Abstract
Rett syndrome (RTT) is a severe neurological disorder caused by mutations in the X-linked MECP2 gene, which encodes a methyl-CpG binding transcriptional repressor. Using the Mecp2-null mouse (an animal model for RTT) and differential display, we found that mice with neurological symptoms overexpress the nuclear gene for ubiquinol-cytochrome c reductase core protein 1 (Uqcrc1). Chromatin immunoprecipitation demonstrated that MeCP2 interacts with the Uqcrc1 promoter. Uqcrc1 encodes a subunit of mitochondrial respiratory complex III, and isolated mitochondria from the Mecp2-null brain showed elevated respiration rates associated with respiratory complex III and an overall reduction in coupling. A causal link between Uqcrc1 gene overexpression and enhanced complex III activity was established in neuroblastoma cells. Our findings raise the possibility that mitochondrial dysfunction contributes to pathology of the Mecp2-null mouse and may contribute to the long-known resemblance between Rett syndrome and certain mitochondrial disorders.
Rett syndrome (RTT) is a profound neurological disorder that almost exclusively affects girls. Approximately 80% of patients possess a mutation in one copy of the X-linked MECP2 gene that is, except in very rare cases, absent in somatic DNA from the parents (1). It follows that MECP2 mutations are the underlying cause of RTT. Mice lacking an intact Mecp2 gene have been generated in several laboratories (5, 10, 37). Without the Mecp2 gene, mice are born, but they acquire neurological symptoms after about 6 weeks and die at about 10 weeks of age (5, 10). Females heterozygous for the disrupted Mecp2 allele are apparently normal for several months, producing multiple litters, but later display symptoms similar to those of the Mecp2-null mice. An important difference is that their health does not progressively decline but remains stable for an apparently normal life span (10). These heterozygous animals are genetically most comparable to RTT patients, and their delayed-onset neurological symptoms and abnormal gait recall the human condition. The Mecp2-null mouse therefore provides an animal model for human RTT with the potential to shed light on its underlying molecular causes (19).
MeCP2 is a nuclear protein that binds preferentially to methylated sites in chromosomal DNA (22, 27, 29) and can function as a DNA methylation-dependent transcriptional repressor (27). Analyses of the Mecp2-null mouse therefore sought genes that might be overexpressed due to an absence of repression by MeCP2. Microarray analysis of brain RNA showed small alterations in the levels of multiple mRNAs when wild-type (wt) and Mecp2-null samples were compared (38). Although the effects were insignificant for individual genes, they became statistically significant when groups of affected genes were considered. Success in finding direct targets of MeCP2 was first achieved via a “candidate gene” approach (6, 24). One of four promoters of the gene for brain-derived neurotrophic factor (Bdnf) was found to bind MeCP2. This promoter is activated by artificial stimulation of cultured murine neurons. Remarkably, MeCP2 becomes phosphorylated upon stimulation and is lost from the promoter, thereby temporarily relieving MeCP2-mediated repression. Recent studies have additionally reported abnormal expression of the Dlx-5 and Dlx-6 (13), UBE3A and GABRB3 (35), and Sgk1 and Fkbp5 (31) genes as a target genes for MeCP2.
In an attempt to detect additional MeCP2-regulated genes and assess their contributions to phenotype, we subjected mRNA from Mecp2-null mouse brains to global analysis of gene expression by using a variant of differential display (17). Populations of mRNAs from mutant and wild-type brains were visualized by amplifying cDNA subsets that were then displayed by gel electrophoresis. Reproducible differences in the intensities of the resulting bands indicate altered expression. Our analysis revealed that the gene encoding a component of the mitochondrial respiratory chain is misregulated in the mutants at a time when symptoms are just beginning. We hypothesized that the phenotype of these mice might involve mitochondrial abnormalities, and we therefore investigated mitochondrial respiration in Mecp2-null mouse brain by using an oxygen electrode. The results showed abnormal respiration in brain mitochondria from mutant symptomatic mice compared to wild-type controls. Blue native gel electrophoresis of respiratory complexes followed by enzymatic staining showed reduced activity in some components of the respiratory chain. Overexpression of the Uqcrc1 gene in cultured cells induced physiological changes in mitochondria that resembled those seen in the Mecp2-null mouse brain. Our data therefore raise the possibility that some of the characteristics of the mouse phenotype, and perhaps of human RTT, are mitochondrial in origin.
MATERIALS AND METHODS
RNA isolation.
RNA was purified from whole brain using TRI reagent (Sigma) according to the manufacturer's recommendations.
ADDER differential display.
Pooled total RNA (three RNA pools, each consisting of three separate RNA preparations; nine brain samples in total) from Mecp2-null male mice (C57/black backcrossed two to five times; average age, 68 days) and the same number of control littermates were used as an input for ADDER (amplification of double-stranded cDNA end restriction fragments) analysis. ADDER was performed as described previously (17) with minor modifications. Briefly, pooled total RNA samples were used for cDNA synthesis, using a poly(A)-annealing oligonucleotide with a biotin label at its 5′ end. cDNA was then adsorbed on magnetic streptavidin beads and cut with MboI (NEB) restriction endonuclease. The resulting DNA ends were ligated to oligonucleotide adaptors and released from beads by AscI (NEB) digestion. cDNA fragments were amplified and used as templates for differential display PCR using primers with different terminal nucleotides (Fig. 1a). Bands with different intensities were cut from the gel, and eluted DNA was PCR amplified using primers with a specific terminal 3′ dinucleotide pair. Reamplified DNA was cloned into a plasmid vector. Restriction analysis of 24 picked clones allowed identification of the misexpressed fragment.
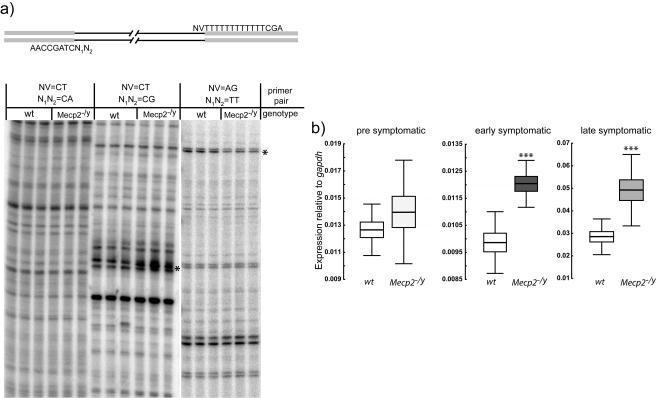
FIG. 1.
Global analysis of gene expression by ADDER reveals misexpressed genes in Mecp2-null mouse brain. (a) Top panel: primers used for differential display PCR. Shaded bars represent constant regions with 3′-terminal varied regions. Variable cDNA sequences are represented by lines. Bottom panel: differential display gels with two bands with altered intensity that were subsequently recovered and identified as Uqcrc1 (asterisk in center panel) and mt-Nd2 (asterisk in right panel). (b) Real-time PCR data showing Uqcrc1 up-regulation in early- and late-symptomatic mice but not in presymptomatic mice. Three pools, each containing RNA from three brains, were analyzed in quadruplicate to generate these data. The horizontal lines represent the means, the boxes delineate the standard errors of the means, and whiskers extend the standard deviations. ***, significant difference (t test, P < 0.001).
