Abstract
The stable accumulation of p53 is detrimental to the cell because it blocks cell growth and division. Therefore, increases in p53 levels are tightly regulated, mainly by its transcriptional target, mdm2, that downregulates p53. Elucidation of new signaling pathways requires the characterization of the members and the nature of their connection. Vaccinia-related kinase 1 (VRK1) contributes to p53 stabilization by partly interfering with its mdm2-mediated degradation, among other mechanisms; therefore, it is likely that some form of autoregulation between VRK1 and p53 must occur. We report here the identification of an autoregulatory loop between p53 and its stabilizing VRK1. There is an inverse correlation between VRK1 and p53 levels in cell lines, and induction of p53 by UV light downregulates VRK1 in fibroblasts. As the amount of p53 protein increases, there is a downregulation of the VRK1 protein level independent of its promoter. This effect is indirect but requires a transcriptionally active p53. The three most common transcriptionally inactive mutations detected in hereditary (Li-Fraumeni syndrome) and sporadic human cancer, p53(R175H), p53(R248W), and p53(R273H), as well as p53(R280K), are unable to induce downregulation of VRK1 protein. The p53 isoforms Δ40p53 and p53β, lacking the transactivation and oligomerization domains, respectively, do not downregulate VRK1. VRK1 downregulation induced by p53 is independent of mdm2 activity and proteasome-mediated degradation since it occurs in the presence of proteasome inhibitors and in mdm2-deficient cells. The degradation of VRK1 is sensitive to chloroquine, an inhibitor of the late endosome-lysosome transport, and to serine protease inhibitors of the lysosomal pathway.
The accumulation of the tumor suppressor protein p53 occurs in response to many different types of cellular stress or DNA damage. The levels of p53 in response to stress present a transient accumulation that may last a few hours (46). This transient p53 accumulation induces a variety of responses designed to protect the cell and allow for damage repair by stopping the cell cycle (1, 28, 38, 43, 57, 61) or inducing apoptosis (41, 52, 62). If the level of p53 is maintained at a high level when unnecessary, there would be a permanent block to cell cycle progression or the cells will die. In any case, the stable and prolonged accumulation of p53 is incompatible with life. Therefore, the p53 molecule has evolved in a way that it is always present at low levels, and when it is stabilized, the accumulation occurs in a transient manner. The normal p53 levels are tightly regulated, and mechanisms aiming at its downregulation are highly developed. This is achieved in part by having a short half-life of about 20 min and, in case of an increase in p53 levels, by the induction of negative regulatory proteins for p53. The best-characterized mechanism is p53 downregulation by one of its transcriptional targets, hdm2/mdm2 (34), so that the induction of mdm2 leads to p53 ubiquitination and degradation by the proteasome (32, 53). The p53 protein is held at low levels under normal growth conditions, and upon stress its level increases rapidly (41) with the subsequent transcriptional activation, including that of hdm2. Hdm2 is a ubiquitin ligase that promotes downregulation of p53, bringing its levels down to basal ones. Therefore, it can be considered that there are waves of p53 accumulation whose magnitude may vary depending on the type of stimulation or the cell type (24). The sequential expression of p53 and mdm2 might also contribute to oscillations in their levels (27). The same occurs with other p53 negative regulators that are also transcriptional targets, as is the case for other ubiquitin ligases such as COP1 (11). The downregulation of positive modulators by p53, such as chk1, has been also reported (15). The p53 mutant (R273H), present in sporadic cancers and in Li-Fraumeni syndrome patients, accumulates in human tumors and has its properties altered with oncogenic potential (25, 39). The p53 mutants detected in many tumors are likely to have altered these autoregulatory mechanisms, but these alterations have not been characterized. Recently, it has been shown that variant p53 proteins lacking either the transactivation (11) or the oligomerization domains (11) but retaining their ability to activate gene transcription can be generated by alternative splicing (8); these variant p53 proteins may have modifications in their regulatory mechanisms.
The human vaccinia-related kinase 1 (VRK1) (26), a member of a new Ser-Thr kinase family in the human kinome (30) has autophosphorylation activity (29, 36) and also phosphorylates different transcription factors such as p53 (4, 29, 58, 23), c-Jun (50), ATF2 (51), and nuclear factor BAF (37). VRK1 is able to induce p53 stabilization by a complex mechanism, a component of which is the phosphorylation of p53 in Thr18 (4, 29), partly preventing its interaction with mdm2 and promoting p300 recruitment that leads to p53 accumulation and transcriptional activation (58). p53 stabilization by VRK1 has been postulated to be a basic control process that operates in cells under normal growth conditions in the absence of or under suboptimal stress and that permits p53 to remain in a readiness state or operate in minor damage responses, such as during replication (58). VRK1 appears to be necessary for a basic control mechanism in cell proliferation since its loss, induced by small interfering RNA (siRNA) and in the absence of stress stimulation, leads to a retardation of cell division and cell death by a mechanism not yet identified (58). In human head and neck squamous cell carcinomas, VRK1 correlates with established proliferation markers, which suggests that VRK1 might be playing a role early in the G1 phase of the cell cycle (47).
We hypothesized that some cross-regulation between VRK1 and p53 proteins must exist so that p53 stabilization can be reversed; this mechanism should involve the inactivation in some way of VRK1 so that its loss will permit the downregulation of p53 by mdm2 or any other mechanism. Elucidation of signaling networks implies the identification of interacting molecules and the characterization of their interaction in order to identify their contribution to different types of biological effects (42). In this report we have identified that the accumulation of p53 is able to induce the downregulation of its stabilizing protein, VRK1, and this process requires the contribution of different p53 domains, does not involve VRK1 transcriptional regulation, and is mediated by the lysosomal pathway of protein degradation.
MATERIALS AND METHODS
Plasmids, antibodies, and reagents.
The VRK1 constructs, pCEFL-HA-VRK1, pCEFL-HA-VRK1(K179E), and pCDNA3.1-VRK1-myc coding either for the wild-type VRK1 or the VRK1(K179E) inactive mutant have been previously described (58). The plasmid pCB6+p53, containing human p53 wild-type cDNA, plasmid pCB6+p53T18D (2), and plasmid pCOC-Mdm2-X2 coding for the Mdm2 protein were from K. Vousden (The Beatson Institute, Glasgow, United Kingdom); the transcriptionally defective mutant p53(R280K) was from A. J. Levine (Rockefeller University, New York) (12); the mutant plasmids pCMV-p53(R175H), pCMV-p53(R248W), and pCMV-p53(R273H) were from Bert Vogelstein (Johns Hopkins University, Baltimore, MD); plasmid pCMV-p53(L22Q, W23S) was from K. Roemer (7); plasmids expressing the p53 isoforms p53β and Δ40p53 (p47) were from J.C. Bourdon (Dundee University, Scotland) (8); and pCMW-p53(L322A) and pCDNA3-p53CΔ60 were from S. Camus (Institute of Molecular and Cellular Biology, Singapore). The plasmid BRR12-ubiquitin-His (pUbiquitin-His) was from S. Lain and D. Lane (Dundee University, Scotland). The p53 siRNA expression plasmid pSUPER.retro.p53 (Oligoengine, Seattle, WA) was used where indicated to suppress the expression of p53. All plasmids used for transfection were endotoxin free and purified with a JetStar Maxi kit from Genomed (Bad Oeynhausen, Germany).
The anti-β-actin antibody was from Sigma (St. Louis, MO). The hemagglutinin (HA) tag was detected with a mouse monoclonal antibody HA-probe (F7) from Covance (Berkeley, Calif.). The p53 protein was detected with a mixture of DO1 antibody (Santa Cruz, CA) and Pab1801 (Santa Cruz, CA) used at 1:500 and 1:1,000, respectively. The p53 isoform, Δ40p53 (p47), lacking the transactivation domain was detected with the CM1 polyclonal antibody at a dilution of 1:20,000 (from A. Craig, Dundee University, Scotland). VRK1 was detected using a rabbit polyclonal antibody (VE1) or a mouse monoclonal antibody (1F6 clone) made against a VRK1 fusion protein. Poly(ADP-ribose) polymerase was determined with a monoclonal antibody from Enzyme Systems Products (Livermore, CA). As secondary antibodies, a goat anti-mouse-horseradish peroxidase or a goat anti-rabbit-horseradish peroxidase (Amersham Pharmacia Biotech) was used at 1:5,000 in Western blotting.
