Abstract
In cancer cells and germ cells, shortening of chromosome ends is prevented by telomerase. Telomerase-deficient cells have a replicative life span, after which they enter senescence. Senescent cells can give rise to survivors that maintain chromosome ends through recombination-based amplification of telomeric or subtelomeric repeats. We found that in Trypanosoma brucei, critically short telomeres are stable in the absence of telomerase. Telomere stabilization ensured genomic integrity and could have implications for telomere maintenance in human telomerase-deficient cells. Cloning and sequencing revealed 7 to 27 TTAGGG repeats on stabilized telomeres and no changes in the subtelomeric region. Clones with short telomeres were used to study telomere elongation dynamics, which differed dramatically at transcriptionally active and silent telomeres, after restoration of telomerase. We propose that transcription makes the termini of short telomeres accessible for rapid elongation by telomerase and that telomere elongation in T. brucei is not regulated by a protein-counting mechanism. Many minichromosomes were lost after long-term culture in the absence of telomerase, which may reflect their different mitotic segregation properties.
Linear chromosomes are capped by telomeres, which are nucleoprotein complexes consisting of tandem DNA repeats and proteins that bind to them. Telomeres serve two major purposes: they protect chromosome ends from the DNA repair machinery that would otherwise recognize them as double-stranded breaks, and, with the help of telomerase, they compensate for the gradual sequence loss that would otherwise arise from the inability of conventional DNA polymerases to replicate chromosome ends (21, 31, 51). Telomerase is present in germ cells, and its activation is a crucial step in tumorigenesis (13, 30). In human and yeast cells, telomerase activity is tightly regulated by proteins that relay information about telomere length (32, 52). The ability of telomerase to extend a telomere is inversely proportional to the initial length of the telomere DNA repeats (36, 37, 56). This protein-counting model explains how short telomeres can be rapidly elongated whereas the length of long telomeres is restricted (37). In human somatic cells, which lack telomerase, telomeres shorten through each replicative cycle (25). This molecular clock limits the proliferation of somatic cells to ∼50 to 60 population doublings (PD), after which they enter a nonreversible growth arrest called senescence. Telomerase-deficient cells occasionally escape senescence or apoptosis. Yeast and human survivor cells acquire the ability to maintain chromosome ends through an alternative lengthening mechanism (ALT) (35, 48). In the absence of telomerase, Saccharomyces cerevisiae can survive by amplification of subtelomeric repeats, recombination among telomeres, or palindrome-based amplification of subtelomeric regions (35, 38). In addition to telomere-telomere recombination-based lengthening, Schizosaccharomyces pombe can overcome the loss of telomeric DNA by chromosome circularization (43).
From 5 to 20% of human cancers lack telomerase activity and use ALT to dramatically increase their telomere length (5). Although telomere-telomere recombination is the major mechanism underlying ALT, fluorescent in situ hybridization of metaphase spreads of human ALT cells reveals that telomeric repeats are undetectable at many chromosome ends (5, 17). It remains unclear whether signal-free ends represent short telomeres, how much telomeric DNA they carry, and whether these ends might be temporarily stabilized until they are able to undergo recombination.
Here we describe the consequences of telomere shortening in Trypanosoma brucei, the causative agent of African sleeping sickness. This parasite persists in the bloodstream of its mammalian host by stochastically changing its surface coat, which consists of a homogeneous layer of variant surface glycoproteins (VSG) (2, 4). VSG genes are dispersed throughout the genome, but the actively transcribed VSG gene is always located in one of ∼20 subtelomeric transcription units called expression sites (ES) (15, 45, 55). Antigenic switching can be achieved by duplicative gene conversion events, in which a transcriptionally silent telomeric or a nontelomeric VSG gene is the donor (28, 49), or by coupled ES silencing and activation, called an in situ switch (28). Despite intensive investigations, the mechanisms that regulate antigenic variation remain elusive, but telomeres are likely to play an important role.
T. brucei has 11 diploid megabase chromosomes (MBC), which contain all the essential genes and most ES; a variable number of intermediate chromosomes (IC), which appear to contain ES and little else; and ∼100 minichromosomes (MC), which consist mainly of 177-bp tandem repeats, silent VSG genes, and telomeres (40, 58). MC are thought to provide a reservoir of VSG genes that can be activated by duplicative transposition. Despite their similarity and number, MC are mitotically stable and faithfully segregate during several years of continuous propagation (1, 60). We previously identified and deleted the gene encoding telomerase reverse transcriptase (TERT) in order to investigate the role that telomeres might play during antigenic variation (16). Here we report the consequences of telomere shortening for different chromosome types. Most interestingly, our results show that telomerase-deficient T. brucei can maintain the length of critically short telomeres by telomere stabilization. MC are lost as telomeres become critically short. By restoration of the TERT gene, we were able to explore the dynamics of telomere elongation at transcriptionally active and silent ES. Our results suggest that T. brucei has evolved a novel way of regulating telomerase-dependent telomere elongation. We propose a model in which transcription modulates telomeric chromatin structure and renders it accessible to the telomerase complex.
MATERIALS AND METHODS
Trypanosome cell lines and complementation constructs.
T. brucei Lister 427, expressing MITat 1.2 (VSG 221), was previously manipulated to constitutively express T7 polymerase and the Tet repressor protein and cultured in HMI-9 medium at 37°C (27, 59). In this cell line, designated “single marker,” both alleles of the TERT open reading frame (ORF) were replaced by genes conferring resistance to hygromycin and puromycin, respectively (16). To complement the TERT deletion phenotype, the T. brucei TERT open reading frame was amplified using primers that introduce NdeI restriction sequences at the 5′ end and HpaI site at the 3′ end of the gene. The PCR product was cloned into pGEM-T Easy (Promega), and clones from independent PCRs were sequenced to exclude the possibility of PCR-derived mutations. To put the ORF under the control of an inducible T7 promoter, the PCR product was released by NdeI/HpaI digestion and cloned into similarly digested pCO57 (this step removes a green fluorescent protein [GFP] cassette from pCO57, a derivative of pLew82 [59]). Proper integration was verified by restriction analysis and sequencing. To introduce a GFP tag at the telomerase C terminus, NdeI sites were introduced by PCR at both ends of the TERT ORF, cloned into pGEM-T Easy, released, and ligated into NdeI-cut and dephosphorylated pCO57. The constructs were linearized by digestion with NotI and transfected into telomerase-deficient cell lines, and integrants were selected with 2.5 μg/ml phleomycin. Upon reintroduction, TERT-expressing clones were verified by Southern blotting (data not shown). Expression of the fusion product was induced by addition of 100 ng/ml doxycycline. DNA was extracted on a weekly basis, and telomere length changes at transcriptionally silent and active ES were monitored by Southern hybridization. To assess whether TERT-GFP is fully functional, its ability to elongate a short telomere was compared with those of a reintroduced wild-type TERT allele and a hemagglutinin-tagged version. Wild-type and TERT-GFP constructs were fully functional, whereas hemagglutinin-TERT did not elongate telomeres (data not shown).
