Abstract
Recruitment of DNA polymerases onto replication origins is a crucial step in the assembly of eukaryotic replication machinery. A previous study in budding yeast suggests that Dpb11 controls the recruitment of DNA polymerases α and ɛ onto the origins. Sld2 is an essential replication protein that interacts with Dpb11, but no metazoan homolog has yet been identified. We isolated Xenopus RecQ4 as a candidate Sld2 homolog. RecQ4 is a member of the metazoan RecQ helicase family, and its N-terminal region shows sequence similarity with Sld2. In Xenopus egg extracts, RecQ4 is essential for the initiation of DNA replication, in particular for chromatin binding of DNA polymerase α. An N-terminal fragment of RecQ4 devoid of the helicase domain could rescue the replication activity of RecQ4-depleted extracts, and antibody against the fragment inhibited DNA replication and chromatin binding of the polymerase. Further, N-terminal fragments of RecQ4 physically interacted with Cut5, a Xenopus homolog of Dpb11, and their ability to bind to Cut5 closely correlated with their ability to rescue the replication activity of the depleted extracts. Our data suggest that RecQ4 performs an essential role in the assembly of replication machinery through interaction with Cut5 in vertebrates.
The initiation of eukaryotic DNA replication involves the assembly of highly conserved replication proteins on replication origins (3, 15, 24). Before the initiation of replication, prereplicative complexes (pre-RCs) are assembled onto the origins at the end of M phase and during G1 phase. The assembly of pre-RCs, referred to as “licensing of origins” (5), is absolutely dependent on the binding of origin recognition complex to origins, which allows the assembly of Cdc6 and Cdt1 and finally leads to the recruitment of minichromosome maintenance proteins (Mcm2 to -7) onto origins (28). At the onset of S phase, the pre-RCs are activated through the action of cyclin-dependent kinase (CDK) and Dbf4-dependent Cdc7 kinase, being converted into the preinitiation complexes, and eventually DNA polymerases are assembled into replication machinery at the origins.
The recruitment of DNA polymerases onto origins is a crucial step in the assembly of replication machinery. A previous study with budding yeast (Saccharomyces cerevisiae) suggests that Dpb11 controls the recruitment of DNA polymerases α and ɛ onto origins (22). Studies with metazoan homologs of Dpb11, in particular with Xenopus laevis Cut5/Mus101, support this view, showing that the metazoan protein is required for the loading of DNA polymerases onto chromatin (8, 33). Further insights into components essential for assembly of the machinery were obtained by Araki and coworkers, who isolated a group of six genes that interact with DPB11, named SLD (“synthetically lethal with dpb11-1”) (12). Proteins Sld1, -4, and -6 are identical to Dpb3 (the third largest subunit of DNA polymerase ɛ), Cdc45, and Rad53 (a checkpoint kinase), respectively. SLD2, -3, and -5 were novel genes and are essential for the growth of yeast. The protein Sld5 is highly conserved among eukaryotes, and yeast and Xenopus Sld5 form a heterotetrameric GINS complex with three Sld5-interacting proteins, Psf1, -2, and -3 (14, 19, 32). This complex is required for the recruitment of Cdc45 onto chromatin, a crucial step in the assembly of DNA polymerases on chromatin (19, 32).
So far only fission yeast homologs of budding yeast Sld2 (also isolated as DRC1 [35], a dosage suppressor of dpb11-1) and Sld3 have been identified (27, 29). Sld3 shows some different behaviors in the cell cycle between fission and budding yeasts (13, 27), but Sld2/Drc1 apparently retains similar functions in the initiation of DNA replication. Both budding and fission yeast Sld2/Drc1 are novel targets of S-phase CDK (S-CDK), and their phosphorylation is essential for chromosomal DNA replication (23, 29). In budding yeast, CDK-dependent phosphorylation of Sld2 is apparently required for physical interaction with Dpb11, and such interaction plays an important role in the assembly of replication machinery (23). Similarly in fission yeast, a mutant of Drc1 lacking putative CDK-dependent phosphorylation sites failed to interact with Cut5, a fission yeast homolog of Dpb11 (29). It is not known, however, whether Sld2/Drc1 is conserved among eukaryotes.
In this study, we isolated Xenopus RecQ4 as a candidate Sld2 homolog. The N-terminal region of RecQ4 shows some sequence similarity with Sld2, and it interacted with Cut5/TopBP1 in egg extracts. The region is required for the initiation of DNA replication, in particular for chromatin binding of DNA polymerase α. In humans, mutations in RecQ4 are responsible for some cases of Rothmund-Thomson syndrome (2, 16, 20), suggesting that RecQ4 is important for preventing genome instability and that mutations in it might lead to cancer or premature aging (1, 7, 9). Here we report that vertebrate RecQ4 performs an essential role in the formation of replication machinery through interaction with Cut5. While this work was in progress, a paper was published suggesting that RecQ4 is required for the initiation of DNA replication in higher eukaryotes (30).
MATERIALS AND METHODS
Cloning and sequencing of Xenopus RecQ4.
We obtained a partial sequence of Xenopus RecQ4 from the expressed sequence tag (EST) database (accession no. BE507000) by a homology search with yeast Sld2/Drc1. The sequence was amplified from Xenopus oocyte cDNA by PCR using a 5′ primer (AGGCATGGAGCGCTATAATGAGG) and a 3′ primer (TCAAGAGGTCGGAAACGAGGAGT). The amplified PCR sequence was used as a probe to screen a λZAP-derived cDNA library of Xenopus oocyte mRNA (18). From 3 × 105 plaques, we isolated three positive clones with a maximum insert size of 4.6 kb. The three clones were sequenced with an automatic DNA sequencer (ABI 311). The full-length Xenopus RecQ4 gene (accession no. AB213025) consists of 4691 bp, containing a 5′ untranslated region of 30 bp, an open reading frame of 4,512 bp, and a 3′ untranslated region of 149 bp.
Protein expression and antibody production.
Recombinant six-His-tagged full-length and N-terminal fragments of Xenopus RecQ4 (corresponding to amino acids 1 to 334, 1 to 596, and 1 to 727) were obtained by using a Bac-to-Bac HT recombinant baculovirus expression system (Invitrogen). The recombinant proteins were expressed in Sf9 insect cells and purified through a Ni-nitrilotriacetic acid column (QIAGEN) following the instructions of the supplier. Recombinant glutathione S-transferase (GST)-Cut5 (corresponding to amino acids 1197 to 1513) and the six-His-tagged N-terminal fragment of RecQ4 (corresponding to amino acids 1 to 596) were expressed in Escherichia coli (lon-minus strain) and purified as described previously (18). Polyclonal rabbit antibodies were raised against the recombinant proteins (Hokudo Inc., Japan) and further affinity purified with recombinant protein immobilized in Affi-Gel 10 (Bio-Rad).
Preparation of Xenopus egg extract and chromatin fractions.
