Abstract
Cytochrome c oxidase (COX) biogenesis requires COX10, which encodes a protoheme:heme O farnesyl transferase that participates in the biosynthesis of heme a. We created COX10 knockout mouse cells that lacked cytochrome aa3, were respiratory deficient, had no detectable complex IV activity, and were unable to assemble COX. Unexpectedly, the levels of respiratory complex I were markedly reduced in COX10 knockout clones. Pharmacological inhibition of COX did not affect the levels of complex I, and transduction of knockout cells with lentivirus expressing wild-type or mutant COX10 (retaining residual activity) restored complex I to normal levels. Pulse-chase experiments could not detect newly assembled complex I, suggesting that either COX is required for assembly of complex I or the latter is quickly degraded. These results suggest that in rapidly dividing cells, complex IV is required for complex I assembly or stability.
Cytochrome c oxidase (COX or complex IV) is the terminal complex of the electron transport chain. COX deficiencies present a wide variety of clinical phenotypes in humans, but their molecular mechanisms are not clear. Complex IV catalyzes the transfer of electrons from reduced cytochrome c to oxygen in a reaction that is coupled to proton pumping across the inner mitochondrial membrane (35). In mammals, COX is a 200-kDa multicomponent protein located in the inner mitochondrial membrane, and it is active as a dimer (34). It is composed of 13 subunits encoded by both the mitochondrial (subunits 1, 2, and 3, which form the catalytic core of the enzyme) and the nuclear genomes (2, 29). The enzyme has two hemes (a and a3), two copper centers (CuA and CuB), and magnesium and zinc ions. Both heme groups and the CuB center are located within the Cox1 subunit, and the CuA site is located within Cox2. Electrons are transferred from reduced cytochrome c to the CuA site, then to heme a, and finally to heme a3-CuB prior to the final transfer to O2 to produce H2O.
The incorporation of heme a in subunit 1 (Cox1), the catalytic site of the complex, requires several factors that modify the protoheme backbone, among which is the product of COX10, a protoheme:heme O farnesyl transferase. Heme a biosynthesis starts with the addition of a farnesyl group at the vinyl group of carbon 2 of heme B to make heme O. This step is catalyzed by Cox10. In the next step, the methyl group on carbon 8 is oxidized to a carboxy methyl group. This reaction is thought to be carried out by Cox15, ferredoxin (Yah1), and ferredoxin reductase (Arh1), which act in concert as a heme monooxygenase (7, 8, 10). The farnesyl chain of heme a is used as a lipophilic anchor holding the heme at the proper position within the oxidase complex (20).
In this study, we analyzed the effect of a COX10 knockout (KO) in the mitochondrial function of mouse skin fibroblasts. Our results showed that mutant cells have not only a COX deficiency but also a marked decrease in the levels of complex I.
MATERIALS AND METHODS
Cell culture.
A primary cell line from skin fibroblasts was established from a mouse homozygous for the floxed COX10 gene (exon 6). loxP sites were introduced into the COX10 gene by homologous recombination of a plasmid containing floxed exon 6 of COX10 in mouse embryonic stem cells. The selection cassette was removed by partial recombination of loxP sites after transient Cre expression (17). Selected embryonic stem cells were injected into blastocysts, and chimera mice were generated. After obtaining germ line transmission of the floxed COX10 allele, we produced homozygous mice (13). A primary cell line from skin fibroblasts was grown from this mouse with floxed COX10. Cells were grown in high-glucose Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, 1 mM pyruvate, 50 μg/ml uridine at 37°C in an atmosphere of 5% CO2. Cells were immortalized by infection with a retrovirus expressing the E6 and E7 genes of type 16 human papilloma virus carrying G418 resistance (kindly provided by E. Shoudbridge) (19). To generate COX10 knockout cells, the immortalized cell line was transfected with the pCre-Hygro plasmid expressing the P1 Cre recombinase (kindly provided by J. Marth). Several clones were obtained by ring cloning and were grown under hygromycin selection. To determine cell growth of the selected clones, 1 × 104 cells were plated in triplicate on 24-well plates, trypsinized, and counted every 24 h.
PCR and Southern blotting.
Total DNA was obtained from cells by phenol-chloroform extraction and isopropanol precipitation. Deletion of exon 6 was detected by PCR and Southern blotting. PCR amplification of the deleted allele was performed within the intronic sequence flanking exon 6. These primers amplify both the deleted and the intact floxed allele. To detect the deleted as well as the floxed allele by Southern blotting, 20 μg of total DNA was digested with BamHI, separated in a 0.7% agarose gel, and transferred to Z-Probe membrane (Bio-Rad, Hercules, CA). The membrane was hybridized with a 32P-radiolabeled COX10 genomic probe (shown in Fig. 1A) using the random Primer Labeling kit from Roche Biomedicals (Indianapolis, IN). Southern blotting to detect mitochondrial DNA (mtDNA) was performed as described above by digesting DNA with SacI or with NheI and hybridizing the blotted membrane to a DNA probe spanning nucleotide positions 5556 to 6268 of the mouse mitochondrial genome.
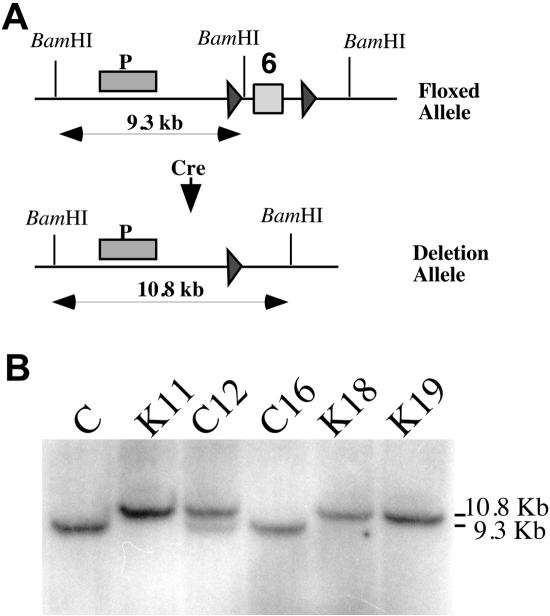
FIG. 1.
Generation of COX10 knockout mouse cells. (A) loxP sites were introduced into COX10 by homologous recombination of a plasmid containing a floxed exon 6 (box marked “6”) in embryonic stem cells. The diagram shows the position of the loxP sites in the gene (black triangles). A primary culture from skin fibroblasts was established from a homozygous mouse with the floxed COX10 and transfected with the pCre-Hygro plasmid to generate KO cells. To generate KO cells, several hygromycin-resistant clones were obtained and characterized. (B) Genetic characterization of the COX10 locus. The parental fibroblast line (C) and five hygromycin-resistant clones were analyzed by Southern blotting to detect the deletion of COX10 exon 6. The deleted allele was detected in the K clones (K11, K18, and K19); the wild-type floxed allele was detected in two C clones (C12 and C16). Clone C12 showed the presence of both the floxed and the deletion allele, indicating that it was heterozygous for the deletion. DNA was digested with BamHI, and the location of the probe is shown as a bar (P) in panel A.
Mitochondrial preparation.
Cells from seven to eight confluent T75 flasks were used for mitochondria isolation by N2 cavitation (16). Cells were trypsinized; resuspended in phosphate-buffered saline (PBS); and washed in 100 mM sucrose, 1 mM EGTA, 20 mM morpholinepropanesulfonic acid, pH 7.4, 0.1% bovine serum albumin (BSA); and centrifuged at 1,000 × g for 5 min. Cell pellets were resuspended in the same buffer containing 10 mM triethanolamine, 5% Percoll, and a protease inhibitor cocktail (Roche Biomedicals). Cells were disrupted by N2 cavitation at 600 lb/in2 for 30 min at 4°C. The osmolarity of the sample was increased to 250 mM sucrose prior to centrifugation at 2,500 × g for 10 min. The supernatant was further centrifuged at 25,000 × g for 10 min to obtain the mitochondrial fraction. Samples were stored at −80°C until needed.