The following protease inhibitors were used: pepstatin for aspartyl proteases; phenylmethylsulfonyl fluoride (PMSF), aprotinin, diisopropylfluorophosphate (DFP), soybean trypsin inhibitor (STI), and leupeptin for serine proteases; iodoacetic acid (IAA) and leupeptin for cysteine proteases; EDTA and 1,10-phenantroline for metaloproteasas; and ALLN, calpain inhibitor III, calpeptin, EST, and PD150606 for calpain proteases (all from Sigma). Chloroquine was from Sigma.
Cell lines and transfections.
The human lung cancer cell line H1299 (p53−/−) was grown in RPMI medium supplemented with 10% fetal calf serum, glutamine, penicillin, and streptomycin in a humidified atmosphere and 5% CO2. HeLa cells, U2OS (p16−/−), and the WS1 normal human fibroblast cell line (ATCC CRL-1502) were grown in Dulbecco's modified Eagle's medium with the same supplements. For transfection experiments, H1299 cells were plated in 60- or 100-mm dishes and transfected with the plasmid indicated in the specific experiments either by the calcium phosphate precipitation method or with JetPI reagent following the manufacturer's instructions (Polytransfection; Illkirch, France). Unless otherwise indicated, the cells were lysed 36 h posttransfection in lysis buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 5 mM EDTA, and 1%Triton X-100, plus protease and phosphatase inhibitors), and 40 μg of whole-cell extract was processed for sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and subjected to immunoblotting with the indicated antibodies. The immortalized fibroblasts from double knockout p53/Mdm2 mice (p53−/−, mdm2−/−) were a gift of G. Lozano (MD Anderson Cancer Center, Houston, TX) and were grown in Dulbecco's modified Eagle's medium with 10% fetal calf serum and antibiotics. Where indicated, the proteasome inhibitor MG132 (Calbiochem) was used at 50 μM for 6 h. Tetracycline was from Sigma (St. Louis, MO) and was used at a concentration of 2 μg/ml. Stimulation of the cells with UV light was done in a Stratalinker from Stratagene (San Diego, CA).
Immunoblotting.
Total protein extracts were quantified using a Bio-Rad protein assay kit. Protein was fractionated in an SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride Immobilon-P membrane (Millipore). The membrane was blocked with TBS-T buffer (25 mM Tris, 50 mM NaCl, 2.5 mM KCl, 0.1% Tween-20) and 5% nonfat milk. Afterwards the membrane was rinsed with TBS-T buffer; and the specific primary antibody (indicated in individual experiments) was added, and the membrane was incubated for 90 min at room temperature. The membrane was rinsed and incubated with a secondary antibody conjugated with peroxidase for 30 min. The membrane was develop for chemiluminescence with ECL reagent (Amersham) and exposed to X-ray films. Quantification was always performed in the linear response range.
Quantitative RT-PCR.
Reverse transcription-PCR (RT-PCR) was performed as previously described (58). Briefly, H1299 cells were cotransfected as described above with the plasmids indicated in the experiment, and total RNA was extracted using an RNAeasy extraction kit from QIAGEN (Hilden, Germany). One hundred nanograms of total RNA was used in a one-step reverse transcription real-time PCR amplification reaction using a Quantitec SYBR Green RT-PCR kit from QIAGEN in an iCycler (Bio-Rad, Hercules, CA). The reaction was analyzed with iCycler software (Bio-Rad). The primers used for VRK1 amplification detection were 5′-CCAACGAGCTGCAAAACCA-3′ and 5′-TGTCATGTAGACCAGACCCCC-3′; for GAPDH amplification detection, the primers were 5′-GGTCTTACTCCTTGGAGGCCATGT-3′ and 5′-ACCTAACTACATGGTTTACATGTT-3′.
RESULTS
The level of VRK1 inversely correlates with p53 levels in cell lines.
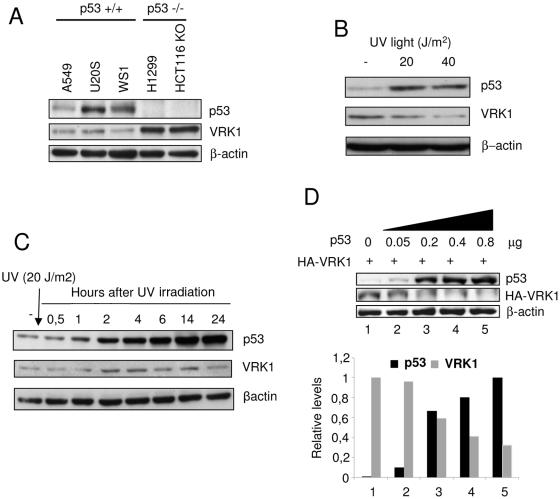
During the course of the work, it was noticed that when a cell had p53 wild-type protein, the level of transfected VRK1 detected was always lower than if the cell line lacked p53. Once it was clear that this effect was not due to transfection efficiency, the correlation with the cell genotype was intriguing and suggested that the explanation might be related to the connection of the two proteins in the pathway (58), with an inverse relation between them. To determine if that might be the case, the level of the two endogenous proteins, VRK1 and p53, was determined in several cell lines (Fig. 1A). The cell lines with wild-type p53 had lower levels of endogenous VRK1. But in the two cell lines lacking p53, such as H1299 or HCT116 (p53 knockout), higher levels of endogenous VRK1 were clearly detected (Fig. 1A). One of the effects of an active VRK1 is to stabilize p53 by increasing its intracellular levels (58), and a stable accumulation of p53 is known to be detrimental to the cell since it prevents cell division and might even trigger an apoptotic response (3, 9, 43). Therefore, we considered the possibility that higher levels of p53 might promote a downregulation of its activator-stabilizer VRK1 protein, thus forming an autoregulatory loop between p53 and VRK1, which might be conceptually similar to the autoregulation between Chk1 and p53 (15) or to the p53-mdm2 autoregulatory loop (34). That interpretation might account for the different levels of VRK1 observed, depending on the p53 genotype of cell lines.
FIG. 1.
Expression of endogenous p53 and VRK1 in tumor cell lines and in UV treated fibroblasts. (A) Expression of VRK1 and p53 in cell lines that were p53 wild type (A549, U2OS, and WS1) or p53−/− (H1299 or HCT116-p53 knockout). VRK1 and p53 were determined by Western blotting, using actin as internal control. (B) Levels of p53 and VRK1 in human fibroblasts treated with the indicated dose of UV light; the extracts were prepared 8 h after treatment and analyzed by Western blotting. (C) The levels of endogenous p53 and VRK1 in WS1 fibroblasts were detected at different times after treatment with 20 J/m2 UV light. The proteins were identified by Western blotting with the corresponding antibodies indicated in the methods section. (D) Normal human fibroblasts, cell line WS1, were transfected with increasing amounts of pCB6+p53 as indicated and 5 μg of pCEFL-HA-VRK1. Cell extracts were prepared 36 h after transfection. The proteins were detected with the corresponding antibodies by Western blotting, and to the right is shown the quantification of the normalized values.
p53 accumulation induced by UV downregulates VRK1.
To determine if the level of p53 might indeed have an inverse correlation with the level of VRK1, we studied the variation in levels of both proteins in the cell response to stimulation with UV irradiation. Human diploid fibroblasts, the WS1 cell line (p53+/+), were treated with two doses of UV light, at 20 J/m2 or 40 J/m2, and cell extracts were analyzed by Western blotting 8 h after irradiation. In both cases the endogenous p53 was stabilized, and there was a reduction of VRK1 levels (Fig. 1B). Next, a time course of the response of WS1 cells irradiated with 20 J/m2 was performed for a period of 24 h. As expected, after UV treatment there was the expected fast stabilization of p53 as a consequence of phosphorylation (46), but the accumulation of p53 continued to increase, reaching significantly much higher levels after 10 h (Fig. 1C), a response typical of fibroblasts (16). After irradiation there was also a transient increase in the level of VRK1 that reached a peak at 4 h, after which it fell even below the initial level. The timing of this reduction in VRK1 levels coincided with the time necessary for p53-dependent de novo gene expression and with the increased level of p53, suggesting that perhaps the accumulated p53 might be implicated in inducing the VRK1 downregulation. Next, we tried to determine if this down-regulatory effect could be reproduced by transfection of p53. To this end, the WS1 fibroblast cell line was transfected with increasing amounts of p53 (plasmid pCB6+p53) and a fixed amount of VRK1 (plasmid pCEFL-HA-VRK1). As the amount of p53 increased, it was accompanied by a reduction in the level of VRK1 protein in these cells (Fig. 1D).
p53 downregulates VRK1 protein level.