Interphase egg extract and demembranated sperm nuclei were prepared as described previously (17). Aphidicolin (Wako) was added to the egg extract at a concentration of 40 μg/ml to inhibit DNA polymerases. GST-p21 (25) and roscovitine (Sigma) were added to the extracts at 50 μg/ml and 200 μM, respectively, to inhibit CDK. Green fluorescent protein-geminin (37) was added at 15 μg/ml to inhibit the licensing reaction. To isolate chromatin fractions, we incubated sperm nuclei in 50 to 100 μl of the egg extracts (4,000 nuclei/μl) for appropriate times at 23°C. The samples were diluted with 10 volumes of extraction buffer (EB; 100 mM KCl, 2.5 mM MgCl2, 50 mM HEPES-KOH, pH 7.5) containing 0.25% NP-40 (Wako) and then centrifuged through a 10% sucrose layer at 10,000 × g for 5 min, and then the pellets were washed with EB. The chromatin-associated proteins were extracted with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer from the pellets and separated from the DNA by passage through a 0.45-μm filter (Millipore; Ultrafree-MC).
Assay for DNA replication activity.
The replication activities of the egg extracts were measured as the incorporation of [α-32P]dCTP into sperm DNA. [α-32P]dCTP was incorporated as described previously (24) except that the autoradiography was quantified by Image Gauge software (Fuji Film). For Cy3-dCTP incorporation, the method was as described below in “Fluorescence microscopy.”
Fluorescence microscopy.
Sperm nuclei (4,000/μl) were incubated in the egg extract containing 10 μM Cy3-dCTP (Amersham Biosciences) for appropriate times at 23°C. The samples were prepared as described previously (26), and fluorescence images were captured with the OpenLab imaging program (Improvision) and analyzed with NIH Image software to estimate the amounts of chromatin-bound proteins and the replication activities of the egg extracts.
Immunodepletion and immunoprecipitation.
Xenopus proteins were immunodepleted as described previously (25) except that rProtein A Sepharose Fast Flow (Amersham Biosciences) was used instead of Affi-Prep protein A matrix, and the antibodies were cross-linked to the rProtein A with dimethyl pimelimidate (Pierce). The following antibodies were used for mock and RecQ4 depletion: mock depletion, control antibody purified from preimmune serum, and RecQ4 depletion, antibody against the N-terminal fragment corresponding to amino acids 1 to 727. The egg extracts were immunoprecipitated as described previously (25).
The pull-down assay of RecQ4 and Cut5 was performed as follows. Recombinant Cut5 (1.5 μg) was incubated with 1.5 μg anti-Cut5 antibody immobilized to rProtein A Sepharose beads (Amersham Biosciences) in 20 μl EB at 4°C for 30 min. The beads were washed three times with 500 μl EB and then further incubated with 1 μg recombinant six-His-tagged RecQ4 fragments in 20 μl EB at 4°C for 90 min. After three washes with EB, the beads were resuspended in SDS-PAGE sample buffer, and solubilized proteins were analyzed by SDS-PAGE. Western blot analysis was performed with the antibodies against six-His tag and Cut5. For phosphatase treatment, the recombinant N2-RecQ4 fragment was treated with 5.6 μg/ml protein phosphatase 1 (Sigma) with or without phosphatase inhibitor (1 μM okadaic acid) at 23°C for 60 min before incubation with the Cut5-conjugated beads. For rephosphorylation, the sample treated with the phosphatase was further incubated with 17 μg/ml cyclinA-Cdk2 (generously provided by T. Masuda-Sasa) at 23°C for 45 min in the presence of 1 μM okadaic acid to prevent the dephosphorylation.
RESULTS AND DISCUSSION
Identification of Xenopus RecQ4 as a candidate homolog of metazoan Sld2/Drc1.
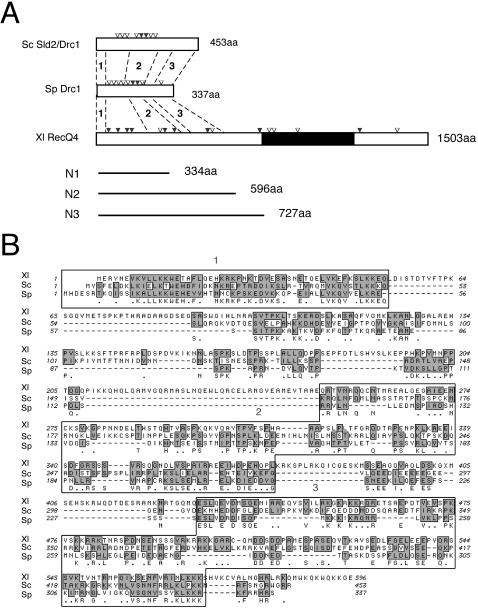
To identify a candidate Xenopus Sld2, we searched EST databases for a Xenopus homolog and obtained a Xenopus EST sequence with a degree of homology (accession no. BE507000). Using the EST sequence as a probe, we screened a Xenopus oocyte cDNA library and isolated a full-length cDNA encoding a putative Xenopus homolog (accession no. AB213025). The open reading frame encodes a protein of 1,503 amino acids, which is more than three times the length of yeast Sld2/Drc1 (Fig. 1A). The N-terminal region of about 50 amino acids shows about 30% identity and 50% similarity with that of yeast Sld2/Drc1, the N-terminal region from 251 to 373 amino acids shows about 20% identity and 30% similarity with the middle regions of yeast proteins, and the region from 430 to 570 amino acids shows about 20% identity and 35% similarity with the C-terminal regions of yeast Sld2/Drc1 (Fig. 1A). The C-terminal region of the Xenopus homolog contains a RecQ helicase domain, which is absent in the yeast Sld2/Drc1 (Fig. 1A). A further database search with the full-length Xenopus sequence revealed that it shares the highest similarity with mammalian RecQ4, whose mutation has been implicated in the generation of some cases of Rothmund-Thomson syndrome in humans (2, 16, 20). Because the sequence shows greater similarity to that of metazoan RecQ4 than to that of yeast Sld2/Drc1, we hereafter call this gene product Xenopus RecQ4.
FIG. 1.
Sequence comparison of Sld2, Drc1, and RecQ4. (A) Schematic structures of Sld2/Drc1 of Saccharomyces cerevisiae (Sc), Schizosaccharomyces pombe (Sp), and Xenopus laevis (Xl) RecQ4. Potential CDK phosphorylation motifs (S/T-P, S/T-P-K/R, and S/T-P-X-K/R) are indicated by open (one motif) and solid (two motifs) inverted triangles. The black region represents the RecQ helicase domain (amino acids 744 to 1086). Bold lines represent N-terminal fragments of Xenopus RecQ4 used in this study. Regions 1, 2, and 3 show the following identity and similarity (% identity/% similarity): Xl RecQ4/Sc Sld2, 30/55, 19/31, and 19/35; Xl RecQ4/Sp Drc1, 35/52, 16/28, and 23/34; and Sc Sld2/Sp Drc1, 29/55, 26/40, and 20/32, respectively. (B) Alignment of N-terminal amino acid sequences of Sc Sld2/Drc1, Sp Drc1, and Xl RecQ4. Identical amino acids are indicated just below the sequence of Sp Drc1, and conserved regions are shaded. Boxed regions show relatively high sequence similarities.
