Abstract
FQR1 is a novel primary auxin-response gene that codes for a flavin mononucleotide-binding flavodoxin-like quinone reductase. Accumulation of FQR1 mRNA begins within 10 min of indole-3-acetic acid application and reaches a maximum of approximately 10-fold induction 30 min after treatment. This increase in FQR1 mRNA abundance is not diminished by the protein synthesis inhibitor cycloheximide, demonstrating that FQR1 is a primary auxin-response gene. Sequence analysis reveals that FQR1 belongs to a family of flavin mononucleotide-binding quinone reductases. Partially purified His-tagged FQR1 isolated from Escherichia coli catalyzes the transfer of electrons from NADH and NADPH to several substrates and exhibits in vitro quinone reductase activity. Overexpression of FQR1 in plants leads to increased levels of FQR1 protein and quinone reductase activity, indicating that FQR1 functions as a quinone reductase in vivo. In mammalian systems, glutathione S-transferases and quinone reductases are classified as phase II detoxification enzymes. We hypothesize that the auxin-inducible glutathione S-transferases and quinone reductases found in plants also act as detoxification enzymes, possibly to protect against auxin-induced oxidative stress.
Auxin plays a central role in the control of plant growth and development. It can stimulate or inhibit cell expansion, stimulate cell division, promote differentiation of vascular tissues, inhibit shoot branching, and promote lateral root formation. Although recent attempts to unravel these signal transduction pathways have been quite successful, it is clear that we have not identified all of the genes that are up-regulated immediately after auxin application.
At present, about a half a dozen families of primary auxin-response genes have been identified. These include the Aux/IAA, SAUR, and GH3 families, members of gene families encoding glutathione S-transferases (GST) and 1-aminocyclopropane-1-carboxylic acid synthases, and NAC1 (for review, see Abel and Theologis, 1996; Xie et al., 2000). Of these, the Aux/IAA family is the most thoroughly studied (Abel et al., 1995). Aux/IAA genes are transcriptional regulators, and gain-of-function mutations in some family members have dramatic effects on plant development (Nagpal et al., 2000; Rogg et al., 2001). Transcription of the Aux/IAA genes is under the control of auxin-response elements (AuxREs; for review, see Guilfoyle et al., 1998).
The GSTs are a distinct class of auxin-responsive genes (for review, see Marrs, 1996). This family of enzymes was named for its ability to conjugate compounds, including xenobiotic compounds, to the tripeptide glutathione. Once such a complex has formed, the glutathione serves as a tag, marking the complex for transport into the vacuole (for review, see Edwards et al., 2000). As a result of this activity, GSTs protect cells from potentially damaging agents.
In mammalian systems, GSTs are classified as phase II detoxifying enzymes. Detoxification of xenobiotic and/or cancer-causing compounds often occurs via two steps. Phase I enzymes catalyze the metabolic activation of the compound. Phase II enzymes then detoxify it, often by catalyzing the conjugation of endogenous metabolites to the xenobiotic (Josephy, 1997). The phase II enzymes also include quinone reductases (QRs) that carry out obligate two-electron reductions of quinones. These reactions may protect cells against oxidative stress by decreasing the rate of formation of semiquinones, which can contribute to the formation of reactive oxygen species (for review, see Ross, 1997).
Expression of mammalian GST and QR genes is coordinately regulated by the cis-acting antioxidant response element (ARE) and can be induced by a wide range of compounds, including some polycyclic aromatic hydrocarbons and oxidants such as hydrogen peroxide (for review, see Rushmore and Pickett, 1993). In plants, auxin inducibility of GST genes is conferred by stress-responsive octapine synthase (ocs) elements (Ulmasov et al., 1994; Chen and Singh, 1999). These ocs elements are similar in sequence to the AREs found in mammalian GST genes and distinct from the AuxREs found in other types of auxin-inducible genes. Until now, the genes encoding GST proteins were the only auxin-regulated genes known to encode phase II enzymes.
Here, we describe the discovery of a primary auxin-response gene that encodes a QR. We identified, cloned, and characterized a novel auxin-responsive cDNA, FQR1, which encodes a flavodoxin-like QR. Accumulation of the corresponding mRNA begins within 10 min of indole-3-acetic acid (IAA) application, and is not inhibited by addition of the protein synthesis inhibitor cycloheximide, demonstrating that FQR1 is a primary auxin-response gene. When expressed in Escherichia coli, the corresponding protein binds FMN, carries out electron transport, and functions as a QR. Overexpression of FQR1 leads to increased levels of QR activity in whole plant extracts, suggesting that FQR1 acts as a QR in planta. This is a novel activity for a protein encoded by an auxin-responsive gene, and its presence raises questions about the extent to which phase II enzymes may be involved in auxin response.
RESULTS
Differential Display Reveals Auxin-Regulated mRNAs
Treating Arabidopsis roots with 1 to 30 μm exogenous IAA causes a number of physiological responses, including reduced rates of root elongation and induction of lateral root formation. We previously established a system for auxin treatment of 7-d-old Arabidopsis seedlings that strongly induces the formation of lateral roots (Laskowski et al., 1995). To determine how that treatment affects gene expression, differential display was performed on total RNA collected from roots 1 h after the initiation of a 15-min treatment with water or 30 μm IAA (Liang and Pardee, 1992). Seventy primer pairs were used to generate 1,412 PCR products. Twenty-one of these products were at least 2-fold induced by the auxin treatment. Based on reproducibility and the magnitude of the response, five of these differentially expressed mRNAs were selected. Their corresponding PCR fragments were isolated from the display gels, re-amplified, and used as probes. The differential expression of one of these mRNAs was confirmed by northern-blot analysis (data not shown). The corresponding cDNA was re-amplified and cloned into a pCRII vector.
FQR1 Has Sequence Similarity to a Family of QRs
A full-length cDNA was obtained by using the PCR-generated fragment as a probe to screen the Arabidopsis library λPRL-2 (Newman et al., 1994). This cDNA clone represents a gene that we named FQR1. The FQR1 cDNA contains a 612-bp open reading frame that encodes a protein with a calculated molecular mass of 21,794 D. This cDNA sequence exactly matches the nucleotide sequence and predicted splice sites for GenBank protein BAA97523.1 (nucleotide AB026634). The gene is located on chromosome 5 immediately 5′ to a hypothetical auxin-responsive-like protein.
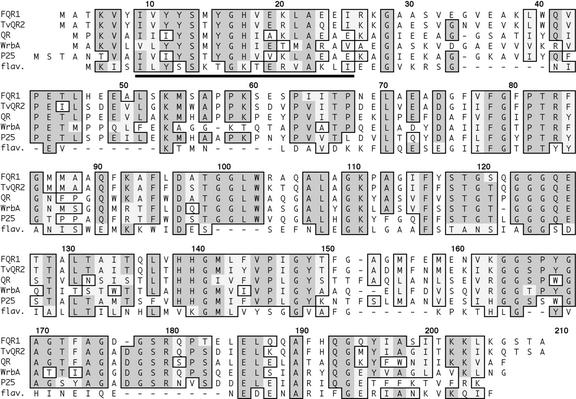
FQR1 is a member of an extensive family of highly conserved proteins found in plants, fungi, archea, and eubacteria (Fig. 1). The founding member of this family is WrbA. FQR1/WrbA family members share sequence motifs that identify them as candidate flavodoxins, which are flavin-binding proteins that function as electron carriers in redox reactions (Grandori and Carey, 1994). The N-terminal region of FQR1 matches the 17-amino acid-long flavodoxin signature sequence [LIV]-[LIVFY]-[FY]-x-[ST]-x-x-[AGC]-x-T-x-x-x-A-x-x-[LIV] (Fig. 1, underlined segment) with one mismatch. The mismatch is shown in the 18th position of the multiple alignment. Here, the expected amino acid in the flavodoxin signature sequence is Thr, whereas FQR1 contains a Val residue. A BLAST search identified three Arabidopsis proteins that share from 48% to 82% amino acid identity with FQR1, indicating that the gene family within Arabidopsis has at least four members. The one family member for which biochemical activity has been determined is a 1,4-benzoquinone reductase from a basidomycete (Brock et al., 1995). However, FQR1/WrbA family members have very little sequence similarity with mammalian QRs.
