Abstract
The fungal secondary metabolite gliotoxin produced by Aspergillus fumigatus has been hypothesized to be important in the development of invasive aspergillosis. In this study, we addressed this hypothesis by disrupting a nonribosomal peptide synthetase (NRPS) (encoded by gliP) predicted to be involved in gliotoxin production. Mutants with a disrupted gliP locus failed to produce gliotoxin, which confirmed the role of the NRPS encoded by gliP in gliotoxin biosynthesis. We found no morphological, developmental, or physiological defects in ΔgliP mutant strains. In addition, disruption of gliP resulted in down regulation of gene expression in the gliotoxin biosynthesis gene cluster, which was restored with addition of exogenous gliotoxin. This interesting result suggests a role for gliotoxin in regulating its own production. Culture filtrates from the ΔgliP mutant were unable to inhibit ionomycin-dependent degranulation of mast cells, suggesting a role for gliotoxin in suppressing mast cell degranulation and possibly in disease development. However, the ΔgliP mutant did not have an impact on survival or tissue burden in a murine inhalational model of invasive aspergillosis. This result suggests that gliotoxin is not required for virulence in an immunosuppressed host with an invasive pulmonary infection.
New medical therapies for life-threatening diseases such as solid-organ transplantation, aggressive cancer therapies, and other immunomodulating therapies have resulted in an increase in invasive fungal infections. In particular, the mortality due to invasive aspergillosis (IA) has increased 357% over the last 25 years (19). IA has become one of the leading causes of death in immunocompromised patients, with mortality rates ranging from 60 to 90% (18, 19). Although IA can be caused by several members of the genus Aspergillus, Aspergillus fumigatus remains the most prevalent causal organism (13, 26).
A. fumigatus is a saprophytic, asexually reproducing fungus that is found in soil and compost piles and primarily functions to recycle carbon and nitrogen throughout the environment (35). Yet the high frequency of A. fumigatus infections strongly suggests that this saprophytic fungus has attributes unique among Aspergillus species that make it an effective opportunistic pathogen. Several physiological and morphological characteristics of A. fumigatus have been hypothesized or demonstrated to be involved in fungal pathogenesis. These include thermotolerant growth, fast growth rates, conidial size and morphology, and the production of cell wall-degrading enzymes and proteases (3, 6, 13, 16).
Like other Aspergillus species, A. fumigatus is known to produce immunosuppressant secondary metabolites, including the epipolythiodioxopiperazine (ETP) toxin gliotoxin (11). Secondary metabolites are compounds produced by many filamentous fungi that are not required for growth but often have important biological activities. For instance, fungal secondary metabolites often have antimicrobial activity, and it is hypothesized that their production is an adaptation that allows fungi to compete with other microorganisms in their ecological niches. In several plant-fungal interactions, secondary metabolites are primary virulence factors that determine outcomes of host-pathogen interactions (25). While several indirect lines of evidence have suggested a role for gliotoxin and other secondary metabolites in invasive fungal infections, no direct evidence has definitively shown their involvement in human-fungal pathogenesis.
With the recent completion of the A. fumigatus genome project, the ability to address the role of secondary metabolites in IA has greatly increased (23). Recently, an ETP toxin and the nonribosomal peptide synthetase (NRPS) involved in its production were identified in the plant pathogen Leptosphaeria maculans (9). With this sequence information, a gene cluster containing an NRPS (gliP) was identified in the genome sequence of A. fumigatus and predicted to be involved in the synthesis of gliotoxin (10). Yet this prediction has not been functionally confirmed.
In this study, we definitively demonstrate that the NRPS gliP is involved in gliotoxin biosynthesis as disruption of the gliP locus eliminated gliotoxin production in A. fumigatus. This mutant was used to address the hypothesis that gliotoxin plays a critical role in the development of IA. Effects of gliotoxin on the expression of the biosynthetic gene cluster involved in its production and on the morphology and physiology of A. fumigatus are also presented.
MATERIALS AND METHODS
Fungal strains, cell lines, media, and growth conditions.
