α-cyano-4-hydroxy-cinnamic acid as a matrix on anchor chip targets (Bruker, Germany). Identification of the proteins was carried out by peptide mass fingerprint data and/or by TOF-TOF PSD. Database searches were performed using the Mascot algorithm (Matrix Science).
RESULTS
Proteomic identification of shedding products from the conditioned medium of cells overexpressing ADAM17.
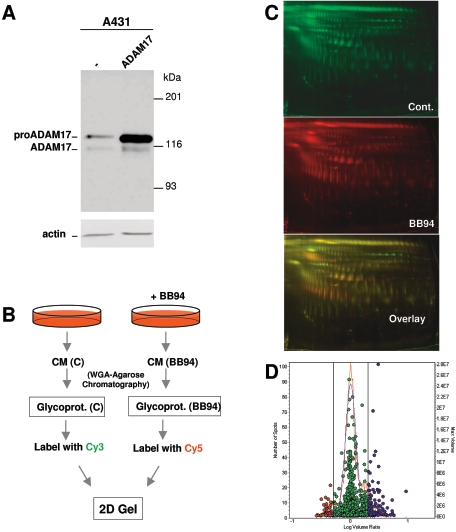
To identify novel metalloprotease substrates, we compared by DIGE the medium conditioned by control cells with that conditioned by cells treated with BB-94, a widely used nonspecific inhibitor that targets MMPs and ADAMs (2). We used A431 cells because they express high levels of the EGFR, making them a good model to study the regulation of ectodomain shedding by this tyrosine kinase. The levels of ADAM17 in A431 cells are low compared to those in a panel of cell lines (see Fig. S1 in the supplemental material); thus, in order to facilitate the identification of ADAM17 substrates, we used permanently transfected A431 cells. Analysis of cell lysates from A431 cells with antibodies against the intracellular domain of ADAM17 revealed two species (Fig. 1A), corresponding to the inactive form of the metalloprotease containing the prodomain (pro-ADAM17) and the processed form of the enzyme (ADAM17). As previously described (31), transfection with the cDNA encoding ADAM17 leads to the predominant accumulation of pro-ADAM17 and to a modest (∼3-fold) increase of the active form of the enzyme (Fig. 1A).
FIG. 1.
Proteomic identification of shedding substrates. A. Expression of ADAM17 in parental and stably transfected A431 cells. Lysates from the indicated cells were analyzed by Western blotting with polyclonal antibodies against the cytoplasmic domain of ADAM17. As a control, the same lysates were analyzed with antiactin antibodies. B. Schematic showing the protocol used. The conditioned media (CM) of A431 cells transfected with TACE and treated with or without BB-94 were subjected to WGA-agarose chromatography. The purified glycoproteins were then labeled with the cyanin dyes Cy3 and Cy5 and analyzed by 2D electrophoresis. C. The 2D gel was scanned with different wavelengths to visualize protein patterns corresponding to proteins labeled with Cy3 and Cy5. D. Experimental (red curve) and normalized model (blue curve) frequency distribution of volume ratios for the spots detected in the fluorescence images of the DIGE experiment. The volume of each individual protein spot, represented as a single data point, is plotted in the right axis. Spots in red represent proteins with a greater-than-1.5-fold decrease in the conditioned medium of cells treated with BB94. Spots in blue represent proteins with a greater-than-1.5-fold increase.
It has been previously shown that the majority of polypeptides secreted by cells correspond to heat shock proteins, actin, and metabolic pathway enzymes (18). Since the extracellular domains of shedding substrates are usually glycosylated, in order to enrich shedding products, we purified glycoproteins by WGA-chromatography as previously described (18) (Fig. 1B). Purified glycoproteins were labeled with the cyanin dyes Cy3 and Cy5 and analyzed by 2D gel electrophoresis (Fig. 1B).
To visualize the protein patterns, the gel was scanned with fluorophore-specific excitation and emission wavelengths to acquire two independent images (Fig. 1C), each corresponding to a separated Cy-labeled probe. Image analysis and determination of significant alterations in protein abundances were performed automatically with the DeCyder software. Only those protein spots exhibiting a decrease in volume ratio exceeding 1.5-fold were selected (Fig. 1D), excised, and subjected to in-gel digestion with trypsin. The resulting peptides were then subjected to peptide mass fingerprinting analysis by matrix-assisted laser desorption ionization mass spectrometry. In a search of the NCBI nonredundant database, peptide mass information identified 32 of the 43 protein spots analyzed, assigning them to seven proteins (Table 1). Several spots corresponded to the TfnR; ALCAM (also known as CD166), a member of the immunoglobulin superfamily with five extracellular immunoglobulin-like domains (28); and Dsg-2, a desmosomal cadherin (17). Only peptides encompassed within the extracellular domains of these three proteins were identified (Fig. 2A), suggesting that the corresponding proteins are indeed products of shedding. The rest of the proteins identified did not correspond to transmembrane proteins, which are the prototypical shedding substrates, and thus were not subsequently analyzed.
TABLE 1.