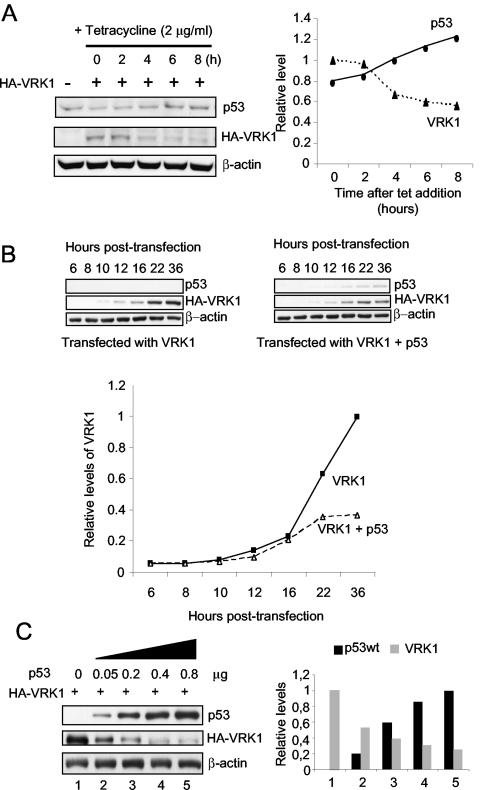
To reproduce this down-regulatory effect in an independent system, a new experiment was designed using a variant of the H1299 (p53−/−, p53Tet inducible) cell line that contains a p53 gene that is inducible by addition of tetracycline to the culture. This cell line has a basal level of p53 expression and thus mimics a normal cell situation. This cell line was transfected with a fixed amount of pCEFL-HA-VRK1, p53 expression was induced by the addition of tetracycline, and the level of VRK1 was determined at different time points. After induction with tetracycline, as the level of p53 increased with time, it was accompanied by a reduction in the level of the VRK1 protein detected with an antibody against the HA epitope tag (Fig. 2A).
FIG. 2.
Downregulation of VRK1 by increasing levels of p53. (A) Inducible endogenous p53 downregulated VRK1. H1299 cells (p53Tet inducible) were transfected with 5 μg of pCEFL-HA-VRK1, and the p53 gene was induced with tetracycline. As the level of p53 increased, there was a noticeable decrease of VRK1 expression levels. Cell extracts were prepared at different time points after induction, and the proteins were detected by immunoblot analysis with the quantification shown in the graph to the right. (B) Time course of p53 and VRK1 protein levels. H1299 cells were transfected with pCEFL-HA-VRK1 (4 μg) with and without pCB6+p53 (200 ng). At different hours after transfection, cell extracts were prepared and analyzed by Western blotting for the amount of each protein. The quantification of the blots is shown in the graph to illustrate the trend in protein levels. The accumulation of p53 interferes with the accumulation of VRK1. The proteins were detected with the corresponding antibodies by Western blotting, and to the right is shown the quantification of the normalized values. (C) H1299 cells (p53−/−) were transfected with a fixed amount (5 μg) of pCEFL-HA-VRK1 and increasing amounts of p53 (wild type) to detect the effect of increasing p53 on VRK1 expressed from different promoters.
To confirm that VRK1 protein downregulation was dependent on the accumulation of p53 protein, a time course experiment for VRK1 expression alone or together with p53 overexpression was performed (Fig. 2B). For this experiment, H1299 (p53−/−) cells were transfected with plasmids pCEFL-HA-VRK1 and pCB6+p53, and at different time points cell extracts were prepared and analyzed by immunoblotting. In the absence of p53, VRK1 protein levels increased constantly, as expected, given the high degree of VRK1 stability. However, in the presence of p53, at first VRK1 increased as a result of de novo protein synthesis; but when there was also a detectable accumulation of p53, VRK1 accumulation slowed down and finally came to a stop, which was particularly noticeable between 24 and 36 h after transfection (Fig. 2B). These data suggested that the effect depends on the concentration of p53 in the cell and was probably due to the modification of gene transcription as a consequence.
To further determine whether such an effect was occurring in a p53 dose-dependent manner and was promoter independent, an experiment was designed to reproduce the previous inducible effect by transfection using the H1299 cell line (p53−/−). This cell line was transfected with increasing amounts of pCB6+p53 and a fixed amount of pCEFL-HA-VRK1 (Fig. 2C), which express VRK1 from a Moloney murine leukemia virus promoter; with pCDNA3.1-VRK1-myc, which expresses VRK1 from a cytomegalovirus promoter; or with pEF1-VRK1, which uses the EF1 gene promoter. In this experiment it was observed that the higher the level of p53, the lower the amount of VRK1 protein detected in cell extracts (Fig. 2C). The effect of p53 on VRK1 was similar independent of the type of promoter from which VRK1 was expressed (not shown). The effect was also confirmed in other tumor cell lines, such as HT144, WS1, A549, and in U2OS (p16−/−), suggesting that the effect is independent of cell type and that p16 is not involved in this process (not shown). These results indicate that the effect is more general and does not depend on the genetic peculiarities of the particular tumor cell line or promoter used.
Contribution of p53 domains to VRK1 protein downregulation.
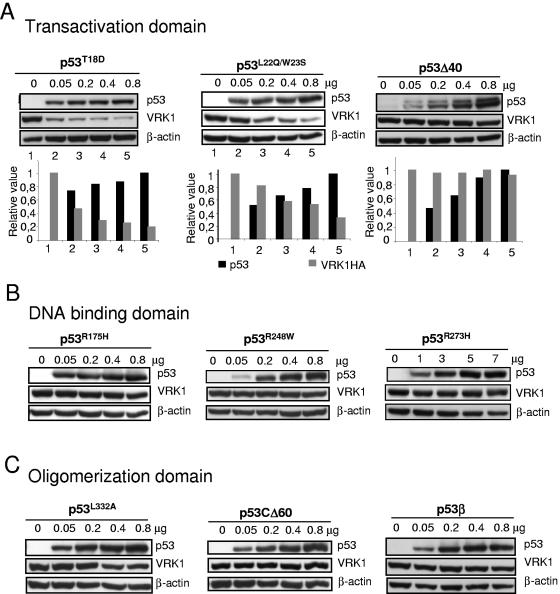
To determine the requirements of the p53 protein required to induce the downregulation of VRK1, the contributions of three main p53 functional regions, the transactivation, DNA binding, and oligomerization domains, were analyzed. The potential contribution of the N-terminal transactivation domain of p53 was studied using three different types of mutants: a phosphorylation mutant that mimics phosphorylation in threonine-18, p53(T18D), the p53(L22Q, W23S) conformational mutant than retains its transcriptional activation role but has different effects on double-strand DNA repair and apoptosis (7, 31), and the isoform Δ40p53, known as p47, that lacks the first 40 amino acids of the transactivation domain (8). This isoform functions as a dominant negative of p53 growth suppression properties (10, 14). The phosphorylation and the conformational mutants were able to induce the downregulation of VRK1 in a dose-dependent manner; however, the Δ40p53, lacking the first forty amino acids, has lost this role (Fig. 3A). These results indicate that the first 40 residues of the transactivation domain are required for this effect and reflect a functional difference in p53 isoforms. Different N-terminal phosphorylation mutants were also assayed with same results as with the p53(T18D) (not shown). p53 interaction with many regulators is mediated by the N-terminal transactivation domain, and it is possible that such interaction is necessary for p53-mediated VRK1 downregulation.
FIG. 3.