Figure 1.
Deduced amino acid sequence of FQR1 and its relationship to the WrbA gene family as aligned by ClustalW in MacVector. The sequences shown in this alignment were selected from those that have a high degree of sequence similarity with FQR1; preference was given to those sequences for which biochemical information is available. FQR1 is displayed on the top line. TvQR2 is quinone oxidoreductase from T. versicolor (accession no. AAG53945; Matvienko et al., 2001), QR is 1,4-benzoquinone reductase from P. chrysosporium (accession no. AAD21025; Akileswaran et al., 1999), WrbA is Trp repressor-binding protein from E. coli (accession no. P30849; Yang et al., 1993), p25 protein is from S. pombe (accession no. P30821; Toda et al., 1992; Turi et al., 1994), and flav. is flavodoxin from C. acetobutylicum (accession no. A38177; Santangelo et al., 1991). Sequence identity with the archeal protein (from Archaeoglobus fulgidus), not shown because of its lower level of similarity, is 32%. Identical residues are indicated with dark gray shading, and similarities are indicated with light gray shading. The N-terminal region matching the flavodoxin signature sequence is underlined.
Recombinant FQR1 Is Associated with a Flavin
FQR1 was expressed in E. coli as an His-tagged fusion protein. When the protein was purified on an Ni-containing column, the FQR1-containing fractions were bright yellow. To determine whether this color might be caused by a bound flavin, we treated the fractions with cold trichloroacetic acid (TCA) to precipitate the protein and separate the flavin. The absorption spectrum of the resulting supernatant was similar to that of flavin standards (Fig. 2A). Fluorescence excitation and emission spectra of the holoenzyme indicate that the protein has characteristics similar to that of known flavoproteins (data not shown).
Figure 2.
FQR1 fusion protein binds a flavin. Protein was isolated from IPTG-induced BL21 (DE3) E. coli harboring the His-tagged FQR1 expression vector and purified on a column containing an Ni2+-charged agarose matrix. A, Flavin was dissociated from purified protein under acidic conditions, and the spectrum of the isolate (dotted line) was compared with that of a flavin (FMN) standard (solid line). B, Thin layer chromatographic analysis of FQR1-associated flavin harvested from an E. coli overexpression system. The isolated protein was dialyzed in phosphate-buffered saline and precipitated using 5% (w/v) TCA. The flavin-containing supernatant was dried, resuspended in 65% (v/v) ethanol, and loaded onto silica plates. Approximately 1 μg of each standard was added. The experiment was performed once each in two different solvent systems. The data shown here are from 3:1:1 butanol:acetic acid:water. Rf, Riboflavin.
To determine the type of flavin bound by the protein, the acid supernatant was concentrated and spotted on a thin-layer chromatography plate. As shown in Figure 2B, the FQR1-associated flavin comigrated with FMN.
FQR1 Functions as an NAD(P)H-Dependent Reductase
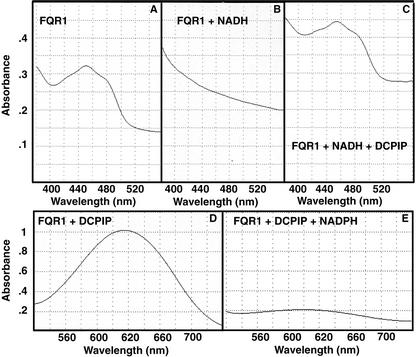
In an in vitro assay, purified recombinant FQR1 functions as an NAD(P)H-dependent reductase. A freshly isolated FQR1 fusion protein has an absorption maximum at about 450 nm (Fig. 3A). Such a spectrum is characteristic of the oxidized form of a flavin. Reduction of the flavin moiety is associated with loss of this absorption maximum (Ghisla, 1980). Addition of an equivalent concentration of NADH results in loss of the peak at 450 nm (Fig. 3B). Subsequent addition of an equivalent concentration of the electron acceptor dichlorophenolindophenol (DCPIP) results in flavin oxidation, as demonstrated by a restoration of the absorption maximum at 450 nm (Fig. 3C). The reduction of DCPIP by FQR1 can also be followed by monitoring its absorption at 600 nm, because the reduced form of DCPIP is colorless rather than blue (Fig. 3, D and E). Rapid reduction of DCPIP requires both FQR1 and a molecule that can donate electrons to FQR1. NADH and NADPH both function in this respect. Denaturing FQR1 by boiling inactivates the reductase activity. As a result of these experiments, we conclude that FQR1 functions as an NAD(P)H-dependent reductase.
Figure 3.
FQR1 functions in electron transport. A, Absorption spectrum of FQR1 fusion protein. B, Absorption spectrum of FQR1 fusion protein from A to which NADH has been added. The loss of A450 is indicative of flavin reduction. C, Absorption spectrum of FQR1 fusion protein-NADH mixture from B to which the electron acceptor DCPIP has been added. At this point, all three components are present at 50 μm. D, Absorption spectrum of FQR1 fusion protein mixed with DCPIP. The high absorbance in the blue spectrum is indicative of the oxidized state of DCPIP. E, Absorption spectrum of FQR1 fusion protein mixed with DCPIP from D to which NADPH has been added. At this point, all three components are present at 50 μm.
FQR1 Is a QR
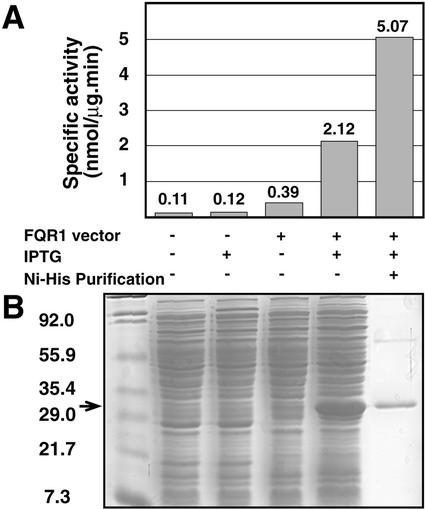
To determine whether FQR1, like its family members, has QR activity, a standard assay for QRs was used (Prochaska and Santamaria, 1988). QR activity is measured as the ability of an enzyme to reduce the quinone substrate menadione to menadiol. Formation of menadiol is evidenced by the subsequent menadiol-induced reduction of 3-(4,5-dimethylthiazo-2-yl)-2,5-diphenyltetrazolium bromide (MTT) to a blue formazan dye. The specific activity of extracts from E. coli cells with and without the FQR1 expression vector were compared. As shown in Figure 4B, the amount of FQR1 protein increased dramatically when cells containing the expression vector were induced with isopropyl-d-thiogalactopyranoside (IPTG). This increase in FQR1 protein level correlates with an increase in the specific activity of the preparation. The specific activity of extracts from IPTG-induced cells that contain the FQR1 expression vector is 2,120 nmol min−1 mg−1, whereas the specific activity of uninduced, vectorless controls is 110 nmol min−1 mg−1 (Fig. 4A). To confirm that the increase in activity seen in induced cells was caused by the increase in FQR1 expression, FQR1 fusion protein was purified by affinity chromatography on Ni-nitrilotriacetic acid agarose beads. The specific activity of the purified fraction is 5,000 nmol min−1 mg−1. In light of these results, we named the protein flavodoxin-like QR (FQR1).
Figure 4.
FQR1 has QR activity. A, Specific activity of FQR1 as determined by the formation of reduced MTT in the QR assay. Numbers for the first four samples reflect the averages of two trials. Protein concentrations for each trial were determined by averaging the results of triplicate Bradford assays. B, Coomassie Brilliant Blue-stained gel showing the relative amount of FQR1 present in the E. coli extracts used in A. The additional lane on the far left contains Mr markers. The arrow indicates the position of the FQR1 fusion protein.