Cytochrome spectra.
The mitochondrial preparation (2 mg), obtained by N2 cavitation, was incubated with 1 M KCl and 20 mM Tris, pH 7.5, in a final volume of 0.5 ml at 4°C. Cytochromes were solubilized by addition of sodium deoxycholate to a final concentration of 1% (vol/vol). Samples were centrifuged at 100,000 × g for 15 min at 4°C. Supernatant was collected and clarified by the addition of 20% potassium cholate to a final concentration of 2%. One aliquot of the preparation was oxidized with potassium ferricyanide, and the other aliquot was reduced with sodium dithionate. Absorbance spectra were determined by scanning the samples at 360 to 650 nm in a Beckman spectrophotometer. The total cytochrome spectrum represents the reduced spectra minus oxidized spectra for each sample.
Cell respiration and enzyme activities.
Oxygen consumption was measured polarographically in 0.3 M mannitol, 10 mM KCl, 5 mM MgCl2, 1 mg/ml BSA, 10 mM KH2PO4, pH 7.4, with a Clark oxygen electrode (Hansatech Instruments, Norfolk, United Kingdom). After measuring intact-cell endogenous respiration, antimycin A was added to block respiration at complex III. Ascorbate-N, N, N′, N′-tetramethyl-p-phenylenediamine (TMPD) was then added to drive complex IV respiration, which was subsequently inhibited with 0.7 mM KCN. Enzyme activities of the different respiratory complexes from isolated mitochondria were analyzed spectrophotometrically as previously described (5).
Immunocytochemistry.
Cells grown on coverslips had their mitochondria labeled with 200 nM MitoTracker (CMXRos; Molecular Probes, Eugene, OR) for 30 min at 37°C. Cells were washed with PBS and fixed with 2% paraformaldehyde in PBS, permeabilized with cold methanol, and then incubated with anti-Cox1-Alexa Fluor 488-conjugated monoclonal antibody (Molecular Probes) for 2 to 4 h. The antibody was used at a concentration of 2 μg/ml in 5% BSA in PBS. Coverslips were mounted with an antifade aqueous mounting gel (Biomeda, Foster City, CA), and the fluorescence was observed in a Carl Zeiss confocal microscope.
Blue native gels.
Blue native polyacrylamide gel electrophoresis (BN-PAGE) for the identification of individual complexes (but not supercomplexes) was performed as described by Nijtmans and colleagues (22). This was accomplished by using dodecylmaltoside instead of digitonin for sample preparations. Approximately 25 to 40 μg of mitochondrial protein was loaded in a 5 to 13% blue native gradient gel. High-molecular-weight markers for native electrophoresis (Amersham Biosciences, Piscataway, NJ) were treated the same as the samples (1.5 M aminocaproic acid, 50 mM Bis-Tris, pH 7.0, lauryl maltoside, and Coomassie brilliant blue G). Lanes of the native gel were cut with a razor and incubated in 1% sodium dodecyl sulfate (SDS), 1% 2-mercaptoethanol for 30 min, and then in 1% SDS for 30 additional minutes. The second-dimension gel (SDS-PAGE) was cast around the gel strip with 10% separating and 4% stacking gels. After native electrophoresis or SDS-PAGE, proteins were transferred to polyvinylidene difluoride (PVDF) membrane for immunoblot analyses as described previously (22).
To perform in-gel activity staining, the blue cathode buffer was replaced by colorless buffer after the gel was run for 3 h at 30 V. The gel was run until the dye front left the gel. For complex I activity, the gel was incubated in 100 mM Tris-HCl, 0.14 mM NADH, 1 mg/ml nitroblue tetrazolium, pH 7.4, for 1 to 3 h at room temperature (18). For complex IV activity stain, gels were incubated as described previously (22, 36) for 2 to 4 h at 37°C. Complex V activity gels were performed as described previously (22), with the following modifications. Gels were washed with distilled water, incubated at room temperature for 1 h with 50 mM glycine, pH 8.6 (4), and then incubated in 35 mM Tris-HCl, 270 mM glycine, 14 mM MgSO4, 0.2% Pb(NO3)2, 5 mM ATP, pH 8.6, overnight at 37°C.
For detection of supercomplexes, isolated mitochondria (200 μg) were resuspended in 0.75 M amino caproic acid and 50 mM Bis-Tris, pH 7.0, and incubated with digitonin (high purity, water soluble; Calbiochem) at a detergent-to-protein ratio of 8:1 (wt/wt) for 20 min on ice. Samples were centrifuged at 20,000 × g for 20 min, and 5% Serva blue was added to the supernatant to achieve a detergent-to-dye ratio of 4:1 (wt/wt). Samples (about 12 to 15 μl) were separated by BN-PAGE in 4 to 10% acrylamide gradient gels. Electrophoresis was performed with blue cathode buffer at 100 V for 1 h and then switched to constant current (15 to 16 mA). The blue cathode was changed to colorless buffer after the dye front reached half of the gel, and electrophoresis was stopped when the dye front left the gel. Proteins were transferred overnight to PVDF membranes as described above. To separate proteins in the second dimension, the procedure described above was followed.
Mitochondrial protein synthesis.
In vivo mitochondrial protein synthesis was determined by labeling cells with [35S]methionine in the presence of emetine, a cytoplasmic protein synthesis inhibitor, as described previously (11).
Inhibition of complex IV activity by KCN.
To determine the concentration of KCN required to inhibit complex IV, total cell respiration was measured with a Clark O2 electrode in the presence of increasing concentrations of KCN. Culture medium containing 0.4 mM KCN was used to completely inhibit COX activity in cultured cells. Medium was replaced everyday to avoid effects of media acidification while inhibiting respiration.
Western blotting.
Mitochondrial samples were separated by SDS-PAGE in 4 to 20% gradient gels and transferred to PVDF membranes. Membranes were blocked with 5% nonfat dry milk in PBS-Tween 20 (0.1%) and blotted with specific antibodies (Molecular Probes) against the complex IV subunits (Cox1, Cox4, Cox5b, and Cox6b), a complex I subunit (Ndufa9), a complex II subunit {succinate dehydrogenase flavoprotein subunit [SDH(Fp)]}, complex III subunits (Uqcrc2 and Uqcrfs1), and a complex V subunit (ATPase-β). Horseradish peroxidase-labeled secondary antibodies were used, and reactions were developed using chemiluminiscence with Superwest signal reagent (Pierce, Rockford, IL). The amount of protein loaded on the gels was normalized using an antibody against voltage-dependent anion channel (VDAC; Molecular Probes, Eugene, OR).
Recombinant lentiviral constructs.
Wild-type COX10 was obtained by PCR amplification of a mouse brain cDNA library and cloned into p156RRLsinPPThCMVMCSpre vector for lentiviral production. Two mutants of COX10 were produced by site-directed mutagenesis using a QuickChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA). Mutant 1 had a C584A point mutation, which corresponds to a C791A (T196K) mutation in human COX10. Mutant 2 had a C671T point mutation, which corresponds to a C878T (P225L) mutation in human COX10. Plasmids were transfected into human kidney 293T cells, and viral particles were produced as described previously (21). The titer of viral preparations was determined by a human immunodeficiency virus type 1 p24 enzyme-linked immunosorbent assay kit (Perkin Elmer Life Sciences, Boston, MA), and 4 × 105 pg of viral particles was added to 2 × 105 K19 cells in a 6-well plate. Cells were expanded and analyzed for oxidative phosphorylation (OXPHOS) complex levels.
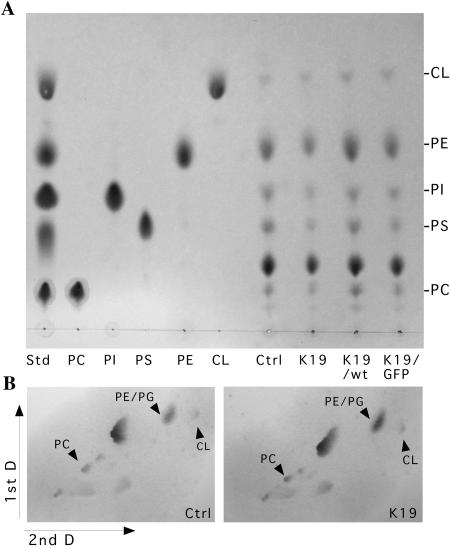
Thin-layer chromatography analysis of phospholipids.