A. fumigatus strain AF293.1 was used to create gliP knockout strain ARC2 (ΔgliP::A. parasiticus pyrG pyrG1) and ectopic transformant strain ARC5 (pyrG1 A. parasiticus pyrG::gliP). A. fumigatus strain AF293.1 is a uracil-auxotrophic (pyrG1) mutant of A. fumigatus strain AF293 (36). A. fumigatus strains were cultivated at 37°C in Czapek Dox broth (Difco Laboratories, Sparks, MD) to measure gliotoxin production and in glucose minimal medium for all other assays as previously described (4, 28). For growth on solid medium, glucose minimal medium with appropriate supplements was used as previously described (28). Rat basophilic leukemia (RBL-2H3) cells were cultured in minimum essential medium (Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum. Cells were trypsinized and replated in 96-well plates prior to experiments.
Construction of gliotoxin-deficient A. fumigatus strain ARC2.
Disruption of the NRPS gene gliP in A. fumigatus strain AF293.1 was accomplished by replacing an ∼3.7-kb internal fragment of the gliP coding region (∼6.4 kb; GenBank accession no. DQ457015) with A. parasiticus pyrG. The disruption construct was created by cloning a sequence homologous to the gliP locus into plasmid pJW24 (donated by Nancy Keller, University of Wisconsin—Madison). 5′ and 3′ gliP homologous sequences, each 1 kb in length, were cloned to flank A. parasiticus pyrG in pJW24. The resulting plasmid, pGLIPKO, was used as a template to amplify the 5.1-kb disruption construct for use in fungal transformation. The primers utilized in vector construction and all of the other primers utilized in this study are presented in Table 1.
TABLE 1.
Primers used in this study
| Primera | Sequence 5′ to 3′ | Function |
|---|---|---|
| 5′GliZ | ACG ACG ATG AGG AAT CGA AC | Real-time PCR of gliotoxin cluster |
| 3′GliZ | TCC AGA AAA GGG AGT CGT TG | Real-time PCR of gliotoxin cluster |
| 5′Glil | AGG CCA TCC TCG TGT GTA AC | Real-time PCR of gliotoxin cluster |
| 3′Glil | GCC GAG GTC TTT GCT GAT AC | Real-time PCR of gliotoxin cluster |
| 5′GliJ | CTC TGA TCG ACG GCC ATA AT | Real-time PCR of gliotoxin cluster |
| 3′GliJ | TCG AGC TGT TGG AGT GTC TG | Real-time PCR of gliotoxin cluster |
| 5′NRPS10 | CAT CGA GGG GCT GGA GGT GAA A | Real-time PCR of gliotoxin cluster |
| 3′NRPS10 | TCG GAT GCT GTC CTC GAT GGT G | Real-time PCR of gliotoxin cluster |
| 5′GliC | ATT GAC CGG GAT GAC ACA TT | Real-time PCR of gliotoxin cluster |
| 3′GliC | ACC GTC GAG GAT TGT ATT GC | Real-time PCR of gliotoxin cluster |
| 5′GliM | CGA TCT GTA CCC CAA CGA GT | Real-time PCR of gliotoxin cluster |
| 3′GliM | TTC TGG AAC TTT GCC AGC TT | Real-time PCR of gliotoxin cluster |
| 5′GliG | GAA ACT GCG CAG CAA CAT TA | Real-time PCR of gliotoxin cluster |
| 3′GliG | TTG GCC ATT TCT CAA ACT CC | Real-time PCR of gliotoxin cluster |
| 5′GliK | CTC ACG GCA TAC AGC GAC TA | Real-time PCR of gliotoxin cluster |