FQR1 Is a Primary Auxin-Response Gene
Preliminary experiments demonstrated that FQR1 mRNA is auxin inducible. Although the amount present in root cultures is higher than in detipped roots from intact plants, the response of FQR1 to auxin treatment is similar in intact and cultured roots (data not shown). Thus, we investigated the response of FQR1 to auxin in roots that had been cultured in liquid Murashige and Skoog medium containing 3% Suc.
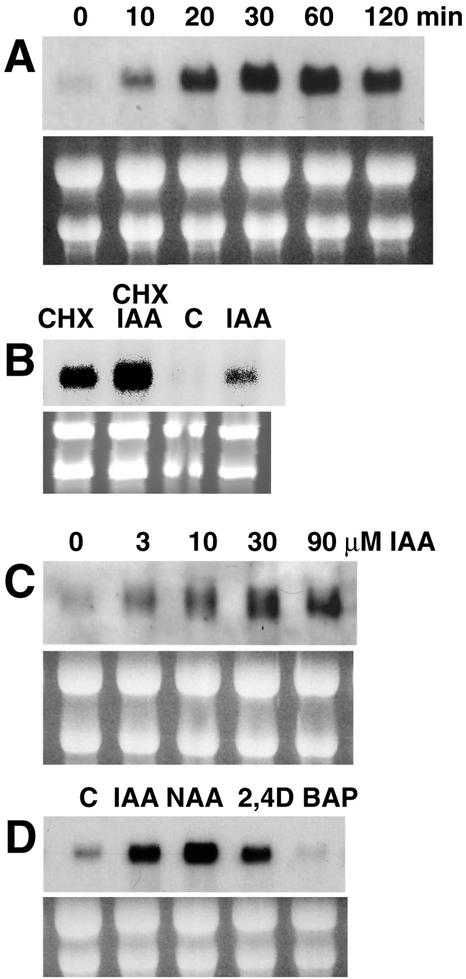
Addition of auxin to cultured roots results in an extremely rapid increase in the amount of accumulated FQR1 mRNA. As shown in Figure 5A, elevated levels of FQR1 mRNA are observed within 10 min of auxin application. The maximal level of mRNA accumulation is approximately 10-fold greater than the level seen in uninduced roots and is reached about 30 min after auxin application. This places FQR1 in a class with the fastest known auxin up-regulated genes (McClure and Guilfoyle, 1987; Abel et al., 1995). When auxin is continually present in the medium, enhanced levels of mRNA are maintained for at least 2 h. Whether these increases in mRNA levels are a result of increased transcription or mRNA stability has not been determined.
Figure 5.
Northern blots were hybridized with probe made from the 550-bp fragment of the FQR1 cDNA. Total RNA (10 μg) was loaded per lane. A, Increase in FQR1 mRNA accumulation with time after auxin treatment. RNA was isolated from cultured roots to which 30 μm IAA was added at t = 0. B, Incubating roots with cycloheximide leads to increased levels of FQR1 mRNA. RNA was extracted from root cultures treated with 10 μm cycloheximide and/or 10 μm IAA for 2 h. “C,” Control; “CHX,” cycloheximide. C, Effect of increasing concentrations of IAA on accumulation of FQR1 mRNA. RNA was isolated from cultured roots 1 h after IAA application. D, Effect of multiple auxin and non-auxin compounds. Hormones were applied to cultured roots for 2 h at a concentration of 30 μm. “C,” Control. rRNA bands from the ethidium bromide-stained gels, shown below each northern blot, served as loading controls.
Whereas the accumulation of mRNA for secondary auxin-response genes requires protein synthesis, primary auxin-response genes are defined by their ability to be induced by auxin in the presence of protein synthesis inhibitors. Induction of FQR1 mRNA by IAA occurs in the presence of the protein synthesis inhibitor cycloheximide (10 μm for 2 h), demonstrating that FQR1 is a primary auxin-response gene (Fig. 5B). Many of the primary auxin-response genes can also be induced by protein synthesis inhibitors alone (Theologis et al., 1985; van der Zaal et al., 1987; Franco et al., 1990). Treating Arabidopsis root cultures with cycloheximide leads to a substantial increase in FQR1 mRNA accumulation (Fig. 5B).
Specificity of FQR1 Induction
To determine whether the response of FQR1 to exogenous IAA could be associated with physiological auxin responses, we examined the effective concentration range of IAA and the variety of molecules that promote the response. Accumulation of FQR1 mRNA is promoted by 1 to 90 μm IAA, the same range of concentrations that is effective in inducing lateral root formation and inhibiting root elongation (Blakely et al., 1988; Fig. 5C). Maximal accumulation, a 13-fold increase over control levels, is reached with 30 μm IAA. Two additional auxins, naphthylacetic acid and 2,4-dichlorophenoxyacetic acid, also induce the response (Fig. 5D), whereas Trp, a molecule that is structurally similar to IAA but does not have auxin activity, fails to induce FQR1 mRNA accumulation. In independent experiments, addition of high concentrations of Trp (50 and 500 μm) to cultured roots for a period of 6 h did not affect the accumulation of this mRNA (data not shown). Incubation in 30 μm cytokinin (6-benzylaminopurine [BAP]) for 2 h also failed to promote FQR1 mRNA accumulation (Fig. 5D). Thus, there is a correlation between the conditions that promote typical auxin responses and the factors that induce FQR1 mRNA accumulation.
Because accumulation of FQR1 mRNA is auxin inducible, we searched the putative promoter region for AuxREs similar to those found in other early auxin-inducible genes. AuxREs are typically composed of pairs of TGTCTC sites, or of a single TGTCTC site together with a second element (Ulmasov et al., 1995). We did not find any obvious TGTCTC-type AuxRE sites, although there are separate TGTCTt sequences at position −1,655 and −990 from the ATG that might have the potential to form the basis for composite elements, if a second, as yet unidentified, element were present. In contrast, there are some close matches for stress-responsive elements. The consensus stress-responsive ocs-like element found in some GSTs is TGACGTAAgcgcTGACGTAA (Ulmasov et al., 1994). Another stress-responsive element is the mammalian ARE. The consensus ARE is two closely spaced TGACNNN followed by a GC box (Dhakshinamoorthy et al., 2000). We searched for elements similar to either of these by looking for the characteristic TGAC motif and identified three sites that have closely spaced repeats of TGAC: TGACcctaccctcgGTCA at −1,026, TTAGTCActaacattagtgttaagatTGACta at −677, and TGACAGCaacatatcaTCA at −489. The presence of these sites raises the possibility that they may confer transcriptional regulation on FQR1 in a manner similar to other genes that contain similar stress-responsive elements.
Accordingly, we tested the specificity of FQR1 induction by examining tissues that were stressed in various ways. Treating root cultures with 5 or 10 mm H2O2 for 30 min or 100 μm CdCl2 for 2 h did not alter the level of FQR1 mRNA accumulation (data not shown). Accumulation of FQR1 mRNA can be detected in shoots as well as in roots and root cultures, and so we investigated the effect of wounding on FQR1 expression in leaves. Removing 50% of the leaf blade of the four largest leaves of 14-d-old plants 6 h before harvest failed to affect the amount of FQR1 mRNA accumulation (data not shown).