Chromatin binding of Xenopus RecQ4 during DNA replication.
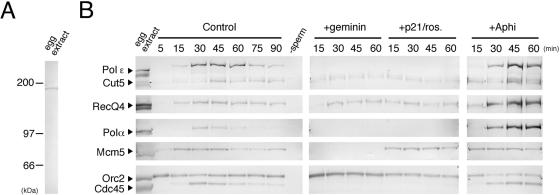
To investigate the function of Xenopus RecQ4 in egg extracts, we prepared three His-tagged N-terminal regions of Xenopus RecQ4 (Fig. 1A, shown as N1, N2, and N3) and the full-length protein. Since the yield of the full-length protein was not sufficient for production of antibody, we produced antibodies against the recombinant N-terminal fragments. The affinity-purified antibodies recognized a single polypeptide in the extracts with an apparent molecular mass of about 185 kDa, closely coinciding with the value calculated from the predicted sequence (Fig. 2A). In the following experiments, we have routinely used the antibody against the N3 fragment, which shows the highest avidity to endogenous RecQ4. The behavior of RecQ4 during DNA replication was then examined in an interphase extract of Xenopus eggs, which enables us to recapitulate the conserved features of eukaryotic DNA replication (26). Using sperm chromatin as templates, we found that the behavior of RecQ4 during DNA replication is similar to that of Cut5. Figure 2B compares the chromatin binding of RecQ4 with that of Cut5 and other replication proteins before and after the initiation of DNA replication. Upon addition of sperm chromatin to the extracts, Orc2, a component of origin recognition complex, bound to chromatin within 5 min, and the replication licensing monitored as chromatin binding of Mcm5 was apparently established within 15 min. After that, nuclear structures were formed around the chromatin within 30 min (data not shown), thus allowing initiation of DNA replication. Chromatin binding of RecQ4 was detected before the initiation of DNA replication, similar to that of Cut5. After the initiation, both RecQ4 and Cut5 behaved similarly to various replication proteins involved in the formation of replication machinery, such as Cdc45 and DNA polymerases α and ɛ. They bound maximally to chromatin at about 30 to 45 min, when replication was fully activated, and gradually dissociated from the chromatin during the progression of DNA replication, whereas Orc2 binding remained constant throughout the incubation. Dissociation of these replication proteins was blocked by inhibiting the activity of the DNA polymerases by aphidicolin, and both RecQ4 and Cut5 were markedly accumulated onto chromatin fractions, as was DNA polymerase α.
FIG. 2.
Chromatin binding of RecQ4 in interphase extracts of Xenopus eggs. (A) Specificity of anti-RecQ4 antibody. Interphase egg extract was resolved by SDS-PAGE and immunoblotted with anti-RecQ4 antibody. (B) Time course of chromatin binding of RecQ4 and other replication proteins under various conditions. Xenopus sperm chromatin was incubated in egg extracts at 23°C in the absence (Control) or the presence of 15 μg/ml green fluorescent protein-geminin (+geminin), 50 μg/ml GST-p21 plus 200 μM roscovitine (+p21/ros.), or 40 μg/ml aphidicolin (+Aphi) for the indicated times. The chromatin fractions isolated by centrifugation and the egg extract (1 μl) were resolved by SDS-PAGE and immunoblotted with antibodies indicated in the figure. Negative control of chromatin fractions was obtained by incubating the extracts in the absence of sperm chromatin (−sperm).
Because RecQ4 bound to the chromatin before the initiation of DNA replication, which requires S-CDK activity, it is apparent that CDK activity was dispensable for its chromatin binding. Figure 2B confirms that RecQ4 bound to chromatin in the presence of CDK inhibitors p21 and roscovitine. In the presence of inhibitors (“+p21/ros.”), and thus in the absence of CDK activity, the chromatin binding of Cdc45 and the polymerases was abolished, whereas the chromatin binding of RecQ4 was fairly constant during incubation and similar to those of Cut5, Mcm5, and Orc2. We further found that RecQ4 was bound to chromatin even though the formation of pre-RC was prevented by the addition of geminin (“+geminin”), an inhibitor of Cdt1 (31, 36), to the extracts. The chromatin binding of RecQ4 was delayed in the presence of geminin relative to that in its absence, and the chromatin binding of RecQ4 at 15 min of incubation was mostly suppressed in the presence of geminin. In a previous report the chromatin binding of RecQ4 in the extracts was apparently suppressed by geminin after 60 min of incubation (30), contradicting our present data showing a low level of chromatin binding of RecQ4 after 60 min of incubation in the presence of geminin. The exact reason for this contradiction is not known, but the delay in binding suggests that pre-RC formation is required for proper chromatin binding of RecQ4. The increased chromatin binding of RecQ4 observed after the initiation of DNA replication was completely suppressed by the addition of CDK inhibitors or geminin. Conversely, these results show that maximal binding of RecQ4 was dependent on pre-RC formation and S-CDK activity, a situation that is similar to that of Cut5 (8).
Xenopus RecQ4 is required for DNA replication in egg extracts.
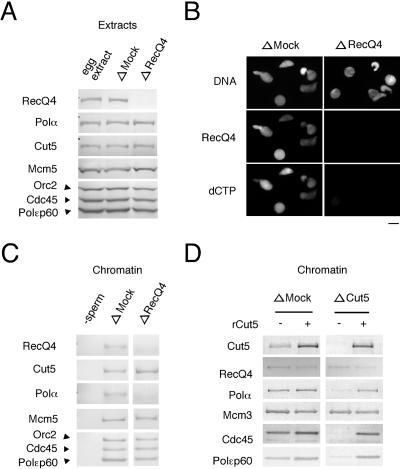
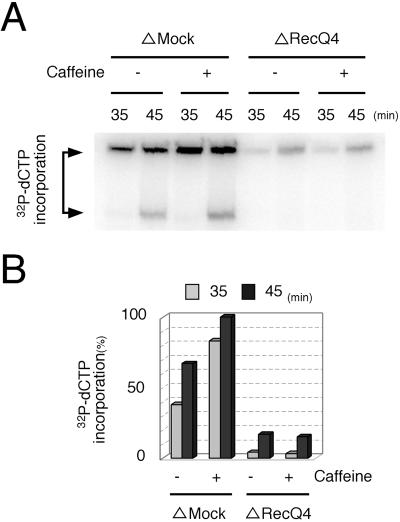
To investigate the involvement of Xenopus RecQ4 in DNA replication, we first compared the replication activities of mock- and RecQ4-depleted egg extracts. Upon treatment of the extracts with anti-RecQ4 antibody-conjugated beads, the replication proteins so far examined in the extracts were not appreciably decreased, but RecQ4 was decreased to a small fraction of mock-depleted extracts (Fig. 3A). Immunofluorescence observation of nuclei formed in treated extracts further showed that RecQ4 was diminished in the nuclei formed in the RecQ4-depleted extracts and that the DNA replication activity measured as the incorporation of fluorescently labeled dCTP into DNA was suppressed in the nuclei formed in RecQ4-depleted extract (Fig. 3B). In contrast, the incorporation of dCTP and localization of RecQ4 coincided in the nuclei formed in mock-depleted extracts (Fig. 3B, compare ΔMock and ΔRecQ4). On depleting RecQ4, we could not detect any appreciable defect in nuclear envelope formation (data not shown), and the morphology of the nuclear chromatin was essentially the same in mock- and RecQ4-depleted extracts (Fig. 3B; also see Fig. 5A). The inhibition of DNA replication by depleting RecQ4 was not due to the activation of checkpoint reactions, which leads to the inhibition of DNA replication (21). Caffeine, which inhibits the activation of the checkpoint, accelerated the replication of mock-depleted extracts but did not show any significant effect on the replication activity of RecQ4-depleted extracts (Fig. 4).
