Abstract
In kinetoplastids spliced leader (SL) RNA is trans-spliced onto the 5′ ends of all nuclear mRNAs, providing a universal exon with a unique cap. Mature SL contains an m7G cap, ribose 2′-O methylations on the first four nucleotides, and base methylations on nucleotides 1 and 4 (AACU). This structure is referred to as cap 4. Mutagenized SL RNAs that exhibit reduced cap 4 are trans-spliced, but these mRNAs do not associate with polysomes, suggesting a direct role in translation for cap 4, the primary SL sequence, or both. To separate SL RNA sequence alterations from cap 4 maturation, we have examined two ribose 2′-O-methyltransferases in Trypanosoma brucei. Both enzymes fall into the Rossmann fold class of methyltransferases and model into a conserved structure based on vaccinia virus homolog VP39. Knockdown of the methyltransferases individually or in combination did not affect growth rates and suggests a temporal placement in the cap 4 formation cascade: TbMT417 modifies A2 and is not required for subsequent steps; TbMT511 methylates C3, without which U4 methylations are reduced. Incomplete cap 4 maturation was reflected in substrate SL and mRNA populations. Recombinant methyltransferases bind to a methyl donor and show preference for m7G-capped RNAs in vitro. Both enzymes reside in the nucleoplasm. Based on the cap phenotype of substrate SL stranded in the cytosol, A2, C3, and U4 methylations are added after nuclear reimport of Sm protein-complexed substrate SL RNA. As mature cap 4 is dispensable for translation, cap 1 modifications and/or SL sequences are implicated in ribosomal interaction.
Most eukaryotic pre-mRNAs acquire a 5′ 7-methyl guanosine (m7G) cap early during transcription elongation by RNA polymerase (pol) II. This m7G cap (cap 0) is attached to the 5′-most base of the mRNA by a 5′-5′ triphosphate linkage that is formed by three separate enzymatic reactions (43). The cap 0 structure has been linked to several posttranscriptional processes, including mRNA stability, intracellular transport, and translation (4). More complex 5′-cap structures that include ribose 2′-O methylations of the first (cap 1) and second (cap 2) nucleotides appear in plants, animals, and viruses (17). In humans the cap 1 ribose 2′-O methylation occurs in the nucleus, while the cap 2 methylation occurs later in the cytoplasm (24, 27, 37). Additional cap modifications have been implicated in translational control in vertebrate cells (23, 24) and may be advantageous for translation of viral mRNAs (13).
Nature's most complex 5′ cap is the hypermethylated cap 4 structure found on substrate spliced leader (SL) RNA in the kinetoplastid protozoa, which include the pathogenic parasites Leishmania major, Trypanosoma brucei and Trypanosoma cruzi. Cap 4 is composed of ribose 2′-O methylations of the first four nucleotides (AACU) with additional base methylations on the first (m26A) and fourth (m3U) positions (5). In kinetoplastids protein-coding genes are transcribed as polycistronic precursors (20) that are resolved into individual mRNAs by 5′-end trans-splicing (5, 15, 36) and 3′-end polyadenylation (28). Thus, SL trans-splicing functions as a trans-capping reaction.
Possible roles for the kinetoplastid cap structure include recognition by, and assembly of, trans-spliceosome components and/or recruitment of ribosomes to the mRNA. Systemic methylation inhibitors in T. brucei led to loss of SL trans-splicing (35, 50), but these deleterious effects were likely downstream of cap 4 formation, as previous data from our laboratory and others have demonstrated that mutations in the SL RNA, which resulted in a spectrum of cap 4 defects, can be trans-spliced in vivo (2, 33, 47, 56). Base substitutions in the cap 4 nucleotides affected both modification of cap 4 and trans-splicing in Leptomonas collosoma (34). In Leishmania tarentolae, mutations in the Sm protein-binding site and stem-loop III of the SL RNA intron resulted in undermethylation of the 5′ cap, inhibition of 3′-end maturation, and loss of trans-splicing (46, 48), while mutation of the SL exon resulted in undermethylated cap 4 that was trans-spliced efficiently (47, 56). Decreased polysome association of mRNA bearing these undermethylated cap 4 structures provided the first evidence that cap 4 and/or primary sequence of the SL may be necessary for translation (56). To determine whether primary SL sequence or cap 4 maturation is responsible for the differential translation, the two potential culprits require study in isolation.
While the mRNA cap-specific 2′-O-methyltransferases (MTases) of human cells have been only partially purified and characterized (27), viral cap 2′-O-MTases (11, 19, 42) have been studied intensively. Position-specific iterative BLAST searches have identified several families of site-specific Rossmann fold 2′-O-MTases (14) that have a highly conserved K-D-K-E tetrad, containing the catalytic core (7). The poxvirus 2′-O-MTase family, including vaccinia virus VP39, grouped with proteins from kinetoplastids (14). Since VP39 contains an m7G cap-binding domain and is known to be a cap 1 cytosolic 2′-O-MTase, these kinetoplastid homologs are excellent candidates for cap 4 methylation of substrate SL, a primary transcript (39) that acquires an m7G cotranscriptionally (32).
Here we report the biological and biochemical characterization of the two kinetoplastid MTases and show their role in cap 4 formation. TbMT417 and TbMT511 were named based on their amino acid sizes. Double-stranded RNA interference (RNAi)-mediated knockdown of TbMT417 and TbMT511 individually and in combination caused no growth defects and revealed incomplete cap 4 profiles on both SL RNA and total mRNA. RNAi-mediated knockdown of TbMT417 affected 2′-O modification of the second adenosine, while knockdown of TbMT511 affected 2′-O formation at positions 3 and 4. Mutagenesis of key elements in both proteins confirmed preference of the m7G cap for substrate recognition. Consistent with observations with L. tarentolae, complete cap 4 on substrate SL RNA was not required for trans-splicing. Cell viability implies that translation is unaffected by incomplete cap 4 maturation. TbMT417 and TbMT511 fusion proteins both localized to the nucleus. SL RNA biogenesis requires cytosolic trafficking (54, 55), and thus the final maturation of cap 4 is performed in the nucleus postimport, consistent with the requirement for Sm protein binding prior to completion of cap 4 formation (33, 46, 53).
MATERIALS AND METHODS
Bioinformatics analyses.
Sequence database searches were carried out with PSI-BLAST (1), using the VP39 sequence as a query. Multiple-sequence alignment between VP39 and its trypanosome homologs found by PSI-BLAST was created based on the results of sequence-structure fitting using protein fold recognition and molecular modeling. Briefly, the sequences of trypanosome candidate cap MTases were aligned with all known protein structures by using the GeneSilico meta-server (25). The VP39 structure was identified as the best template. Starting from several alternative alignments to VP39 proposed by different methods, we generated refined models of TbMT417 and TbMT511 by the “FRankenstein's monster” procedure (21), namely, by iterating three-dimensional model building, model evaluation/scoring via COLORADO3D (40), recombination of best-scoring fragments of models, and local shifting of the alignment to VP39 in the poorly scoring regions. This procedure optimized the sequence-structure compatibility in the regions that could be aligned to VP39. The remaining loops and terminal extensions, which had no counterparts in the VP39 structure, were modeled de novo using the ROSETTA method (45) and combined with the homology-modeled core as described previously (22). Finally, spatial superposition of the structures of VP39 and the modeled trypanosome proteins was used to generate the structure-based sequence alignment.
RNAi construction Cloning.
The full genomic coding sequence was amplified for TbMT417 (accession no. Tb11.02.2500) using primers MT417Hind-F (GGAAGCTTATGAACTTTCCGGATAGTAGG) and MT417Xba-R (GGTCTAGACGGTGCACGATCAGAGGC). The first 1,279 nucleotides (nt) of the TbMT511-coding region (accession no. Tb09.211.3130) were amplified using oligonucleotides MT426-Xba-F (GGTCTAGAATGTATCCTAGTCTTTTTAAG) and MT426 Xho/Xba-R (GGCTCGAGTCTAGAGTGCTTCAGTACGAGGGC). Each product was cloned using the TOPO-TA system (Invitrogen), digested with HindIII and XbaI, and subcloned into the pZJM (51) vector. The vectors were sequenced to verify proper insertion and named pZJMTbMT417 and pZJMTbMT511. For the double-knockdown RNAi construction, the same 1,279-nt region was amplified using TbMT511-SpeI (CCTTTGCTCGTGTGCACTAGTATGTATC) and MT426 Xho/Xba-R (GGCTCGAGTCTAGAGTGCTTCAGTACGAGGGC), cloned into TOPO vector, and subcloned into the pZJMMT417 vector digested with SpeI and XhoI, leaving the first 718 nt of the TbMT417 coding sequence followed by the TbMT511 fragment.
RNAi strains and cell culture.