FQR1 Protein Is Present throughout the Plant
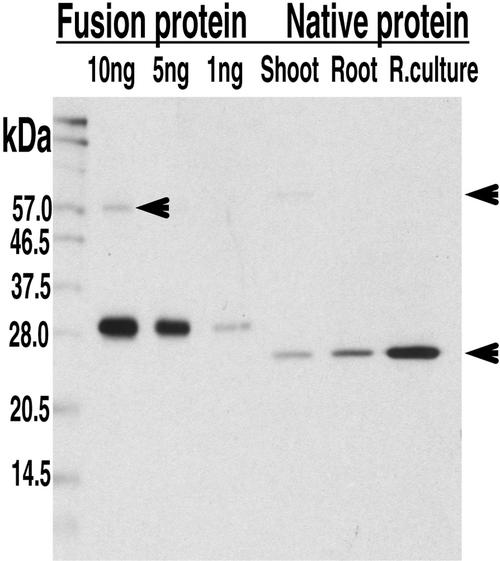
To determine where FQR1 is expressed in the plant, polyclonal antibodies were raised to the His-tagged FQR1 fusion protein. A 1:10,000 dilution of the resulting serum detects both the purified fusion protein and a single group of native proteins (Fig. 6). Because the antibody is polyclonal and two members of the FQR1 family in Arabidopsis that share 84% and 62% amino acid identity with FQR1 have masses only 0.5 and 0.9 kD larger than FQR1, we cannot rule out the possibility that the single band of native proteins recognized by the antibody includes these family members. The apparent masses of the fusion protein (about 30 kD) and the native protein (about 25 kD) are somewhat larger than their predicted masses (27 and 22 kD, respectively, based on the apoprotein with a single FMN). In some experiments, a protein with an estimated molecular mass of 62 kD is also apparent. Homodimerization or multimerization of proteins in this family has been frequently reported (Brock et al., 1995; Grandori et al., 1998; Uetz. et al., 2000), as has binding to both related family members and to proteins of distinct sequence (Yang et al., 1993; Uetz et al., 2000).
Figure 6.
Western blot showing that anti-FQR1 antibody recognizes both fusion and native proteins. Lanes 1, 2, and 3: 10, 5, and 1 ng of the FQR1 fusion protein, respectively; lanes 4, 5, and 6, 8 μg of total protein extract from wild-type shoots, roots, and root cultures. As expected, the molecular mass of the fusion protein is approximately 5 kD more than the native protein. The blot was incubated with a 1:10,000 dilution of immune serum. Incubation with preimmune serum did not reveal any bands. The lower arrow indicates the native protein. The positions of comparatively faint bands of larger molecular mass are indicated with the upper arrows.
The protein(s) recognized by the anti-FQR1 antibody is present in both roots and shoots. The higher levels of protein accumulation in root cultures as compared with roots, and in roots as compared with shoots are consistent with the patterns of FQR1 mRNA accumulation. We did not detect reliable changes in the level of FQR1 protein accumulation in response to auxin application. In two of four experiments, a slight increase was seen in the range from 4 to 12 h after a pulse of auxin was given to roots grown on agar plates, but these increases were not consistent (data not shown). These data may reflect an actual lack of auxin-induced FQR1 accumulation. Alternatively, they could reflect a modest increase in FQR1 accumulation against a relatively high background of constitutive expression on the part of related family members.
Overexpression of FQR1 Increases the Total QR Activity in Plant Extracts
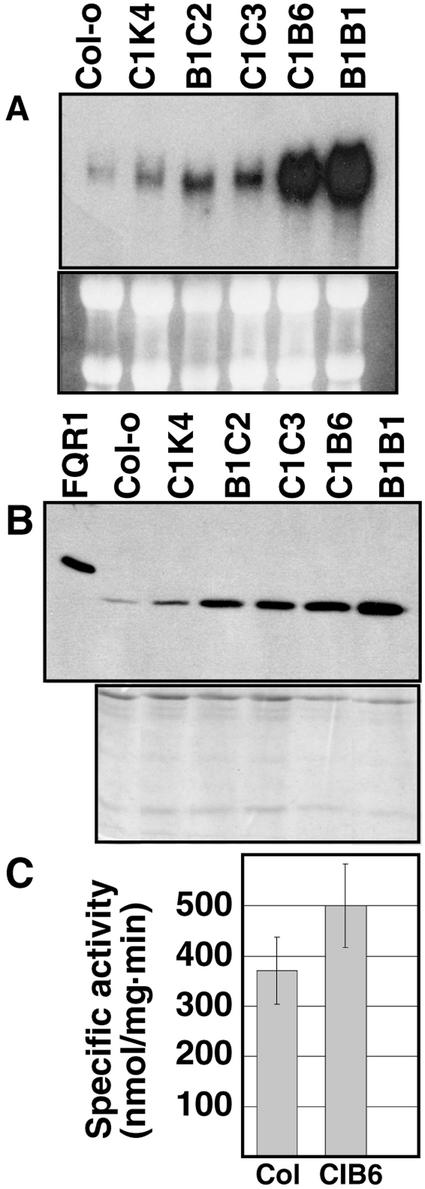
Arabidopsis plants were transformed with a cauliflower mosaic virus (CaMV) 35S::FQR1 construct, and five distinct lines that overexpress FQR1 were established. Northern-blot analysis confirmed that these plants have elevated levels of FQR1 mRNA (Fig. 7A). The transformed plants were grown in soil, and their phenotype was observed through several generations. We did not notice a morphological difference between soil-grown wild-type and transformed plants. Measurements of root length and number of lateral roots per centimeter, two features known to be regulated by auxin, were compared for IAA-treated and untreated seedlings in all of the lines tested. No correlation was observed between either of these features and the degree of FQR1 overexpression (data not shown).
Figure 7.
Levels of FQR1 mRNA, FQR1 protein, and QR activity in untreated plants transformed with CaMV35S::FQR1 as compared with wild type. All five of the transformed lines (C1K4, B1C2, C1C3, CIB6, and B1B1) carry the same construct. A, Northern blot probed with FQR1 cDNA. Samples of leaf and stem tissue were taken from 21-d-old plants. RNA (10 μg) was loaded per lane. rRNA bands shown below the northern blot served as loading controls. B, Western blot shows that FQR1 protein levels correlate with FQR1 mRNA abundance. Lane 1, 10 ng of His-tagged FQR1 purified from E. coli. Lane 2 to 7, 8 μg of protein extract from 2-week-old soil-grown plants. The Coomassie Brilliant Blue-stained gel, shown below the western blot, was loaded with equivalent amounts of the samples used in the western blot and served as a loading control. C, Specific activity of extracts from CIB6 transgenic plants (n = 5 isolations) is greater than that of wild-type (Col-0) plants (n = 6). The error bars represent sds. Results are statistically significant at P = 0.01.
These data raise the question as to whether the plants with elevated levels of FQR1 mRNA have elevated levels of FQR1 protein. Western-blot analysis indicates that the level of FQR1 protein accumulation in these lines does vary in accord with the relative amount of FQR1 mRNA, although the magnitude of the increase in protein levels is less than that of the mRNA (Fig. 7B). To determine whether the increase in FQR1 protein levels corresponds to an increase in QR activity, QR assays were run on extracts from wild-type and CIB6 plants. On average, the specific activity of extracts from seedlings of the CIB6 line was 34% greater than the specific activity of extracts from wild-type seedlings (Fig. 7C). These data suggest that production of FQR1 results in an increase in in planta QR activity.
DISCUSSION
FQR1 Is an FMN-Binding QR
The data presented here demonstrate that FQR1 is an FMN-binding QR. FQR1 belongs to a family of highly conserved flavin-binding reductases, the founding member of which is WrbA from E. coli. All of the studies of FQR1/WrbA family enzymes point to their playing a role in the detoxification of xenobiotics or oxidized metabolic products. The FQR1/WrbA family member isolated from the white rot fungus Phanerochaete chrysosporium is a 1,4-benzoquinone reductase (QR). This FMN-binding protein can be reduced by NADH and NADPH and transfers electrons to a variety of substrates. The most efficient electron acceptor identified is 2,6-dimethoxybenzoquinone (DMBQ), a compound commonly found in plants (Handa et al., 1983; Brock and Gold, 1996). Expression of the enzyme is induced by a variety of aromatic compounds, including 2-methoxybenzoquinone and vanillic acid, a product of lignin degradation (Akileswaran et al., 1999). Because P. chrysosporium actively degrades lignin, the fungus is likely to encounter elevated levels of oxidized metabolic products such as these. It is hypothesized that QR may play a role in protecting the fungus from toxic levels of quinones that may result from breakdown of the plant cell wall.