FIG. 3.
Effect of RecQ4 depletion from the egg extract on DNA replication and chromatin binding of DNA polymerase α. (A) Depletion of RecQ4 from the extracts. The egg extracts were treated with preimmune or anti-RecQ4 antibodies conjugated to protein A beads. Untreated egg extract and mock- and RecQ4-depleted extracts (1 μl) were resolved by SDS-PAGE and immunoblotted with the antibodies indicated in the figure. (B) Immunofluorescent detection of nuclear RecQ4. Xenopus sperm chromatin was incubated in mock- and RecQ4-depleted extracts for 40 min at 23°C. The samples were treated with 0.1% NP-40 and then fixed with 3.7% formaldehyde in EB. Nuclear localization of RecQ4 was visualized with rabbit anti-RecQ4 antibody followed by Alexa 488-labeled anti-rabbit immunoglobulin G. DNA replications were monitored as the incorporation of Cy3-dCTP into DNA, and DNA was visualized with Hoechst 33258 dye. Fluorescence images were captured with the OpenLab imaging program (Improvision). (C) Chromatin binding of Cut5 in the absence of RecQ4. Xenopus sperm chromatin was incubated in mock- or RecQ4-depleted extracts for 40 min at 23°C. Isolated chromatin fractions were resolved by SDS-PAGE and immunoblotted with the antibodies indicated. (D) Chromatin binding of RecQ4 in the absence of Cut5. Xenopus sperm chromatin was incubated in mock- or Cut5-depleted extracts with (+) or without (−) recombinant Cut5 for 40 min at 23°C.
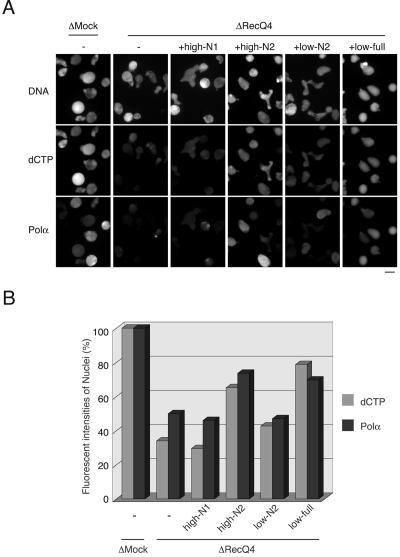
FIG. 5.
Rescue of DNA replication activity and nuclear localization of DNA polymerase α by recombinant RecQ4 in RecQ4-depleted extracts. (A) Sperm chromatin was incubated in mock (ΔMock)- or RecQ4 (ΔRecQ4)-depleted extracts in the absence (−) or presence of full-length RecQ4 (+low-full, 2 μg/ml), N2-RecQ4 (+low-N2, 0.8 μg/ml; +high-N2, 16 μg/ml), or N1-RecQ4 (+high-N1, 9 μg/ml) at 23°C for 40 min. The samples were treated for 2 min with 0.1% Triton X-100 in EB containing an additional 100 mM NaCl and then fixed with 3.7% formaldehyde. Nuclear localization of DNA polymerase α was visualized with rabbit anti-polymerase α antibody followed by Alexa 488-labeled anti-rabbit immunoglobulin G. DNA replication was monitored as the incorporation of Cy3-dCTP into DNA, and DNA was visualized with Hoechst 33258 dye. (B) The fluorescent intensity of each nucleus was quantified using NIH Image software, and average intensities of over 40 nuclei were normalized by the intensities of the nuclei from mock-depleted extract.
FIG. 4.
Effect of caffeine on DNA replication in mock- or RecQ4-depleted extracts. (A) Xenopus sperm chromatin was incubated in mock- (ΔMock) or RecQ4-depleted (ΔRecQ4) extracts with (+) or without (−) 5 mM caffeine for indicated times at 23°C. DNA replication activity was monitored as the incorporation of [α-32P]dCTP into DNA. At the times indicated, DNA was subjected to agarose gel electrophoresis and then autoradiographed. [α-32P]dCTP incorporation was carried out as described previously (24) except that the autoradiography was quantified by Image Gauge software (Fuji Film). (B) The amount of 32P incorporated into DNA in panel A was quantified, taking the highest value of the mock-depleted extracts as 100%.
Examination of chromatin fractions isolated after nuclear formation further confirmed that the nuclear formation and consequent activation of CDK were intact in RecQ4-depleted extracts. Figure 3C shows that the chromatin binding of both Cdc45 and DNA polymerase ɛ, which depends on CDK activity (26) and thus happens only after nuclear formation, was not affected by the depletion of RecQ4, but the chromatin binding of RecQ4 was diminished in the depleted extracts. Intriguingly, the depletion of RecQ4 markedly suppressed the chromatin binding of DNA polymerase α without affecting the binding of Cdc45 (Fig. 3C) and GINS (data not shown), both of which are required for polymerase binding (19, 32). The suppression of the chromatin binding of DNA polymerase α was further confirmed by fluorescence microscopic observation of nuclei formed in the depleted extracts (Fig. 5). These results clearly show that depletion of RecQ4 led to the inhibition of the chromatin binding of DNA polymerase α, which eventually led to the inhibition of DNA replication in the extracts.
In accord with the chromatin binding of Cdc45 in the absence of RecQ4, Cut5 was bound to chromatin at a similar level in both the presence and the absence of RecQ4 in the extracts (Fig. 3C). We have previously reported that the chromatin binding of Cut5 consists of two distinct modes, and a low level of binding in the absence of CDK activity (CDK-independent binding) is sufficient for DNA replication and thus for the binding of Cdc45 (8). Therefore, RecQ4 is apparently dispensable for the CDK-independent binding of Cut5. Nevertheless, the mode of chromatin binding of RecQ4 is similar to that of Cut5 (Fig. 2), suggesting that Cut5 plays a role in the chromatin binding of RecQ4. Thus, we examined whether Cut5 is required for the chromatin binding of RecQ4. Unexpectedly, RecQ4 was bound to chromatin at a similar level in the absence of Cut5, although it was lower than that observed with mock-depleted extracts. In contrast, the polymerases and Cdc45 were not bound to chromatin in the absence of endogenous Cut5, and recombinant Cut5 could rescue the chromatin binding of the polymerases and Cdc45 without affecting the binding of RecQ4 (Fig. 3D). Since Cut5 is required for the initiation, we could not test the functional relevance of a low level of RecQ4 binding in DNA replication, but these results show that RecQ4 can bind to chromatin in the absence of Cut5.