Proteins identified by mass spectrometry
| Protein | Accession no. | No. of spots | Avg (SD) fold decrease |
|---|---|---|---|
| Ribosomal protein RS.40 K | A31233 | 1 | 3.9 |
| Transferrin receptor | A93343 | 5 | 2.7 (0.23) |
| Extracellular matrix protein 1 | U65938 | 7 | 2.5 (0.40) |
| ALCAM | I39428 | 8 | 2.4 (0.22) |
| Annexin V | A67023 | 1 | 2.1 |
| β-Hexosaminidase alpha chain | A23561 | 3 | 1.6 (0.04) |
| Desmoglein 2 | S38673 | 7 | 1.6 (0.14) |
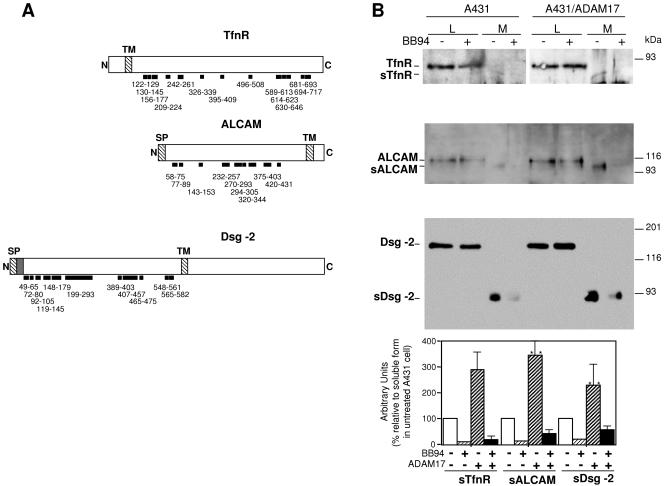
FIG. 2.
Analysis of the shedding of TfnR, ALCAM, and Dsg-2 in parental A431 cells and in the same cells transfected with ADAM17. A. Schematics showing the N terminus (N), signal peptide (SP), transmembrane region (TM), and C terminus (C) of the TfnR, ALCAM, and Dsg-2. The prodomain of Dsg-2, processed by furin-like proprotein convertases, is shown as a shaded box following the SP. The thick lines represent the peptides observed by mass spectrometry analysis and the numbers their location in each protein. B. Parental A431 cells or the same cells permanently transfected with ADAM17 were incubated with or without BB-94. Cells were washed, lysed, and cell lysates (L) analyzed by Western blotting with antibodies against the extracellular domains of TfnR, ALCAM, and Dsg-2 as indicated. Conditioned media (M) were concentrated by WGA-agarose chromatography and analyzed similarly. The protein loading corresponding to cell lysates was normalized with antiactin. The protein loading corresponding to medium samples was normalized by Ponceau red staining of the blots. These controls are not shown to simplify the figure. The averages from three independent experiments ± standard deviations are shown. **, Student's t test values (P < 0.01) comparing soluble material produced by A431 to that produced by parental ADAM17 cells.
To validate the results obtained by DIGE, we monitored the shedding of TfnR, ALCAM, and Dsg-2 by analyzing the levels of these proteins in lysates and conditioned media from parental and transfected A431 cells by Western blotting. The shedding of TfnR has been partially characterized; previous reports have shown that the TfnR is expressed as a cell surface ∼90-kDa transmembrane protein that can be shed, generating a ∼80-kDa soluble form (13, 19). In agreement with these results and with the analysis by DIGE, a ∼80-kDa species can be detected in the medium conditioned by A431 overexpressing ADAM17. The shedding of ALCAM has not been reported yet. In cell lysates we detected the expected ∼110-kDa species (28). The electrophoretic migration of the soluble form of ALCAM, present in the conditioned medium of A431/ADAM17 cells, corresponds to the expected molecular mass (∼100 kDa) of nearly the entire extracellular domain (Fig. 2A), arguing that it is a shedding product. The shedding of Dsg-2 is largely uncharacterized. A recent report shows that Dsg-2 is expressed as three cell-associated forms: a ∼160-kDa form, representing the full-length molecule, and fragments of ∼100 and ∼60 kDa (22). In addition, a soluble ∼60-kDa fragment accumulates in the extracellular medium of cells expressing Dsg-2 (22); since the production of this soluble fragment is lowered by metalloprotease inhibitors, it likely represents a shedding product (22). In lysates from A431/ADAM17 cells, we detected a main product likely corresponding to full-length Dsg-2 (Fig. 2B). In overexposed Western blots we also detected additional products likely corresponding to the 100-kDa fragment (data not shown; see also Fig. S2 in the supplemental material). In agreement with the results of Lorch et al. (22), in the conditioned medium we detected soluble Dsg-2 (sDsg-2), albeit of a higher molecular size (∼90 kDa) than that detected by those authors. Treatment with BB-94 largely prevented the accumulation of the soluble forms of the three proteins analyzed, confirming that they are shedding products.
