Abstract
The importance of polarized growth for fungi has elicited significant effort directed at better understanding underlying mechanisms of polarization, with a focus on yeast systems. At sites of tip growth, multiple protein complexes assemble and coordinate to ensure that incoming building material reaches the appropriate destination sites, and polarized growth is maintained. One of these complexes is the polarisome that consists of Spa2, Bud6, Pea2, and Bni1 in Saccharomyces cerevisiae. Filamentous hyphae differ in their development and life style from yeasts and likely regulate polarized growth in a different way. This is expected to reflect on the composition and presence of protein complexes that assemble at the hyphal tip. In this study we searched for polarisome homologues in the model filamentous fungus Aspergillus nidulans and characterized the S. cerevisiae Spa2 and Bud6 homologues, SpaA and BudA. Compared to the S. cerevisiae Spa2, SpaA lacks domain II but has three additional domains that are conserved within filamentous fungi. Gene replacement strains and localization studies show that SpaA functions exclusively at the hyphal tip, while BudA functions at sites of septum formation and possibly at hyphal tips. We show that SpaA is not required for the assembly or maintenance of the Spitzenkörper. We propose that the core function of the polarisome in polarized growth is maintained but with different contributions of polarisome components to the process.
The defining feature of filamentous fungi is the ability to form highly polarized hyphae capable of invading and exploring new territory. The pivotal events that enable the formation of polarized hyphae are the specification and stabilization of an axis of cell polarity. Despite recent progress, the molecular mechanisms underlying these events remain poorly understood in the filamentous fungi (22). With the completion of several fungal genome sequencing projects and the development of tools that facilitate the construction of null mutants, one approach to addressing this problem is to characterize annotated homologues of gene products known to be important for polarized growth in the yeast Saccharomyces cerevisiae. This approach is based on the premise that S. cerevisiae provides a strong paradigm for the basic mechanisms involved in the establishment and maintenance of hyphal polarity.
During the vegetative cycle in S. cerevisiae, the axial (i.e., Bud3, Bud4, and Bud10) and bipolar (Bud8, Bud9, Rax1, and Rax2) landmark proteins direct the establishment of polarity (6, 14, 31). They generate positional information that is recognized by the Ras-related GTPase Bud1 and its associated regulators, Bud2 and Bud5. Activated Bud1 then triggers assembly of the polarity establishment machinery, in which the Rho-related GTPase Cdc42 plays a central role. In its GTP-bound active state, Cdc42 interacts with downstream effectors that include the p21-activated kinases Ste20 and Cla4 (46). In addition, activated Cdc42 recruits several protein complexes to polarization sites. These include the following: (i) the exocyst, which is required for the localized fusion of vesicles with the plasma membrane, (ii) the Arp2/3 complex, which nucleates branched actin filaments that form a cortical actin network involved in endocytosis, and (iii) the polarisome, which controls the formation of actin cables that support the transport of exocytic vesicles (61).
The polarisome was identified in S. cerevisiae as a 12S multiprotein complex that contains Spa2, Pea2, and Bud6/Aip3 and interacts with Bni1 (54). All polarisome components localize to the tips of growing buds and shmoos during polarized tip growth and the mother-bud neck during cytokinesis. Bni1 (bud neck involved) is a formin with multiple domains that controls the localized assembly of actin cables (12, 48, 49) at these sites. A bni1 deletion mutant is viable but has a spherical shape and a wider bud neck due to defects in polarization (42). The proline-rich FH1 domain of Bni1 is responsible for binding profilin, a protein that sequesters ATP-bound G-actin and thereby locally increases the concentration of actin monomer. The FH2 domain then catalyzes the addition of these monomers to the barbed end of nascent actin filaments (38). The C-terminal Dia autoregulatory domain of Bni1 binds in an intramolecular fashion to the N-terminal Rho-binding domain, thereby inhibiting the actin-filament nucleation function of Bni1 (38). Autoinhibition is released upon the binding of a GTP-bound Rho-GTPase with the Rho-binding domain. The Rho-GTPases implicated in the activation of Bni1 include Cdc42 (11), Rho1 (30), as well as Rho3 and Rho4 (9). The less conserved FH3 domain is proposed to be important for the localization of Bni1 (45).
Spa2 (spindle pole associated) is a proposed scaffold protein that was originally identified as a protein that interacts with human anti-spindle pole autoantibodies (56). Spa2 deletion mutants form rounder cells and exhibit a significantly reduced capacity to generate pheromone-induced shmoos (15). As a scaffold protein, Spa2 interacts with numerous proteins, including the other polarisome components (i.e., Bni1, Bud6/Aip3, and Pea2), members of mitogen-activated protein kinase pathways (i.e., Ste11, Ste7, Mkk1, Mkk2, and Slt2) (54, 60), and actin-interacting proteins (i.e., Myo2, Myo1, and Pan1) (55). It is important to note that Spa2 and Bni1 have homologues, Sph1 (47) and Bnr1 (26), respectively, with whom they share overlapping functions. For example, the spa2Δ sph1Δ double deletion mutant is viable with a slightly exacerbated phenotype compared to that of either of the single deletion mutants (47), while the bni1Δ bnr1Δ double disruption mutant shows a temperature-sensitive phenotype and is not viable at the restrictive temperature (26).
Two parallel screens, one for mutants defective in the bipolar pattern of bud site selection (65) and one for actin-interacting proteins (2), led to the identification of Bud6/Aip3 (bud site selection/actin-interacting protein). Besides the defect in bipolar budding, bud6 deletion mutants have rounder vegetative cells than the wild type does, a wider bud neck and bud scar, reduced growth rate and decreased viability, a variation in cell size, and a delay in cytokinesis due to a defect in spindle disassembly (2). On the ultrastructural level, bud6 mutants possess fewer secretory vesicles, form abnormal septa, and display abnormal actin bars in nuclei. The Bud6-green fluorescent protein (GFP) localization pattern was shown to be independent of actin and septins (2). Bud6 interacts with actin through its AIP3 domain, as well as with Pea2, Spa2, and Bni1 (37, 54).
The morphology of fungal hyphae is far more complex that that of yeast cells or even pseudohyphae (19, 57). Although core functions involved in the establishment and maintenance of cell polarity are most likely conserved, the need to support long-range transport along multiple polarity axes must impose additional requirements upon filamentous fungi. This alone indicates that it may not be possible to generalize mechanisms that control polarization and that the presence of equivalent polarization complexes in yeasts and filamentous fungi needs to be tested. The polarisome is a good model to start with. Aspergillus nidulans has a single formin named SepA (septation defective) with conserved formin family domains (21, 36). SepA is an essential gene, and temperature-sensitive sepA mutants lack septa, possess abnormally wide hyphae that undergo abnormal apical branching, and display restricted colonial growth. SepA-GFP localizes to the hyphal tip as a narrow crescent subtended by a dot and also forms a constricting ring at sites of septum formation (52). The colocalization of SepA with actin at both hyphal tips and septation sites suggests a likely role in regulating the dynamics of actin structures (52).
A structure that is present at the hyphal tip in fungi that form only true filaments, including A. nidulans, is the Spitzenkörper (3, 24). The Spitzenkörper is an accumulation of vesicles and other cell components that is present only at actively growing hyphal tips. It is not clear whether the function of the Spitzenkörper overlaps, and to what extent, with the function of the polarisome in filamentous fungi (24, 61).
To date, the only filamentous fungi in which Spa2 and Bud6 homologues have been characterized are Candida albicans and Ashbya gossypii (8, 29, 66). Moreover, C. albicans Spa2 and Bud6 likely associate in a polarisome equivalent (8). However, because these fungi are closely related to S. cerevisiae (i.e., they are all members of the subphylum Saccharomycotina), it is not clear that they are representative of the broader spectrum of filamentous fungi. Accordingly, in this study, we characterize the Spa2 and Bud6 homologues in the model filamentous fungus Aspergillus nidulans and explore their role in a putative filamentous fungal polarisome.
MATERIALS AND METHODS
Strains and growth conditions.
A list of yeast strains used in this study is presented in Table 1. Media were made by the method of Kafer (28). Strains were grown at 28°C unless indicated otherwise.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| A28 | pabaA6 biA1 | FGSC (accession no. A28) |
| GR5 | pyrG89 wA3 pyroA4 | FGSC (accession no. A773) |
| UI224 | pyrG89 yA2 argB2 | 51 |
| TN02A3 | pyrG89 argB2 pyroA4 ΔnkuA::argB | 39 |
| ASH630 | pyrG89 wA3 sepA1 | Lab collection |
| AKS70 | pyrG89 yA2 pabaA1 sepA::gfp::pyr-4 | 52 |
| ACP115 | pyrG89 wA3 pyroA4 tpmA::gfp::pyr-4 | 43 |
| AHS11 | pyrG89 wA3 pyroA4 tubA::gfp::pyr-4 | Lab collection |
| AAV1 | pyrG89 spaA::gfp::pyr-4 wA3 pyroA4 | This study |
| AAV2 | pyrG89 wA3 pyroA4 alcA(p)::gfp::budA::pyr-4 | This study |
| AAV9 | pyrG89 ΔspaA::pyr-4 wA3 pyroA4 | This study |
| AAV11 | pyrG89 ΔspaA::pyr-4 wA3 pyroA4 | This study |
| AAV12 | pyrG89 yA2 spaA::gfp::argB argB2 | This study |
| AAV21 | pyrG89 wA3 pyroA4 ΔbudA::pyro-A4 | This study |
| AAV64 | pyrG89 wA3 argB2 pyroA4 | This study |
| AAV65 | pyrG89 yA2 argB2 pyroA4 | This study |
| AAV85 | pyrG89 ΔspaA::pyr-4 wA3 pyroA4 tubA::gfp::pyr-4 | This study |
| AAV86 | pyrG89 ΔspaA::pyr-4 wA3 pyroA4 tpmA::gfp::pyr-4 | This study |
| AAV87 | pyrG89 sepA::gfp::pyr-4 ΔspaA::pyr-4 wA3 pyroA4 | This study |
Bioinformatic analysis of SpaA and BudA.