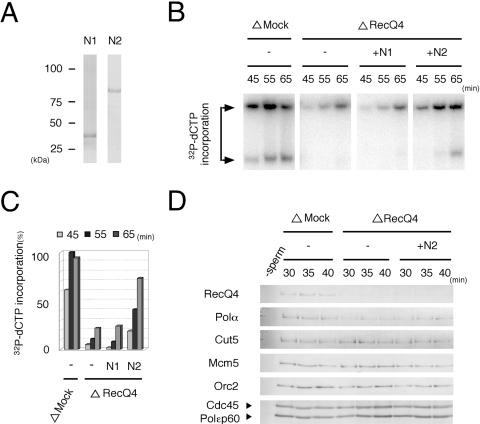
We next examined whether recombinant RecQ4 proteins could rescue the replication activity of RecQ4-depleted extracts (Fig. 5). The activity of RecQ4-depleted extracts was decreased to about one-third of that of mock-depleted extracts (Fig. 5B). The addition of full-length RecQ4 at concentrations of about 20% of the endogenous protein level, estimated to be about 10 μg/ml, could efficiently rescue the replication activity (“+low-full”). The N2 fragment, covering most of the putative CDK phosphorylation sites, could partially rescue the activity at a similar concentration (“+low-N2”) and at four times the endogenous level could rescue the activity as much as the full-length protein could (“+high-N2”). The N1 fragment, which is about half the size of N2 but contains only the N-terminal Sld2-similar regions, could not rescue the replication activity at four times the endogenous level (“+high N1”). To compare more precisely how N1 and N2 rescue the replication activity, we measured the time course of incorporation of 32P-labeled dCTP into sperm DNA (Fig. 6B). Again, we found that N2 but not N1 could rescue the replication activity, to as much as 70% of that of the mock-depleted extracts (Fig. 6B). These results indicate that the defect in the replication activity of RecQ4-depleted extracts was due to the depletion of RecQ4 from the extracts, and the N-terminal region devoid of the helicase domain was responsible for the replication activity.
FIG. 6.
Rescue of DNA replication activity of RecQ4-depleted extracts by N-terminal fragments of RecQ4. (A) SDS-polyacrylamide gel showing recombinant N-terminal fragments of Xenopus RecQ4. N1- and N2-RecQ4 were expressed in Sf9 cells using a baculovirus expression system. The proteins purified through Ni-nitrilotriacetic acid resin were resolved by SDS-PAGE and visualized with Coomassie blue staining. (B and C) Rescue of DNA replication by N-terminal fragments of RecQ4. Sperm chromatin was incubated in mock- (ΔMock) or RecQ4-depleted (ΔRecQ4) extracts in the absence (−) or presence of N2-RecQ4 (+N2, 16 μg/ml) or N1-RecQ4 (+N1, 9 μg/ml) at 23°C. DNA replication activity was monitored as the incorporation of [α-32P]dCTP into DNA. At the indicated times, DNA was isolated, electrophoresed in an agarose gel, and autoradiographed. The bar graph (C) shows the amount of 32P incorporated into DNA, taking the highest value of the mock-depleted extracts as 100%. (D) Recovery of chromatin binding of DNA polymerase α by N-terminal fragments of RecQ4 in RecQ4-depleted extracts. Xenopus sperm chromatin was incubated in mock (ΔMock)- or RecQ4 (ΔRecQ4)-depleted extracts in the absence (−) or presence of N2-RecQ4 (+N2, 16 μg/ml) for the indicated times at 23°C. Isolated chromatin fractions were resolved by SDS-PAGE and immunoblotted with the indicated antibodies.
In addition to rescuing DNA replication, the N2 fragment could rescue the chromatin binding of DNA polymerase α in the RecQ4-depleted extracts. As shown in Fig. 5A, the chromatin localization of the polymerase observed under fluorescent microscopy was decreased by the depletion of RecQ4 from the extracts. The replication activity and the chromatin localization of the polymerase were resumed by the addition of a high concentration of N2 or a low concentration of full-length RecQ4, but not by a high concentration of the N1 fragment (Fig. 5). The chromatin binding of the polymerase was further confirmed by immunoblotting analysis of the chromatin fractions (Fig. 6D). The chromatin binding of polymerase α but not Cdc45 or polymerase ɛ was diminished in RecQ4-depleted extracts, and the high concentration of N2 could partially rescue the polymerase binding (Fig. 6D). The chromatin binding of Cdc45 and polymerase ɛ was essentially unaffected by depletion or addition of recombinant RecQ4 proteins to the extracts. These results therefore show that RecQ4 is required for the chromatin binding of DNA polymerase α.
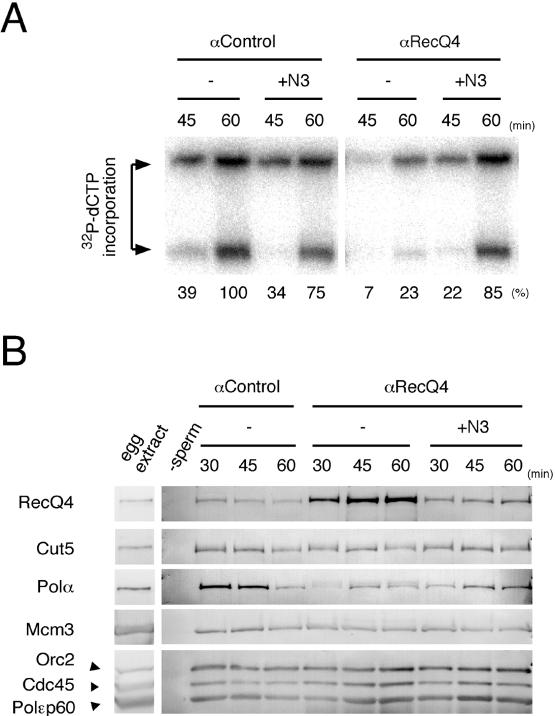
We further examined the potential role of the N-terminal region of RecQ4 in DNA replication by examining the effect of the antibody against N3, which was used for the immunodepletion, during replication. The activity in the presence of anti-RecQ4 antibody was decreased to about 20% of that in the presence of control antibody (Fig. 7A). A similar inhibition of DNA replication was observed with the antibody against N1 or N2 (data not shown), and the inhibition was diminished by preincubating the antibody with an excess molar ratio of the N3. The replication activity in the presence of the fragment was essentially the same in the presence of the control and anti-RecQ4 antibodies (Fig. 7A). The immunoblotting of the chromatin fractions again showed that the chromatin binding of DNA polymerase α was markedly decreased upon the addition of anti-RecQ4 but not control antibody, and the preincubation of the antibody with the N3 recovered the chromatin binding of the polymerase. The chromatin binding of Cdc45, which is required for the chromatin binding of the polymerase, was not affected by the presence of the antibody, but the chromatin binding of RecQ4 was increased upon the addition of anti-RecQ4 antibody. Since RecQ4 was not detected in the chromatin fractions in the absence of sperm chromatin and in the presence of the antibody (data not shown), the increase in RecQ4 was not due to coprecipitation of RecQ4-antibody complex with the chromatin fractions; this suggests that the antibody stabilizes the chromatin binding of RecQ4.
