Abstract
RNA polymerases can be shared by a particular group of genes in a transcription “factory” in nuclei, where transcription may be coordinated in concert with the distribution of coexpressed genes in higher-eukaryote genomes. Moreover, gene expression can be modulated by regulatory elements working over a long distance. Here, we compared the conformation of a 130-kb chromatin region containing the mouse α-globin cluster and their flanking housekeeping genes in 14.5-day-postcoitum fetal liver and brain cells. The analysis of chromatin conformation showed that the active α1 and α2 globin genes and upstream regulatory elements are in close spatial proximity, indicating that looping may function in the transcriptional regulation of the mouse α-globin cluster. In fetal liver cells, the active α1 and α2 genes, but not the inactive ζ gene, colocalize with neighboring housekeeping genes C16orf33, C16orf8, MPG, and C16orf35. This is in sharp contrast with the mouse α-globin genes in nonexpressing cells, which are separated from the congregated housekeeping genes. A comparison of RNA polymerase II (Pol II) occupancies showed that active α1 and α2 gene promoters have a much higher RNA Pol II enrichment in liver than in brain. The RNA Pol II occupancy at the ζ gene promoter, which is specifically repressed during development, is much lower than that at the α1 and α2 promoters. Thus, the mouse α-globin gene cluster may be regulated through moving in or out active globin gene promoters and regulatory elements of a preexisting transcription factory in the nucleus, which is maintained by the flanking clustered housekeeping genes, to activate or inactivate α-globin gene expression.
The human genome project has revealed that there are about 32,000 protein-encoding genes, which are distributed in 22 pairs of autosomes and 2 sex chromosomes. The genome-wide bioinformatic analysis of chromosomal gene expression profiles and chromosomal gene positions reveals that genes are not randomly distributed in the genome. Moreover, highly expressed genes, especially housekeeping genes, are often clustered within specific chromosomal regions (9, 25), suggesting that their chromosomal locations may be related to their transcriptional activities and inherent regulatory properties (42, 44). The estimation of the total number of nascent transcripts, active polymerase molecules, and number of transcription sites within a cell suggests that many different templates may be attached in a “cloud” of loops around a site; each site, or transcription “factory,” would contain approximately 30 active polymerases and associated transcripts (22). Active RNA polymerases are concentrated in these discrete “factories,” where they work together on the transcription of particular groups of genes (34, 35, 39). However, little is known about the functional correlation between the clustered gene order and their transcription. It is also unclear how these clustered genes are spatially organized in nuclei with respect to transcription and how they, as individual transcription units, are regulated in a transcription “factory.”
It has been shown that higher-eukaryote genomes contain many important distant regulatory elements (e.g., enhancer, silencer, and insulator/boundary elements), which often work over a long distance up to thousands of kilobases to control gene expression (11, 15, 26, 29, 30, 38). Looping, linking, and tracking models have been proposed to explain how distantly positioned enhancers might function (7). Much of our knowledge about the functional mechanism of distant enhancers has come from studies on the locus control region (LCR) of the β-globin gene cluster. By employing two independent techniques, chromosome conformation capture (3C) and RNA tagging and recovery of associated proteins, it has been shown that looping is important for long-range activation of β-globin genes (8, 10, 40). Multiple flanking hypersensitive sites (HSs) can interact with active β-like globin promoters to establish a unique structure, dubbed the active chromatin hub (ACH), by looping out the interval regions (40). Further investigations have defined the functions of some related HSs and transcription factors in building β-ACH (13, 33). However, it remains unclear whether the looping mechanism is operative in other long-range gene activation events.
The mouse α-globin cluster and its flanking housekeeping genes provide another model system for addressing the above questions. Although both α- and β-globins are expressed in a tissue- and development-specific manner, activated by distant enhancers, and presumed to have a common origin (20), they display remarkable differences in chromatin environments and regulatory features in mammals (2, 6, 18). The mouse β-globin gene cluster possesses an independent chromatin domain which is confined by two sides of the condensed chromatin domain in erythroid cells and is protected from the transgenic position effect (26). In contrast, the mouse α-globin locus is embedded in a GC-rich region neighbored by a cluster of housekeeping genes and cannot avoid the transgenic position effect (19). Interestingly, its distant major regulatory element, HS26 (corresponding to HS40 in humans), is located in an intron of the housekeeping gene C16orf35 (5, 16). However, why this distant regulatory element can regulate α-globin expression without being interfered with by neighboring genes remains unknown. Moreover, whether the expressing genes of the α-globin locus interact with the flanking housekeeping genes is yet to be determined.
In this study, we investigated the chromatin conformation of the mouse α-globin locus and its flanking housekeeping genes by 3C assay and the occupancy of RNA polymerase II (Pol II) across the whole region by chromatin immunoprecipitation (ChIP) assay in mouse erythroid cells (14.5-day-postcoitum [dpc] fetal liver cells) and nonerythroid cells (14.5 fetal brain cells). The upstream regulatory elements of the mouse α-globin locus are found to be in close proximity to the development-specifically activated α1 and α2 genes in fetal liver cells. Remarkably, the active globin genes in expressing cells colocalize with upstream housekeeping genes, while in nonexpressing cells, the silenced mouse α-globin genes are separated from the congregated housekeeping genes. A comparison of the occupancies of RNA Pol II showed that the active α1 and α2 globin gene promoters have much higher RNA Pol II occupancy in fetal liver than in brain. The RNA Pol II occupancy at the developmentally repressed ζ gene promoter is much lower than that at the active α1 and α2 promoters in liver cells. However, the RNA Pol II occupancies at housekeeping genes are similar in fetal liver and brain. These data indicate that the mouse α-globin gene cluster may be regulated through recruitment of active globin genes and regulatory elements to a shared nuclear subcompartment which is occupied by the flanking colocalized housekeeping genes.
MATERIALS AND METHODS
NcoI digestion efficiency tested by Southern blotting.
At 14.5 dpc, fetal liver and brain cells were treated as described in the following chromosome conformation capture procedure (with 2% formaldehyde) except for the ligation step. The cross-linked DNA and non-cross-linked genomic DNA digested by NcoI were purified and analyzed by 1% agarose electrophoresis to compare their digestion efficiencies. The 15 μg purified DNA was analyzed by Southern blotting to compare the cleavage of the different restriction sites. The following probes were used: HS8, a 502-bp PCR fragment, which hybridizes to a 2.2-kb NcoI HS8 fragment; ζ, a 485-bp PCR fragment, which hybridizes to a 2.2-kb NcoI ζ fragment; α1, a 388-bp PCR fragment, which hybridizes to a 2.2-kb NcoI α1 fragment; α2, a 420-bp PCR fragment, which hybridizes to a 1.5-kb NcoI α2 fragment. The primers for amplifying the probes are as follows: HS8-L, GATCTACAGACTGCCCTCCCAAGTC; HS8-R, TATAAAGTGCTTTCCCTCACCAGGG; ζ-L, CATAGCCATTTGTTGCCAATCAGTG; ζ-R, GGGCTTCATAGTGAGACCGCA TC; α1-L, TGCTCACATCCATTCAGACACAGAC; α1-R, AAGGTTGGGACAAGTACAGTTAGGG; α2-L, GCTGCCCTTCCCTCATCCTCTG; α2-R, AAATCCGGTTGTTACTTGATCATGC.
