Abstract
Seed oils of a number of Asteraceae and Euphorbiaceae species are enriched in 12-epoxyoctadeca-cis-9-enoic acid (vernolic acid), an unusual 18-carbon Δ12-epoxy fatty acid with potential industrial value. It has been previously demonstrated that the epoxy group of vernolic acid is synthesized by the activity of a Δ12-oleic acid desaturase-like enzyme in seeds of the Asteraceae Crepis palaestina and Vernonia galamensis. In contrast, results from metabolic studies have suggested the involvement of a cytochrome P450 enzyme in vernolic acid synthesis in seeds of the Euphorbiaceae species Euphorbia lagascae. To clarify the biosynthetic origin of vernolic acid in E. lagascae seed, an expressed sequence tag analysis was conducted. Among 1,006 randomly sequenced cDNAs from developing E. lagascae seeds, two identical expressed sequence tags were identified that encode a cytochrome P450 enzyme classified as CYP726A1. Consistent with the seed-specific occurrence of vernolic acid in E. lagascae, mRNA corresponding to the CYP726A1 gene was abundant in developing seeds, but was not detected in leaves. In addition, expression of the E. lagascae CYP726A1 cDNA in Saccharomyces cerevisiae was accompanied by production of vernolic acid in cultures supplied with linoleic acid and an epoxy fatty acid tentatively identified as 12-epoxyoctadeca-9,15-dienoic acid (12-epoxy-18:2Δ9,15) in cultures supplied with α-linolenic acid. Consistent with this, expression of CYP726A1 in transgenic tobacco (Nicotiana tabacum) callus or somatic soybean (Glycine max) embryos resulted in the accumulation of vernolic acid and 12-epoxy-18:2Δ9,15. Overall, these results conclusively demonstrate that Asteraceae species and the Euphorbiaceae E. lagascae have evolved structurally unrelated enzymes to generate the Δ12-epoxy group of vernolic acid.
12-Epoxyoctadeca-cis-9-enoic acid (vernolic acid) is a C18 fatty acid that is structurally distinct from other plant fatty acids by the presence of an epoxy group between its Δ12 and Δ13 carbon atoms. This unusual fatty acid is enriched in the seed oils of several Asteraceae genera, including Stokesia, Vernonia, and Crepis (Gunstone, 1954; Badami and Patil, 1981). Vernolic acid is also a major component of the seed oils of certain Euphorbiaceae species such as Euphorbia lagascae (Kleiman et al., 1965) and Bernardia pulchella (Spitzer et al., 1996). In the seed oils of these plants, vernolic acid can compose 50% to 90% (w/w) of the total fatty acids.
Vegetable oils that contain vernolic acid have a number of potential industrial applications because of the unique chemical properties associated with the Δ12-epoxy group. Vernolic acid-enriched seed oils, for example, can be used as plasticizers of polyvinyl chloride, a market that is currently served by petroleum-derived compounds such as phthalates and to a lesser extent by chemically epoxidized soybean and linseed oil (Perdue et al., 1986; Budziszewski et al., 1996). In addition, the ability of the epoxy group to crosslink makes vernolic acid-containing oils useful in adhesives and coating materials such as paint (Perdue et al., 1986). Furthermore, vernolic acid can be used as a precursor of monomeric components of nylon-11 and nylon-12 (Ayorinde et al., 1989, 1997).
The epoxy group of vernolic acid has been shown to result from the insertion of an oxygen atom at the Δ12 double bond of linoleic acid bound to phosphatidylcholine (PC) in seeds of E. lagascae and Vernonia galamensis (Bafor et al., 1993; Liu et al., 1998). However, previous studies have indicated that this activity is catalyzed by divergent classes of enzymes in seeds of E. lagascae and Asteraceae species that accumulate vernolic acid (Bafor et al., 1993; Lee et al., 1998). In the case of E. lagascae seed, metabolic studies have suggested that a cytochrome P450-type enzyme is involved in the formation of the epoxy group of vernolic acid (Bafor et al., 1993). This conclusion is supported by the ability of carbon monoxide to strongly inhibit epoxygenase activity in microsomes from E. lagascae seed (Bafor et al., 1993). This activity is also partially inhibited by cytochrome P450 reductase antibodies (Bafor et al., 1993). In contrast, studies with microsomes from seeds of the Asteraceae species Crepis palaestina have suggested that the epoxy group of vernolic acid is formed by a fatty acid desaturase-type enzyme rather than by a cytochrome P450 (Lee et al., 1998). In seed microsomes of this plant, epoxygenase activity is inhibited by cyanide, but is relatively insensitive to carbon monoxide and cytochrome P450 reductase antibodies (Lee et al., 1998). The involvement of a desaturase-type enzyme in vernolic acid synthesis in Asteraceae species was confirmed by the identification of cDNAs for Δ12-oleic acid desaturase (FAD2)-related enzymes from seeds of C. palaestina and V. galamensis (Hitz, 1998; Lee et al., 1998). Expression of these cDNAs in transgenic plants resulted in the accumulation of vernolic acid (Hitz, 1998; Lee et al., 1998).
The role of a cytochrome P450 epoxygenase in vernolic acid synthesis in E. lagascae seed has yet to be conclusively demonstrated by the identification and transgenic expression of a corresponding cDNA. In this study, we have reexamined the biosynthetic origin of vernolic acid in E. lagascae by analysis of expressed sequence tags (ESTs) from developing seeds of this plant. Using this strategy, we have identified a cDNA for a cytochrome P450 enzyme that generates Δ12-epoxy fatty acids, including vernolic acid, when expressed in yeast (Saccharomyces cerevisiae) and transgenic plant cells.