A second member of the FQR1/WrbA gene family, TvQR2, was isolated from the parasitic plant Triphysaria versicolor (Matvienko et al., 2001). In this case, it is hypothesized that the encoded enzyme is involved in detoxification of quinones, such as DMBQ, that are released by the roots of neighboring plants. This hypothesis is supported by the observation that DMBQ induces TvQR2 expression in both parasitic and nonparasitic plants (Matvienko et al., 2001).
A homologous gene in Schizosaccharomyces pombe was identified based on its ability to be regulated by Pap1, an AP-1 like transcription factor that is activated by oxidative stress (Toda et al., 1992). Treating S. pombe with 2 mm hydrogen peroxide for 6 h increases the amount of p25 protein in wild-type cells, although not in pap1− cells (Kudo et al., 1999). Overexpression of this protein is observed in a brefeldin A-resistant mutant. This indicates that p25 might play a role in protection against brefeldin A. If so, however, it does not act alone, because overexpression of the protein is not sufficient to confer brefeldin A resistance (Turi et al., 1994). The protein encoded by a similar gene in Saccharomyces cerevisiae (YDR032C) also shows induction with hydrogen peroxide (Godon et al., 1998). These data strongly suggest that p25 may serve as a cellular response to oxidative stress.
Members of the FQR1/WrbA Enzyme Family Are Distantly Related to Mammalian QRs
Although several members of the FQR1/WrbA enzyme family have QR activity, their amino acid sequences are quite different from the well known FAD-binding QRs, such as the mammalian NQO1. Initial studies of the sequence and predicted structure of WrbA family members lead to the proposal that members of this family were novel flavodoxins (Grandori and Carey, 1994): FMN-binding proteins that function in electron transport (for review, see Simondsen and Tollin, 1980). Subsequent studies substantiate the prediction that FQR1/WrbA family members bind FMN (Brock et al., 1995; Grandori et al., 1998). For FQR1, thin-layer chromatography analysis of the cofactor associated with the His-tagged protein demonstrates that, when expressed in E. coli, the flavin associated with FQR1 is FMN. Thus, both the primary sequence and cofactor bound distinguish the WrbA family members from the mammalian QRs. Given this low degree of conservation, it is perhaps surprising that enzymes in the FQR1 family have QR activity. This apparent anomaly can be resolved by considering the three-dimensional structures of the enzymes. The catalytic domain of mammalian QR1 is more similar to the FMN-containing flavodoxin from Clostridium MP, a close relative of FQR1, than to other FAD-containing enzymes (for review, see Bianchet et al., 1999). Thus, FQR1 belongs to a family of proteins that share amino acid similarity with flavodoxins and similar three-dimensional folds with QRs. Because all of the members of the WrbA family that have been biochemically characterized are known to bind FMN and have QR activity, we suggest that the family be renamed the flavodoxin-like QRs to better reflect the activities of these proteins.
Mammalian QR Protects against Chemotoxicity
QRs have been extensively studied in mammalian systems. NQO1 is an FAD-binding cytosolic enzyme that carries out obligate two-electron reductions of quinones. This enzyme plays a role in protection against oxidative stress and xenobiotic toxicity (for review, see Ross et al., 2000). This protection may be carried out through the reduction of specific substrates, such as coenzyme Q (Beyer et al., 1996), or more generally by carrying out the two-electron reduction of a quinone to a hydroquinone. This later reduction effectively prevents the one-electron reduction of a quinone to a semiquinone. Semiquinones are potentially damaging because they may pass a single electron to oxygen, thereby generating highly destructive reactive oxygen species (for review, see Ross, 1997). Loss of QR function is not lethal. NQO1 knockout mice are viable but have an increased NADH/NAD ratio and fail to gain weight at the rate that wild-type mice do (Gaikwad et al., 2001). The observation that NQO1 knockouts are viable is mirrored by work with members of the FQR1/WrbA family. Knockouts of FQR1/WrbA family members from E. coli, S. pombe, and S. cerevisiae are viable, and, when grown in standard laboratory conditions, result in organisms that are morphologically indistinguishable from wild type (Toda et al., 1992; Yang et al., 1993; Winzeler et al., 1999).
FQR1 Is a Novel Primary Auxin-Response Gene
Accumulation of FQR1 mRNA is one of the fastest known auxin responses. Significantly enhanced levels of FQR1 mRNA are observed within 10 min of IAA application, the earliest time tested. Half-maximal induction is reached about 20 min after auxin application and steady state is reached in 30 min. These times are directly comparable to those of the three fastest members of the PS-IAA4/5 family in Arabidopsis. Accumulation of IAA1, IAA2, and IAA5 mRNA begins 6 to 8 min after auxin application and reaches a maximal, steady-state level approximately 30 min after application (Abel et al., 1995). Transcriptional activation of the SAUR genes is only slightly faster. Increased levels of SAUR mRNAs are first detectable 2.5 to 5 min after the application of 50 μm 2,4-dichlorophenoxyacetic acid, half-maximal induction is reached 10 min after application, and the steady state is reached at 60 min (McClure and Guilfoyle, 1987).
As described in “Results,” FQR1 mRNA accumulation is increased by treatment with auxin in the presence of the protein synthesis inhibitor cycloheximide and by cycloheximide alone. Cycloheximide inducibility is a common feature of many of the early auxin-regulated genes and can be interpreted as evidence in favor of the existence of a short-lived transcriptional repressor (for review, see Abel and Theologis, 1996). If such a repressor is functioning in this case, it presumably binds to a site other than the consensus AuxRE, because a TGTCTC-type AuxRE was not observed in the putative promoter region. DMBQ inducibility of TvQR2, the T. versicolor homolog, is also a primary response. In that case, however, a relatively short (40 min) treatment with cycloheximide alone did not suffice to induce the gene (Matvienko et al., 2001).
Although FQR1 is clearly a primary auxin-response gene, it is not clear that the induction of FQR1 is specific to auxin. The apparent lack of an AuxRE combined with the presence of a putative stress-responsive element suggests that the induction of FQR1 could be mediated as a cellular stress response. Although the experiments here did not identify effective inducers other than cycloheximide and auxin, it should be noted that only a limited number of agents were tried, and thus, no general conclusion about the nature of the induction can be reached. It is interesting to note that mRNA corresponding to the mammalian QR NQO1 accumulates in response to treatment with polycyclic aromatic hydrocarbons, as well as oxidants such as hydrogen peroxide, and hypoxia (Ross, 1997). This induction is mediated by cis-acting AREs that have consensus elements similar to those found in the putative promoter of FQR1 (Rushmore et al., 1991).
CONCLUSIONS
FQR1 is a novel primary auxin-response gene that encodes a protein with QR activity. Although the QR activity of FQR1 suggests a possible role in protecting against oxidative stress, the in vivo function of FQR1 is not yet known. Identification of its in vivo reaction partners is an exciting avenue for future research. It is intriguing that QRs and GSTs, plant enzymes with functions that match two of the many mammalian phase II enzymes, are auxin responsive, and it will be interesting to learn whether any other plant homologs of the mammalian phase II enzymes are also auxin responsive.
MATERIALS AND METHODS
Plant Tissue
Seeds of Arabidopsis ecotype Columbia-0 were surface sterilized, vernalized in the dark for 1 week at 4°C, mixed with 0.1% agarose, and plated on 0.8% agar containing Murashige and Skoog salts, vitamins, and 3% Suc. Plates were wrapped with Parafilm (American National Can, Chicago), vertically oriented, and placed in a 22°C chamber. Plates containing 7-d-old plants were placed horizontally and flooded for 15 min with approximately 25 mL of sterile water with or without IAA. For differential display, plants were rinsed with fresh sterile water once after treatment. Roots cut just below the hypocotyl junction under sterile conditions were frozen in liquid nitrogen.