3C.
The 3C procedure was used as previously described (12, 40) with a few modifications to determine the spatial organization of the 130-kb chromatin region containing the mouse α-globin locus and several flanking housekeeping genes. Cells from fetal liver and fetal brain (both from 14.5-dpc embryos) were isolated and passed through a cell strainer cap to obtain a homogeneous single-cell suspension. Per experiment, equivalent cells from 5 fetal livers and 10 fetal brains (approximately 1 × 108 cells) were resuspended in 100 ml of Dulbecco's minimal essential medium supplemented with 10% fetal calf serum. The samples were cross-linked by 2% formaldehyde for 10 min at room temperature and then quenched by the addition of glycine to 0.125 M. Cells were harvested and washed twice using ice-cold 1× phosphate-buffered saline and then lysed in ice-cold lysis buffer (10 mM Tris, pH 8.0, 10 mM NaCl, 0.2% NP-40) containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, and 1 μg/ml pepstatin A) for 10 min. Nuclei were harvested and washed once using ice-cold 1× phosphate-buffered saline and then resuspended in 1× NcoI restriction buffer (500 μl per 1 × 107 nuclei) containing 0.3% sodium dodecyl sulfate (SDS) and incubated at 37°C for 30 min with constant shaking. Triton X-100 was added to 1.8% and shaked at 37°C for 30 min to sequester the SDS. The 1 × 107 cross-linked nuclei were digested with 750 U NcoI (New England Biolabs) at 37°C for 16 to 20 h with constant shaking. The restriction enzyme was inactivated by the addition of SDS to 1.6% and incubated at 65°C for 20 min. The reaction was diluted by 15-fold with 1× T4 ligase buffer. Triton X-100 was added to 1% and shaked at 37°C for 30 min. Chromatin DNAs were ligated using 100 U T4 ligase (New England Biolabs) for 4 to 5 h at 16°C followed by 30 min at room temperature. Then, 5 M NaCl was added to a final concentration of 200 mM. The samples were incubated at 65°C for 4 h to reverse the cross-linking. Next, 40 μl (10 mg/ml) of proteinase K was added, and the samples were incubated overnight at 45°C. The DNA was purified by phenol-chloroform/isoamyl alcohol (25:24:1) extraction and ethanol precipitation and used for the PCR.
To calibrate the differences in quality and quantity of template, the ligation frequencies were normalized between the analyzed pairs (with sites about 7.6 kb apart) to those detected between two restriction fragments in the Errc3 locus. Errc3 encodes a subunit of the basal transcription factor TFIIH and was used to normalize the differences of DNA amounts between two tissues (32). To compare the intensities obtained with different primer sets in a quantitative manner, a control template containing all possible ligation products in equimolar amounts was made to correct for the PCR amplification efficiency of each set. For this purpose, two bacterial artificial chromosome (BAC) clones spanning the analyzed loci were used. For the mouse α-globin locus, a 187-kb BAC (no. RP23-130H16; CHORI BACPAC Resources) was used. In addition, a 233-kb BAC containing the mouse Errc3 locus (no. RP23-148C24; CHORI BACPAC Resources) was used. The mouse globin BAC was mixed with the Errc3 BAC in equimolar amounts. Then, the mixed BAC DNA templates were digested and ligated. The ligated mixture was diluted to the appropriate concentration, and then the digested and ligated genomic DNA was added and used as a control sample. For quantitative PCR analysis of the ligation products, the linear range of amplification was determined for the fetal liver and brain samples by serial dilution. An appropriate amount of DNA within the linear range (typically 200 to 300 ng of DNA for both liver and brain) was subsequently applied for the experiments. The linear range of control sample was also determined by serial dilution of random ligation mix made in the same DNA amount (300 ng) by the mixture of the digested and ligated genomic DNA. The 5′ side of the restriction fragment was used to design the primer unless this coincided with the repetitive DNA sequences or was not compatible with the paired primer. Primer sequences for the tested restriction fragments were as follows: I, GATGTCTTGTGGTTTTTGATGTGG; II, GATGTCTTGTGGTTTTTGATGTGG; III, GATGTCTTGTGGTTTTTGATGTGG; IV, TGCGGTTCCTGTGAAATGGTG; V, AATGGTGTCTCCACGGGTTGT; VI, GGAAACAACCTCCCTTCTGCC; VII, CAGGACCTTTTAATGGCGAATG; VIII, TCATAATAGGTGGGTGGAATCAG; IX, GGTATGGACTTGGTTCTGTAGC; X, TCTTGAAAATATTAGGCTGAAGGC; XI, CACACTGGGTCGGAAGTTGAG; XII, AAAATATGGGCCAGCAAGACGG; XIII, ACAGTGCAGGTCTGGATACAAG; XIV, CAGTACCGTCCTGGAACATGG; XV, TGGAGGATGCGGCAGTTAGTG; XVI, AAATGATGCGGGACAGTTCG; XVII, CACAGAACTGGGCAATTTATATC; AGCAGCATCATACTCCAGGTAAACA; XIX, TTTGTCTGTCCTCGGCTTGGG. Errc3-1 (CTATATTCAACTGCTGTTCCCATG) and Errc3-2 (TTCTACCAGCAGATCCGTATTCC) are the primers from two NcoI restriction fragments of the Errc3 gene used for correction control amplification. All the test primer pairs were verified by amplifying the control sample and sequencing the PCR products. All the PCRs were done at least three times with the following PCR protocol: 4-min heat start at 94°C and 35 cycles of denaturation at 94°C for 40 s, annealing at 58°C for 40 s, and extension at 72°C for 40 s, and then further extension at 72°C for 10 min and holding at 4°C. All PCR products were run on 2% agarose gels and quantified on an UVIscan imager (UVI). An example of PCR-amplified ligation products on 2% agarose gel and the equation used to calculate ligation frequency are shown in Fig. 3A. The correction method is the same as that given in the work of Dekker et al. and Tolhuis et al. (12, 40). The correction equation is given in Fig. 3A. The calculation gives a relative ligation frequency for each tissue, since it corrects for the differences in PCR amplification efficiencies, amounts of templates, and sizes of PCR products (40).
