Abstract
The hexose-proton symporter HUP1 shows a spotty distribution in the plasma membrane of the green alga Chlorella kessleri. Chlorella cannot be transformed so far. To study the membrane localization of the HUP1 protein in detail, the symporter was fused to green fluorescent protein (GFP) and heterologously expressed in Saccharomyces cerevisiae and Schizosaccharomyces pombe. In these organisms, the HUP1 protein has previously been shown to be fully active. The GFP fusion protein was exclusively targeted to the plasma membranes of both types of fungal cells. In S. cerevisiae, it was distributed nonhomogenously and concentrated in spots resembling the patchy appearance observed previously for endogenous H+ symporters. It is documented that the Chlorella protein colocalizes with yeast proteins that are concentrated in 300-nm raft-based membrane compartments. On the other hand, it is completely excluded from the raft compartment housing the yeast H+/ATPase. As judged by their solubilities in Triton X-100, the HUP1 protein extracted from Chlorella and the GFP fusion protein extracted from S. cerevisiae are detergent-resistant raft proteins. S. cerevisiae mutants lacking the typical raft lipids ergosterol and sphingolipids showed a homogenous distribution of HUP1-GFP within the plasma membrane. In an ergosterol synthesis (erg6) mutant, the rate of glucose uptake was reduced to less than one-third that of corresponding wild-type cells. In S. pombe, the sterol-rich plasma membrane domains can be stained in vivo with filipin. Chlorella HUP1-GFP accumulated exactly in these domains. Altogether, it is demonstrated here that a plant membrane protein has the property of being concentrated in specific raft-based membrane compartments and that the information for its raft association is retained between even distantly related organisms.
A large volume of recent studies suggest that the plasma membranes of mammalian and fungal cells are laterally subcompartmented. One type of microdomain, enriched in sterols, sphingolipids, and specific proteins, comprises lipid rafts (1, 11, 46). In mammalian cells, the sizes of individual rafts have been estimated to be 25 to 70 nm in diameter. Individual rafts cannot be resolved in living cells by light microscopy, which indicates that the distance between the rafts is on the same order of magnitude as their size (29, 35). A patchy distribution of raft markers in mammalian cells is light-microscopically visible only after treatment with antibodies or cross-linkers (15). Due to their insolubility in mild nonionic detergents (1% Triton X-100 at 4°C), rafts are operationally defined as detergent-resistant membranes (DRM) (6). After density gradient centrifugation of mild detergent-treated membranes, raft-associated proteins are found floating in the low-density fraction due to the lipids remaining attached to the proteins (1, 6).
In the yeast Saccharomyces cerevisiae, large raft-based membrane microdomains or, more likely, raft clusters can be visualized in living cells (27). As judged from detergent solubilities and from the difference in the membrane patterns, two raft-based membrane compartments (RMCs) can be distinguished. One (RMC P) is occupied by the proton ATPase Pma1p (2, 22, 27), and the other (RMC C) is known to contain the two H+ symporters Can1p and Fur4p and a membrane protein of unknown function, Sur7p (26, 27, 55). The two compartments (RMC P and RMC C) have been estimated to cover about 80% of the cell surface of yeast cells (26), which implies that many more proteins are expected to be associated with each of them.
For plant cells, the occurrence of rafts has not been firmly established; so far, solely the existence of mild detergent-insoluble proteins from tobacco leaf plasma membranes and Arabidopsis membranes has been reported (4, 28, 33).
For characterizations of plant membrane proteins, especially their transport functions, yeast has been an extremely helpful model organism in the past (13, 49). Once the Chlorella hexose-H+ symporter, the HUP1 protein, and the related transporter STP1 from Arabidopsis had been functionally expressed in yeast cells (40, 41), and once specific yeast mutants were complemented with corresponding plant genes coding for transporters and ion channels (37, 43, 45), these methods became standard procedures, leading to the detection and characterization of hundreds of plant transporters (13, 49). It is not implausible, therefore, to look for a potential raft association of plant membrane proteins with the help of yeast cells.
For this study, we used HUP1, the first eukaryotic H+ symporter to be described and cloned (19, 42), as a model protein. The HUP1 protein is a member of the major facilitator superfamily of transport proteins with 12 putative transmembrane helices and most likely is active as a monomer. We constructed a HUP1-green fluorescent protein (HUP1-GFP) fusion and expressed it in S. cerevisiae. As observed by confocal microscopy, the protein clearly exhibited a patchy distribution in the plasma membrane and colocalized with the endogenous yeast raft proteins constituting RMC C. Lipid analysis of HUP1p purified to homogeneity from yeast membranes revealed that the protein is specifically associated with ergosterol, phosphatidylethanolamine, and phosphatidylcholine (PC) (38). In yeast mutants deficient in ergosterol biosynthesis, HUP1-GFP is distributed homogenously in the plasma membrane. In addition, we demonstrate that HUP1-GFP expressed in Schizosaccharomyces pombe concentrates in the sterol-rich end zones of this fission yeast. With the use of an anti-HUP1 protein antibody, a patchy distribution of the transporter can also be seen in Chlorella cross sections. The data can be taken as evidence that at least this plant protein associates and concentrates within lipid raft-based membrane compartments.
MATERIALS AND METHODS
Strains and growth conditions.
The baker's yeast and fission yeast strains used for this study are listed in Table 1. Chlorella kessleri was grown in mineral medium and induced for d-glucose uptake as previously described (50). Plasmid amplification was carried out in the Escherichia coli host strain DH5α. The bacterial strains were incubated in 2TY medium (1% tryptone, 1.6% yeast extract, 0.5% NaCl) supplemented with ampicillin (100 mg/ml) for the selection of transformants. Yeast wild-type strains were grown in rich medium (for S. cerevisiae, YPD [2% peptone, 1% yeast extract, 2% glucose]; for S. pombe, YE [1% yeast extract, 3% glucose]), while transformants were selected in yeast minimal medium (0.67% Difco yeast nitrogen base without amino acids, 2% glucose, and essential amino acids). pVT100-U and YEplac181 transformants were selected on uracil- and leucine-free media, respectively. For construction of the hxt1-7Δ erg6Δ strain, genomic DNA of the erg6 mutant was used as a template for PCR to amplify the erg6Δ::kanMX4 open reading frame. Therefore, primers were used which anneal from 198 bp to 224 bp upstream and 178 bp to 205 bp downstream of the ERG6 open reading frame. The PCR product was directly transformed into hxt1-7Δ cells (36).
TABLE 1.