RESULTS
Identification of a Novel Cytochrome P450 cDNA from E. lagascae Seed ESTs
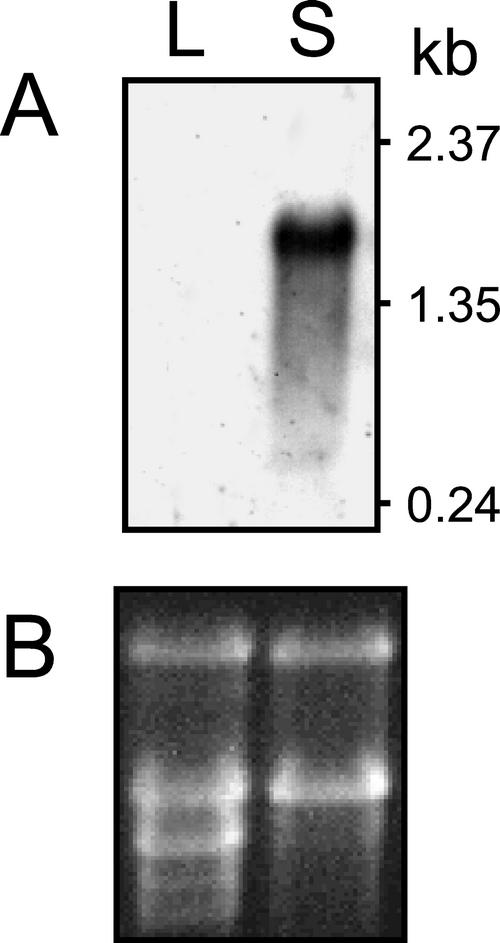
An EST approach was undertaken to determine the genetic basis for the biosynthesis of the Δ12-epoxy group of vernolic acid in E. lagascae seeds. Nucleotide sequences were obtained from the 5′ ends of 1,006 cDNAs that were chosen randomly from a library derived from developing E. lagascae seed. Based on the demonstrated pathway of vernolic acid synthesis in Asteraceae seeds (Hitz, 1998; Lee et al., 1998) and the proposed pathway in E. lagascae seeds (Bafor et al., 1993), homology comparisons of ESTs focused on those encoding polypeptides related to Δ12-oleic acid desaturases (FAD2) and cytochrome P450 enzymes. The pool of ESTs included one partial cDNA for a FAD2-type enzyme that was most similar to Δ12-desaturases rather than to functionally divergent forms of FAD2 such as fatty acid hydroxylases and epoxygenases. In addition, four ESTs were identified with homology to the cytochrome P450 superfamily. Two of these ESTs contained identical nucleotide sequences and encoded full-length polypeptides (GenBank accession no. AF406732). The enzyme encoded by this cDNA class was designated CYP726A1 by the cytochrome P450 nomenclature committee (Dr. David R. Nelson, University of Tennessee Health Science Center, Memphis; http://drnelson.utmem.edu/CytochromeP450.html) based on amino acid sequence properties described below. Northern analysis indicated that mRNA for the CYP726A1 gene is abundant in developing seeds of E. lagascae but is not detectable in leaves (Fig. 1). This expression profile is consistent with the seed-specific occurrence of vernolic acid in E. lagascae plants (E.B. Cahoon, unpublished data). Based on this, the two ESTs corresponding to the CYP726A1 cDNA were chosen for further characterization, and no additional studies were conducted with the two remaining cytochrome P450 ESTs or with the partial FAD2 EST.
Figure 1.
Northern-blot analysis of CYP726A1 gene expression in E. lagascae. A radiolabeled probe derived from the CYP726A1 cDNA was hybridized to 10 μg of total RNA isolated from leaves (L) and developing seeds (S) of E. lagascae as shown in A. The ethidium bromide-stained gel corresponding to the northern blot is shown in B.
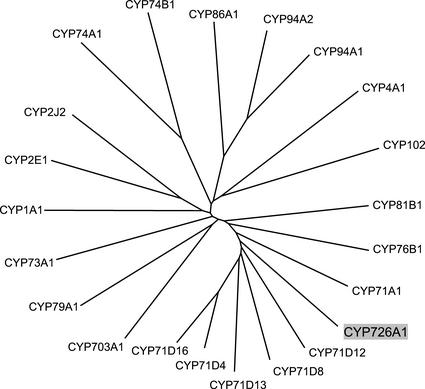
The E. lagascae CYP726A1 cDNA encodes a 500-amino acid polypeptide with a predicted Mr of 56,460 D (Fig. 2). The primary structure of this polypeptide has features that are distinctive for cytochrome P450 enzymes. These include an I helix that agrees with the consensus sequence (A/G) GX(D/E) T(T/S) for group A cytochromes P450, as well as a heme-binding domain that differs from the consensus sequence PFG(A/S/V) GRRXC(P/A/V) G in one position (Fig. 2; Durst and Nelson, 1995). Based on amino acid sequence identity with known cytochromes P450, the E. lagascae polypeptide was grouped by the P450 nomenclature committee into a new family, as indicated by the “CYP726” designation. Classification as a new family typically signifies that a cytochrome P450 shares <40% amino acid sequence identity with members of existing P450 families (Werck-Reichart and Feyereisen, 2000). However, the E. lagascae CYP726A1 does share 40% to 41% amino acid sequence identity with at least several members of the family CYP71 subfamily D, including soybean CYP71D8 and potato (Solanum tuberosum) CYP71D4 (Fig. 3). Neither of these polypeptides has any reported function. The few functionally characterized members of the CYP71D subfamily include tabersonine 16-hydroxylase (CYP71D12; Schröder, et al., 1999), (-)-4S-limoene hydroxylases (CYP71D13, CYP71D15, and CYP71D18; Lupien, et al., 1999), and cembratriene-ol hydroxylase (CYP71D16; Wang, et al., 2001). It is interesting that the E. lagascae CYP726A1 is more distantly related (13%–25% identity) to known eukaryotic fatty acid-metabolizing enzymes such as human arachidonic acid epoxygenases (CYP1A1, 19% identity; CYP2J2, 17% identity), the Arabidopsis fatty acid ω-hydroxylase (CYP86A1, 15% identity), and the Jerusalem artichoke (Helianthus tuberosus) fatty acid in-chain hydroxylase (CYP81B1, 25% identity; Fig. 3).
Figure 2.
The deduced amino acid sequence of the E. lagascae CYP726A1. The amino acid sequence corresponding to the conserved I helix motif is underlined, and the sequence corresponding to heme-binding region is highlighted in gray.
Figure 3.
Phylogenetic comparison of the E. lagascae CYP726A1 with selected members of the cytochrome P450 superfamily. The unrooted phylogenetic tree shown was generated by use of the neighbor-joining algorithm (Saitou and Nei, 1987). The P450s represented (and corresponding GenBank accession numbers) are as follows: CYP1A1, human (Homo sapiens) arachidonic acid epoxygenase (K03191); CYP2E1, Mus musculus lauric acid ω-1 hydroxylase (X62595); CYP2J2, human arachidonic acid epoxygenase (U37143); CYP94A2, common vetch (Vicia sativa) medium chain fatty acid hydroxylase (AF092917); CYP94A1, common vetch fatty acid ω-hydroxylase (AF030260); CYP86A1, Arabidopsis fatty acid ω-hydroxylase (X90458); CYP4A1, Rattus norvegicus fatty acid ω-hydroxylase (M57718); CYP74B1, Capsicum annuum fatty acid hydroperoxide lyase (U51674); CYP74A1, flax (Linum usitatissimum) allene oxide synthase (U00428); CYP102, Bacillus megaterium fatty acid in-chain hydroxylase (J04832); CYP73A1, Jerusalem artichoke (Helianthus tuberosus) cinnamate 4-hydroxylase (Z17369); CYP79A1, Sorghum bicolor Tyr N-hydroxylase (U32624); CYP703A1, Petunia × hybrida lauric acid monooxygenase (AB006790); CYP76B1, Jerusalem artichoke 7-ethoxycoumarin O-deethylase (Y09920); CYP81B1, Jerusalem artichoke fatty acid in-chain hydroxylase (AJ000477); CYP71A1, avocado (Persea americana) monoterpene monooxygenase (M32885); CYP71D8, soybean elicitor-induced P450 (unknown function) (O81974); CYP71D13, Mentha sp. (-)-(4S)-limonene-3-hydroxylase (AF124816); CYP71D4, potato fungus-induced P450 (unknown function) (AJ296346); and CYP71D12, Catharanthus roseus tabersonine 16-hydroxylase (AJ238612); and CYP71D16, tobacco cembratriene-ol hydroxylase (AF166332). The E. lagascae CYP726A1 (AF406732) is highlighted in gray.