When plants were grown for differential display, the agar plates also contained 0.1 mg L−1 ampicillin and 0.01 mg L−1 benomyl, a fungicide. To promote root growth, the plates were kept dark for the first 3 d. To increase the probability of identifying an auxin-regulated gene involved in meristem formation, the terminal 0.5 cm (approximate) of the root was cut off and left out of the sample.
To generate conveniently large quantities of tissue for use in northern-blot analysis, Arabidopsis roots were also grown in liquid culture as described by Williams and Sussex (1995). Each compound under study was added to the liquid culture, resulting in continuous stimulation rather than a pulse.
Plants for the wounding experiment were grown in soil for 14 d. To wound the plants, one-half of each of the largest four leaves was cut away with scissors. If the plant did not have four visible leaves, three were used instead. Shoot samples were harvested 6 h after the leaves were cut.
Hormone Preparation
Hormones were prepared as 3 mm stocks and filter sterilized before use. The auxin stock was prepared by dissolving 0.026 g of IAA in 0.5 mL of ethanol, which was then added a drop at a time to 49.5 mL of water. Cytokinin (BAP) was dissolved in 0.5 mL of 1 m NaOH and diluted to 25 mL with water.
Differential Display
RNA was isolated in Kirby buffer according to the procedure of Leutwiler et al. (1986). For differential display, a subsequent 30-min treatment with RNase-free DNase I, a phenol:chloroform extraction, and an ethanol precipitation were performed. Differential display was carried out as described by Liang and Pardee (1992) with modifications. Total RNA (0.4 μg) from detipped roots was mixed with 40 pmol of oligo(dT) primer, left at 70°C for 10 min, and then placed on ice. Reverse transcription proceeded for 1 h at 37°C with Superscript enzyme and reagents suggested by the manufacturer (Invitrogen, Carlsbad, CA) in a final volume of 40 μL. PCR reactions contained 2 μL of the cDNA reaction, 20 pmol of anchor primer (T12GC) primer, 10 pmol of arbitrarily chosen 10-mer primer, 2 μm deoxyribonucleotide triphosphates, 0.5 μL of Taq polymerase, and 10 μCi of 35S in buffer (final buffer concentrations: 50 mm KCl, 10 mm Tris-HCl, pH 8.3, 1.5 mm MgCl2, 0.001% [w/v] gelatin). PCR was carried out as described by Liang and Pardee (1992) with an annealing temperature of 40°C (2 min at 94°C, followed by 40 cycles of 30 s at 94°C, 1 min at 40°C, 30 s at 72°C, followed by 5 min at 72°C). Reactions were run on 5% polyacrylamide sequencing gels until the xylene cyanol dye marker reached the end of the gel to retain DNA fragments larger than about 125 bp. After PCR amplification, cDNA fragments from the bands of interest were cloned using the TA cloning kit (Invitrogen, Carlsbad, CA).
Northern Analysis
The cloned PCR-amplified fragment of FQR1 served as the template for the probes used in northern-blot analysis. Some experiments were carried out with the Illuminator nonradioactive detection kit (Stratagene, La Jolla, CA) according to manufacturer's directions. Results from these experiments were confirmed with 32P-labeled random-primed probes. RNA was fractionated on a 1.2 or 1.5% formaldehyde gel, and inspection of the ethidium bromide-stained RNA indicated that equal amounts were loaded in each well. Gels were transferred overnight to a Zetaprobe membrane (Bio-Rad, Hercules, CA) following the manufacturer's protocol for alkaline transfer. After transfer, gels were examined on a UV light box to make sure that no ethidium bromide-stained products remained. Baked membranes were prehybridized (0.25 m sodium phosphate, pH 7.5, 7% SDS, 1% bovine serum albumin, 1 mm EDTA, pH 8) at 65°C for at least 1 h. Following overnight hybridization, blots were washed at 65°C for 1 h in 1× SSC plus 0.5% SDS and then for an additional 1 h in 0.5× SSC plus 0.5% SDS. Results were visualized either via exposure to autoradiographic film or with a PhosphorImager. Specific numbers for fold-induction by IAA were calculated by dividing the PhosphorImager-determined density of the induced band by the density of the control.
Screening
The λPRL-2 cDNA library was constructed by Thomas Newman from a mixture of shoots and cultured roots and was supplied by the Arabidopsis Biological Resource Center at Ohio State University (Columbus, OH). Plaques were transferred to a noncharged nylon membrane, and the baked blots were subjected to hybridization according to the directions supplied with the Illuminator nonradioactive detection kit (Stratagene).
Sequencing
Sequencing reactions were carried out with the Sequenase, version 2.0, kit (United States Biochemical, Cleveland) according to manufacturer's directions. Custom primers were obtained from the DNA-sequencing facility at the University of California (Berkeley).
Transgenic Plants
A CaMV35S::FQR1 construct was made by digesting FQR1 with XhoI and BamHI and cloning it into the 4K1 shuttle vector (Peralta et al., 1986). This plasmid was subsequently digested with EcoRI and HindIII, cloned into the SLJ7292 binary vector (Jones et al., 1992), and introduced into Agrobacterium tumefaciens via a triparental mating. Arabidopsis plants were transformed via vacuum infiltration, and T1 seed was grown on kanamycin. Resistant plants were selected, transferred to soil, and allowed to self. Seeds of these T2 plants were grown on kanamycin, and those that gave rise to 100% kanamycin-resistant progeny were selected and served as the founders for the lines used in all subsequent experiments. The CIB6 line is available from Arabidopsis Biological Resource Center (stock no. CS3902).
To compare the morphology of 35S::FQR1 plants to wild-type plants, seeds of each type were cold treated for 1 week at 4°C, and then grown to maturity in soil at 23°C (16 h light/8 h dark). Roots of 7-d-old seedlings grown on sterile agar plates were assessed via manual measurements of root length. The number of lateral roots emerging from the epidermis was determined using a dissecting microscope.
Antibody Production
The entire open reading frame of the FQR1 cDNA was excised with NcoI and BamHI, and the resulting fragment was ligated into the pET-30a (+) vector (Novagen, Madison, WI) so as to add an N-terminal extension encoding a 4,872 D peptide including a 6 His tag, thrombin, an S Tag, and an enterokinase cleavage site. DH5α cells and, subsequently, BL21 (DE3) cells were transformed with this plasmid. FQR1 was purified by inducing expression in BL21 cells with 0.4 mm IPTG for 3 h, 20 min and then by following protein purification steps outlined in the pET system manual (Novagen). Eluted fractions were dialyzed overnight against phosphate-buffered saline. Antibody production was performed by Millbrook Farm (Amherst, MA). The first injection consisted of 230 μg of purified His-tagged protein mixed 1:1 with Freund's adjuvent; subsequent injections included the same amount of protein mixed with incomplete Freund's adjuvant.
Western Blots
Plant tissue was ground with a mortar and pestle in liquid nitrogen. Grinding buffer (3 mL of 0.1 m Tris-HCl, pH 8.0, 0.01 m MgCl2, 40 mm β-mercaptoethanol, and 18% Suc) was added per gram fresh weight of tissue. Ground tissue was filtered through cheesecloth and spun at 10,000 rpm in a microfuge for 5 min at 4°C. The protein concentration in the supernatant was determined using the Sigma Micro Protein Determination Kit (Sigma Diagnostics, St. Louis). Samples were heated at 100°C for 4 to 5 min, and 8 μg of protein from each plant extract was loaded on a 12% acrylamide gel with a 4% stacking gel. After transfer to a supported nitrocellulose membrane, the membrane was blocked overnight at 4°C in TTBS (20 mm Tris, pH 7.6, 137 mm NaCl) plus 0.1% (v/v) Tween 20 and 5% (w/v) dry milk, washed three times in TTBS, and incubated with a 1:10,000 dilution of serum. After an additional three washes in TTBS, detection was performed using LumiGLO (New England Biolabs, Beverly, MA) following the manufacturer's directions.