Yeast strains used for this study
| Strain | Genotype | Source or reference |
|---|---|---|
| Saccharomyces cerevisiae strains | ||
| SEY6210 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 | S. Emr |
| SEY6210a | MATaura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 | J. Stolz |
| BY4742 | MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0 trp1-Δ901 bar1-1 | C. Brachmann |
| erg6 mutant | BY4742 except erg6Δ::kanMX4 | Euroscarf |
| erg24 mutant | BY4742 except erg24Δ::kanMX4 | Euroscarf |
| lcb1-100 (RH3804) mutant | MATα lcb1-100 leu2 trp1 ura3 lys2 bar1 | H. Riezman |
| Sur7mRFP (KM153) | SEY6210a except SUR7::mRFP::kanMX4 | K. Malinska |
| Pma1dsRed (KM) | SEY6210 except PMA1::dsRed::kanMX4 | K. Malinska |
| hxt1-7Δ (RE700) mutant | MATahxtΔ::HIS3::Δhxt4 hxt5::LEU2 hxt2Δ::HIS3 hxt3Δ::LEU2::Δhxt6 hxt7::HIS3 ura3-52 his3-11,15 leu2-3,112 | 36 |
| hxt1-7Δ erg6Δ mutant | hxt1-7Δ except erg6Δ::kanMX4 | This study |
| Schizosaccharomyces pombe strain | ||
| FY254 | h−ura4-D18 leu1-32 can1-1 ade6-M210 | 23 |
In liquid media, more distinct patchy patterns of HUP1-GFP were obtained at low glucose concentrations (0.2%). Due to the temperature-sensitive phenotype of the lcb1-100 mutant, this strain was incubated at 25°C. During the experiment with lcb1-100/pHUP1GFP, the cells in logarithmic growth phase (optical density at 600 nm, 0.8) were shifted to 30°C for 4 h. All other yeast strains were incubated at 30°C.
Plasmids. (i) pVT100-U HUP1GFP.
The HUP1 cDNA was amplified by PCR from plasmid pTF201 (41), using a 5′ primer containing a HindIII restriction site and a 3′ primer containing a BamHI site. The PCR product was subcloned into the 2μm vector pVT100-U-GFP (J. Stolz, unpublished). In this vector, the gene encodes HUP1 C-terminally fused to GFP and is under the control of a constitutive ADH1 promoter.
(ii) YEplac181 HUP1GFP.
The HUP1-GFP DNA was cut from pVT100-U HUP1GFP, including its ADH1 promoter and terminator, as SphI-HindIII and HindIII-MfeI fragments. In a three-way ligation, these fragments were subcloned into the 2μm vector YEplac181 cut with SphI and EcoRI.
Isolation of lipid rafts.
For the isolation of lipid rafts according to the methods of Bagnat et al. (2) and Malinska et al. (27), crude membranes corresponding to 200 μg protein were incubated in 300 μl cold TNE buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA) containing protease inhibitors (Roche Diagnostics GmbH, Mannheim, Germany) and 1% Triton X-100 for 30 min on ice. Subsequently, the samples were overlaid with an Optiprep (Nycomed) step gradient and centrifuged for 3 h at 45,000 rpm in a Beckman SW60 rotor at 4°C. After centrifugation, six equal fractions were collected, and the proteins were immunodetected.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblot analysis.
Protein fractions from the density gradient were incubated with sample buffer at 37°C for 10 min, loaded into a 10% polyacrylamide gel, and finally blotted onto a polyvinylidene difluoride membrane. For immunodetection of HUP1-GFP, Pma1p, Gas1p, and Hxt1p, the membrane was incubated with rabbit polyclonal antibodies. The detection of primary antibodies was carried out using a peroxidase-conjugated immunoglobulin G anti-rabbit antibody and a chemiluminescence detection system (Pierce Biotechnology Inc., Rockford, IL).
Lipid analysis.
Lipids from the top and bottom fractions of the Optiprep gradient were extracted with 3 ml of chloroform-methanol (2:1). After centrifugation, the lower, organic phase was washed with 10% methanol, and the lipid-containing chloroform phase was evaporated by a stream of nitrogen. The dried lipids were dissolved in 8 μl toluene and separated by one-dimensional high-performance thin-layer chromatography with five stepwise developments, as described by Ruiz and Ochoa (39). Pure lipids dissolved in toluene were spotted onto thin-layer chromatography plates and served as standards. The lipids were detected by charring.
Filipin staining of sterols in S. pombe cells.
Wild-type cells were grown in rich medium and prepared for microscopy during early logarithmic growth phase. Just before concentration of the cells by a brief centrifugation, filipin (Sigma) dissolved in dimethyl sulfoxide was added at a final concentration of 5 μg/ml.
Transport assay.
For testing of glucose uptake, yeast cells were grown on minimal medium supplemented with the required amino acids and 2% maltose as a carbon source. The cells from mid-exponential growth phase were collected, washed three times in 50 mM potassium phosphate buffer, pH 6.3, and resuspended in the same buffer to a final optical density of 1.0. A mixture of radioactive and nonradioactive glucose (specific activity, 2.5 μCi/μmole) was added to aliquots of 0.33 ml of the yeast suspension to a final concentration of 75 μM. Samples of 100 μl were withdrawn at intervals, diluted in 2 ml of water, filtered with 0.8-μm-pore-size cellulose-acetate filters (Schleicher & Schuell), and washed with 2 ml of water. The radioactivity was determined by scintillation counting.
Isolation of membranes from Chlorella cells.
Cells were harvested, resuspended in TNE buffer containing protease inhibitors, and homogenized with glass beads in a ribolyser. The homogenate was subjected to differential centrifugation at 20,000 × g for 30 min and subsequently at 100,000 × g for 1 h (microsomes). The microsomal pellet was resuspended in TNE buffer and used for the isolation of DRMs.
Light microscopy.
Chlorella cells induced for glucose uptake were washed and pelleted; they were then fixed, further treated, and antibody stained as described previously (48). Fixation was carried out with 0.1% glutaraldehyde and 6% formaldehyde; the cell pellets were dehydrated with increasing concentrations of ethanol and embedded in methacrylate. Two-micrometer ultrathin microtome sections were stained with an anti-HUP1 antibody.
Yeast cells were concentrated for microscopy by a short centrifugation step. Immobilization of the cells was achieved with 1% agarose. Microscopy of specimens was carried out with an LSM 510 META confocal microscope (Carl Zeiss, Jena, Germany). Fluorescence signals of GFP (excitation at 488 nm, Ar laser) were detected using a 505- to 550-nm-band-pass emission filter; for monomeric red fluorescent protein (mRFP) signals (excitation at 543 nm, HeNe laser), a 585-nm-long-pass emission filter was used. In cases of double labeling, the specimens were scanned sequentially to avoid any cross talk of fluorescence channels.
For visualization of filipin-stained cells, a Zeiss Axioskop equipped with an HBO 50 Hg lamp and a 3CCD color video camera (Sony) was used. Filipin was detected with the following equipment: a UV-G365 filter, an FT395 dichroic mirror, and an LP420 emission light filter.