Expression of E. Lagascae CYP726A1 in Yeast
The E. lagascae CYP726A1 was expressed in yeast WHT1 cells to determine its activity. The WHT1 cell line is engineered to coexpress a plant NADPH-cytochrome P450 reductase for enhancement of recombinant cytochrome P450 activity (Jung et al., 2000). In vitro fatty acid epoxygenase assays were initially conducted with microsomes isolated from Gal-induced cells transformed with the expression vector alone or with the CYP726A1 cDNA in the expression vector linked to the GAL1 promoter. The substrate used in these assays was sn-1-palmitoyl/sn-2-[1-14C]linoleoyl-PC, as linoleic acid-containing species of PC have been shown to function as substrates in the measurement of epoxygenase activity in microsomes of developing E. lagascae seed (Bafor et al., 1993). With this substrate, epoxygenase activity was detected in microsomes of cells transformed with the CYP726A1 cDNA, albeit at low levels (approximately 0.2 pmol min−1 mg−2 protein), but was absent from microsomes of cells transformed with the expression vector alone.
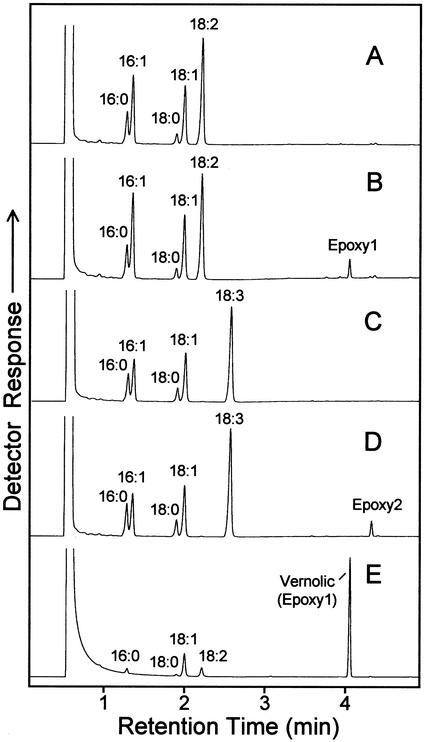
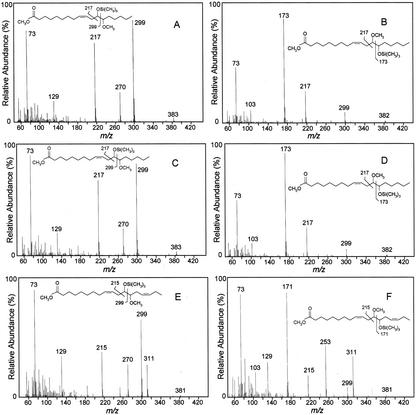
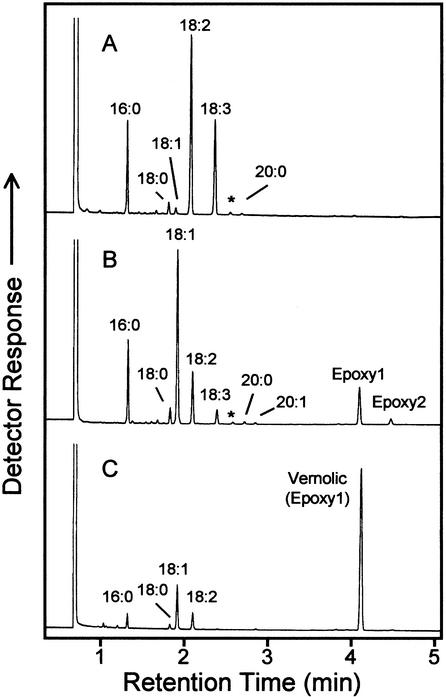
No further attempts were made to optimize the assay conditions or to use alternative substrates. Instead, recombinant WHT1 cells containing the CYP726A1 cDNA linked to the GAL1 promoter were examined for their ability to produce epoxy fatty acids in vivo. For these experiments, cells were grown with Gal induction in media lacking exogenous fatty acids or in media containing linoleic acid (18:2Δ9,12) or α-linolenic acid (18:3Δ9,12,15). The fatty acid content of these cells was then examined by gas chromatography (GC) and GC-mass spectrometry (MS) for the presence of epoxy fatty acids. No epoxy fatty acids were detected when cells were grown without exogenous fatty acids (results not shown). However, when linoleic acid was included in the media, a novel fatty acid was detected in cells expressing CYP726A1 that was absent from vector control cells. The methyl ester of this fatty acid, which is indicated as “Epoxy1” in Figure 4B, displayed the same GC retention time as the methyl ester of vernolic acid (Fig. 4E). In addition, acidic methanol derivatives of this fatty acid methyl ester displayed mass spectra (Fig. 5, C and D) identical to those of similarly prepared derivatives of methyl vernolic acid (Fig. 5, A and B). Based on these data, Epoxy1 in Figure 5B is identified as the methyl ester of vernolic acid. This finding thus demonstrates that the E. lagascae CYP726A1 functions as a Δ12-linoleic acid epoxygenase in the biosynthesis of vernolic acid.
Figure 4.
GC analysis of fatty acid methyl esters prepared form yeast cells expressing the E. lagascae CYP726A1 cDNA in media supplemented with linoleic (B) or α-linolenic acid (D). Gas chromatograms in A and C show fatty acid methyl esters from yeast cells harboring the expression vector without cDNA insert and grown in the presence of exogenous linoleic and α-linolenic acids, respectively. The gas chromatogram in E corresponds to fatty acid methyl esters prepared from E. lagascae seeds. Based on mass spectral analyses shown in Figure 5, the peak labeled Epoxy1 in B is identified as the methyl ester of vernolic acid, and the peak labeled Epoxy2 in D is tentatively identified as the methyl ester of 12-epoxyoctadeca-9,15-dienoic acid. Other labeled peaks correspond to methyl esters of the following fatty acids: 16:0, palmitic acid; 16:1, palmitoleic acid; 18:0, stearic acid; 18:1, oleic acid; 18:2, linoleic acid; and 18:3, α-linolenic acid.
Figure 5.
MS of derivatives of methyl esters of vernolic acid (A and B) and epoxy fatty acids from yeast cells expressing the E. lagascae CYP726A1 cDNA in media supplemented with linoleic acid (C and D) or α-linolenic acid (E and F). Epoxy fatty acid methyl esters were first reacted in acidic methanol, which generates a 12-hydroxy/13-methoxy product and a 13-methoxy/12-hydroxy product from opening of the epoxy ring. The two products obtained from a given epoxy fatty acid methyl ester were then converted to trimethylsilyl derivatives to produce the mass spectra shown. The mass spectra in A and B were obtained from derivatives of methyl vernolic acid from E. lagascae seed. The mass spectra in C and D were obtained from derivatives of the fatty acid methyl ester identified as “Epoxy1” in Figure 4B. The mass spectra in E and F were obtained from derivatives of the fatty acid methyl ester identified as “Epoxy2” in Figure 4D. As indicated, the mass spectra in E and F are consistent with derivatives of methyl 12-epoxyoctadeca-9,15-dienoate.