| 3′GliK | ATA ATC CAA CCG AGC CAC TG | Real-time PCR of gliotoxin cluster |
| 5′GliA | TTT GCG ATC AAC GAA CTC TG | Real-time PCR of gliotoxin cluster |
| 3′GliA | CCC TTG ACG GAC TGG AAG TA | Real-time PCR of gliotoxin cluster |
| 5′GliN | GCA AGA GGT GCA AGA GAA GG | Real-time PCR of gliotoxin cluster |
| 3′GliN | GGA TCG GAT CAA AGT CCT CA | Real-time PCR of gliotoxin cluster |
| 5′GliF | GGG GGC CGA TAA TAT CAA CT | Real-time PCR of gliotoxin cluster |
| 3′GliF | AAG ATG GCC AAT CCA CCA TA | Real-time PCR of gliotoxin cluster |
| 5′GliT | ACT CCA CCA TCC AGT TCC AG | Real-time PCR of gliotoxin cluster |
| 3′GliT | TCC GAG TAT CCC TCG ATG TC | Real-time PCR of gliotoxin cluster |
| 5′AfactinII | TCA CTG CCC TTG CTC CCT CGT C | Real-time PCR of gliotoxin cluster |
| 3′AfactinII | GCA CTT GCG GTG AAC GAT CGA A | Real-time PCR of gliotoxin cluster |
| 5′SalIOL1GliP | TCG TCG ACG ATC GAG GAA CAG ACC CAT C | GliP disruption construct |
| 3′EcoRIOL2GliP | GCG AAT TCC GCA AGC TCT GTG AAG ATG A | GliP disruption construct |
| 5′XbaIOL3GliP | TCT CTA GAC GAT TGA TCA TTC CCG TCT C | GliP disruption construct |
| 3′SacIOL4GliP | GTG AGC TCC TCT GCA TCA TGC CAT CAG T | GliP disruption construct |
| 5′GlioPReplace | GGA TGC GAG TCG GTG TAA CT | Amplifies 0.7-kb internal region of gliP |
| 3′GlioPReplace | ATC TCT TGG GCG ACA TTG AC | Amplified 0.7-kb internal region of gliP |
| 5′GliPKOLF | TGG CTC GCT GCA CGC GTA GAC | Check homologous recombination left (5′) |
| 3′PyrGKOLF | TAG GGT ACC TGT CCG CGC GGG G | Check homologous recombination left (5′) |
| 5′PyrGKORT | TGG CGA CCA CAC CCG TCC TGT G | Check homologous recombination right (3′) |
| 3′GliPKORT | GCC ATA TGA AGC ATC GAC TTA CC | Check homologous recombination right (3′) |
Gli RT primers are from reference 10. Primers 5′NRPS10 and 3′NRPS10 are for gliP.
Creation of fungal protoplasts and polyethylene glycol-mediated transformation of A. fumigatus were performed as previously described (5). Briefly, 2 μg of the gliPKO PCR-generated disruption construct was incubated on ice for 50 min with 1 × 107 fungal protoplasts in a total volume of 100 μl. Transformants were initially screened by PCR to identify potential homologous recombination events at the gliP locus. PCR was performed with primers designed to amplify a 700-bp internal region of the gliP locus (5′GliPReplace and 3′GliPReplace), which should be absent in gliP disruptant strains. In addition, PCR was performed with primers designed to amplify only the disrupted gliP locus (5′GlipKOLF and 3′PyrGKOLF; 5′PyrGKORT and 3′GliPKORT). Homologous recombination was confirmed by Southern analysis with the digoxigenin labeling system (Roche Molecular Biochemicals, Mannheim, Germany) as previously described (7). To eliminate the chance of heterokaryons, each transformant was streaked with sterile toothpicks a minimum of two times to obtain colonies from single spores.
Determination and quantification of gliotoxin production.