The open reading frames AN3815.2 and AN1324.2 were retrieved from the Aspergillus nidulans Database (http://www.broad.mit.edu/annotation/fungi/aspergillus/) and the NCBI (National Center for Biotechnology Information) database (http://www.ncbi.nlm.nih.gov) by BLASTP, using the Saccharomyces cerevisiae YLL021W (spa2) and YLR319C (bud6) open reading frame translations from the Saccharomyces Genome Database (http://www.yeastgenome.org) as queries (1). On the basis of existing homologies, AN3815.2 and AN1324.2 were subsequently renamed spaA and budA, respectively. Both Fungal Genome Initiative (http://www.broad.mit.edu/annotation/fungi/fgi/) and NCBI databases were searched for SpaA and BudA homologues in filamentous fungi. MacVector 7.0 software (Accelrys, Madison, WI) was used for sequence analysis, pairwise and multiple sequence alignments. Conserved domains were identified in the Conserved Domain Database (CDD) at the NCBI website (33) using RPS-BLAST (34). ETANDEM software from the European Molecular Biology Open Source Software Suite (EMBOSS) program package (developed by the European Molecular Biology Laboratory [EMBL] [http://bioweb.pasteur.fr/seqanal/interfaces/etandem.html]) was used for identifying nucleotide tandem repeats.
spaA and budA gene replacements.
The spaA gene replacement was constructed by fusion PCR (64). The spaA gene upstream and downstream sequences were amplified using genomic DNA from strain A28 as the template (Table 1), and the pyr-4 gene of Neurospora crassa was amplified from plasmid pRG3 (62). A list of all primers used in this study is presented in Table 2. Primers spaA-10F and spaA-10R were used to amplify a 2,019-bp sequence upstream of spaA, primers pyr-4-1F and pyr-4-1R were used to amplify the pyr-4 cassette, and spaA-11F and spaA-11R were used to amplify a 2,037-bp sequence downstream of spaA. Amplification of individual fragments was performed with the High Fidelity PCR system (Roche Diagnostics Corporation, Indianapolis, IN) on a Px2 Hybaid thermal cycler or Eppendorf Mastercycler gradient. The amplification products were purified using the QIAquick gel extraction kit (QIAGEN Inc., Valencia, CA), or the GeneClean II kit (Q-Biogene, Irvine, CA). The fusion PCR was performed in a total volume of 10 μl by using the Long Template PCR system (Roche) and cycling conditions recommended by Yang et al. (64), with adjusted annealing temperatures. The spaA gene replacement was cloned into the pCR2.1-TOPO vector (Invitrogen Corporation, Carlsbad, CA) and digested with KpnI, which cuts once into the multicloning site of the vector. The linear plasmid was transformed into strain GR5 (Table 1), and transformants were tested for homologous integration of the gene replacement construct by PCR as outlined by Yang et al. (64). Primer pair spaA-12F and pyr-4-2R was used for the upstream region, and primer pair pyr-4-3F and spaA-13R was used for the downstream region. In addition, to confirm the absence of spaA, two primer pairs were used: spaA-12F and spaA-13R, which amplified a shorter fragment in the event of a homologous spaA gene replacement fragment integration, and spaA-7F and spaA-7R, which amplified the spaA gene from “start” to one codon before “stop.”
TABLE 2.
Primers used in this study
| Primer | DNA sequencea | Purpose |
|---|---|---|
| spaA-10F | GGGTGCTATGCTTGACTTACTAGGCGTC | spaA gene replacement |
| spaA-10R | CCGCACAGATGCGTAAGGAGATCTGCTAGAAAAGTGTCGTTCCAATCCAC | spaA gene replacement |
| pyr-4-1F | GTGGATTGGAACGACACTTTTCTAGCAGATCTCCTTACGCATCTGTGCGG | spaA gene replacement |
| pyr-4-1R | CTAATCTGATGAACTGGCATGGAGGACTATGCATCAGAGCAGATTGTACTG | spaA gene replacement |
| spaA-11F | CAGTACAATCTGCTCTGATGCATAGTCCTCCATGCCAGTTCATCAGATTAG | spaA gene replacement |
| spaA-11R | ATTTGTATCTTCATCGGAGCCTTCCTGG | spaA gene replacement |
| spaA-12F | CGGTCCCCTTGTTATTGATGG | spaA gene replacement integration verification |
| pyr-4-2R | GGCTGTTGTGATTTGCGTTTG | spaA gene replacement integration verification |
| pyr-4-3F | CGAAAGCACGACAGAGGAAGAAG | spaA gene replacement integration verification |
| spaA-13R | CCGTCCAAGCCTTATTGAACAG | spaA gene replacement integration verification |
| budA-12F | CACAGGTACCAGGAACACAAGTCGGTCATGGG | budA gene replacement |
| budA-12R | CACAACTAGTCGGACCTTAACGAAAGACAGCAC | budA gene replacement |
| pyroA-3F | CACAACTAGTGGTTGTTGCCAAGCATGATCG | budA gene replacement |
| pyroA-3R | CACAGAATTCTCTGATGCCAGCCTCTGAAGAC | budA gene replacement |
| budA-13F | CACAGAATTCGCTTGTATGCAAAACAGCCAGG | budA gene replacement |
| budA-13R | CACAGGGCCCGAGAAGATCAGGAAAGGACTGCC | budA gene replacement |
| budA-7F | GAGTGGATGAGGAGAGTATGCCAG | budA gene replacement integration verification |
| budA-7R | GGGGAAATAAATGTCAGGTGAAGG | budA gene replacement integration verification |
| budA-8F | TGGCTAAGCGTGGGTGGTAATAC | budA gene replacement integration verification |
| budA-8R | AGAAGGGAGGGTTTGAATGACC | budA gene replacement integration verification |
| sepG1004for | GAATTCGTCACGAAGTTCCAACTT | spaA::GFP construct |
| sepG1004rev | GGATCCTCGGAAGTCATCGTCCTC | spaA::GFP construct |
| budA-1F | GGCGCGCCGATGACTTCACAGTCTTCTGCAC | alcAp::budA::gfp construct |
| budA-1R | TTAATTAAGGGCATCGTTTTCTCTGAA | alcAp::budA::gfp construct |
| GFPe-1F | GGACGACGGGAACTACAAGA | alcAp::budA::gfp construct integration verification |
| budA-3R | TGTGGAATTGTGAGCGGATA | alcAp::budA::gfp construct integration verification |
| spaA-7F | CACCATGTCGCCCGTCTCCGTAGAC | GATEWAY spaA::gfp entry construct |
| spaA-7R | TCGGAAGTCATCGTCCTCGG | GATEWAY spaA::gfp entry construct |
| budA-4F | CACCATGACTTCACAGTCTTCTGCACC | GATEWAY budA::gfp entry construct |
| budA-4R | ATTCTCGGGGTCTGTTTTATGGC | GATEWAY budA::gfp entry construct |
All sequences are written in the 5′ to 3′ direction. The underlined sequences in these primers are added restriction enzyme site nucleotide extensions or extensions that are complementary to the corresponding primers.
The budA gene replacement was constructed in several steps. First, a 2,171-bp sequence upstream of budA was amplified using primers budA-12F and budA-12R, and a 2,097-bp sequence downstream was amplified using primers budA-13F and budA-13R. Plasmid pAVT24 containing the pCR2.1-TOPO vector and a pyroA cassette from wild-type strain A28 was used as a template to amplify 1,780 bp of the pyroA marker. The pyroA-3 fragment included 573 bp of upstream sequence and was amplified with primers pyroA-3F and pyroA-3R. The downstream budA fragment was ligated into the EcoRI-ApaI site of the pCR2.1-TOPO vector, yielding pAV19. Next, the pyroA-containing fragment was ligated into the SpeI-EcoRI site of pAV19, yielding pAV20. The upstream budA fragment was then integrated at the KpnI-SpeI site of pAV20, giving pAV21. The gene replacement was cut out with ApaI and KpnI, gel extracted and purified, and transformed into strain GR5. Homologous integration by double recombination was verified by PCR the same way as in the spaA gene replacement, using primer pair budA-7F and budA-7R and primer pair budA-8F and budA-8R, as well as primer pair budA4F and budA-4R, which amplified the budA gene from “start” to one codon before “stop.” The gene replacement construct from pAV21 was also transformed into strain TN02A3 (39). Ten transformants were crossed with strains AAV64 and AAV65 (Table 1), and progeny were scored on complete medium and minimal medium lacking pyridoxine to confirm the presence of the pyroA-marked gene replacement.