Bal 31 digestion.
Bal 31 digestion was performed as described previously (41). Briefly, 100 μg of DNA in 500 μl volume were incubated at 30°C with 5 units of Bal 31. One-hundred-microliter samples were removed periodically, added to 2 μl 0.5 M EDTA, incubated at 65°C for 10 min, and stored at −20°C until DNA was extracted by phenol-chloroform-isoamyl-alcohol, ethanol precipitated, and dissolved in 10 mM Tris (pH 7.4), 0.1 mM EDTA. DNA was digested with EcoRI and loaded on an 0.8% agarose gel. Southern blotting and hybridization procedures were performed as described previously (16).
Telomere tailing procedure and PCR amplification.
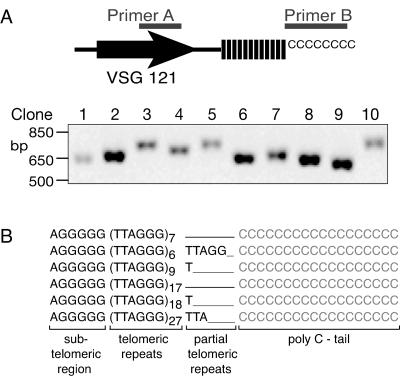
Telomere tailing and PCR amplification were carried out as described elsewhere (19), with minor modifications. To increase the efficiency of telomere tailing, terminal restriction fragments (TRF) corresponding to the size of stabilized VSG 121 telomere (∼1.6 kb) were isolated from an 0.8% agarose gel. Approximately 100 ng of TRF in One-Phor-All buffer (Pharmacia), 1mΜ dCTP, and 5 U of terminal deoxynucleotidyl transferase (Amersham Biosciences) was incubated for 30 min at 37°C. The efficiency of the tailing reaction was verified by adding radiolabeled dCTP to the reaction mix and running the products on agarose gels (data not shown). The tailing reactions were PCR amplified using 0.75 μM primer A (5′-ACACAGGCGAACCAGAATGACGCTGCAGCCAAAGCA-3′), 1.0 μM primer B (CCCCC)3, 320 μM deoxynucleoside triphosphates, 10× Taq polymerase buffer, and 2.5 U of Taq polymerase by incubating for 2 min at 94°C, followed by 45 cycles of 45 seconds at 94°C, 45 seconds at 65°C, and 70 seconds at 72°C. PCR-amplified products were separated on a 1.2% agarose gel, and nontailed TRF were amplified in parallel as a negative control. Specific products were gel purified and subcloned into pGEM-T (Promega). Transformed bacteria (Escherichia coli DH 10B) were grown at 30°C to minimize recombination events. Restriction digests were performed as described by the manufacturer (New England Biolabs). Sequencing reactions were carried out by GeneWiz Inc. (North Brunswick, NJ).
RAGE.
DNA agarose plugs were prepared as described by Navarro and Cross (45). Briefly, 2 × 108 cells were purified from the blood of infected mice, resuspended in 0.5 ml L-buffer (LB) (0.1 M EDTA [pH 8.0], 10 mM Tris-HCl [pH 7.6], 20 mM NaCl), and incubated for 10 min at 42°C. A 0.5-ml quantity of 1.6% low-temperature-gelling agarose (Sigma) in LB was added to 0.5 ml cells, mixed, and poured into plug molds (Bio-Rad Laboratories). Plugs were treated in 3 ml LB with 1 mg/ml proteinase K at 50°C for 2 days. After two washes for 15 min with LB, proteinase K treatment and washing were repeated. Plugs were embedded in 0.8% agarose in 0.5× Tris-borate-EDTA gel. Chromosome-sized DNA was separated by rotating agarose gel electrophoresis (RAGE) (Stratagene, Inc.) under conditions described previously (44): a linear ramp of 100 to 300 s for 10 h at 120 V and then 1,000 to 2,500 s for 80 h at 50 V. Gels were dried at room temperature and hybridized as described above. Separation of MC and IC took place at a rotation angle of 120° in 0.5× Tris-borate-EDTA at 12°C. The program consisted of one ramped pulse time of 60 to 20 s at 110 V for 60 h (Luisa Figueiredo, unpublished data). For in-gel hybridization with radiolabeled telomeric oligonucleotides, gels were dried at room temperature, denatured, and neutralized. In-gel hybridization was preformed as described previously (16). Alternatively, DNA was blotted onto an N+ Hybond membrane, UV cross-linked, and hybridized. Signals were quantified using a phosphorimager screen and ImageQuant software (Molecular Dynamics).
To quantify the extent of MC loss, gels were run in duplicate: gel 1 was in-gel hybridized using a radiolabeled (TTAGGG)4 probe, whereas gel 2 was blotted onto Hybond N+ membrane and sequentially hybridized with probes for the 50-bp repeat, 177-bp repeat, and individual VSG genes; 177- and 50-bp probes were obtained by PCR amplification from genomic DNA as previously described (18). Hybridized membranes were exposed to phosphorimager screens, and signal intensities of individual bands were quantified using ImageQuant software. To analyze the reduction in MC 177-bp signal, 50-bp repeat and VSG probes were used to normalize loading.
Slot blot hybridization.
A 0.4- to 0.8-μg quantity of DNA from wild-type and long-term-cultured TERT-deficient strains was added to 6× SSC (900 mM NaCl, 90 mM sodium citrate, pH 7.0), boiled for 10 min, and placed on ice. Duplicate samples (200 and 400 μl) of denatured DNA were applied to a vacuum manifold device containing a 6× SSC-equilibrated Hybond N+ membrane, which was subsequently UV cross-linked and hybridized with 177-bp and 50-bp repeat probes. Signal intensities were quantified using phosphorimager screens.
RESULTS
Long-term consequences of telomerase deletion.