Czapek Dox broth was used to determine the production of gliotoxin in liquid broth cultures. Freshly harvested A. fumigatus conidia from wild-type strain AF293, ΔgliP mutant strain ARC2, or ectopic transformant strain ARC5 were inoculated into 50 ml of Czapek Dox broth and incubated for defined periods of time at 37°C while shaking at 200 rotations/min. Following growth, mycelia were separated from culture filtrates (CFs) by vacuum filtration. Mycelial mats were lyophilized overnight and weighed to determine the dry weight of fungal tissue. CFs were extracted 2× with equal volumes of high-performance liquid chromatography (HPLC) grade chloroform, and the organic layers were combined and dried to completeness on a roto-evaporator (Brinkmann-Eppendorf, Hamburg, Germany). Dried extracts were resuspended in 1 ml of HPLC grade methanol. Gliotoxin production was measured by reverse-phase HPLC on a Waters HPLC system with a UV light detector (Waters Corporation, Milford, MA). A Zorbax SB-C18 reverse-phase column (5 μm, 4.6 by 250 mm) was used (Agilent Technologies, Palo Alto, CA). Twenty microliters of the methanol-resuspended culture extracts was injected onto the column and eluted isocratically with 0.1% (vol/vol) trifluoroacetic acid-65% (vol/vol) HPLC grade water-34.9% (vol/vol) HPLC grade acetonitrile as previously described (27). The pressure in the column ranged from 1,400 to 1,800 lb/in2, and the flow rate was 1.00 ml min−1. Gliotoxin was detected at a wavelength of 264 nm. A standard curve of peak area versus gliotoxin concentration was constructed with gliotoxin (Sigma Aldrich, St. Louis, MO) standards ranging from 25 μg/ml to 1 mg/ml. Quantification of gliotoxin production by A. fumigatus strains was done by measuring CFs from cultures grown at 37°C for 72 h in Czapek Dox broth. The amount of gliotoxin produced by each Aspergillus strain is presented as milligrams of toxin produced per gram of dried fungal tissue.
Determination of spore morphology, colony growth, and sporulation.
Scanning electron microscopy was used to assess the surface morphology of conidia from A. fumigatus strains AF293, ARC2, and ARC5. Sporulation and colony growth were quantified as previously described (19). Briefly, 10-μl aliquots containing 1 × 107 conidia from freshly harvested glucose minimal medium plates were placed in the center of glucose minimal medium and Sabouraud's agar plates. Diameters of three colonies per A. fumigatus strain were measured once daily over a period of 5 days. The average change in colony diameter per 24 h of growth was calculated for three independent cultures. Sporulation was quantified by harvesting conidia from 5-day-old glucose minimal medium plates inoculated as for colony growth determination. Conidia were harvested with 20 ml of sterile 0.05% Tween 80, filtered through two layers of miracloth (EMD Biosciences, La Jolla, CA), and quantified with a hemacytometer. Conidia were harvested from three independent cultures, and the mean and standard deviation of the total number of conidia collected from the plate are reported.
Mast cell ionomycin chase β-hexosaminidase release assay.
Hindrance of degranulation of RBL cells was determined by measuring β-hexosaminidase release after pretreatment with fungal CF and stimulation with a calcium ionophore. Fungal CFs were prepared in Czapek Dox broth as described for gliotoxin production and sterilized by filtering through a 0.22-μm Millipore Express Plus membrane (Millipore Corporation, Billerica, MA). Briefly, monolayers of RBL cells (5 × 104) were exposed to 0.1 ml of fungal CF in Czapek Dox broth for 120 min. Filtrates were removed before stimulation with 0.1 ml of 1 μM ionomycin in Tyrode's buffer for 60 min. The cells were lysed with 0.1% Triton in Tyrode's buffer. Supernatants and total cell lysates were incubated with 3.4 mg/ml p-nitrophenyl-N-acetyl-β-d-galactosaminide in 0.1 M sodium citrate buffer (pH 4.5) at 37°C for 60 min. Addition of 0.1 M carbonate buffer (pH 10.0) terminated the reaction. Release of the product p-nitrophenol was evaluated by measuring optical absorbance at 405 nm. Results are expressed as the percentage of β-hexosaminidase released into the medium versus the total. The experiments were performed in quadruplicate wells for replicates 1 and 2 and triplicate wells for replicate 3. The variables are presented as the mean ± the standard error of the mean, and comparisons between groups were made by analysis of variance, followed by a multiple-comparison Tukey test. The analyses were done with the statistical package S-PLUS 7.0 for Windows (Insightful Corp., Seattle, WA), and a significance level of P < 0.05 was accepted.
Gene expression assays.