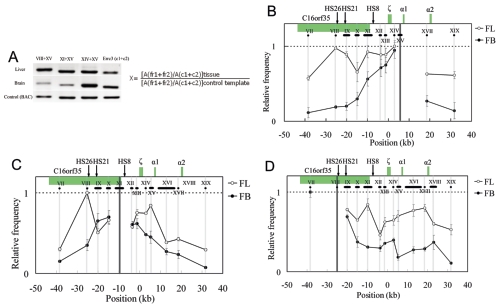
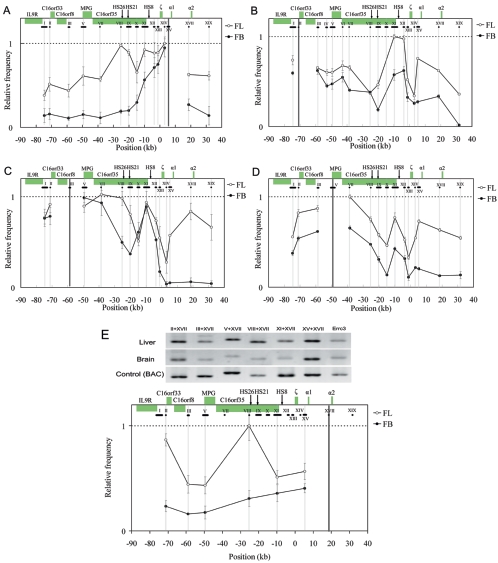
FIG. 3.
Spatial organization of the mouse α-globin locus. An example of PCR-amplified ligation products on 2% agarose gel and the equation used to calculate the ligation frequency of two given fragments, such as 1 and 2, are shown in panel A. Cross-linking patterns of the fragments XV close to the α1 gene (B), XI containing part of HS8 (C), and VIII containing HS26 (D) respectively, in fetal liver (unfilled circles) and brain (filled circles) are shown. The equation in panel A is used to calculate the relative ligation frequency (X). A(fr1+fr2): peak area (determined with UVI scan) of PCR signal obtained with two-test fragment ligation products. A(c1+c2): peak area of PCR signal obtained with two-Errc3 cut fragment (not linked) ligation products. A(fr1+fr2) and A(c1+c2) are determined for both tissues (fetal liver and brain) and the control sample (see Methods). In Fig. 3B to D, the highest relative ligation frequency value was set to 1, and then the relative values are plotted on the y axis for various test fragment sets. The horizontal black lines below the locus indicate the size and position of the analyzed RFs. The vertical gray lines show the central position of the analyzed RFs, and the black line represents the central position of the fixed RFs: XV (B), XI (C), and VIII (D). The x axis shows the distances relative to the ζ-globin mRNA cap site. Error bars represent the standard errors of the means.
ChIP assay.
ChIP analysis was carried out for mouse 14.5-dpc fetal liver and brain cells. In brief, isolated, 1% formaldehyde-cross-linked cells, as described above, were lysed in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris, pH 8.1) containing protease inhibitors and sonicated on ice until cross-linked chromatin DNA was sheared to an average length of 0.5 to 1 kb. The sonicated cell supernatant (1 × 107 cells per experiment) was diluted 10-fold in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.1, 167 mM NaCl). The precleared chromatin using protein A-agarose (Upstate) was incubated without antibody or with the anti-RNA Pol II (H-224) antibody (Santa Cruz Biotechnology) or immunoglobulin G (Upstate), respectively, overnight at 4°C. Immunoprecipitates were recovered by incubation with protein A-agarose (Upstate) for 2 h at 4°C, followed by low-speed centrifugation. The washed pellets were reverse cross-linked by incubation for 6 h at 65°C and digested with proteinase K (Roche) for 12 to 14 h at 45°C. The DNA was extracted with phenol-chloroform/isoamyl alcohol (25:24:1), precipitated with ethanol, and used for PCR analysis. Meanwhile, 1/10 of the sonicated cell supernatant was also reverse cross-linked, and DNA was purified and used for input to correct for the differences in PCR amplification efficiencies and DNA amounts. The primers used for ChIP analysis are available by request. The linear range of ChIP sample and input were determined by serial dilution. An appropriate DNA amount was used for the quantitative PCR analysis. All PCRs were done at least three times with the following PCR protocol: 4-min heat start at 94°C and 35 cycles of denaturation at 94°C for 40 s, annealing at 55°C for 40 s, and extension at 72°C for 40 s, and then further extension at 72°C for 10 min and holding at 4°C. All PCR products were resolved by 2% agarose gel electrophoresis, revealed by staining with ethidium bromide, and then quantified on an UVI. All the ChIP analyses were performed three times independently. GAPDH, which is ubiquitously expressed in all tissue cells, was used as a positive control to correct the differences in DNA amounts of two tissues. RNA Pol II intensity at MyoD, which is not expressed in the tested tissue cells, showed the background occupancy of RNA Pol II. An example of PCR-amplified ChIP products on 2% agarose gel and the equation used to calculate the relative intensity of RNA pol II at DNA are shown in Fig. 5A. This calculation gives RNA Pol II intensity on each site, since it corrects for the differences in PCR amplification efficiencies, amounts of templates, and the sizes of PCR products by input control samples and GAPDH.
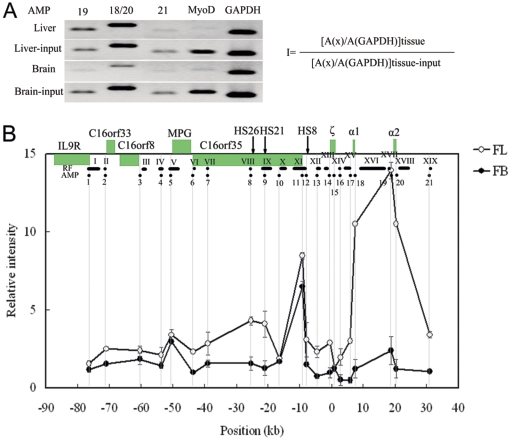
FIG. 5.
Occupancies of RNA Pol II at hypersensitive sites and gene promoters in the 130-kb chromatin region analyzed with ChIP assay. An example of PCR-amplified ChIP and input products on 2% agarose gel and the equation used to calculate the relative intensity of RNA Pol II at a given site are shown in panel A. The occupancy pattern of RNA Pol II across the 130-kb chromatin region is shown in panel B. The equation in panel A is used to calculate RNA Pol II intensity at a given site (I). A(x): peak area (determined with UVI scan) of PCR signal obtained with a certain site in the locus. A(GAPDH): peak area of PCR signal obtained with the GAPDH gene promoter. A(x) and A(GAPDH) are determined for both ChIP and input samples of two tissues. The relative intensity is plotted on the y axis for each test site using a scale relative to MyoD. The black dots below the locus indicate the analyzed amplicons (AMP) in ChIP. The gray vertical lines indicate the positions of the amplicons. The x axis shows the distances relative to the ζ-globin mRNA cap site. Error bars represent the standard errors of the means.