Reverse transcription and real-time PCR analysis.
The RNA pools were used for cDNA synthesis as described elsewhere (18). Real-time PCR analysis was performed using an iCycler real-time PCR machine (Bio-Rad). Four parallel reactions were carried out for each cDNA pool with IQ SYBR green Supermix (Bio-Rad) or homemade mix (0.5× SYBR green [Molecular Probes], 10 nM fluorescein [Sigma], 1× PCR buffer with MgCl2 [2 mM final concentration; Roche], and 200 μM deoxynucleoside triphosphates [ABgene] with 1 U FastStart Taq polymerase [Roche]). Results were always displayed relative to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) cDNA. Primers used for this analysis are shown in Table 1.
TABLE 1.
Primers used for real-time PCR analysis
| Gene accession no. | Gene | Forward primer | Reverse primer |
|---|---|---|---|
| XM_146892 | GAPDH gene | TACCCCCAATGTGTCCGTCG | CCTGCTTCACCACCTTCTTG |
| NM_025407 | Uqcrc1 | ACGGTGGGAGTGTGGATTGAC | CATTGCCAGGCCGATTCTTTG |
| AK087448 | Unknown, cDNA | GCATATCAAGTGCATATCAAGTG | CTTAAATTAGTTTGCTTTGCTTG |
| BC028971 | Gtl2/Meg3 | TGCAGCGGAGAGCCATAAATAAC | ACATCGCCTCCCTCCCTCGTG |
| AK011516 | Hist1h2bc | GATACTAGCAGATTAACCACCAT | TTCTTATCACAAATTTCTACAGT |
| AK034339 | Unknown, cDNA | GCCTATCATGCAGACCACAG | GCTCGCAGGTAAGGATGTAG |
| AU018611 | mt-Nd2 | GGGCATGAGGAGGACTTAACCAAAC | TGAGGTTGAGTAGAGTGAGGGATGG |
| NM_175092 | Rhof | GTCCCAAGCCCACTGTTTCTG | TTGATGCCTGTGTTCTCCTGATAG |
| AK049648 | Unknown, cDNA | AGTTCCAGAATAACCGCTCTCC | CTCTCTCACCCATCTGATACCTTAG |
| AK029199 | Cdon | TCCCAGTGGTAGCCTCTTATCC | ATTGGTGCCACACTGTCCTTG |
| BC025130 | Ccl19 | GAGCCCTGTGTCTTGAGTAAAG | ACTTGGCTGGGTTAGGTCTG |
| BC058513 | Snrp70 | GCTGACTGGTGGGAGTGTGAG | TGCCATCTGCGTGCTTGTAAG |
Chromatin immunoprecipitation.
Three whole brains (from adult mice age 2 to 5 months) provided sufficient input material for six or seven immunoprecipitations. Flash-frozen brains were ground in liquid nitrogen, and the powder was poured into fixation solution (1% formaldehyde in phosphate-buffered saline). Fixation was continued for 15 min at room temperature and quenched with 0.125 M (final concentration) glycine solution. Pelleted cells were washed with phosphate-buffered saline and homogenized in a Dounce homogenizer. After centrifugation, cells were resuspended in 9 ml of cell lysis buffer (0.2% NP-40, 10 mM NaCl, 10 mM Tris-HCl [pH 8], Complete [Roche] protease inhibitors). Lysate was triturated through a 25-gauge needle to remove lumps and incubated for 15 min with an additional 6 ml of lysis buffer. Nuclei were then harvested by spinning at 4,000 × g for 5 min and resuspended in 3.6 ml of nucleus lysis buffer (50 mM Tris-HCl, 10 mM EDTA, 1% sodium dodecyl sulfate [SDS], protease inhibitors). Nuclei were lysed for 10 min at room temperature and diluted with 2.2 ml of immunoprecipitation dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris HCl [pH 8], protease inhibitors). Chromatin was sonicated twice for 4 min with a Branson Sonifier 250 (duty cycle 60; output, 6) to obtain chromatin DNA fragments of about 200 bp on average. Chromatin was then cleared by centrifugation, diluted five times, precleared with protein A-Sepharose (Amersham), and subjected to overnight immunoprecipitation with rabbit polyclonal antibody 674 against MeCP2 (28) and rabbit polyclonal histone H3 dimethyl K9 antibody (Abcam). Antibody precipitates were bound to protein A-Sepharose for 1 h. Washes were performed once with TSEI (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl [pH 8]), four times with TSEII (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 500 mM NaCl, 20 mM Tris-HCl [pH 8]), once with buffer III (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl [pH 8]), and three times with Tris-EDTA. Antibody precipitates were then extracted twice with extraction solution (1% SDS, 0.1 M sodium hydrocarbonate), and cross-links were reversed overnight at 65°C. DNA was purified with the QIAGEN PCR purification kit and eluted in 50 μl of elution buffer (QIAGEN). The final eluate (2 μl) was used for PCRs. Uqcrc1 promoter PCR was done with primers uq1pd (CTTCTGTGTCTCCATTTCCCAAG) and uq1pr (TCTGTGCAAGAAGGTGTCCAC). Bdnf pIII primers were as described previously (6).
Mitochondrial isolation and respiration measurements.
Isolated mitochondria were prepared from whole brains of wt and Mecp2-null mice as described previously (20) with modifications (2). Briefly, brains were homogenized in 5 ml of isolation buffer (0.25 M sucrose, 10 mM MOPS [morpholinepropanesulfonic acid], 1 mM EGTA, 1 mg/ml bovine serum albumin, pH 7.4) before dilution to 10 ml with isolation buffer and centrifugation at 2,000 × g for 3 min at 4°C. The pellet was discarded and the supernatant respun. Following removal of the pellet, the supernatant was spun at 12,500 × g for 8 min. The resultant pellet was resuspended in 3% Ficoll in isolation buffer and was layered upon 6% Ficoll in isolation buffer. The gradient was spun at 11,500 × g for 30 min. The pellet was resuspended in 10 ml of isolation buffer with 5 mg of digitonin/g initial brain weight before centrifugation at 11,500 × g for 10 min, removal of supernatant, resuspension in 10 ml isolation buffer, and a further centrifugation at 11,500 × g. The final pellet was collected, resuspended in 1 ml isolation buffer, and kept on ice.
Measurements of oxygen consumption were carried out in a polarographic oxygen electrode (Rank Brothers Ltd.) thermostatted to 30°C. Small quantities (50 or 100 μl) of mitochondrial suspension were added to 3 ml of respiration buffer (25 mM sucrose, 75 mM mannitol, 95 mM KCl, 20 mM Tris-HCl [pH 7.4], 5 mM KH2PO4, 50 μM EDTA, 1 mg/ml bovine serum albumin) and a flat baseline acquired prior to addition of substrates. Complex I substrates (pyruvate and malate at final concentrations of 5 mM and 2.5 mM, respectively) were used to assay respiratory chain activity through complexes I, III, and IV. The addition of 8 μM rotenone to inhibit complex I activity, followed by the addition of succinate to a final concentration of 15 mM, allowed the measurement of respiratory activity via complexes II, III, and IV. Complex IV activity was measured following the addition of N,N,N′,N′-tetramethylphenylenediamine (TMPD) and ascorbate to final concentrations of 80 μM and 10 mM, respectively, while the respiratory chain was blocked at complex III with 50 nM myxathiazol. The TMPD auto-oxidation rate was found to be the same in the presence or absence of the complex IV inhibitor KCN. TMPD rates presented were all subjected to an auto-oxidation rate subtraction. Sufficient ADP was added to allow the adequate measurement of state 3 respiration. The uncoupling protonophore carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) was added to a final concentration of 1.6 μM. Respiration data were normalized to protein concentrations, which were determined by Peterson's modification of the assay of Lowry et al. (32).