As expected, parental cells produced significantly lower levels of the soluble extracellular domains of the proteins analyzed (Student's t test, P < 0.01); in fact, the extracellular domains of TfnR and ALCAM were barely detectable in the conditioned medium of parental A431 cells (Fig. 2B), probably because of the low affinity of the antibodies available. Thus, these results indicate that DIGE is an appropriate technique to identify shedding products; using this technique, we have identified TfnR, ALCAM, and Dsg-2 as shedding substrates that can be cleaved by overexpressed ADAM17.
Shedding of ALCAM and Dsg-2 in cells genetically deficient for ADAM17.
The analysis of transfected A431 indicates that when overexpressed, ADAM17 can act on the three molecules identified by DIGE. Since the shedding of TfnR has already been partially characterized (13, 19), we focused on the analysis of ALCAM and Dsg-2. To further explore the role of ADAM17 in the shedding of these molecules, we used M2, a mutant cell line derived from CHO cells which expresses only inactive pro-ADAM17 (Fig. 3A) (9, 31). Western blot analysis with specific antibodies did not reveal any specific band corresponding to Dsg-2 (data not shown), indicating that CHO cells do not express this molecule or, alternatively, that the antibodies used do not cross-react with the hamster ortholog. In contrast, several bands were detected with the antibody against ALCAM in CHO cells (Fig. 3B). Thus, we analyzed the shedding of endogenous ALCAM and transfected Dsg-2.
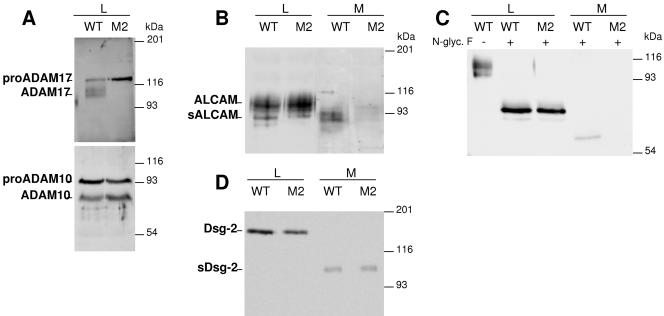
FIG. 3.
Analysis of the shedding of ALCAM and Dsg-2 in cells genetically deficient for ADAM 17. A. Wild-type CHO cells (WT) or M2 mutant cells were lysed and cell lysates analyzed by Western blotting with antibodies against ADAM17 or ADAM10 as indicated. B. Lysates (L) from wild-type CHO cells or M2 cell mutants and medium samples (M) obtained after 48 h of serum starvation were concentrated by WGA-agarose chromatography and analyzed by Western blotting with antibodies against the extracellular domain of ALCAM. C. Cell lysates and medium samples obtained as for panel B were treated without or with N-glycosidase F as indicated and analyzed by Western blotting as for panel B. D. WT CHO or M2 cells were transiently transfected with the cDNA encoding human Dsg-2. Cell lysates or conditioned media from transfected cells were analyzed by Western blotting with antibodies against the extracellular domain of Dsg-2.
Soluble ALCAM is readily detected in the conditioned medium of wild-type CHO cells. In contrast, the release of anti-ALCAM immunoreactive species to the medium conditioned by M2 cells was severely diminished, showing that the shedding of ALCAM is impaired in the mutant cells. Transfection of ADAM17 into M2 cells restored the ability to cleave ALCAM (data not shown). In contrast to the discrete band detected in A431 cells (Fig. 2B), the anti-ALCAM antibody stains several species in lysates from CHO cells (Fig. 3B). To determine whether these species correspond to differentially glycosylated forms of ALCAM, we repeated the experiment, treating proteins from cell lysates and conditioned media with N-glycosidase F. In deglycosylated protein samples one major band is detected (Fig. 3C), confirming the specificity of the different bands detected with anti-ALCAM. Thus, these results show that ADAM17 is largely responsible for the shedding of ALCAM in CHO cells.
Transfection of Dsg-2 leads to the generation of the expected cell-associated and soluble forms of the cell adhesion molecule. Unexpectedly, and in contrast with the results obtained for ALCAM, the shedding of Dsg-2 was not affected in M2 cells (Fig. 3D). This result indicates that ADAM17 has a role in the shedding of Dsg-2 only when overexpressed and shows that other proteases can act on the cell adhesion molecule.
Shedding of Dsg-2 in cells genetically deficient for ADAM10.
ADAM17 is most similar to ADAM10, and both metalloproteases are involved in protein ectodomain shedding. In fact, in some instances, such as in the case of the β-amyloid precursor protein (10), both ADAM17 and ADAM10 act on the same substrate. To determine the expression of ADAM10 in CHO cells, we analyzed cell lysates from CHO and M2 cells by Western blotting with polyclonal antibodies against ADAM10 previously used in our lab (9). The bands detected were identified as pro-ADAM10 and ADAM10 devoid of prodomain because they are also stained by a different anti-ADAM10 antibody and because of their shift in electrophoretic migration upon reduction (data not shown), a previously described hallmark of ADAMs (see, for example, reference 20).