Molecular manipulations were done using standard protocols (50). The transformation method was based on the protocol by Oakley and Osmani (40).
GFP fusion constructs.
Plasmid pCP33 was obtained by ligating a 1,004-bp C-terminal spaA fragment into the EcoRI-BamHI site of pCP19 (43) using primers sepG1004for and sepG1004rev. Strain GR5 was transformed with pCP33, yielding strain AAV1 that expresses a SpaA-GFP fusion protein (Table 1).
Another C-terminal SpaA-GFP fusion was made by utilizing the GATEWAY system. The full-length spaA without the stop codon was amplified using primers spaA-7F and spaA-7R and ligated into the pENTR/D-TOPO vector using the pENTR Directional TOPO cloning kit (Invitrogen Corporation, Carlsbad, CA), yielding plasmid pAV11. pAV11 was transformed into One Shot ccdB Survival T1 phage-resistant cells (Invitrogen Corporation, Carlsbad, CA), the plasmid was isolated with the QIAprep Spin Miniprep kit (QIAGEN Inc., Valencia, CA), and transferred into the pMT-sGFP destination vector (58) using the GATEWAY LR Clonase enzyme mix according to the manufacturer's instructions (Invitrogen Corporation, Carlsbad, CA). The resulting plasmid pAV15 was transformed into strain UI224, yielding strain AAV12 containing the SpaA-GFP fusion protein (Table 1).
An N-terminal BudA-GFP fusion was made by ligating an 1,194-bp amino-terminal fragment of budA (from “start” using primers budA1F and budA1R) at the AscI-PacI site of the pMCB17apx vector (10, 13), resulting in plasmid pAV2. Strain GR5 was transformed with pAV2, resulting in strain AAV2 containing the fusion construct. Homologous integration of the fusion construct was confirmed with primers GFPe-1F and budA-3R (Table 2).
To select for spontaneous mutations in the pyr-4 gene that disrupt the function of its product, orotidine 5′-monophosphate decarboxylase, strain AAV9 was plated on plates containing 5.74 mM (1g/liter) fluoroorotic acid (U.S. Biological, Swampscott, MA). The spaA gene replacement was confirmed by PCR. The resulting strain AAV11 was used for transformations with plasmids pLO72 containing a tubA fragment fused to gfp (provided by B. Oakley [41]), pCP32 containing a tpmA fragment fused to gfp (43), and pKES59 containing a sepA fragment fused to gfp (52). Transformations yielded strains AAV85 expressing TubA-GFP, AAV86 expressing TpmA-GFP, and AAV87 expressing SepA-GFP.
Cell wall, nuclear, and membrane staining.
For observing nuclear distribution and cell wall deposition, hyphae were fixed using a modified standard protocol (23) [fixing solution contained 3.7% formaldehyde, 25 mM EGTA, 50 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), and 0.5% dimethyl sulfoxide] for 20 min and then stained with staining solution containing both 273 nM fluorescent brightener 28 (Sigma-Aldrich Corporation, St. Louis, MI) and 160 nM Hoechst 33258 (Molecular Probes, Eugene, OR). Membranous material was visualized with FM4-64 (Molecular Probes, Eugene, OR) based on the protocol by Peñalva (44). Conidiospores were grown on coverslips in YGV medium with appropriate supplements for 10 to 12 h and exposed to medium containing 32.92 μM FM4-64 for 2 min (28). The hyphae on coverslips were then transferred to medium without FM4-64 to recover for 1.5 to 4 h, and live growing cells were observed.
Microscopy.
Digital images of plates were collected with an Olympus C-3020ZOOM digital camera. Differential interference contrast (DIC) and fluorescent images were collected with either an Olympus BX51 microscope with a reflected fluorescence system fitted with a Photometrics CoolSnap HQ camera or an Olympus Fluoview confocal laser-scanning microscope. Images were processed with IPLab Scientific Image Processing 3.5.5 (Scanalytics Inc., Fairfax, VA) and Adobe Photoshop 6.0 (Adobe Systems Incorporated, San Jose, CA).
RESULTS
Molecular characterization of SpaA and BudA.
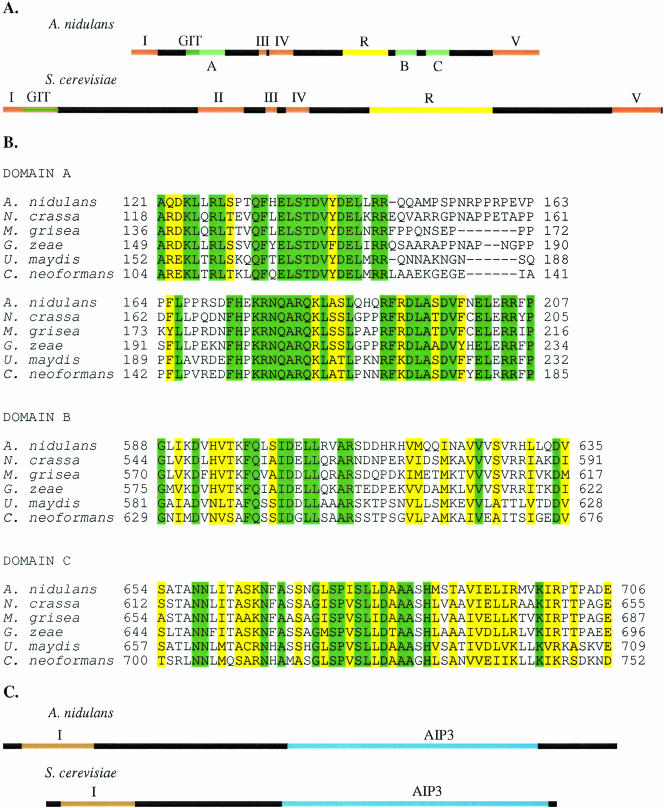
A search for S. cerevisiae Spa2 homologues in A. nidulans yielded a single gene that was renamed spaA. spaA is 2,840 nucleotides long and contains three exons and two introns (Fig. 1A). It translates to a protein 906 amino acids long, with a molecular mass of 101.24 kDa. Out of five Spa2 domains identified in S. cerevisiae (47), only domain II is not present in SpaA. Domain I exhibits 20% similarity but lacks the Spa2 direct repeats. Domains III, IV, and V show 35%, 37%, and 40% similarity, respectively. A nine-amino-acid repeat characterized in Spa2 (15) is also present in SpaA, but the segment (R) is shorter and possesses fewer repeats. A search of the Conserved Domain Database revealed that SpaA contains the GIT domain, a helical motif in the GIT family of ADP-ribosylation factor GTP-activating proteins. A comparison of the SpaA amino acid sequence with the sequences of Spa2 homologues from five other Ascomycetes and Basidiomycetes (Neurospora crassa, Magnaporthe grisea, Gibberella zeae, Ustilago maydis, and Cryptococcus neoformans) showed that there are three SpaA domains conserved among filamentous fungi (Fig. 1B). Spa2 domain A is located between domains I and III and contains the GIT domain and extends towards the carboxy terminus. The amino acid sequences exhibit 60% similarity among the tested sequences. Domains B and C are positioned between the nine-amino-acid repeat sequence and domain V and exhibit 51% and 68% similarity, respectively. According to the CDD, domain C is a part of a slightly larger region that exhibits weak similarity with an uncharacterized integral membrane protein conserved domain, COG5594.1. The Ashbya gossypii AgSpa2 homologue exhibits more similarity with the Spa2 homologues of filamentous fungi than the S. cerevisiae Spa2 does but exhibits less similarity than the Spa2 homologues of filamentous fungi show among themselves. In addition, both spaA and spa2 have tandem repeats in their DNA sequences (29). All these characteristics together suggest that, while some functions of SpaA may be conserved between diverse filamentous and nonfilamentous fungi, SpaA may also have roles unique for filamentous fungi.
FIG. 1.
Homology domains in SpaA and BudA. (A) (Top) Map of A. nidulans SpaA domains. Regions of homology with S. cerevisiae (orange), regions with repeats (yellow), and regions of homology with filamentous fungi (light green) are indicated. The GIT domain is shown in dark green. The domains in SpaA have the following spans: domain I, amino acids 1 to 59; domain III, amino acids 284 to 300; domain IV, amino acids 307 to 360; domain V, amino acids 802 to 906; domain R (9 amino acid repeat), amino acids 470 to 572; GIT, amino acids 121 to 151, domain A, amino acids 121 to 207; domain B, amino acids 588 to 634; and domain C, amino acids 654 to 706. Domain C overlaps with a slightly larger region (amino acids 640 to 745) of COG 5594.1. (Bottom) A map of S. cerevisiae Spa2 domains is shown for comparison. (B) Alignment of domains A, B, and C in five filamentous fungi. Identical (green) and similar (yellow) amino acids are indicated. (C) (Top) Map of A. nidulans BudA domains. Regions of homology (orange and blue) are shown. Domain I spans amino acids 30 to 146, and AIP3 spans amino acids 454 to 853. (Bottom) A map of S. cerevisiae Bud6 domains is shown for comparison.