RESULTS
Overall experimental system for comparative 3C.
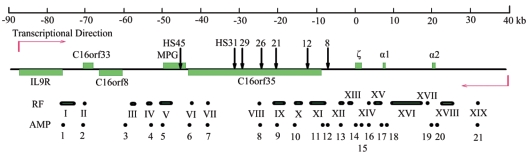
The mouse α-globin gene cluster, which resides in the telomeric region of chromosome 11, consists of one embryonic and two fetal/adult genes organized in the 5′-ζ-α1-α2-3′ orientation (24). The erythroid cell-specific HSs have been identified at −26 kb (HS26), −21 kb (HS21), and −8 kb (HS8) upstream of the mouse ζ globin mRNA cap site (5). There are other HSs, which are defined at −12, −29, −31, and −45 kb in a mouse erythroleukemia cell line (3). Several housekeeping genes have been identified upstream of mouse α-globin genes, including C16orf35, MPG, C16orf8, and C16orf33 (16). The MPG gene encodes N-methylpurine-DNA glycosylase, which removes a diverse group of damaged bases from DNA, including cytotoxic and mutagenic alkylation adducts of purines (16). So far, no function has been annotated to the C16orf8 gene. Conserved domain analysis showed that it is homologous to some members of the rhomboid family. C16orf33 encodes the U11/U12 snRNP 25,000-molecular-weight protein, a component of spliceosome. The function of C16orf35, which is expressed in a wide range of cell lines and tissues, is still unknown (16, 43). HS31, HS29, HS26, HS21, and HS12 are positioned in the transcription unit of C16orf35. Similarly, HS45 is located in MPG, and HS8 is close to the C16orf35 promoter. As illustrated in Fig. 1, the chromatin region described above spans 130 kb. Here, we chose the mouse definitive erythroid cells/14.5-dpc fetal liver cells and nonerythroid cells/14.5-dpc fetal brain cells for comparative 3C analysis. In the 3C assay, NcoI was chosen to digest the cross-linked DNA-protein complexes, since the fragments after cut separate most of the gene promoters and HSs within the 130-kb chromatin region (with the exception of HS8 and C16orf35) (Fig. 1). We checked the digestion efficiencies for the cross-linked chromatin from fetal liver and brain cells. Electrophoresis showed that the digestion efficiencies of cross-linked DNA from fetal liver and brain were similar (Fig. 2A). Southern blot analysis demonstrated that the digestion efficiencies were comparable between a hypersensitive site, HS8, a nonexpressing gene (ζ), and two expressing genes (α1 and α2) in fetal liver cells, indicating that the local chromatin configuration has no effect on the digestion efficiency (Fig. 2B). Digestion efficiencies are also comparable among these sites in fetal brain cells (data not shown). Incomplete cuts were observed in cross-linked samples, which may be due to formaldehyde treatment (32). In addition, quantitative PCR was also used to test the digestion efficiency. Similar efficiencies were obtained at different sites for both fetal liver and brain cells (data not shown).
FIG. 1.
Schematic presentation of the mouse α-globin locus and the flanking region. Black arrows indicate erythroid cell-specific hypersensitive sites, and green boxes indicate genes. The transcription direction toward centromere (above the line) or telomere (below the line) is indicated by red arrows. A set of NcoI restriction fragments (RF) used for 3C analysis and amplicons (AMP) used for ChIP analysis are shown below the locus. RFs are indicated by roman numerals, and AMPs are shown in arabic numerals. Distances are in kilobases, counting from the ζ-globin mRNA cap site.
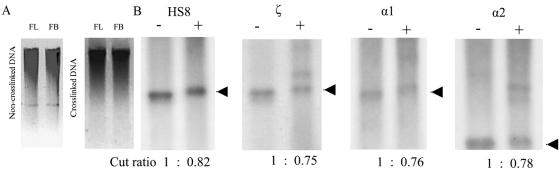
FIG. 2.
NcoI digestion efficiency, tested by Southern blots. (A). Electrophoresis of the non-cross-linked genomic DNA (left panel) and cross-linked DNA-protein complex (right panel) digested by NcoI shows similar patterns for fetal liver (FL) and brain (FB) cells. (B) Southern blotting shows that the NcoI digestion efficiencies of the fragments containing HS8, ζ, α1, and α2 are comparable in definitive erythrocytes (liver cells). − indicates genomic DNA not treated with formaldehyde; + indicates cross-linked DNA treated with formaldehyde. Arrowheads indicate cut fragments. The cut ratios of the cut fragments from non-cross-linked genomic DNA and formaldehyde-cross-linked DNA are showed at the bottom.
The distant regulatory elements of the mouse α-globin locus come in close proximity to active globin genes through looping.
The comparative 3C assay of expressing and nonexpressing cells (fetal liver and brain cells), both from 14.5-dpc fetal embryos, was performed to analyze the spatial organization across the whole mouse α-globin gene cluster in order to determine the particular chromatin conformation. The Errc3 gene, encoding a subunit of the basal transcription factor TFIIH, was used to normalize the differences of DNA amounts in two tissue cells, since its transcription level is similar in two tissues (data not shown). Fragment XV is the closest to the α1 gene. The spatial organization between XV and the fragments VIII and XI, which contain HS26 and HS8, respectively, was examined first. Figure 3A shows that PCR bands> from ligations between VIII and XV and between XI and XV are much weaker than the PCR band from the ligation between XIV and XV in brain samples, while these bands appear similar in the liver, indicating that distance may affect ligation frequency in the brain but not in the liver. To extensively study the chromatin conformation in the α-globin locus, the ligation frequencies within this region were examined and quantitated with a fixation of fragments XV, XI, and VIII (Fig. 3B, C, and D, respectively). In the nonexpressing-tissue brain cells, when the fragment XV, closest to the α1-globin gene, is fixed, the ligation frequency gradually decreased with fragments (from XIV to VII) located farther away toward its 5′ end on the linear genomic DNA (Fig. 3B). Similar results were obtained for the middle fragment XI (Fig. 3C). When the 5′ side fragment VIII was fixed, it also exhibited a roughly similar pattern (Fig. 3D). Although the more distant fragments XIV and XVIII show small, elevated ligation frequencies, they are much lower than that of the closest fragment, IX (Fig. 3D). Such a correlation between distance in space and distance in the genomic location indicates that there is no particular spatial organization across the mouse α-globin locus in nonexpressing brain cells.