Spectrophotometric Measurements
Cold TCA was added to FQR1 purified from Escherichia coli to a final concentration of 5% (w/v) TCA. The mixture was left on ice in the dark for 10 to 30 min and then centrifuged for 5 min at room temperature. Absorbance measurements of the supernatant were made in a LambdaBio20 spectrophotometer (PerkinElmer, Shelton, CT).
Electron Transport Assays
FQR1 fusion protein was freshly prepared on a column with an Ni2+-charged agarose matrix, and the concentration of individual fractions was determined by the Lowry method (Protein Assay Kit, Sigma). Protein preparations were either used immediately or after overnight dialysis against 50 mm Tris buffer. Purified protein, NAD(P)H, and electron acceptors were added to a cuvette such that the final concentration of all components was 50 μm after the last addition was made. Absorbance spectra were taken immediately after each new addition.
QR Assay
E. coli transformed with the FQR1-coding expression vector were grown to an A600 of 0.8 to 1.2, at which time one-half of the cultures were treated with IPTG (400 μm final concentration) for 3 to 4 h. Cells were chilled on ice for 10 min and centrifuged at 3,500 rpm for 12 min. After freezing, the resulting pellets were resuspended in 50 mm Tris-HCl, pH 8, sonicated, and centrifuged at 13,500 rpm in a cold microfuge for 15 min. The resulting supernatant was diluted to varying concentrations (usually 1:20) in 50 mm Tris-HCl and used in the assay as described by Prochaska and Santamaria (1988). A 22-mL reaction mixture was generated with 1.1 mL of 0.5 m Tris-HCl, pH 7.4, 14.3 mg of bovine serum albumin, 146.7 μL of 1.5% Tween 20, 14.7 μL of 7.5 mm FAD, 146.7 μL of 150 mm Glc 6-phosphate, 13.2 μL of 50 mm NADP, and 6.6 mg of MTT. Each sample was mixed with 800 μL of the reaction mixture and 2 μL of Glc 6-phosphate dehydrogenase, and the total volume was increased to 995 μL with water. To initiate the reaction, 5 μL of 10 mm menadione was added. The absorption of the reaction mix at 610 nm was measured continuously over a period of 10 min.
For the QR assays on plant extracts, seeds were grown on vertically oriented Murashige and Skoog plates with 1.5% (w/v) Suc for 11 d in a 22°C to 24°C growth chamber, with 16 h of 60 μE m−2 s−1 light from fluorescent and incandescent bulbs. Entire seedlings were collected, frozen, and ground with about 530 μL of extraction buffer (100 mm Tris acetate, pH 8, 100 mm potassium acetate, pH 8, 2 mm EDTA, 5 mm DTT, 250 mm sodium ascorbate, 10% [v/v] glycerol) per g fresh weight of tissue. This extract was centrifuged in a microfuge for 8 min at full speed. All procedures were carried out in the cold. Supernatants were diluted 1:40 or 1:45 and added to the QR assay mixture described above lacking menadione. These samples were incubated at room temperature for about 35 min to allow the endogenous, QR-independent reduction of MTT to formazan dye to subside. After confirming that the menadione-independent activity in each sample had reached minimal levels, menadione was added and formazan dye formation was recorded over a period of 10 min. Protein concentrations for each sample were calculated as the average concentration obtained from three Bradford assays prepared using Bio-Rad reagent.
ACKNOWLEDGMENTS
We thank Winslow R. Briggs for helpful comments, Mary E. Williams for technical advice, and Debbie Shen for help screening the cDNA library.
Footnotes
This work was supported in part by the National Science Foundation (grant no. IBN–920597/DIR–9104374). Support for K.A.D. was provided by Williams College and a Molecular and Cellular Biology Training Grant Award, University of California-Davis. Support for M.A.G. was provided by a Howard Hughes Medical Institute grant to Williams College.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010581.
LITERATURE CITED
- Abel S, Nguyen MD, Theologis A. The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol. 1995;251:533–549. doi: 10.1006/jmbi.1995.0454. [DOI] [PubMed] [Google Scholar]
- Abel S, Theologis A. Early genes and auxin action. Plant Physiol. 1996;111:9–17. doi: 10.1104/pp.111.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akileswaran L, Brock BJ, Cereghino JI, Gold MH. 1,4-Benzoquinone reductase from Phanerochaete chrysosporium: cDNA cloning and regulation of expression. Appl Environ Microbiol. 1999;65:415–421. doi: 10.1128/aem.65.2.415-421.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer RE, Segura-Aguilar J, DiBernaardo S, Cavazzoni M, Fato R, Fiorentini D, Galli MC, Setti M, Landi L, Lenaz G. The role of DT-diaphorase in the maintenance of the reduced antioxidant form of coenzyme Q in the membrane systems. Proc Natl Acad Sci USA. 1996;93:2528–2532. doi: 10.1073/pnas.93.6.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchet MA, Foster C, Faig M, Talalay P, Amzel LM. Structure and mechanism of cytosolic quinone reductases. Biochem Soc Trans. 1999;27:610–615. doi: 10.1042/bst0270610. [DOI] [PubMed] [Google Scholar]
- Blakely LM, Blakely RM, Colowit PM, Elliott DS. Experimental studies on lateral root formation in radish seedling roots: II. Analysis of the dose-response to exogenous auxin. Plant Physiol. 1988;87:414–419. doi: 10.1104/pp.87.2.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock BJ, Gold MH. 1,4-Benzoquinone reductase from the basidiomycete Phanerochaete chrysosporium: spectral and kinetic analysis. Arch Biochem Biophys. 1996;331:31–40. doi: 10.1006/abbi.1996.0279. [DOI] [PubMed] [Google Scholar]
- Brock BJ, Rieble S, Gold MH. Purification and characterization of a 1,4-benzoquinone reductase from the basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 1995;61:3076–3081. doi: 10.1128/aem.61.8.3076-3081.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Singh KB. The auxin, hydrogen peroxide and salicylic acid induced expression of the Arabidopsis GST6 promoter is mediated in part by an ocs element. Plant J. 1999;19:667–677. doi: 10.1046/j.1365-313x.1999.00560.x. [DOI] [PubMed] [Google Scholar]
- Dhakshinamoorthy S, Long DJ, II, Jaiswal AK. Antioxidant regulation of genes encoding enzymes that detoxify xenobiotics and carcinogens. Curr Top Cell Regul. 2000;36:201–216. doi: 10.1016/s0070-2137(01)80009-1. [DOI] [PubMed] [Google Scholar]
- Edwards R, Dixon DP, Walbot V. Plant glutathione S-transferases: enzymes with multiple functions in sickness and in health. Trends Pharmacol Sci. 2000;5:193–198. doi: 10.1016/s1360-1385(00)01601-0. [DOI] [PubMed] [Google Scholar]
- Franco AR, Gee MA, Guilfoyle TJ. Induction and superinduction of auxin-responsive messenger RNA with auxin and protein synthesis inhibitors. J Biol Chem. 1990;265:15845–15849. [PubMed] [Google Scholar]
- Gaikwad A, Long DJ, II, Stringer JL, Jaiswal AK. In vivo role of NAD(P)H:quinone oxidoreductase 1 (NQO1) in the regulation of intracellular redox state and accumulation of abdominal adipose tissue. J Biol Chem. 2001;276:22559–22564. doi: 10.1074/jbc.M101053200. [DOI] [PubMed] [Google Scholar]
- Ghisla S. Fluorescence and optical characteristics of reduced flavins and flavoproteins. Methods Enzymol. 1980;66:360–373. doi: 10.1016/0076-6879(80)66481-7. [DOI] [PubMed] [Google Scholar]
- Godon C, Liagniel G, Lee J, Buhler J-M, Kieffer S, Perrot M, Boucherie H, Toledano MB, Labarre J. The H2O2 stimulation in Saccharomyces cerevisiae. J Biol Chem. 1998;273:22480–22489. doi: 10.1074/jbc.273.35.22480. [DOI] [PubMed] [Google Scholar]