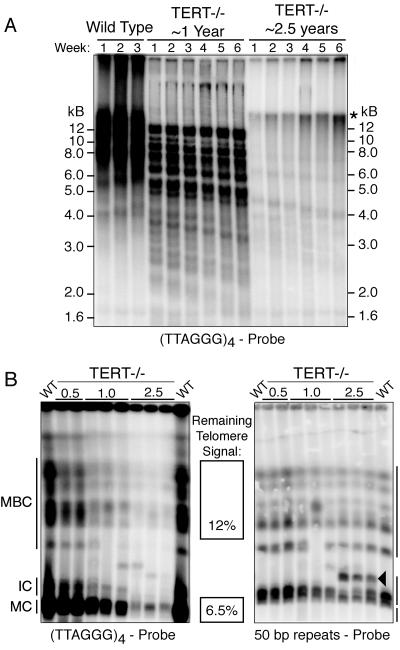
Deletion of the TERT gene leads to progressive shortening of the T. brucei telomere tract, at a rate of 3 to 6 bp/population doubling (16). The average telomere of the Lister 427 laboratory strain used in these studies is ∼15 kb (41). To monitor long-term telomere shortening and its consequences, several independent TERT−/− clones were continuously cultured without any selection regimen for ∼2.5 years. Telomeres of wild-type T. brucei grow at a constant rate of 7 to 9 bp/PD with no apparent limit during continuous propagation in the laboratory (3, 46). After ∼1 year in culture, telomeres of a TERT-deficient clone shortened significantly, and a dramatic reduction of telomere signal was evident after ∼2.5 years of continuous propagation (Fig. 1A). The six strong bands in Fig. 1A represent classes of telomeres of similar length and not six individual telomeres. A slowly migrating band also appeared in extensively propagated TERT-deficient strains (Fig. 1A). As judged by two-dimensional gel electrophoresis with DNA from human ALT cells as a positive control, this band does not appear to represent circular double-stranded DNA (data not shown). In-gel hybridization of native agarose gels, using strand-specific probes, excluded the possibility that this DNA is single stranded, and preliminary experiments showed that the band is susceptible to Bal 31 digestion, indicating that it is either nicked or terminal (data not shown). We have been thwarted in all our attempts to determine the origin and significance, if any, of this band.
FIG. 1.
Telomere loss in telomerase-deficient T. brucei. (A) Lanes 1 to 3 show wild-type telomeres. Due to their abundance and length, they are not well resolved on this gel. Telomerase-deficient clones exhibit gradual telomere shortening at a rate of 3 to 6 bp/PD during 6-week time courses after 1 or 2.5 years in continuous culture. DNA was digested with MboI and AluI and hybridized using a (TTAGGG)4 probe. Equal loading was ensured by fluorimetric quantification of DNA prior to loading. The asterisk indicates the appearance of a new heterogeneous band in TERT-deficient cells. (B) Telomere loss on different chromosome types. MBC were separated from IC and MC. Left panel: telomeric signal decreased in clones cultured for 0.5, 1.0, and 2.5 years. Right panel: a duplicate gel hybridized with a 50-bp probe as a loading control. The arrowhead marks rearrangements among IC. WT, wild type.
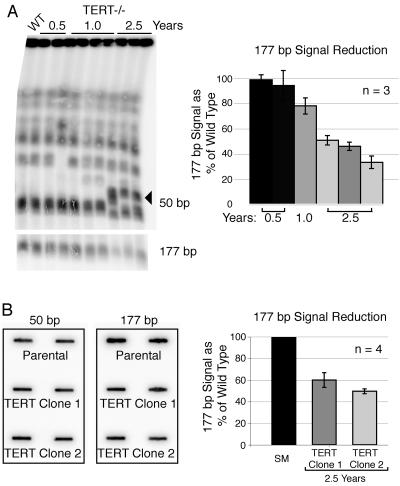
To measure the decline in telomere signal in different chromosome classes, gels were dried and probed with a radiolabeled (TTAGGG)4 probe (Fig. 1B). To ensure equal loading, the telomeric signal was normalized to a 50-bp repeat signal, which is uniquely present in long arrays upstream of ES promoters (Fig. 1B, right panel). Comparable results were obtained when a single-copy gene was used as a loading control (data not shown). Quantitative analysis of these blots revealed that the telomere signal on MBC declined to ∼12% of its initial intensity after ∼2.5 years in culture. Surprisingly, the telomere signal on MC reproducibly decreased to 6.5% (±1.2%) (Fig. 1B, middle panel). We observed one example of a rearrangement among MCB, which is consistent with the very infrequent rearrangements observed many years ago and with the fact that the diploid chromosomes of T. brucei can differ greatly in size, due to variations in long chromosome-internal repetitive regions, transposon-like sequences, and other pseudogenes that accumulate in large subtelomeric regions and possibly facilitate recombination (39, 42). We also noticed DNA rearrangements within the IC region of the gel (Fig. 1B, left panel). By separation of individual IC and hybridization with IC-specific genes, we confirmed the origin of these rearrangements to IC (data not shown). These changes could be the result of chromosome fusions or rearrangements. However, since we have very little understanding of IC function or relevance, we did not investigate these events further. To investigate what could account for this difference of telomere loss between MBC and MC and to study the effects of telomere loss on MC stability, we analyzed the karyotype of TERT-deficient cell lines in more detail by hybridization to a 177-bp repeat motif that is unique to MC. The 177-bp repeat signal decreased to approximately half of its initial intensity, relative to 50-bp and MBC-internal (data not shown) loading controls, over the course of ∼2.5 years (Fig. 2A). These results were reproducible for independently propagated clones.
FIG. 2.
Loss of minichromosomes in telomerase-deficient T. brucei. (A) Left panel: whole chromosomes, separated by RAGE and analyzed by Southern blotting. Upper panel: 50-bp repeat probe serves as loading control (the arrowhead indicates IC rearrangements). Lower panel: the same gel upon rehybridization with MC-specific 177-bp repeat probe (only the MC part of the gel is shown). Right panel: phosphorimager quantification of MC signal. The 177-bp signal was normalized to the 50-bp signal; bars represent the average 177-bp signal from three independent experiments (standard deviation is indicated) with up to three clones. (B) Left panel: slot blots using the 50-bp repeat probe as loading control and the 177-bp probe to confirm MC loss in long-term-cultured TERT−/− strains (clones 1 and 2). Right panel: quantified results of four experiments.
If MC had fused to each other, the 177-bp signal could have been redistributed over the gel and thereby reduced in the designated MC region. Although we did not observe any obvious chromosome fusions, we also quantified MC in unfractionated DNA by “slot blotting” (Fig. 2B). Quantitative phosphorimager analysis for four independent experiments confirmed that the 177-bp MC signal was reduced to ∼50% of its initial intensity (Fig. 2B). We attribute the decline of the 177-bp signal to loss of MC, which can also be detected by ethidium bromide staining (data not shown). This interpretation is consistent with the 50% reduction in TTAGGG signal on MC compared to MBC.
No significant rearrangements were detected in MBC using 50-bp repeat, VSG gene, and chromosome-internal hybridization probes (Fig. 1 and 2 and data not shown). Despite dramatic telomere shortening, MC loss, and rearrangements among IC in long-term-cultured telomerase-deficient strains, we could not detect any growth or viability problems, as judged by growth curves and plating assays (data not shown).