Expression of genes in the predicted gliotoxin biosynthesis cluster was assessed by real-time reverse transcriptase PCR (RT-PCR). Expression of genes in the gliotoxin biosynthesis cluster (GenBank accession no. AY83877) was examined in Czapek Dox broth cultures after 48 h of growth at 37°C. In addition, to confirm the regulation of the gliotoxin cluster by the presence or absence of gliotoxin, gliotoxin at final concentrations of 500 ng/ml, 5 μg/ml, and 20 μg/ml was added after 24 h to the ARC2 culture, which was incubated for an additional 24 h. Total RNA was extracted from the Czapek Dox broth cultures with the Ambion RNAqueous total RNA isolation kit (Ambion, Austin, TX). Total RNA was treated with Ambion Turbo DNA-free DNase I (Ambion, Austin, TX) to remove contaminating genomic DNA. A total of 500 ng of DNase-treated total RNA from each A. fumigatus strain was reverse transcribed with Superscript III RT (Invitrogen, Carlsbad, CA). Real-time RT-PCR was conducted with 20-μl reaction volumes with the iQ SYBR green supermix (Bio-Rad Laboratories, Hercules, CA), 2 μl of a 1:5 dilution of first-strand cDNA, and 0.4 μl of each 10 μM primer stock. No-RT controls were used to confirm elimination of contaminating genomic DNA. Real-time PCR was performed with an iQ Cycler real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA). PCRs for each gene were done in triplicate, and data were analyzed with the gene expression software included with the iQ Cycler system. Melt curve analysis was performed after the PCR was complete to confirm the absence of nonspecific amplification products. The fold change in gliotoxin cluster gene expression between A. fumigatus wild-type strain AF293 and the gliotoxin mutant ARC2, with or without addition of gliotoxin, was calculated by using 2−ΔΔCt, and all values were normalized to the expression of the A. fumigatus actin gene.
Murine virulence assays.
Outbred ICR (Harlan Sprague-Dawley) male mice (19 to 21 g in size) were used to assess the role of gliotoxin in fungal virulence. Mice were housed five per cage and had access to food and water ad libitum. All animal research procedures were approved by the Institutional Animal Care and Use Committee at the University of Texas Health Science Center at San Antonio. Mice were immunosuppressed with intraperitoneal (i.p.) injections of cyclophosphamide at 250 mg/kg and with cortisone acetate injected subcutaneously (s.c.) at 250 mg/kg 2 days prior to infection. On day 3 postinfection, repeat injections were given with cyclophosphamide (200 mg/kg i.p.) and cortisone acetate (250 mg/kg s.c.). Mice were given ceftazidime (50 mg/kg s.c.) daily beginning 2 days prior to infection.
Twenty mice per A. fumigatus strain (AF293, gliotoxin-deficient mutant ARC2, or ectopic mutant ARC5) were infected with an acrylic aerosol chamber that allows simultaneous challenge of 50 to 60 mice. In this acrylic chamber, A. fumigatus conidia were nebulized at 109 CFU/ml of fluid for 1 h. Infection inoculum was prepared by growing the A. fumigatus isolates on potato dextrose agar plates at 37°C for 7 days. Conidia were harvested by washing the plate surface with sterile phosphate-buffered saline-0.1% Tween 80, dislodging conidia by gentle rubbing with a sterile glass rod. The resultant conidial suspension was centrifuged at 15,000 × g (Sorvall SS-34 rotor) without braking and adjusted to the desired concentration of 109 conidia/ml by hemacytometer count. Mice were immunosuppressed, infected, and observed for survival for 14 days after A. fumigatus challenge. Any animals showing distress were immediately sacrificed and recorded as deaths within 24 h. Lungs from all mice sacrificed during the experiment were removed for fungal burden assessment.
For tissue burden studies, additional mice were immunosuppressed as described above, infected, and sacrificed at set time points of 1 h and day 7 after A. fumigatus challenge. When mice were sacrificed, serum and lungs were removed on that day. Tissues were aseptically removed, weighed, and homogenized with an overhead stirrer in 2 ml of sterile saline with gentamicin at 2.5 mg/liter and chloramphenicol at 400 mg/liter. Tissue homogenates were quantitatively cultured by serial dilution plating on potato dextrose agar incubated at 37°C overnight. Statistical analysis of tissue burdens among the three fungal strains was performed with a Kruskal-Wallis test and Dunn's multiple-comparison test. A log rank test was used for survival comparisons. Statistical significance was defined as P < 0.05.