The S. cerevisiae Bud6 also has a single homologue in A. nidulans. It is 3,013 nucleotides long, contains two exons and one intron (Fig. 1C), and translates to a 981-amino-acid protein with a molecular mass of 107.54 kDa. Compared to the S. cerevisiae Bud6, it shares the AIP3 domain at the carboxy terminus that is important for interactions with actin and another highly homologous domain at the amino terminus (I). An alignment of BudA with Bud6 homologues of five other fungi from the Ascomycetes and Basidiomycetes (N. crassa, G. zeae, U. maydis, C. neoformans, and A. gossypii) showed that both these domains are conserved among filamentous fungi.
A spaA gene replacement strain has a polarity maintenance defect.
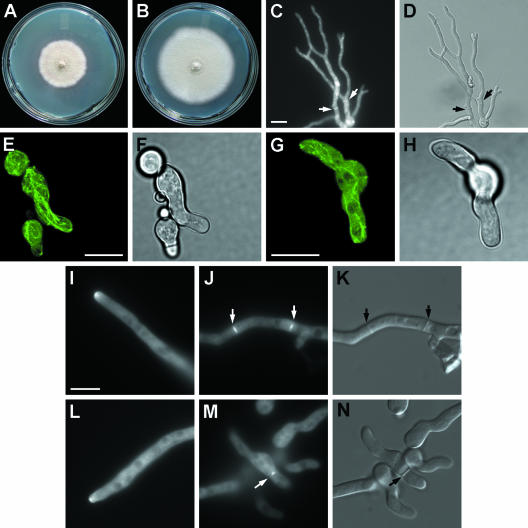
To deduce the role of SpaA in polarized growth, we constructed the spaA gene replacement strain AAV9 (Table 1). This strain has a lower growth rate than the wild type does, and its colonies are more compact (Fig. 2A and B). Moreover, this phenotype segregated as a single gene mutation when AAV9 was backcrossed with the wild type. In wild-type A. nidulans strains, new branches predominantly emerge at a certain distance from the hyphal tip, generating a lateral branch. Dichotomous branching, in which a new branch is formed by splitting the existing tip in two, is a rare occurrence in the wild type. After 13 h of growth on YGV medium with appropriate supplements at 42°C, 99.7% of GR5 hyphae branched laterally (see Fig. 7J and K), and only 1.9% branched dichotomously. In contrast, in the spaA gene replacement strain AAV9, 63.7% branched laterally and 70.6% branched dichotomously under the same conditions (Fig. 2C and D). Data from two separate experiments were compiled, and only hyphae with at least one branch were scored (sample size n = 200). The switch in the branching pattern suggests a role for SpaA at the hyphal tip. Nuclear and cell wall staining of the spaA gene replacement strain AAV9 showed that nuclear distribution and cell wall deposition are not disrupted (Fig. 2C and D). The spaA gene replacement strain expressing GFP-labeled tubulin, AAV85, displayed a relatively normal distribution of microtubules at the hyphal tip (Fig. 2E to H). However, we cannot exclude the possibility that there are differences within other regions of the hyphae. Therefore, SpaA does not affect nuclear structure, nuclear distribution, cell wall structure, cell wall deposition, or hyphal tip microtubule distribution. Actin cables were visualized by GFP-labeled tropomyosin in strain AAV86. The general distribution of actin cables at the hyphal tip and at septum sites (43) is maintained. However, the GFP signal was generally less intense and faded more rapidly than in the wild type (Fig. 2I to N). This suggests that actin cables were less stable than in the wild type. Strain AAV9 generates conidiophores with conidia and produces cleistothecia with ascospores in crosses, indicating that elimination of SpaA does not disrupt the asexual or sexual cycle.
FIG. 2.
Phenotype of the spaA gene replacement strain AAV9. (A) Colony morphology of AAV9 after 7 days of growth on MNV medium (28) at 42°C. (B) Colony morphology of wild-type control strain GR5 after 7 days of growth on MNVUU medium at 42°C. (C) Nuclear (stained with Hoechst 33258) and cell wall distribution (stained with fluorescent brightener 28) of AAV9 after 13 h of growth on YGV medium at 28°C. Bar = 10 μm and applies to the images in panels C and D. (D) DIC image of hyphae from panel C. Arrows indicate positions of septa. (E) Microtubule distribution in AAV9 after 10 h of growth on YGV medium at 28°C. Bar = 10 μm and applies to the images in panels E and F. (F) DIC image of germ tubes from panel E. (G) Microtubule distribution in GR5 after 10 h of growth on YGVUU at 28°C. Bar = 10 μm and applies to the images in panels G and H. (H) DIC image of germ tubes from panel G. (I) Actin cable distribution at hyphal tip in AAV9 after 15 h of growth on YGV at 28°C. Bar = 10 μm and applies to the images in panels I to N. (J) Actin cable distribution at septation sites in AAV9 hypha. (K) DIC image of hypha from panel J. (L) Actin cable distribution at the hyphal tip of GR5 after 16 h of growth on YGVUU at 28°C. (M) Actin cable distribution at the septation site of GR5 germ tube after 12 h of growth. (N) DIC image of the germ tube from panel M. Arrows indicate positions of septa. Images in panels E to H were captured by using a confocal laser-scanning microscope.
FIG. 7.
BudA-GFP distribution in strain AAV2. (A) BudA-GFP signal at septation sites under inducing conditions. (B) DIC image of panel A. (C) Merged image of the images shown in panels A and B. Images in panels A to C were captured by using a confocal laser-scanning microscope. The inserts show an enlarged segment of a hypha containing a septum (marked with asterisks). (D and F) BudA-GFP signal at septation sites under inducing conditions. (E) DIC image of panel D. (G) DIC image of panel F. (H) Fluorescent image of cell wall and nuclear staining under repressing conditions in strain AAV2 under repressing conditions. (I) DIC image of panel H. Dichotomous branching can be seen. (J) Fluorescent image of cell wall and nuclear staining of GR5 under the same conditions as those for strain AAV2 in the images shown in panels H and I. (K) DIC image corresponding to lateral branching (J) can be seen. Bars = 10 μm in all images and apply to the image they are on and the corresponding DIC image. The arrows and arrowheads indicate the positions of septa.
SpaA-GFP localizes to hyphal tips.
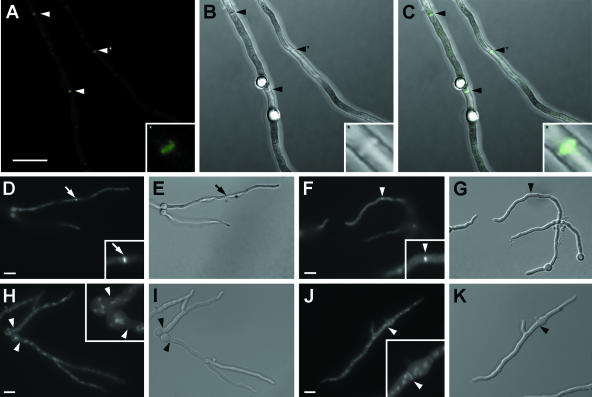
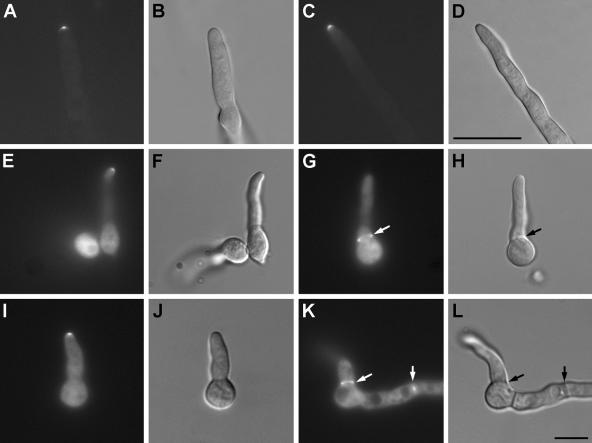
To localize SpaA, we constructed strain AAV1 that expresses a C-terminal SpaA-GFP fusion protein. The phenotypes of the strains harboring these fusions are indistinguishable from the phenotype of the wild-type strain. SpaA-GFP localizes to hyphal tips, but not to septation sites (Fig. 3A to K). At hyphal tips in the mid-focal plane, SpaA-GFP can be seen as a narrow crescent with a slightly rounded central segment. Strain AAV12, which contains a C-terminal SpaA-GFP fusion protein constructed using the GATEWAY technology, shows the same distribution (58). This indicates that the function of SpaA is localized at hyphal tips. As SpaA is present at sites of polarized growth, we looked at conidiophores and conidiospore formation to see whether the same mechanism involved in polarity maintenance during vegetative growth is present during conidiogenesis. Single time points were observed during conidiogenesis, and SpaA-GFP localized to the apex of phialides, but not to the conidiophore vesicle or spores (Fig. 3L and M).
FIG. 3.