Telomere length stabilization in the absence of telomerase.
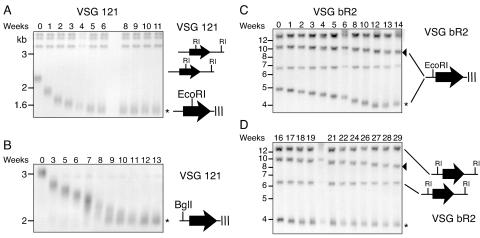
We monitored several MBC telomeres for ∼2.5 years. Once they became exceptionally short, the fate of individual telomeres was followed by hybridization to unique telomeric VSG genes during further propagation. VSG 121 and bR2 telomeres were chosen for analysis because they had become particularly short. There are three copies of VSG 121, one of which is telomeric. The telomeric restriction fragment, released by digestion with EcoRI, gradually shortened during the first 2 weeks of observation (Fig. 3A). Between weeks 3 and 5, the rate of shortening decreased, and the length of this restriction fragment ultimately stabilized (weeks 6 to 11). The length of the VSG 121 chromosome end remained stable during several additional months of continuous culture (data not shown). We confirmed this result by analyzing the VSG 121 telomere in a second independent clone, using a different restriction enzyme to liberate the terminal fragment (Fig. 3B).
FIG. 3.
Telomere length stabilization at dramatically shortened VSG 121 and VSG bR2 telomeres during continuous culture of telomerase-deficient clones (1 week corresponds to ∼24 population doublings). Bands corresponding to chromosome-internal copies of VSG 221 and bR2 do not change in size. (A and B) Stabilization of the VSG 121 terminal restriction fragment (asterisk) liberated by EcoRI (A) or BglI (B) digestion in two clones after 6 or 10 weeks of culture (chromosome-internal BglI fragments migrate higher and are not shown). (C) EcoRI liberates the two telomere-linked copies of VSG bR2. One copy is located in an ES with a long telomere (arrowhead), and the second copy is located in an ES with a shorter telomere, whose length stabilized after 12 weeks (asterisk). (D) The population from panel C was cloned at week 14, and one clone was propagated for an additional 13 weeks. The short telomere (asterisk) remained stable throughout the remaining 13 weeks, whereas the longer terminal restriction fragment continuously shortened (arrowhead).
To exclude the possibility that stabilization was occurring exclusively at the VSG 121 telomere, we analyzed a silent ES telomere harboring VSG bR2 (Fig. 3C and D). There are four copies of VSG bR2, two of which are ES-linked. Nontelomeric VSG bR2 restriction fragments did not change in size but the ES-linked fragments gradually shortened. One ES-linked copy of VSG bR2 had a short telomere that allowed good resolution of size changes on an agarose gel. This telomere progressively shortened up to week 12 and then stabilized. At week 14, a clone was picked out of the population and kept in culture for an additional 13 weeks (Fig. 3D). The VSG bR2 band was initially sharp (week 16). Over time, the average length of the terminal fragment did not change significantly (compare Fig. 3C and D), but the band became more heterogeneous. Upon further culturing, telomere stabilization also occurred on chromosome arms harboring VSG 224 and VSG 1.8 (data not shown).
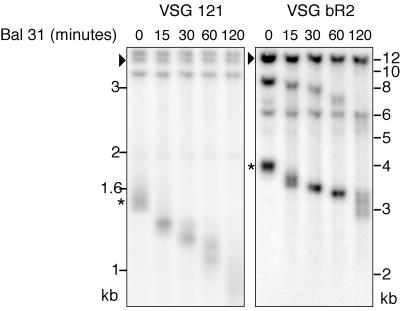
To confirm that the stabilized short chromosome ends remained terminal and did not fuse with other chromosome arms, we treated the DNA with the exonuclease Bal 31, after which the DNA was cut to liberate terminal restriction fragments that could be visualized with VSG gene probes. Telomere-linked copies of VSG 121 and VSG bR2, regardless of whether they were stabilized or not, were highly susceptible to Bal 31 treatment (Fig. 4), indicating that they remained terminal. Nontelomeric copies were unaffected by Bal 31. Telomerase-deficient S. pombe can survive by circularizing its chromosome (43). Circularized chromosomes remain trapped in the slot during RAGE separation. Using VSG 121 and bR2 as hybridization probes, we verified that the chromosomes harboring these VSG genes remained linear and that VSG 121 and bR2 did not recombine onto other chromosomes (data not shown).
FIG. 4.
Short stabilized restriction fragments are terminal. Genomic DNA was digested with exonuclease Bal 31. Stabilized terminal restriction fragments containing VSG 121 (left panel) or VSG bR2 (right panel) were liberated by EcoRI digestion and were susceptible to Bal 31 digestion (asterisk), whereas chromosome-internal copies (arrowheads) were not.
Taken together, telomere length analyses of different chromosome arms over several months indicated that, upon reaching a critical length, terminal restriction fragments were stabilized by a telomerase-independent mechanism. This mechanism seems to act exclusively at critically short telomeres, since longer telomeres continued to progressively shorten. Such a distinction has not been observed in other species in the absence of telomerase.
Cloning and sequencing of short stabilized telomeres.
Up to this point, we did not know whether short stabilized chromosome arms still contained telomeric repeats or whether they were maintained by amplification of subtelomeric sequences. To address this question, the short stabilized VSG 121 chromosome end was cloned and sequenced using a telomere tailing method (19). Poly(C) tails were added to purified terminal restriction fragments by using terminal transferase. VSG 121 and poly(G) primers were used to PCR amplify the region encompassing the 3′ end of the VSG 121 ORF, the subtelomeric region, and the end of the chromosome. PCR products were ligated into a cloning vector and analyzed by restriction digestion and sequencing. The size of the cloned PCR products (Fig. 5A) matched the size range observed on Southern blots and in the uncloned PCR products (data not shown). Sequencing (Fig. 5B) revealed terminal TTAGGG repeat tracts that ranged from 41 to 200 bp, matching the size heterogeneity observed by Southern blotting and PCR amplification (Fig. 3 and data not shown). The end of the sequence varied among individual clones. By adapter cloning of telomere ends, it was previously shown that T. brucei telomeres terminate in TTAGGG (11). This and other work suggested that the templating region of telomerase RNA ends in AATCCC-5′ (6). To verify that no changes occurred in the subtelomeric region, we PCR amplified this region by using VSG 121 and (CCCTAA)5 primers. Comparison of wild-type and mutant cells showed no amplifications or rearrangements in the ∼200-bp subtelomeric region between the VSG gene and the TTAGGG repeat tract. In conclusion, our results indicate that a potentially novel mechanism maintains 40 to 200 bp of terminal TTAGGG repeats in the absence of telomerase.