Since ADAM10 is expressed by wild-type CHO cells as well as M2 mutants (Fig. 3A), this ADAM could be responsible for the shedding of Dsg-2. Thus, we analyzed the possible role of ADAM10 in the shedding of Dsg-2 by using MEFs derived from ADAM10 knockout mice. As expected, while no detectable ADAM10 species are expressed by knockout cells, the expression of ADAM17 remains unaltered (Fig. 4A). The shedding of Dsg-2 in MEFs from wild-type mice was normal. In contrast, no soluble form could be detected in the conditioned medium of ADAM10−/− cells, even in transfectants expressing high levels of Dsg-2 (Fig. 4B). These results indicate the requirement of ADAM10 for the shedding of the desmosomal cadherin and that the levels of ADAM17 expressed in knockout MEFs cannot compensate for the deficiency of ADAM10. To confirm the requirement of ADAM10, we cotransfected Dsg-2 with this metalloprotease in knockout MEFs. ADAM10 expression restored the generation of soluble Dsg-2 (Fig. 4B), showing the role of this metalloprotease in the shedding of Dsg-2.
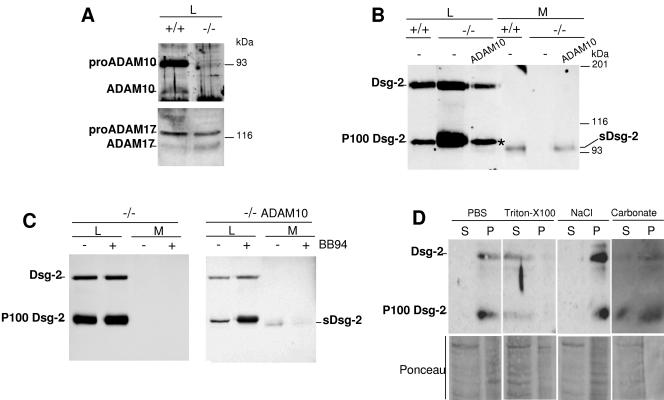
FIG. 4.
Analysis of the shedding of Dsg-2 in cells genetically deficient for ADAM10. A. MEFs from control mice (+/+) or ADAM10 knockout mice (−/−) were lysed and analyzed by Western blotting with polyclonal antibodies against ADAM10 or ADAM17. B. +/+ or −/− MEFs were transiently cotransfected with the cDNA encoding Dsg-2 and a control DNA (−) or the cDNA encoding ADAM10. Cell lysates (L) or conditioned media (M) obtained as for Fig. 3 from transfected cells were analyzed by Western blotting with antibodies against Dsg-2. C. −/− MEFs were transiently cotransfected with the cDNA encoding Dsg-2 and a control DNA (−/−) or ADAM10 (−/− ADAM10), treated with or without BB-94, and lysed. Cell lysates and medium samples were obtained and analyzed as for panel B. D. Microsomal fractions from ADAM10−/− MEFs permanently transfected with Dsg-2 were extracted with PBS, 1% Triton X-100, 1 M NaCl, or 0.1 M Na2CO3 (pH 12.0). Similar proportions of each fraction were analyzed by Western blotting with antibodies against Dsg-2. The efficiency of the extraction was verified by Ponceau red staining of total proteins.
In cell lysates from MEFs, in addition to the ∼160-kDa full-length form of Dsg-2 a ∼100-kDa species is apparent (Fig. 4B, P100 Dsg-2). In fact, in ADAM 10−/− MEFs this ∼100-kDa Dsg-2 species is predominant (Fig. 4B and C). Since the electrophoretic migration of the ∼100-kDa form is similar to that of sDsg-2, it could represent a shedding product that remains bound to cells. To test this possibility, we treated ADAM10−/− MEFs with BB-94. As shown in Fig. 4C, the metalloprotease inhibitor does not affect the levels of the P100 Dsg-2 (Fig. 4C, left), arguing that it is the product of a protease not inhibited by BB-94. Interestingly, transfection with ADAM10 decreased the levels of the P100 Dsg-2 band (Fig. 4C, right). Furthermore, treatment of transfected knockout cells with BB-94 upregulates P100 Dsg-2 (Fig. 4C, right). Collectively, these results indicate that Dsg-2 can be a substrate of two types of proteolytic activities, ADAM17 and ADAM10, which shed the ectodomain of Dsg-2, and of an unknown protease not inhibitable by BB-94, which leads to the generation of a cell-associated fragment (P100 Dsg-2).
To further characterize the association of P100 Dsg-2 with cells, we performed fractionation experiments. Upon extraction with high salt or high pH, all of the P100 Dsg-2 cofractionated with membranes, strongly indicating that this Dsg-2 species is an integral membrane protein. This result, along with the electrophoretic migration of P100 Dsg-2, indicates that the cleavage that generates P100 Dsg-2 occurs after the transmembrane and removes most of the cytoplasmic domain.
EGF concomitantly upregulates ADAM17 and the shedding of Dsg-2.