Electron microscopy.
Transmission electron microscopy was performed using standard techniques. Mitochondrial samples were fixed overnight in 2.5% glutaraldehyde and stained in 2% (wt/vol) osmium tetroxide for 1 h (33). Sections were examined with a Philips CM120 Biotwin transmission electron microscope. Mitochondrial shape and size and density of cristae were scored before revealing the genotypes.
Bisulfite DNA modification.
Bisulfite DNA modification was performed using standard procedures. Briefly, 1 μg of whole-brain genomic DNA was digested with BamHI (NEB) restriction endonuclease, denatured at 100°C for 5 min, and incubated with 0.3 M NaOH (final concentration) for 20 min. After addition of 10 volumes of bisulfite/hydroquinone mix (0.51 g/ml sodium hydrogensulfite [Aldrich, no. 24 397-3), 0.11 g/ml hydroquinone (Sigma, no. H9003), and 0.4 M NaOH), the solution was overlaid with mineral oil and incubated for 5 h at 55°C. Bisulfite-treated DNA was precipitated with isopropanol (with 50 μg of glycogen), desulfonated with NaOH (0.3 M final concentration) for 15 min at 37°C, purified, and amplified by PCR. Primers for amplification (ubisd [AAATTATTTTTATATTGTTTTTTTT] and ubisr [AAACCCTTCATCTAATCCCATCTA]) were designed with Methprimer (23). PCR-amplified DNA was cloned using a TOPO cloning kit (Invitrogen). Data in the paper were derived from sequencing 14 or 15 clones.
Blue native electrophoresis and histochemical staining.
Blue native electrophoresis was performed as described elsewhere (36). Briefly, to solubilize respiratory complexes, equal protein amounts of purified mitochondrial sample were supplied with 3.5 volumes of buffer C (1 M aminohexanoic acid, 50 mM BisTris-HCl [pH 7.0]) and 1 volume of 10% dodecylmaltoside. The samples were centrifuged at 100,000 × g for 15 min. The supernatant protein concentration was measured using a Bio-Rad Dc kit. The samples were then supplemented with 1/10 volume of 5% Serva Blue G in 1 M aminohexanoic acid and loaded (90 μg of protein per lane) on a gradient (5 to 13%) native polyacrylamide gel. Catalytic staining was performed as described previously (39). Briefly, to stain for complex I (NADH) activity, the gel was incubated in a solution containing 2 mM Tris-HCl (pH 7.4), 0.1 mg/ml NADH, and 2.5 mg/ml nitroblue tetrazolium on a shaking platform until the gel developed maximal staining. Complex IV activity was evaluated in 25 ml of solution containing 12.5 mg 3,3′-diaminobenzidine tetrahydrochloride, 0.05 M phosphate buffer (pH 7.4), 20 μg/ml catalase, 25 mg cytochrome c, and 1.87 mg sucrose. After staining, the gels were fixed in 50% methanol and 10% acetic acid for 15 min.
Production and analysis of Uqcrc1-overexpressing cells.
N2A cells were cultured in Dulbecco modified Eagle medium plus 10% fetal calf serum plus nonessential amino acids (Invitrogen). The mouse Uqcrc1 coding sequence was PCR amplified from cDNA (Clontech MARATHON Ready mouse 11-day embryo cDNA) and cloned into pBABE Puro (EcoRI, SalI) plasmid. The resulting clone was confirmed by sequencing and transfected into the Phoenix Eco retrovirus packaging cell line by using calcium phosphate. Viral supernatant was collected 2 days after transfection and used to infect N2A cells in the presence of 4 μg/ml Polybrene. Puromycin (3 μg/ml) was applied 48 h after infection and maintained during cell culture. The medium was replaced with puromycin-free medium 24 h before respiration measurements. For Western blotting, cells were boiled in SDS loading buffer and proteins were resolved on a 10% SDS-polyacrylamide gel. Uqcrc1 and porin proteins were detected simultaneously using mouse monoclonal antibodies (anti-Uqcrc1, Molecular Probes A21362; antiporin, Molecular Probes A31855).
Measurements of mitochondrial respiration in neuroblastoma cells.
Mitochondrial oxygen consumption was measured in permeabilized neuroblastoma (N2A) cells essentially by the method of Hofhaus et al. (12). Cells were harvested by trypsinization, diluted 1:5 in medium A (250 mM sucrose, 10 mM MgCl2, 20 mM HEPES [pH 7.1]), and spun down at 250 × g for 2 min. Recovered cells were resuspended in 1 ml medium A plus 1 ml of 200 μg/ml digitonin. Cells were incubated at 37°C for 1 min prior to a 1/10 dilution in medium A and spun down at 250 × g for 3 min. The final pellet was resuspended in 1 ml medium A and triturated using five passes through a fire-polished glass pipette to separate the cells. Cell counts were performed in triplicate for each sample, using a hemocytometer. Cell viability was tested using trypan blue. Typically >95% of cells stained with trypan blue following digitonin permeabilization. Permeabilized cells (4 × 106) were introduced into the electrode chamber to a final volume of 3 ml in N2A respiration buffer (medium A supplemented with 1 mM ADP and 2 mM KH2PO4). Following acquisition of a stable baseline, the complex I substrates pyruvate and malate were added (to concentrations of 5 mM and 2.5 mM, respectively) before complex I was inhibited by addition of rotenone (2 μM). The complex II substrate succinate was then added (15 mM) and respiration measured before maximal inhibition of the chain at complex III by using myxathiazol (50 nM). Finally, the complex IV substrate TMPD (80 μM) was added together with ascorbate (10 mM) and a rate determined prior to complete inhibition of the chain at complex IV by addition of KCN (1 mM).
RESULTS
Detection of misregulated genes by differential display.
Mecp2-null mice are normal at birth but develop symptoms at around 6 weeks of age, leading to death after approximately 10 weeks (5, 10). As the time of onset of these phenotypic effects shows considerable variability among individuals, we selected mice according to symptom stage rather than chronological age, using the criteria shown in Table 2. In order to maximize our ability to detect alterations in the expression of low-abundance transcripts, we compared mRNA populations in mutant and wt brains by using a variant of differential display called ADDER (17). We detected as broad a range of mRNAs as possible by using multiple primer sets covering all possible nucleotide combinations flanking the 3′ poly(A) and 5′ adaptors of the mRNA. ADDER is reported to detect transcripts present at ∼10 copies per cell and to be sensitive to small differences in RNA levels (17). We performed ADDER on three pools (three brains per pool) of total RNA from brains of “late-symptomatic” Mecp2-null mice (Table 2), using as controls three equivalent pools from brains of wt littermates. In total, we used 192 primer combinations, theoretically allowing detection of about 10,000 RNA species. We observed 39 bands that were more intense in Mecp2-null samples than in the wt and 11 bands that were less intense. Bands corresponding to 36 nonrepetitive genes were recovered and tested for aberrant gene expression by quantitative real-time PCR. Significant deregulation of gene expression in late-symptomatic brains was verified for 12 genes (Table 3). We next asked whether these genes are aberrantly expressed in Mecp2-null mice that had only recently begun to display symptoms (“early-symptomatic” mice [Table 2]). Quantitative PCR on three pools of Mecp2-null brains (three brains per pool) revealed 3 out of the 12 genes to be misexpressed in brains of early-symptomatic mice, in comparison with age-matched controls (t test, P ≤ 0.05). All of the identified genes were expressed apparently normally in presymptomatic Mecp2-null brain RNA (Table 3).