Implication of p53 protein domains in the induction of VRK1 downregulation. (A) Role of the transactivation domain. The effects of the phosphorylation mimicking mutant p53(T18D), the conformational double mutant p53(L22Q, W23S), and the p53 isoform lacking the transactivation domain (Δ40p53) were tested in combination with pCEFL-HA-VRK1 (5 μg). The Δ40p53 isoform was detected with the CM1 polyclonal antibody. At the bottom is shown the quantification of the blots to illustrate the changes in both proteins. (B) Contribution of the p53 DNA binding domain. Lack of effect of the most common p53 transcriptional mutants, p53(R175H), p53(R248W), and p53(R273H) on the level of VRK1 protein expressed from plasmid pCEFL-HA-VRK1 (5 μg). Cell extracts were prepared 36 h after transfection, and the levels of both proteins were determined by Western blotting. The transfected VRK1 was detected with an antibody specific for the HA epitope. (C) Contribution of the p53 oligomerization domain. The conformational mutant p53(L322A) and isoforms lacking the C-terminal region p53β and p53CΔ60 were studied.
To ascertain if the DNA binding domain of p53 was a requirement for induction of VRK1 downregulation, several mutants in this domain were studied. The three most common naturally occurring p53 mutants in human cancer, both sporadic and hereditary, were studied. The conformational mutant p53(R175H) and the DNA contact mutants p53(R248W) and p53(R273H) (55) account for approximately one-third of all p53 mutations in human cancer (6, 56). A p53 mutant [p53(R280K)] engineered to prevent binding to DNA and without effects on other p53 properties was also studied (12). All of these p53 mutants were unable to induce a downregulation of VRK1 protein levels (Fig. 3B). This lack of effect indicated that the action of p53 on VRK1 levels requires the integrity of the DNA binding domain and is therefore probably mediated by a DNA-bound form of p53. This result opens the possibility of the effect being mediated by a p53-dependent induction or repression of a gene not yet identified.
Finally, the potential contribution of the p53 oligomerization domain was analyzed. The C terminus is necessary for the anti-growth arrest and anti-apoptotic effects of p53 (54). Three different p53 variants in this domain were used: the p53(L332A) conformational mutant that has a defective oligomerization, pCDNA3-p53CΔ60 (lacking the last 60 residues), and the p53β isoform lacking the oligomerization domain (8). The loss of this domain resulted in the loss of the effect on VRK1, while the conformational mutant p53(L332A) was slightly less efficient in inducing it (Fig. 3C). These data suggested that oligomerization of p53 is necessary for the induction of VRK1 protein downregulation and also indicated a functional difference between p53 isoforms. This further supports the idea of a transcriptionally active p53 role in VRK1 downregulation.
p53 does not affect VRK1 transcription.
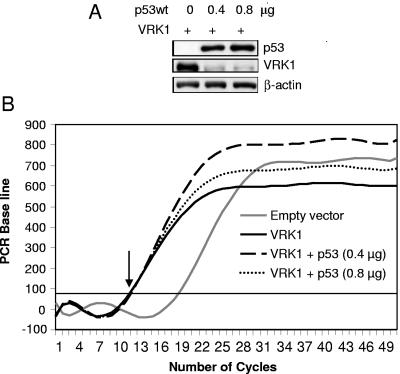
There is the possibility that p53 might affect the expression of the VRK1 gene and contribute in that way to VRK1 regulation. However, the previous observations in which the effect was detected by expressing VRK1 from different promoters do not support this possibility. To further rule out this possibility, mRNA levels were determined by quantitative RT-PCR under conditions where p53 was overexpressed by transfection with three different amounts (0.2, 0.4, and 0.8 μg) of plasmid pCB6+p53. A similar result was obtained with the three doses of p53, and results of the latter two are shown in Fig. 4. These concentrations of p53 induced a strong downregulation of VRK1 protein levels as shown by immunoblot analysis (Fig. 4A); therefore, at these points or earlier, there should also be a decrease in VRK1 RNA if this effect is mediated by downregulation of the promoter. The level of p53 protein did not appear to have an effect on VRK1 transcription (Fig. 4B), since the levels of VRK1 mRNA, both endogenous and transfected, were not significantly affected by p53, as shown by the start of amplification at the same cycle in both samples (indicated by an arrow in the figure) at a time when there is already an important reduction in VRK1 protein level. This effect was observed even when both proteins were expressed from the same cytomegalovirus promoter; therefore, the mechanism cannot be due to a p53 effect on the type of promoter used to express VRK1.
FIG. 4.
The p53 protein does not affect VRK1 gene transcription levels. (A) Levels of p53 and VRK1 protein at the time point used for RNA determination. The levels of VRK1 protein in cells transfected with either 0.4 or 0.8 μg of plasmid pCB6+p53 are shown. The extracts were prepared 36 h after transfection. The proteins were detected by immunoblotting. (B) The levels of VRK1 mRNA, whether after p53 overexpression or not, were determined by RT-PCR as described in Materials and Methods. The results obtained in cells transfected with 0.4 (p53) and 0.8 (p53) μg of plasmid pCB6+p53 are shown.
VRK1 downregulation induced by p53 protein is abrogated by p53 siRNA.
To further establish the dependence of VRK1 downregulation on p53 levels, a different approach was used based on the use of siRNA. For this experiment a fixed level of p53 protein expression was selected, the one corresponding to 200 ng of pCB6+p53, and this p53 protein was eliminated with vector-delivered short hairpin RNA (shRNA) specific for p53 (plasmid pSUPER.retro.p53). The p53 protein was effectively eliminated, and in this situation the level of VRK1 was near its control level (Fig. 5A, top). The p53 shRNA does not affect VRK1 (Fig. 5A, bottom). Next, to show the dependence on p53 levels an experiment was designed in such a way that p53 was first increased in a concentration-dependent manner, and at the high point, it was reduced by shRNA also in a concentration-dependent way. In this experiment the levels of p53 and VRK1 behave inversely, establishing that cycles of fluctuations of these two proteins can occur based on the p53 protein expression levels (Fig. 5B).
FIG. 5.
p53 siRNA blocks the downregulation of VRK1 in H1299 cells. (A) H1299 cells were transfected with pCB6+p53 and an siRNA specific for p53 (pSUPER.retro.p53) as well as pCEFL-HA-VRK1 (5 μg) in different combinations to show that the level of the cotransfected p53 expressed from plasmid pCB6+p53 is downregulated. The cell extracts were prepared 36 h after transfection and analyzed by Western blotting with the corresponding antibodies. (B) H1299 cells were transfected with pCB6+p53 and 5 μg of pCEFL-HA-VRK1. The increase in p53 protein, as expected, downregulated the level of VRK1, but when cells were cotransfected with increasing amounts of siRNA specific for p53, expression plasmid pSUPER.retro.p53, the downregulation of VRK1 was reduced. The graph shows the quantification of results for both proteins.
The downregulation of VRK1 by p53 is mediated by an mdm2-independent mechanism.
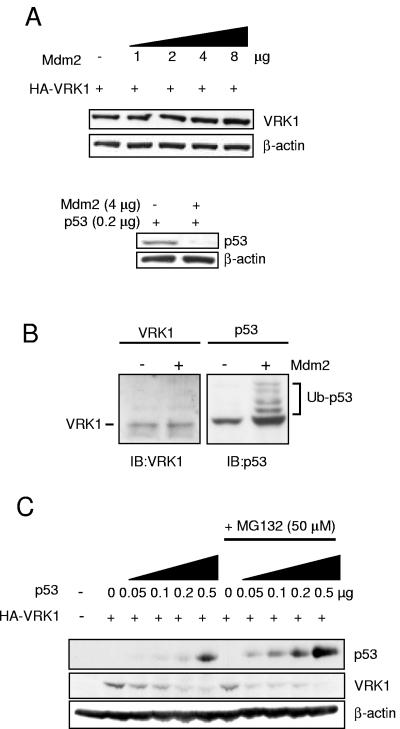
A likely mechanism by which p53 is able to downregulate VRK1 might be the result of a proteolytic activity that is activated, directly or indirectly, following the activation of p53-dependent transcription. In this context the most likely mechanism by which VRK1 might be downregulated as a consequence of p53 transcription is by the action of a ubiquitin ligase, such as that encoded by mdm2/hdm2, which represents the proteasome pathway of protein degradation. Furthermore, the mdm2 gene is a p53 transcriptional target, and this protein promotes p53 degradation, following ubiquitination, by the proteasome (35). Therefore, it could be postulated that VRK1 could also be downregulated by mdm2 or another ubiquitin ligase. To determine if mdm2 might be implicated in the downregulation of VRK1 by p53, different experiments were performed. First, H1299 cells were transfected with increasing amounts of plasmid pCOC-mdm2. The increasing levels of Mdm2 protein by itself did not affect the amount or migration in the SDS-polyacrylamide gel electrophoresis gel of VRK1 protein detected in the cells by Western blotting (FIG. 6A, top blots), indicating that it is not ubiquitinated by Mdm2. As a control of the mdm2 ability to induce the degradation of target proteins by ubiquitination, H1299 cells were cotransfected in parallel with p53 in the presence or absence of mdm2 to show that mdm2 did indeed downregulate p53 in this cell line (Fig. 6A, bottom blots).