FIG. 7.
Inhibition of DNA replication and chromatin binding of DNA polymerase α by anti-RecQ4 antibody. (A) Xenopus sperm chromatin was incubated in egg extract containing 72.5 μg/ml of preimmune antibody (αControl) or anti-RecQ4 (αRecQ4) antibody in the absence (−) or presence of N-terminal 727-amino-acid fragments of RecQ4 (+N3, 82.7 μg/ml). DNA replication activity was monitored as the incorporation of [α-32P]dCTP into DNA. At the indicated times, DNA was isolated, electrophoresed in an agarose gel, and autoradiographed. The amount of 32P incorporated into DNA was quantified, taking the highest value of the control extracts as 100%. (B) Effect of anti-RecQ4 antibody on the chromatin binding of various replication proteins. Xenopus sperm chromatin was incubated in egg extract as in panel A, and the chromatin fractions were collected at the indicated times. Chromatin fractions and 1 μl of egg extract were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. Negative control of chromatin fractions was obtained by incubating the extracts in the absence of sperm chromatin (−sperm).
Interaction between Xenopus RecQ4 and Cut5.
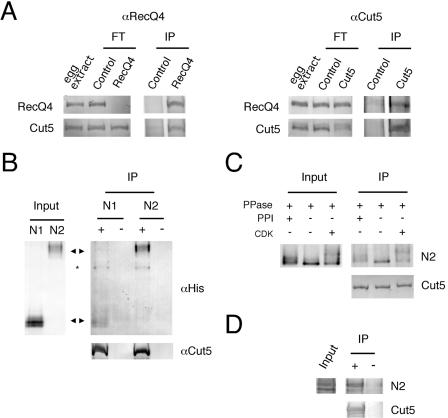
To obtain further insight into the role of RecQ4 in DNA replication, we explored proteins interacting with RecQ4 in the extracts. By examining the immunoprecipitates of egg extracts with anti-RecQ4 antibody, we found that Cut5 was coprecipitated with RecQ4 (Fig. 8A), but other replication proteins, namely, Mcm2 to -7, Cdc45, and DNA polymerases, were not coprecipitated (data not shown). In addition, the antibody against Cut5 could immunoprecipitate RecQ4 with Cut5 from the extracts (Fig. 8A). These results show that Xenopus RecQ4 could form a complex with Cut5 in the extracts. This feature is reminiscent of yeast Sld2/Drc1, which interacts with Dpb11 (12).
FIG. 8.
Physical interaction between RecQ4 and Cut5. (A) Coimmunoprecipitation of RecQ4 and Cut5. The egg extracts (80 μl) were incubated with preimmune control immunoglobulin G (Control), anti-RecQ4 (RecQ4), or anti-Cut5 (Cut5) antibody-conjugated protein A beads. The untreated 1 μl of egg extract, flowthrough extracts (FT), and the proteins bound to the beads (IP) were resolved by SDS-PAGE and blotted onto nitrocellulose membrane for immunostaining. (B) Physical interaction of RecQ4 and Cut5 in vitro. Recombinant N1- and N2-RecQ4 fragments were incubated with anti-Cut5 antibody (−) or the antibody prebound with Cut5 (+) conjugated to protein A beads, and proteins bound to the antibody were pulled down with the beads. Input fragments (Input) and immunoprecipitated fragments (IP) were resolved by SDS-PAGE and blotted onto a nitrocellulose membrane for immunostaining. Arrowheads indicate the position of each fragment. *, degradation product of Cut5. (C) Effect of phosphorylation of RecQ4 on the interaction with Cut5. Recombinant N2-RecQ4 fragment was treated with 5.6 μg/ml protein phosphatase 1 with (+) or without (−) phosphatase inhibitor (1 μM okadaic acid). For rephosphorylation, the sample treated with the phosphatase was further incubated with 17 μg/ml cyclin A/Cdk2 in the presence of 1 μM okadaic acid to prevent the dephosphorylation. The samples were then incubated with Cut5 prebound to anti-Cut5 antibody. Both input (Input) and immunoprecipitated N2 fragments (IP) were resolved by SDS-PAGE and blotted onto a nitrocellulose membrane for immunostaining. (D) Physical interaction of unphosphorylated RecQ4 and Cut5 in vitro. The experiment was carried out as described for panel B except that the N2 fragment expressed in E. coli was used instead of the protein expressed in Sf9 cells.
Since the N-terminal region of RecQ4 has similarity to Sld2 and is essential for DNA replication, we next examined whether the N-terminal fragments of RecQ4 could physically interact with Cut5. Figure 8B shows that N2, which could rescue the replication activity of the RecQ4-depleted extracts, was efficiently pulled down by Cut5 in vitro, but only a small amount of N1 could be pulled down by Cut5, even at twice the concentration of N2. Weak interaction of N1 with Cut5 in vitro coincides with the poor ability of the N1 fragment to rescue the replication activity of the depleted extracts (Fig. 6; see also reference 30), raising a possibility that physical interaction of RecQ4 with Cut5 plays an important role in its activity.
Previous studies of the interaction between Dpb11/Cut5 and Sld2/Drc1 showed that CDK-dependent multiple phosphorylation of Sld2/Drc1 is required for efficient interaction with Dpb11/Cut5 (23, 29). The recombinant RecQ4 fragments used in the present study were apparently phosphorylated, because protein bands resolved by SDS-PAGE had a smear appearance, which was abolished by treating the protein samples with protein phosphatase (Fig. 8C). In addition, N2 and N1 fragments could be efficiently phosphorylated with cyclin A/Cdk2 in vitro (data not shown). We therefore examined whether the protein phosphorylation is required for the interaction of RecQ4 and Cut5 in vitro. Initially, we used alkaline phosphatase to dephosphorylate the recombinant N2 fragments, and the amount of N2 pulled down by Cut5 was substantially decreased by the phosphatase treatment. But we were unable to rephosphorylate the N2 fragments once they were dephosphorylated by alkaline phosphatase at alkaline pH (data not shown). Thus, we used protein phosphatase 1 to dephosphorylate the fragment at neutral pH. Figure 8C shows that the treatment with the protein phosphatase diminished the smear-phosphorylated bands on SDS-PAGE, but the amount of proteins pulled down by Cut5 was not decreased by the dephosphorylation. In addition, rephosphorylation with cyclin A-Cdk2 did not increase the amount of proteins pulled down by Cut5. To further examine the effect of protein phosphorylation, we prepared a recombinant N2 fragment with bacteria for the pull-down assay. As shown in Fig. 8D, bacterially produced protein did not show any smear bands on SDS-PAGE, but it was efficiently pulled down with Cut5. Accordingly, these data suggest that protein phosphorylation was not required for the interaction of Cut5 and RecQ4 in vitro.