RESULTS
Expression of HUP1-GFP in S. cerevisiae.
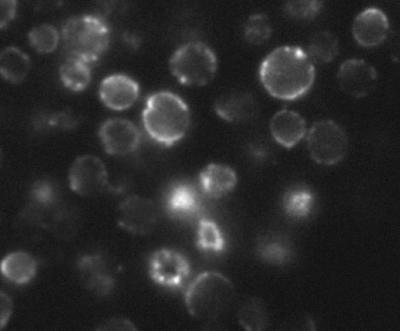
When the expression of the HUP1 transport protein in Chlorella was checked with a polyclonal antibody (48), it was localized in the plasma membrane. It was also noticed, however, that the antibody staining of the plasma membrane was not homogenous, but rather showed a patchy distribution (Fig. 1). This is reminiscent of similar nonhomogenous distributions of several endogenous transport proteins in yeast. H+ symporters such as Can1p and Fur4p are localized in 300-nm patches, as described recently (26, 27). On the other hand, the hexose facilitator Hxt1p is homogenously distributed within the plasma membrane. The Chlorella hexose-H+ symporter, the HUP1 protein, is fully functional when expressed in S. cerevisiae or in S. pombe (38, 40). The question arose as to whether this heterologous H+ symporter joins the specific membrane compartment of endogenous symporters in the yeast plasma membrane.
FIG. 1.
Cross sections of Chlorella kessleri cells stained with anti-HUP1 antibody (for details, see Materials and Methods).
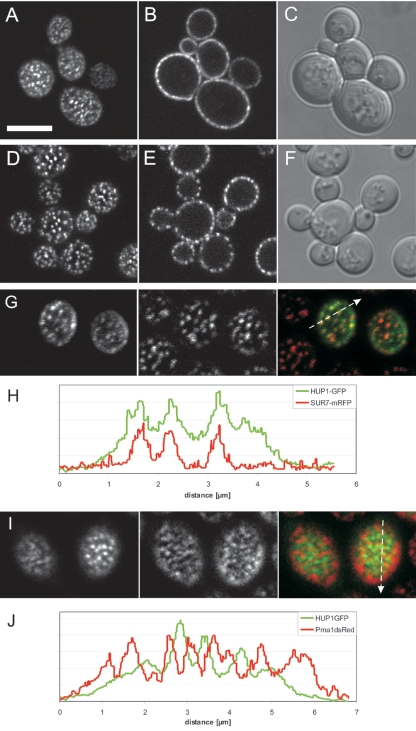
A HUP1-GFP fusion construct was prepared and expressed in S. cerevisiae. The GFP fusion protein was functional and catalyzed the uptake of [14C]glucose (see below and Fig. 7). As evident from a confocal cross section, HUP1-GFP was localized almost exclusively in the plasma membrane (Fig. 2B). The patchiness of its distribution can be clearly seen in cell surface views (Fig. 2A). Due to the thickness of the confocal “slice,” the nonhomogenous distribution cannot be resolved as clearly in cross sections as in surface views. However, the patchiness is easily observable in surface views as well as cross sections of yeast cells expressing endogenous membrane proteins such as Sur7-GFP (Fig. 2D and E). Sur7p, a protein of unknown function, contains three transmembrane domains and is localized in the plasma membrane (55). Recently, it has been shown that Sur7p colocalizes with Can1p and Fur4p within the 300-nm patches (26). To see whether the heterologous HUP1 protein is housed in the same compartment (RMC C), the localization of Sur7-mRFP and HUP1-GFP in the same cell was investigated. Figure 2G and H demonstrate clearly that the two proteins colocalize. Additionally, we observed that Sur7-mRFP is more highly concentrated within the patches than is HUP1-GFP. This is most clearly seen when the ratios of maxima to minima are compared in the fluorescence intensity profile (Fig. 2H). On the other hand, the yeast plasma membrane H+-ATPase Pma1p forms a network-like subcompartment clearly excluding the Can1, Fur4, and Sur7 protein patches (26, 27). When HUP1-GFP was expressed in cells together with Pma1-dsRed, these two proteins also excluded each other (Fig. 2I and J).
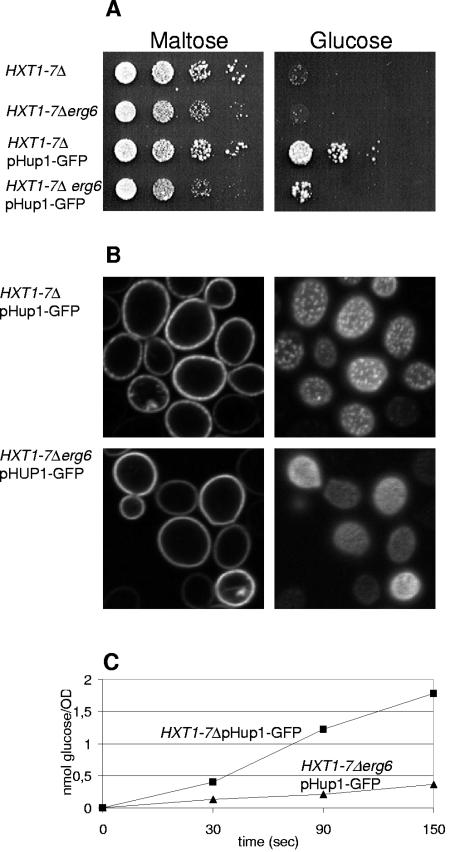
FIG. 7.
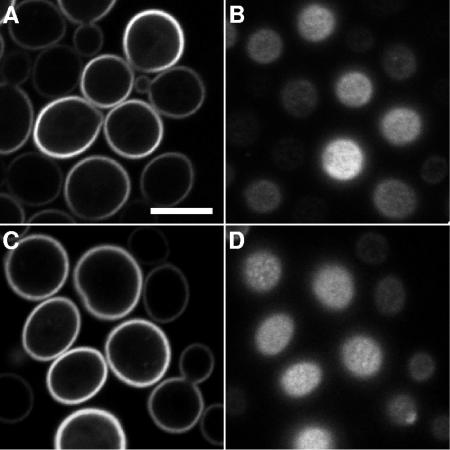
Ergosterol is required for optimal glucose uptake and growth of S. cerevisiae hxt1-7Δ mutants. (A) The S. cerevisiae mutant lacking seven hexose facilitator genes (36) grows on maltose (left) and can grow on glucose only when transformed with the gene for Chlorella HUP1-GFP (right). Optimal growth also requires an intact ERG6 gene (compare rows 3 and 4). The individual strains were grown in SD medium, diluted in water, and spotted in dilution steps of 10 on plates containing either 2% maltose or 2% glucose. (B) HUP1-GFP is expressed in untransformed cells or in erg6 mutant cells of the hxt1-7Δ strain. In both types of cells, the HUP1-GFP is targeted to the plasma membrane; 300-nm clusters are not formed in the erg6 mutant cells. (C) Rates of glucose uptake in the absence of ERG6 were considerably decreased.