Procyclic T. brucei strain 29-13 (52) containing a T7 RNA pol and the tetracycline (Tet) repressor was grown at 27°C in SM medium supplemented with 10% fetal bovine serum in the presence of hygromycin (50 μg/ml) and G418 (15 μg/ml). Log-phase cultures (approximately 5 × 106 cells/ml) were used for transfection as described previously (51).
RNAi was performed using the pZJM system (51). The pZJMTbMT417, pZJMTbMT511, and pZJMTbMT417-MT511 constructions were transfected into T. brucei 29-13 cells by electroporation as described previously (55), and drug-resistant T. brucei was cloned by limiting dilution in SM medium under the selection pressure of G418 (15 μg/ml) (ICN), hygromycin (50 μg/ml) (PGC Scientific), and Zeocin (20 μg/ml) (Invitrogen). RNAi was induced by addition of 100 ng/ml Tet to each clonal pZJM cell line at 1 × 106 cells/ml, and growth was assayed using a Coulter Counter (Beckman/Coulter).
RNA analysis.
Total cell RNA was isolated with the TRIzol reagent (Invitrogen) as described previously (55). High-resolution acrylamide RNA blotting, RNA primer extension using Moloney murine leukemia virus reverse transcriptase (RT), and DNA sequencing reactions were performed as described previously (46, 47, 55). It should be noted that the RT used for primer extension was critical for obtaining an accurate result; Superscript RT (Invitrogen) appears to have an inefficient single-base terminal transferase activity that confounds data interpretation, while Moloney murine leukemia virus RT (Gibco) gave extensions that ran true to the sequencing ladder. Low-resolution formaldehyde-agarose RNA blots were generated as described previously (47). Low-resolution formaldehyde-agarose blots were hybridized with [α-32P]CTP-incorporated random hexamer probes made using Ready-To-Go DNA labeled beads (Amersham Biosciences) and exposed using PhosphorImager (Amersham Biosciences) cassettes. For the trans-splicing assay, mRNA was isolated using the MicroPoly(A)Purist kit (Ambion), primer extension was performed using the γ-32P-end-labeled oligonucleotide TbWTexon (CAATATAGTACAGAAACTG), and samples were verified to be free of substrate SL RNA contamination using the intron-specific primer TbSL40 (CTACTGGGAGCTTCTCATAC).
Cap analysis.
mRNA from each cell line was purified using the MicroPoly(A)Purist mRNA purification kit (Ambion), starting with 10 μg of total RNA. To label the cap structure, 300 ng of mRNA was decapped with tobacco acid pyrophosphatase (TAP) (5 U; Roche) for 1 h at 37°C in the provided reaction buffer. The α phosphate was removed using HK phosphatase (2 U; Epicentre) for 1 h at 30°C. The RNA was extracted twice with phenol, ethanol precipitated, and γ-32P end labeled with T4 polynucleotide kinase. The radiolabeled RNA was digested in 50 mM ammonium acetate (pH 4.5) and 2 mM EDTA with 40 U/ml RNase T2 at 37°C for 12 h. The digestion products were resolved on 25% acrylamide-8 M urea gels and exposed to PhosphorImager (Amersham) cassettes.
TbMT417 and TbMT511 overexpression and purification.
To produce histidine (His)-tagged TbMT417 and TbMT511 proteins, genes encoding TbMT417 and TbMT511 were cloned in the pET28a expression vector (Novagen). For overexpression, the recombinant plasmids were individually transformed into Escherichia coli BL21(DE3)LysS (Novagen). TbMT417 fusion protein was obtained in soluble form in cell lysates from cultures growing overnight at 24°C in the presence of 2% ethanol and 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). TbMT511 fusion protein was mostly insoluble and present in inclusion bodies. Inclusion bodies were obtained and purified from harvested cells and solubilized in buffer A containing 50 mM 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS) (pH 11.0), 0.3% N-laurylsarcosine, and 0.1 mM dithiothreitol (DTT) at a final protein concentration of 2 to 5 mg/ml (31). The solubilized protein was then extensively dialyzed against buffer B containing 20 mM Tris (pH 7.9), 50 mM NaCl, and 0.1 mM DTT.
The His-tagged proteins were purified by Ni column chromatography with His-Bind resin (Novagen) according to the manufacturer's protocol. Solubilized proteins were loaded onto a 1-ml His-Bind column. After the column was washed thoroughly with buffer C (20 mM Tris [pH 7.9], 500 mM NaCl, and 80 mM imidazole), the His-tagged proteins were eluted with buffer C containing 500 mM imidazole.
AdoMet-Binding Assay.
Recombinant TbMT417 and TbMT511 proteins were incubated in 20-μl reaction mixtures containing 25 mM MOPS [3-(N-morpholino)propanesulfonate] (pH 7.0), 100 mM NaCl, 2 mM DTT, 30 mM EDTA, and 1.6 M S-[methyl-3H]adenosyl-l-methionine (AdoMet) for 60 min at 37°C. Following incubation, the reaction mixtures were chilled in ice for 5 min and then transferred onto a Parafilm strip, which was again placed on ice. The binding reaction mixtures were irradiated with UV light (Stratalinker; Stratagene) for 1 min at a distance of 9 cm from the light source. The samples were combined with one-third volume of 4× Laemmli buffer (26) and heated at 100°C for 3 min before being loaded onto a 0.5-mm-thick, 15-cm-long 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel. Electrophoresis was carried out in TGS buffer (27 mM Tris, 187 mM glycine, 0.1% SDS) at 200 V for 45 min. Gels were fixed and soaked in 1 M sodium salicylic acid (pH 6.0) for 30 min before drying and exposure to X-ray film at −80°C.
m7G cap-binding assay.
Gel shift assays were performed by using nucleotide 32P-labeled RNA probes generated by in vitro transcription with the T7 Maxiscript kit (Ambion). Plasmid pBluescript SK(−) digested with HindIII was used as template DNA. To prepare transcripts with m7G cap at the 5′ terminus, in vitro transcription reactions were carried out with 1 mM monomethylated cap analog (m7G[5′]ppp[5′]G) and 1/10 of the usual concentration of GTP (0.1 mM). Reaction products were gel purified.
Formation of RNA-protein complex was observed by the relative shift in mobility of the radiolabeled probe when electrophoresed through a polyacrylamide gel. Following incubation of the binding reaction mixtures at 28°C for 30 min, the samples were size separated by electrophoresis in a 5% (60:1 acrylamide-bisacrylamide) polyacrylamide gel (2.5 h at 4°C and 150 V in 0.5× Tris-borate-EDTA) that had been prerun for 45 min under the same running conditions. The gels were dried and exposed to a PhosphorImager screen (Amersham Biosciences).
Subcellular localization.
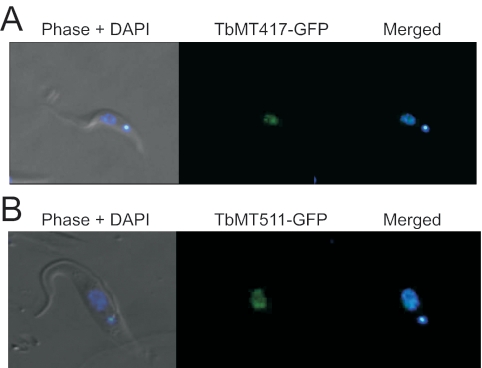
Two green fluorescent protein (GFP) fusion constructions were made using the pNICK2 vector (a derivative of pLEW100 containing different restriction sites; provided by N. Downey, Luther College, Decorah, Iowa). A 5′ HindIII::TbMT417::XbaI fragment containing the full open reading frame of TbMT417 lacking a stop codon was amplified with Pfu Turbo DNA pol (Stratagene) and subcloned into the HindIII and XbaI sites of pNICK2, creating an in-frame C-terminal GFP fusion of TbMT417 with a 2-amino-acid (aa) (SR) linker. A 5′ HindIII::TbMT511::XbaI fragment containing a 1.2-kb portion of TbMT511 and lacking a stop codon was amplified with Pfu Turbo DNA pol and subcloned into the HindIII and XbaI sites of pNICK2, producing an in-frame C-terminal fusion of TbMT511-GFP with a 2-aa (SR) linker. The TbMT511-GFP construction contains the first 426 aa of TbMT511. Both plasmids were sequenced in both directions. Ten micrograms of each plasmid was linearized at the NotI site and electroporated into KH4A YTAT T. brucei (obtained from Kent Hill, UCLA) as described above. Stable transfectants were selected with 50 μg/ml hygromycin, and 24 clonal cell lines were made by limiting dilution. These clonal lines were rinsed and washed once in 1× phosphate-buffered saline containing 100 ng of DAPI (4′,6′-diamidino-2-phenylindole) (Sigma)/ml for 5 min at room temperature. Live cells were mounted on microscope slides and visualized through the indicated filters (see Fig. 9) (Chroma) with a Zeiss Axiocam compound fluorescence microscope fitted with a Zeiss 63× objective and a Zeiss Axiocam digital camera. Digital images were taken with Zeiss Axiovision software and compiled for publication with Adobe Photoshop 7.0 (Adobe).