- Grandori R, Carey J. Six new candidate members of the alpha/beta twisted open-sheet family detected by sequence similarity to flavodoxin. Protein Sci. 1994;3:2185–2193. doi: 10.1002/pro.5560031204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori R, Khalifah P, Boice JA, Fairman R, Giovanielli K, Carey J. Biochemical characterization of WrbA, founding member of a new family of multimeric flavodoxin-like proteins. J Biol Chem. 1998;273:20960–20966. doi: 10.1074/jbc.273.33.20960. [DOI] [PubMed] [Google Scholar]
- Guilfoyle T, Hagen G, Ulmasov T, Murfett J. How does auxin turn on genes? Plant Physiol. 1998;118:341–347. doi: 10.1104/pp.118.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa SS, Kinghorn AG, Cordell GA, Farnsworth NR. Plant anticancer agents. XXVI. Constituents of Peddies Fischeri. J Nat Prod. 1983;46:248–250. doi: 10.1021/np50026a020. [DOI] [PubMed] [Google Scholar]
- Jones JDG, Shlumukov L, Carland F, English J, Scofield SR, Bishop GJ, Harrison K. Effective vectors for transformation, expression of heterologous genes, and assaying transposon excision in transgenic plants. Transgen Res. 1992;1:285–297. doi: 10.1007/BF02525170. [DOI] [PubMed] [Google Scholar]
- Josephy DP, with Mannervik B, de Montellano PO. Molecular Toxicology. Oxford: Oxford University Press; 1997. [Google Scholar]
- Kudo N, Taoka H, Toda T, Yoshida M, Horinouchi S. A novel nuclear export signal sensitive to oxidative stress in the fission yeast transcription factor Pap1. J Biol Chem. 1999;274:15151–15158. doi: 10.1074/jbc.274.21.15151. [DOI] [PubMed] [Google Scholar]
- Laskowski MJ, Williams ME, Nusbaum HC, Sussex IM. Formation of lateral root meristems is a two-stage process. Development. 1995;121:3303–3310. doi: 10.1242/dev.121.10.3303. [DOI] [PubMed] [Google Scholar]
- Leutwiler LS, Meyerowitz EM, Tobin EM. Structure and expression of three light-harvesting chlorophyll a/b-binding protein genes in Arabidopsis thaliana. Nucleic Acids Res. 1986;14:4051–4064. doi: 10.1093/nar/14.10.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- Marrs KA. The functions and regulation of glutathione S-transferases in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:127–158. doi: 10.1146/annurev.arplant.47.1.127. [DOI] [PubMed] [Google Scholar]
- Matvienko M, Wojtowicz A, Wrobel R, Jamison D, Goldwasser Y, Yoder JI. Quinone oxidoreductase message levels are differentially regulated in parasitic and non-parasitic plants exposed to allelopathic quinones. Plant J. 2001;25:375–387. doi: 10.1046/j.1365-313x.2001.00971.x. [DOI] [PubMed] [Google Scholar]
- McClure B, Guilfoyle T. Characterization of a class of small auxin-inducible soybean polyadenylated RNAs. Plant Mol Biol. 1987;9:611–623. doi: 10.1007/BF00020537. [DOI] [PubMed] [Google Scholar]
- Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed J. AXR2encodes a member of the Aux/IAA protein family. Plant Physiol. 2000;123:563–574. doi: 10.1104/pp.123.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman T, DeBruijn FJ, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M et al. Genes galore: a summary of methods for accessing results from large-scale partial sequencing of anonymous ArabidopsiscDNA clones. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta EG, Hellmiss R, Ream W. Overdrive, a T-DNA transmission enhancer on the A. tumefascienstumor-inducing plasmid. EMBO J. 1986;5:1137–1142. doi: 10.1002/j.1460-2075.1986.tb04338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochaska HJ, Santamaria AB. Direct measurement of NAD(P)H:quinone reductase from cells cultured in microtiter wells: a screening assay for anticarcinogenic enzyme inducers. Anal Biochem. 1988;169:328–336. doi: 10.1016/0003-2697(88)90292-8. [DOI] [PubMed] [Google Scholar]
- Rogg LE, Lasswell J, Bartel B. A gain-of-function mutation in IAA28suppresses lateral root development. Plant Cell. 2001;13:465–480. doi: 10.1105/tpc.13.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross D. Quinone reductases. In: Guengrich FP, editor. Comprehensive Toxicology. Vol. 3. New York: Pergamon; 1997. pp. 179–197. [Google Scholar]
- Ross D, Kepa JK, Winski SL, Beall HD, Anwar A, Siegel D. NAD(P)H:quinone oxidoreductase 1 (NQO1): chemoprotection, bioactivation, gene regulation, and genetic polymorphisms. Chem Biol Interact. 2000;129:77–97. doi: 10.1016/s0009-2797(00)00199-x. [DOI] [PubMed] [Google Scholar]
- Rushmore TH, Morton MR, Pickett CB. The antioxidant responsive element: activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J Biol Chem. 1991;266:11632–11639. [PubMed] [Google Scholar]
- Rushmore TH, Pickett CB. Glutathione S-transferases, structure, regulation, and therapeutic implications. J Biol Chem. 1993;268:11475–11478. [PubMed] [Google Scholar]
- Santangelo JD, Jones DT, Woods DR. Metronidazole activation and isolation of Clostridium acetobutylicumelectron transport genes. J Bacteriol. 1991;173:1088–1095. doi: 10.1128/jb.173.3.1088-1095.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simondsen RP, Tollin G. Structure-function relations in flavodoxins. Mol Cell Biochem. 1980;33:13–24. doi: 10.1007/BF00224568. [DOI] [PubMed] [Google Scholar]
- Theologis A, Huynh TV, Davis RW. Rapid induction of specific mRNAs by auxin in pea epicotyl tissue. J Mol Biol. 1985;183:53–68. doi: 10.1016/0022-2836(85)90280-3. [DOI] [PubMed] [Google Scholar]
- Toda T, Shimanuki M, Saka Y, Yamano H, Adachi Y, Shirakawa M, Kyogoku Y, Yanagida M. Fission yeast pap1-dependent transcription is negatively regulated by an essential nuclear protein, crm1. Mol Cell Biol. 1992;12:5474–5484. doi: 10.1128/mcb.12.12.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turi TG, Webster P, Rose JK. Brefeldin A sensitivity and resistance in Schizosaccharomyces pombe: isolation of multiple genes conferring resistance. J Biol Chem. 1994;269:24229–24236. [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield T, Judson R, Knoght J, Lockshon D, Narayan V, Srinivasan M, Pochart P et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle T. The ocs element in the soybean GH2/4promoter is activated by both active and inactive auxin and salicylic acid analogues. Plant Mol Biol. 1994;26:1055–1064. doi: 10.1007/BF00040688. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Liu ZB, Hagen G, Guilfoyle TJ. Composite structure of auxin response elements. Plant Cell. 1995;7:1611–1623. doi: 10.1105/tpc.7.10.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zaal EJ, Memelink J, Mennes AM, Quint A, Libbenga KR. Auxin-induced mRNA species in tobacco cell cultures. Plant Mol Biol. 1987;10:145–157. doi: 10.1007/BF00016152. [DOI] [PubMed] [Google Scholar]
- Williams ME, Sussex I. Developmental regulation of ribosomal protein L16 genes in Arabidopsis thaliana. Plant J. 1995;8:65–76. doi: 10.1046/j.1365-313x.1995.08010065.x. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H et al. Functional characterization of the S. cerevisiaegenome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Xie Q, Frugis G, Colgan D, Chua N. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000;14:3024–3036. doi: 10.1101/gad.852200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Ni L, Somerville RL. A stationary-phase protein of Escherichia colithat affects the mode of association between the trp repressor protein and operator-bearing DNA. Proc Natl Acad Sci USA. 1993;90:5796–5800. doi: 10.1073/pnas.90.12.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]