FIG. 2.
Saccharomyces cerevisiae (SEY6210/pHUP1GFP) expressing the Chlorella glucose-H+ symporter HUP1-GFP. The top row of images shows a confocal surface view (A), a cross section (B), and a differential interference contrast (DIC) image (C). The S. cerevisiae membrane protein Sur7-mRFP localizes to 300-nm patches of the raft-based membrane compartment C (RMC C) (26). The second row of images shows a confocal surface view (D), a cross section (E), and a DIC image (F). HUP1-GFP colocalizes with Sur7-mRFP (G and H). Confocal surface views in panel G are as follows: left, HUP1-GFP; middle, Sur7-mRFP; and right, merge. (H) Fluorescence intensity profile of patches scanned as indicated in the merged image. HUP1-GFP does not colocalize with Pma1-dsRed (I and J). Confocal surface views in panel I are as follows: left, HUP1-GFP; middle, Pma-dsRed; right, merge. (J) Fluorescence intensity profile of a cell coexpressing HUP1-GFP and Pma1-dsRed. HUP1-GFP is concentrated in areas where H+-ATPase is minimal. Size bar, 5μm.
Detergent resistance of HUP1.
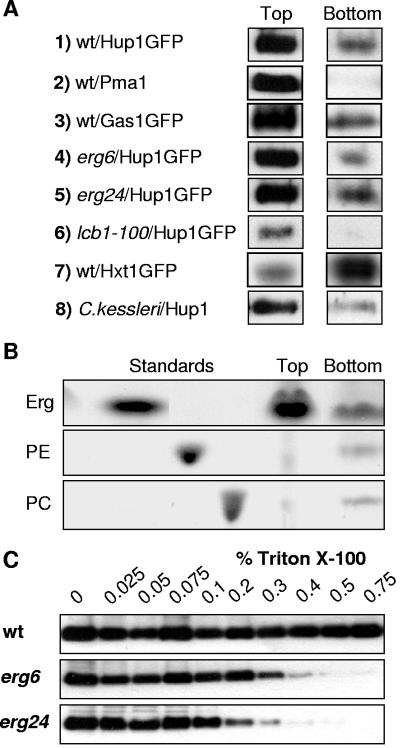
The most commonly applied criterion for raft association of membrane proteins is their detergent insolubility. To test the solubility of the HUP1 protein, yeast membranes containing HUP1-GFP were treated with 1% Triton X-100 at 4°C and fractionated on an Optiprep density gradient. After centrifugation, six equal fractions from top to bottom were tested for their content of HUP1-GFP. The protein distributions in the top and bottom fractions are shown in Fig. 3A. More than 70% of the total HUP1 protein was found floating in the top fraction (Fig. 3A, panel 1), indicating its association with lipid rafts. A similar distribution was observed when HUP1 was extracted from Chlorella kessleri membranes (Fig. 3A, panel 8). In contrast Hxt1p, a yeast transporter which was previously shown to be distributed homogenously within the plasma membrane (27), remained in the bottom fraction of the gradient (>80%) (Fig. 3A, panel 7), whereas the well-established raft proteins Pma1p and Gas1p (2) were detected predominantly in the top fraction (Fig. 3A, panels 2 and 3).
FIG. 3.
Properties of detergent-resistant membranes. (A) Yeast membranes containing HUP1-GFP were solubilized with Triton X-100, and the membrane proteins were separated in a density gradient (see Materials and Methods). The top fraction (floating proteins and rafts) and the bottom fraction (soluble proteins not associated with lipids) were immunoanalyzed as described in Materials and Methods. Besides HUP1, Pma1p, Gas1p, and Hxt1p (panels 1 to 3 and 7), HUP1-containing membranes of three mutants (panels 4 to 6) and of Chlorella cells (panel 8) were tested. (B) Lipids from the top and bottom fractions were extracted and analyzed by thin-layer chromatography (Erg, ergosterol; PE, phosphatidylethanolamine; PC, phosphatidylcholine). (C) HUP1-GFP was extracted from membranes of wild-type and erg mutant cells with Triton X-100. Membranes were extracted with the indicated concentrations of Triton X-100, followed by high-speed centrifugation. The amount of HUP1 protein that remained associated with the membrane pellet was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
The lipids from the top and bottom fractions were extracted with chloroform-methanol, and the extracts were separated by thin-layer chromatography. The top fraction contained the majority of ergosterol (Fig. 3B). Based on the amount of PC present in each fraction, the enrichment of ergosterol in the top fraction was estimated to be >10-fold. As determined by mass spectrometry (B. Brügger et al., unpublished data), the ratio of doubly unsaturated to singly unsaturated fatty acids of PC in the two fractions also agrees with the expectation of an increased association of unsaturated or less saturated fatty acids with nonraft fractions. The ratios of doubly to singly unsaturated fatty acids determined for PC from the bottom and top fractions were 2.13 and 1.23, respectively. Thus, the enrichment of HUP1 protein in the top fraction of an Optiprep gradient is accompanied by a greater abundance of ergosterol and an increase in the saturation of the accompanying lipids.
Requirement of ergosterol and sphingolipids for the formation of raft-based clusters.
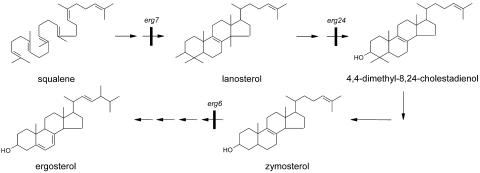
Yeast membrane proteins such as Pma1p and Can1p require the presence of ergosterol and sphingolipids within the plasma membrane to behave as raft proteins (2, 27). To see whether the lateral membrane distribution of the heterologous protein HUP1 was affected in mutants lacking ergosterol or sphingolipids, the corresponding mutant strains were transformed with the HUP1-GFP-encoding plasmid. As shown in Fig. 4A and B, targeting of the protein to the plasma membrane in the erg6 mutant was hardly affected, but the protein was almost homogenously distributed and not concentrated in the typical 300-nm patches. Similar pictures were obtained with the erg24 mutant (data not shown) and the lcb1-100 mutant (Fig. 4C and D). Thus, the formation of raft-based membrane compartments responsible for the patchy pattern obviously requires both ergosterol and sphingolipids. Testing the detergent solubility of the HUP1 protein expressed in erg mutants and its separation by density gradient centrifugation showed, however, that there was no significant change in the top-to-bottom distribution of the protein (Fig. 3A, panels 5 to 7). By this criterion, the HUP1 proteins in erg6 and erg24 mutants still had raft properties, although the formation of the 300-nm patches was greatly suppressed (Fig. 4A and B). Therefore, it has to be assumed that the patches represent raft clusters. The ergosterol biosynthetic intermediates lanosterol and zymosterol, which accumulate in erg24 and erg6 mutants, respectively (Fig. 5) (14, 32), obviously enable the yeast cell to form rafts—most likely with the typical dimensions known for mammalian cells (35)—but they no longer allow raft clustering. To characterize the raft association of the HUP1 protein in more detail, we used the method of protein extraction by increasing the concentration of Triton X-100 at 20°C (10). The HUP1 protein from wild-type membranes treated with 0.75% detergent was still insoluble, whereas 0.3 to 0.4% Triton X-100 was sufficient to solubilize HUP1 completely from the membranes of the erg mutants (Fig. 3C).