FIG. 9.
TbMT417 and TbMT511 localize to the nucleoplasm. A. Clonal YTAT cell lines expressing the pNICKTbMT417-GFP construct were stably transfected to constitutively express the TbMT417-GFP fusion protein. The DAPI stain for DNA served as markers for the nucleus and kinetoplastid. B. Clonal YTAT cell lines expressing the pNICKTbMT511-GFP construct were stably transfected to constitutively express the TbMT417-GFP fusion protein.
RESULTS
Identification of two potential cap-specific ribose 2′-O-MTases in T. brucei.
The separation of SL mutation from reduced cap 4 formation in an otherwise wild-type background will allow evaluation of their respective roles in SL RNA biogenesis and function. Two potential trypanosome 2′-O-MTases were identified in a search for Rossmann fold class of MTases of the RrmJ/fibrillarin superfamily that are characterized by the conserved tetrad of residues K-D-K-E (14).
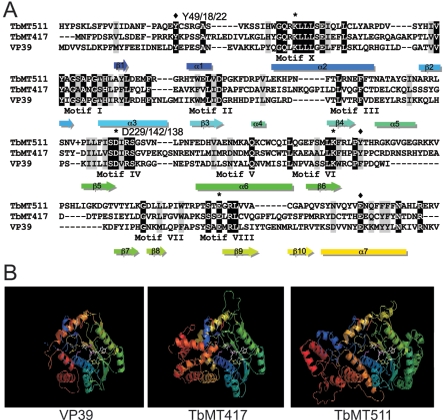
The first, designated TbMT417 (GeneDB no. Tb11.02.2500), was a predicted 47.5-kDa protein with 23% identity and 43% similarity to VP39. The second, designated TbMT511 (Tb09.211.3130), was a predicted 57.4-kDa protein with 39% identity and 52% similarity to VP39. Both enzymes have been examined elsewhere (2, 3). Homologs of VP39 were found in other poxviruses, and TbMT417 and TbMT511 homologs were identified in L. major (LmjF24.0110 and LmjF35.3380) and T. cruzi (Tc00.1047053509877.20, Tc00.1047053507669.180, Tc00.1047053503983.29, Tc00.1047053507003.70, Tc00.1047053507011.210), but no other relatives were found in other organisms. The kinetoplastid proteins were more similar to each other than either the vertebrate or invertebrate poxvirus clades (see Fig. S1 in the supplemental material). These two T. brucei proteins were assayed at the biological and biochemical levels. Both kinetoplastid proteins are larger than the prototype VP39, possessing extensions at both the amino and carboxy termini and an insertion of 13 or 17 aa between strand β6 and strand β7 of the Rossmann fold (Fig. 1A). TbMT511 contains an additional 69-aa insertion between helix α5 and strand β5 of the Rossmann fold and a 16-aa insertion between helix α7 and helix α8 that is present (11 aa) in the Chordopoxviridae of vertebrates but absent from TbMT417 and the Entomopoxviridae of invertebrates. Comparison of the core regions of the three proteins revealed the conserved K-D-K-E motif typical for 2′-O-ribose MTases (TbMT417, K40-D142-K182-E227; TbMT511, K66-D229-K266-E315) (Fig. 1A). TbMT511 had a second, lower-confidence variant (K66-D188-K266-E315) identified in the course of our analysis (data not shown). Along with the perfect conservation of the catalytic residues, aromatic residues and glutamic acid corresponding to a Y-Y-E motif of VP39 were found in both TbMT417 (Y18-Y187-E253) and TbMT511 (Y49-Y271-E335). The coplanar arrangement of the aromatic residues constitutes the m7G cap-binding domain of VP39 as determined from the crystal structure (19) and the cap-binding domains of the unrelated proteins CBC20, eukaryotic initiation factor 4E, and influenza virus RNA pol subunit PB2 (13).
FIG. 1.
Conserved motifs in the T. brucei cap-specific 2′-O-methyltransferases. A. Alignment of amino acid sequences of TbMT417 and TbMT511 with the core region of vaccinia virus VP39. ⧫, conserved aromatic residues and the ancillary E predicted to be involved in interactions with the m7G cap. *, amino acids in the conserved catalytic triad K-D-K. Numbers represent the relative positions of key residues in TbMT511, TbMT417, and VP39. B. Three-dimensional structure models of VP39, TbMT417, and TbMT511. The guanine base of the cap moiety, the methylated ribose, and AdoMet are indicated in white. The cap-binding Y residues are shown in red, the K-D-K catalytic triad is shown in magenta, and the AdoMet-binding residues (Y and D) are shown in yellow.
Structure modeling of the TbMT417 and TbMT511 sequences on the coordinates of the VP39 crystal structure revealed a similar topology of the larger proteins with the VP39 core in the conserved regions (Fig. 1B). The amino- and carboxy-terminal sequences and internal insertions in the kinetoplastid proteins were modeled de novo, and the corresponding structures have lower confidence than the homology-modeled core. The locations of the kinetoplastid-specific insertions are shown relative to the topological diagram (12, 19) of the consensus Rossmann fold (see Fig. S2 in the supplemental material). The conservation of characteristic additional secondary structures present in VP39, the Y-Y-E triad of residues for cap binding and the K-D-K-E tetrad characteristic of a 2′-O-MTase, in the two kinetoplastid proteins strongly suggests functions that are necessary for cap 4 MTases.
TbMT417 performs cap 2 methylation.
To determine if TbMT417 plays a role in cap 4 formation of SL RNA in T. brucei, we performed inducible RNAi knockdown of the TbMT417 mRNA. Given the unique nature of the structure and the importance placed on cap 4, we anticipated that elimination of a cap 4 modification enzyme would be lethal. Our initial assay to assess cap 4 formation was primer extension using primers that are specific for SL intron or exon sequences, depending on the RNA population to be queried (either substrate SL RNA or substrate SL RNA and mRNA, respectively). A limitation of this assay is the high efficiency of extension termination in the presence of modified nucleotides; whereas we had believed that a certain level of readthrough takes place, nucleotide titration studies show that termination is efficient and is not sensitive to fluctuations in nucleotide concentration. Thus, downstream methylations may occur independently of upstream methylations, blocking detection by primer extension of missing methylations 5′ of the termination event. Alternatively, depending on the substrate specificity of a given MTase, the loss of one modification may interrupt subsequent methylations if, for example, the missing methylation is required for substrate SL 5′ end recognition by a subsequent methyltransferase.
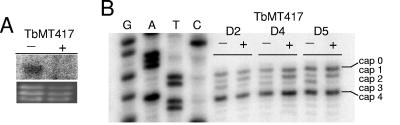
Stably transfected clonal cell lines harboring a Tet-inducible TbMT417 RNAi construction displayed no detrimental effect on growth up to 9 days postinduction (data not shown). The efficiency of the RNAi knockdown was assayed by RNA blotting, demonstrating reduction of the specific TbMT417 mRNA by the second day (Fig. 2A). The viability of the cells in the absence of TbMT417 indicated that TbMT417 is not an essential protein. To assay the effects of TbMT417 knockdown on SL RNA cap structure, primer extensions were performed on substrate SL using total RNA extracted from uninduced and Tet-induced cells and a primer specific to the intron (Fig. 2B). In induced samples the primer extension band that comigrated with A2 in a parallel DNA sequence ladder increased in intensity from day 2 to day 5, indicating accumulation of cap 1 intermediates. A simultaneous reduction in intensity of the band comigrating with nucleotide C3, corresponding to cap 2, was observed relative to that in uninduced samples. The shift in extension pattern implies a defect in 2′-O methylation of the second adenosine nucleotide, i.e., reduction of cap 2. The primer extension band comigrating with A5, attributed to modifications on the fourth nucleotide, was not significantly affected. Thus, the cap 2 modification decreased by TbMT417 knockdown was not necessary for one or both of the modifications of position 4. The U1 snRNA shares six of the first seven nucleotides with SL at its 5′ end and has equivalent modifications through cap 2 (41); thus, we assayed for the presence of the cap 2 modification by primer extension for U1 snRNA in the pZJMTbMT417 cell line. No changes were detected in the U1 snRNA primer extension pattern in the induced-RNAi samples (data not shown), suggesting that TbMT417 modifies the SL RNA specifically.
FIG. 2.
TbMT417 modifies the second position of SL RNA. A. RNA was collected at 2 days from pZJMTbMT417 cells grown in the presence (+) and absence (−) of Tet. The RNA was run on a 1.1% formaldehyde agarose gel, blotted, and probed for the presence of TbMT417 mRNA. The ethidium bromide stain of the rRNAs is a loading control. B. Primer extension analysis using total RNA collected from pJZMTbMT417 RNAi inductions at days 2, 4, and 5. Extension products from the TbSL40 primer were separated on a 6% acrylamide-8 M urea gel and run next to a cognate sequence ladder.