Supplementation of the growth media with α-linolenic acid resulted in the production of a second novel fatty acid in cells expressing the CYP726A1 cDNA under control of the GAL1 promoter (Fig. 4D). The methyl ester of this fatty acid, which is labeled as “Epoxy2” in Figure 4D, displayed a longer retention time than that of methyl vernolic acid. In addition, the mass spectra of acidic methanol derivatives of Epoxy2 were consistent with those arising from a C18 fatty acid methyl ester containing a Δ12-epoxy group flanked by two double bonds (Fig. 5, E and F). Given that the precursor of this product is 18:3Δ9,12,15, Epoxy2 is thus tentatively identified as the methyl ester of 12-epoxyoctadeca-9,15-dienoic acid (12-epoxy-18:2Δ9,15). This result demonstrates that the E. lagascae CYP726A1 is also able to catalyze the Δ12-epoxidation of α-linolenic acid.
Under the growth conditions described, amounts of Δ12-epoxy fatty acids accumulated by cells transformed with the CYP726A1 cDNA ranged from approximately 1% to 5% (w/w) of the total fatty acid content of cultures supplied with linoleic acid or α-linolenic acid. Examination of the major phospholipid classes of cultures supplemented with linoleic acid indicated that vernolic acid is enriched in PC. Relative amounts of vernolic acid in PC were nearly 2-fold higher than that in phosphatidylinositol and 4-fold higher than that in phosphatidylethanolamine (results not shown).
Expression of the E. lagascae CYP726A1 in Transgenic Tobacco Callus
To further characterize its activity, the E. lagascae CYP726A1 was expressed in tobacco callus under control of the cauliflower mosaic virus 35S promoter. A similar plant expression system has been previously used to characterize other unusual fatty acid biosynthetic enzymes, including the coriander Δ4-desaturase (Cahoon et al., 1992). In callus expressing CYP726A1, two novel fatty acids were detected, which are indicated as “Epoxy1” and “Epoxy2” in Figure 6B. Epoxy1 displayed the same GC retention time as the methyl ester of vernolic acid (Fig. 6), and the mass spectra of derivatives of Epoxy1 were identical to those obtained from similar derivatives of the vernolic acid methyl ester (results not shown). Epoxy1 is thus identified as the methyl ester of vernolic acid. Epoxy2 in Figure 6B displayed the same GC retention time and mass spectral fragmentation as the methyl ester of the epoxy fatty acid formed from α-linolenic acid in recombinant yeast (Figs. 4 and 5). This fatty acid methyl ester is thus tentatively identified as methyl 12-epoxy-18:2Δ9,15. Overall, these results confirm those obtained from yeast expression studies described above that indicated that CYP726A1 functions as a Δ12-epoxygenase.
Figure 6.
GC analysis of fatty acid methyl esters prepared from tobacco callus transformed with the expression vector lacking cDNA insert (A) or from tobacco callus expressing the E. lagascae CYP726A1 cDNA under control of the cauliflower mosaic virus 35S promoter (B). Shown in C is a gas chromatogram of fatty acid methyl esters prepared from E. lagascae seed. Based on mass spectral analyses, the peak in B labeled Epoxy1 corresponds to the methyl ester of vernolic acid, and the peak labeled Epoxy2 is tentatively identified as the methyl ester of 12-epoxyoctadeca-9,15-dienoic acid. Peaks labeled with asterisks correspond to phytol.
Δ12-Epoxy fatty acids detected in callus expressing the E. lagascae CYP726A1 accounted for more than 15% (w/w) of the total fatty acid content (Table I). This included approximately 13% (w/w) in the form of vernolic acid. In addition, the Δ12-epoxy fatty acids were found in all of the major phospholipid classes (i.e. PC, phosphatidylethanolamine, and phosphatidylinositol; results not shown). The relative amounts of epoxy fatty acids were nearly evenly distributed among each of these lipid classes and were approximately equal to that in the total lipid extract. Interestingly, the accumulation of epoxy fatty acids was accompanied by large alterations in the unsaturated fatty acid content of the transgenic callus. Most notably, the relative amount of oleic acid increased from approximately 2.7% (w/w) in the vector control callus to more than 40% (w/w) of the total fatty acids in CYP726A1-expressing callus (Table I).
Table I.
Fatty acid composition of tobacco callus transformed with the expression vector only (p35S) or with the E. lagascae CYP726A1 cDNA (+E. lagascae CYP726A1) under control of the cauliflower mosaic virus 35S promoter
| Fatty Acid | p35S (vector control) (n = 3) | +E. lagascae CYP726A1 (n = 3) |
|---|---|---|
| wt% of total fatty acids | ||
| 16:0 | 18.8 ± 1.6 | 15.9 ± 0.9 |
| 18:0 | 4.0 ± 0.7 | 4.5 ± 0.7 |
| 18:1 | 2.7 ± 0.9 | 43.2 ± 2.7 |
| 18:2 | 46.4 ± 5.0 | 14.9 ± 1.7 |
| 18:3 | 27.1 ± 5.4 | 4.6 ± 0.5 |
| Vernolic | N.D.a | 13.3 ± 0.7 |
| 12-Epoxy-18:2 | N.D. | 2.2 ± 0.2 |
| Otherb | ≤1.0 | ≤1.5 |
Results were obtained by measurement of three callus samples from independent transformations ± sd.
N.D., Not detected.
Includes 20:0, 20:1, 22:0, and 22:1.
Expression of the E. lagascae CYP726A1 in Somatic Soybean Embryos
To further characterize its activity, the E. lagascae CYP726A1 was expressed in somatic soybean embryos under control of a strong seed-specific promoter. Somatic soybean embryos are enriched in triacylglycerols and provide a model system for predicting the functions of transgenes in developing soybean seeds (Kinney, 1996). As with the transgenic tobacco callus, expression of CYP726A1 in soybean somatic embryos was accompanied by the accumulation of two epoxy fatty acids, both of which were absent from untransformed embryos (Table II). These fatty acids were identified as vernolic acid and 12-epoxy-18:2Δ9,15 on the basis of GC retention times of their methyl esters and mass spectra of derivatives produced by reaction in acidic methanol (data not shown). The Δ12-epoxy fatty acids composed nearly 8% (w/w) of the total fatty acids of the transgenic embryos. This level of accumulation was accompanied by only a small decrease in linoleic acid and α-linolenic acid content and a slight increase in oleic acid content relative to that of untransformed embryos.
Table II.
Fatty acid composition of untransformed somatic soybean embryos (Untransformed) or transgenic somatic soybean embryos expressing the E. lagascae CYP726A1 cDNA (+E. lagascae CYP726A1)
| Fatty Acid | Untransformed (n = 3) | +E. lagascae CYP726A1 (n = 3) |
|---|---|---|
| wt% of total fatty acids | ||
| 16:0 | 14.2 ± 0.2 | 13.4 ± 0.6 |
| 18:0 | 3.4 ± 0.5 | 3.9 ± 1.0 |
| 18:1a | 8.7 ± 0.7 | 10.5 ± 1.9 |
| 18:2 | 53.7 ± 3.9 | 46.9 ± 1.9 |
| 18:3 | 19.1 ± 3.2 | 17.1 ± 2.1 |
| Vernolic | N.D.b | 5.4 ± 0.4 |
| 12-Epoxy-18:2 | N.D. | 1.8 ± 0.1 |
| Otherc | ≤0.9 | ≤1.0 |
Values were obtained from three separate measurements (± sd) of single embryos.