Nucleotide sequence accession number.
The nucleotide sequence of the updated gliP sequence reported here has been deposited in GenBank under accession number DQ457015.
RESULTS
Disruption of the gliP-encoded NRPS.
Sequencing of the gliP cDNA revealed a 30-nucleotide discrepancy with respect to the previously published sequence. Our cDNA clone had an additional 30 nucleotides starting at position 613 in the cDNA sequence, bringing the total length of the gliP cDNA to 6,408 bp. This result is likely due to misannotation of the intron's splice site in the computer-based gene prediction algorithm. This updated gliP sequence was deposited in GenBank under accession number DQ457015.
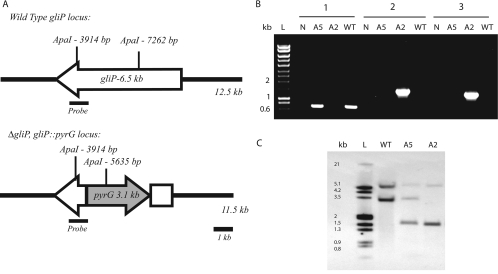
To definitively study the role of gliotoxin in A. fumigatus physiology and pathogenesis, we created a mutation in the gliP NRPS gene predicted to be involved in gliotoxin biosynthesis (Fig. 1A). Transformation of A. fumigatus protoplasts with the gliPKO PCR disruption construct yielded 25 transformants. Transformant ARC2 had no detectable PCR amplicon with primers (5′GlioPReplace and 3′GlioPReplace) designed to amplify the targeted replacement region of the gliP locus (Fig. 1B). ARC2 had detectable PCR amplicons with primers (5′GliPKOLF and 3′PyrGKOLF; 5′PyrGKORT and 3′GliPKORT) designed to amplify the predicted ΔgliP disruption locus after homologous recombination (Fig. 1B). These results suggested that the gliP locus had been precisely replaced with the ΔgliP::pyrG mutant allele in transformant ARC2.
FIG. 1.
Disruption of A. fumigatus NRPS gene gliP. (A) Representation of the gliP locus in wild-type and ΔgliP mutant strains. In ΔgliP mutant strain ARC2, a 3.8-kb internal section of the gliP coding region was replaced with the 3.1-kb pyrG gene from A. parasiticus. The ApaI restriction enzyme was used to digest chromosomal DNA, which was probed with the 1.0-kb probe as indicated. (B) PCR confirmation of gliP gene disruption. Lanes: L, DNA ladder; N, water control; A5, ectopic transformant strain ARC5; A2, ΔgliP mutant strain ARC2; WT, wild-type strain AF293. Group 1, amplification of internal gliP amplicon absent in ΔgliP mutant strain ARC2 because this sequence was replaced with pyrG via homologous recombination. Groups 2 and 3, 5′ and 3′ flanking regions, respectively. Amplification products in ΔgliP mutant strain ARC2 indicate successful homologous recombination at the gliP locus. (C) Southern analysis of A. fumigatus transformants. Lanes are the same as those in panel B.
By Southern analysis with ApaI-digested genomic DNA (Fig. 1A and C), we confirmed disruption of the gliP-encoded NRPS in transformant ARC2. Transformant ARC2 has a single insertion of the disruption construct into the gliP locus replacing the 3.7-kb internal fragment of the gliP-encoded NRPS, as indicated by the absence of the 3.3-kb wild-type band and the presence of a single 1.7-kb disruption construct band. Transformant ARC5 had a single ectopic integration of the disruption construct into the A. fumigatus genome, as indicated by the single 1.7-kb band corresponding to the disruption construct. This transformant was used as a gliP wild-type control since it complemented the pyrG auxotrophic defect in the recipient AF293.1 strain.
Measurement of gliotoxin production.