FIG. 5.
Terminal sequence of short stabilized telomeres. (A) Terminal restriction fragments were C tailed, PCR amplified, and cloned. Size variation among the cloned products reflected the heterogeneity observed on Southern blots and in the uncloned PCR product. (B) Six representative sequences showed 7 to 27 perfect hexameric repeats; the terminal nucleotide varied.
Rates of telomere elongation at transcriptionally silent and active ES.
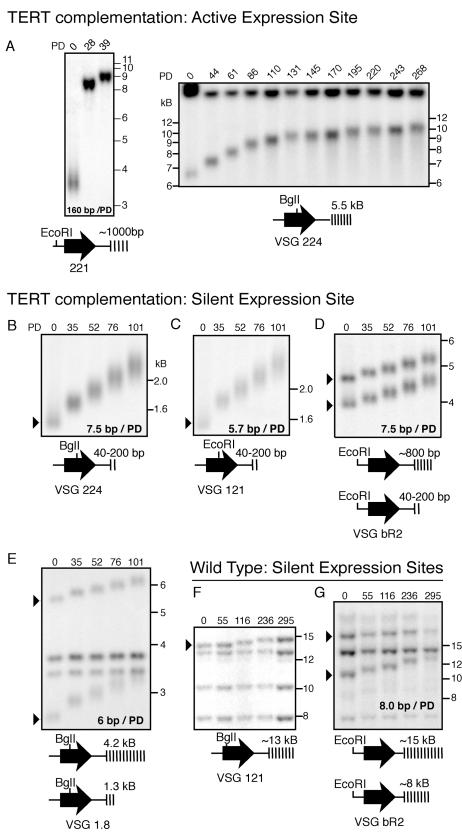
Previous studies showed that an artificially seeded short telomere at the transcriptionally active ES was rapidly elongated by telomerase (29). To determine the rate of elongation at naturally shortened stabilized telomeres and to verify that these short telomeres could be extended by telomerase, we restored TERT to cells that had been in culture for ∼2.5 years and studied the dynamics of telomere elongation. As shown in Fig. 6A, a short 3.5-kb terminal restriction fragment containing the actively transcribed VSG 221 and less than 1 kb of telomeric repeats was rapidly elongated. The rate of elongation, measured from the initial day of complementation to the first possible time point at which sufficient DNA could be obtained from the cells (PD 28), was ∼160 bp/PD. This experiment was repeated with six independently complemented clones with similar results, so we are confident the rapid elongation is a consequence of telomerase restoration rather than random recombination in individual clones. When the elongated telomere reached 8 to 10 kb, the rate of elongation decreased to ∼6 to 8 bp/PD (Fig. 6A, right panel). Although the resolving power of an 0.8% agarose gel is limited as telomere restriction fragments became large, the elongation rate clearly remained at ∼6.5 bp/PD for the last 7 weeks of the time course. When telomerase was restored to clones having different telomere lengths at the active ES, the rate of telomere elongation was inversely related to initial telomere length.
FIG. 6.
Telomere elongation dynamics at transcriptionally active and silent ES telomeres of various lengths after restoration of telomerase. The initial telomere length and restriction fragments used are schematically represented under each panel. (A) Left panel: a short transcriptionally active VSG 221 ES telomere was rapidly elongated. As the telomere reached a length of ∼8.5 kb, the rate of elongation decreased. Right panel: elongation of a 6.5-kb active VSG 224 ES telomere. During the initial ∼90 population doublings, the rate of telomere elongation was ∼25 bp/PD, but it subsequently decreased to ∼6.5 bp/PD. (B to D) Elongation of three short stabilized silent ES telomeres (VSG 224, VSG 121, and VSG bR2) after telomerase restoration. (E) Two silent ES-linked copies of VSG 1.8, with very different initial telomere lengths, are elongated at the same rate. (F and G) Long VSG bR2 and VSG 121 telomeres elongate at a constant rate in wild-type T. brucei.
In contrast, the rate of elongation at transcriptionally silent telomeres was only 6 to 8 bp/PD when telomerase was restored, regardless of their initial length. Telomeres harboring VSG 224, VSG 121, and one copy of VSG bR2 were short (40 to 200 bp) and stable, prior to reintroduction of telomerase (Fig. 6B, C, and D). The same extension rate was observed at the larger of the two VSG bR2 telomeres (Fig. 6D) and at two longer telomeres of ∼4.2 kb and 1.3 kb at the VSG 1.8 ES (Fig. 6E).
Over 20 years ago, it was observed that T. brucei telomeres of 5 to 9 kb grow at a constant rate (3, 46). We wondered whether, in accordance with the conventional protein-counting model of telomere length homeostasis, elongation would cease when a telomere reached a specific length. To address this question, we turned to extensively propagated wild-type T. brucei and found that silent ES telomeres longer than 10 or 15 kb continue to grow at a rate of 6 to 8 bp/PD (Fig. 6F and G).
In conclusion, very short transcriptionally silent and active ES telomeres have very different elongation dynamics. Short active ES telomeres are very rapidly elongated by telomerase. As telomere length increases, the rate of elongation decreases to 6 to 8 bp/PD, as at silent ES.
DISCUSSION
We monitored telomere loss, in the absence of TERT, at several MBC and IC during ∼2.5 years. Shortening of nontranscribed telomeres occurred at a constant rate of 4 to 6 bp/PD, and we did not observe any telomere-telomere recombination. Telomere shortening led to the loss of many apparently nonessential MC and to rearrangements among IC. MBC remained stable. Several possibilities could account for these differences. Loss of MBC, which contain essential genes, would presumably be lethal. IC appear only to contain ES, which can be active or silent, so rearrangements or loss may be tolerated to some degree. It is possible that MC with very short telomeres could be more susceptible to nucleolytic degradation. MBC and MC exhibit different positional dynamics during mitosis (12, 18, 24). Although segregation of MC normally occurs with high fidelity, as shown by their stable inheritance over 5 years of propagation (1), it remains unclear how the large number of MC are faithfully replicated and segregated. We speculate that MC might be attached to the mitotic spindle through their telomeres. As a consequence of dramatic telomere shortening, MC could be missegregated and give rise to cells lacking a particular MC or containing extra copies. Cells harboring extra MC might require slightly more time to replicate. During 2.5 years, cells that lost a minichromosome could have a minute replication advantage and become dominant within a population. A comparable situation has been observed between different types of survivors in telomerase-deficient S. cerevisiae. Type I survivors, which maintain their chromosome ends by extensive amplification of subtelomeric Y′ elements, were outgrown by type II (telomere-telomere recombination) survivors, presumably because the extensive amplifications (10% increase in genome size) increased the time needed for replication (53).