Analysis of the mouse α-globin locus in the expressing tissue, 14.5-dpc fetal liver cells, shows a significantly distinct spatial organization (Fig. 3B to D). When fragment XV, which is very close to the active α1 gene, was used as the fixed fragment, the distant fragment VIII containing HS26, fragment IX containing HS21, and fragment XI containing half of HS8 showed highly elevated ligation frequencies in fetal liver compared to brain, and their ligation frequencies were higher than their interval fragment X (Fig. 3B). When fragment XI containing half of HS8 was used as the fixed fragment, a much higher ligation frequency with HS26 in liver than in brain was observed (Fig. 3C). In addition, the ligation frequency between XI and XV (close to the α1 gene) forms a peak in liver but not in brain, confirming the results obtained when the fragment XV is fixed (Fig. 3B). When the fragment containing HS26 is fixed, it shows much higher ligation frequencies with its downstream fragments (X to XIX) in erythroid cells than those in nonerythroid cells (Fig. 3D). In particular, one peak of high ligation frequency with fragment XI (containing half of HS8) and one broad plateau of high ligation frequency with the region harboring active α1 and α2 genes (fragments XIV to XVII) stand out in liver cells compared to those in brain cells (Fig. 3D). The above data gave reciprocal confirmation that as with the β-globin gene cluster (32, 40), in expressing cells, the distant regulatory elements of the mouse α-globin locus, even though they are located in or close to the transcription unit of other genes, may come in physical proximity to the active α-globin genes through looping out the interval DNA regions.
Coexpressing housekeeping genes colocalize with active α-globin genes through looping.
Distinct from the mouse β-globin gene cluster, which possesses an independent chromatin domain sided by two condensed chromatin domains in erythroid cells (26), the α-globin locus is neighbored by a group of housekeeping genes (19). Since both α-globin genes and the nearby housekeeping genes are expressed in definitive erythroid cells, we investigated their spatial relationships. In the distant 5′ 80-kb region of the α1 gene, there are four genes: C16orf33, C16orf8, MPG, and C16orf35, which have been identified as housekeeping genes (16). When the ligation frequency of fragment XV near the α1 gene versus other sites in the 130-kb region was determined, surprisingly, this fragment showed two- to threefold-higher ligation frequencies with the upstream regions containing four clustered housekeeping genes in definitive erythroid cells (14.5-dpc fetal liver cells) than those in nonerythroid cells (14.5-dpc fetal brain cells) (Fig. 4A). In fetal brain cells, fragment XV exhibited decreased ligation frequencies with more distant loci (Fig. 4A). To examine if the α-globin genes colocalize with the upstream housekeeping genes in expressing cells but not in nonexpressing cells, the relative ligation frequencies of fragments II, III, and V containing the promoters of the C16orf33, C16orf8, and MPG genes (Fig. 4B, C, and D, respectively) were measured versus other fragments across the 130-kb region. They all showed two significant peak regions of high ligation frequencies in definitive erythroid cells: one with fragment XI containing the C16orf35 promoter and the other with fragments XV and XVII, which are close to the active α1 and α2 genes, respectively. The ligation frequencies of these peak regions with the fixed fragments were higher than those of the interspace regions between them (Fig. 4B to D). As illustrated in Fig. 4B to D, the fixed fragments II, III, and V, containing the promoters of the C16orf33, C16orf8, and MPG genes, respectively, showed lower ligation frequencies with fragment XIV, which is close to the ζ gene, than with its nearby fragments XI, XV, and XVII, which are close to the C16orf35, α1, and α2 genes, respectively, in expressing liver cells. These results suggest that in erythroid cells the active α1 and α2 globin genes, but not the development-specifically repressed ζ gene, colocalize with the upstream promoters of housekeeping genes C16orf33, C16orf8, and MPG through looping (Fig. 4B, C, and D, respectively). However, in nonerythroid cells, only one peak region of high ligation frequency was present, which is the fragment XI containing the C16orf35 gene promoter (Fig. 4B to D). The other peak seen in liver cells with fragments XV and XVII, close to the active α1 and α2 genes, does not exist in brain (Fig. 4B to D). These data indicate that the repressed nonexpressing α-globin genes are not in close proximity with these housekeeping genes in brain. When the fragment XVII close to α2 was fixed, ligation frequencies with VIII (HS26) and XI (HS8) were greater in the liver than in the brain (Fig. 4E). Similar to the results on fragment XV close to the α1 gene (Fig. 3A), the ligation frequencies of fragment XVII close to the α2 gene with fragments V (MPG), III (C16orf8), and II (C16ord33) are also greater in the liver than in the brain (Fig. 4E). In addition, ligation frequency between HS26 and α2 is much higher than that between HS8 and α2 in liver cells. Taken together, in fetal liver, both the active α1- and α2-globin genes, but not the inactive ζ-globin gene, colocalize with the neighboring housekeeping genes C16orf33, C16orf8, MPG, and C16orf35 by forming many transcriptional loops.
FIG. 4.
Spatial organization of the 130-kb chromatin region containing the α-globin locus and the flanking housekeeping genes. The cross-linking patterns of the fragments XV (A), II (B), III (C), V (D), and XVII (E) next to promoters of α1, C16orf33, C16orf8, MPG, and α2, respectively, are shown. All symbols and color patterns are the same as in Fig. 3.
The occupancy of RNA Pol II at active α-globin genes is highly enriched.
The special chromatin conformation of these clustered genes may be primarily related to their transcription status. It has been found that RNA polymerase II (Pol II) associates with regulatory elements in both LCR and promoters to modulate the transcription of β-globin genes (23). In order to examine the correlation of RNA Pol II loading with transcription regulation, the occupancies of RNA Pol II at regulatory elements and gene promoters were measured in the whole 3C-analyzed 130-kb chromatin region by ChIP. An example of PCR-amplified ChIP sample and input control sample and the equation to calculate the relative intensity of RNA Pol II are shown in Fig. 5A. The universally expressed GAPDH gene promoter was used as a positive control to correct the difference in DNA templates of two tissues. The MyoD promoter was used as a negative control to show the background occupancy of RNA Pol II on DNA (Fig. 5A), since it is repressed in both liver and brain cells (23). The relative intensity of each site is shown on Fig. 5B using a scale relative to MyoD. In definitive erythroid cells, the highest occupancies of RNA Pol II were noted at the mouse α1 and α2 promoters, about 13- to 15-fold relative to the MyoD promoter (Fig. 5B). In contrast to the high level of expression of α1 and α2, the expression from the embryonic ζ promoter appeared low due to its low occupancy by RNA Pol II (Fig. 5B) (24). There was more than an 8- to 10-fold and approximately a 4- to 5-fold enrichment at the C16orf35 and MPG promoters, respectively, compared to MyoD. Moreover, there was relatively little enrichment at the C16orf33 and C16orf8 gene promoters, yet the levels were still higher than that at the MyoD promoter (Fig. 5B). However, in nonerythroid cells, the intensity of RNA Pol II at the mouse α1 and α2 gene promoters remarkably decreased, whereas there was a similar occupancy pattern at the housekeeping gene promoters to that in erythroid cells (Fig. 5B). The different enrichments of RNA Pol II at these housekeeping gene promoters may be correlated to their different expressions in cells.