SpaA-GFP distribution in strain AAV1. (A) SpaA-GFP fluorescence, (B) DIC, and (C) merged images of a 1-day-old hyphal tip. Images in panels A to C were captured by using the confocal laser-scanning microscope. Bar = 5 μm and applies to the images in panels A to C. All remaining bars are 10 μm and should be applied to the image they are in and the corresponding DIC image. SpaA-GFP fluorescence at the hyphal tips of (D and E) 12-hour-old germlings and the corresponding inserts with enlarged hyphal tips, (F) 12-hour-old germlings that do not have any signal at a septation site (white arrow points to the septum position), and (G)14-hour-old hyphal tips. (H to K) DIC images of images in panels D to G, respectively. In panel J, the black arrow points to the septum position. (L) SpaA-GFP fluorescence and (M) DIC image of strain AAV1 conidiophore. GFP signal is visible only at the tips of phialides (white arrows).
The Spitzenkörper colocalizes with SpaA-GFP at the hyphal tip.
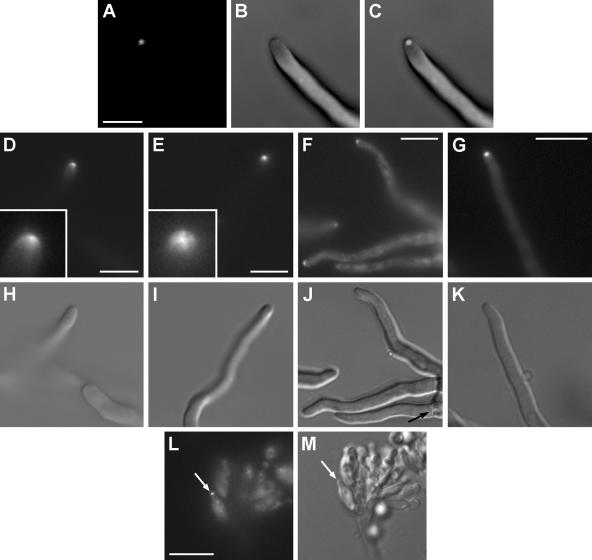
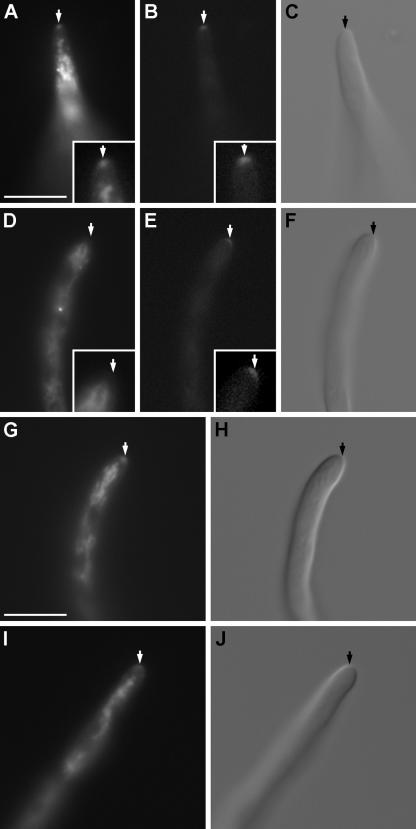
FM4-64 staining of strain AAV1 in which SpaA-GFP is in a wild-type background revealed that the Spitzenkörper is present and that it colocalizes with SpaA-GFP at the hyphal tip (Fig. 4A to C). The distributions of the Spitzenkörper and SpaA-GFP, however, are not identical. The Spitzenkörper forms a round-shaped distinct dot adjacent to the apex, while SpaA-GFP occupies the same region but is less confined and extends to various extents subcortically and radially in the form of a crescent. In addition, the Spitzenkörper disappears within 10 min of observation, but SpaA-GFP persists (Fig. 4D to F). We did not observe the opposite, i.e., the Spitzenkörper persisting and the SpaA-GFP signal disappearing. The persistence of the SpaA-GFP signal most likely reflects the persistence of SpaA at the apical cortex, but we cannot exclude the possibility that the FM4-64 signal is simply less stable and bleaches out faster than the GFP signal does. The localization and difference in the persistence at the hyphal tip together suggest that besides being a part of the Spitzenkörper, SpaA-GFP has additional localization sites and consequently could have functions distinct from Spitzenkörper function. To test this, we stained the spaA gene replacement strain AAV9 with FM4-64. The Spitzenkörper was present in growing hyphal tips of AAV9 (Fig. 4G and H). The Spitzenkörper in the wild-type strain GR5 is presented in Fig. 4I and J for comparison. This suggests that SpaA is not required for the assembly and maintenance of the Spitzenkörper. However, the mechanism and components involved in the switch from lateral to dichotomous branching are still not clear, and therefore, it is possible that SpaA modifies the branching pattern through Spitzenkörper function.
FIG. 4.
Spitzenkörper distribution in live cells of the SpaA-GFP-expressing strain AAV1, the spaA gene replacement strain AAV9, and the wild-type strain GR5. (A) A round Spitzenkörper is present at the hyphal tip of AAV1. Bar = 10 μm and applies to the images in panels A to F, I, and J. (B) The SpaA-GFP signal in the same hypha as in panel A corresponds with the Spitzenkörper distribution and extends radially in the shape of a crescent. (C) A DIC image of the same hypha as in panels A and B. (D) A hyphal tip that does not show the Spitzenkörper, but (E) does show the characteristic SpaA-GFP distribution at the hyphal tip. (F) A DIC image of the hypha from panels D and E. (G) The Spitzenkörper at a hyphal tip of AAV9. Bar = 10 μm, and also applies to the image in panel H. (H) A DIC image of the same hypha as in panel G. (I) The Spitzenkörper at the tip of a GR5 hypha. (J) A DIC image of the same hypha as in panel I. Arrows indicate the apical points of a hyphae. The inserts in panels A, B, D, and E show magnified hyphal tips.
SpaA and SepA localization is partially independent.
As both SepA and SpaA could function together, we looked at the distribution of SpaA-GFP in the sepA1 mutant strain ASH630. ASH630 has a point mutation in the FH2 domain of sepA that causes substitution of a hydrophilic amino acid for a hydrophobic amino acid and presumably results in impaired protein-protein interactions at this domain (52). SpaA was present at hyphal tips of ASH630 (Fig. 5A and B), suggesting that SepA function is not required for the deposition of SpaA at the hyphal tip of this mutant. The same distribution is present in the wild type (Fig. 5C and D).
FIG. 5.
(A) SpaA-GFP signal at a hyphal tip of strain ASH630. (B) DIC image of hypha from panel A. (C) SpaA-GFP signal at a hyphal tip of strain GR5. (D) DIC image of the hypha from C. Bar = 20 μm and applies to the images in panels A to D. (E) SepA-GFP signal in a 13.5-hour-old hyphal tip of the spaA gene replacement strain AAV9. (F) DIC image of the hypha from panel E. (G) SepA-GFP signal at a septum in spaA gene replacement strain AAV9 hypha. (H) DIC image of the hypha from panel G. (I) SepA-GFP signal at the hyphal tip of a 14-hour-old AKS70 hypha. (J) DIC image of the hypha from panel I. (K) SepA-GFP signal at a septum in an AKS70 hypha. (L) DIC image of the hypha from panel K. Bar = 10 μm and applies to images in panels E to L. The arrows indicate the positions of septa.
Next, we looked at the distribution of SepA-GFP at the tips of vegetative hyphae of the spaA gene replacement strain. SepA-GFP still localizes to the tip and septum sites when SpaA is absent (Fig. 5E to H). The same distribution of SepA-GFP is present in a wild-type background (Fig. 5I to L). This indicates that, unlike S. cerevisiae, SpaA is not required for localization of SepA to the hyphal tip.
budA gene replacement.