FIG. 4.
HUP1-GFP expressed in ergosterol erg6 mutant (A and B) and sphingolipid biosynthesis lcb1-100 mutant (C and D). Confocal cross sections (A and C) and surface views (B and D) are shown. Controls corresponding to the wild type are shown in Fig. 2A and B. Size bar, 5μm.
FIG. 5.
Pathway of ergosterol biosynthesis in S. cerevisiae. Only the erg7 mutation, blocking the conversion of squalene epoxide via ring closures to lanosterol, is lethal.
Expression of HUP1-GFP in S. pombe.
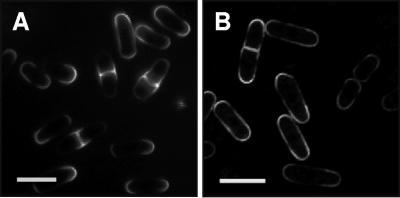
In S. pombe cells, the highest concentration of sterols has been detected in the caps of the rod-shaped cells and, shortly before cell division, also in membranes of the division plane (52) (Fig. 6A). To check whether HUP1-GFP localizes to “S. cerevisiae-type” 300-nm patches in S. pombe or whether the HUP1 protein is targeted to and ends up within the sterol-rich zones of S. pombe, the cells were transformed with a HUP1-GFP construct and analyzed by confocal microscopy. As shown in Fig. 6B, the GFP fluorescence was strongest in the cap regions of S. pombe and also appeared in the division plane shortly before cytokinesis. These data indicate that the HUP1 protein by itself does not induce the formation of the 300-nm RMCs. Nevertheless, the HUP1 protein concentrates in sterol-rich membrane domains in S. pombe.
FIG. 6.
HUP1-GFP expressed in Schizosaccharomyces pombe (FY254/pHUP1-GFP). Sterol-rich membrane domains were visualized with filipin (A; fluorescence microscope, UV light) and HUP1-GFP localization (B; confocal microscope). Size bars, 10 μm.
Decreased transport activity of HUP1 in ergosterol-depleted cells.
A most important question is, of course, whether the localization pattern of the HUP1 protein (in RMC C or more evenly distributed, as in erg mutants) affects the activity of the protein. To this end, the ERG6 gene was deleted from a yeast strain lacking seven endogenous hexose transporters, the hxt1-7Δ mutant (36). The hxt1-7Δ and hxt1-7Δ erg6Δ mutants were transformed with a plasmid encoding the HUP1-GFP fusion and grown on maltose. As documented in Fig. 7A, the HUP1-GFP protein enabled growth on glucose, but only very poorly in the erg6 mutant background.
To analyze the poor growth of erg6 mutants expressing HUP1 in more detail, we performed uptake experiments with [14C]glucose. As shown in Fig. 7C, the initial rate of [14C]glucose uptake into erg6 mutant cells also amounted to only about 20% that of ERG6-carrying cells. In both types of cells, the targeting of HUP1-GFP to the plasma membrane was not affected. No internal fluorescence is visible in either confocal cross section (Fig. 7B). The amounts of HUP1 protein expressed differed at most by a factor of 2, as estimated from the fluorescence intensities (Fig. 7B; also compare Fig. 2 and 4) and from Western blots (data not shown). Thus, we postulate that localization within the raft clusters significantly increases the catalytic activity of this transporter.
DISCUSSION
Lipid rafts were originally postulated to constitute a mechanism for protein sorting and trafficking in animal cells, especially polarized epithelial cells (6, 47). The sorting and transport of apical (but not basolateral) membrane proteins were impaired in cells with reduced cholesterol contents (17). Also, for S. cerevisiae, the existence of rafts was first documented in relation to the biosynthetic delivery of proteins to the plasma membrane (2).
The raft concept has raised enormous attention, and many publications have related lipid rafts to various biological functions, such as signaling, cell adhesion, and docking of pathogens (for reviews see references 5, 12, and 59). The most convincing examples concerning raft functions have been reported for hematopoietic cells. For example, in T-cell signaling, tyrosine phosphorylation of the LAT protein (linker for activation of T cells) is essential. If a palmitoylation site of the LAT protein is mutated, the nonpalmitoylated protein still localizes to the plasma membrane but not to DRMs. As a consequence, this protein is not tyrosine phosphorylated and does not function in signaling anymore (24, 57). In general, the function of rafts is thought to be brought about in any of the following three ways: (i) an increased concentration of proteins in rafts could facilitate homomeric and heteromeric interactions, (ii) the lateral compartmentation of the plasma membrane might separate signaling components until a signal allows them to come together, or (iii) the different lipid environment as such may affect the functions of membrane proteins. Activities of many membrane proteins are affected specifically by phospholipids and sterols (for a review, see reference 31).
In comparison with the intense interest in rafts of mammalian and yeast cells, surprisingly little attention has been paid to potential lipid rafts of plant membranes. Although the typical plant sterols sitosterol and stigmasterol promote raft domain formation in model membranes (54), evidence for the existence of such domains in biological membranes of plants is still scarce. Tobacco leaves and Arabidopsis callus tissue represent the only plant tissues for which the existence of detergent-resistant membranes has been reported (4, 28, 33). Based on their occurrence in low-buoyant-density fractions after mild detergent treatment and centrifugation, a number of glycosylphosphatidylinositol-anchored proteins associated with DRMs have been identified. Whether the detergent insolubility and density gradient data reflect the true organization of lipids in living cells or whether rafts and their association with defined proteins arise as an artifact during detergent extraction is, however, still under debate (16). Agreement exists only in stating that detergent resistance as a sole criterion for raft association may be insufficient (59).
The visualization of rafts or raft-based clusters in living cells of S. cerevisiae and S. pombe (3, 27, 52) represents an additional phenomenon indicating the existence of rafts, at least for fungal cells. Valdez-Taubas and Pelham (51) argue that the raft-like domains in polarized fungal cells arise simply due to polarized exocytosis and slow lateral diffusion. However, raft-based membrane compartmentation documented by patchy, nonpolarized distributions of membrane proteins is shown here and has been reported previously for vegetatively growing S. cerevisiae cells (27).