TABLE 2.
Classification of Mecp2-null mice according to the manifestation of phenotype
| Symptom stage | Manifestation of phenotype
|
Age (days)a | ||||
|---|---|---|---|---|---|---|
| Clasping | Inertia | Tremor | Weight loss | LOCb | ||
| Presymptomatic | − | − | − | − | − | ∼30 |
| Early symptomatic | −/+ | + | + | − | − | ∼55 |
| Late symptomatic | + | + | + | + | + | ∼70 |
Average age of mice displaying the symptoms.
LOC, loss of condition.
TABLE 3.
Genes found by a variant of differential display and confirmed by real-time PCR
| Gene accession no. | Gene (protein or comment) | Valuea for the following symptom stage and strain:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Presymptomatic
|
Early symptomatic
|
Late symptomatic
|
||||||||||||||
| wt
|
Null
|
P | wt
|
Null
|
P | wt
|
Null
|
P | ||||||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |||||
| NM_025407 | Uqcrc1(ubiquinol-cytochrome c reductase core protein 1) | 1.00 | 0.15 | 1.10 | 0.30 | 0.32 | 1.00 | 0.12 | 1.22 | 0.09 | <0.001 | 1.00 | 0.28 | 1.73 | 0.56 | <0.001 |
| AK087448 | Unknown (contains SAM domain) | 1.00 | 0.16 | 0.96 | 0.12 | 0.28 | 1.00 | 0.28 | 0.69 | 0.10 | 0.004 | |||||
| BC028971 | Gtl2/Meg3 (imprinted, maternally expressed) | 1.00 | 0.42 | 1.14 | 0.37 | 0.42 | 1.00 | 0.22 | 1.69 | 0.33 | <0.001 | |||||
| AK011516 | Hist1h2bc (histone 1 H2bc) | 1.00 | 0.37 | 1.00 | 0.21 | 0.95 | 1.00 | 0.37 | 0.82 | 0.25 | 0.17 | 1.00 | 0.34 | 0.44 | 0.16 | <0.001 |
| AK034339 | Unknown (similar to esterase/lipase/thioesterase family members) | 1.00 | 0.25 | 1.05 | 0.20 | 0.49 | 1.00 | 0.23 | 1.37 | 0.50 | 0.039 | 1.00 | 0.26 | 1.35 | 0.47 | 0.038 |
| AU018611 | mt-Nd2 (NADH dehydrogenase 2) | 1.00 | 0.16 | 1.14 | 0.30 | 0.16 | 1.00 | 0.44 | 0.73 | 0.25 | 0.08 | 1.00 | 0.17 | 0.6 | 0.08 | <0.001 |
| NM_175092 | Rhof (ras homolog gene family, member f) | 1.00 | 0.65 | 1.44 | 0.64 | 0.12 | 1.00 | 0.33 | 0.67 | 0.29 | 0.02 | |||||
| AK049648 | Unknown | 1.00 | 0.15 | 1.13 | 0.32 | 0.26 | 1.00 | 0.46 | 2.07 | 0.96 | 0.002 | 1.00 | 0.34 | 1.39 | 0.34 | 0.01 |
| AK029199 | Cdon (cell adhesion molecule-related/down-regulated by oncogenes) | 1.00 | 0.25 | 0.89 | 0.23 | 0.31 | 1.00 | 0.16 | 1.39 | 0.23 | <0.001 | |||||
| BC025130 | Ccl19 (chemokine [C-C motif] ligand 19) | 1.00 | 0.40 | 1.00 | 0.30 | 0.96 | 1.00 | 0.21 | 2.69 | 0.71 | <0.001 | |||||
| BC058513 | Snrp70 (U1 small nuclear ribonucleoprotein polypeptide A) | 1.00 | 0.42 | 1.40 | 0.59 | 0.08 | 1.00 | 0.38 | 1.44 | 0.57 | 0.04 | |||||
Mean values are normalized against the GAPDH gene and expressed relative to wt values. Significance was determined by the Student t test. Boldface indicates genes that are significantly up-regulated (P < 0.05).
The three genes that were misexpressed in early symptomatic animals were considered more likely to be primary contributors to the observed pathology than those expressed only in terminally ill animals. One of the three genes is a putative member of the esterase/lipase/thioesterase gene family. This gene is expressed in several tissues and has 43% amino acid sequence identity with arylacetamide deacetylase, which plays a role in lipid metabolism. A second gene was of unknown function, as no annotated relatives could be detected. In this study, we focused on the third early misexpressed gene, which encodes Uqcrc1, a core subunit of complex III which is part of the mitochondrial electron transport chain. Interestingly, the gene for NADH dehydrogenase subunit 2 (mt-Nd2), another component of the mitochondrial electron transport chain (complex I), was found among the 11 genes that were misregulated in late-symptomatic animals. Uqcrc1, a nucleus-encoded gene, was significantly up-regulated in early- and late-symptomatic brains, whereas mitochondrially encoded mt-Nd2 was significantly down-regulated in late-symptomatic, but not early-symptomatic, brains (Fig. 1a and b; Table 3). We have failed to detect any difference between the amounts of Uqcrc1 protein present in the brains of Mecp2-null (early- and late-symptomatic) mice and wt littermates by Western blotting (data not shown; see Discussion).
There are several possible reasons why the set of genes detected by differential display did not overlap with those reported in previous studies. Theoretically ADDER permits expression analysis of about 80% of polyadenylated transcripts, but in practice this number is considerably lower. Our best estimate was that we examined expression of up to 10,000 transcription units, which is around half of the total number of expressed sequences in mouse brain and may therefore exclude some reported targets. An additional limitation of the technique is that the PCR amplification step may be unrepresentative, leaving some genes undetected. The microarray experiment performed by Nuber et al. (31) utilized an array of 13,000 genes (about 60% of the expected total), and changes of less than twofold were disregarded. As most of the genes found by ADDER differential display are misregulated less than twofold, they may be missed by the microarray data analysis. Reexamining the microarray data set, we found that Gtl2/Meg3 shows up-regulation of 1.6-fold. This gene therefore is found in both analyses.
MeCP2 binds to the Uqcrc1 promoter in vivo.