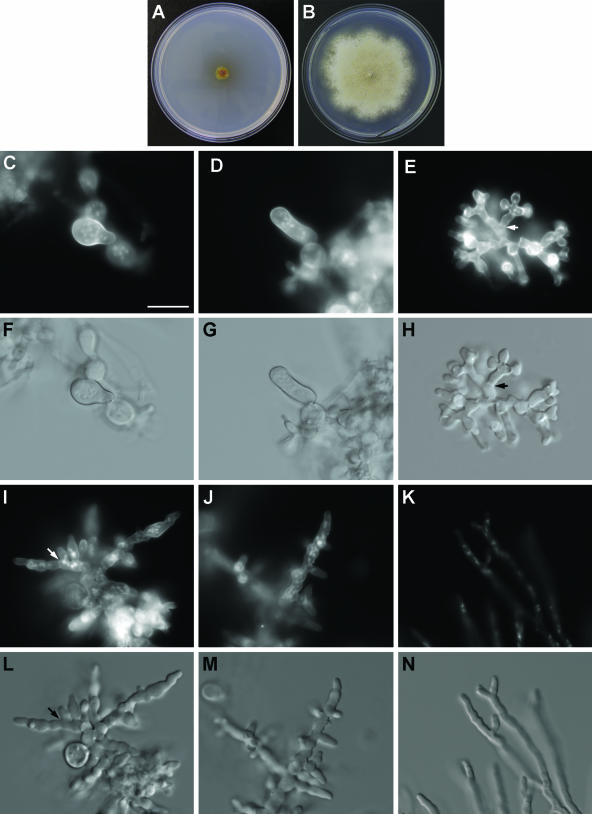
Linearized ΔbudA::pyroA fragment was transformed into strain GR5. Out of 47 tested transformants that grew on selective plates, all had the wild-type phenotype. Diagnostic PCR (64) indicated that eight transformants contained both the ΔbudA::pyroA gene replacement fragment and the endogenous budA gene. This would be possible only if the transformants were heterokaryotic. However, even homokaryotic colonies obtained after the plating of uninucleate conidiospores possessed both the replacement and wild-type alleles of budA. The presence of wild-type budA and ΔbudA::pyroA in the same nucleus can be explained if the absence of budA is lethal or near lethal. In this case, the selection for viability or improved growth may have resulted in retention of a short budA-containing chromosomal fragment, resulting in a disomic state. The principle of using a duplicated fragment that contains the wild-type version of an essential gene to shelter a lethal knockout underlies a proposed strategy for observing the phenotypes of lethal mutations in N. crassa (35). To eliminate the possible presence of extrachromosomal fragments, we backcrossed the homokaryotic strains with strain GR5 that was used in the original transformation, and 50 to 100 progeny were tested for each cross. There were three categories of colony phenotypes in the progeny: about half of them had the wild-type phenotype, another half were slightly slower growing and more compact, and two were very slow growing, with an irregular colony front and without any conidia. Diagnostic PCR indicated that only the two very slowly growing progeny had the ΔbudA::pyroA gene replacement fragment and no endogenous budA gene. One of two strains, AAV21, was examined further. To confirm the ΔbudA::pyroA gene replacement phenotype, we additionally transformed a strain that favors homologous DNA integration, TN02A3 (39), with the ΔbudA::pyroA gene replacement construct. Eighteen transformants were tested by diagnostic PCR, and 10 contained both the ΔbudA::pyroA gene replacement fragment and endogenous budA. All 10 of these transformants were crossed with appropriate strains (see Materials and Methods), and their progeny were scored for the AAV21 phenotype. Out of seven successful outcrosses, three yielded progeny that clearly showed the same phenotype as that of AAV21.
In the absence of BudA, the hyphal growth rate is dramatically reduced compared to that of the wild type (Fig. 6A and B). The colonies are compact and do not develop conidiophores, indicating that the asexual cycle is disrupted. We could not obtain spores to follow the germination sequence, but within the growing colony we could identify large round cells that may be rare conidiospores. These cells were much larger than wild-type conidia and contained a large number of nuclei. The only two germinating spores we could find are presented in Fig. 6C, D, F, and G. We encountered a range of different hyphal morphologies and branching types in strain AAV21. The most common was a slowly growing dichotomously branched appearance which had an increased branching frequency and wider hyphal diameter compared to the wild type. The hyphae also had an increased number of nuclei per hyphal diameter, and septa were present (Fig. 6E and H). Other regions had slow growth, a lateral branching pattern, an increased hyphal diameter with irregular constrictions, and an increased number of nuclei per hyphal diameter (Fig. 6I, J, L, and M). In all these cases, the increased hyphal diameter and the meandering appearance indicated a difficulty in maintaining polarity. Finally, there were regions where the growth rate and hyphal diameter were closer to those of the wild type, but the branching pattern was dichotomous (Fig. 6K and N). AAV21 colonies do not form cleistothecia, indicating that the sexual cycle is disrupted.
FIG. 6.
Phenotype of budA gene replacement strain AAV21. (A) Growth of the budA gene replacement strain AAV21 on MNUU medium at 28°C for 8 days. (B) Growth of strain GR5 on MNVUU medium at 28°C for 8 days. (C to N) Cell wall and nuclear distribution within AAV21 spores and hyphae after growth at 28°C for 2 days and the corresponding DIC images. (C) Large multinucleate spore with an emerging germ tube. (D) Wide potato-shaped multinucleate germling. (E) Multinucleate dichotomously branched hyphae with increased branching frequency and increased hyphal diameter. (F) DIC image of panel C. (G) DIC image of panel D. (H) DIC image of panel E. (I and J) Hyphae with an increased hyphal diameter and irregular nuclear distribution. (K) Dichotomous branching pattern of hyphae with regular nuclear distribution. (L) DIC image of panel I. (M) DIC image of panel J. (N) DIC image of panel K. Bar = 10 μm and applies to all images. Arrows indicate the positions of septa.
BudA localizes to sites of septum formation.
We constructed strain AAV2 in which the expression of budA::gfp is controlled by the alcA inducible promoter. Expression of a gene by this promoter is repressed on glucose-containing medium, partially induced on glycerol-containing medium, and overexpressed on ethanol-containing medium (62). On inducing medium, the BudA-GFP signal was visible only at sites of septum formation (Fig. 7A to G), not at hyphal tips or any other hyphal structures. On repressing medium, the branching pattern was predominantly dichotomous, and although septa were present, they did not stain as strongly and were not as abundant as in the wild type (Fig. 7H to K). At 12 h, germ tubes with more than eight nuclei were scored for the presence or absence of septa. At this stage, a septum should be present at the base of the germ tube. In the wild type, 98.5% of germlings had septa compared to only 46.5% of AAV2 germlings (n = 200; data from two separate experiments were compiled for each strain). Under repressing conditions, the hyphae of AAV2 appeared wider than those of the wild type. The absence of BudA did not affect the distribution of nuclei.
DISCUSSION
In A. nidulans, the single formin homologue, SepA, localizes to the hyphal tip in the form of a cortical crescent and a subjacent apical dot (52). The Spitzenkörper, a structure present exclusively in hyphal tips of actively growing hyphae (16, 24), is also visualized as an apical dot (44). These distributions, together with the function of both structures in hyphal morphogenesis (24), strongly suggest that SepA is a component of the Spitzenkörper. In yeasts, formins associate with proteins such as Spa2 and Bud6 to form a complex called the polarisome that is necessary for normal morphogenesis (46). The association of formins with other proteins has not been explored in filamentous fungi, raising the question of whether a functional polarisome exists in this group. Another unknown is the relationship between polarisome homologues and the Spitzenkörper in filamentous fungi. The absence of the Spitzenkörper in the growing tips of budding yeast cells (8) would render any connection of this kind unique for filamentous fungi. As the first step in answering these questions, we used a reverse genetic approach to functionally characterize the A. nidulans Spa2 and Bud6 homologues, SpaA and BudA.
Our bioinformatic analysis of SpaA showed that although some domains are conserved among both yeasts and filamentous fungi (i.e., domains I, III, IV, V, and GIT), SpaA possesses other domains that are conserved in filamentous fungi but not in S. cerevisiae (i.e., domains A, B, and C). Because SpaA does not contain transmembrane domains, the similarity of domain C to a conserved domain present in membrane proteins is particularly interesting, suggesting a possible function at the plasma membrane that could be shared with membrane proteins. Hyphal tips are enriched for membrane-associated signaling proteins, such as various Rho-GTPases (4, 18). Some of these signaling proteins, such as RacI, are present in filamentous fungi but not in S. cerevisiae (5, 7), making them potential candidates for interaction with domain A, B, or C. BudA, on the other hand, is conserved and has two regions of homology with other fungal Bud6 homologues, an N-terminal homology domain, and an AIP3 homology domain implicated in actin binding (2).
The dichotomous branching pattern of the spaA gene replacement mutant and SpaA-GFP localization to the tip suggest that SpaA functions solely at the tip. This and the increase in apical branching associated with the loss of SpaA indicate a possible role for SpaA in the stabilization of hyphal polarity axes. In S. cerevisiae, Spa2 is distributed both at bud and shmoo tips, as well as at mother-bud necks (15). Deletion of S. cerevisiae Spa2 results in a greatly reduced capacity to form shmoos and in disruption of the polarized growth machinery, including mislocalization of actin (15) and the secretion marker Sec4 (53). In Candida albicans, a facultative fungal human pathogen that generates yeast, pseudohyphal, and true hyphal cells, the Spa2 homologue localizes to a crescent adjacent to the plasma membrane in the tips of true hyphae (8, 66). The presence of Spa2 at septation sites is rare (66), and even then it is not clear whether the authors were assessing Spa2 distribution in true hyphae or pseudohyphae (for classification of C. albicans cell types, see reference 57), as shallow constrictions at septation sites were visible. C. albicans Spa2 deletion results in broader germ tubes with a less polarized appearance (8). In A. gossypii, a filamentous fungus closely related to S. cerevisiae, A. gossypii Spa2 (AgSpa2) localizes both to hyphal tips and to septation sites (29). On the basis of the slower growth and persistent dichotomous branching of the AgSpa2 deletion strain, the authors propose that AgSpa2 determines the area of growth at the hyphal tip and organizes the incorporation of exocytic vesicles at the hyphal tip. Nevertheless, the unifying feature of Spa2 in all yeasts appears to be its involvement in morphogenetic events at growing tips and at septation sites. Notably, the role of Spa2 in A. nidulans, a representative of the euascomycete filamentous fungi, differs from the yeasts in two important respects. First, SpaA function appears to be exclusively limited to polarization events at the hyphal tip. We were unable to detect an obvious role for SpaA in any aspect of septum formation. Second, SpaA is not required for SepA localization, as SepA is present at both hyphal tips and septa in the spaA gene replacement strain. By contrast, formin localization at S. cerevisiae bud sites and C. albicans hyphal tips is largely dependent on the presence of Spa2 (42).