DISCUSSION
One of the existing questions in transcriptional regulation is how distant cis-regulatory elements modulate their target genes. There is increasing evidence that looping is an important mechanism by which distant cis-acting genomic elements and their associated genes are brought together in long-range communications. Some examples, such as the boundary element scs in Drosophila, the β-globin gene cluster, the Ifng gene region, and the Vκ gene and its enhancer and boundary element (4, 14, 27, 40), show similar looping mechanisms. The study of the spatial organization of a 200-kb region spanning the mouse β-globin locus showed that distant HSs are in close proximity with β-globin genes through looping (40). It was proposed that the three-dimensional clustering of HSs and active globin genes forms an active chromatin hub (ACH) to regulate gene expression in the β-globin locus (40). In addition, this erythroid cell-specific β-ACH may also be developmentally regulated through association with different active genes during different development stages (10, 32). However, distinct from the β-globin cluster, which is encompassed by a cluster of suppressed olfactory receptor genes (26), the mouse α-globin locus is neighbored by a cluster of housekeeping genes and its distant enhancer is located in the transcription unit of a housekeeping gene (19). So it was hypothesized that they may form a conformation different from that of the β-globin gene cluster to avoid the interference of the flanking genes. Unexpectedly, we found that, similar to the β-globin gene cluster, the upstream regulatory elements of the mouse α-globin gene locus are also congregated together with the downstream active α1 and α2 globin genes in expressing cells (Fig. 3 and 4). We therefore speculate that the mouse α-globin gene locus may also form an erythroid cell-specific ACH-like structure (α-ACH) in the nucleus which is similar to β-ACH. However, this conformation cannot be observed in mouse nonerythroid cells where the globin gene expression is suppressed (Fig. 3 and 4). Hence, it will be of interest to investigate whether the conformation of the mouse α-globin gene locus is similar to that of the β-globin locus in the mode of development-coupled transcription regulation. These results may also suggest possible roles of the looping structures in interactions among spatially positioned cis-acting elements in the coordinated transcription regulation.
The important roles of DNA-protein and protein-protein interactions in the formation of looping structures have previously been investigated in the pairing of two sides of the boundary elements, scs and scs′, in Drosophila and the building of β-ACH for the β-globin gene cluster (4, 13). The above-described spatially assembled complexes also most likely take place through dynamic looping in conjunction with the recruitment of trans-acting factors. It has also been established that the hematopoietic transcription factors GATA-1 and NF-E2 largely contribute to α-globin gene activation (1, 28), but how these factors act in the α-globin chromatin environment is unclear. The measurement of the enrichment of erythroid cell-specific activators, GATA-1 and NF-E2, at the mouse α-globin locus revealed strong interactions of GATA-1 with HS26, HS8, active α1 and α2 gene promoters, and NF-E2 with HS26 in induced murine erythroleukemia cells (1), suggesting that these activators may have potential roles in mediating the long-range activation of mouse α-globin genes through looping. In β-globin gene transcription activation, it has been proposed that the interaction of NF-E2, which specifically binds to HS2 of LCR, with the promoter regions in which its binding sequences are not present may result from the interactions with other proteins, such as TAFII130 (36). Moreover, it has been documented that GATA-1-NF-E2 cooperativity is required for RNA Pol II loading at the mouse adult β-major promoter, probably through a looping structure (23). Here, we also observed the erythroid cell-specific high occupancy of RNA Pol II at HS26 and active α1 and α2 genes. While studying the occupancy pattern of the NF-E2p45 subunit across the mouse α-globin locus, we found that NF-E2p45 has the strongest interaction with HS26 and relatively lower occupancy at α1 and α2 in liver cells, but there is no NF-E2p45 occupancy at these sites in brain cells (G. L. Zhou and D. P. Liu, unpublished data). These data imply that NF-E2 and RNA Pol II may be involved in the establishment of mouse α-ACH. Recently, it was also found that GATA-1 and FOG-1 are essential anchors for a tissue-specific chromatin loop at the mouse β-globin locus (41). In mouse definitive erythroid cells, the fetal and adult α1 and α2 gene promoters are highly enriched with histone acetylation and H3-K4 dimethylation. However, the embryonic ζ gene promoter is development-specifically deacetylated at histones H3 and H4 and lacks active H3-K4 dimethylation (17). Therefore, it is of further interest to determine how RNA Pol II, GATA-1, and NF-E2 cooperate with other proteins and how these interactions are coordinated to mediate α-ACH formation and influence histone modification during erythroid development.
The expression of mouse α-globin genes may harmonize with the expression of neighboring housekeeping genes that overlap with their distant regulatory elements. There is a cluster of housekeeping genes upstream of the mouse α-globin gene locus where every two adjacent genes have opposite transcriptional direction alternatively (Fig. 1). The genome-wide bioinformatic analysis indicated that the highly expressed genes, especially those housekeeping genes, which are often clustered in specific chromosomal regions, may be accommodated to their transcription regulation (9, 25). To explore how the mouse α-globin locus is coordinated with the flanking, ubiquitously expressed housekeeping genes, we analyzed the extended 130-kb region for chromatin conformation and RNA Pol II occupancy at HSs and promoters in erythroid and nonerythroid cells. The results show that in erythroid cells the coexpressed globin genes and housekeeping gene promoters are congregated together through the formation of many transcriptional loops (Fig. 4 and 6). In nonerythroid cells, the coexpressed housekeeping gene promoters still colocalize, but the tissue-specific, silenced globin genes are excluded (Fig. 4 and 6). Hence, it is possible that both the clustered gene order on the chromosome and the spatial congregation in the nucleus might favor cotranscription. In nonerythroid cells, their spatial assembly excludes the tissue-specific α-globin gene locus probably silenced through an independently suppressed chromatin configuration. The results may also suggest that the assembled flanking housekeeping genes may maintain a preexisting structural subcompartment in the nucleus to favor their ubiquitous expression. The recruitment of active α-globin gene promoters and the regulatory elements in this preexisting structural subcompartment may result in globin gene expression in erythroid cells, and the exclusion from this subcompartment in nonerythroid cells may inactivate globin gene expression. It is interesting that active α1 and α2 globin gene promoters have a much higher RNA Pol II enrichment in fetal liver than in brain. The RNA Pol II occupancy at the ζ gene promoter, which was specifically repressed during development, is much lower than that at the α1 and α2 promoters (Fig. 5). However, the RNA Pol II occupancies at the housekeeping genes are similar in fetal liver and brain. The different intensities of RNA Pol II at these gene promoters may be correlated to their different levels of transcription. It has been observed that RNA Pol II could be concentrated in the distinct “transcription factories” specializing in the transcription of many different templates (21, 22, 35). Thus, it is likely that the spatial colocalization of the transcribing genes within this region can lead to the formation of a putative transcription “factory” and concentrate RNA polymerase II. The recently observed phenomena that the distantly separated (even on different chromosomes) active genes dynamically colocalize to shared sites of ongoing transcription in nuclei lend further support for this possibility (31, 37). It was suggested that such an organization may favor transcription via efficient utilization of RNA Pol II and trans-acting factors and may also allow the coupling of different nuclear processes (e.g., pre-mRNA splicing, DNA repair, and DNA replication) in three-dimensional space.