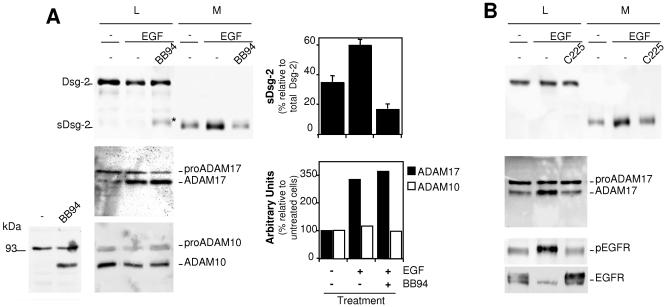
It has been recently shown that inhibitors of the EGFR increase the levels of cell-associated Dsg-2 fragments (22), opening the possibility that EGFR signaling regulates the shedding of Dsg-2. On the other hand, we have recently shown that prolonged EGFR activation leads to the upregulation of the expression of ADAM17 (B. Santiago-Josefat et al., submitted for publication). Thus, we analyzed the effect of EGF on the shedding of Dsg-2. As shown in Fig. 5A, treatment with EGF leads to an increase in the levels of sDsg-2. The effect of EGF can be largely prevented by BB-94, indicating that the activation of the EGFR leads to the upregulation of the shedding of Dsg-2. In agreement with the results shown in Fig. 4C, treatment with BB-94 leads to an increase in the levels of P100 Dsg-2 (Fig. 5A, asterisk).
FIG. 5.
Analysis of the shedding of Dsg-2 in cells treated with EGF. A. Parental A431 cells were treated with or without EGF and BB-94 as indicated and lysed. Cell lysates (L) and medium samples (M) were analyzed by Western blotting with antibodies against Dsg-2, ADAM17, or ADAM10 as indicated. Note that when a lysis buffer without BB-94 is used, only pro-ADAM10 is detected in cell lysates from A431 cells (bottom left). Addition of BB-94 allows the detection of mature ADAM10 (bottom left), and thus a buffer containing this metalloprotease inhibitor was used in the right bottom panel. Western blots were quantified, and the averages from three independent experiments ± standard deviations or the averages from two experiments are shown. B. A431 cells were treated with or without EGF and C225 (a monoclonal antibody that blocks the activation of the EGFR) for 48 h. Cell lysates and medium samples were analyzed by Western blotting with antibodies against Dsg-2, ADAM17, phospho-EGFR, or EGFR as indicated.
To determine the protease most likely involved in the shedding of Dsg-2, we analyzed the levels of ADAM17 and ADAM10 in the same cell lysates. In agreement with recent results (Santiago-Josefat et al., submitted), mature ADAM17 is upregulated by EGF treatment (Fig. 5A). In contrast to the results shown in Fig. 3A and 4A, analysis of A431 cell lysates with antibodies against ADAM10 showed only one band corresponding to pro-ADAM10 (Fig. 5A, bottom left). Addition of BB94 to the lysis buffer leads to the detection of mature ADAM10 in the lysates of A431 cells (Fig. 5A, bottom left). This result indicates that A431 cells contain a metalloprotease inhibited by BB94 that degrades ADAM10. Therefore, to analyze ADAM10 in A431 cells, we added BB94 to the lysis buffer. Under these conditions, both forms of ADAM10 are readily detected (Fig. 5A, bottom center), and they remain largely unchanged in cells treated with EGF. Thus, these results strongly suggest that ADAM17 is involved in the upregulation of the shedding of Dsg-2.
As a control of the effect of EGF, we repeated the experiment in the presence of C225, an antibody directed against the extracellular domain of the EGFR that prevents the ligand-mediated activation of the receptor (32). The blockade of EGFR activation by C225 also resulted in the concomitant blockade of the increase in the shedding of Dsg-2 and in the upregulation of ADAM17, further linking both processes. As expected, C225 prevented the increase in the phosphorylation of the EGFR induced by EGF, as judged by immunoblotting with anti-phospho-EGFR (Fig. 5B). Note that, as previously described (32), the long-term treatment with EGF used in this experiment leads to the downmodulation of the receptor (Fig. 5B), which is also prevented by treatment with C225 (Fig. 5B).
DISCUSSION
Increasing evidence shows that the function of extracellular metalloproteases goes far beyond the degradation of the extracellular matrix (24). In addition, they regulate several aspects of the cellular behavior; some of them, such as cell proliferation and migration, are clearly related to the development of cancer. Recent results show that, despite the negative outcome of clinical trials performed with nonspecific MPIs (12), certain metalloproteases can still be considered potential targets of tumor therapy (see, for example, reference 7). However, the inhibition of others may favor tumor development (4). Thus, it appears mandatory to characterize in detail the functions of individual metalloproteases and define which ones should be inhibited. Consequently, the functional characterization of individual metalloproteases, through the identification of their repertoire of substrates, i.e., their degradome (21), remains a current challenge, which is complicated by the fact that different metalloproteases can act on one substrate. Furthermore, metalloproteases are frequently overexpressed in tumors, and overexpressed metalloproteases may have an expanded repertoire of substrates.