Includes primarily oleic acid (18:1Δ9) and lesser amounts of cis-vaccenic acid (18:1Δ11; 1%–2% of the total fatty acids).
N.D., Not detected.
Includes 20:0, 20:1, and 22:0.
DISCUSSION
We have successfully used an EST strategy to identify a cytochrome P450 cDNA (designated CYP726A1) that corresponds to a gene that is highly expressed in E. lagascae seed. We have also shown that the expression of this cDNA in yeast and transgenic plants is accompanied by the production of Δ12-epoxy fatty acids, including vernolic acid. This finding confirms results from previous metabolic studies with E. lagascae seed that suggested the involvement of a cytochrome P450 in vernolic acid synthesis based on the sensitivity of epoxygenase activity to carbon monoxide (Bafor et al., 1993). A cytochrome P450-mediated pathway thus contrasts with the route of vernolic acid synthesis in the Asteraceae C. palaestina and V. galamensis. In seeds of these plants, the Δ12-epoxy group of vernolic acid has instead been shown to result from the activity of a Δ12-oleic acid desaturase related enzyme (Hitz, 1998; Lee et al., 1998). Therefore, our results, together with those from Asteraceae species, conclusively demonstrate that the Δ12-epoxygenase activity associated with vernolic acid synthesis can be mediated by structurally unrelated heme- and non-heme-containing enzymes.
Overall, the in vivo properties displayed by the E. lagascae CYP726A1 are in general agreement with the in vitro activity reported for the fatty acid epoxygenase in E. lagascae seed microsomes (Bafor et al., 1993). For example, CYP726A1 is able to epoxidize linoleic acid or α-linolenic acid when expressed in yeast or transgenic plant cells. This finding is consistent with results from in vitro assays of E. lagascae seed microsomes that indicate that linoleic acid and α-linoleic acid function equally well as epoxygenase substrates (Bafor et al., 1993). In addition, the detection of vernolic acid primarily in PC of yeast cells that express CYP726A1 is consistent with the demonstration that linoleic acid bound to PC can serve as a substrate for the epoxygenase in E. lagascae seed microsomes (Bafor et al., 1993).
It is interesting that the accumulation of epoxy fatty acids in transgenic tobacco callus expressing CYP726A1 was accompanied by a >15-fold increase in oleic acid content relative to callus transformed with only the expression vector. A similar but less dramatic phenomenon has been previously observed with the transgenic expression of divergent forms of the Δ12-oleic acid desaturase (FAD2) that produce unusual fatty acids with modifications at the Δ12 position, including hydroxylation, epoxidation, and double bond conjugation (Broun and Somerville, 1997; Broun et al., 1998; Cahoon et al., 1999; Singh et al., 2001). One hypothesis that has been proposed to explain this phenotype is that the unusual fatty acid products inhibit the activity of the native FAD2 and thus effectively block the conversion of oleic acid to linoleic acid (Broun and Somerville, 1997; Cahoon et al., 2001). This hypothesis may also account for the high oleic acid content of the tobacco callus transformed with the E. lagascae CYP726A1 cDNA. Tobacco callus contains only small amounts of triacylglycerol (E.B. Cahoon, unpublished data). As a result, nearly all of the epoxy fatty acids accumulate in phospholipids, including PC, the primary substrate for the Δ12 desaturation of oleic acid by FAD2 (Shanklin and Cahoon, 1998). The epoxy fatty acid products accumulated in PC may then act as inhibitors of the native FAD2 activity. In such a metabolic scenario, inhibition of FAD2 activity would be further accentuated by the inability of tobacco callus cells to partition epoxy fatty acids out of phospholipids and into triacylglycerols. It should be noted that a similar increase in oleic acid content was not detected in soybean somatic embryos that express the E. lagascae CYP726A1. This may be in part due to the lower levels of epoxy fatty acids that accumulate in this tissue compared with tobacco callus. Although the fatty acid content of lipid classes was not measured, the soybean somatic embryos may also more efficiently remove epoxy fatty acids from PC for eventual sequestration in triacylglycerol.
Of note, the primary structure of the E. lagascae CYP726A1 is not closely related to any known fatty acid-modifying cytochrome P450 enzyme, including mammalian arachidonic acid epoxygenases (e.g. CYP2J2 and CYP2J6). Instead, this polypeptide shares the highest identity (≤41% identity) with members of the CYP71D subfamily. The few functionally characterized CYP71D enzymes catalyze the hydroxylation of small molecules such as mono- and diterpenes and alkaloids (e.g. CYP71D4, CYP71D12, and CYP71D16; Lupien et al., 1999; Schröder et al., 1999; Wang et al., 2001). In this regard, there was no evidence of fatty acid hydroxylase activity (e.g. ricinoleic acid accumulation) in yeast or transgenic plant cells upon expression of the E. lagascae CYP726A1. Quite remarkable is the fact that the E. lagascae CYP726A1 shares <12% amino acid sequence identity with FAD2-type Δ12-epoxygenases from C. palaestina (Lee et al., 1998) and V. galamensis (Hitz, 1998), yet these enzymes catalyze the same reaction. With more detailed structural characterization, it will be interesting to determine if the E. lagascae CYP726A1 and the non-heme fatty acid epoxygenases of Asteraceae species have similar active sites. Additional substrate feeding studies using the yeast system described here may also be useful for determining whether these heme and non-heme epoxygenases share identical mechanisms for positioning the placement of epoxy groups in fatty acids.
The E. lagascae CYP726A1 described here provides an additional tool for the production of industrially valuable epoxy fatty acids in transgenic oilseeds. One novel approach for future studies may be to coexpress CYP726A1 and a fatty acid desaturase-type Δ12-epoxygenase to see whether the combined activities of these enzymes result in enhanced production of epoxy fatty acids. Additional metabolic factors will undoubtedly be required to achieve high levels of epoxy fatty acid accumulation in transgenic oilseeds. These factors will likely include enzymes that maintain efficient flux of epoxy fatty acids between PC and triacylglycerol, the sites of epoxy fatty acid synthesis and deposition (as described in detail by Voelker and Kinney [2001]).
MATERIALS AND METHODS
cDNA Library Construction and EST Analysis
Total RNA was isolated from developing seeds of Euphorbia lagascae using the method described by Jones et al. (1995). Enrichment of poly(A)+ RNA from the E. lagascae total RNA and subsequent cDNA library construction were conducted as previously described (Cahoon et al., 1999). The resulting library consisted of cDNA inserts cloned 5′→3′ into the EcoRI/XhoI sites of pBluescript II SK+ and was maintained in Escherichia coli DH10B cells (Invitrogen, Carlsbad, CA).