Isolated mitochondria (0.5 mg to 1 mg) were dried in a speed vacuum, and lipids were extracted in 0.5 ml chloroform-methanol (2:1) by homogenization of the sample. After centrifugation, supernatant was collected and the pellet was extracted again. Both extracts were combined and washed three times with a 0.1 volume of 0.9% NaCl (14). The solvent was evaporated in a speed vacuum, and extracted lipids were resuspended in 50 to 100 μl of chloroform. For one-dimension thin-layer chromatography (TLC), samples and phospholipid standards (Sigma, St. Louis, MO) were applied to silica plates and separated in a tank previously equilibrated in chloroform-ethanol-water-triethylamine (30:35:7:35) as described previously (31). Plates were dried in a fume hood and then sprayed with molybdenum blue reagent (Sigma) to detect phospholipids. For two-dimension TLC, lipids extracted from 1 mg mitochondria were applied to the silica plate and separated in the first dimension with chloroform-methanol-water-ammonium hydroxide (70:30:3:6.6), dried for 45 min, and then separated in the second dimension using chloroform-methanol-water (65:35:5) as a solvent (23). Plates were dried completely and sprayed with molybdenum blue.
Pulse-chase labeling.
Cells were incubated with 40 μg/ml chloramphenicol in complete medium overnight. The next day the cells were washed with regular and then with methionine-free media (with dialyzed serum, pyruvate, and uridine) for 1 h. Fresh methionine-free medium with 100 μg/ml cycloheximide was added to the cells for 10 min, followed by the addition of 630 μCi of [35S]methionine (Trans35S label; MP Biomedicals, Irvine, CA). Cells were incubated for 2 h at 37°C. After a PBS wash, chase was accomplished with complete medium. Cells were harvested by trypsinization and counted at different time points. Cells were washed in PBS, and 2.5 × 106 cells were resuspended in 200 μl of PBS and solubilized with 70 μl of 8 mg/ml digitonin (Sigma, St. Louis, MO) in PBS. Samples were incubated on ice for 10 min, 1 ml cold PBS was added, and then the solution was centrifuged for 5 min at 20,000 × g. The pellet was washed with PBS and resuspended in 100 μl of 1.5 M aminocaproic acid, 50 mM Bis-Tris, pH 7.4. OXPHOS complexes were solubilized with 5 μl of 10% lauryl maltoside and incubated on ice for 10 min. Samples were centrifuged at 20,000 × g for 30 min at 4°C, and 10 μl of sample buffer (750 mM aminocaproic acid, 50 mM Bis-Tris, pH 7.0, 50 mM EDTA, 5% Serva G) was added to supernatant. Samples were loaded into a 4 to 10% acrylamide BN gel.
RESULTS
Creation and characterization of COX10 knockout cells.
An immortalized skin fibroblast cell line was established from a homozygous mouse for the floxed COX10 gene (exon 6; see Fig. 1A and reference 13). To generate COX10 knockout cells, the immortalized cell line was transfected with a plasmid expressing the P1 Cre recombinase (pCre-Hygro), and several hygromycin-resistant clones were obtained. PCR analysis of clones C12 and C16 showed the presence of the intact floxed allele using specific primers flanking the loxP sites. In contrast, we were unable to detect the floxed allele in clones K11, K18, and K19. Using the same primers, we were able to detect the band corresponding to the deleted allele in K11, K18, and K19. These results were confirmed by Southern blotting (Fig. 1B). Digestion of DNA with BamHI shows the floxed allele in C, C12, and C16 (approximately 9.3 kb) and the deleted allele (10.8 kb; the deletion of the region flanked by the loxP sites results in the loss of a BamHI site, therefore the resulting band is larger than the floxed one) in K11, K18, and K19. C12 was heterozygous for the deleted and floxed alleles (Fig. 1B).
COX10 knockout fibroblasts lack COX activity and heme a.
The growth characteristics of the COX10 KO cells were determined in high-glucose, uridine-supplemented medium. The doubling times were the following: K11, 42.2 h; C12, 31.2 h; C16, 24 h; K18, 37.9 h; and K19, 38.0 h. Therefore, “K” clones grew slower than “C” clones. Clones K11, K18, and K19 acidified the media faster, suggesting increased lactate production. We measured total cell respiration and observed that K11, K18, and K19 were unable to respire, whereas C12 and C16 respired at levels similar to those of the parental control (Fig. 2A). When ascorbate/TMPD was used to reduce cytochrome c, the electron donors for COX, K11, K18, and K19 were unable to consume oxygen (Fig. 2A).
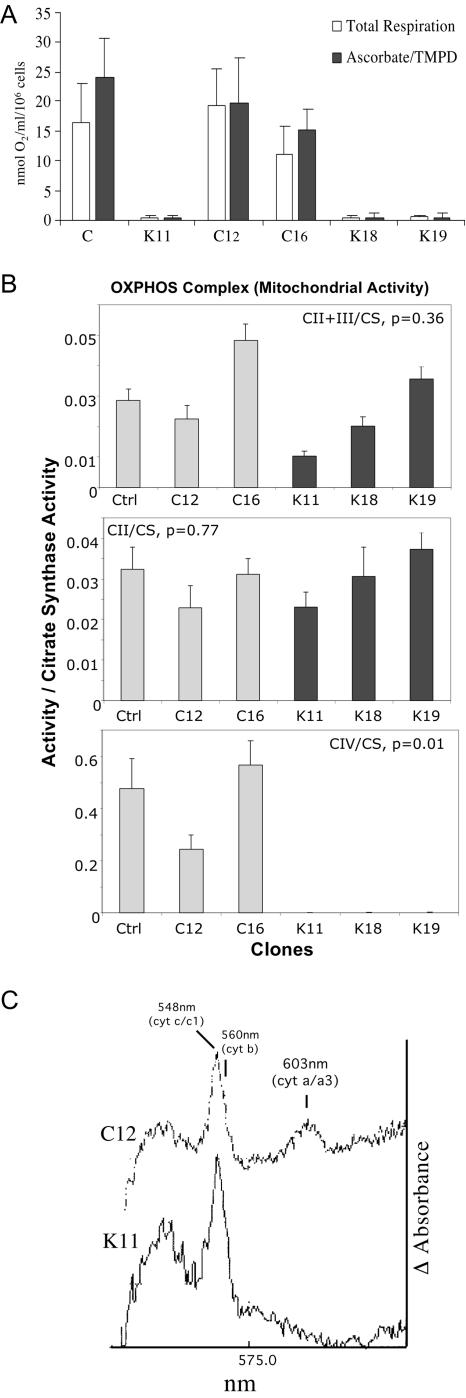
FIG. 2.
Function of mitochondrial enzymes in COX10 KO cells. (A) Total cell respiration of COX10 KO cells was determined by the rate of oxygen consumption on a Clark O2 electrode. Knockout clones (K11, K18, and K19) did not respire, either with endogenous substrates or with ascorbate-TMPD, which donates electrons directly to complex IV. (B) Respiratory complex activities of COX10 KO cells. Enzymatic activities were measured spectrophotometrically in isolated mitochondria. Specific activities, initially obtained as micromoles/minute/milligram, are expressed as ratios to citrate synthase activity. Error bars represent standard deviations of at least six independent measurements. (C) Cytochrome spectra of COX10 KO cells. The differential oxidized spectra minus reduced spectra for C12 and K11 mitochondrial cytochromes are shown. The COX10 KO clone K11 lacked the cytochrome a and a3 peak at 603 nm. cyt, cytochrome; Ctrl, control.