FIG. 6.
Mdm2 is not implicated in p53 downregulation of VRK1. (A) Mdm2 protein overexpression does not produce VRK1 degradation by itself. H1299 cells were transfected with 5 μg of pCDNA-VRK1 expression plasmid alone or together with increasing amounts of plasmid encoding pCOC-Mdm2 where indicated. One microgram of plasmid (pUbiquitin-His) encoding ubiquitin was added in all cases. Also 0.2 μg of pCB6+p53 plasmid was cotransfected with Mdm2-encoding plasmid under similar conditions (lower panel). Whole-cell extracts were prepared 36 h after transfection and analyzed by Western blotting with the corresponding antibody. (B) Detection of ubiquitination in p53 but not VRK1 in H1299 cells. The cells were transfected with plasmid pCOC-Mdm2 (4 μg), pCEFL-HA-VRK1 (3 μg), or pCB6+p53 (1 μg) in the presence of 1 μg of pUbiquitin-His. The cell extracts were analyzed by immunoblotting with an anti-VRK1 antibody or a mix of antibodies for p53 to detect their potential change in migration in the gel if there was ubiquitination. (C) VRK1 downregulation by p53 is independent of Mdm2 protein and proteasome-mediated degradation. Mouse embryo fibroblasts derived from double knockout mice (p53−/− mdm2−/−) were cotransfected with the indicated plasmids pCEFL-HA-VRK1 (3 μg) or pCB6+p53. Where indicated, the proteasome inhibitor MG132 was added at 50 μM. Cells were processed as described in panel A, and levels of p53 and VRK1 proteins were detected.
To confirm that VRK1 is not ubiquitinated, we performed an experiment under conditions where protein ubiquitination can be detected using p53 as a positive control. The H1299 cell was transfected with pCOC-mdm2 and either pCB6+p53 or pCEFL-HA-VRK1 in the presence of pUbiquitin-His, so that ubiquitin is not limiting, and of the proteasome inhibitor MG132 to prevent degradation of the proteins by the proteasome and permit accumulation of ubiquitinated proteins. The change in migration of ubiquitinated p53 was clearly detected in the form of a p53 ladder, but there was no change in the migration in the gel of the VRK1 protein (Fig. 6B), suggesting that ubiquitinated forms of this protein do not exist under these conditions.
To further rule out any participation of mdm2, murine embryo fibroblasts deficient in p53 and mdm2 derived from double-knockout mice were used (35). These fibroblasts were transfected with increasing amounts of pCB6+53 and a fixed amount of pCEFL-HA-VRK1. The downregulation of VRK1 was also achieved (Fig. 6C) indicating that mdm2 was not necessary for this effect. Next, to rule out the possibility that the effect is not mediated by any other ubiquitin ligase and degradation in the proteasome, a similar experiment was performed in the presence of the proteasome inhibitor MG132. The proteasome inhibitor was able to maintain even higher levels of p53 (Fig. 6C), indicating that even in the absence of mdm2 there was some proteasome-mediated degradation of p53; but the effect of p53 levels on VRK1 was always observed, and its magnitude was even larger in the presence of MG132 as the levels of p53 were higher (Fig. 6C), probably resulting from blocking the effect of other ubiquitin-ligases. These experiments suggested that the downregulation of VRK1 was mediated by an mdm2- and proteasome-independent mechanism. p53 degradation by mdm2 partially prevented the downregulation of VRK1, not by a direct Mdm2 effect but as a consequence of reduced p53 levels (not shown).
Downregulation of VRK1 is sensitive to inhibition of endosome-lysosome traffic and lysosomal protease inhibitors.
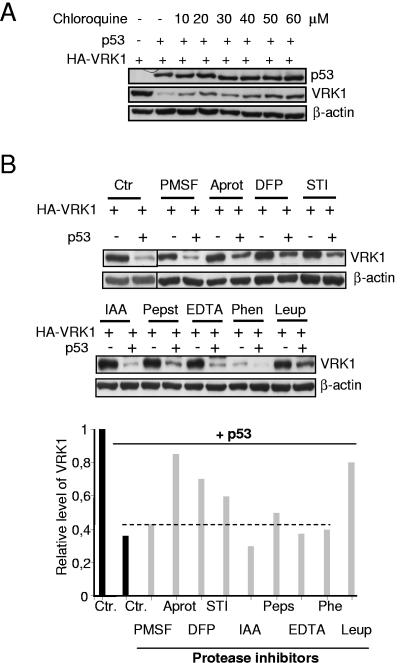
The alternative pathway of protein degradation is represented by the lysosomal pathway, mainly composed of acidic serine and cysteine proteases (17). In this pathway the proteins have to be transported from late endosomes to the lysosomes, where they are degraded by lysosomal serine proteases that function at an acidic pH. Therefore, it was decided to test the potential implication of this route by using a variety of endosome-lysosome traffic and protease inhibitors that affect different types of intracellular proteases. The movement of late endosomes to lysosomes can be experimentally inhibited by chloroquine (18, 65). Therefore, the sensitivity of VRK1 downregulation to inhibitors of the transport system was tested by performing a dose-response analysis with chloroquine. As the amount of chloroquine was increased in the range of 10 to 60 μM, there was an inhibition of p53-induced downregulation of VRK1 (Fig. 7A); at higher concentrations chloroquine was toxic to the cells. These data indicated that VRK1 enters the late-endosome to lysosome system, and thus, partial protection was conferred by chloroquine.
FIG. 7.
VRK1 downregulation occurs in the lysosomal pathway. (A) Sensitivity of VRK1 downregulation to the inhibition of the late endosome to lysosome traffic by chloroquine. H1299 cells were transfected with pCB6+p53 and pCEFL-HA-VRK1. Chloroquine at the indicated concentrations was added 12 h after transfection, and cell extracts were prepared for immunoblot analysis 36 h after transfection. (B) Effect of protease inhibitors on the downregulation of VRK1 induced by p53. H1299 cells were transfected with pCB6+p53 and pCEFL-HA-VRK1, and different protease inhibitors were added to the culture 18 h after transfection. The inhibitors were used at the specified concentrations for 5 h. The serine protease inhibitors were the following: 1 mM PMSF, 10 μl/ml aprotinin (Aprot), 0.1 mM DFP, and 10 μl/ml STI. The following protease inhibitors were also used: for serine and cysteine, 50 μM leupeptin (Leup); for cysteine, 50 μM IAA; and for aspartyl, 1 μM pepstatin (Peps). As metalloprotease inhibitors, 5 mM EDTA and 5 mM 1,10-phenantroline (Phen) were used. Calpain inhibitors were also tested with no effect. The cells were collected 23 h after transfection and lysed for determination of VRK1 and p53 levels by Western blotting. Ctr, controls without inhibitors.
Next, the sensitivity to a variety of protease inhibitors was also determined. The protease inhibitors tested were pepstatin for aspartyl proteases; PMSF, aprotinin, DFP, STI, and leupeptin for serine proteases; IAA and leupeptin for cysteine proteases; EDTA and 1,10-phenantroline for metaloproteasas; and ALLN, calpain inhibitor III, calpeptin, EST, and PD150606 for calpain proteases. Because these inhibitors are in general toxic, the cells 18 h after transfection were treated for 5 h with the corresponding inhibitor, washed, and incubated for an additional 5 h before the extracts were prepared. Cells were lysed and analyzed by immunoblotting for the corresponding proteins (Fig. 7B). Most of the inhibitors of serine proteases (aprotinin, leupeptin, and DFP) or cysteine proteases such as leupeptin partially prevented the downregulation of VRK1. Leupeptin is furthermore an established inhibitor of the lysosomal pathway of protein degradation (44). None of the calpain inhibitors affected the downregulation (not shown). The possible activation of caspases was also determined by the analysis of poly(ADP-ribose) polymerase proteolytic cleavage, which was not affected in these experiments, suggesting that there was no activation of the caspase pathway (not shown).