Short MBC telomeres stabilized as they reached a length of ∼40 to 200 bp. Bal 31 digestion confirmed that the remaining telomeric repeats were indeed terminal and that stabilized chromosomes did not fuse, suggesting that as little as 40 bp of telomeric repeats is sufficient to stabilize a chromosome end. The terminal nucleotide varied between different clones, which differs from the situation in wild-type cells (11). Telomere stabilization appears to be sufficient to ensure genomic integrity of MBC in a large majority of cells, as there was no discernible impact on long-term population growth. Nevertheless, TERT−/− T. brucei might have an enhanced propensity to undergo homologous recombination, similarly to sir/tlc-deficient yeast cells that become survivors without going through crisis (34). In Drosophila, chromosome end protection depends upon heterochromatin protein 1, which binds chromosome ends in a sequence-independent manner. It has been suggested that the epigenetic state of the chromatin rather than a particular sequence mediates end protection (33). It is possible that protection of short silent ES telomeres is in part mediated by the epigenetic state of its chromatin, but we have no information on this.
We did not detect extrachromosomal telomeric circles, which could explain why short telomeres remained stable during several months of continuous culture and did not undergo the dramatic telomere elongation observed in human ALT cells and type II S. cerevisiae survivors (10, 48, 53).
Immortal human ALT cells are characterized by increased telomere length and telomere heterogeneity and by the presence of extrachromosomal telomeric circles and ALT-associated PML bodies (5, 8, 26, 57). There is evidence that telomere-telomere recombination accounts for the increase in telomere length in ALT cells (17). It has also been suggested that extrachromosomal telomeric circles could serve as a template for rapid rolling-circle-based elongation of telomeres (14, 17, 54). However, telomere fluorescent in situ hybridization on metaphase nuclei of ALT cell lines shows many chromosome arms with little or no telomeric signal (47). The lack of subtelomeric marker genes in ALT cells has hampered efforts to follow the fate of individual telomeres. A recent report identified a human ALT cell line that maintains short yet stable telomeres in the absence of characteristic ALT markers (7). In this cell line, the average telomere length was ∼5 kb, as assessed by telomere blotting of RsaI/HinfI-digested genomic DNA. The precise subtelomeric locations of the RsaI and HinfI sites are unknown, but the size of individual telomeres could be shorter than 5 kb. Could the mechanism that stabilized telomere length in this cell line be similar to the mechanism that stabilizes T. brucei telomeres as short as 40 bp?
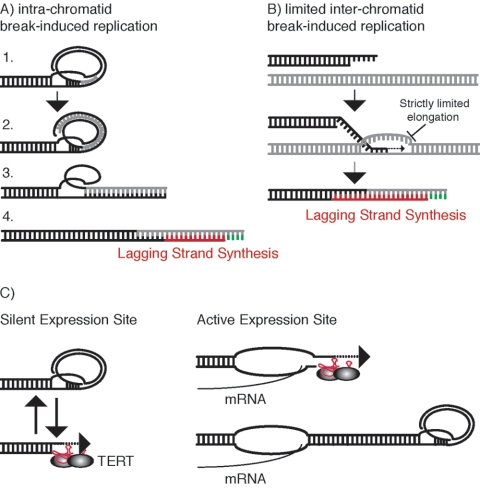
We propose a speculative model that would restrict the extent of telomere elongation and lead to the telomere stabilization that we observe. Telomeres of human, mouse, plants, and T. brucei can invade upstream tandem repeats, giving rise to a loop-like structure called the t-loop (9, 22, 23, 41). During telomere stabilization, a conventional DNA polymerase could extend the invading 3′ telomere terminus (intrastrand break-induced replication) (Fig. 7A). The displaced strand would give rise to a bubble. Subsequent unwinding of the downstream double-stranded region, realignment, and lagging-strand synthesis would complete replication. The extent of elongation would be limited by the size of the loop, which in turn is limited by the length of the telomere tract. Elongation could thus be restricted and lead to telomere stabilization. Alternatively, interstrand break-induced replication, in conjunction with machinery that tightly controls elongation to 40 to 200 bp, could facilitate telomere stabilization in the absence of telomerase (Fig. 7B).
FIG. 7.
(A) Speculative model for telomerase-independent telomere stabilization by intrastrand break-induced replication. Conventional DNA polymerases could extend the invaded 3′ end, using the C-rich strand as a template. Extension might be limited by the 5′ end of the telomere terminus. Subsequent unwinding and strand displacement, followed by lagging-strand synthesis (in red), could result in a limited extension of short telomeres. The RNA primer is shown in green. (B) An alternative model for stabilization by interstrand break-induced replication in conjunction with some mechanism that tightly limits elongation of the 3′ terminus to 40 to 200 bp. (C) Model for telomere length regulation in T. brucei. Left panel: silent ES telomeres. Telomere termini are tucked away, forming a t-loop structure (41). During some particular stage of the cell cycle, t-loops are resolved, and telomerase accesses the short 3′ overhangs and adds 6 bp ending in TTAGGG (11). Elongation occurs at the same rate regardless of the initial telomere length. Right panel: short active ES telomere. VSG gene transcription extends into the telomere, disrupts telomere structure (such as a t-loop), and allows telomerase unrestricted access to extend the 3′ end. As the telomere length increases, transcription does not reach the telomere end, allowing the assembly of a telomerase-inaccessible structure. As a consequence, the extension rate decreases to that of transcriptionally silent telomeres.
Short stabilized telomeres in telomerase-deficient T. brucei enabled us to study telomere length regulation upon reintroduction of telomerase. Telomere length regulation has been studied in numerous organisms. In human cells, telomere length homeostasis is regulated by a negative feedback loop, executed through numerous telomere-binding proteins that relay information about telomere length to the telomere terminus (51). How these proteins accomplish this task remains unclear, but recent work suggested that chromatin remodeling (or t-loop resolution) could regulate telomerase access to telomere termini (52). However, this protein-counting model does not appear to apply to T. brucei, where transcriptionally silent telomeres were elongated at the same rate, regardless of their initial telomere length. Furthermore, during in vitro culture, telomere homeostasis is not achieved even at long telomeres, which appear to grow indefinitely. This result implies that the negative feedback loop that regulates telomere length in other organisms is absent from transcriptionally silent telomeres in T. brucei.
In contrast to silent ES, a short active ES telomere was elongated at ∼160 bp/PD. As the length of an active ES telomere increased, the rate of elongation decreased until it matched the rate at silent ES. The only known difference between active and silent ES is transcriptional activity, and it has been shown that transcription extends into telomeric repeats (50). Very recently, a direct correlation between the strength of an inserted subtelomeric promoter and the rate of telomerase-dependent extension at the adjacent short seeded telomere was demonstrated (20). Based on all these observations, we propose a model in which transcription disrupts telomeric chromatin and enhances telomerase access and elongation (Fig. 7C). After rapid elongation, transcription no longer reaches the telomere terminus, which can now reassemble into a t-loop, rendering the 3′ end inaccessible during most of the cell cycle. The elongation dynamics now match those of transcriptionally silent ES.