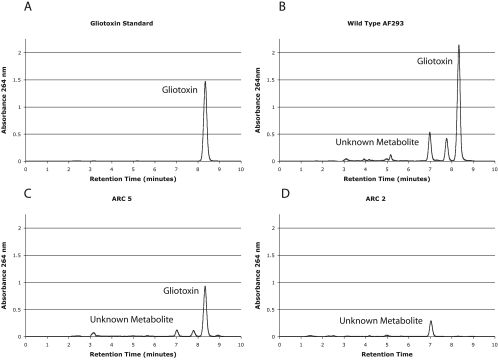
Previous bioinformatic and gene expression analyses suggested that gliP encodes the NRPS responsible for gliotoxin production in A. fumigatus (9, 10). To test this hypothesis, HPLC was used to determine if disruption of the gliP-encoded NRPS eliminates gliotoxin production. HPLC analysis of culture extracts from wild-type strain AF293 and transformation control strain ARC5 revealed the presence of a peak with a retention time (8.3 min) identical to that of a gliotoxin standard (Fig. 2A to C). This peak at 8.3 min is absent in the ΔgliP mutant ARC2, indicating that disruption of the gliP-encoded NRPS eliminated gliotoxin production (Fig. 2D). This result definitively confirms that gliP encodes the NRPS responsible for gliotoxin production. Quantification of gliotoxin production in strains ARC5 and AF293 revealed that the ectopic transformant ARC5 still produced wild-type levels of gliotoxin (13.1 ± 1.71 mg of toxin/g of fungus versus 10.96 ± 1.96 mg of toxin/g of fungus, respectively) and that the transformation procedure itself did not disturb or alter gliotoxin biosynthesis. We observed an additional peak (retention time of 7.05 min) with unknown identity produced in equal concentrations by all three strains.
FIG. 2.
HPLC analysis of the A. fumigatus strains used in this study. Chloroform extracts of CFs from Czapek Dox broth cultures grown for 72 h at 37°C. Gliotoxin and unknown metabolite peaks are identified. Panels: A, gliotoxin standard, 1 mg/ml; B, wild-type AF293; C, ectopic transformant ARC5; D, ΔgliP mutant strain ARC2 (lacks the peak at approximately 8.3 min, which demonstrates elimination of gliotoxin production in this mutant).
Phenotypic characteristics.
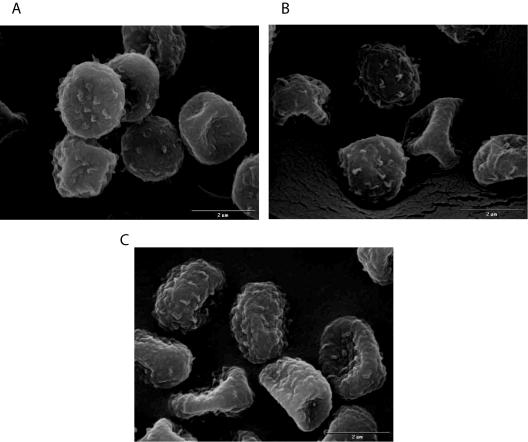
No detectable morphological or development differences were observed among strains ARC2, ARC5, and AF293. Conidia of ARC2 and ARC5 displayed normal pigmentation in culture, and scanning electron microscopy revealed no structural defects (Fig. 3). Conidiation was not affected in the gliotoxin-deficient mutant ARC2 or the ectopic transformant ARC5 (Table 2). Colony growth, as measured by the rate of increasing colony diameter, was not affected in any of the strains, regardless of the medium type (glucose minimal medium versus Sabouraud's medium), at 37°C (Table 2). Thus, we conclude that elimination of gliotoxin production has no effect on the overall growth and physiology of A. fumigatus.
FIG. 3.
Scanning electron microscopy of conidia from the A. fumigatus strains used in this study. Panels: A, ectopic transformant ARC5; B, ΔgliP mutant strain ARC2; C, wild-type strain AF293. No change in conidial surface morphology is apparent.
TABLE 2.