Essential role of Xenopus RecQ4 in assembly of the replication machinery.
Xenopus RecQ4 was required for the initiation of DNA replication in egg extracts. RecQ4 was apparently required for the loading of polymerase α but not of Cdc45. Efficient rescue of the replication activity of the depleted extracts by the N-terminal fragment of RecQ4, devoid of the helicase domain, further suggests that the N-terminal region of RecQ4, which shows sequence similarity to that of yeast Sld2/Drc1, retains a function distinct from that of the helicase activity. Previous studies of RecQ4 mutation in patients with Rothmund-Thomson syndrome indicate that the helicase domain is not essential for viability: that is, patients homozygous for the RecQ4 mutation in the helicase domain show severe defects in terms of the syndrome phenotype, but these defects are not acutely lethal to embryonic development (2, 16, 20). On the other hand, studies of RecQ4 knockout in mice suggest that RecQ4 is essential for embryonic development (10, 11). If we take these results into account, the N-terminal region of RecQ4 has an essential function in DNA replication. During the course of our study, Ashok Venkitaraman (Cancer Research UK) and his colleagues reported that human and Xenopus RecQ4 are essential for chromosomal DNA replication and showed that mutation in the helicase domain of RecQ4 led to loss of its activity in rescuing DNA replication in RecQ4-depleted extracts (30). They further showed that a short N-terminal fragment consisting of 118 amino acids could partially rescue the replication activity of the depleted extracts. Poor rescue by the short fragment is consistent with our finding that N2 but not N1 could efficiently rescue the activity and suggests that the fragment is not properly folded in the absence of the C-terminal region. We cannot exclude the possibility that RecQ helicase activity is also required for efficient DNA replication. In fact, the full-length recombinant protein was more efficient in the rescue experiments than the N2 fragment (Fig. 5).
Previous studies in budding yeast have demonstrated that Sld2/Drc1 has an essential role in the initiation of DNA replication and is one of the target proteins of S-CDK (23, 29). However, the precise role of Sld2 in the initiation reaction has not yet been elucidated. Based on the sequence similarity, we have identified RecQ4 as a putative homolog of Sld2/Drc1. The following results support the view that RecQ4 is a functional homolog of Sld2 in vertebrates. (i) The N-terminal region of Xenopus RecQ4 has sequence similarity with that of Sld2/Drc1. In addition, we found the enrichment of putative CDK phosphorylation sites in this region. Human, Xenopus, and mouse RecQ4s contain 9, 15, and 7 SP motifs in the N-terminal region, respectively. (ii) The N-terminal region of RecQ4 has an essential role in replication, as described above. (iii) RecQ4 physically interacts with Cut5 in vitro and in vivo. In budding yeast, Sld2 was initially identified as an interactor with Dpb11 (12). Although the interaction between Sld2 and Dpb11 is controlled by CDK-dependent phosphorylation, overproduction of both proteins in yeast cells leads to coprecipitation, even in G1 phase (H. Araki [NIG Japan], personal communication). Therefore, the interaction with Cut5/Dpb11 is a conserved feature of Sld2/Drc1 homologs, and our data further suggest that RecQ4 plays an essential role in DNA replication through interaction with Cut5.
We found that the phosphorylation of RecQ4 is dispensable for its interaction with Cut5 in vitro and presumably in Xenopus egg extracts, contradicting the idea that RecQ4 is a functional homolog of Sld2/Drc1. However, such a difference may be explained by the functional diversity of a replication protein with less-conserved structures. Further study of the physiological role of phosphorylation of RecQ4 should help us to define whether RecQ4 is a functional homolog of Sld2/Drc1 or not.
A remaining question concerns the function of RecQ4 in DNA replication. Our data showed that it is required for the chromatin binding of DNA polymerase α. An independent study of human and Xenopus RecQ4 reported that the chromatin binding of replication protein A (RPA) was inhibited in RecQ4-depleted extracts (30). Since RPA is required for the chromatin binding of polymerase and is tightly bound to chromatin only after the unwinding of DNA (26, 34), these results suggest that RecQ4 is required for the unwinding of DNA. Intriguingly, we found that the chromatin binding of RecQ4 was increased by the antibody treatment, which inhibited the binding of the polymerase. Similarly, we found that the chromatin binding of RecQ4 was increased by depleting RPA from the extracts, which also inhibited the polymerase binding (unpublished data). These results suggest that RecQ4 is accumulated on chromatin when the chromatin loading of the polymerase and possibly the unwinding reaction have been suppressed. It is therefore possible that the dissociation of RecQ4 is required for promoting the loading of the polymerase and the unwinding reaction and that RecQ4 associates with chromatin only transiently during the initiation reaction. The present study and a previous one (30) showed that RecQ4 plays an essential role at the last stage of the assembly of replication machinery. In addition to its replication function, recent reports suggest that RecQ helicase Sgs1 is required for stabilizing DNA polymerases at stalled replication forks (4, 6), also implicating a crucial function of RecQ helicase in the checkpoint response. The marked accumulation of chromatin-bound RecQ4 in the presence of aphidicolin suggests its role in the S-phase checkpoint. Further examination of the intrinsic function of the helicase domain of RecQ4 should give us an insight into the molecular mechanisms involved in the generation of the Rothmund-Thomson syndrome phenotype.
ADDENDUM
During the revision of the manuscript, Macris et al. published a paper (20a) reporting that human RecQ4 has an ATPase activity stimulated by single-stranded DNA and single-stranded DNA annealing activity that is inhibited by RPA. Thus, our finding of accumulation of RecQ4 in the absence of RPA or in the presence of the antibody suggests that RecQ4 plays an important role in the early step of the DNA unwinding reaction.
Acknowledgments
We thank Ashok Venkitaraman and Hiroyuki Araki for communicating their results to us before publication.
This work was supported by Grants-in-Aid for Scientific Research on Priority Area (A) from the Ministry of Education, Science, Sports and Culture, Japan.