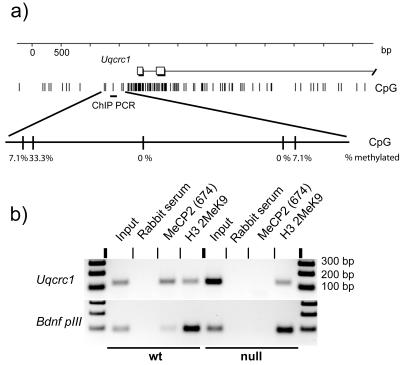
The delayed onset of Uqcrc1 up-regulation may mean that misexpression is an indirect consequence of the absence of MeCP2. However, the amount of MeCP2 in neurons is known to increase dramatically as neurons mature (15, 26), and therefore some genes that are not initially affected by MeCP2 may come under its regulatory influence as MeCP2 becomes more abundant. The increase in the concentration of MeCP2 in murine brain occurs progressively during postnatal life as synaptogenesis proceeds and neurons mature (16, 26). If Uqcrc1 is a direct target, MeCP2 should bind in the vicinity of its promoter. This prediction was verified by chromatin immunoprecipitation, which showed that an anti-MeCP2 antibody could precipitate a DNA region near the Uqcrc1 transcription start site (Fig. 2). This region was not detected when Mecp2-null brain nuclei were subjected to immunoprecipitation, thereby eliminating the possibility of nonspecific cross-reaction of the antibody with other nuclear components (Fig. 2b). A known MeCP2 binding site, promoter III of the Bdnf gene (6, 24), served as a positive control for the immunoprecipitation reaction. To control for the equivalence of Mecp2-null brain chromatin preparations, we found that an antibody against dimethylated lysine 9 of histone H3 gave identical recovery of the Uqcrc1 promoter region in Mecp2-null and wt brain nuclei. We also tested for the presence of methyl-CpG sites in the region that appeared to bind MeCP2 by using bisulfite sequencing. One of a group of CpGs flanking the Uqcrc1 promoter CpG island was found to be 33% methylated in total brain DNA (Fig. 2a). We conclude that MeCP2 is associated with the Uqcrc1 gene in brain and may directly influence its expression, although we cannot exclude the possibility that Uqcrc1 overexpression is an indirect consequence of MeCP2 deficiency.
FIG. 2.
MeCP2 binds the promoter region of the Uqcrc1 gene. (a) Uqcrc1 promoter map showing CpG frequency, methylation status, and the region used for PCR amplification of immunoprecipitated DNA. (b) Chromatin immunoprecipitation reveals MeCP2 bound to the Uqcrc1 promoter in wt mouse brain but not in Mecp2-null mouse brain. Bdnf promoter III is a positive control. DNA is specifically immunoprecipitated with MeCP2 antibody but not with rabbit serum. No DNA is precipitated from Mecp2-null mouse brain by anti-MeCP2 antibody. Chromatin quality is similar in wt and Mecp2-null mouse brains, because anti-dimethyl H3 K9 antibody is able to precipitate similar amounts of DNA.
Mitochondrial abnormalities in the Mecp2-null mouse brain.
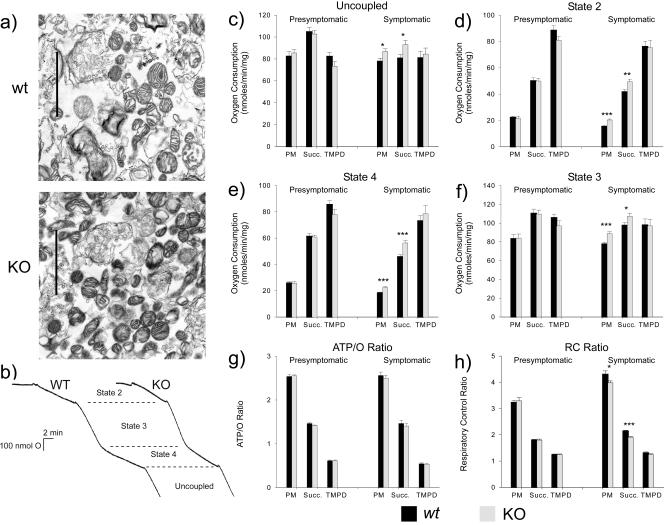
We speculated that the abnormal expression of Uqcrc1 in symptomatic Mecp2-null mice might affect mitochondrial morphology and/or physiology. Initial examination of purified mitochondria by electron microscopy did not reveal gross structural differences between wt and Mecp2-null mice (Fig. 3a). We therefore performed a polarographic oxygen electrode study to evaluate the activities of different complexes within the electron transport chain (Fig. 3b). The protonophore FCCP uncouples the activity of complexes I to IV from the rate-limiting electrochemical proton gradient, thereby permitting maximal activity. Symptomatic Mecp2-null mitochondrial samples consistently showed increased uncoupled respiration rates when substrates were fed into the respiratory chain upstream of complex III (Fig. 3c, PM and Succ.) but not with a substrate entering downstream of complex III (Fig. 3c, TMPD). This difference was not seen in brain mitochondria from presymptomatic mice (Fig. 3c, left panel). The data suggest that in symptomatic Mecp2-null mice the maximal capacity of the respiratory chain upstream of complex IV is increased. This is confirmed by the observation that differences in state 3 respiration rates between symptomatic Mecp2-null and wt mice were also observed only for substrates that feed in to the respiratory chain upstream of complex IV (Fig. 3f).
FIG. 3.
Investigation of the respiratory chain in mitochondria isolated from whole brains of wt and Mecp2-null mice. (a) Electron micrographs showing isolated mitochondria from a symptomatic Mecp2-null (KO) mouse and an age-matched wt littermate. Bar, 2 μm. (b) Typical output traces from a polarographic oxygen electrode. Initially mitochondria consume very little oxygen, but following the addition of substrates, oxygen consumption is moderately increased (state 2). Addition of ADP (state 3) permits rapid respiration, during which the proton gradient is relieved via ATP synthase. Following phosphorylation of all the ADP present, respiration slows again (state 4). At this point, mitochondria were directly uncoupled via the addition of the protonophore FCCP, allowing the proton pumps to run freely. (c to h) Each pair of bars shows respiration rates using one of three different substrates: pyruvate plus malate (PM), succinate (Succ.), or TMPD (see Materials and Methods). Panels c, d, e, and f compare respiration rates during the different respiratory states from brain mitochondria isolated from presymptomatic Mecp2-null animals (light bars; n = 8 runs with separate mitochondrial preparations derived from four animals) and age-matched wt littermate controls (dark bars). Data from symptomatic Mecp2-null mouse mitochondria (n = 16 runs with preparations from eight animals) compared to wt controls are shown at the right in each panel. Significantly increased respiration rates were observed in mitochondria from symptomatic Mecp2-null animals with complex I substrates (PM) and the complex II substrate (Succ.) for all respiratory states. All comparisons were tested with the t test. *, P < 0.05; **, P < 0.01; ***, represent P < 0.001. (g) Calculated ATP/O ratios for presymptomatic and postsymptomatic Mecp2-null animals and wt controls. No significant differences were observed with any of the substrates. (h) Calculated respiratory control (RC) ratios for presymptomatic and postsymptomatic Mecp2-null animals and wt controls. Significant decreases were observed in symptomatic Mecp2-null animals compared to wt controls for complex I (P = 0.05) and complex II (P < 0.001) substrates only. Error bars indicate standard deviations.