Our results do not fully address the relationship between the Spa2 homologue and Spitzenkörper, but they do provide some informative insights. We already know that SepA colocalizes with the Spitzenkörper but that it also forms a membrane crescent. As in C. albicans (8), SpaA may overlap with the Spitzenkörper to some degree and regulate its function, even if different binding partners are involved. However, in C. albicans, the deletion of SpaA results in the loss of a visible Spitzenkörper, while this is not the case in A. nidulans. Although the presence of the Spitzenkörper in the spaA gene replacement strain shows that SpaA is not required for the establishment and maintenance of the Spitzenkörper, we cannot exclude the possibility that the absence of SpaA causes the switch from a lateral to a dichotomous branching pattern by altering the function of the Spitzenkörper. Alternatively, the colocalization of SpaA and the Spitzenkörper in wild-type hyphae may simply be a result of different independent complexes acting simultaneously at the site of active hyphal tip growth (61). In this case, localization of SpaA at the Spizenkörper may merely reflect transport of SpaA to destination sites at the hyphal tip. We know that components of the secretory pathway mediate the localization of polarisome components in yeasts (27), and the same may apply for A. nidulans. Although a more detailed study at higher resolution needs to be done to confirm a more cortical position of SpaA, our results support the view that SpaA as a part of the putative polarisome acts, at least partially, independently of the Spitzenkörper.
Whereas the deletion of bud6 in S. cerevisiae or Schizosaccharomyces pombe has no adverse affect on colony formation (17, 65), the A. nidulans budA gene replacement caused a severe growth defect. This appears to be largely attributable to the failure to establish and maintain stable axes of hyphal polarity. Because we could not detect BudA localization at hyphal tips, which may be due to a weak or unstable signal, the molecular function of BudA at this site remains elusive. However, the significant overlap in polarity defects caused by budA and sepA deletion mutants (52) is consistent with the possibility that BudA may regulate SepA function at hyphal tips.
The modest septation defect observed in the repressed alcA(p)::gfp::budA strain implies that, like Bud6 in S. cerevisiae and S. pombe, both yeasts, BudA also has a role in septum formation. Moreover, the transient presence of BudA at septation sites, where both SepA and actin cables can be found, suggests that BudA may function as a part of the cytokinetic actin ring (20). Indeed, this may be a shared function common to all ascomycete Bud6 homologues. Although the role of BudA at septation sites is likely to promote actin ring formation (see below) (38), we cannot rule out the possibility that BudA has an additional function that promotes SepA recruitment to the septation site. In particular, recent results from S. cerevisiae show that Bud6 acts downstream of the septins to mediate the formation of a membrane barrier at the mother-bud neck (32). This barrier enables the localized accumulation of factors required for cytokinesis and septum assembly. The A. nidulans septin AspB also localizes to septation sites, where it ultimately forms a double ring that sandwiches the cytokinetic actin ring (63). Accordingly, BudA may act downstream of the septins to coordinate the localization of SepA and actin at septation sites in A. nidulans (20, 52).
Our results raise important questions concerning the conservation of the polarisome. The range of polarisome gene conservation within the A. nidulans genome, from highly conserved (BudA) to partially conserved (SpaA) to not present at all (Pea2 [22, 59]), initially suggested that it may have an altered function relative to S. cerevisiae and related filamentous hemiascomycetes. Our characterization of SpaA provides some support for this notion. In particular, SpaA functions exclusively at hyphal tips, where we propose that it acts as a scaffold to promote the formation of a multiprotein complex required for the organization of the cytoskeleton and/or localized vesicle trafficking. Moreover, this complex may not include the formin SepA. Instead, like the mammalian GIT domain proteins (25), SpaA may regulate hyphal tip organization by association with alternative partners, such as the paxillins, ADP-ribosylation factors, or p21-activated kinases. There may also be additional fungal-specific partners that bind to SpaA via domain A, B, or C. On the other hand, our results suggest that BudA may function like yeast Bud6 as an actin monomer-binding protein that generates a localized pool of actin monomer to promote microfilament formation (38). In this context, the formin-Bud6 interaction may constitute the conserved core of the fungal polarisome.
To conclude, we suggest that the basic function of the putative polarisome in polarized growth is preserved in filamentous fungi, albeit with a modified set of components that have altered contributions to the polarisome function.
Acknowledgments
This study was supported with funds provided by the Nebraska Research Foundation.
We thank Berl R. Oakley (The Ohio State University) for plasmid pLO72. We also thank Claire L. Pearson for constructing plasmid pCP33.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amberg, D. C., J. E. Zahner, J. W. Mulholland, J. R. Pringle, and D. Botstein. 1997. Aip3p/Bud6p, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol. Biol. Cell 8:729-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartnicki-García, S., E. Hegert, and G. Gierz. 1990. A novel computer model for generating cell shape: application to fungal morphogenesis, p. 43-60. In P. J. Kuhn, A. P. J. Trinci, M. J. Jung, M. W. Goosey, and L. G. Coping (ed.), Biochemistry of cell walls and membranes in fungi. Springer-Verlag, Berlin, Germany.
- 4.Boyce, K. J., M. J. Hynes, and A. Andrianopoulos. 2001. The CDC42 homolog of the dimorphic fungus Penicillium marneffei is required for correct cell polarization during growth but not development. J. Bacteriol. 183:3447-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyce, K. J., M. J. Hynes, and A. Andrianopoulos. 2003. Control of morphogenesis and actin localization by the Penicillium marneffei RAC homologue. J. Cell Sci. 116:1249-1260. [DOI] [PubMed] [Google Scholar]
- 6.Chant, J. 1999. Cell polarity in yeast. Annu. Rev. Cell Dev. Biol. 15:3365-3391. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C., and M. Dickman. 2004. Dominant active Rac and dominant negative Rac revert the dominant active Ras phenotype in Colletotrichum trifolii by distinct signalling pathways. Mol. Microbiol. 51:1493-1507. [DOI] [PubMed] [Google Scholar]
- 8.Crampin, H., K. Finley, M. Gerami-Nejad, H. Court, C. Gale, J. Berman, and P. Sudbery. 2005. Candida albicans hyphae have a Spitzenkörper that is distinct from the polarisome found in yeast and pseudohyphae. J. Cell Biol. 118:2935-2947. [DOI] [PubMed] [Google Scholar]
- 9.Dong, Y., D. Pruyne, and A. Bretscher. 2003. Formin-dependent actin assembly is regulated by distinct modes of Rho signaling in yeast. J. Cell Biol. 161:1081-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Efimov, V. P. 2003. Roles of NUDE and NUDF proteins of Aspergillus nidulans: insights from intracellular localization and overexpression effects. Mol. Biol. Cell 14:871-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evangelista, M., K. Blundell, M. S. Longtine, C. J. Chow, N. Adames, J. R. Pringle, M. Peter, and C. Boone. 1997. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science 276:118-122. [DOI] [PubMed] [Google Scholar]
- 12.Evangelista, M., S. Zigmond, and C. Boone. 2003. Formins: signaling effectors for assembly and polarization of actin filaments. J. Cell Sci. 116:2603-2611. [DOI] [PubMed] [Google Scholar]
- 13.Fernández-Ábalos, J. M., H. Fox, C. Pitt, B. Wells, and J. H. Doonan. 1998. Plant-adapted green fluorescent protein is a versatile vital reporter for gene expression, protein localization and mitosis in the filamentous fungus, Aspergillus nidulans. Mol. Microbiol. 27:121-130. [DOI] [PubMed] [Google Scholar]
- 14.Fujita, A., M. Lord, T. Hiroko, T. Chen, C. Oka, Y. Misumi, and J. Chant. 2004. Rax1, a protein required for the establishment of the bipolar budding pattern in yeast. Gene 327:161-169. [DOI] [PubMed] [Google Scholar]
- 15.Gehrung, S., and M. Snyder. 1990. The SPA2 gene of Saccharomyces cerevisiae is important for pheromone-induced morphogenesis and efficient mating. J. Cell Biol. 111:1451-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Girbardt, M. 1957. Der Spitzenkörper von Polystictus versicolor. Planta 50:47-59. [Google Scholar]
- 17.Glynn, J. M., H. J. Lustig, A. Berlin, and F. Chang. 2001. Role of bud6p and tea1p in the interaction between actin and microtubules for the establishment of cell polarity in fission yeast. Curr. Biol. 11:836-845. [DOI] [PubMed] [Google Scholar]
- 18.Guest, G. M., X. Lin, and M. Momany. 2004. Aspergillus nidulans RhoA is involved in polar growth, branching, and cell wall synthesis. Fungal Genet. Biol. 41:13-22. [DOI] [PubMed] [Google Scholar]
- 19.Harris, S. D. 1997. The duplication cycle in Aspergillus nidulans. Fungal Genet. Biol. 22:1-12. [DOI] [PubMed] [Google Scholar]