REFERENCES
- 1.Bachrati, C. Z., and I. D. Hickson. 2003. RecQ helicase: suppressors of tumorigenesis and premature aging. Biochem. J. 374:577-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balraj, P., P. Concannon, R. Jamal, A. Beghini, T. S. Hoe, A. S. Khoo, and L. Volpi. 2002. An unusual mutation in RECQ4 gene leading to Rothmund-Thomson syndrome. Mutat. Res. 508:99-105. [DOI] [PubMed] [Google Scholar]
- 3.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. [DOI] [PubMed] [Google Scholar]
- 4.Bjergbaek, L., J. A. Cobb, M. Tsai-Pflugfelder, and S. M. Gasser. 2005. Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance. EMBO J. 24:405-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blow, J. J., and B. Hodgson. 2002. Replication licensing—defining the proliferative state? Trends Cell Biol. 12:72-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cobb, J. A., L. Bjergbaek, K. Shimada, C. Frei, and S. M. Gasser. 2003. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 22:4325-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furuichi, Y. 2001. Premature aging and predisposition to cancers caused by mutations in RecQ family helicases. Ann. N. Y. Acad. Sci. 928:121-131. [DOI] [PubMed] [Google Scholar]
- 8.Hashimoto, Y., and H. Takisawa. 2003. Xenopus Cut5 is essential for a CDK-dependent process in the initiation of DNA replication. EMBO J. 10:2526-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hickson, I. 2003. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer 3:169-178. [DOI] [PubMed] [Google Scholar]
- 10.Hoki, Y., R. Araki, A. Fujimori, T. Ohhata, H. Koseki, R. Fukumura, M. Nakamura, H. Takahashi, Y. Noda, S. Kito, and M. Abe. 2003. Growth retardation and skin abnormalities of the Recql4-deficient mouse. Hum. Mol. Genet. 12:2293-2299. [DOI] [PubMed] [Google Scholar]
- 11.Ichikawa, K., T. Noda, and Y. Furuichi. 2002. Preparation of the gene targeted knockout mice for human premature aging diseases, Werner syndrome, and Rothmund-Thomson syndrome caused by the mutation of DNA helicases. Nippon Yakurigaku Zasshi 119:219-226. [DOI] [PubMed] [Google Scholar]
- 12.Kamimura, Y., H. Masumoto, A. Sugino, and H. Araki. 1998. Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol. Cell. Biol. 18:6102-6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamimura, Y., Y. S. Tak, A. Sugino, and H. Araki. 2001. Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J. 20:2097-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanemaki, M., A. Sanchez-Diaz, A. Gambus, and K. Labib. 2003. Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature 423:720-724. [DOI] [PubMed] [Google Scholar]
- 15.Kearsey, S. E., and S. Cotterill. 2003. Enigmatic variations: divergent modes of regulating eukaryotic DNA replication. Mol. Cell 12:1067-1075. [DOI] [PubMed] [Google Scholar]
- 16.Kitano, S., A. Shimamoto, M. Goto, R. W. Miller, W. A. Smithson, N. M. Lindor, and Y. Furuichi. 1999. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat. Genet. 22:82-84. [DOI] [PubMed] [Google Scholar]
- 17.Kubota, Y., and H. Takisawa. 1993. Determination of initiation of DNA replication before and after nuclear formation in Xenopus egg cell free extracts. J. Cell Biol. 123:1321-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubota, Y., S. Mimura, S.-I. Nishimoto, H. Takisawa, and H. Nojima. 1995. Identification of the yeast MCM3-related protein as a component of Xenopus DNA replication licensing factor. Cell 81:601-609. [DOI] [PubMed] [Google Scholar]
- 19.Kubota, Y., Y. Takase, Y. Komori, Y. Hashimoto, T. Arata, Y. Kamimura, H. Araki, and H. Takisawa. 2003. A novel ring-like complex of Xenopus proteins essential for the initiation of DNA replication. Genes Dev. 17:1141-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindor, N. M., Y. Furuichi, S. Kitao, A. Shimamoto, C. Arndt, and S. Jalal. 2000. Rothmund-Thomson syndrome due to RECQ4 helicase mutations: report and clinical and molecular comparisons with Bloom syndrome and Werner syndrome. Am. J. Med. Genet. 90:223-228. [DOI] [PubMed] [Google Scholar]
- 20a.Macris, M. A., L. Krejci, W. Bussen, A. Shimamoto, and P. Sung. 2006. Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair 5:172-180. [DOI] [PubMed] [Google Scholar]
- 21.Marheineke, K., and O. Hyrien. 2004. Control of replication origin density and firing time in Xenopus egg extracts. J. Biol. Chem. 270:28071-28081. [DOI] [PubMed] [Google Scholar]
- 22.Masumoto, H., A. Sugino, and H. Araki. 2000. Dpb11 controls the association between DNA polymerases α and ɛ and the autonomously replicating sequence region of budding yeast. Mol. Cell. Biol. 20:2809-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masumoto, H., S. Muramatsu, Y. Kamimura, and H. Araki. 2002. S-CDK-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature 415:651-655. [DOI] [PubMed] [Google Scholar]
- 24.Méndez, J., and B. Stillman. 2003. Perpetuating the double helix: molecular machines at eukaryotic DNA replication origins. Bioessays 25:1158-1167. [DOI] [PubMed] [Google Scholar]
- 25.Mimura, S., and H. Takisawa. 1998. Xenopus Cdc45-dependent loading of DNA polymerase α onto chromatin under the control of S-phase CDK. EMBO J. 17:5699-5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mimura, S., T. Masuda, T. Matsui, and H. Takisawa. 2000. Central role for Cdc45 in establishing an initiation complex of DNA replication in Xenopus egg extracts. Genes Cells 5:439-452. [DOI] [PubMed] [Google Scholar]
- 27.Nakajima, R., and H. Masukata. 2002. SpSld3 is required for loading and maintenance of SpCdc45 on chromatin in DNA replication in fission yeast. Mol. Biol. Cell 13:1462-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishitani, H., and Z. Lygerou. 2002. Control of DNA replication licensing in a cell cycle. Genes Cells 7:523-534. [DOI] [PubMed] [Google Scholar]
- 29.Noguchi, E., P. Shanahan, C. Noguchi, and P. Russell. 2002. CDK phosphorylation of Drc1 regulates DNA replication in fission yeast. Curr. Biol. 12:599-605. [DOI] [PubMed] [Google Scholar]
- 30.Sangrithi, M. N., J. A. Bernal, M. Madine, A. Philpott, J. Lee, W. G. Dunphy, and A. R. Venkitaraman. 2005. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell 121:887-898. [DOI] [PubMed] [Google Scholar]
- 31.Tada, S., A. Li, D. Maiorano, M. Mechali, and J. J. Blow. 2001. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat. Cell Biol. 3:107-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takayama, Y., Y. Kamimura, M. Okawa, S. Muramatsu, A. Sugino, and H. Araki. 2003. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 17:1153-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Hatten, R. A., A. V. Tutter, A. H. Holway, A. M. Khederian, J. C. Walter, and W. M. Michael. 2002. The Xenopus Xmus101 protein is required for the recruitment of Cdc45 to origins of DNA replication. J. Cell Biol. 159:541-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walter, J., and J. Newport. 2000. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol. Cell 5:617-627. [DOI] [PubMed] [Google Scholar]
- 35.Wang, H., and S. J. Elledge. 1999. DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 96:3824-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wohlschlegel, J. A., B. T. Dwyer, S. K. Dhar, C. Cvetic, J. C. Walter, and A. Dutta. 2000. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 290:2309-2312. [DOI] [PubMed] [Google Scholar]
- 37.Yoshida, K., H. Takisawa, and Y. Kubota. 2005. Intrinsic nuclear import activity of geminin is essential to prevent re-initiation of DNA replication in Xenopus eggs. Genes Cells 10:63-73. [DOI] [PubMed] [Google Scholar]