EST analysis of the E. lagascae developing seed cDNA library was performed as previously described (Cahoon et al., 1999) to generate approximately 500 bp of partial nucleotide sequence from the 5′ ends of 1,006 random cDNAs. Putative identities of these cDNAs were determined by comparison of their partial sequences with translated sequences in public databases using the National Center for Biotechnology Information BLASTX program (Altschul et al., 1990). From the EST population, two identical full-length cDNAs encoding a polypeptide (designated CYP726A1) related to known cytochromes P450 were chosen for functional characterization. Nucleotide sequences were determined for both strands of the cDNAs by dye-terminator sequencing as previously described (Cahoon et al., 1999).
Expression of E. lagascae CYP726A1 in Yeast (Saccharomyces cerevisiae)
The coding sequence of the E. lagascae CYP726A1 cDNA was amplified by PCR using the Advantage-GC cDNA polymerase kit (CLONTECH, Palo Alto, CA) and the primer pair 5′-TCAAGGAGAAAAAACCCCGGATCCATGGAGCAGAAAAATCTCTCTTTTCCG-3′ (sense) and 5′- GGCCAGTGAATTGTAATACGACTCACTATAGGGCG-3′ (antisense). Besides homology to the cytochrome P450 coding region, these primers share homology with the vector pRS315 (Sikorski and Hieter, 1989) that was modified as previously described (Jung et al., 2000) by the insertion of bidirectional GAL1/GAL10 promoters. The resulting amplification product was hybridized to the digested vector, and was transformed into yeast strain WHT1 where the expression plasmid is formed by gap repair (Hua et al., 1997). Proper integration of the CYP726A1 open reading frame downstream of the GAL1 promoter was confirmed by partial sequencing of the resulting plasmid. The WHT1 cell line used in these studies was engineered as previously described (Jung et al., 2000) to coexpress NADPH-cytochrome P450 reductase from Jerusalem artichoke (Hasenfratz, 1992) for enhancement of recombinant cytochrome P450 activity.
Microsomes were prepared from WHT1 cells transformed with the expression vector lacking insert or with the expression vector containing CYP726A1 cDNA. Growth conditions and methods for microsome preparation were as previously described (Pompon, et al., 1996; Jung, et al., 2000). Using the isolated microsomes, fatty acid epoxygenase assays were conducted in a 100-μL reaction volume and consisted of 80 mm K2HPO4 (pH 8.0), 20% (w/v) Suc, 10 mm NADPH, and 10 μm l-1-palmitoyl-2-[1-14C]linoleoyl-PC (58 mCi mmol−1; PerkinElmer Life Sciences, Boston). The PC substrate was solubilized in 12.5 mm CHAPS {3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid} and was added to the assay in a 12-μL volume. Reactions were started with the addition of 500 μg of microsomal protein and were conducted for 1 h at room temperature. Reactions were stopped with the addition of 1.5 mL of 1:2 (v/v) chloroform:methanol and they were then partitioned into organic and aqueous phases with the addition of 0.5 mL of chloroform and 0.8 mL of water. The organic layer was recovered and dried under nitrogen. Fatty acid methyl esters were prepared from the organic extract by incubation in 1% (w/v) sodium methoxide in methanol for 20 min at room temperature and were then extracted as previously described (Cahoon et al., 2001). Radiolabeled methyl esters of epoxy fatty acid reaction products were separated from nonepoxy fatty acid methyl esters by development on silica gel 60 thin-layer chromatography (TLC) plates (Whatman, Clifton, NJ) in hexane:ethyl ether (80:20, v/v). Reaction products were detected by phosphorimaging and were quantified by liquid scintillation counting.
Fatty Acid Analysis of E. lagascae CYP726A1 expressed in Yeast
Yeast WHT1 colonies containing the pRS315-derived expression plasmid (described above) with or without the CYP726A1 cDNA were grown for 3 d at 30°C in media lacking Leu (0.17% [w/v] yeast nitrogen base without amino acids [Difco, Detroit], 0.5% [w/v] ammonium sulfate, 0.01% [w/v] adenine, 0.07% [w/v] CSM-Leu [Bio 101, Vista, CA], and 0.2% [w/v] Tergitol NP-40 [Sigma, St. Louis]) and were supplemented with glycerol and Glc to final concentrations of 5% (v/v) and 0.5% (w/v), respectively. Cells were then washed twice in the medium described above that contained 2% (w/v) Gal in place of glycerol and Glc as the carbon source. The washed cells were then diluted to OD600 ≈ 0.4 in media consisting of 1% (w/v) yeast extract, 2% (w/v) peptone, 2% (w/v) Gal, 0.01% (w/v) adenine, and 0.2% (w/v) Tergitol NP-40. In some experiments (as described below), the media was supplemented with linoleic acid or α-linolenic acid at a final concentration of 0.45 mm. The cultures were maintained with shaking (250 rpm) at 28°C and were grown to OD600 ≈ 12. Cells from 3-mL cultures were collected by centrifugation and the cell pellets were then dried under vacuum. Fatty acid methyl esters were prepared by transesterification of the dried cell pellet in 1% (w/v) sodium methoxide in methanol and analyzed by GC as previously described (Cahoon et al., 2001). The retention times of putative epoxy fatty acid methyl esters in cell extracts were compared with that of vernolic acid methyl ester prepared from E. lagascae seed. More detailed structural characterization of epoxy fatty acid methyl esters in yeast extracts was conducted using GC-MS as described below.
For analysis of the fatty acid content of phospholipid classes, 10-mL cultures were grown as described above and harvested by centrifugation. The recovered cell pellets were dried under vacuum and then resuspended in 3-mL of chloroform:methanol (1:2, v/v). To aid in cell disruption, 250 mg of acid-washed glass beads (0.5 mm in diameter) were added and the cells were vortexed for 2 min. After 1 h of incubation at room temperature, 1 mL of chloroform and 1.9 mL of water were added to the extract. Following mixing and centrifugation, the lower organic layer was recovered, dried under nitrogen, and resuspended in chloroform:methanol (6:1, v/v). The phospholipid classes were then separated by development of the lipid extract on silica gel silica gel 60 TLC plates (Whatman) in chloroform:methanol: 14.8 m ammonium hydroxide (65:35:4, v/v). Phospholipid bands were visualized by light staining with iodine vapors and were identified by comigration with standards. Fatty acid methyl esters were prepared from the separated phospholipid classes by incubation of TLC scrapings in 1% (w/v) sodium methoxide in methanol and were analyzed by GC as previously described (Cahoon et al., 2001).
Expression of E. lagascae CYP726A1 in Tobacco Callus
The coding sequence of the E. lagascae CYP726A1 cDNA was amplified by PCR using Pfu polymerase (Stratagene, La Jolla, CA) and the oligonucleotide primers: 5′-gcggccgcgaattcGGAAAATGGAGCAGAAAAATC-3′ (sense) and 5′-gcggccgcggatccTTAGAACATCGTTAATTAAAG-3′ (antisense). (Note: The bases in lowercase indicate added restriction sites for cloning of the PCR product.) The amplification product was first subcloned into pCR-Script AMP (Stratagene) and was then transferred as an EcoRI/BamHI fragment into the corresponding sites of vector pART7 (Gleave, 1992). The resulting plasmid contained the open reading frame of the E. lagascae CYP726A1 cDNA flanked at its 5′ end by the cauliflower mosaic virus 35S promoter and at its 3′ end by the transcription termination portion of the octopine synthase gene. This plant expression cassette was moved as a NotI fragment into the binary vector pART27 (Gleave, 1992) to generate the plasmid pElCYP-35S. A vector control was prepared by insertion of the promoter and termination elements from pART7 into the NotI site of pART27. The resulting plasmid was designated p35S. Plasmids pElCYP-35S and p35S were then transformed into the Agrobacterium tumefaciens strain LBA4404 by electroporation. Cultures derived from these cells were used for transformation of tobacco (Nicotiana tabacum cv Xanthi) leaf discs according to the method described by Rogers, et al. (1986). Transgenic callus derived from these transformations was selected by kanamycin resistance, and expression of the E. lagascae CYP726A1 cDNA derived transgene in this tissue was confirmed by northern-blot analysis.