The primer extension analysis suggests that TbMT417 is a 2′-O-MTase involved in methylation of nucleotide A2 of the SL RNA, the absence of which is not lethal to T. brucei. The detection of accumulated cap 1 extension products despite the relative stability of the +5 cap 4 modification indicated that the intermediate population was skewed by the knockdown of TbMT417, while at least one if not both of the subsequent methylations at position 4 was not affected by the absence of the cap 2 methylation. The cap 3 methylation does not show any accumulation in either the noninduced or induced RNA population, possibly due to the lack of termination ability caused by that modification or to an extreme transience as an intermediate prior to position 4 methylation. A parallel study using knockout of the identical gene demonstrated the same phenotypes described here (3). A stable cap 3 intermediate has not been seen in mutagenesis or termination studies (32, 46, 47).
TbMT511 knockdown results in accumulation of cap 2.
To investigate the properties of the related protein TbMT511, RNAi and primer extension analyses were performed. The identification of two potential catalytic tetrads by sequence analysis gives this enzyme increased possibilities for activity.
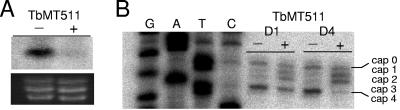
As observed with the TbMT417 RNAi cell lines, the growth rate of Tet-induced clonal TbMT511 RNAi cell lines did not diverge from that of noninduced controls (data not shown). RNA blotting demonstrated knockdown of the TbMT511 transcript at day 4 (Fig. 3A), confirming that RNAi had been successful and indicating that TbMT511 was also not an essential protein, consistent with other studies (2). To test whether TbMT511 plays a role in cap 4 modification, the substrate SL RNA primer extension profile was determined for total RNA (Fig. 3B). Induced samples showed an increase in bands comigrating with A2 and C3 and a marked reduction of the mature A5 termination, corresponding to a buildup of cap 1 and cap 2 intermediates coupled with a reduction of position 4 modification. A band corresponding to cap 3 was not observed.
FIG. 3.
TbMT511 removal leads to cap 2 accumulation. A. RNA was collected after 4 days from pZJMTbMT511 cells grown in the presence (+) and absence (−) of Tet. The RNA was run on a 1.1% formaldehyde agarose gel, blotted, and probed for the presence of TbMT511 mRNA. The ethidium bromide stain of the rRNAs is a loading control. B. Primer extension analysis using total cell RNA collected from clonal pJZMTbMT511 RNAi cell lines at days 1 and 4 after Tet induction. Extension products were separated on a 6% acrylamide-8 M urea gel and run next to the cognate sequence ladder.
The accumulation of cap 2 intermediates is consistent with a buildup of the TbMT511 substrate, suggesting that cap 3 methylation is the modification performed by this enzyme. The reduction of U4 termination products could reflect an interruption in the production of recognizable cap 3 substrate created by the action of TbMT511 or could be a direct effect in the unusual case that this single enzyme catalyzes methylations on both C3 and U4. Furthermore, the second methylation on U4 must also be factored into the altered pattern, as this base modification can also terminate primer extensions (6). The TbMT511 RNAi results are consistent with the phenotype observed in the double knockout of the same gene, in which a residual termination at position 4 is observed but not identified as a base or sugar modification (2).
Double RNAi knockdown of TbMT417 and TbMT511 is not lethal.
Since TbMT417 and TbMT511 generate different nonessential modifications within the cap 4 of SL RNA, we determined the effect of knockdown of both putative 2′-O-MTases simultaneously.
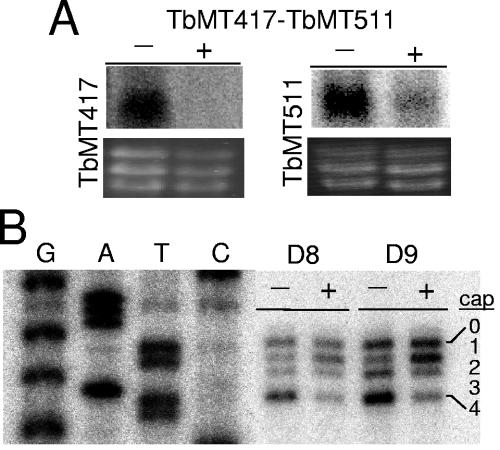
RNAi for the double knockdown was performed with the full coding region of TbMT417 and a partial region of TbMT511. No difference was seen in cell growth of the induced and uninduced cell lines monitored through day 9 (data not shown), indicating that the combined action of these enzymes was not required for cell viability. RNA blots probed for each of the T. brucei MTases showed efficient knockdown of the TbMT417 mRNA and substantial reduction of the TbMT511 mRNA by day 5 (Fig. 4A). Primer extension analysis of substrate SL RNA from total RNA preparations revealed an increase in A2 and a decrease in bands comigrating with nucleotides C3 and A5, corresponding, respectively, to cap 2 and cap 4 (Fig. 4B). This pattern reflected an additive combination of the two individual RNAi phenotypes.
FIG. 4.
Double knockdown of TbMT417 and TbMT511 produces an additive phenotype. A. RNA was collected after 5 days from pZJMTbMT417-TbMT511 cells grown in the presence (+) and absence (−) of Tet. The RNA was run on a 1.1% formaldehyde agarose gel, blotted, and probed. The ethidium bromide stain of the rRNAs is included as a loading control. B. Primer extension analysis using total RNA collected from clonal pJZMTbMT417-TbMT511 RNAi cell lines at days 8 and 9 after Tet induction. Extension products were separated on a 6% acrylamide-8 M urea gel and run next to the cognate sequence ladder.
With the simultaneous reduction of two potential SL cap 4 2′-O-MTases, the overall profile of the substrate SL RNA population begins to resemble the profile seen in the Sm complex disruption RNAi studies (33, 53), i.e., the majority species at cap 1. In this population, the Sm complex formation should not be affected, and thus the complete trafficking cycle can take place, leading to the potential for trans-splicing of these cap 1 substrates. In order to assess the potential of these substrates for participation in trans-splicing, we examined the cap structure of the mRNA population in induced cell lines by primer extension using an SL exon-specific primer.
trans-splicing of SL with reduced cap 4 methylations.
The lack of growth defects in any of the three RNAi lines generated with T. brucei MTases indicated that key pathways such as trans-splicing and translation are largely unperturbed by the absence of mature cap 4 structures on the SL RNA. However, other dynamics could be at play, including differential trans-splicing or mRNA stability. To determine whether undermethylated SL RNA was trans-spliced in induced RNAi cells, the primer extension assay was applied to poly(A)+-selected RNA, using the SL exon as a general mRNA extension primer (56).
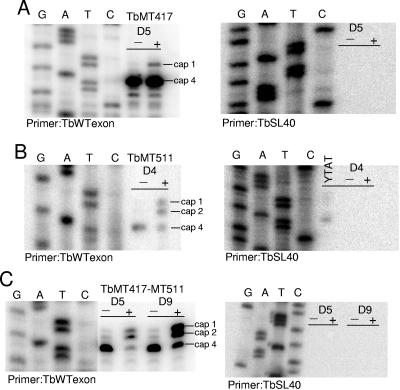
Primer extension analysis of pZJMTbMT417 knockdown mRNA samples produced a prominent band at A5, the corresponding termination position for the mature cap 4, and a minor band at A2, the cap 1 intermediate detected in the substrate SL population (Fig. 5A). The substrate SL extension profile indicated that modification at nucleotide 4 can proceed in the absence of cap 2, and thus it is possible that the A2 position in some proportion of the cap 4-terminating transcripts is unmodified but cannot be detected by this assay. The mRNA from pZJMTbMT511-induced cells showed three extension stops of roughly equal intensity corresponding to cap 1, cap 2, and cap 4 products (Fig. 5B). The primer extension assay for trans-splicing of SL in double-knockdown cells revealed a pattern of extension stops that reflected more closely the effects observed for TbMT511 (Fig. 5C). The cap 2 products may be present due to residual activity of TbMT417 or may be created by a lower-efficiency redundant enzyme. Control extensions with SL RNA intron probe were performed to detect substrate SL RNA contamination in the poly(A)+ preparations and confirmed that the exon-specific extension products were due to mRNA templates. In contrast to the extensions with substrate SL RNA, primer extension from mRNA did not yield a band comigrating with nucleotide A1, or cap 0; the cap 0 intermediate is a transient form present only on SL RNA that is nascent or has yet to exit the nucleus (55).
FIG. 5.
Undermethylated SLs are trans-spliced onto mRNA. In the left panels, poly(A)+-selected mRNA was extended using TbWTexon, complementary to the exon sequence, and separated on a 6% acrylamide-8 M urea gel and run next to the cognate sequence ladder. In the right panels poly(A)+-selected mRNA was extended with TbSL40 to control against contamination by substrate SL RNA. A. Induced (+) and uninduced (−) mRNAs from pZJMTbMT417 cells. B. Induced and uninduced mRNAs from pZJMTbMT511 cells. C. Induced and uninduced mRNAs from pZJMTbMT417-TbMT511 cells.