The nonhomogenous distribution of the HUP1 symporter protein detected by specific antibodies at the Chlorella cell surface resembled the 300-nm patches of Can1-GFP, Fur4-GFP, and Sur7-GFP observed in S. cerevisiae. Since yeast has turned out to be an ideal host for the functional expression of plant transporters (41, 42; for review, see references 13 and 49), we expected that these heterologous membrane proteins might become compartmentalized within the fungal plasma membrane. Assuming that the membrane patches visible in Chlorella are raft-type subdomains, and with respect to the fact that the HUP1 protein does show detergent resistance when extracted from Chlorella membranes (Fig. 3A, panel 8), we checked whether HUP1-GFP joins one of the raft compartments detected in the plasma membrane of S. cerevisiae (26).
As shown in Fig. 2, HUP1-GFP colocalizes with Sur7-mRFP (Fig. 2G and H). This demonstrates that the Chlorella symporter localizes in yeast subdomain RMC C, which also houses Can1p, Fur4p, and Sur7p. It is notable that, in contrast to Sur1-mRFP, HUP1-GFP is also present to a minor extent in the non-raft-cluster area, which possibly indicates that the heterologous protein does not carry the optimal sorting information required for yeast raft or raft cluster association. This conclusion is derived from the overall background of HUP1-GFP in the nonraft area, which is higher than that of Sur7-mRFP (Fig. 2G and H). Also, less Can1-GFP is present outside the 300-nm patches (27). As a member of the raft cluster compartment RMC C, the hexose-H+ symporter of Chlorella is distinctly separated from the H+-ATPase of RMC P (Fig. 2G). This clear separation of a membrane area responsible for the current generated by proton export from an area occupied by proton importers (Can1p, Fur4p, and HUP1) may be of functional relevance. The alkaline and acid banding of Chara described by Lucas and Smith (25) may thus be a much more general phenomenon and valid even at a nanoscale. This and other functional aspects of the subcompartmentation of transport proteins in the plasma membrane will obviously be the most interesting aspects to be worked on in the future.
The results indicate that sterols are important for the activity of the HUP1 protein (Fig. 7). Whether the clustering of the transporter within the membrane or the presence of a critical amount of a specific sterol is decisive for the effect observed is still an open question. The possibility that ergosterol is a more potent cofactor for the activity of the HUP1 protein than zymosterol (Fig. 5) cannot be excluded. These two possibilities, however, may simply represent two sides of the same coin.
In the raft field, a committed discussion is taking place regarding whether it is primarily a lipid-phase separation that drives raft formation with the raft proteins subsequently concentrating in their favorite lipid phases. Alternatively, membrane proteins with specific lipid shells may initiate the formation of smaller and larger rafts depending on the extent of fusion of these protein-lipid units (59). The expression of HUP1-GFP in S. pombe cells characterized by a specific pattern of sterol-rich plasma membrane domains (Fig. 6) clearly shows that the heterologous protein joins the sterol-rich zones of the host organism. At first sight, this seems to support the idea of lipid-phase separation as the primary cause of raft formation. It has to be realized, however, that these large raft-based membrane domains in S. pombe contain various endogenous membrane proteins and that heterologous HUP1-GFP most likely simply joins these rafts, independent of the mechanism by which they arose.
Additional evidence for the raft association of HUP1-GFP was obtained by detergent extraction and by studies of mutants with defects in sterol and ceramide synthesis. HUP1-GFP behaves to a large extent as a detergent-resistant membrane protein; 70 to 80% of the total HUP1 protein was found in thetop fraction of the density gradient (Fig. 3A, panel 1). The S. cerevisiae erg6 and erg24 mutants are unable to synthesize ergosterol, the main sterol, which amounts to >50% of the lipid components of the plasma membrane (58). In contrast to the case in wild-type cells, the fluorescence pattern of HUP1-GFP expression was quite homogenous in both mutants, and the same was observed for the lcb1-100 mutant (Fig. 4C and D), which contains only 4% of the wild-type content of sphingolipids at the nonpermissive temperature (56). Further support for the postulated raft association of HUP1 is given by the fact that the purified protein is associated with lipids that are typical for rafts. A biotinylation domain-tagged HUP1 protein purified by affinity chromatography to the extent that only a single band could be seen on a silver-stained sodium dodecyl sulfate gel (8) contained approximately three molecules of ergosterol per protein molecule (38). The fact that sterols represent the major component of lipids specifically associated with the protein not only supports a raft association of HUP1 protein but also strengthens the old observation that interference of sterols by nystatin changes the kinetic behavior of the Chlorella HUP1 transport protein in vivo and in vitro (18, 30).
A frequently applied procedure for testing whether biological phenomena are related to the existence of rafts in mammalian cells is a manipulation of the sterol content in membranes. Reduction of the sterol content can be achieved either by interfering with sterol biosynthesis (lovostatin) or by extracting the sterols from intact cells with methyl-β-cyclodextrin (34). However, possible pitfalls such as changes in membrane permeability or in the membrane's Nernst potential due to a decreased cholesterol content are generally neglected, and the functions of lipid rafts may therefore be considerably overrated (12, 29). The same skepticism may hold when considering the decreased transport activities in the erg mutants observed in this study.
Only recently, several studies have dealt with mutants defective in sterol biosynthesis in Arabidopsis (7, 9, 44, 53). The phenotypes also showed severe developmental defects in mutants where the sitosterol branch of the pathway (and not the campesterol/brassinolide one) was inhibited (7). The tendency to discuss these results and related ones (20, 21) in connection with a potential role of lipid rafts urgently demands more evidence of their existence in plants. The facts presented here demonstrate that a plasma membrane protein of the lower plant Chlorella possesses all the crucial features for localizing in rafts. The approach described may be a generally applicable and convenient method to learn about lipid requirements and potential raft properties of other plant plasma membrane proteins.
Acknowledgments
The expert technical assistance of Ingrid Fuchs is gratefully acknowledged. We thank Britta Brügger, Heidelberg, Germany for the mass spectrometric analysis of lipids, Laura Popolo, Milan, Italy, Ramon Serrano, Valencia, Spain, and Eckhard Boles, Frankfurt, Germany, for Gas1p, Pma1p, and Hxt1p antibodies, Jan Malinsky and Katka Malinska, Prague, Czech Republic, for helpful discussions and advice, and Ruth Stadler, Erlangen, Germany, for Fig. 1.
The financial support of DFG (Schwerpunkt 1108) and of Fonds der Chemischen Industrie is acknowledged. M. Opekarova was supported by LC545 funds.