In order to identify novel metalloprotease substrates we compared by DIGE the extracellular proteomes of cells treated with or without BB-94, a hydroxamic acid-derived nonspecific metalloprotease inhibitor. We used cells overexpressing ADAM17 because of the potential interest of this metalloprotease. ADAM17 sheds the extracellular domains of several ligands of the EGFR (26), some of which are inactive when membrane anchored (8). Blockage of this metalloprotease causes the inhibition of the EGFR (8, 14), a tyrosine kinase inhibited by several anticancer drugs currently used clinically (5). However, ADAM17 acts on dozens, perhaps hundreds, of cell surface molecules (2). Therefore, a detailed definition of the degradome of ADAM17 is required before it is considered further as a therapeutic target.
The results obtained by DIGE show that the shedding of TfnR, ALCAM, and Dsg-2 is upregulated in A431 cells transfected with ADAM17, arguing that, when overexpressed, this metalloprotease can act on them. The impaired shedding of ALCAM in mutant cells defective in ADAM17 activity confirms a central role of this metalloprotease in the shedding of the cell adhesion molecule. In contrast, further characterization of the shedding of Dsg-2 in the same cells clearly shows the participation of another metalloprotease(s). ADAM10 seems to play a major role, since no shedding of Dsg-2 is apparent in MEFs from knockout mice. Thus, both ADAM10 and, when overexpressed, ADAM17 seem to act on Dsg-2.
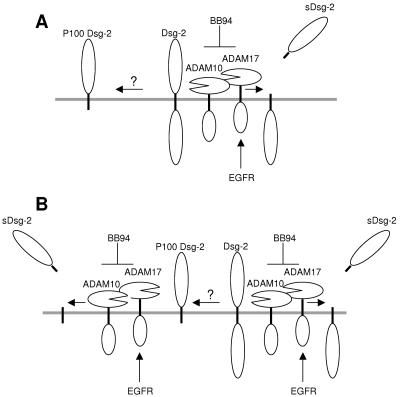
P100 Dsg-2 is a Dsg-2 species with a slightly slower electrophoretic mobility than that of sDsg-2. Biochemical fractionation indicates that P100 Dsg-2 is an integral membrane protein and, thus, that it contains the transmembrane domain. This Dsg-2 fragment is particularly apparent in ADAM10−/− MEFs or in cells treated with BB-94. A hypothetical competition between two types of proteases, i.e., on one hand ADAM10 and -17 and on the other hand an unknown proteolytic activity endowed with the ability to cleave the intracellular domain of Dsg-2, would explain these results (Fig. 6A). Alternatively, P100 Dsg-2 could also be a substrate of ADAM10 and ADAM17 (Fig. 6B). Inhibition of ADAM10 (by genetic ablation) or of both ADAM10 and ADAM17 (by BB-94) would increase the availability of Dsg-2 to be cleaved by the unknown protease (Fig. 6A) and prevent the shedding of the ectodomain of P100 Dsg-2, leading to its accumulation (Fig. 6B).
FIG. 6.
Hypothetical models for the generation of different Dsg-2 species. A. Schematic showing the shedding of Dsg-2 by ADAM10 and ADAM17. Activation of the EGFR leads to the upregulation of ADAM17 and the increase of the production of sDsg-2. Inhibition of ADAM10 and ADAM17 leads to the increase in the availability of Dsg-2 and the cleavage by an unidentified proteolytic activity that generates cell-associated P100 Dsg-2. B. Both full-length Dsg-2 and P100 Dsg-2 could be substrates of ADAM10 and ADAM17.
The γ-secretase complex cleaves transmembrane proteins within the transmembrane domain, releasing the cytoplasmic moiety (1, 15). Presenilins are the catalytic core of the γ-secretase complex (1, 15). Treatment with L685,458, a γ-secretase inhibitor that efficiently prevents the γ-secretase site cleavage of APP (see Fig. S2A, right, in the supplemental material), had no effect on the generation of P100 Dsg-2 (see Fig. S2A, left, in the supplemental material). Furthermore, using cells derived from Presenilin-1 knock out mice, which display a highly reduced γ-secretase activity, we did not detect a reduction in the levels of P100 Dsg-2 (see Fig. S2B in the supplemental material). Inhibitors of calpains or caspase-3 that prevent the release of the intracellular domains of certain transmembrane proteins (see, for example, references 16 and 23) or proteasome inhibitors were also without effect (see Fig. S2C in the supplemental material), indicating that these proteases do not significantly contribute to the generation of P100 either. Thus, the mechanism responsible for the production of P100 Dsg-2 does not seem to be mediated by these proteolytic activities and awaits further characterization.
Based on published results (17, 22, 28), it is expected that an increased level of ALCAM and Dsg-2 shedding will result in decreased cell-cell adhesion. Since both molecules support cell-cell adhesion by establishing homo- or heterotypic interactions, a decreased level of full-length ALCAM and Dsg-2 at the cell surface will reduce cell-cell contacts. In addition, soluble forms of these cell adhesion molecules, encompassing most of the extracellular domain, may act as dominant-negative forms competing for the interaction between full-length forms located in adjacent cells.