We also determined the enzymatic activity of respiratory complexes in isolated mitochondria from the different clones (Fig. 2B). As expected, the K clones, which were unable to respire, were completely deficient in COX activity. The combined activity of complexes II and III or the activity of isolated complex II (normalized to citrate synthase [CS]) was variable but not significantly altered (Fig. 2B).
We analyzed the relative levels of total cytochromes by differential absorption spectra. Figure 2C shows a differential spectra of mitochondria isolated from clones K11 and C12. We could not detect the absorption peak at 603 nm, characteristic of cytochrome aa3 in clone K11. The absorption peaks corresponding to cytochromes c and c1 were clearly visible in K11. Mitochondria from clones K18 and K19 also had undetectable cytochrome aa3 (data not shown). These results indicate that the deletion of exon 6 of COX10 causes a specific impairment of cytochrome aa3 synthesis but does not affect the synthesis of other cytochromes.
Cox1 is synthesized but is highly unstable in the absence of Cox10.
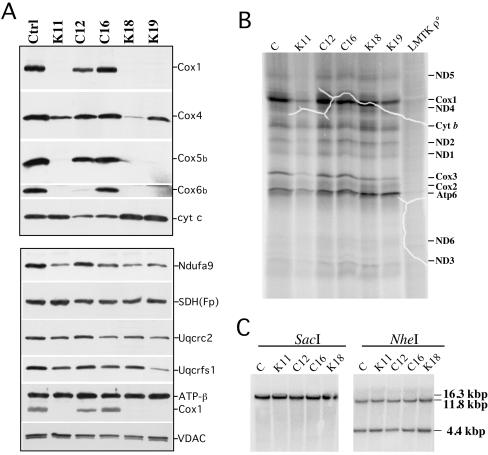
The steady-state levels of representative subunits of the respiratory complexes in all the clones are shown in Fig. 3A. Cox1, Cox5b, and Cox6b were undetectable or barely detectable in the COX10 KO cells, whereas Cox4 was significantly reduced in the K clones compared to controls. Surprisingly, the Cox6b immunodetectable material in C12 was reduced, even though in every other aspect this clone behaved as a control. Cytochrome c levels were increased in K18 and K19 compared to controls. The protein levels of other respiratory complex subunits are also shown in Fig. 3A. The levels of Ndufa9 (complex I) and Uqcrc2 (complex III) were relatively decreased in K11, K18, and K19. In addition, the iron sulfur protein of complex III (Uqcrfs1) was slightly decreased, especially in K19. The levels of SDH(Fp) (complex II) and ATPase-β (complex V) were similar in all the clones. Antibodies against VDAC1, an outer mitochondrial membrane protein, were used to verify the amount of mitochondrial protein loaded in the gels.
FIG. 3.
Synthesis and steady-state levels of OXPHOS proteins in COX10 KO cells. (A) We analyzed the steady-state levels of several subunits of cytochrome oxidase by Western blotting, including Cox1, Cox4, Cox5b, Cox6b, and cytochrome c. The lower part of the panel shows the steady-state levels of subunits of complex I (Ndufa9), complex II [SDH(Fp)], complex III (Uqcrc2 and Uqcrfs1), and complex V (ATPase-β). Protein loading was analyzed using an antibody against VDAC1. (B) [35S]methionine-labeled mitochondrial proteins (60 μg) were separated on SDS-15% PAGE. Fluorographic bands were assigned as described previously (11). LMTK ρ°, mouse cell line devoid of mtDNA. (C) Southern analyses of total DNA digested with SacI or with NheI. The membrane was hybridized with a 32P-labeled mitochondrial probe (mouse mtDNA nucleotide positions 5556 to 6268). Ctrl, control; cyt, cytochrome.
We next performed mitochondrial protein synthesis experiments to assess if newly synthesized mtDNA-coded polypeptides, particularly Cox1, could be detected in the K clones. Figure 3B shows a fluorography of the pulse-labeled mitochondrial proteins separated by SDS-PAGE. Bands corresponding to the different mtDNA-coded COX subunits can be observed in all the clones. Cox1 was readily detectable, although in the K clones the ratio of Cox1 to cytochrome b (or to ND1) was decreased by approximately 20% (P < 0.02). This small decrease is likely related to Cox1 instability due to impaired maturation. Conversely, the levels of Atp6 were relatively increased in the K mutants, possibly because of increased levels of complex V.
We also analyzed the mtDNA by Southern blotting to test if differences observed in the steady-state levels of proteins could be the result of damage to the mtDNA. Figure 3C shows that there were no differences in mtDNA levels between the clones.
Mitochondria morphology is altered in COX10 KO fibroblasts.
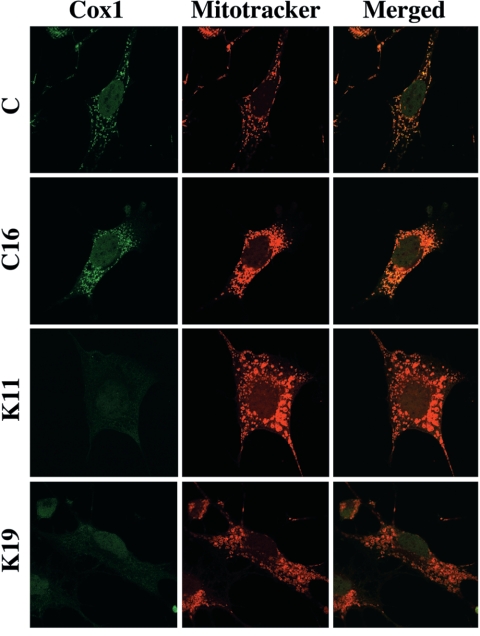
Immunocytochemistry experiments using either a Cox1 monoclonal antibody or MitoTracker CMX revealed the typical filamentous network of mitochondria in control clones (Fig. 4). Immunocytochemistry of K clones showed no detectable Cox1 staining. Staining of clones K11 and K19 with MitoTracker showed abnormal mitochondrial morphology. In these clones, mitochondria were rounder and appeared larger than in the controls (Fig. 4).
FIG. 4.
Mitochondrial morphological abnormalities in COX10 KO cells. Cell lines were immunostained for Cox1 and MitoTracker red. Cox1 colocalized with MitoTracker in control cells but was absent in KO clones. Control cells displayed a typical mitochondrial filamentous network, whereas COX10 KO cells contained round enlarged mitochondria.
Steady-state levels of respiratory complex I are affected by the lack of complex IV in COX10 KO fibroblasts.
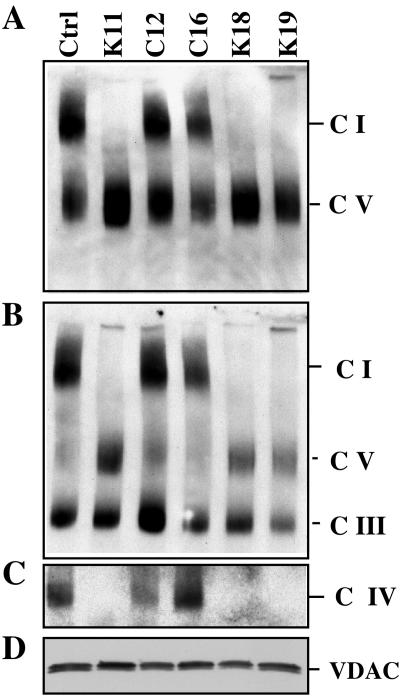
To assess the steady-state levels of fully assembled oxidative phosphorylation (OXPHOS) complexes, cell lines were analyzed by blue native gel electrophoresis followed by Western blotting. Mitochondria were solubilized with lauryl maltoside in conditions that preserve the individual complexes but not supercomplexes. Figure 5C shows that clones K11, K18, and K19 lacked complex IV. Surprisingly, they had markedly reduced levels of fully assembled complex I (Fig. 5A). The levels of complex III, determined by using an antibody against Uqcrc2, revealed a small decrease on the K clones relative to the C clones (Fig. 5B).
FIG. 5.