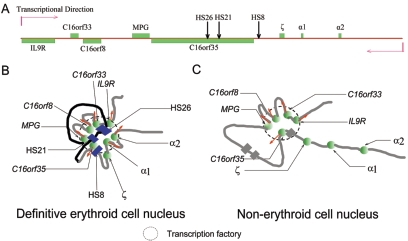
FIG. 6.
Schematic representation of the linear structure in genome (A) and the putative three-dimensional structure of a large chromatin region in definitive erythroid cells (B) and nonerythroid cells (C). In definitive erythroid cells, active α-globin genes are in close proximity with the upstream HSs to form an α-ACH, similar to β-ACH. Moreover, mouse α-ACH colocalizes with the upstream housekeeping genes, which may form a subcompartment in the nucleus like a “transcription factory” where the concentrated RNA polymerase may be shared by a particular group of genes (B). In nonerythroid cells (C), the repressed mouse α-globin genes are separated from the congregated housekeeping genes, which retain a “transcription factory” shared by these housekeeping genes. The symbols in panel A are as described in the legend to Fig. 1. In panels B and C, the green spheres represent protein complexes on gene promoters. The gray and black strings represent chromatin. The red arrows indicate transcription direction. In panel B, the blue boxes represent protein complexes on distant erythroid-specific HSs. In panel C, the gray boxes show the positions of erythroid cell-specific HSs.
In summary, active α-globin genes and their distant cis-regulatory elements are recruited to a preexisting structural subcompartment in erythroid cells, which forms a transcription factory shared by the flanking colocalized housekeeping genes; but in nonerythroid cells the silenced α-globin genes are excluded from this factory. Moreover, the expressing α-globin genes are highly occupied by RNA Pol II. Our results provide evidence for a possible correlation between spatial organization of genomic elements and transcription regulation. Further investigations are required to characterize how a putative transcription “factory” among these genes is established and how spatially colocalized cis elements regulate transcription in this transcription factory in the nucleus.
Acknowledgments
We thank K. E. Cullen, J. Dekker, B. Tolhuis, and R. J. Palstra for the advice and protocol of 3C technology. We also thank Jennifer P. Pons for reading and proofing the manuscript.
This work was supported by grants from the National Natural Science Foundation of China (grant numbers 30393110 and 30421003).
REFERENCES
- 1.Anguita, E., J. Hughes, C. Heyworth, G. A. Blobel, W. G. Wood, and D. R. Higgs. 2004. Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J. 23:2841-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anguita, E., C. A. Johnson, W. G. Wood, B. M. Turner, and D. R. Higgs. 2001. Identification of a conserved erythroid specific domain of histone acetylation across the alpha-globin gene cluster. Proc. Natl. Acad. Sci. USA 98:12114-12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anguita, E., J. A. Sharpe, J. A. Sloane-Stanley, C. Tufarelli, D. R. Higgs, and W. G. Wood. 2002. Deletion of the mouse alpha-globin regulatory element (HS -26) has an unexpectedly mild phenotype. Blood 100:3450-3456. [DOI] [PubMed] [Google Scholar]
- 4.Blanton, J., M. Gaszner, and P. Schedl. 2003. Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev. 17:664-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouhassira, E. E., M. F. Kielman, J. Gilman, M. F. Fabry, S. Suzuka, O. Leone, E. Gikas, L. F. Bernini, and R. L. Nagel. 1997. Properties of the mouse alpha-globin HS-26: relationship to HS-40, the major enhancer of human alpha-globin gene expression. Am. J. Hematol. 54:30-39. [DOI] [PubMed] [Google Scholar]
- 6.Brown, K. E., S. Amoils, J. M. Horn, V. J. Buckle, D. R. Higgs, M. Merkenschlager, and A. G. Fisher. 2001. Expression of alpha- and beta-globin genes occurs within different nuclear domains in haemopoietic cells. Nat. Cell Biol. 3:602-606. [DOI] [PubMed] [Google Scholar]
- 7.Bulger, M., and M. Groudine. 1999. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 13:2465-2477. [DOI] [PubMed] [Google Scholar]
- 8.Bulger, M., and M. Groudine. 2002. TRAPping enhancer function. Nat. Genet. 32:555-556. [DOI] [PubMed] [Google Scholar]
- 9.Caron, H., B. van Schaik, M. van der Mee, F. Baas, G. Riggins, P. van Sluis, M. C. Hermus, R. van Asperen, K. Boon, P. A. Voute, S. Heisterkamp, A. van Kampen, and R. Versteeg. 2001. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science 291:1289-1292. [DOI] [PubMed] [Google Scholar]
- 10.Carter, D., L. Chakalova, C. S. Osborne, Y. F. Dai, and P. Fraser. 2002. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 32:623-626. [DOI] [PubMed] [Google Scholar]
- 11.Cullen, K. E., M. P. Kladde, and M. A. Seyfred. 1993. Interaction between transcription regulatory regions of prolactin chromatin. Science 261:203-206. [DOI] [PubMed] [Google Scholar]
- 12.Dekker, J., K. Rippe, M. Dekker, and N. Kleckner. 2002. Capturing chromosome conformation. Science 295:1306-1311. [DOI] [PubMed] [Google Scholar]
- 13.Drissen, R., R. J. Palstra, N. Gillemans, E. Splinter, F. Grosveld, S. Philipsen, and W. de Laat. 2004. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 18:2485-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eivazova, E. R., and T. M. Aune. 2004. Dynamic alterations in the conformation of the Ifng gene region during T helper cell differentiation. Proc. Natl. Acad. Sci. USA 101:251-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farrell, C. M., A. G. West, and G. Felsenfeld. 2002. Conserved CTCF insulator elements flank the mouse and human beta-globin loci. Mol. Cell. Biol. 22:3820-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flint, J., C. Tufarelli, J. Peden, K. Clark, R. J. Daniels, R. Hardison, W. Miller, S. Philipsen, K. C. Tan-Un, T. McMorrow, J. Frampton, B. P. Alter, A. M. Frischauf, and D. R. Higgs. 2001. Comparative genome analysis delimits a chromosomal domain and identifies key regulatory elements in the alpha globin cluster. Hum. Mol. Genet. 10:371-382. [DOI] [PubMed] [Google Scholar]
- 17.Fu, X. H., D. P. Liu, X. B. Tang, G. Liu, X. Lv, Y. J. Li, and C. C. Liang. 2005. A conserved, extended chromatin opening within alpha-globin locus during development. Exp. Cell Res. 309:174-184. [DOI] [PubMed] [Google Scholar]