Fatty acid compositional analyses were performed using transgenic callus that had been maintained on kanamycin selection plates for 3 to 4 weeks. Approximately 250 mg of callus was homogenized in 1 mL of 1% (w/v) sodium methoxide in methanol, and the resulting fatty acid methyl esters were extracted and analyzed by GC as previously described (Cahoon et al., 2001). Fatty acid analysis of phospholipids from callus samples was conducted using lipid extraction and TLC procedures described above.
Expression of E. lagascae CYP726A1 in Somatic Soybean (Glycine max) Embryos
The coding sequence of the E. lagascae CYP726A1 cDNA was amplified by PCR and the product was subcloned as described above for tobacco callus expression. The amplification product was subsequently cloned as a NotI fragment into the plant expression vector pKS123 to generate pKR31. pKS123 contains the promoter of the gene for the α′-subunit of β-conglycinin (Doyle et al., 1986), which confers strong seed-specific expression of transgenes. This vector is identical to the previously described pKS67 (Cahoon et al., 1999) except that it contains two AscI restriction sites that flank the promoter and termination elements of the plant expression cassette.
pKR31 was subsequently introduced into somatic embryos of soybean (cv A2872) using the particle bombardment method of transformation (Finer and McMullen, 1991). Selection and propagation of transgenic somatic embryos was conducted as previously described (Finer and McMullen, 1991; Cahoon et al., 1999). Expression of the E. lagascae CYP726A1 transgene was confirmed by PCR using sequence specific primers and first strand cDNA prepared from total RNA isolated from transgenic somatic soybean embryos. The fatty acid composition of single transgenic embryos was determined by GC as previously described (Cahoon et al., 2001). The results reported were from transformation event MSE578-6-9.
Preparation and GC-MS Analysis of Epoxy Fatty Acid Derivatives
Epoxy fatty acid methyl esters from yeast and transgenic plant tissue was characterized by MS following acid derivatization, which provides for more diagnostic fragmentation patterns (Kleiman and Spencer, 1973). Acidic reaction of methyl esters of Δ12 epoxy fatty acids (e.g. vernolic acid) results in the opening of the epoxy ring and the generation of two products: a Δ12-hydroxy/Δ13-methoxy derivative and a Δ12-methoxy/Δ13-hydroxy derivative.
Fatty acid methyl esters from sodium methoxide transesterification of yeast or plant tissues (as described above) were heated at 70°C in 1 mL of 2.5% (v/v) sulfuric acid in methanol for 20 min. After cooling, 1 mL of water was added, and fatty acid methyl esters were extracted with 2 mL of hexane. The fatty acid methyl esters were then dried under nitrogen and subsequently reacted with 0.5 mL of the silylating reagent bis-(trimethylsilyl) trifluoroacetamide:trimethylchlorosilane (99:1, v/v) (Supelco, Bellefonte, PA) at 50°C for 30 min to generate trimethylsilyl ether derivatives of hydroxyl groups.
The derivatized fatty acid methyl esters were dried under nitrogen and were resuspended in hexane for analysis using a gas chromatograph (HP6890; Hewlett-Packard, Palo Alto, CA) interfaced with a mass selective detector (HP5973; Hewlett-Packard) operating at an electron impact ionization potential of 70 eV. The epoxy fatty acid methyl ester derivatives were partially resolved using a 30-m × 0.25-mm i.d. HP-INNOWax column (Hewlett-Packard) with the oven temperature programmed from 185°C (5-min hold) to 240°C (10-min hold) at 7.5°C min−1.
Northern-Blot Analysis of CYP726A1 Expression in E. lagascae
Total RNA was extracted from leaves and developing seeds of E. lagascae using Trizol (Invitrogen) according to the manufacturer's protocol. Northern analysis of 10 μg of total RNA from each tissue was conducted as previously described (Cahoon et al., 2001). The hybridization probe was prepared from the open reading frame of the E. lagascae CYP726A1 cDNA by random hexamer priming.
ACKNOWLEDGMENTS
We thank Dr. Maureen Dolan and colleagues of DuPont Genomics for cDNA library sequencing. We also thank Christine Howells and George Cook (DuPont) for preparation of transgenic somatic soybean embryos, Kevin Stecca (DuPont) for providing vector pKS123, and Rebecca Cahoon (DuPont) for critical reading of the manuscript.
Footnotes
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010768.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Meyers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ayorinde FO, Nana EY, Nicely PD, Woods AS, Price EO, Nwaonicha CP. Synthesis of 12-aminododecanoic and 11-aminoundecanoic acids from vernolic acid. J Am Oil Chem Soc. 1997;74:531–538. [Google Scholar]
- Ayorinde FO, Powers FT, Streete LD, Shepard RL, Tabi DN. Synthesis of dodecanedioic acid from Vernonia galamensisoil. J Am Oil Chem Soc. 1989;66:690–692. [Google Scholar]
- Badami RC, Patil KB. Structure and occurrence of unusual fatty acids in minor seed oils. Prog Lipid Res. 1981;19:119–153. doi: 10.1016/0163-7827(80)90002-8. [DOI] [PubMed] [Google Scholar]
- Bafor M, Smith MA, Jonsson L, Stobart K, Stymne S. Biosynthesis of vernoleate (cis-12-epoxyoctadeca-cis-9-enoate) in microsomal preparations from developing endosperm of Euphorbia lagascae. Arch Biochem Biophys. 1993;303:145–151. doi: 10.1006/abbi.1993.1265. [DOI] [PubMed] [Google Scholar]
- Broun P, Boddupalli S, Somerville C. A bifunctional oleate 12-hydroxylase:desaturase from Lesquerella fendleri. Plant J. 1998;13:201–210. doi: 10.1046/j.1365-313x.1998.00023.x. [DOI] [PubMed] [Google Scholar]
- Broun P, Somerville C. Accumulation of ricinoleic, lesquerolic, and densipolic acids in seeds of transgenic Arabidopsisplants that express a fatty acyl hydroxylase cDNA from castor bean. Plant Physiol. 1997;113:933–942. doi: 10.1104/pp.113.3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budziszewski GJ, Croft KPC, Hildebrand DF. Uses of biotechnology in modifying plant lipids. Lipids. 1996;31:557–569. doi: 10.1007/BF02523826. [DOI] [PubMed] [Google Scholar]