Steady-state levels of respiratory holocomplexes in COX10 KO cells. A Western blot of one-dimension BN-PAGE was probed with monoclonal antibodies against (A) the Ndufa9 subunit of complex I (C I) and ATPase-β of complex V, (B) Uqcrc2 of complex III, (C) and Cox1 of complex IV. The same samples were subjected to SDS-PAGE, and VDAC was immunodetected as a loading control (D). Ctrl, control.
Samples of lauryl maltoside-solubilized mitochondria, obtained by BN-PAGE, were also analyzed in a second dimension by SDS-PAGE. We could observe that the levels of complex I were markedly reduced, confirming the observations in the one-dimension BN-PAGE. No subcomplexes were observed with anti-complex I antibodies (data not shown).
The decrease in complex I steady-state levels is not due to a decrease in COX activity.
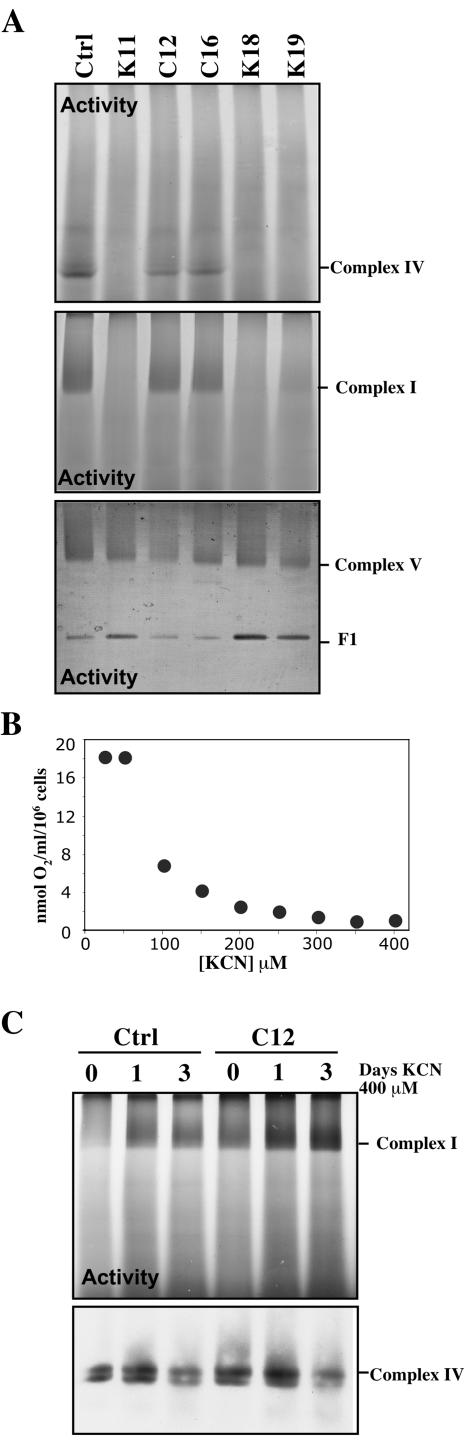
In-gel activities of complexes I, IV, and V were also obtained. As expected, complex IV activity was absent in the KO clones. Complex I activity was markedly decreased, whereas complex V activity was slightly higher (Fig. 6A). Although the absence of complex IV affected the levels of complex I, it was unclear if the effect was caused by the physical absence of COX or because of the COX enzyme deficiency. To address this issue, we treated control cells with different concentrations of KCN. At 200 μM KCN we observed more than 95% inhibition of cell respiration in mouse control fibroblasts (Fig. 6B). To ensure complete inhibition of complex IV, control cells were grown in 400 μM KCN for 1 and 3 days and analyzed by BN-PAGE. There was no decrease in complex I during this period (Fig. 6C, upper gel), suggesting that, at least in this time frame, lack of COX activity does not impact complex I levels. COX was still present in these treated cells (Fig. 6C, lower gel).
FIG. 6.
Complex I is not destabilized by decreased COX enzyme activity. (A) In-gel activity for complexes I, IV, and V showed a severe defect in complex I in the K clones. F1 is the dissociated F1 component of complex V. (B) The concentration of KCN required for the complete inhibition of COX activity in a control cell was determined to be 250 to 300 μM. Cells were grown in the presence of 400 μM KCN for different times (1 or 3 days), and enzymatic activities of complex I were determined after BN-PAGE. Panel C shows complex I in-gel activity stain (upper) of control and C12 cells at 0, 1, and 3 days of KCN treatment. The lower part of panel C shows a Western blot demonstrating that although the activity of COX was inhibited by KCN, complex IV was still present. Ctrl, control.
Introduction of wild-type or point mutant forms of COX10 in COX10 KO fibroblasts restores complex I levels.
We took advantage of the KO cells to assess if COX10 mutants identified in patients with mitochondrial diseases could restore COX activity. The wild-type mouse COX10 cDNA as well as two different mutants carrying point mutations analogous to those found in patients were introduced into a lentivirus expression system. Mutant 1 had a C584A point mutation which corresponds to a C791A (T196K) mutation in human COX10. Mutant 2 had a C671T point mutation which corresponds to a C878T (P225L) mutation in human COX10 (3).
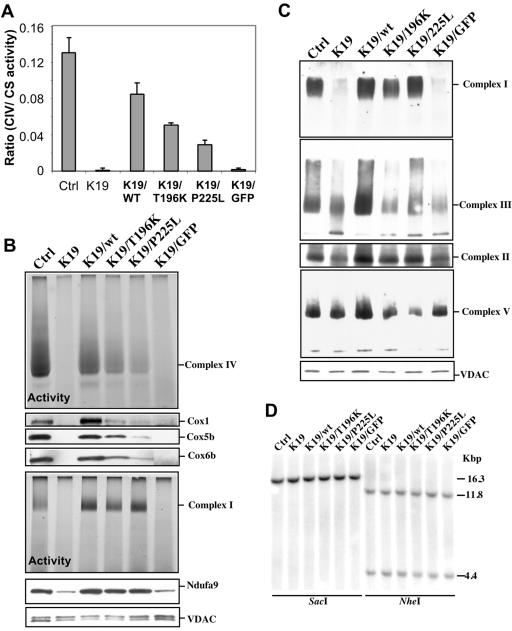
Clone K19 was transduced with the recombinant lentiviruses, after which cells were expanded and COX and citrate synthase (CS) activities measured in homogenates. COX/CS activity was restored to levels that were 65% of that of control cells when wild-type COX10 was used. Cells transduced with the T196K mutant had 59%, whereas the P225L mutant had 34%, of the COX/CS activity observed in the cells transduced with wild-type COX10 (Fig. 7A). Mitochondria were prepared from these cells and analyzed by BN-PAGE. Figure 7B shows that the wild-type COX10 restored COX activity in K19. The mutant forms of COX10 also restored COX activity, albeit at a lower level. The steady-state levels of COX subunits correlated with the levels of COX activity (Fig. 7B). Infection of clone K19 with a green fluorescent protein-containing recombinant virus had no effect on its COX activity.
FIG. 7.
Levels of complexes IV and I are restored by transduction of COX10 KO fibroblasts with recombinant COX10. The COX-deficient clone K19 was infected with recombinant lentivirus carrying the wild-type and two mutant forms of COX10. The mutations corresponded to alterations in the human COX10 gene observed in patients with mitochondrial disorders. (A) Infection with wild-type COX10 restored complex IV (CIV) activity to 65% of control levels, whereas the mutants had a less potent effect. (B) Similar results were observed by BN in-gel COX activity and Western blotting for COX subunits. (C) The steady-state levels of complexes I, III, IV, and V are shown by BN-PAGE Western blots. (D) The mtDNA levels in the different COX10-transduced lines are shown. Ctrl, control; GFP, green fluorescent protein; wt, wild type.