Under physiological conditions, the proton translocation associated with complexes I, III, and IV works against an electrochemical proton gradient, providing a mechanism for respiratory control. The coupled states 2 and 4 represent this condition. Increases in state 2 and 4 oxygen consumption were observed between symptomatic Mecp2-null and wt mitochondria when substrates were fed in at complexes I and II but not when these were bypassed by addition of a complex IV substrate (Fig. 3d and e). Again, these differences were not seen in presymptomatic mice.
Measurement of the amount of oxygen consumed during the conversion of a known amount of ADP (during state 3) allows the calculation of the ATP/O ratio. This relates the stoichiometries of protons translocated per electron flowing down the respiratory chain to the number of protons flowing through the ATP synthase per ATP molecule produced. No significant differences were observed in any of the measured ATP/O ratios (Fig. 3g), implying that the respiration rate differences reported above were not the result of a change in the respiratory chain proton pumps or the efficiency of ATP synthase.
Our results show that respiration rates for symptomatic Mecp2-null animals are significantly increased relative to those for wt controls for all substrates that feed in upstream of complex IV. Examination of the respiratory control ratios indicates that the rates of coupled respiration have increased proportionally more than the uncoupled rates (Fig. 3h). This probably indicates an increase in the non-Ohmic proton conductance across the mitochondrial inner membrane (30). To maintain the proton gradient against this background “leak,” the electron transport chain works faster and therefore consumes more oxygen. Thus, mitochondria from symptomatic mutants appear to have an overall greater respiratory capacity (Fig. 3c) but also appear to work less efficiently (Fig. 3e and h).
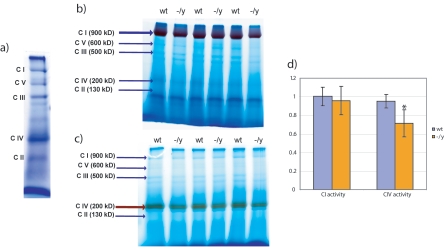
Complexes I and II transfer electrons independently to complex III via a common pool of ubiquinone. Despite the independent nature of the electron paths, the observed genotype-specific respiratory effects were similar with substrates specific for either complex. The common link between the two pathways is complex III. We could not rule out the possibility, however, that the observations are due to elevated activities of both complexes I and II. We tested this possibility for complex I by using blue native electrophoresis, which allows activity-based visualization of each respiratory complex (36). Coomassie blue staining of respiratory complexes from wt and Mecp2-null mouse brains showed apparently normal levels (Fig. 4a and data not shown). We next assayed the enzymatic activities of complexes I and IV in the gel by using histochemical staining for NADH dehydrogenase and cytochrome oxidase (39). Complexes I and IV each produced a single band of the expected size (Fig. 4b and c). Samples derived from three wild-type and mutant littermate pairs were quantified using densitometry. The results showed that complex I activity was indistinguishable between wt and mutant brain mitochondria (t test. P > 0.05). This result argues against the possibility that complex I is responsible for the increase in respiration rates and therefore further implicates complex III. As there is currently no in-gel assay for complex III activity, this hypothesis was not directly testable. An interesting by-product of the blue native electrophoresis analysis was the discovery of a reproducible decrease in the activity of complex IV (t test, P < 0.001) in samples derived from Mecp2-null mouse brains (Fig. 4d).
FIG. 4.
Analysis of the respiratory complexes by blue native electrophoresis. (a) Coomassie blue-stained blue native electrophoresis gel shows resolved mitochondrial respiratory complexes. (b and c) Enzymatic staining of complexes I and IV, respectively. Samples from littermates are beside each other. The stained band is highlighted with thicker arrows. (d) Statistical analysis of the enzymatic staining. Three experiments were done with three pairs of mice, and data were evaluated using the t test. Complex IV staining in Mecp2-null mice is significantly lower (P < 0.001), while complex I staining is the same (P = 0.38). Error bars show standard deviations.
Uqcrc1 overexpression causes abnormal mitochondrial respiration in N2A cells.
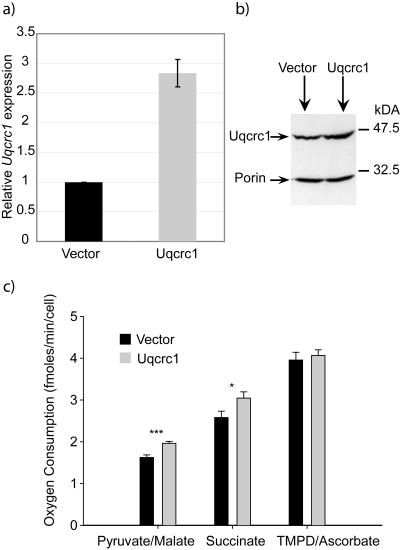
Multiple events could potentially cause the mitochondrial respiration abnormalities in Mecp2-null mouse brain described above. The simplest hypothesis, however, is that overexpression of Uqcrc1 alone is responsible for the measured increase in oxygen consumption with complex I and II substrates. To test this hypothesis, we overexpressed Uqcrc1 in neuroblastoma cells. The Uqcrc1 gene was introduced into mouse N2A cells by using retroviral infection, and infected cells were maintained and expanded by selecting for the vector-carried puromycin resistance gene. Stable Uqcrc1-overexpressing cell lines produced 2.8 times more RNA and 1.6 times more Uqcrc1 protein than the control cells infected with vector only (Fig. 5a and b). Permeabilized cell suspensions of Uqcrc1-overexpressing N2A cells and control cells were then used for respiration analysis with a Clarke oxygen electrode as described above. When substrates upstream of complex III were used (Fig. 5c, pyruvate/malate and succinate), we observed a significant increase in the uncoupled respiration rate in the Uqcrc1-overexpressing cell line (t test, Ppy/mal < 0.001 and Psucc < 0.05; n = 19). Respiration rates with a complex IV substrate (Fig. 5c, TMPD/ascorbate) resulted in similar rates of oxygen consumption in both cell lines (t test, P > 0.05). Thus, artificial overexpression of Uqcrc1 strikingly reproduces the data obtained with isolated uncoupled mitochondria from the brains of Mecp2-null mice discussed above (compare Fig. 3 and 5).
FIG. 5.
Overexpression of Uqcrc1 in an N2A cell line causes increased mitochondrial respiration. (a) Real-time PCR analysis of Uqcrc1 mRNA expression relative to GAPDH. Uqcrc1 expression in control cell line was normalized to 1. (b) Representative Western blot of whole-cell lysates from control and Uqcrc1-overexpressing N2A cells. Porin serves as a loading control. Statistical analysis of duplicate experiments indicates 1.6-fold overexpression of Uqcrc1 protein. (c) Oxygen electrode data for the ADP-uncoupled respiration rates of permeabilized control and Uqcrc1-overexpressing cells in the presence of pyruvate/malate, succinate, or TMPD/ascorbate as respiratory substrates. Significance was tested using the t test. **, P < 0.01; *, P < 0.05. Error bars indicate standard deviations.
DISCUSSION
Relationship of Uqcrc1 overexpression to mitochondrial abnormalities.