- Cahoon EB, Carlson TJ, Ripp KG, Schweiger BJ, Cook GA, Hall SE, Kinney AJ. Biosynthetic origin of conjugated double bonds: production of fatty acid components of high-value drying oils in transgenic somatic soybean embryos. Proc Natl Acad Sci USA. 1999;96:12935–12940. doi: 10.1073/pnas.96.22.12935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon EB, Ripp KG, Hall SE, Kinney AJ. Formation of conjugated Δ8, Δ10-double bonds by Δ12-oleic acid desaturase-related enzymes: biosynthetic origin of calendic acid. J Biol Chem. 2001;276:2637–2643. doi: 10.1074/jbc.M009188200. [DOI] [PubMed] [Google Scholar]
- Cahoon EB, Shanklin J, Ohlrogge JB. Expression of a coriander desaturase results in petroselinic acid production in transgenic tobacco. Proc Natl Acad Sci USA. 1992;89:11184–11188. doi: 10.1073/pnas.89.23.11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Schuler MA, Godette WD, Zenger V, Beachy RN, Slightom JL. The glycosylated seed storage proteins of Glycine max and Phaseolus vulgaris: structural homologies of genes and proteins. J Biol Chem. 1986;261:9228–9238. [PubMed] [Google Scholar]
- Durst F, Nelson DR. Diversity and evolution of plant P450 and P450-reductases. Drug Metab Drug Interact. 1995;12:189–206. doi: 10.1515/dmdi.1995.12.3-4.189. [DOI] [PubMed] [Google Scholar]
- Finer JJ, McMullen MD. Transformation of soybean via particle bombardment of embryonic suspension culture tissue. In Vitro Cell Dev Biol. 1991;27:175–182. [Google Scholar]
- Gleave AP. A versatile binary vector system with a T-DNA organizational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- Gunstone FD. Fatty acids: part II: the nature of the oxygenated acid present in Vernonia anthelmintica(Willd.) seed oil. J Chem Soc (London) 1954;May:1611–1616. [Google Scholar]
- Hasenfratz MP. Clonage de la NADPH-cytochrome P450 reductase et d'une proteine calnexine-like chez Helianthus tuberosus. PhD thesis. Strasbourg, France: Universite Louis Pasteur; 1992. [Google Scholar]
- Hitz WD, inventor December 8, 1998. Fatty acid modifying enzymes from developing seeds of Vernonia galamensis. U.S. Patent No. 5,846,784
- Hua SB, Qiu M, Chan E, Zhu L, Luo Y. Minimum length of sequence homology required for in vivocloning by homologous recombination in yeast. Plasmid. 1997;38:91–96. doi: 10.1006/plas.1997.1305. [DOI] [PubMed] [Google Scholar]
- Jones A, Davies HM, Voelker TA. Palmitoyl-acyl carrier protein (ACP) thioesterase and the evolutionary origin of plant acyl-ACP thioesterases. Plant Cell. 1995;7:359–371. doi: 10.1105/tpc.7.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung W, Yu O, Lau S-MC, O'Keefe DP, Odell J, Fader G, McGonigle B. Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat Biotechnol. 2000;18:208–212. doi: 10.1038/72671. [DOI] [PubMed] [Google Scholar]
- Kinney AJ. Development of genetically engineered soybean oils for food applications. J Food Lipids. 1996;3:273–292. [Google Scholar]
- Kleiman R, Smith CR, Jr, Yates SG. Search for new industrial oils: fifty-eight Euphorbiaceae oils, including one rich in vernolic acid. J Am Oil Chem Soc. 1965;42:169–172. [Google Scholar]
- Kleiman R, Spencer GF. Gas chromatography-mass spectrometry of methyl esters of unsaturated oxygenated fatty acids. J Am Oil Chem Soc. 1973;50:31–38. [Google Scholar]
- Lee M, Lenman M, Banas, Bafor M, Singh S, Schweizer M, Nilsson R, Liljenberg C, Dahlqvist A, Gummeson P-O et al. Identification of non-heme diiron proteins that catalyze triple bond and epoxy group formation. Science. 1998;280:915–918. doi: 10.1126/science.280.5365.915. [DOI] [PubMed] [Google Scholar]
- Liu L, Hammond EG, Nikolau BJ. In vivo studies of the biosynthesis of vernolic acid in the seed of Vernonia galamensis. Lipids. 1998;33:1217–1221. doi: 10.1007/s11745-998-0326-3. [DOI] [PubMed] [Google Scholar]
- Lupien S, Karp F, Wildung M, Croteau R. Regiospecific cytochrome P450 limonene hydroxylases from mint (Mentha) species: cDNA isolation, characterization, and functional expression of (-)-4S-limonene-3-hydroxylase and (-)-4S-limonene-6-hydroxylase. Arch Biochem Biophys. 1999;368:181–192. doi: 10.1006/abbi.1999.1298. [DOI] [PubMed] [Google Scholar]
- Perdue RE, Jr, Carlson KD, Gilbert MG. Vernonia galamensis, potential new crop source of epoxy fatty acid. Econ Bot. 1986;40:54–68. [Google Scholar]
- Pompon D, Louerat B, Bronne A, Urban P. Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 1996;272:51–64. doi: 10.1016/s0076-6879(96)72008-6. [DOI] [PubMed] [Google Scholar]
- Rogers SG, Horsch RB, Fraley RT. Gene transfer in plants: production of transformed plants using Ti plasmid vectors. Methods Enzymol. 1986;118:627–640. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: anew method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schröder G, Unterbusch E, Kaltenbach M, Schmidt J, Strack D, De Luca V, Schröder J. Light-induced cytochrome P450-dependent enzyme in indole alkaloid biosynthesis: tabersonine 16-hydroxylase. FEBS Lett. 1999;458:97–102. doi: 10.1016/s0014-5793(99)01138-2. [DOI] [PubMed] [Google Scholar]
- Shanklin J, Cahoon EB. Desaturation and related modifications of fatty acids. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:611–641. doi: 10.1146/annurev.arplant.49.1.611. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Thomaeus S, Lee M, Stymne S, Green A. Transgenic expression of a Δ12-epoxygenase gene in Arabidopsisseeds inhibits accumulation of linoleic acid. Planta. 2001;212:872–879. doi: 10.1007/s004250000456. [DOI] [PubMed] [Google Scholar]
- Spitzer V, Aitzemuller K, Vosmann K. The seed oil of Bernardia puchella(Euphorbiaceae)-a rich source of vernolic acid. J Am Oil Chem Soc. 1996;73:1733–1735. [Google Scholar]
- Voelker T, Kinney AJ. Variations in the biosynthesis of seed-storage lipids. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:335–361. doi: 10.1146/annurev.arplant.52.1.335. [DOI] [PubMed] [Google Scholar]
- Wang E, Wang R, DeParasis J, Loughrin JH, Gan S, Wagner GJ. Suppression of a P450 hydroxylase gene in plant trichome glands enhances natural-product-based aphid resistance. Nat Biotechnol. 2001;19:371–374. doi: 10.1038/86770. [DOI] [PubMed] [Google Scholar]
- Werck-Reichart D, Feyereisen R. Cytochromes P450: a success story. Genome Biol. 2000;1:3003.1–3003.9. doi: 10.1186/gb-2000-1-6-reviews3003. [DOI] [PMC free article] [PubMed] [Google Scholar]