- 20.Harris, S. D. 2001. Septum formation in Aspergillus nidulans. Curr. Opin. Microbiol. 4:736-739. [DOI] [PubMed] [Google Scholar]
- 21.Harris, S. D., L. Hamer, K. E. Sharpless, and J. E. Hamer. 1997. The Aspergillus nidulans sepA gene encodes an FH1/2 protein involved in cytokinesis and the maintenance of cellular polarity. EMBO J. 16:3474-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris, S. D., and M. Momany. 2004. Polarity in filamentous fungi: moving beyond the yeast paradigm. Fungal Genet. Biol. 41:391-400. [DOI] [PubMed] [Google Scholar]
- 23.Harris, S. D., J. L. Morrell, and J. E. Hamer. 1994. Identification and characterization of Aspergillus nidulans mutants defective in cytokinesis. Genetics 136:517-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris, S. D., N. D. Read, R. W. Roberson, B. Shaw, S. Seiler, M. Plamann, and M. Momany. 2005. Spitzenkörper meets polarisome: microscopy, genetics, and genomics coverage. Eukaryot. Cell 4:225-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoefen, R. J., and B. C. Berk. 2006. The multifunctional GIT family of proteins. J. Cell Sci. 119:1469-1475. [DOI] [PubMed] [Google Scholar]
- 26.Imamura, H., K. Tanaka, T. Hihara, M. Umikawa, T. Kamei, K. Takahashi, T. Sasaki, and Y. Takai. 1997. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 16:2745-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin, H., and D. C. Amberg. 2000. The secretory pathway mediates localization of the cell polarity regulator Aip3p/Bud6p. Mol. Biol. Cell 11:647-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kafer, E. 1977. Meiotic and mitotic recombination in Aspergillus, and its chromosomal aberrations. Adv. Genet. 19:33-131. [DOI] [PubMed] [Google Scholar]
- 29.Knechtle, P., F. Dietrich, and P. Philippsen. 2003. Maximal polar growth potential depends on the polarisome component AgSpa2 in the filamentous fungus Ashbya gossypii. Mol. Biol. Cell 14:4140-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kohno, H., K. Tanaka, A. Mino, M. Umikawa, H. Imamura, T. Fujiwara, Y. Fujita, K. Hotta, H. Qadota, T. Watanabe, Y. Ohya, and Y. Takai. 1996. Bni1 implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 15:6060-6068. [PMC free article] [PubMed] [Google Scholar]
- 31.Lord, M., F. Inose, T. Hiroko, T. Hata, A. Fujita, and J. Chant. 2002. Subcellular localization of Axl1, the cell type-specific regulator of polarity. Curr. Biol. 12:1347-1352. [DOI] [PubMed] [Google Scholar]
- 32.Luedke, C., S. B. Frei, I. Sbalzarini, H. Schwarz, A. Spang, and Y. Barral. 2005. Septin-dependent compartmentalization of the endoplasmic reticulum during yeast polarized growth. J. Cell Biol. 169:897-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CCD: a conserved domain database for protein classification. Nucleic Acids Res. 33:D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marchler-Bauer, A., and S. H. Bryant. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327-W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Metzenberg, R. L. 2005. Construction of minimally-sheltered knockout mutants of Neurospora crassa. Fungal Genet. Newslett. 52:11-13. [Google Scholar]
- 36.Morris, R. N. 1976. Mitotic mutants of Aspergillus nidulans. Genet. Res. 26:237-254. [DOI] [PubMed] [Google Scholar]
- 37.Moseley, J. B., and B. L. Goode. 2005. Differential activities and regulation of S. cerevisiae formins Bni1 and Bnr1 by Bud6. J. Biol. Chem. 280:28023-28033. [DOI] [PubMed] [Google Scholar]
- 38.Moseley, J. B., I. Sagot, A. L. Manning, Y. Xu, M. J. Eck, D. Pellman, and B. L. Goode. 2004. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol. Cell. Biol. 15:896-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nayak, T., E. Szewczyk, C. E. Oakley, A. Osmani, L. Ukil, S. L. Murray, M. J. Hynes, S. A. Osmani, and B. R. Oakley. 2006. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 173:1557-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oakley, B. R., and S. A. Osmani. 1993. Cell cycle analysis using the filamentous fungus Aspergillus nidulans, p. 27-142. In P. Fantes and R. Brooks (ed.), The cell cycle: a practical approach. Academic Press, London, United Kingdom.
- 41.Osmani, A. H., J. Davies, C. E. Oakley, B. R. Oakley, and S. A. Osmani. 2003. TINA interacts with the NIMA kinase in Aspergillus nidulans and negatively regulates astral microtubules during metaphase arrest. Mol. Biol. Cell 14:3169-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ozaki-Kuroda, K., Y. Yamamoto, H. Nohara, M. Kinoshita, T. Fujiwara, K. Irie, and Y. Takai. 2001. Dynamic localization and function of Bni1p at the sites of directed growth in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:827-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pearson, C. L., K. Xu, K. E. Sharpless, and S. D. Harris. 2004. MesA, a novel fungal protein required for the stabilization of polarity axes in Aspergillus nidulans. Mol. Biol. Cell 15:3658-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peñalva, M. A. 2005. Tracing the endocytic pathway of Aspergillus nidulans with FM4-64. Fungal Genet. Biol. 42:963-975. [DOI] [PubMed] [Google Scholar]
- 45.Petersen, J., O. Nielsen, R. Egel, and I. M. Hagan. 1998. FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J. Cell Biol. 141:1217-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pruyne, D., and A. Bretscher. 2000. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 113:365-375. [DOI] [PubMed] [Google Scholar]
- 47.Roemer, T., L. Vallier, Y. J. Sheu, and M. Snyder. 1998. The Spa2-related protein, Sph1p, is important for polarized growth in yeast. J. Cell Sci. 111:479-494. [DOI] [PubMed] [Google Scholar]
- 48.Sagot, I., S. K. Klee, and D. Pellman. 2002. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat. Cell Biol. 4:42-50. [DOI] [PubMed] [Google Scholar]
- 49.Sagot, I., A. A. Rodal, J. Moseley, B. L. Goode, and D. Pellman. 2002. An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol. 4:626-631. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 51.Semighini, C. P., M. R. Van Zeska Kress Fagundes, J. C. Ferreira, R. C. Pascon, M. H. De Souza Goldman, and G. H. Goldman. 2003. Different roles of the Mre11 complex in the DNA damage response in Aspergillus nidulans. Mol. Microbiol. 48:1693-1709. [DOI] [PubMed] [Google Scholar]
- 52.Sharpless, K. E., and S. D. Harris. 2002. Functional characterization and localization of the Aspergillus nidulans formin SEPA. Mol. Biol. Cell 13:469-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sheu, Y. J., Y. Barral, and M. Snyder. 2000. Polarized growth controls cell shape and bipolar bud site selection in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:5235-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheu, Y. J., B. Santos, N. Fortin, C. Costigan, and M. Snyder. 1998. Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol. Cell. Biol. 18:4053-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shih, J. L., S. L. Reck-Peterson, R. Newitt, M. S. Mooseker, R. Aebersold, and I. Herskowitz. 2005. Cell polarity protein Spa2p associates with proteins involved in actin function in Saccharomyces cerevisiae. Mol. Biol. Cell 16:4595-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Snyder, M. 1989. The SPA2 protein of yeast localizes to sites of cell growth. J. Cell Biol. 108:1419-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sudbery, P., N. Gow, and J. Berman. 2004. The distinct morphogenetic states of Candida albicans. Trends Microbiol. 12:317-324. [DOI] [PubMed] [Google Scholar]
- 58.Toews, M. W., J. Warmbold, S. Konzack, P. Rischitor, D. Veith, K. Vieken, C. Vinuesa, H. Wei, and R. Fischer. 2004. Establishment of mRFP1 as a fluorescent marker in Aspergillus nidulans and construction of expression vectors for high throughput protein tagging using recombination in vitro (GATEWAY). Curr. Genet. 45:383-389. [DOI] [PubMed] [Google Scholar]
- 59.Valtz, N., and I. Herskowitz. 1996. Pea2 protein of yeast is localized to sites of polarized growth and is required for efficient mating and bipolar budding. J. Cell Biol. 135:725-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Drogen, F., and M. Peter. 2002. Spa2 functions as a scaffold-like protein to recruit the Mpk1p MAP kinase module to sites of polarized growth. Curr. Biol. 12:1698-1703. [DOI] [PubMed] [Google Scholar]
- 61.Virag, A., and S. D. Harris. 2006. The Spitzenkörper—a molecular perspective. Mycol. Res. 110:4-13. [DOI] [PubMed] [Google Scholar]
- 62.Waring, R. B., G. S. May, and N. R. Morris. 1989. Characterization of an inducible expression system in Aspergillus nidulans using alcA and tubulin-coding genes. Gene 79:119-130. [DOI] [PubMed] [Google Scholar]
- 63.Westfall, P. J., and M. Momany. 2002. Aspergillus nidulans septin AspB plays pre- and postmitotic roles in septum, branch and conidiophore development. Mol. Biol. Cell 13:110-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang, L., L. Ukil, A. Osmani, F. Nahm, J. Davies, C. P. C. De Souza, X. Dou, A. Perez-Balaguer, and S. A. Osmani. 2004. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot. Cell 3:1359-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zahner, J. E., H. A. Harkins, and J. R. Pringle. 1996. Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 16:1857-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng, X. D., Y. M. Wang, and Y. Wang. 2003. CASPA2 is important for polarity establishment and maintenance in Candida albicans. Mol. Microbiol. 49:1391-1405. [DOI] [PubMed] [Google Scholar]