The mRNA cap phenotypes of these three RNAi lines lend further support to previous observations that undermethylated SL RNA is efficiently trans-spliced and also attest to the stability of the undermethylated transcripts (47, 56). As the identities of the cap 1 MTases have not been revealed, those are the only remaining modifications that might influence either process directly.
SL substrate requirements vary for each MTase.
As detailed previously, the methylation pattern 5′ of a modified position cannot be assessed by primer extension, and thus we applied an assay to examine the mRNA population from the 5′ end. To examine the methylation status of positions 5′ of primer extension termination stops, digestion with RNase T2 was performed on mRNA preparations that were decapped, dephosphorylated, and γ-32P labeled such that their 5′ ends, represented predominantly by the SL, were labeled at A1. RNase T2 has been used in the study of cap 4 to great advantage (2, 32, 50). RNase T2 cleaves unmethylated RNA completely but is inhibited by the presence of 2′-O-methyl groups, such that mature cap 4 results in the generation of a 5-nt product. For the TbMT417 knockdown mRNA population, the mRNA cap distribution showed predominantly cap 4 by primer extension and did not reflect the substrate SL pattern of equal proportions of cap 2 and cap 4. In the case of TbMT511 knockdown, the absence of a primer extension termination corresponding to the cap 3 product could be due to a nucleotide specificity issue rather than an actual loss of the cap 3 methylation. The RNase T2 assay will clarify both situations.
RNase T2-resistant products for induced and uninduced pZJMTbMT417 and pZJMTbMT511 cells were generated from poly(A)+ RNA preparations after m7G cap removal using TAP, α-phosphate removal, and 32P labeling with polynucleotide kinase. The digestion products were resolved by electrophoresis through a sequencing gel (Fig. 6). Two controls were run, including a treatment without TAP to visualize product contributions from uncapped RNA species copurifying in the poly(A)+ RNA preparations and a mock TAP/RNase T2 digestion using the wild-type T. brucei YTAT strain as a positive control for the mature cap 4 digestion pattern. Size markers to approximate the migration of the cap 4 ladder were generated by performing a mild alkali hydrolysis on a 10-nt RNA oligonucleotide with the unmethylated SL sequence that had been 5′-32P labeled with polynucleotide kinase. These controls indicated that the cap 4 product was clearly visible, migrating just ahead of the cognate size marker in the ladder, and that the poly(A)+ RNA preparations contained additional uncapped RNA species that could be 32P labeled with polynucleotide kinase. In the case of TbMT417 poly(A)+ RNA, the RNA from Tet-induced cells showed a substantial decrease in the RNase T2-resistant cap 4 band relative to that in the uninduced control, revealed a band corresponding to a cap 2 product as judged by the size markers, and demonstrated that the cap 4 structure in the mRNA is largely incomplete. Definitive conclusions about any cap 1 products in the TbMT417 sample were not possible because the signal was masked by a comigrating band in the control sample without TAP; however, a suggestive relative increase in the band designated 1 in Fig. 6 may be the anticipated corollary to the cap 1 primer extension product. The TbMT511 Tet-induced poly(A)+ RNA samples revealed a significant reduction of the RNase T2-resistant cap 4 product and accumulation of the anticipated cap 2 product; significantly, a faint shadow corresponding to the cap 3 product was also seen (band 2 in Fig. 6). The presumptive cap 1 product showed a decrease rather than an increase. These results must be viewed in light of the limitation that the RNAi effect is a knockdown rather than a total elimination of the targeted enzyme, and thus some residual activity may account for the presence of the wild-type bands.
FIG. 6.
RNase T2 digestion shows the lack of mature cap 4 on mRNA. Poly(A)+-selected RNA was treated with TAP and HK phosphatase to remove the 5′ cap and phosphates, 5′-32P labeled by polynucleotide kinase, and digested with RNase T2. The samples were from wild-type cells (YTAT) and cells uninduced (−) or induced (+) for RNAi against TbMT417 (day 5) and TbMT511 (day 4). A ladder of alkali-treated RNA (-OH) for the corresponding sequence was coelectrophoresed as a size marker. −TAP represents a control sample that was not treated with TAP. Bands 1 and 2 are discussed in the text.
The combined approaches of primer extension and the RNase T2 assay yielded a more complete picture of the two T. brucei MTase RNAi phenotypes. TbMT417 catalyzes the cap 2 2′-O methylation, the absence of which may retard but does not inhibit subsequent cap methylations at position 4. The majority of the TbMT417 mRNA population has at least one of the two methylations at position 4; however, the cap 2 methylation is largely absent in these molecules. TbMT511 is responsible for the cap 3 2′-O methylation, the absence of which has pronounced consequences for both base and ribose methylations at position 4, as well as contributing to a backlog of precursors in the form of cap 1 and cap 2. Whether TbMT511 also affects 2′-O methylation of position 4 remains an open question. All of these variants in cap structure are trans-spliced and stable in the mRNA population, reinforcing the conclusion that only cap 1 might hold sway over either of these processes. The m7G cap is known to be critical for SL stability (39, 47) and for recognition of mRNA by the translation machinery (16).
Recombinant TbMT417 and TbMT511 proteins bind AdoMet.
The aspartic acid residue (D138) of the K-D-K-E functional tetrad of vaccinia virus VP39 is crucial for MTase activity and for binding methyl group donor AdoMet (42, 44). Inspection of the VP39, TbMT417, and TbMT511 protein sequences and structures (Fig. 1) suggested the presence of similar conserved aspartic residues in the T. brucei MTases. To determine if the same catalytic centers are conserved, TbMT511 and TbMT417 were tested for AdoMet binding.
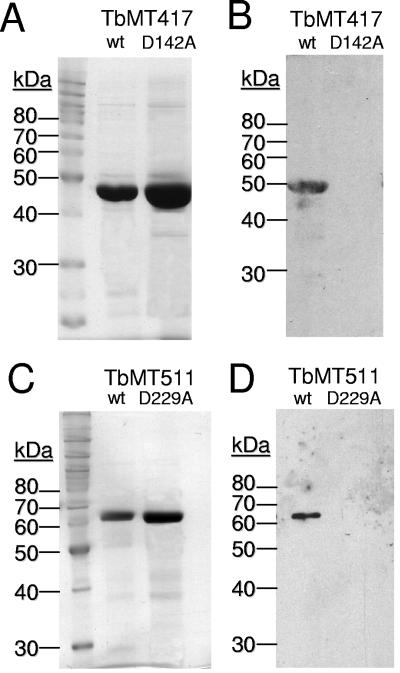
His-tagged TbMT417 and TbMT511 were constructed for ease of biochemical purification. Substitution mutations D142A and D229A were engineered into TbMT417 and TbMT511, respectively, to challenge the assignment of these residues in the functional tetrad. Both wild-type and mutated proteins were expressed and purified from E. coli. In a photo-cross-linking experiment, wild-type proteins bound radiolabeled AdoMet (Fig. 7A and C), whereas the mutated TbMT417 D142A and TbMT511 D229A did not (Fig. 7B and D), confirming the role of the aspartic acid residues in binding AdoMet. Intriguingly, mutation of TbMT511 D188A, targeting the secondary potential motif, also caused loss of the ability to bind substrate (data not shown).
FIG. 7.
Recombinant TbMT417 and TbMT511 bind AdoMet. A. SDS-polyacrylamide gel electrophoresis analysis of purified wild-type (wt) and D142A-mutated TbMT417 recombinant protein. B. Fluorograph to detect the 3H-labeled AdoMet-binding ability of wild-type and mutated recombinant TbMT417. C. SDS-polyacrylamide gel electrophoresis analysis of purified wild-type and D229A-mutated MT511 recombinant protein. D. Fluorograph of SDS-polyacrylamide gel to detect the UV-cross-linking of 3H-labeled AdoMet to wild-type and mutated recombinant TbMT511.
The anticipated AdoMet binding results obtained with the wild-type recombinant proteins support the predicted MTase activity, allowing the pursuit of further biochemical characterization. As proper folding of the mutated protein cannot be confirmed, the negative findings obtained for the engineered tetrad mutations will be validated by future in vivo experiments to test hypotheses on TbMT511 function.
TbMT511 and TbMT417 affinity for m7G cap structure.
A second relevant characteristic of VP39 is an interaction with the m7G cap via an aromatic amino acid sandwich. The homolog alignments predicted conserved tyrosine residues in both TbMT417 and TbMT511 proteins corresponding to Y22 of VP39. Gel shift assays were performed using 32P-labeled, in vitro-transcribed Bluescript RNA with or without the m7G cap structure to evaluate cap-specific binding properties of the recombinant T. brucei MTase proteins. Competition studies were performed to assess substrate specificity, while mutagenesis studies addressed the identification of the key tyrosine residue in each T. brucei MTase.