REFERENCES
- 1.Bagnat, M., A. Chang, and K. Simons. 2001. Plasma membrane proton ATPase Pma1p requires raft association for surface delivery in yeast. Mol. Biol. Cell 12:4129-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagnat, M., S. Keranen, A. Shevchenko, and K. Simons. 2000. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA 97:3254-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagnat, M., and K. Simons. 2002. Cell surface polarization during yeast mating. Proc. Natl. Acad. Sci. USA 99:14183-14188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borner, G. H., D. J. Sherrier, T. Weimar, L. V. Michaelson, N. D. Hawkins, A. Macaskill, J. A. Napier, M. H. Beale, K. S. Lilley, and P. Dupree. 2005. Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 137:104-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, D. A., and E. London. 2000. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J. Biol. Chem. 275:17221-17224. [DOI] [PubMed] [Google Scholar]
- 6.Brown, D. A., and J. K. Rose. 1992. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68:533-544. [DOI] [PubMed] [Google Scholar]
- 7.Carland, F. M., S. Fujioka, S. Takatsuto, S. Yoshida, and T. Nelson. 2002. The identification of CVP1 reveals a role for sterols in vascular patterning. Plant Cell 14:2045-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caspari, T., I. Robl, J. Stolz, and W. Tanner. 1996. Purification of the Chlorella HUP1 hexose-proton symporter to homogeneity and its reconstitution in vitro. Plant J. 10:1045-1053. [DOI] [PubMed] [Google Scholar]
- 9.Diener, A. C., H. Li, W. Zhou, W. J. Whoriskey, W. D. Nes, and G. R. Fink. 2000. Sterol methyltransferase 1 controls the level of cholesterol in plants. Plant Cell 12:853-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Draeger, A., S. Wray, and E. B. Babiychuk. 2005. Domain architecture of the smooth-muscle plasma membrane: regulation by annexins. Biochem. J. 387:309-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edidin, M. 2003. Lipids on the frontier: a century of cell-membrane bilayers. Nat. Rev. Mol. Cell Biol. 4:414-418. [DOI] [PubMed] [Google Scholar]
- 12.Edidin, M. 2003. The state of lipid rafts: from model membrane to cells. Annu. Rev. Biophys. Biomol. Struct. 32:257-283. [DOI] [PubMed] [Google Scholar]
- 13.Frommer, W. B., and O. Ninnemann. 1995. Heterologous expression of genes in bacterial, fungal, animal, and plant cells. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46:419-444. [Google Scholar]
- 14.Gaber, R. F., D. M. Copple, B. K. Kennedy, M. Vidal, and M. Bard. 1989. The yeast gene ERG6 is required for normal membrane function but is not essential for biosynthesis of the cell-cycle-sparking sterol. Mol. Cell. Biol. 9:3447-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harder, T., P. Scheiffele, P. Verkade, and K. Simons. 1998. Lipid domain structure of the plasma membrane revealed by patching of membrane components. J. Cell Biol. 141:929-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heerklotz, H. 2002. Triton promotes domain formation in lipid raft mixtures. Biophys. J. 83:2693-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keller, P., and K. Simons. 1998. Cholesterol is required for surface transport of influenza virus hemagglutinin. J. Cell Biol. 140:1357-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komor, B., E. Komor, and W. Tanner. 1974. Transformation of a strictly coupled active transport system into a facilitated diffusion system by nystatin. J. Membr. Biol. 17:231-238. [DOI] [PubMed] [Google Scholar]
- 19.Komor, E. 1973. Proton-coupled hexose transport in Chlorella vulgaris. FEBS Lett. 38:16-18. [DOI] [PubMed] [Google Scholar]
- 20.Lalanne, E., D. Honys, A. Johnson, G. H. Borner, K. S. Lilley, P. Dupree, U. Grossniklaus, and D. Twell. 2004. SETH1 and SETH2, two components of the glycosylphosphatidylinositol anchor biosynthetic pathway, are required for pollen germination and tube growth in Arabidopsis. Plant Cell 16:229-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavy, M., K. Bracha-Drori, H. Sternberg, and S. Yalovsky. 2002. A cell-specific, prenylation-independent mechanism regulates targeting of type II RACs. Plant Cell 14:2431-2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, M. C., S. Hamamoto, and R. Schekman. 2002. Ceramide biosynthesis is required for the formation of the oligomeric H+-ATPase Pma1p in the yeast endoplasmic reticulum. J. Biol. Chem. 277:22395-22401. [DOI] [PubMed] [Google Scholar]
- 23.Liang, D. T., J. A. Hodson, and S. L. Forsburg. 1999. Reduced dosage of a single fission yeast MCM protein causes genetic instability and S phase delay. J. Cell Sci. 112:559-567. [DOI] [PubMed] [Google Scholar]
- 24.Lin, J., A. Weiss, and T. S. Finco. 1999. Localization of LAT in glycolipid-enriched microdomains is required for T cell activation. J. Biol. Chem. 274:28861-28864. [DOI] [PubMed] [Google Scholar]
- 25.Lucas, W. J., and E. A. Smith. 1973. The formation of alkaline and acid regions at the surface of Chara corallina cells. J. Exp. Bot. 24:1-14. [Google Scholar]
- 26.Malinska, K., J. Malinsky, M. Opekarova, and W. Tanner. 2004. Distribution of Can1p into stable domains reflects lateral protein segregation within the plasma membrane of living S. cerevisiae cells. J. Cell Sci. 117:6031-6041. [DOI] [PubMed] [Google Scholar]
- 27.Malinska, K., J. Malinsky, M. Opekarova, and W. Tanner. 2003. Visualization of protein compartmentation within the plasma membrane of living yeast cells. Mol. Biol. Cell 14:4427-4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mongrand, S., J. Morel, J. Laroche, S. Claverol, J. P. Carde, M. A. Hartmann, M. Bonneu, F. Simon-Plas, R. Lessire, and J. J. Bessoule. 2004. Lipid rafts in higher plant cells. Purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J. Biol. Chem. 279:36277-36286. [DOI] [PubMed] [Google Scholar]
- 29.Munro, S. 2003. Lipid rafts: elusive or illusive? Cell 115:377-388. [DOI] [PubMed] [Google Scholar]
- 30.Opekarova, M., and W. Tanner. 1994. Nystatin changes the properties of transporters for arginine and sugars. An in vitro study. FEBS Lett. 350:46-50. [DOI] [PubMed] [Google Scholar]
- 31.Opekarova, M., and W. Tanner. 2003. Specific lipid requirements of membrane proteins—a putative bottleneck in heterologous expression. Biochim. Biophys. Acta 1610:11-22. [DOI] [PubMed] [Google Scholar]