In summary, our results show that proteomic analysis by DIGE is a useful method to identify shedding substrates. We have identified two novel substrates of ADAM17, Dsg-2 and ALCAM, both of which are adhesion molecules that mediate cell-cell interactions. Presumably, shedding of these cell adhesion molecules weakens cell-cell adhesion. In tumor cells, which frequently overexpress ADAM17 (8) and overactivated EGFR (32), the shedding of Dsg-2 and ALCAM is expected to be higher than in normal cells. Thus, ADAM17 may contribute to the loss of cell adhesion, a key characteristic of cancer cells (17), and therefore its inhibition may be beneficial for tumor treatment.
Supplementary Material
Acknowledgments
We thank Juan Carlos Rodriguez-Manzaneque for critical reading of the manuscript.
This work was supported by grants from Fundació “La Caixa” (BM 05-208-00) and from the European Comission (FP6-503228) to J.A. and from Fondo de Investigaciones Sanitarias (FIS PI040793) to F.C. The Proteomics Laboratory is a member of the National Spanish Institute for Proteomics (PROTEORED). J.J.B.-S. is supported by the AGAUR (Agencia de Gestió d'Ajuts Universitaris i de Recerca). B.S.-J. and C.E. are postdoctoral fellows from the Vall d'Hebron University Hospital Research Institute and Juan de la Cierva program (Spanish Ministry of Education), respectively.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Annaert, W., and B. De Strooper. 2002. A cell biological perspective on Alzheimer's disease. Annu. Rev. Cell Dev. Biol. 18:25-51. [DOI] [PubMed] [Google Scholar]
- 2.Arribas, J., and A. Borroto. 2002. Protein ectodomain shedding. Chem. Rev. 102:4627-4638. [DOI] [PubMed] [Google Scholar]
- 3.Baker, A. H., D. R. Edwards, and G. Murphy. 2002. Metalloproteinase inhibitors: biological actions and therapeutic opportunities. J. Cell Sci. 115:3719-3727. [DOI] [PubMed] [Google Scholar]
- 4.Balbin, M., A. Fueyo, A. M. Tester, A. M. Pendas, A. S. Pitiot, A. Astudillo, C. M. Overall, S. D. Shapiro, and C. Lopez-Otin. 2003. Loss of collagenase-2 confers increased skin tumor susceptibility to male mice. Nat. Genet. 35:252-257. [DOI] [PubMed] [Google Scholar]
- 5.Baselga, J., and C. L. Arteaga. 2005. Critical update and emerging trends in epidermal growth factor receptor targeting in cancer J. Clin. Oncol. 23:2445-2459. (Erratum, 23:6281.) [DOI] [PubMed] [Google Scholar]
- 6.Blobel, C. P. 2005. ADAMs: key components in EGFR signalling and development. Nat. Rev. Mol. Cell. Biol. 6:32-43. [DOI] [PubMed] [Google Scholar]
- 7.Boire, A., L. Covic, A. Agarwal, S. Jacques, S. Sherifi, and A. Kuliopulos. 2005. PAR1 is a matrix metalloprotease-1 receptor that promotes invasion and tumorigenesis of breast cancer cells. Cell 120:303-313. [DOI] [PubMed] [Google Scholar]
- 8.Borrell-Pagès, M., F. Rojo, J. Albanell, J. Baselga, and J. Arribas. 2003. TACE is required for the activation of the EGFR by TGF-alpha in tumors. EMBO J. 22:1114-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borroto, A., S. Ruiz-Paz, T. Villanueva de la Torre, M. Borrell-Pages, A. Merlos-Suarez, A. Pandiella, C. P. Blobel, J. Baselga, and J. Arribas. 2003. Impaired trafficking and activation of TACE in cell mutants defective in protein ectodomain shedding. J. Biol. Chem. 278:25933-25939. [DOI] [PubMed] [Google Scholar]
- 10.Buxbaum, J. D., K. N. Liu, Y. Luo, J. L. Slack, K. L. Stocking, J. J. Peschon, R. S. Johnson, B. J. Castner, D. P. Cerretti, and R. A. Black. 1998. Evidence that tumor necrosis factor alpha converting enzyme is involved in regulated alpha-secretase cleavage of the Alzheimer amyloid protein precursor. J. Biol. Chem. 273:27765-27767. [DOI] [PubMed] [Google Scholar]
- 11.Coughlin, S. R. 2000. Thrombin signalling and protease-activated receptors. Nature 407:258-264. [DOI] [PubMed] [Google Scholar]
- 12.Coussens, L. M., B. Fingleton, and L. M. Matrisian. 2002. Matrix metalloproteinase inhibitors and cancer—trials and tribulations. Science 295:2387-2392. [DOI] [PubMed] [Google Scholar]
- 13.Dassler, K., M. Kaup, R. Tauber, and H. Fuchs. 2003. Mutational suppression of transferrin receptor shedding can be compensated by distinct metalloproteases acting on alternative sites. FEBS Lett. 536:25-29. [DOI] [PubMed] [Google Scholar]