DISCUSSION
VRK1 is a nuclear kinase with p53 among its targets (29); therefore, in this report we have attempted to characterize new aspects of their interaction in the context of a novel signaling pathway. This is particularly important since VRK1 is likely to play an important role in the control of normal cell proliferation (58). Because VRK1 induces an accumulation of p53, it is expected that the levels and activity of VRK1 will be tightly regulated. Accumulation of p53 can have a deleterious effect on the cell since it can stop cell division and induce apoptosis. Once the biological effect of p53 has been achieved, it is reasonable to expect that the stabilizing factor will return to basal levels in some way; otherwise, there would be a permanent activation and accumulation of p53. One of the mechanisms by which VRK1 contributes to p53 stabilization is by phosphorylation of Thr18 that prevents the p53-Mdm2 interaction (19, 22, 48) and promotes p300 recruitment and transcriptional activation (58). It was expected that a p53 stabilizing protein, such as VRK1, should be the target of a down-regulatory process. A downregulation of another p53 positive modulator, the Chk1 protein, has already been identified (15). In this report we have shown that increasing the level of intracellular p53 led to the downregulation of VRK1 levels. The inverse correlation between VRK1 and p53 observed in cell lines can also be induced in vivo under conditions in which a stable accumulation of p53 can be obtained, as is the case of stimulation of fibroblasts with UV light that induces senescence (60), but other ways to induce the effect more effectively may be found. Senescence in fibroblasts is already known to be accompanied by Thr18 phosphorylation and accumulation of p53 (13). The inverse correlation can be reproduced by transfection in different cell types, making it more amenable for its characterization.
The contribution of p53 to VRK1 regulation is an intermediate step in the process for which it is absolutely essential, the structural maintenance of p53 molecules, since alterations in each domain affect the downregulation of VRK1. This requirement is important because it reflects a functional difference between normal p53 and p53 mutated in human cancer, either sporadic or hereditary, or p53 isoforms. The response is affected by the transactivation domain of p53 since the effect is lost in the case of Δ40p53, an isoform lacking the first 40 amino acids, which have a dominant negative role and counteract growth suppression (10, 14). However, the conformational mutant p53(L22Q, W23S), which affects some response elements but not others (7, 31, 45, 59), still induces downregulation. Many p53 regulators interact with p53 through this N-terminal domain. Therefore, it is likely that some p53 interaction not yet identified could have a role in the VRK1 downregulation induced by p53. The contribution of the integrity of the DNA-binding domain is demonstrated by the loss of effect when common mutations, either conformational or with loss of DNA contact, are present. This might have consequences in tumors bearing these mutations. Some conformational mutations are not active and contribute to tumor development by routes currently under characterization since their pattern of tumor formation is different from that induced by structural mutants affecting contact with DNA (25, 39). It is important to note that this lack of effect occurs with the most common p53 mutations, detected in both sporadic tumors and hereditary Li-Fraumeni syndrome (40). The dependence on the conservation of the oligomerization domain is further consistent with the requirement for the integrity of p53 as the formation of a functional tetramer is also important for the effect. All these data point to a potentially important functional consequence for VRK1 downregulation since it does not occur when p53 isoforms lacking transactivation or oligomerization domains are expressed (8, 10). The p53 gene is mutated in more than half of human tumors (63), and the mutations are concentrated in the DNA-binding domain affecting the transcriptional role of p53. In tumors with p53 mutations, the mechanism responsible for VRK1 negative regulation is probably not induced, leading to a more stable VRK1 protein or even higher levels. Since VRK1 activity is correlated with cell proliferation, these tumor cell lines are likely to have a higher potential to divide, but whether the process is successful will also depend on other cell properties. The exact implication, if any, of this mechanism in cancer progression has yet to be elucidated.
The results with the different p53 mutants indicate that the mechanism appears to be either a direct consequence of a gene regulated by p53 or an interaction of p53 with some regulator that includes binding to DNA. The regulated phenomenon is not the expression of VRK1 itself, since the effect is independent of the type of promoter from which VRK1 was expressed, either endogenous or from different types of plasmids. The genes induced by p53 in the H1299 cell line have been partially characterized by microarray analysis (20, 33, 49, 64), but a clear candidate gene cannot be identified as a potential mediator of the effect reported. VRK1 and other p53 activators act by increasing p53 stability, with the consequent rise in p53 levels. Several mechanisms ensure that p53 accumulation is transient, and they include the transcriptional activation of negative modulators, as is the case for mdm2, and the downregulation of positive modulators, as is the case of VRK1, through the transcription of other p53-dependent genes as an intermediate step. It may be either a protease that can degrade VRK1 or a protein that somehow modifies VRK1 stability, either by interacting with it or by a covalent modification. Both may also affect its kinase activity, which in some way can influence VRK1 stability by making it more susceptible to enter the proteolytic degradation via the lysosome. The inactive kinase VRK1(K179E), although less stable, was also equally downregulated by p53 (unpublished results).
Intracellular proteins can be degraded by one of the two alternative pathways, the proteasome or the lysosome. VRK1 lacks PEST sequences that would make a protein susceptible to proteasome-mediated degradation (5, 21). Mechanistically, the VRK1 downregulation induced by p53 is independent of a proteasome-mediated pathway, and mdm2 is not implicated since the downregulation was detected in mdm2-deficient cells and was also insensitive to proteasome inhibitors. However, the downregulation is sensitive to some serine protease inhibitors, suggesting that the final step is executed by a member this protease family. In this group is included leupeptin, the prototype inhibitor of the lysosome-mediated degradation pathway (44). Therefore, it is highly likely that this is the pathway responsible for the effect, but how VRK1 is targeted for degradation is not known. This would be the step that requires p53-dependent transcription to control the targeting of VRK1 for lysosomal degradation.
In conclusion, in this report we have identified and characterized a possible autoregulatory loop between the tumor suppressor p53 protein and its stabilizing protein, the VRK1 kinase; this regulation, as an intermediate step, depends on the p53 DNA-binding and transactivation domain and is not mediated by mdm2. This downregulation as final step requires the transport of VRK1 from late endosomes to lysosomes and the subsequent degradation mediated by lysosomal serine proteases. The identification of this new regulatory circuit opens up new possibilities to better understand the regulation of cell proliferation in higher eukaryotes.
Acknowledgments
This work was funded by grants from Ministerio de Educación y Ciencia (SAF2004-02900), Fundación de Investigación Médica MM, Junta de Castilla y León (SAN-SA04/05 and CSI05A05), and Fundación Memoria Samuel Solórzano Barruso. A.V. and S.B. were supported by a fellowship from Ministerio de Educación y Ciencia. F.M.V. was supported by fellowships from Fundación Ramón Areces and Asociación Española contra el Cáncer.