- 32.Parks, L. W., and W. M. Casey. 1995. Physiological implications of sterol biosynthesis in yeast. Annu. Rev. Microbiol. 49:95-116. [DOI] [PubMed] [Google Scholar]
- 33.Peskan, T., M. Westermann, and R. Oelmuller. 2000. Identification of low-density Triton X-100-insoluble plasma membrane microdomains in higher plants. Eur. J. Biochem. 267:6989-6995. [DOI] [PubMed] [Google Scholar]
- 34.Pike, L. J. 2003. Lipid rafts: bringing order to chaos. J. Lipid Res. 44:655-667. [DOI] [PubMed] [Google Scholar]
- 35.Pralle, A., P. Keller, E. L. Florin, K. Simons, and J. K. Horber. 2000. Sphingolipid-cholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells. J. Cell Biol. 148:997-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reifenberger, E., K. Freidel, and M. Ciriacy. 1995. Identification of novel HXT genes in Saccharomyces cerevisiae reveals the impact of individual hexose transporters on glycolytic flux. Mol. Microbiol. 16:157-167. [DOI] [PubMed] [Google Scholar]
- 37.Riesmeier, J. W., L. Willmitzer, and W. B. Frommer. 1992. Isolation and characterization of a sucrose carrier cDNA from spinach by functional expression in yeast. EMBO J. 11:4705-4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robl, I., R. Grassl, W. Tanner, and M. Opekarova. 2000. Properties of a reconstituted eukaryotic hexose/proton symporter solubilized by structurally related non-ionic detergents: specific requirement of phosphatidylcholine for permease stability. Biochim. Biophys. Acta 1463:407-418. [DOI] [PubMed] [Google Scholar]
- 39.Ruiz, J. I., and B. Ochoa. 1997. Quantification in the subnanomolar range of phospholipids and neutral lipids by monodimensional thin-layer chromatography and image analysis. J. Lipid Res. 38:1482-1489. [PubMed] [Google Scholar]
- 40.Sauer, N., T. Caspari, F. Klebl, and W. Tanner. 1990. Functional expression of the Chlorella hexose transporter in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 87:7949-7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sauer, N., K. Friedlander, and U. Graml-Wicke. 1990. Primary structure, genomic organization and heterologous expression of a glucose transporter from Arabidopsis thaliana. EMBO J. 9:3045-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sauer, N., and W. Tanner. 1989. The hexose carrier from Chlorella. cDNA cloning of a eucaryotic H+-cotransporter. FEBS Lett. 259:43-46. [DOI] [PubMed] [Google Scholar]
- 43.Schachtman, D. P., J. I. Schroeder, W. J. Lucas, J. A. Anderson, and R. F. Gaber. 1992. Expression of an inward-rectifying potassium channel by the Arabidopsis KAT1 cDNA. Science 258:1654-1658. [DOI] [PubMed] [Google Scholar]
- 44.Schrick, K., S. Fujioka, S. Takatsuto, Y. D. Stierhof, H. Stransky, S. Yoshida, and G. Jurgens. 2004. A link between sterol biosynthesis, the cell wall, and cellulose in Arabidopsis. Plant J. 38:227-243. [DOI] [PubMed] [Google Scholar]
- 45.Sentenac, H., N. Bonneaud, M. Minet, F. Lacroute, J. M. Salmon, F. Gaymard, and C. Grignon. 1992. Cloning and expression in yeast of a plant potassium ion transport system. Science 256:663-665. [DOI] [PubMed] [Google Scholar]
- 46.Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature 387:569-572. [DOI] [PubMed] [Google Scholar]
- 47.Simons, K., and G. van Meer. 1988. Lipid sorting in epithelial cells. Biochemistry 27:6197-6202. [DOI] [PubMed] [Google Scholar]
- 48.Stadler, R., K. Wolf, C. Hilgarth, W. Tanner, and N. Sauer. 1995. Subcellular localization of the inducible Chlorella HUP1 monosaccharide-H+ symporter and cloning of a Co-induced galactose-H+ symporter. Plant Physiol. 107:33-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanner, W., and T. Caspari. 1996. Membrane transport carriers. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47:595-626. [DOI] [PubMed] [Google Scholar]
- 50.Tanner, W., and N. Sauer. 1989. Uptake of sugars and amino acids by Chlorella. Methods Enzymol. 174:390-402. [Google Scholar]
- 51.Valdez-Taubas, J., and H. R. Pelham. 2003. Slow diffusion of proteins in the yeast plasma membrane allows polarity to be maintained by endocytic cycling. Curr. Biol. 13:1636-1640. [DOI] [PubMed] [Google Scholar]
- 52.Wachtler, V., S. Rajagopalan, and M. K. Balasubramanian. 2003. Sterol-rich plasma membrane domains in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 116:867-874. [DOI] [PubMed] [Google Scholar]
- 53.Willemsen, V., J. Friml, M. Grebe, A. van den Toorn, K. Palme, and B. Scheres. 2003. Cell polarity and PIN protein positioning in Arabidopsis require sterol methyltransferase 1 function. Plant Cell 15:612-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu, X., R. Bittman, G. Duportail, D. Heissler, C. Vilcheze, and E. London. 2001. Effect of the structure of natural sterols and sphingolipids on the formation of ordered sphingolipid/sterol domains (rafts). Comparison of cholesterol to plant, fungal, and disease-associated sterols and comparison of sphingomyelin, cerebrosides, and ceramide. J. Biol. Chem. 276:33540-33546. [DOI] [PubMed] [Google Scholar]
- 55.Young, M. E., T. S. Karpova, B. Brugger, D. M. Moschenross, G. K. Wang, R. Schneiter, F. T. Wieland, and J. A. Cooper. 2002. The Sur7p family defines novel cortical domains in Saccharomyces cerevisiae, affects sphingolipid metabolism, and is involved in sporulation. Mol. Cell. Biol. 22:927-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zanolari, B., S. Friant, K. Funato, C. Sutterlin, B. J. Stevenson, and H. Riezman. 2000. Sphingoid base synthesis requirement for endocytosis in Saccharomyces cerevisiae. EMBO J. 19:2824-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, W., J. Sloan-Lancaster, J. Kitchen, R. P. Trible, and L. E. Samelson. 1998. LAT: the ZAP-70 tyrosine kinase substrate that links T cell receptor to cellular activation. Cell 92:83-92. [DOI] [PubMed] [Google Scholar]
- 58.Zinser, E., C. D. Sperka-Gottlieb, E. V. Fasch, S. D. Kohlwein, F. Paltauf, and G. Daum. 1991. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J. Bacteriol. 173:2026-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zurzolo, C., G. van Meer, and S. Mayor. 2003. The order of rafts. Conference on microdomains, lipid rafts and caveolae. EMBO Rep. 4:1117-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]