Wild-type TbMT417 shifted both uncapped and capped T7 transcripts (Fig. 8A, lanes 2 and 5, respectively). Binding experiments performed with a fivefold excess of nonlabeled, uncapped transcript resulted in a marked reduction of binding to the labeled uncapped RNA (Fig. 8A, lane 3). However, binding to capped RNA was still present (lane 6), indicating that the protein binds to the capped RNA probe with higher specificity. An apparent decrease in binding to the capped RNA substrate was seen in presence of uncapped competitor RNA, but this can be attributed to incomplete capping of the labeled substrate. To further probe the specificity for binding the cap structure, substitution mutations were made in the conserved tyrosine residues. Mutations Y18A and Y49A were introduced into TbMT417 and TbMT511, respectively. TbMT417Y18A mutated protein was unable to bind capped RNA probe even when used in a 10-fold excess to wild-type (lanes 3 and 4). Similar cap binding was observed with wild-type TbMT511 (Fig. 8C, lane 2) but not with the Y49A-mutated protein (lanes 3 and 4).
FIG. 8.
TbMT417 and TbMT511 bind the mRNA m7G cap. A. Purified TbMT417 protein was incubated with 32P-labeled, uncapped RNA probe alone (lane 2) and in the presence of a fivefold molar excess of unlabeled uncapped RNA transcript (lane 3) as a competitor. Similarly, radiolabeled RNA with m7G cap at the 5′ end (lane 4) was used as substrate for binding to TbMT417 alone (lane 5) or in the presence of a fivefold molar excess of cold uncapped competitor RNA (lane 6). B. Binding experiments were performed with m7G-capped RNA as substrate (lane 1), using 50 ng of purified wild-type (WT) TbMT417 (lane 2) or 50 ng (lane 3) or 250 ng (lane 4) of a mutated form (Y18A) of TbMT417. C. Same as B, but purified wild-type (lane 2) or mutated Y49A forms (lanes 3 and 4) of recombinant TbMT511 were used in the binding experiments.
The basic binding studies performed here confirm two of the predicted features of the T. brucei VP39 homologs, identifying key amino acid residues in both proteins and paving the way for more in-depth studies. The interaction with the methyl donor AdoMet fortifies the MTase assignment, while the binding preference for m7G-capped transcripts is consistent with the structure of substrate SL RNA. Further studies examining the relative effects on catalysis of methylations at cap 4 sites will reveal the specific substrate requirements of these specialized enzymes.
TbMT417 and TbMT511 localize to the nucleoplasm.
Maturation of the complete cap 4 requires trafficking of the SL RNA through the cytoplasm for the acquisition of Sm proteins (33, 53). Methylations beyond cap 1 occur in an Sm-dependent manner either in the cytosol or postimport in the nucleus. In vitro cap 4 modification has been demonstrated in T. brucei, requiring preassembly of substrate SL in RNPs (49). We assayed the intracellular localization of TbMT417 and TbMT511 to determine the spatial location of these downstream effector proteins. A cotranscriptional cap 4 formation model has been proposed (32); it fails to address the issue of intron mutations affecting Sm complex formation, although the data from Mair et al. (32) demonstrate that the Sm-binding site represents a clear boundary that must be crossed transcriptionally for SL RNA cap formation beyond cap 2.
Wild-type YTAT T. brucei was transfected with a plasmid encoding GFP fusion proteins for TbMT417 or TbMT511. Microscopic observation of the cells under UV illumination showed that the TbMT417 and TbMT511 fusion proteins localized to the nucleoplasm as judged by colocalization with the DAPI-stained nuclear DNA (Fig. 9). Both MTase-GFP fusion proteins showed some clustering in discrete spots within the overall nuclear staining.
SL RNAs receive their TbMT417 and TbMT511 cap modifications within the nucleus. The various cap intermediates accumulating upon perturbation of SL cytoplasmic trafficking indicate that TbMT417 and TbMT511 act on SL after import into the nucleus: SL RNA blocked before export from the nucleus by leptomycin B accumulates SL methylated at cap 0 (55), and SL RNA blocked in the cytosol by knockdown of Sm proteins also accumulates the cap 1 (33, 53), as does Sm-binding-site-mutagenized SL (46).
DISCUSSION
In order to differentiate between the biological effects of SL primary sequence and cap structure we have identified and partially characterized two enzymes involved in modifications of the unique kinetoplastid cap 4 structure, both in vitro and in vivo. TbMT417 and TbMT511 show conservation of residues involved in the ribose 2′-O-MTase activity (7) and those involved in cap binding in the vaccinia virus cap 1 methyltransferase VP39 (38). TbMT417 is a 2′-O-MTase that modifies the second adenosine residue of SL RNA, while knockdown of TbMT511 produces modification defects at cap position 3 and cap position 4. Neither of the MTases was required for cell viability, casting doubt on an essential role for all cap 4 modifications in trans-splicing and translation. The methylations at position 1 must be queried to settle these issues. Recombinant MTases were shown to bind the AdoMet cofactor and m7G cap-containing transcripts as predicted. The nonessential nature of the MTases will facilitate future studies on the efficiency of translation of undermethylated mRNA and on the determinants of T. brucei MTase substrate specificity.
The role of the cap 4 modifications and the identification of critical SL features have been addressed by different approaches and have resulted in mutually exclusive conclusions. A major question remaining from the SL mutagenesis studies is the relative effect of cap 4 methylation versus that of changing the primary SL sequence of cap 4 structure. In studies where the first four nucleotides of the SL are changed (34, 47), it is not possible to distinguish between these two variables. Mutagenesis of domains outside cap 4 could be attributed to other processing defects such as the loss of Sm complex formation (30, 34, 46). The effects due to base changes can be eliminated by modulation of the modifying enzymes as described here. SL sequences mutated at positions 10 to 19 and 20 to 29 fail to associate with polysomes in L. tarentolae (56). In these cases, the cap 4 primary sequence is unchanged, but the SLs are both undermethylated and trans-spliced. No alteration of U1 snRNA cap structure was seen in TbMT417 knockdown, despite the sequence identity of the 5′-most four nucleotides of U1 snRNA and SL RNA, indicating that this identical modification is performed by a separate, and possibly redundant, enzyme that may recognize SL at low efficiency.
The order of substrate SL RNA 3′ trimming and 5′ methylation events has not yet been determined. Whether the T. brucei MTases make up a complex of proteins involved in processing can be addressed in colocalization studies, including use of the nuclear 3′-polishing enzyme TbSNIP (54). Since knockdown of TbSNIP did not affect cap 4, the formation of the 3′ end may be independent of the 5′-end modifications and linked only in that both processing events require cytoplasmic trafficking. Knockdown of TbMT417 did not affect the primer extension stop corresponding to cap 4, indicating that ribose and/or base methylation of A4 is not dependent on cap 2 modification. Knockdown of TbMT511 resulted in a defect of cap 3 and undermethylation of cap 4, either directly or indirectly. Further experiments are required to distinguish whether one or both 2′-O modifications are performed by TbMT511 and whether there is an interdependence of the two. In vivo SL RNA cap 4 modifications are added sequentially in a 5′-to-3′ direction (32). Synthesis up to cap 2 was observed on artificially truncated transcripts of 67 and 111 nt; complete cap 4 was not observed until synthesis of nt 127 (32), which is also consistent with a dependence on the Sm-binding site. No cap 3 product was described, supporting the transience of this intermediate.
Transcription terminator inhibition studies led to the proposal of a cotranscriptional model for formation of the cap 4 structure (32). However, several lines of evidence indicate that later steps in the modification of the SL RNA are posttranscriptional and require cytosolic trafficking. Numerous exon and intron mutants in L. tarentolae are not truncated but are undermethylated (46, 47), suggesting that the primary sequence as well as secondary structure influences the recognition of SL RNA as a substrate for catalysis (nt 10 to 29) or result in subcellular mislocalization due to structural issues (intron structures stem-loop II, the Sm-binding site, and stem-loop III). Nuclear retention of newly synthesized substrate SL by the lethal inactivation of exportin 1 (XPO1/CRM1) leads to the accumulation of unmethylated (cap 0) SL RNA (55). The lethal downregulation of two components of the Sm complex resulted in accumulation of undermethylated (cap 1) substrate SL in the cytosol (33, 53). Modification of caps 2, 3, and 4 depends on export from the nucleus and association with the Sm protein complex in the cytosol, but where these methylations occur was not clear. Localization of TbMT417 and TbMT511 to the nucleoplasm dictates that their cap modifications occur in the nucleus after import from the cytosol. Nuclear MTase localization is also consistent with cotranscriptional addition of cap 4 (32); however, a further prediction would be for the MTases to colocalize to a single SL transcription center, likely focused in a region adjacent to the nucleolus, as observed in colocalization studies of the SL genes with RNA polymerase II in T. cruzi (10). We see no such localization for TbMT417 or TbMT511.