In conclusion, we speculate that T. brucei telomeres play an important role in a variety of cellular processes, including chromosome segregation, genome integrity, and the regulation of VSG gene switching. Furthermore, due to numerous unique subtelomeric VSG genes, T. brucei telomerase-deficient mutants offer a unique opportunity to study the molecular mechanism underlying telomere stabilization, which may enhance our understanding of telomere maintenance in some cancerous human cells.
Acknowledgments
We are grateful to Titia de Lange and Joachim Lingner for helpful discussions and protocols, to Jenny Li for technical assistance with growing trypanosomes in animals, to Sean Rooney for human ALT DNA, to Luisa Figueiredo and Bibo Li for inspiring discussions and probes, and to Veena Mandava and Christian Janzen for discussion and critical reading of the manuscript.
This work was supported by NIH grant AI 50614.
REFERENCES
- 1.Alsford, S., B. Wickstead, K. Ersfeld, and K. Gull. 2001. Diversity and dynamics of the minichromosomal karyotype in Trypanosoma brucei. Mol. Biochem. Parasitol. 113:79-88. [DOI] [PubMed] [Google Scholar]
- 2.Barry, J. D., and R. McCulloch. 2001. Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Adv. Parasitol. 49:1-70. [DOI] [PubMed] [Google Scholar]
- 3.Bernards, A., P. A. Michels, C. R. Lincke, and P. Borst. 1983. Growth of chromosome ends in multiplying trypanosomes. Nature 303:592-597. [DOI] [PubMed] [Google Scholar]
- 4.Borst, P., and S. Ulbert. 2001. Control of VSG gene expression sites. Mol. Biochem. Parasitol. 114:17-27. [DOI] [PubMed] [Google Scholar]
- 5.Bryan, T. M., A. Englezou, J. Gupta, S. Bacchetti, and R. R. Reddel. 1995. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 14:4240-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cano, M. I., J. M. Dungan, N. Agabian, and E. H. Blackburn. 1999. Telomerase in kinetoplastid parasitic protozoa. Proc. Natl. Acad. Sci. USA 96:3616-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerone, M. A., C. Autexier, J. A. Londono-Vallejo, and S. Bacchetti. 2005. A human cell line that maintains telomeres in the absence of telomerase and of key markers of ALT. Oncogene 24:7893-7901. [DOI] [PubMed] [Google Scholar]
- 8.Cesare, A. J., and J. D. Griffith. 2004. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol. Cell. Biol. 24:9948-9957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cesare, A. J., N. Quinney, S. Willcox, D. Subramanian, and J. D. Griffith. 2003. Telomere looping in P. sativum (common garden pea). Plant J. 36:271-279. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Q., A. Ijpma, and C. W. Greider. 2001. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 21:1819-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiurillo, M. A., and J. L. Ramirez. 2002. Characterization of Leishmania major Friedlin telomeric terminus. Mem. Inst. Oswaldo Cruz 97:343-346. [DOI] [PubMed] [Google Scholar]
- 12.Chung, H. M., C. Shea, S. Fields, R. N. Taub, L. H. Van der Ploeg, and D. B. Tse. 1990. Architectural organization in the interphase nucleus of the protozoan Trypanosoma brucei: location of telomeres and mini-chromosomes. EMBO J. 9:2611-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Lange, T. 1994. Activation of telomerase in a human tumor. Proc. Natl. Acad. Sci. USA 91:2882-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Lange, T. 2004. T-loops and the origin of telomeres. Nat. Rev. Mol. Cell. Biol. 5:323-329. [DOI] [PubMed] [Google Scholar]
- 15.De Lange, T., and P. Borst. 1982. Genomic environment of the expression-linked extra copies of genes for surface antigens of Trypanosoma brucei resembles the end of a chromosome. Nature 299:451-453. [DOI] [PubMed] [Google Scholar]
- 16.Dreesen, O., B. Li, and G. A. M. Cross. 2005. Telomere structure and shortening in telomerase-deficient Trypanosoma brucei. Nucleic Acids Res. 33:4536-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunham, M. A., A. A. Neumann, C. L. Fasching, and R. R. Reddel. 2000. Telomere maintenance by recombination in human cells. Nat. Genet. 26:447-450. [DOI] [PubMed] [Google Scholar]
- 18.Ersfeld, K., and K. Gull. 1997. Partitioning of large and minichromosomes in Trypanosoma brucei. Science 276:611-614. [DOI] [PubMed] [Google Scholar]
- 19.Forstemann, K., M. Hoss, and J. Lingner. 2000. Telomerase-dependent repeat divergence at the 3′ ends of yeast telomeres. Nucleic Acids Res. 28:2690-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glover, L., and D. Horn. 2006. Repression of polymerase I-mediated gene expression at Trypanosoma brucei telomeres. EMBO Rep. 7:93-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greider, C. W., and E. H. Blackburn. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43:405-413. [DOI] [PubMed] [Google Scholar]
- 22.Griffith, J. D., L. Comeau, S. Rosenfield, R. M. Stansel, A. Bianchi, H. Moss, and T. de Lange. 1999. Mammalian telomeres end in a large duplex loop. Cell 97:503-514. [DOI] [PubMed] [Google Scholar]
- 23.Groff-Vindman, C., A. J. Cesare, S. Natarajan, J. D. Griffith, and M. J. McEachern. 2005. Recombination at long mutant telomeres produces tiny single- and double-stranded telomeric circles. Mol. Cell. Biol. 25:4406-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gull, K., S. Alsford, and K. Ersfeld. 1998. Segregation of minichromosomes in trypanosomes: implications for mitotic mechanisms. Trends Microbiol. 6:319-323. [DOI] [PubMed] [Google Scholar]
- 25.Harley, C. B., A. B. Futcher, and C. W. Greider. 1990. Telomeres shorten during ageing of human fibroblasts. Nature 345:458-460. [DOI] [PubMed] [Google Scholar]
- 26.Henson, J. D., A. A. Neumann, T. R. Yeager, and R. R. Reddel. 2002. Alternative lengthening of telomeres in mammalian cells. Oncogene 21:598-610. [DOI] [PubMed] [Google Scholar]
- 27.Hirumi, H., and K. Hirumi. 1989. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 75:985-989. [PubMed] [Google Scholar]
- 28.Horn, D., and G. A. M. Cross. 1997. Analysis of Trypanosoma brucei VSG expression site switching in vitro. Mol. Biochem. Parasitol. 84:189-201. [DOI] [PubMed] [Google Scholar]