The data presented here show that MeCP2 binds to the promoter of the Uqcrc1 gene in vivo and that Uqcrc1 mRNA expression is elevated in brains of Mecp2-null mice that have acquired neurological symptoms. Uqcrc1 up-regulation correlates positively with symptom severity and with a significant increase in mitochondrial respiratory capacity and a reduction in respiratory efficiency. The defect appears to be associated with respiratory complex III, which contains the Uqcrc1 protein, as increased respiration was seen when appropriate substrates were provided to either complex I or complex II. Blue native electrophoresis showed normal levels of complex I activity in the mutant, indicating that this complex is not responsible for the observed increase in respiration. As these findings implicated complex III as the source of enhanced respiratory activity, we asked whether overexpression of Uqrcrc1 alone is sufficient to increase mitochondrial respiration. Indeed, a cell line overexpressing Uqcrc1 showed increased respiration rates with substrates upstream of complex IV. This argues that the mitochondrial respiratory abnormalities seen in Mecp2-null brain are a consequence of Uqcrc1 overexpression.
Although the evidence strongly supports a causal relationship between Uqcrc1 overexpression and abnormalities in mitochondrial respiration, we have not been able to detect reliable increases in brain Uqcrc1 protein due to the ∼1.7-fold increase in Uqcrc1 mRNA in the symptomatic Mecp2-null brain. Two possible explanations for this discrepancy can be considered: (i) Uqcrc1 may be subject to tight posttranscriptional regulation, for example, by feedback modulation of mRNA translation efficiency or by degradation of excess unassembled protein, or (ii) Western blotting may be insensitive to quantitative changes of such low magnitude. The finding that a 2.8-fold up-regulation of Uqcrc1 mRNA in N2A cells led to only a 1.6-fold up-regulation of protein is relevant. A comparable ratio of excess mRNA to excess protein in the brain samples would lead to a 1.2-fold increase in Uqcrc1 protein, which would be difficult to detect by Western blotting.
While the molecular origin of these respiratory abnormalities remains unclear, we speculate that the exquisite sensitivity of mitochondrial assembly to the stoichiometry of its constituent proteins is responsible. For example, increased amounts of Uqcrc1 may destabilize complex III assembly, leading to faster electron transfer or perhaps transfer by Uqcrc1 alone. Another possibility is that Uqcrc1 interacts with a complex III inhibitor, which would be titrated by excess Uqcrc1. In the structure of complex III (cytochrome bc1), Uqcrc1 is orientated towards the matrix, which might allow interactions of this kind (14).
Our studies uncovered other mitochondrial defects in the Mecp2-null mouse brain: (i) significantly elevated coupled respiration rates, suggesting increased proton conductance across the inner mitochondrial membrane; (ii) reduced activity of cytochrome oxidase, implying a defect in complex IV; and (iii) down-regulation of mRNA for NADH dehydrogenase, a mitochondrially encoded component of complex I, in late symptomatic animals. The observed increase in coupled respiration of up to 30% indicates a loss of respiratory control which may have a significant impact on mitochondrial efficiency in the Mecp2-null mice. The other changes are subtle (<2-fold) but may reflect suboptimal mitochondrial performance in brains of symptomatic Mecp2-null mice. Whether these effects are related to Uqcrc1 overexpression or arise independently is not currently known.
Mitochondrial abnormalities and RTT.
The notion that mitochondrial abnormalities play a role in RTT predates the discovery of the genetic origin of the condition. With the realization that MeCP2, a nuclear transcriptional repressor, is mutated in RTT, the mitochondrial link became less compelling. It was recently noted, however, that a patient with symptoms normally associated with mitochondrial disorders (hypotonia, small stature, developmental delay, and a slight decrease in respiratory chain enzyme activity) harbored mutations in the MECP2 gene (11). This overlap between symptoms of RTT and mitochondrial disorders recalls early reports of structural abnormalities (4, 8, 9, 34) and defects in the electron transport chain (7, 8) in mitochondria from skin and muscle biopsies of RTT patients. Moreover, about half of RTT patients were reported to have elevated levels of circulatory lactic or pyruvic acid, which might be caused by defects in the efficiency of the respiratory chain and urea cycle complexes, both of which are mitochondrial (21, 25).
In humans many disorders affecting the brain are due to mutations in nuclear or mitochondrially encoded components of the mitochondrion, sometimes resulting in increased oxidative stress or induction of neuronal apoptosis. As Rett syndrome is not a neurodegenerative disorder (3), any contribution of mitochondrial dysfunction to RTT symptoms may take the form of chronic mitochondrial underperformance, rather than catastrophic failure leading to neuronal death. The changes that we have observed in the mouse model of RTT appear to meet this criterion, as they are small in magnitude and may therefore compromise brain function without precipitating cellular death. Future work will probe the involvement of MeCP2 in the expression of proteins that are targeted to the mitochondrion and will test the hypothesis that aberrant mitochondrial function as seen in mice has a role to play in RTT.
Acknowledgments
We thank Jim Selfridge and Hannah Moore for assistance and advice, Bird laboratory members and Cathy Abbott for critical comments on the manuscript, and John Findlay for assistance in using the electron microscope.
This research was funded by the Wellcome Trust, the Rett Syndrome Research Foundation, and Jeans for Genes (Rett Syndrome United Kingdom). S.K. was a Darwin Trust Scholar and now holds an RSRF fellowship. A.P. was supported by a BBSRC studentship.