The protein fold recognition analysis and modeling of the trypanosomal MTases identified putative 2′-O-MTase and cap-binding residues that have been challenged experimentally. By analogy to Y22 and F180 in VP39, we have shown that aromatic residues Y18 in TbMT417 and Y49 in TbMT511 are required for specific cap binding. Based on D138 in VP39 we have confirmed that D142 in TbMT417 and D229 in TbMT511 are required for AdoMet binding. VP39 is a cap 1 MTase for which the structure and function relationships have been determined by crystal structure analysis (19). In contrast, TbMT417 and TbMT511 have specificity for the cap 2 nucleotide and for the cap 3, and possibly cap 4, nucleotides, respectively. The single insert within the Rossmann fold of TbMT417 and the three inserts within TbMT511 may alter the juxtaposition of the cap-binding site and the AdoMet-binding/catalytic sites relative to the RNA chain, providing a spatial explanation for substrate selectivity. Furthermore, the identification of a potential secondary TbMT511 AdoMet-binding tetrad configuration, the disruption of which abolished AdoMet binding, leaves the door open for the unprecedented situation in which a single enzyme might catalyze the methylation of two adjacent ribose moieties.
The identification of the two kinetoplastid MTases by sequence similarity and iterative motif searching has evolutionary and functional implications. The identification of a family of MTases found only in poxviruses and kinetoplastids is extraordinary. While it cannot be ruled out that the conserved features of the MTase catalytic triad and the coplanar cap-binding domain may have evolved independently by convergent evolution, phylogenetic analysis supports a monophyletic origin for the genes in both groups of organisms (14).
It is possible that this gene family may have evolved by horizontal gene transfer between pathogens of a common host(s) without the direct involvement of the host's genome. Furthermore, the similarity between the two kinetoplastid genes suggests that they evolved from each other by a duplication event after the acquisition of the original gene in an ancestral kinetoplastid. Insertion-deletion (indel) differences seen in TbMT417 and TbMT511 relative to VP39 are prevalent in other conserved kinetoplastid proteins (8). Indels may confer pathogeneic properties (9) or may allow interactions with other proteins. In the case of the kinetoplastid VP39 homologs, both indels of TbMT511 are contained within the Rossmann fold and could alter the shape of the RNA-binding pocket, effectively extending the path on the surface that connects the MTase active site relative to the cap-binding site. Since the poxvirus life cycle and VP39 localization are cytosolic, a further property acquired in the kinetoplastid homologs is a nuclear localization signal. The most common of these signals consist of monopartite or bipartite runs of basic amino acids within a 7- to 20-aa stretch. A concentration of arginine and lysine residues is seen in the carboxy-terminal region of TbMT417 and the amino-terminal region of TbMT511.
The elaborate structure of the hypermethylated cap 4 and elements in the SL exon sequence may both contribute to the process of translation in the kinetoplastids, beyond the simple possession of an m7G cap. Consistent with a role for cap 4 in translation, the L. major cytoplasmic cap-binding protein eukaryotic initiation factor 4E bound preferentially chemically synthesized cap 4 over m7G (29). Structural studies of the T. cruzi 80S ribosome show that a kinetoplastid-specific insertion near the mRNA exit channel may function in SL RNA exon recognition by sequence or cap structure (18). Neither our dual T. brucei MTase knockdown studies nor TbMT511 chromosomal knockout (2, 3) revealed growth phenotypes, implicating the primary SL exon sequences, especially in the region from position 10 to 28, and focusing attention on the three methylations of the first nucleotide. SL exon mutagenesis studies at the single and dinucleotide levels in L. tarentolae are under way to delimit the sequences involved in substrate recognition and/or ribosomal interaction. Identification of novel MTases associating with known enzymes forming potential SL processing centers or continued mining of the trypanosome genome project databases will eventually lead to the identification of the additional T. brucei MTase candidates, which can then be tested directly.
Supplementary Material
Acknowledgments
This work was supported by NIH grant AI034056. J.R.Z was supported by NSF Louis Stokes Alliance for Minority Participation Award HRD-0115115:3 and USPHS National Research Service Award GM07104. G.M.Z was supported by NIH Microbial Pathogenesis training grant 2-T32-AI-07323.
We thank Paul Gershon, Robert Hitchcock, Sean Thomas, and Scott Westenberger for stimulating discussions; Nick Downey for the GFP fusion plasmid; Kent Hill for cells and the use of his fluorescence microscope; and Ana Sanchez and Jennifer Wu for help with early RNAi experiments.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arhin, G. K., H. Li, E. Ullu, and C. Tschudi. 2006. A protein related to the vaccinia virus cap-specific methyltransferase VP39 is involved in cap 4 modification in Trypanosoma brucei. RNA 12:53-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arhin, G. K., E. Ullu, and C. Tschudi. 2006. 2′-O-Methylation of position 2 of the trypanosome spliced leader cap 4 is mediated by a 48kDa protein related to vaccinia virus VP39. Mol. Biochem. Parasitol. 147:137-139. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee, A. K. 1980. 5′-terminal cap structure in eucaryotic messenger ribonucleic acids. Microbiol. Rev. 44:175-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bangs, J. D., P. F. Crain, T. Hashizume, J. A. McCloskey, and J. C. Boothroyd. 1992. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J. Biol. Chem. 267:9805-9815. [PubMed] [Google Scholar]
- 6.Basturea, G. N., K. E. Rudd, and M. P. Deutscher. 2006. Identification and characterization of RsmE, the founding member of a new RNA base methyltransferase family. RNA 12:426-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bujnicki, J. M., and L. Rychlewski. 2001. Reassignment of specificities of two cap methyltransferase domains in the reovirus lambda 2 protein. Genome Biol. 2:research 0038.1-research0038.6. [DOI] [PMC free article] [PubMed]
- 8.Cherkasov, A., S. J. Lee, D. Nandan, and N. E. Reiner. 2006. Large-scale survey for potentially targetable indels in bacterial and protozoan proteins. Proteins 62:371-380. [DOI] [PubMed] [Google Scholar]
- 9.Cherkasov, A., D. Nandan, and N. E. Reiner. 2005. Selective targeting of indel-inferred differences in spatial structures of highly homologous proteins. Proteins 58:950-954. [DOI] [PubMed] [Google Scholar]
- 10.Dossin, F. M., and S. Schenkman. 2005. Actively transcribing RNA polymerase II concentrates on spliced leader genes in the nucleus of Trypanosoma cruzi. Eukaryot. Cell 4:960-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egloff, M.-P., D. Benarroch, B. Selisko, J.-L. Romette, and B. Canard. 2002. An RNA cap (nucleoside-2′-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 21:2757-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fauman, E. B., R. M. Blumenthal, and X. Cheng. 1999. Structure and evolution of AdoMet-dependent methyltransferases, p. 1-38. In X. Cheng and R. M. Blumenthal (ed.), S-adenosylmethionine-dependent methyltransferases: structures and functions. World Scientific Publishing, Singapore.