The reintroduction of COX10 also restored complex I activity (Fig. 7B) and the steady-state levels of fully assembled complex I (Fig. 7C). Although the restoration of COX activity did not reach the levels of the control cells, the levels of complex I in the lentivirus-transfected cells (wild type or mutants) were equal to or higher than those in the controls. The steady-state level of Ndufa9 subunit was also increased in the COX10-transduced cells (lower part of Fig. 7B). A smaller increase in complexes II and III occurred, but no consistent changes in complex V were observed (Fig. 7C). Southern blotting showed no alterations in mtDNA levels in K19- or COX10-repopulated lines (Fig. 7D).
Lack of complex IV destabilizes supercomplexes but does not alter the cardiolipin content.
By treating mitochondria with digitonin, followed by one-dimension and two-dimension BN-PAGE, we observed that control cells have a supercomplex composed of complexes I, III, and IV (Fig. 8A and B). The concentration of digitonin was titrated, but at digitonin:protein dilutions of less than 4:1 (wt/wt) no clear bands were observed. Most experiments were performed with digitonin at 8:1 (wt/vol). The abundance of the supercomplex harboring complexes I, III, and IV was relatively low, but it was previously described that COX dissociates relatively easily from supercomplexes during extraction (27), so it is possible that a higher percentage of COX is present in supercomplexes in vivo. On the other hand, most of complexes I and III were present in a stable I/III supercomplex (Fig. 8A). In COX10 KO cells, not only did the supercomplex containing complexes I, III, and IV disappear (Fig. 8C) but the supercomplex made of complexes I and III also was markedly decreased (Fig. 8A and C), which is not surprising considering the drastic reduction in complex I. A possible mechanism for the complex I reduction would be the increased instability of complex I that is not assembled into supercomplexes. Cardiolipin has been shown to be essential for supercomplex formation (15). Therefore, we tested if the lack of COX altered cardiolipin levels. Thin-layer chromatography experiments with two different solvent systems showed that the levels of cardiolipin were not markedly altered in a COX-deficient clone (Fig. 9).
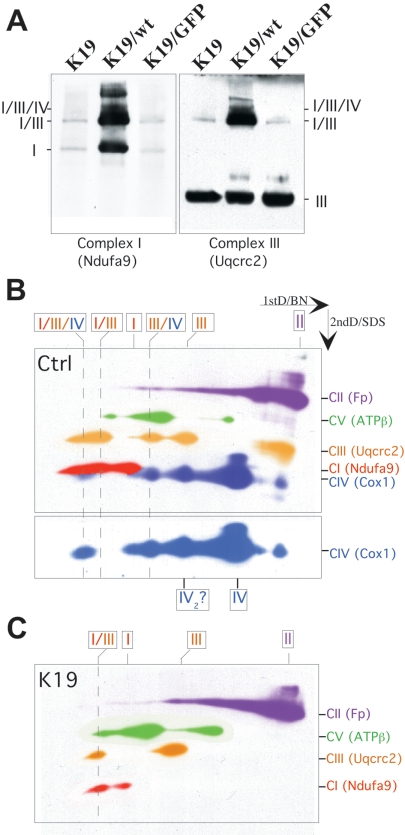
FIG. 8.
Lack of COX does not prevent complex I and III association. (A) Western one-dimension BN-PAGE showed that COX10 KO cells (clone K19 or K19 transduced with lentivirus-green fluorescent protein [GFP]) had decreased levels of both complex I and I/III supercomplex. Clone K19 transduced with a lentivirus expressing the wild-type COX10 cDNA had most of complexes I and III as a stable I/III supercomplex. (B) Two-dimension (2ndD) BN Western blots were performed sequentially in the same membrane. Pseudocolors identifying the different complexes were added for clarity. This experiment showed the presence of I/III/IV and I/III supercomplexes in control cells (B). Because Cox1 migrates close to Ndufa9, we added an isolated exposure of a Cox1 Western blot to the bottom of panel B. (C) Using different antibodies on 2D gels showed that the I/III/IV supercomplex was absent in KO clones and the I/III supercomplex was markedly reduced. wt, wild type; Ctrl, control; CI, complex I.
FIG. 9.
Cardiolipin levels are not markedly altered by COX deficiency. (A) One-dimension TLC comparing the phospholipids composition of two COX-competent cell lines (control [Ctrl] and K19/wild type [wt]) and two COX-deficient lines (K19 and K19/green fluorescent protein [GFP]). (B) Separation of cardiolipin by two-dimension TLC using a different solvent system (see Materials and Methods). The identity of cardiolipin was established not only by analyzing standards on two dimensions but also by the analysis of a yeast mutant in cardiolipin synthesis (data not shown). PI, phosphatidylinositol; PA, phosphatidic acid; PS, phosphatidylserine; PG, phosphatidylglycerol; PC, phosphatidylcholine; PE, phosphatidylethanolamine; CL, cardiolipin. PDME (phosphatidyldimethylethanolamine) was used as a two-dimension standard, but this phospholipid was not detected in the cells. Std, standard; D, dimension.
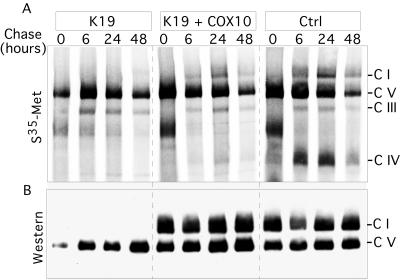
Complex I assembly cannot be detected in the absence of complex IV.
To determine if the reduction of complex I was associated with postassembly degradation, we labeled COX10 KO and control cells with [35S]methionine in the presence of a cytosolic protein synthesis inhibitor (cycloheximide). Cells were labeled for 2 h and chased for 6, 24, and 48 h in the absence of cycloheximide. Finally, OXPHOS complexes were separated in BN-PAGE and subjected to autoradiography (Fig. 10A). The labeling of complex IV can be observed in both controls, although lower levels of COX are present in the K19 cell line transduced with wild-type COX10 (see Fig. 7A). We could not observe newly synthesized assembled complex I in the K19 cell line, suggesting that the absence of COX either affects complex I assembly or causes it to be extremely unstable. The steady-state levels of complex I and complex V were estimated in the same samples by Western blot analysis (Fig. 10B). In agreement with the reintroduction of COX10 mutants into K19 (Fig. 7), very low levels of COX are sufficient for the synthesis/stability of complex I.
FIG. 10.
Newly assembled complex I is undetectable in cells with COX deficiency. A COX-deficient cell line (K19) and two controls were pulse labeled for 2 h with Trans35S label and chased for the indicated times as described in Materials and Methods. (A) Autoradiogram of a BN gel (4 to 10% acrylamide gel). (B) The same samples analyzed by Western blotting using antibodies to Ndufa9 and ATPase-β. S35-Met, [35S]methionine; C I, complex I; Ctrl, control.
DISCUSSION
Effect of COX10 deletion on COX assembly.
Mutations in COX10 have been associated with several mitochondrial diseases (3, 12, 32). Determination of the steady-state levels of COX subunits in tissues from patients with COX10 mutations showed a significant decrease in Cox3 and Cox6c (∼50% of control) and a severe decrease of Cox2 subunits (<3%) (32). In fibroblasts from the same patient, the steady-state levels of Cox1 were decreased by 35 to 40%, Cox4 by 60%, Cox5b by 25%, and Cox6b by 50%. Our study of the COX10 KO fibroblasts showed undetectable levels of Cox1 and Cox6b, barely detectable levels of Cox5b, and a significant reduction of Cox4. The difference observed between our COX10 KO fibroblasts and the patient's fibroblasts could be explained by the differences in the residual COX activity. Because there is essentially no COX assembled in our KO cells, the unassembled subunits are likely to be degraded rapidly, although some subunits, such as Cox4, are more stable than others. This conclusion is supported by our experiments where mutant COX10 was reintroduced into the KO cells and subunit stability correlated with the presence of the complex. The observation that COX activity was restored in the KO cells transduced with the mutant COX10 alleles in an infant with a severe metabolic disorder correlated well with the activities observed in the patient fibroblasts. Lentivirus expressing the T196K COX10 restored the activity to 59% of that of the wild-type COX10, whereas the P225L COX10 restored it to 34% of that of the wild type. Fibroblasts from the patient harboring both mutant alleles (compound heterozygous) had 40% of the COX activity observed in controls (3).