REFERENCES
- 1.Amir, R. E., I. B. van den Veyver, I., M. Wan, C. Q. Tran, U. Francke, and H. Y. Zoghbi. 1999. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 23:185-188. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, M. F., and N. R. Sims. 2000. Improved recovery of highly enriched mitochondrial fractions from small brain tissue samples. Brain Res. Brain Res. Protoc. 5:95-101. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong, D. D. 2002. Neuropathology of Rett syndrome. Ment. Retard. Dev. Disabil. Res. Rev. 8:72-76. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong, D. D. 1992. The neuropathology of the Rett syndrome. Brain Dev. 14(Suppl.):S89-S98. [PubMed] [Google Scholar]
- 5.Chen, R. Z., S. Akbarian, M. Tudor, and R. Jaenisch. 2001. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat. Genet. 27:327-331. [DOI] [PubMed] [Google Scholar]
- 6.Chen, W. G., Q. Chang, Y. Lin, A. Meissner, A. E. West, E. C. Griffith, R. Jaenisch, and M. E. Greenberg. 2003. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science 302:885-889. [DOI] [PubMed] [Google Scholar]
- 7.Coker, S. B., and A. R. Melnyk. 1991. Rett syndrome and mitochondrial enzyme deficiencies. J. Child Neurol. 6:164-166. [DOI] [PubMed] [Google Scholar]
- 8.Dotti, M. T., L. Manneschi, A. Malandrini, N. De Stefano, F. Caznerale, and A. Federico. 1993. Mitochondrial dysfunction in Rett syndrome. An ultrastructural and biochemical study. Brain Dev. 15:103-106. [DOI] [PubMed] [Google Scholar]
- 9.Eeg-Olofsson, O., A. G. al Zuhair, A. S. Teebi, and M. M. al Essa. 1989. Rett syndrome: genetic clues based on mitochondrial changes in muscle. Am. J. Med. Genet. 32:142-144. [DOI] [PubMed] [Google Scholar]
- 10.Guy, J., B. Hendrich, M. Holmes, J. E. Martin, and A. Bird. 2001. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat. Genet. 27:322-326. [DOI] [PubMed] [Google Scholar]
- 11.Heilstedt, H. A., M. D. Shahbazian, and B. Lee. 2002. Infantile hypotonia as a presentation of Rett syndrome. Am. J. Med. Genet. 111:238-242. [DOI] [PubMed] [Google Scholar]
- 12.Hofhaus, G., R. M. Shakeley, and G. Attardi. 1996. Use of polarography to detect respiration defects in cell cultures. Methods Enzymol. 264:476-483. [DOI] [PubMed] [Google Scholar]
- 13.Horike, S., S. Cai, M. Miyano, J. F. Cheng, and T. Kohwi-Shigematsu. 2005. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat. Genet. 37:31-40. [DOI] [PubMed] [Google Scholar]
- 14.Iwata, S., J. W. Lee, K. Okada, J. K. Lee, M. Iwata, B. Rasmussen, T. A. Link, S. Ramaswamy, and B. K. Jap. 1998. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science 281:64-71. [DOI] [PubMed] [Google Scholar]
- 15.Jung, B. P., D. G. Jugloff, G. Zhang, R. Logan, S. Brown, and J. H. Eubanks. 2003. The expression of methyl CpG binding factor MeCP2 correlates with cellular differentiation in the developing rat brain and in cultured cells. J. Neurobiol. 55:86-96. [DOI] [PubMed] [Google Scholar]
- 16.Kishi, N., and J. D. Macklis. 2004. MECP2 is progressively expressed in post-migratory neurons and is involved in neuronal maturation rather than cell fate decisions. Mol. Cell Neurosci. 27:306-321. [DOI] [PubMed] [Google Scholar]
- 17.Kornmann, B., N. Preitner, D. Rifat, F. Fleury-Olela, and U. Schibler. 2001. Analysis of circadian liver gene expression by ADDER, a highly sensitive method for the display of differentially expressed mRNAs. Nucleic Acids Res. 29:E51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kriaucionis, S., and A. Bird. 2004. The major form of MeCP2 has a novel N-terminus generated by alternative splicing. Nucleic Acids Res. 32:1818-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kriaucionis, S., and A. Bird. 2003. DNA methylation and Rett syndrome. Hum. Mol. Genet. 12(Spec. No. 2):R221-R227. [DOI] [PubMed] [Google Scholar]
- 20.Lai, J. C., J. M. Walsh, S. C. Dennis, and J. B. Clark. 1977. Synaptic and non-synaptic mitochondria from rat brain: isolation and characterization. J. Neurochem. 28:625-631. [DOI] [PubMed] [Google Scholar]
- 21.Lappalainen, R., and R. S. Riikonen. 1994. Elevated CSF lactate in the Rett syndrome: cause or consequence? Brain Dev. 16:399-401. [DOI] [PubMed] [Google Scholar]
- 22.Lewis, J. D., R. R. Meehan, W. J. Henzel, I. Maurer-Fogy, P. Jeppesen, F. Klein, and A. Bird. 1992. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 69:905-914. [DOI] [PubMed] [Google Scholar]
- 23.Li, L. C., and R. Dahiya. 2002. MethPrimer: designing primers for methylation PCRs. Bioinformatics 18:1427-1431. [DOI] [PubMed] [Google Scholar]
- 24.Martinowich, K., D. Hattori, H. Wu, S. Fouse, F. He, Y. Hu, G. Fan, and Y. E. Sun. 2003. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science 302:890-893. [DOI] [PubMed] [Google Scholar]
- 25.Matsuishi, T., F. Urabe, H. Komori, Y. Yamashita, E. Naito, Y. Kuroda, M. Horikawa, and E. Ohtaki. 1992. The Rett syndrome and CSF lactic acid patterns. Brain Dev. 14:68-70. [DOI] [PubMed] [Google Scholar]
- 26.Mullaney, B. C., M. V. Johnston, and M. E. Blue. 2004. Developmental expression of methyl-CpG binding protein 2 is dynamically regulated in the rodent brain. Neuroscience 123:939-949. [DOI] [PubMed] [Google Scholar]
- 27.Nan, X., F. J. Campoy, and A. Bird. 1997. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell 88:471-481. [DOI] [PubMed] [Google Scholar]
- 28.Nan, X., H. H. Ng, C. A. Johnson, C. D. Laherty, B. M. Turner, R. N. Eisenman, and A. Bird. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393:386-389. [DOI] [PubMed] [Google Scholar]
- 29.Nan, X., P. Tate, E. Li, and A. Bird. 1996. DNA methylation specifies chromosomal localization of MeCP2. Mol. Cell. Biol. 16:414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicholls, D. G., and S. J. Ferguson. 2002. Bioenergetics 3. Academic Press, San Diego, Calif.
- 31.Nuber, U. A., S. Kriaucionis, T. C. Roloff, J. Guy, J. Selfridge, C. Steinhoff, R. Schulz, B. Lipkowitz, H. H. Ropers, M. C. Holmes, and A. Bird. 2005. Up-regulation of glucocorticoid-regulated genes in a mouse model of Rett syndrome. Hum. Mol. Genet. 14:2247-2256. [DOI] [PubMed] [Google Scholar]
- 32.Peterson, G. L. 1977. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal. Biochem. 83:346-356. [DOI] [PubMed] [Google Scholar]
- 33.Rajapakse, N., K. Shimizu, M. Payne, and D. Busija. 2001. Isolation and characterization of intact mitochondria from neonatal rat brain. Brain Res. Brain Res. Protoc. 8:176-183. [DOI] [PubMed] [Google Scholar]
- 34.Ruch, A., T. W. Kurczynski, and M. E. Velasco. 1989. Mitochondrial alterations in Rett syndrome. Pediatr. Neurol. 5:320-323. [DOI] [PubMed] [Google Scholar]
- 35.Samaco, R. C., A. Hogart, and J. M. LaSalle. 2005. Epigenetic overlap in autism-spectrum neurodevelopmental disorders: MECP2 deficiency causes reduced expression of UBE3A and GABRB3. Hum. Mol. Genet. 14:483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schagger, H. 1995. Quantification of oxidative phosphorylation enzymes after blue native electrophoresis and two-dimensional resolution: normal complex I protein amounts in Parkinson's disease conflict with reduced catalytic activities. Electrophoresis 16:763-770. [DOI] [PubMed] [Google Scholar]
- 37.Shahbazian, M., J. Young, L. Yuva-Paylor, C. Spencer, B. Antalffy, J. Noebels, D. Armstrong, R. Paylor, and H. Zoghbi. 2002. Mice with truncated MeCP2 recapitulate many Rett syndrome features and display hyperacetylation of histone H3. Neuron 35:243-254. [DOI] [PubMed] [Google Scholar]
- 38.Tudor, M., S. Akbarian, R. Z. Chen, and R. Jaenisch. 2002. Transcriptional profiling of a mouse model for Rett syndrome reveals subtle transcriptional changes in the brain. Proc. Natl. Acad. Sci. USA 99:15536-15541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zerbetto, E., L. Vergani, and F. Dabbeni-Sala. 1997. Quantification of muscle mitochondrial oxidative phosphorylation enzymes via histochemical staining of blue native polyacrylamide gels. Electrophoresis 18:2059-2064. [DOI] [PubMed] [Google Scholar]