- 13.Fechter, P., and G. G. Brownlee. 2005. Recognition of mRNA cap structures by viral and cellular proteins. J. Gen. Virol. 86:1239-1249. [DOI] [PubMed] [Google Scholar]
- 14.Feder, M., J. Pas, L. S. Wyrwicz, and J. M. Bujnicki. 2003. Molecular phylogenetics of the RrmJ/fibrillarin superfamily of ribose 2′-O-methyltransferases. Gene 302:129-138. [DOI] [PubMed] [Google Scholar]
- 15.Freistadt, M. S., G. A. M. Cross, and H. D. Robertson. 1988. Discontinuously synthesized mRNA from Trypanosoma brucei contains the highly methylated 5′ cap structure, m7GpppA*A*C(2′-O)mU*A. J. Biol. Chem. 263:15071-15075. [PubMed] [Google Scholar]
- 16.Furuichi, Y., A. LaFiandra, and A. J. Shatkin. 1977. 5′-Terminal structure and mRNA stability. Nature 266:235-239. [DOI] [PubMed] [Google Scholar]
- 17.Furuichi, Y., and A. J. Shatkin. 2000. Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 55:135-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao, H., M. J. Ayub, M. J. Levin, and J. Frank. 2005. The structure of the 80S ribosome from Trypanosoma cruzi reveals unique rRNA components. Proc. Natl. Acad. Sci. USA 102:10206-10211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hodel, A. E., P. D. Gershon, X. Shi, and F. A. Quiocho. 1996. The 1.85 Å structure of vaccinia protein VP39: a bifunctional enzyme that participates in the modification of both mRNA ends. Cell 85:247-256. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, P. J., J. M. Kooter, and P. Borst. 1987. Inactivation of transcription by UV irradiation of T. brucei provides evidence for a multicistronic transcription unit including a VSG gene. Cell 51:273-281. [DOI] [PubMed] [Google Scholar]
- 21.Kosinski, J., I. A. Cymerman, M. Feder, M. A. Kurowski, J. M. Sasin, and J. M. Bujnicki. 2003. A “FRankenstein's monster” approach to comparative modeling: merging the finest fragments of Fold-Recognition models and iterative model refinement aided by 3D structure evaluation. Proteins 53(Suppl. 6):369-379. [DOI] [PubMed] [Google Scholar]
- 22.Kosinski, J., M. J. Gajda, I. A. Cymerman, M. A. Kurowski, M. Pawlowski, M. Boniecki, A. Obarska, G. Papaj, P. Sroczynska-Obuchowicz, K. L. Tkaczuk, P. Sniezynska, J. M. Sasin, A. Augustyn, J. M. Bujnicki, and M. Feder. 2005. FRankenstein becomes a cyborg: the automatic recombination and realignment of Fold-Recognition models in CASP6. Proteins 61(Suppl. 7):106-113. [DOI] [PubMed] [Google Scholar]
- 23.Kuge, H., G. G. Brownlee, P. D. Gershon, and J. D. Richter. 1998. Cap ribose methylation of c-mos mRNA stimulates translation and oocyte maturation in Xenopus laevis. Nucleic Acids Res. 26:3208-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuge, H., and J. D. Richter. 1995. Cytoplasmic 3′ poly(A) addition induces 5′ cap ribose methylation: implications for translational control of maternal mRNA. EMBO J. 14:6301-6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurowski, M. A., and J. M. Bujnicki. 2003. GeneSilico protein structure prediction meta-server. Nucleic Acids Res. 31:3305-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Langberg, S. R., and B. Moss. 1981. Post-transcriptional modifications of mRNA. Purification and characterization of cap I and cap II RNA (nucleoside-2′-)-methylatransferases from HeLa cells. J. Biol. Chem. 256:10054-10060. [PubMed] [Google Scholar]
- 28.LeBowitz, J. H., H. Q. Smith, L. Rusche, and S. M. Beverley. 1993. Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev. 7:996-1007. [DOI] [PubMed] [Google Scholar]
- 29.Lewdorowicz, M., Y. Yoffe, J. Zuberek, J. Jemielity, J. Stepinski, R. Kierzek, R. Stolarski, M. Shapira, and E. Darzynkiewicz. 2004. Chemical synthesis and binding activity of the trypanosomatid cap-4 structure. RNA 10:1469-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lücke, S., G. L. Xu, Z. Palfi, M. Cross, V. Bellofatto, and A. Bindereif. 1996. Spliced leader RNA of trypanosomes: in vivo mutational analysis reveals extensive and distinct requirements for trans splicing and cap 4 formation. EMBO J. 15:4380-4391. [PMC free article] [PubMed] [Google Scholar]
- 31.Mahmood, R., B. Mittra, J. C. Hines, and D. S. Ray. 2001. Characterization of the Crithidia fasciculata mRNA cycling sequence binding proteins. Mol. Cell. Biol. 21:4453-4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mair, G., E. Ullu, and C. Tschudi. 2000. Cotranscriptional cap 4 formation on the Trypanosoma brucei spliced leader RNA. J. Biol. Chem. 275:28994-28999. [DOI] [PubMed] [Google Scholar]
- 33.Mandelboim, M., S. Barth, M. Biton, X.-H. Liang, and S. Michaeli. 2003. Silencing of Sm proteins in Trypanosoma brucei by RNAi captured a novel cytoplasmic intermediate in SL RNA biogenesis. J. Biol. Chem. 278:51469-51478. [DOI] [PubMed] [Google Scholar]
- 34.Mandelboim, M., C. L. Estraño, C. Tschudi, E. Ullu, and S. Michaeli. 2002. On the role of exon and intron sequences in trans-splicing utilization and cap 4 modification of the trypanosomatid Leptomonas collosoma SL RNA. J. Biol. Chem. 277:35210-35218. [DOI] [PubMed] [Google Scholar]
- 35.McNally, K. P., and N. Agabian. 1992. Trypanosoma brucei spliced-leader RNA methylations are required for trans splicing in vivo. Mol. Cell. Biol. 12:4844-4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perry, K. L., K. P. Watkins, and N. Agabian. 1987. Trypanosome mRNAs have unusual “cap 4” structures acquired by addition of a spliced leader. Proc. Natl. Acad. Sci. USA 84:8190-8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perry, R. P., and D. E. Kelley. 1976. Kinetics of formation of 5′ terminal caps in mRNA. Cell 8:433-442. [DOI] [PubMed] [Google Scholar]
- 38.Quiocho, F. A., G. Hu, and P. D. Gershon. 2000. Structural basis of mRNA recognition by proteins. Curr. Opin. Struct. Biol. 10:78-86. [DOI] [PubMed] [Google Scholar]
- 39.Saito, R. M., M. G. Elgort, and D. A. Campbell. 1994. A conserved upstream element is essential for transcription of the Leishmania tarentolae mini-exon gene. EMBO J. 13:5460-5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sasin, J. M., and J. M. Bujnicki. 2004. COLORADO3D, a web server for the visual analysis of protein structures. Nucleic Acids Res. 32:W586-W589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schnare, M. N., and M. W. Gray. 1999. A candidate U1 small nuclear RNA for trypanosomatid protozoa. J. Biol. Chem. 274:23691-23694. [DOI] [PubMed] [Google Scholar]
- 42.Schnierle, B. S., P. D. Gershon, and B. Moss. 1994. Mutational analysis of a multifunctional protein, with mRNA 5′ cap-specific (nucleoside-2′-)-methyltransferase and 3′-adenylyltransferase stimulatory activities, encoded by vaccinia virus. J. Biol. Chem. 269:20700-20706. [PubMed] [Google Scholar]
- 43.Shatkin, A. J., and J. L. Manley. 2000. The ends of the affair: capping and polyadenylation. Nat. Struct. Biol. 7:838-842. [DOI] [PubMed] [Google Scholar]
- 44.Shi, X., T. G. Berhardt, S.-M. Wang, and P. D. Gershon. 1997. The surface region of the bifunctional vaccinia RNA modifying protein VP39 that interfaces with poly(A) polymerase is remote from the the RNA binding cleft used for its mRNA 5′ cap methylation function. J. Biol. Chem. 272:23292-23303. [DOI] [PubMed] [Google Scholar]
- 45.Simons, K. T., C. Kooperberg, E. Huang, and D. Baker. 1997. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J. Mol. Biol. 268:209-225. [DOI] [PubMed] [Google Scholar]
- 46.Sturm, N. R., and D. A. Campbell. 1999. The role of intron structures in trans-splicing and cap 4 formation for the Leishmania spliced leader RNA. J. Biol. Chem. 274:19361-19367. [DOI] [PubMed] [Google Scholar]
- 47.Sturm, N. R., J. Fleischmann, and D. A. Campbell. 1998. Efficient trans-splicing of mutated spliced leader exons in Leishmania tarentolae. J. Biol. Chem. 273:18689-18692. [DOI] [PubMed] [Google Scholar]
- 48.Sturm, N. R., M. C. Yu, and D. A. Campbell. 1999. Transcription termination and 3′-end processing of the spliced leader RNA in kinetoplastids. Mol. Cell. Biol. 19:1595-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ullu, E., and C. Tschudi. 1995. Accurate modification of the trypanosome spliced leader cap structure in a homologous cell-free system. J. Biol. Chem. 270:20365-20369. [DOI] [PubMed] [Google Scholar]
- 50.Ullu, E., and C. Tschudi. 1991. trans splicing in trypanosomes requires methylation of the 5′ end of the spliced leader RNA. Proc. Natl. Acad. Sci. USA 88:10074-10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, Z., J. C. Morris, M. E. Drew, and P. T. Englund. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275:40174-40179. [DOI] [PubMed] [Google Scholar]
- 52.Wirtz, E., S. Leal, C. Ochatt, and G. A. M. Cross. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89-101. [DOI] [PubMed] [Google Scholar]
- 53.Zeiner, G. M., S. Foldynová, N. R. Sturm, J. Lukes, and D. A. Campbell. 2004. SmD1 is required for spliced leader RNA biogenesis. Eukaryot. Cell 3:241-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeiner, G. M., R. A. Hitchcock, N. R. Sturm, and D. A. Campbell. 2004. 3′-end polishing of the kinetoplastid spliced leader RNA is performed by SNIP, a 3′→5′ exonuclease with a motley assortment of small RNA substrates. Mol. Cell. Biol. 24:10390-10396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeiner, G. M., N. R. Sturm, and D. A. Campbell. 2003. Exportin 1 mediates nuclear export of the kinetoplastid spliced leader RNA. Eukaryot. Cell 2:222-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zeiner, G. M., N. R. Sturm, and D. A. Campbell. 2003. The Leishmania tarentolae spliced leader contains determinants for association with polysomes. J. Biol. Chem. 278:38269-38275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.