We observed the synthesis of mtDNA-coded subunits in KO clones, including Cox1. These data indicate that the lack of heme a incorporation in apo-Cox1 does not affect translation of the full-length protein but renders it very unstable and susceptible to degradation. A recent report showed that the presence of fully assembled COX has a positive effect on Cox1 synthesis in budding yeast (6). Although we cannot discard such a mechanism, we believe that the small decrease observed in pulse-labeled Cox1 (20%) is more likely a result of lack of maturation and fast degradation.
Studies performed by Nijtmans and colleagues in human cells suggested a COX assembly pathway involving four intermediates, S1 to S4 (22). COX biosynthesis starts with the incorporation of heme a into Cox1 to form the subcomplex S1. This step is required prior to the incorporation of Cox4 to form S2. We were unable to detect assembly intermediates of complex IV in COX10 KO cells using a Cox1 antibody. Williams et al. showed that fibroblasts of a patient with a COX10 missense mutation also did not accumulate assembly intermediates (33). Likewise, Coenen et al. showed the absence of assembly intermediates in cells from another patient with a missense COX10 mutation (12), reflecting the important role of Cox1 in initiating assembly.
Complex I assembly/stability is affected by the lack of COX.
Unexpectedly, the lack of Cox10 had a strong effect on the levels of complex I. Similarly, mammalian cells harboring mutations in the apocytochrome b gene that preclude the assembly of complex III have a greatly compromised complex I stability (1, 26). Mutations in some complex I subunits also affected the levels of complex III (30). The existence of a “respirasome” has been proposed by Schagger and Pfeiffer (27), where respiratory complexes, including complexes I, III, and IV, are assembled into supercomplexes. More recently, these associations have been confirmed by single-particle image analysis (25). It has been proposed that the presence of supercomplexes provides a more efficient system by promoting substrate channeling as well as complex stability (9, 26). Recently, Stroh and colleagues purified a complete “respirasome” from Paracoccus denitrificans using chromatography and observed a stoichiometry of complexes I, III, and IV of 1:4:4. They also found that complex I stability was impaired by the absence of complex III or IV (28).
Two respiratory supercomplexes were described in human skeletal muscle 2, a smaller one (S) composed of complexes I and III and a larger one (L) composed of complexes I, III, and IV (27). In control individuals, the levels of the L supercomplex were higher than those of the S supercomplex. However, Surf1 mutations associated with partial COX activity had a reversed ratio of S/L supercomplex (26). There was no evidence that complex I was affected in the respirasome in analyses of muscle from this patient with a partial COX deficiency. On the other hand, they showed that a reduction in complex III abundance in muscle from two different patients had a marked negative effect on complex I levels. More recently, we observed that a mouse conditional knockout of COX10 in skeletal muscle was not associated with a decrease in complex I levels (13). Moreover, cells from patients with COX10 missense mutations also have normal complex I levels (3). An important difference is that both the skeletal muscle COX10 KO (13) as well as previously published patient cells with COX deficiency (3) have residual COX activity. In contrast, our “K” fibroblasts are null mutants and are devoid of COX. Our cell lines with reduced levels of COX had normal or higher than normal levels of complex I, indicating that very small amounts of COX can promote complex I synthesis/stability.
What causes the decrease in the steady-state levels of complex I in our COX10 KO cells? A simple model, where COX would protect complex I from degradation by direct physical interaction, is not supported by the observation that a reduction in complex I levels occurs only when the levels of COX are essentially zero. Our results with KCN inhibition, as well as published data on cells growing on antimycin A (1), argue against the scenario where OXPHOS function, at least in the short term, is necessary to stabilize the complexes.
We found that besides the disappearance of I/III/IV supercomplexes in the COX10 KO cells, the I/III supercomplexes were also markedly decreased. Because previous (1, 26) data showed that complex I is unstable if not associated with complex III, it was possible that the disruption of this interaction could lead to the decrease in complex I. However, two pieces of evidence suggest this is not the case: (i) the ratio of I/III to I is not changed in the KO cells, even though both are markedly reduced, and (ii) pulse-chase experiments could not detect complex I formation, suggesting that lack of COX impaired complex I assembly or that the latter is rapidly degraded.
Previous work with Saccharomyces cerevisiae showed that cardiolipin and supercomplexes are interdependent (15). The lack of COX blocks proton pumping, which would affect the mitochondrial pH. In yeast, the mitochondrial pH has been shown to alter cardiolipin synthesis (15), which in itself impairs supercomplex formation (15). However, we were not able to detect changes in cardiolipin levels in COX-deficient cells. Therefore, we believe that the decrease in complex I is related to a direct effect of the lack of complex IV, probably during assembly.
The mechanism which most fit the data is that complex I assembly requires the presence of COX, even in very small amounts. Recently, Rocher et al. reported that a subassembly of complex I interacts with complexes III and IV (24). They found that the complex I 17-kDa subunit, which has been implied in the membrane arm assembly pathway (30), may need to be associated with complexes III and IV in order to associate with the ND6 subunit. Such an assembly pathway would fit our data as well as the observation that reintroduction of COX led to a higher than normal level of complex I, possibly because of the accumulation of assembly intermediates.
In summary, ours and previous observations underscore the concept that oxidative phosphorylation complexes are not independent entities but rather are parts of a physically and functionally interconnected system.
Acknowledgments
We are grateful to Jamey D. Marth (University of California at San Diego) for the targeting construct. We also thank Antoni Barrientos (University of Miami) for help with the cytochrome spectra determination and both A. Barrientos and Sion L. Williams for critical comments.
This work was supported by Public Health Service grants NS041777 and GM55766 to C.T.M. and by a Muscular Dystrophy Association Research Development Grant to F.D. H.F. is a recipient of the Lois Pope LIFE Fellowship.