- 29.Horn, D., C. Spence, and A. K. Ingram. 2000. Telomere maintenance and length regulation in Trypanosoma brucei. EMBO J. 19:2332-2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, N. W., M. A. Piatyszek, K. R. Prowse, C. B. Harley, M. D. West, P. L. Ho, G. M. Coviello, W. E. Wright, S. L. Weinrich, and J. W. Shay. 1994. Specific association of human telomerase activity with immortal cells and cancer. Science 266:2011-2015. [DOI] [PubMed] [Google Scholar]
- 31.Lingner, J., T. R. Hughes, A. Shevchenko, M. Mann, V. Lundblad, and T. R. Cech. 1997. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276:561-567. [DOI] [PubMed] [Google Scholar]
- 32.Loayza, D., and T. De Lange. 2003. POT1 as a terminal transducer of TRF1 telomere length control. Nature 423:1013-1018. [DOI] [PubMed] [Google Scholar]
- 33.Louis, E. J. 2002. Are Drosophila telomeres an exception or the rule? Genome Biol. 3:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowell, J. E., A. I. Roughton, V. Lundblad, and L. Pillus. 2003. Telomerase-independent proliferation is influenced by cell type in Saccharomyces cerevisiae. Genetics 164:909-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lundblad, V. 2002. Telomere maintenance without telomerase. Oncogene 21:522-531. [DOI] [PubMed] [Google Scholar]
- 36.Marcand, S., V. Brevet, and E. Gilson. 1999. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 18:3509-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marcand, S., E. Gilson, and D. Shore. 1997. A protein-counting mechanism for telomere length regulation in yeast. Science 275:986-990. [DOI] [PubMed] [Google Scholar]
- 38.Maringele, L., and D. Lydall. 2004. Telomerase- and recombination-independent immortalization of budding yeast. Genes Dev. 18:2663-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melville, S. E., C. S. Gerrard, and J. M. Blackwell. 1999. Multiple causes of size variation in the diploid megabase chromosomes of African trypanosomes. Chromosome Res. 7:191-203. [DOI] [PubMed] [Google Scholar]
- 40.Melville, S. E., V. Leech, M. Navarro, and G. A. Cross. 2000. The molecular karyotype of the megabase chromosomes of Trypanosoma brucei stock 427. Mol. Biochem. Parasitol. 111:261-273. [DOI] [PubMed] [Google Scholar]
- 41.Munoz-Jordan, J. L., G. A. M. Cross, T. de Lange, and J. D. Griffith. 2001. t-loops at trypanosome telomeres. EMBO J. 20:579-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Myler, P. J., R. F. Aline, Jr., J. K. Scholler, and K. D. Stuart. 1988. Multiple events associated with antigenic switching in Trypanosoma brucei. Mol. Biochem. Parasitol. 29:227-241. [DOI] [PubMed] [Google Scholar]
- 43.Nakamura, T. M., J. P. Cooper, and T. R. Cech. 1998. Two modes of survival of fission yeast without telomerase. Science 282:493-496. [DOI] [PubMed] [Google Scholar]
- 44.Navarro, M., and G. A. M. Cross. 1996. DNA rearrangements associated with multiple consecutive directed antigenic switches in Trypanosoma brucei. Mol. Cell. Biol. 16:3615-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Navarro, M., and G. A. M. Cross. 1998. In situ analysis of a variant surface glycoprotein expression-site promoter region in Trypanosoma brucei. Mol. Biochem. Parasitol. 94:53-66. [DOI] [PubMed] [Google Scholar]
- 46.Pays, E., M. Laurent, K. Delinte, N. Van Meirvenne, and M. Steinert. 1983. Differential size variations between transcriptionally active and inactive telomeres of Trypanosoma brucei. Nucleic Acids Res. 11:8137-8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perrem, K., L. M. Colgin, A. A. Neumann, T. R. Yeager, and R. R. Reddel. 2001. Coexistence of alternative lengthening of telomeres and telomerase in hTERT-transfected GM847 cells. Mol. Cell. Biol. 21:3862-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reddel, R. R. 2003. Alternative lengthening of telomeres, telomerase, and cancer. Cancer Lett. 194:155-162. [DOI] [PubMed] [Google Scholar]
- 49.Robinson, N. P., N. Burman, S. E. Melville, and J. D. Barry. 1999. Predominance of duplicative VSG gene conversion in antigenic variation in African trypanosomes. Mol. Cell. Biol. 19:5839-5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rudenko, G., and L. H. Van der Ploeg. 1989. Transcription of telomere repeats in protozoa. EMBO J. 8:2633-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smogorzewska, A., and T. de Lange. 2004. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73:177-208. [DOI] [PubMed] [Google Scholar]
- 52.Teixeira, M. T., M. Arneric, P. Sperisen, and J. Lingner. 2004. Telomere length homeostasis is achieved via a switch between telomerase-extendible and nonextendible states. Cell 117:323-335. [DOI] [PubMed] [Google Scholar]
- 53.Teng, S. C., and V. A. Zakian. 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:8083-8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tomaska, L., M. J. McEachern, and J. Nosek. 2004. Alternatives to telomerase: keeping linear chromosomes via telomeric circles. FEBS Lett. 567:142-146. [DOI] [PubMed] [Google Scholar]
- 55.Vanhamme, L., P. Poelvoorde, A. Pays, P. Tebabi, H. Van Xong, and E. Pays. 2000. Differential RNA elongation controls the variant surface glycoprotein gene expression sites of Trypanosoma brucei. Mol. Microbiol. 36:328-340. [DOI] [PubMed] [Google Scholar]
- 56.van Steensel, B., and T. de Lange. 1997. Control of telomere length by the human telomeric protein TRF1. Nature 385:740-743. [DOI] [PubMed] [Google Scholar]
- 57.Wang, R. C., A. Smogorzewska, and T. de Lange. 2004. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119:355-368. [DOI] [PubMed] [Google Scholar]
- 58.Weiden, M., Y. N. Osheim, A. L. Beyer, and L. H. Van der Ploeg. 1991. Chromosome structure: DNA nucleotide sequence elements of a subset of the minichromosomes of the protozoan Trypanosoma brucei. Mol. Cell. Biol. 11:3823-3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wirtz, E., S. Leal, C. Ochatt, and G. A. M. Cross. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89-101. [DOI] [PubMed] [Google Scholar]
- 60.Zomerdijk, J. C., R. Kieft, and P. Borst. 1992. A ribosomal RNA gene promoter at the telomere of a mini-chromosome in Trypanosoma brucei. Nucleic Acids Res. 20:2725-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]