REFERENCES
- 1.Agarwal, M. L., W. R. Taylor, M. V. Chernov, O. B. Chernova, and G. R. Stark. 1998. The p53 network. J. Biol. Chem. 273:1-4. [DOI] [PubMed] [Google Scholar]
- 2.Ashcroft, M., M. H. Kubbutat, and K. H. Vousden. 1999. Regulation of p53 function and stability by phosphorylation. Mol. Cell. Biol. 19:1751-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balint, E. E., and K. H. Vousden. 2001. Activation and activities of the p53 tumour suppressor protein. Br. J. Cancer 85:1813-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barcia, R., S. Lopez-Borges, F. M. Vega, and P. A. Lazo. 2002. Kinetic properties of p53 phosphorylation by the human vaccinia-related kinase 1. Arch. Biochem. Biophys. 399:1-5. [DOI] [PubMed] [Google Scholar]
- 5.Beinke, S., M. J. Robinson, M. Hugunin, and S. C. Ley. 2004. Lipopolysaccharide activation of the TPL-2/MEK/extracellular signal-regulated kinase mitogen-activated protein kinase cascade is regulated by IκB kinase-induced proteolysis of NF-κB1 p105. Mol. Cell. Biol. 24:9658-9667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Béroud, C., and T. Soussi. 2003. The UMD-p53 database: new mutations and analysis tools. Hum. Mutation 21:176-181. [DOI] [PubMed] [Google Scholar]
- 7.Boehden, G. S., N. Akyuz, K. Roemer, and L. Wiesmuller. 2003. p53 mutated in the transactivation domain retains regulatory functions in homology-directed double-strand break repair. Oncogene 22:4111-4117. [DOI] [PubMed] [Google Scholar]
- 8.Bourdon, J. C., K. Fernandes, F. Murray-Zmijewski, G. Liu, A. Diot, D. P. Xirodimas, M. K. Saville, and D. P. Lane. 2005. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. [DOI] [PMC free article] [PubMed]
- 9.Canman, C., T. M. Gilmer, S. B. Coutts, and M. B. Kastan. 1995. Growth factor modulation of p53-mediated growth arrest versus apoptosis. Genes Dev. 9:600-611. [DOI] [PubMed] [Google Scholar]
- 10.Courtois, S., G. Verhaegh, S. North, M. G. Luciani, P. Lassus, U. Hibner, M. Oren, and P. Hainaut. 2002. DeltaN-p53, a natural isoform of p53 lacking the first transactivation domain, counteracts growth suppression by wild-type p53. Oncogene 21:6722-6728. [DOI] [PubMed] [Google Scholar]
- 11.Dornan, D., I. Wertz, H. Shimizu, D. Arnott, G. D. Frantz, P. Dowd, K. O'Rourke, H. Koeppen, and V. M. Dixit. 2004. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 429:86-92. [DOI] [PubMed] [Google Scholar]
- 12.Epstein, C. B., E. F. Attiyeh, D. A. Hobson, A. L. Silver, J. R. Broach, and A. J. Levine. 1998. p53 mutations isolated in yeast based on loss of transcription factor activity: similarities and differences from p53 mutations detected in human tumors. Oncogene 16:2115-2122. [DOI] [PubMed] [Google Scholar]
- 13.Ferbeyre, G., E. de Stanchina, A. W. Lin, E. Querido, M. E. McCurrach, G. J. Hannon, and S. W. Lowe. 2002. Oncogenic ras and p53 cooperate to induce cellular senescence. Mol. Cell. Biol. 22:3497-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh, A., D. Stewart, and G. Matlashewski. 2004. Regulation of human p53 activity and cell localization by alternative splicing. Mol. Cell. Biol. 24:7987-7997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottifredi, V., O. Karni-Schmidt, S.-Y. Shieh, and C. Prives. 2001. p53 down-regulates CHK1 through p21 and the retinoblastoma protein. Mol. Cell. Biol. 21:1066-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gudkov, A. V., and E. A. Komarova. 2003. The role of p53 in determining sensitivity to radiotherapy. Nat. Rev. Cancer 3:117-129. [DOI] [PubMed] [Google Scholar]
- 17.Hunziker, W., and H. J. Geuze. 1996. Intracellular trafficking of lysosomal membrane proteins. Bioessays 18:379-389. [DOI] [PubMed] [Google Scholar]
- 18.Ignatiuk, A., J. P. Quickfall, A. D. Hawrysh, M. D. Chamberlain, and D. H. Anderson. 2006. The smaller isoforms of ankyrin 3 bind to the p85 subunit of phosphatidylinositol 3′-kinase and enhance platelet-derived growth factor receptor down-regulation. J. Biol. Chem. 281:5956-5964. [DOI] [PubMed] [Google Scholar]
- 19.Jabbur, J. R., A. D. Tabor, X. Cheng, H. Wang, M. Uesugi, G. Lozano, and W. Zhang. 2002. Mdm-2 binding and TAF(II)31 recruitment is regulated by hydrogen bond disruption between the p53 residues Thr18 and Asp21. Oncogene 21:7100-7113. [DOI] [PubMed] [Google Scholar]
- 20.Kannan, K., N. Amariglio, G. Rechavi, and D. Givol. 2000. Profile of gene expression regulated by induced p53: connection to the TGF-β family. FEBS Lett. 470:77-82. [DOI] [PubMed] [Google Scholar]
- 21.Katagiri, C., K. Masuda, T. Urano, K. Yamashita, Y. Araki, K. Kikuchi, and H. Shima. 2005. Phosphorylation of Ser-446 determines stability of MKP-7. J. Biol. Chem. 280:14716-14722. [DOI] [PubMed] [Google Scholar]
- 22.Kussie, P. H., S. Gorina, V. Marechal, B. Elenbaas, J. Moreau, A. J. Levine, and N. P. Pavletich. 1996. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 274:948-953. [DOI] [PubMed] [Google Scholar]
- 23.Kwon, S. Y., Y. J. Choi, T. H. Kang, K. H. Lee, S. S. Cha, G. H. Kim, H. S. Lee, K. T. Kim, and K. J. Kim. 2005. Highly efficient protein expression and purification using bacterial hemoglobin fusion vector. Plasmid 53:274-282. [DOI] [PubMed] [Google Scholar]
- 24.Lahav, G., N. Rosenfeld, A. Sigal, N. Geva-Zatorsky, A. J. Levine, M. B. Elowitz, and U. Alon. 2004. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat. Genet. 36:147-150. [DOI] [PubMed] [Google Scholar]
- 25.Lang, G. A., T. Iwakuma, Y. A. Suh, G. Liu, V. A. Rao, J. M. Parant, Y. A. Valentin-Vega, T. Terzian, L. C. Caldwell, L. C. Strong, A. K. El-Naggar, and G. Lozano. 2004. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 119:861-872. [DOI] [PubMed] [Google Scholar]
- 26.Lazo, P. A., F. M. Vega, and A. Sevilla. 2005. Vaccinia-related kinase-1. AfCS Nature Molecule Pages [Online.] doi: 10.1038/mp.a003025.01. [DOI]
- 27.Lev Bar-Or, R., R. Maya, L. A. Segel, U. Alon, A. J. Levine, and M. Oren. 2000. Generation of oscillations by the p53-Mdm2 feedback loop: a theoretical and experimental study. Proc. Natl. Acad. Sci. USA 97:11250-11255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine, A. 1997. p53, the cellular gatekeeper for growth and division. Cell 88:323-331. [DOI] [PubMed] [Google Scholar]
- 29.Lopez-Borges, S., and P. A. Lazo. 2000. The human vaccinia-related kinase 1 (VRK1) phosphorylates threonine-18 within the mdm-2 binding site of the p53 tumour suppressor protein. Oncogene 19:3656-3664. [DOI] [PubMed] [Google Scholar]
- 30.Manning, G., D. B. Whyte, R. Martinez, T. Hunter, and S. Sudarsanam. 2002. The protein kinase complement of the human genome. Science 298:1912-1934. [DOI] [PubMed] [Google Scholar]
- 31.Matas, D., A. Sigal, P. Stambolsky, M. Milyavsky, L. Weisz, D. Schwartz, N. Goldfinger, and V. Rotter. 2001. Integrity of the N-terminal transcription domain of p53 is required for mutant p53 interference with drug-induced apoptosis. EMBO J. 20:4163-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michael, D., and M. Oren. 2003. The p53-Mdm2 module and the ubiquitin system. Semin. Cancer Biol. 13:49-58. [DOI] [PubMed] [Google Scholar]