REFERENCES
- 1.Acin-Perez, R., M. P. Bayona-Bafaluy, P. Fernandez-Silva, R. Moreno-Loshuertos, A. Perez-Martos, C. Bruno, C. T. Moraes, and J. A. Enriquez. 2004. Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol. Cell 13:805-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, S., A. T. Bankier, B. G. Barrell, M. H. de Bruijn, A. R. Coulson, J. Drouin, I. C. Eperon, D. P. Nierlich, B. A. Roe, F. Sanger, P. H. Schreier, A. J. Smith, R. Staden, and I. G. Young. 1981. Sequence and organization of the human mitochondrial genome. Nature 290:457-465. [DOI] [PubMed] [Google Scholar]
- 3.Antonicka, H., S. C. Leary, G. H. Guercin, J. N. Agar, R. Horvath, N. G. Kennaway, C. O. Harding, M. Jaksch, and E. A. Shoubridge. 2003. Mutations in COX10 result in a defect in mitochondrial heme A biosynthesis and account for multiple, early-onset clinical phenotypes associated with isolated COX deficiency. Hum. Mol. Genet. 12:2693-2702. [DOI] [PubMed] [Google Scholar]
- 4.Appleby, R. D., W. K. Porteous, G. Hughes, A. M. James, D. Shannon, Y. H. Wei, and M. P. Murphy. 1999. Quantitation and origin of the mitochondrial membrane potential in human cells lacking mitochondrial DNA. Eur. J. Biochem. 262:108-116. [DOI] [PubMed] [Google Scholar]
- 5.Barrientos, A., L. Kenyon, and C. T. Moraes. 1998. Human xenomitochondrial cybrids. Cellular models of mitochondrial complex I deficiency. J. Biol. Chem. 273:14210-14217. [DOI] [PubMed] [Google Scholar]
- 6.Barrientos, A., A. Zambrano, and A. Tzagoloff. 2004. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 23:3472-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barros, M. H., C. G. Carlson, D. M. Glerum, and A. Tzagoloff. 2001. Involvement of mitochondrial ferredoxin and Cox15p in hydroxylation of heme O. FEBS Lett. 492:133-138. [DOI] [PubMed] [Google Scholar]
- 8.Barros, M. H., and A. Tzagoloff. 2002. Regulation of the heme A biosynthetic pathway in Saccharomyces cerevisiae. FEBS Lett. 516:119-123. [DOI] [PubMed] [Google Scholar]
- 9.Bianchi, C., M. L. Genova, G. Parenti Castelli, and G. Lenaz. 2004. The mitochondrial respiratory chain is partially organized in a supercomplex assembly: kinetic evidence using flux control analysis. J. Biol. Chem. 279:36562-36569. [DOI] [PubMed] [Google Scholar]
- 10.Brown, K. R., B. M. Allan, P. Do, and E. L. Hegg. 2002. Identification of novel hemes generated by heme A synthase: evidence for two successive monooxygenase reactions. Biochemistry 41:10906-10913. [DOI] [PubMed] [Google Scholar]
- 11.Chomyn, A. 1996. In vivo labeling and analysis of human mitochondrial translation products. Methods Enzymol. 264:197-211. [DOI] [PubMed] [Google Scholar]
- 12.Coenen, M. J., L. P. Van Den Heuvel, C. Ugalde, M. Ten Brinke, L. G. Nijtmans, F. J. Trijbels, S. Beblo, E. M. Maier, A. C. Muntau, and J. A. Smeitink. 2004. Cytochrome c oxidase biogenesis in a patient with a mutation in COX10 gene. Ann. Neurol. 56:560-564. [DOI] [PubMed] [Google Scholar]
- 13.Diaz, F., C. K. Thomas, S. Garcia, D. Hernandez, and C. T. Moraes. 2005. Mice lacking COX10 in skeletal muscle recapitulate the phenotype of progressive mitochondrial myopathies associated with cytochrome c oxidase deficiency. Hum. Mol. Genet. 14:2737-2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Folch, J., M. Lees, and G. H. Sloane Stanley. 1957. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226:497-509. [PubMed] [Google Scholar]
- 15.Gohil, V. M., P. Hayes, S. Matsuyama, H. Schagger, M. Schlame, and M. L. Greenberg. 2004. Cardiolipin biosynthesis and mitochondrial respiratory chain function are interdependent. J. Biol. Chem. 279:42612-42618. [DOI] [PubMed] [Google Scholar]
- 16.Gottlieb, R. A., and S. Adachi. 2000. Nitrogen cavitation for cell disruption to obtain mitochondria from cultured cells. Methods Enzymol. 322:213-221. [DOI] [PubMed] [Google Scholar]
- 17.Hennet, T., F. K. Hagen, L. A. Tabak, and J. D. Marth. 1995. T-cell-specific deletion of a polypeptide N-acetylgalactosaminyl-transferase gene by site-directed recombination. Proc. Natl. Acad. Sci. USA 92:12070-12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung, C., C. M. Higgins, and Z. Xu. 2000. Measuring the quantity and activity of mitochondrial electron transport chain complexes in tissues of central nervous system using blue native polyacrylamide gel electrophoresis. Anal. Biochem. 286:214-223. [DOI] [PubMed] [Google Scholar]
- 19.Lochmuller, H., T. Johns, and E. A. Shoubridge. 1999. Expression of the E6 and E7 genes of human papillomavirus (HPV16) extends the life span of human myoblasts. Exp. Cell Res. 248:186-193. [DOI] [PubMed] [Google Scholar]
- 20.Mogi, T., K. Saiki, and Y. Anraku. 1994. Biosynthesis and functional role of haem O and haem A. Mol. Microbiol. 14:391-398. [DOI] [PubMed] [Google Scholar]
- 21.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 22.Nijtmans, L. G., N. S. Henderson, and I. J. Holt. 2002. Blue native electrophoresis to study mitochondrial and other protein complexes. Methods 26:327-334. [DOI] [PubMed] [Google Scholar]
- 23.Poorthuis, B. J., P. J. Yazaki, and K. Y. Hostetler. 1976. An improved two dimensional thin-layer chromatography system for the separation of phosphatidylglycerol and its derivatives. J. Lipid Res. 17:433-437. [PubMed] [Google Scholar]
- 24.Rocher, C., W. Fan, E. Ruiz-Pesini, and D. C. Wallace. 2005. The early stage of complex I assembly is linked with complex III and IV supercomplex. Mitochondrial Physiol. 10.9:63-64. [Online.] http://www.mitophysiology.org/index.php?mip2005_abstracts. [Google Scholar]
- 25.Schafer, E., H. Seelert, N. H. Reifschneider, F. Krause, N. A. Dencher, and J. Vonck. 20March2006, posting date. Architecture of active mammalian respiratory chain supercomplexes. J. Biol. Chem. [Online] doi: 10.1074/jbc.MS13525200. [Epub ahead of print.] [DOI] [PubMed]
- 26.Schagger, H., R. de Coo, M. F. Bauer, S. Hofmann, C. Godinot, and U. Brandt. 2004. Significance of respirasomes for the assembly/stability of human respiratory chain complex I. J. Biol. Chem. 279:36349-36353. [DOI] [PubMed] [Google Scholar]
- 27.Schagger, H., and K. Pfeiffer. 2000. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19:1777-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stroh, A., O. Anderka, K. Pfeiffer, T. Yagi, M. Finel, B. Ludwig, and H. Schagger. 2004. Assembly of respiratory complexes I, III, and IV into NADH oxidase supercomplex stabilizes complex I in Paracoccus denitrificans. J. Biol. Chem. 279:5000-5007. [DOI] [PubMed] [Google Scholar]
- 29.Tsukihara, T., H. Aoyama, E. Yamashita, T. Tomizaki, H. Yamaguchi, K. Shinzawa-Itoh, R. Nakashima, R. Yaono, and S. Yoshikawa. 1996. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science 272:1136-1144. [DOI] [PubMed] [Google Scholar]
- 30.Ugalde, C., R. J. Janssen, L. P. van den Heuvel, J. A. Smeitink, and L. G. Nijtmans. 2004. Differences in assembly or stability of complex I and other mitochondrial OXPHOS complexes in inherited complex I deficiency. Hum. Mol. Genet. 13:659-667. [DOI] [PubMed] [Google Scholar]
- 31.Vaden, D. L., V. M. Gohil, Z. Gu, and M. L. Greenberg. 2005. Separation of yeast phospholipids using one-dimensional thin-layer chromatography. Anal. Biochem. 338:162-164. [DOI] [PubMed] [Google Scholar]
- 32.Valnot, I., J. C. von Kleist-Retzow, A. Barrientos, M. Gorbatyuk, J. W. Taanman, B. Mehaye, P. Rustin, A. Tzagoloff, A. Munnich, and A. Rotig. 2000. A mutation in the human heme A:farnesyltransferase gene (COX10) causes cytochrome c oxidase deficiency. Hum. Mol. Genet. 9:1245-1249. [DOI] [PubMed] [Google Scholar]
- 33.Williams, S. L., I. Valnot, P. Rustin, and J. W. Taanman. 2004. Cytochrome c oxidase subassemblies in fibroblast cultures from patients carrying mutations in COX10, SCO1, or SURF1. J. Biol. Chem. 279:7462-7469. [DOI] [PubMed] [Google Scholar]
- 34.Yoshikawa, S. 1997. Beef heart cytochrome c oxidase. Curr. Opin. Struct. Biol. 7:574-579. [DOI] [PubMed] [Google Scholar]
- 35.Zaslavsky, D., and R. B. Gennis. 2000. Proton pumping by cytochrome oxidase: progress, problems and postulates. Biochim. Biophys. Acta 1458:164-179. [DOI] [PubMed] [Google Scholar]
- 36.Zerbetto, E., L. Vergani, and F. Dabbeni-Sala. 1997. Quantification of muscle mitochondrial oxidative phosphorylation enzymes via histochemical staining of blue native polyacrylamide gels. Electrophoresis 18:2059-2064. [DOI] [PubMed] [Google Scholar]