- 18.Hardison, R. 1998. Hemoglobins from bacteria to man: evolution of different patterns of gene expression. J. Exp. Biol. 201:1099-1117. [DOI] [PubMed] [Google Scholar]
- 19.Higgs, D. R., J. A. Sharpe, and W. G. Wood. 1998. Understanding alpha globin gene expression: a step towards effective gene therapy. Semin. Hematol. 35:93-104. [PubMed] [Google Scholar]
- 20.Hosbach, H. A., T. Wyler, and R. Weber. 1983. The Xenopus laevis globin gene family: chromosomal arrangement and gene structure. Cell 32:45-53. [DOI] [PubMed] [Google Scholar]
- 21.Jackson, D. A., A. B. Hassan, R. J. Errington, and P. R. Cook. 1993. Visualization of focal sites of transcription within human nuclei. EMBO J. 12:1059-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson, D. A., F. J. Iborra, E. M. Manders, and P. R. Cook. 1998. Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol. Biol. Cell 9:1523-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, K. D., J. A. Grass, M. E. Boyer, C. M. Kiekhaefer, G. A. Blobel, M. J. Weiss, and E. H. Bresnick. 2002. Cooperative activities of hematopoietic regulators recruit RNA polymerase II to a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. USA 99:11760-11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leder, A., D. Swan, F. Ruddle, P. D'Eustachio, and P. Leder. 1981. Dispersion of alpha-like globin genes of the mouse to three different chromosomes. Nature 293:196-200. [DOI] [PubMed] [Google Scholar]
- 25.Lercher, M. J., A. O. Urrutia, and L. D. Hurst. 2002. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat Genet. 31:180-183. [DOI] [PubMed] [Google Scholar]
- 26.Li, Q., K. R. Peterson, X. Fang, and G. Stamatoyannopoulos. 2002. Locus control regions. Blood 100:3077-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, Z., and W. T. Garrard. 2005. Long-range interactions between three transcriptional enhancers, active Vκ gene promoters, and a 3′ boundary sequence spanning 46 kilobases. Mol. Cell. Biol. 25:3220-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loyd, M. R., Y. Okamoto, M. S. Randall, and P. A. Ney. 2003. Role of AP1/NFE2 binding sites in endogenous alpha-globin gene transcription. Blood 102:4223-4228. [DOI] [PubMed] [Google Scholar]
- 29.Mutskov, V. J., C. M. Farrell, P. A. Wade, A. P. Wolffe, and G. Felsenfeld. 2002. The barrier function of an insulator couples high histone acetylation levels with specific protection of promoter DNA from methylation. Genes Dev. 16:1540-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nobrega, M. A., I. Ovcharenko, V. Afzal, and E. M. Rubin. 2003. Scanning human gene deserts for long-range enhancers. Science 302:413. [DOI] [PubMed] [Google Scholar]
- 31.Osborne, C. S., L. Chakalova, K. E. Brown, D. Carter, A. Horton, E. Debrand, B. Goyenechea, J. A. Mitchell, S. Lopes, W. Reik, and P. Fraser. 2004. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 36:1065-1071. [DOI] [PubMed] [Google Scholar]
- 32.Palstra, R. J., B. Tolhuis, E. Splinter, R. Nijmeijer, F. Grosveld, and W. de Laat. 2003. The beta-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 35:190-194. [DOI] [PubMed] [Google Scholar]
- 33.Patrinos, G. P., M. de Krom, E. de Boer, A. Langeveld, A. M. Imam, J. Strouboulis, W. de Laat, and F. G. Grosveld. 2004. Multiple interactions between regulatory regions are required to stabilize an active chromatin hub. Genes Dev. 18:1495-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pombo, A., D. A. Jackson, M. Hollinshead, Z. Wang, R. G. Roeder, and P. R. Cook. 1999. Regional specialization in human nuclei: visualization of discrete sites of transcription by RNA polymerase III. EMBO J. 18:2241-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pombo, A., E. Jones, F. J. Iborra, H. Kimura, K. Sugaya, P. R. Cook, and D. A. Jackson. 2000. Specialized transcription factories within mammalian nuclei. Crit. Rev. Eukaryot. Gene Expr. 10:21-29. [PubMed] [Google Scholar]
- 36.Sawado, T., K. Igarashi, and M. Groudine. 2001. Activation of beta-major globin gene transcription is associated with recruitment of NF-E2 to the beta-globin LCR and gene promoter. Proc. Natl. Acad. Sci. USA 98:10226-10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spilianakis, C. G., M. D. Lalioti, T. Town, G. R. Lee, and R. A. Flavell. 2005. Interchromosomal associations between alternatively expressed loci. Nature 435:637-645. [DOI] [PubMed] [Google Scholar]
- 38.Spitz, F., F. Gonzalez, and D. Duboule. 2003. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell 113:405-417. [DOI] [PubMed] [Google Scholar]
- 39.Szentirmay, M. N., and M. Sawadogo. 2000. Spatial organization of RNA polymerase II transcription in the nucleus. Nucleic Acids Res. 28:2019-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tolhuis, B., R. J. Palstra, E. Splinter, F. Grosveld, and W. de Laat. 2002. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10:1453-1465. [DOI] [PubMed] [Google Scholar]
- 41.Vakoc, C. R., D. L. Letting, N. Gheldof, T. Sawado, M. A. Bender, M. Groudine, M. J. Weiss, J. Dekker, and G. A. Blobel. 2005. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol. Cell 17:453-462. [DOI] [PubMed] [Google Scholar]
- 42.Versteeg, R., B. D. van Schaik, M. F. van Batenburg, M. Roos, R. Monajemi, H. Caron, H. J. Bussemaker, and A. H. van Kampen. 2003. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Res. 13:1998-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Will, C. L., C. Schneider, M. Hossbach, H. Urlaub, R. Rauhut, S. Elbashir, T. Tuschl, and R. Luhrmann. 2004. The human 18S U11/U12 snRNP contains a set of novel proteins not found in the U2-dependent spliceosome. RNA 10:929-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamashita, T., M. Honda, H. Takatori, R. Nishino, N. Hoshino, and S. Kaneko. 2004. Genome-wide transcriptome mapping analysis identifies organ-specific gene expression patterns along human chromosomes. Genomics 84:867-875. [DOI] [PubMed] [Google Scholar]