- 33.Mirza, A., Q. Wu, L. Wang, T. McClanahan, W. R. Bishop, F. Gheyas, W. Ding, B. Hutchins, T. Hockenberry, P. Kirschmeier, J. R. Greene, and S. Liu. 2003. Global transcriptional program of p53 target genes during the process of apoptosis and cell cycle progression. Oncogene 22:3645-3654. [DOI] [PubMed] [Google Scholar]
- 34.Moll, U. M., and O. Petrenko. 2003. The MDM2-p53 interaction. Mol. Cancer Res. 1:1001-1008. [PubMed] [Google Scholar]
- 35.Montes de Oca Luna, R., D. S. Wagner, and G. Lozano. 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378:203-206. [DOI] [PubMed] [Google Scholar]
- 36.Nichols, R. J., and P. Traktman. 2004. Characterization of three paralogous members of the mammalian vaccinia related kinase family. J. Biol. Chem. 279:7934-7946. [DOI] [PubMed] [Google Scholar]
- 37.Nichols, R. J., M. S. Wiebe, and P. Traktman. 2006. The vaccinia-related kinases phosphorylate the N terminus of BAF, regulating its interaction with DNA and its retention in the nucleus. Mol. Biol. Cell 17:2451-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okorokov, A. L. 2003. p53 in a crosstalk between DNA repair and cell cycle checkpoints. Cell Cycle 2:233-235. [PubMed] [Google Scholar]
- 39.Olive, K. P., D. A. Tuveson, Z. C. Ruhe, B. Yin, N. A. Willis, R. T. Bronson, D. Crowley, and T. Jacks. 2004. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 119:847-860. [DOI] [PubMed] [Google Scholar]
- 40.Olivier, M., D. E. Goldgar, N. Sodha, H. Ohgaki, P. Kleihues, P. Hainaut, and R. A. Eeles. 2003. Li-Fraumeni and related syndromes: correlation between tumor type, family structure, and TP53 genotype. Cancer Res. 63:6643-6650. [PubMed] [Google Scholar]
- 41.Oren, M. 2003. Decision making by p53: life, death and cancer. Cell Death Differ. 10:431-442. [DOI] [PubMed] [Google Scholar]
- 42.Papin, J. A., T. Hunter, B. O. Palsson, and S. Subramaniam. 2005. Reconstruction of cellular signalling networks and analysis of their properties. Nat. Rev. Mol. Cell. Biol. 6:99-111. [DOI] [PubMed] [Google Scholar]
- 43.Prives, C., and P. A. Hall. 1999. The p53 pathway. J. Pathol. 187:112-126. [DOI] [PubMed] [Google Scholar]
- 44.Qin, H., Q. Shao, S. A. Igdoura, M. A. Alaoui-Jamali, and D. W. Laird. 2003. Lysosomal and proteasomal degradation play distinct roles in the life cycle of Cx43 in gap junctional intercellular communication-deficient and -competent breast tumor cells. J. Biol. Chem. 278:30005-30014. [DOI] [PubMed] [Google Scholar]
- 45.Roemer, K., and N. Mueller-Lantzsch. 1996. p53 transactivation domain mutant Q22, S23 is impaired for repression of promoters and mediation of apoptosis. Oncogene 12:2069-2079. [PubMed] [Google Scholar]
- 46.Saito, S., H. Yamaguchi, Y. Higashimoto, C. Chao, Y. Xu, A. J. Fornace, Jr., E. Appella, and C. W. Anderson. 2003. Phosphorylation site interdependence of human p53 post-translational modifications in response to stress. J. Biol. Chem. 278:37536-37544. [DOI] [PubMed] [Google Scholar]
- 47.Santos, C. R., M. Rodriguez-Pinilla, F. M. Vega, J. L. Rodriguez-Peralto, S. Blanco, A. Sevilla, A. Valbuena, T. Hernandez, A. J. van Wijnen, F. Li, E. de Alava, M. Sanchez-Cespedes, and P. A. Lazo. 2006. VRK1 signaling pathway in the context of the proliferation phenotype in head and neck squamous cell carcinoma. Mol. Cancer Res. 4:177-185. [DOI] [PubMed] [Google Scholar]
- 48.Schon, O., A. Friedler, M. Bycroft, S. Freund, and A. Fersht. 2002. Molecular mechanism of the interaction between MDM2 and p53. J. Mol. Biol. 323:491-501. [DOI] [PubMed] [Google Scholar]
- 49.Scian, M. J., K. E. R. Stagliano, M. A. Ellis, S. Hassan, M. Bowman, M. F. Miles, S. P. Deb, and S. Deb. 2004. Modulation of gene expression by tumor-derived p53 mutants. Cancer Res. 64:7447-7454. [DOI] [PubMed] [Google Scholar]
- 50.Sevilla, A., C. R. Santos, R. Barcia, F. M. Vega, and P. A. Lazo. 2004. c-Jun phosphorylation by the human vaccinia-related kinase 1 (VRK1) and its cooperation with the N-terminal kinase of c-Jun (JNK). Oncogene 23:8950-8958. [DOI] [PubMed] [Google Scholar]
- 51.Sevilla, A., C. R. Santos, F. M. Vega, and P. A. Lazo. 2004. Human vaccinia-related kinase 1 (VRK1) activates the ATF2 transcriptional activity by novel phosphorylation on Thr-73 and Ser-62 and cooperates with JNK. J. Biol. Chem. 279:27458-27465. [DOI] [PubMed] [Google Scholar]
- 52.Shen, Y., and E. White. 2001. p53-dependent apoptosis pathways. Adv. Cancer Res. 82:55-84. [DOI] [PubMed] [Google Scholar]
- 53.Shirangi, T. R., A. Zaika, and U. M. Moll. 2002. Nuclear degradation of p53 occurs during down-regulation of the p53 response after DNA damage. FASEB J. 16:420-422. [DOI] [PubMed] [Google Scholar]
- 54.Sigal, A., D. Matas, N. Almog, N. Goldfinger, and V. Rotter. 2001. The C terminus of mutant p53 is necessary for its ability to interfere with growth arrest or apoptosis. Oncogene 20:4891-4898. [DOI] [PubMed] [Google Scholar]
- 55.Sigal, A., and V. Rotter. 2000. Oncogenic mutations of the p53 tumor suppressor: the demons of the guardian of the genome. Cancer Res. 60:6788-6793. [PubMed] [Google Scholar]
- 56.Soussi, T., C. Ishioka, M. Claustres, and C. Beroud. 2006. Locus-specific mutation databases: pitfalls and good practice based on the p53 experience. Nat. Rev. Cancer 6:83-90. [DOI] [PubMed] [Google Scholar]
- 57.Taylor, W. R., and G. R. Stark. 2001. Regulation of the G2/M transition by p53. Oncogene 20:1803-1815. [DOI] [PubMed] [Google Scholar]
- 58.Vega, F. M., A. Sevilla, and P. A. Lazo. 2004. p53 Stabilization and accumulation induced by human vaccinia-related kinase 1. Mol. Cell. Biol. 24:10366-10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Venot, C., M. Maratrat, V. Sierra, E. Conseiller, and L. Debussche. 1999. Definition of a p53 transactivation function-deficient mutant and characterization of two independent p53 transactivation subdomains. Oncogene 18:2405-2410. [DOI] [PubMed] [Google Scholar]
- 60.von Zglinicki, T., G. Saretzki, J. Ladhoff, F. d'Adda di Fagagna, and S. P. Jackson. 2005. Human cell senescence as a DNA damage response. Mech. Ageing Dev. 126:111-117. [DOI] [PubMed] [Google Scholar]
- 61.Vousden, K. H. 2002. Activation of the p53 tumor suppressor protein. Biochim. Biophys. Acta 1602:47-59. [DOI] [PubMed] [Google Scholar]
- 62.Wahl, G. M., and A. M. Carr. 2001. The evolution of diverse biological responses to DNA damage: insights from yeast and p53. Nat. Cell Biol. 3:E277-286. [DOI] [PubMed] [Google Scholar]
- 63.Walker, D. R., J. P. Bond, R. E. Tarone, C. C. Harris, W. Makalowski, M. S. Boguski, and M. S. Greenblatt. 1999. Evolutionary conservation and somatic mutation hotspot maps of p53: correlation with p53 protein structural and functional features. Oncogene 18:211-218. [DOI] [PubMed] [Google Scholar]
- 64.Wang, L., Q. Wu, P. Qiu, A. Mirza, M. McGuirk, P. Kirschmeier, J. R. Greene, Y. Wang, C. B. Pickett, and S. Liu. 2001. Analyses of p53 target genes in the human genome by bioinformatic and microarray approaches. J. Biol. Chem. 276:43604-43610. [DOI] [PubMed] [Google Scholar]
- 65.Xiao, K., D. F. Allison, M. D. Kottke, S. Summers, G. P. Sorescu, V. Faundez, and A. P. Kowalczyk. 2003. Mechanisms of VE-cadherin processing and degradation in microvascular endothelial cells. J. Biol. Chem. 278:19199-19208. [DOI] [PubMed] [Google Scholar]