- 14.Dong, J., L. K. Opresko, P. J. Dempsey, D. A. Lauffenburger, R. J. Coffey, and H. S. Wiley. 1999. Metalloprotease-mediated ligand release regulates autocrine signaling through the epidermal growth factor receptor. Proc. Natl. Acad. Sci. USA 96:6235-6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fortini, M. E. 2002. Signalling: gamma-secretase-mediated proteolysis in cell-surface-receptor signalling. Nat. Rev. Mol. Cell. Biol. 3:673-684. [DOI] [PubMed] [Google Scholar]
- 16.Franco, S. J., and A. Huttenlocher. 2005. Regulating cell migration: calpains make the cut. J. Cell Sci. 118:3829-3838. [DOI] [PubMed] [Google Scholar]
- 17.Getsios, S., A. C. Huen, and K. J. Green. 2004. Working out the strength and flexibility of desmosomes. Nat. Rev. Mol. Cell. Biol. 5:271-281. [DOI] [PubMed] [Google Scholar]
- 18.Guo, L., J. R. Eisenman, R. M. Mahimkar, J. J. Peschon, R. J. Paxton, R. A. Black, and R. S. Johnson. 2002. A proteomic approach for the identification of cell-surface proteins shed by metalloproteases. Mol. Cell Proteomics 1:30-36. [DOI] [PubMed] [Google Scholar]
- 19.Kaup, M., K. Dassler, C. Weise, and H. Fuchs. 2002. Shedding of the transferrin receptor is mediated constitutively by an integral membrane metalloprotease sensitive to tumor necrosis factor alpha protease inhibitor-2. J. Biol. Chem. 277:38494-38502. [DOI] [PubMed] [Google Scholar]
- 20.Krätzschmar, J., L. Lum, and C. P. Blobel. 1996. Metargidin, a membrane anchored metalloproteases-disintegrin protein with an RGD integrin binding sequence. J. Biol. Chem. 271:4593-4596. [DOI] [PubMed] [Google Scholar]
- 21.Lopez-Otin, C., and C. M. Overall. 2002. Protease degradomics: a new challenge for proteomics. Nat. Rev. Mol. Cell. Biol. 3:509-519. [DOI] [PubMed] [Google Scholar]
- 22.Lorch, J. H., J. Klessner, J. K. Park, S. Getsios, Y. L. Wu, M. S. Stack, and K. J. Green. 2004. Epidermal growth factor receptor inhibition promotes desmosome assembly and strengthens intercellular adhesion in squamous cell carcinoma cells. J. Biol. Chem. 279:37191-37200. [DOI] [PubMed] [Google Scholar]
- 23.Marambaud, P., J. Shioi, G. Serban, A. Georgakopoulos, S. Sarner, V. Nagy, L. Baki, P. Wen, S. Efthimiopoulos, Z. Shao, T. Wisniewski, and N. K. Robakis. 2002. A presenilin-1/gamma-secretase cleavage releases the E-cadherin intracellular domain and regulates disassembly of adherens junctions. EMBO J. 21:1948-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mott, J. D., and Z. Werb. 2004. Regulation of matrix biology by matrix metalloproteinases. Curr. Opin. Cell Biol. 16:558-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Overall, C. M., and C. Lopez-Otin. 2002. Strategies for MMP inhibition in cancer: innovations for the post-trial era. Nat. Rev. Cancer 2:657-672. [DOI] [PubMed] [Google Scholar]
- 26.Sahin, U., G. Weskamp, K. Kelly, H. M. Zhou, S. Higashiyama, J. Peschon, D. Hartmann, P. Saftig, and C. P. Blobel. 2004. Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J. Cell Biol. 164:769-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins on silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 28.Swart, G. W. M. 2002. Activated leukocyte cell adhesion molecule (CD166/ALCAM): developmental and mechanistic aspects of cell clustering and cell migration. Eur. J. Cell Biol. 8:313-321. [DOI] [PubMed] [Google Scholar]
- 29.Tam, E. M., C. J. Morrison, Y. I. Wu, M. S. Stack, and C. M. Overall. 2004. Membrane protease proteomics: isotope-coded affinity tag MS identification of undescribed MT1-matrix metalloproteinase substrates. Proc. Natl. Acad. Sci. USA 101:6917-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van den Bergh, G., and L. Arckens. 2004. Fluorescent two-dimensional difference gel electrophoresis unveils the potential of gel-based proteomics. Curr. Opin. Biotechnol. 15:38-43. [DOI] [PubMed] [Google Scholar]
- 31.Villanueva de la Torre, T., J. J. Bech-Serra, S. Ruiz-Paz, J. Baselga, and J. Arribas. 2004. Inactivating mutations block the tumor necrosis factor-alpha-converting enzyme in the early secretory pathway. Biochem. Biophys. Res. Commun. 314:1028-1035. [DOI] [PubMed] [Google Scholar]
- 32.Yarden, Y., and M. X. Sliwkowski. 2001. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell. Biol. 2:127-137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.