Growth and conidiation of A. fumigatus strains used in this studya
| A. fumigatus strain | Conidiation on glucose minimal medium (mean no. of conidia harvested/ culture ± SD) | Mean diam increase (cm)/ 24 h ± SD
|
|
|---|---|---|---|
| Growth on Sabouraud's agar | Growth on glucose minimal medium | ||
| AF293 | 2.87 × 1010 ± 1.31 × 1010 | 2.13 ± 0.13 | 1.30 ± 0.07 |
| ARC5 | 2.67 × 1010 ± 2.40 × 109 | 2.13 ± 0.32 | 1.31 ± 0.24 |
| ARC2 | 2.87 × 1010 ± 1.68 × 109 | 2.13 ± 0.22 | 1.32 ± 0.04 |
Growth for all experiments was conducted at 37°C.
Expression of gliotoxin biosynthesis gene cluster.
Genes involved in the production of fungal secondary metabolites are often physically located together in the genome in gene clusters. We used real-time RT-PCR to examine the expression of genes in the gliotoxin biosynthesis cluster in the wild-type and gliP mutant strains (Table 3). These data demonstrated a significant reduction at 48 h of incubation in the expression of all genes in the gliotoxin biosynthesis cluster in ΔgliP strain ARC2 relative to that in wild-type strain AF293 (Table 3). As expected, expression of gliP was eliminated (0.00007) in strain ARC2 compared to that in strain AF293. Expression of the predicted gliotoxin transporter gene gliA and the transcription factor gene gliZ was also virtually eliminated.
TABLE 3.
Analysis of gliotoxin biosynthesis cluster gene expression in ΔgliP mutant strain ARC2a
| Gene | Product | ARC2 | ARC2 + 500 ng/ml gliotoxin | ARC2 + 5 μg/ml gliotoxin | ARC2 + 20 μg/ml gliotoxin | Wild type + 20 μg/ml gliotoxin |
|---|---|---|---|---|---|---|
| gliA | MFS transporter | 0.00 ± 0.00 | 0.01 ± 0.00 | 0.33 ± 0.05 | 2.10 ± 0.27 | 1.37 ± 0.69 |
| gliC | Cytochrome P450 monooxygenase | 0.29 ± 0.01 | 0.57 ± 0.05 | 0.79 ± 0.07 | 1.29 ± 0.41 | 1.04 ± 0.49 |
| gliF | Unknown | 0.11 ± 0.01 | 0.55 ± 0.10 | 0.39 ± 0.07 | 2.30 ± 0.52 | 1.28 ± 0.63 |
| gliG | Glutathione S-transferase | 0.04 ± 0.01 | 0.05 ± 0.15 | 0.10 ± 0.3 | 3.18 ± 0.62 | 1.01 ± 0.50 |
| gliI | Aminocyclopropane carboxylate synthase | 0.30 ± 0.04 | 2.96 ± 0.24 | 1.02 ± 0.15 | 2.70 ± 1.10 | 1.22 ± 0.63 |
| gliJ | Dipeptidase | 0.45 ± 0.02 | 1.23 ± 0.27 | 0.66 ± 0.09 | 1.18 ± 0.84 | 2.12 ± 1.20 |
| gliK | Unknown | 0.09 ± 0.03 | 0.34 ± 0.03 | 0.15 ± 0.02 | 3.43 ± 0.51 | 1.57 ± 0.75 |
| gliM | O-Methyltransferase | 0.03 ± 0.00 | 0.27 ± 0.09 | 0.19 ± 0.05 | 4.81 ± 0.72 | 1.22 ± 0.63 |
| gliN | Methyltransferase | 0.16 ± 0.00 | 1.00 ± 0.15 | 0.65 ± 0.14 | 2.70 ± 1.28 | 1.85 ± 0.88 |
| gliP | Peptide synthetase | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 1.88 ± 0.91 |
| gliT | Thioredoxin reductase | 0.09 ± 0.00 | 0.85 ± 0.47 | 0.32 ± 0.21 | 2.7 ± 1.34 | 0.75 ± 0.61 |
| gliZ | Zinc finger transcription factor | 0.03 ± 0.00 | 0.12 ± 0.02 | 0.14 ± 0.06 | 4.29 ± 0.24 | 1.82 ± 1.18 |
The wild-type strain and ΔgliP mutant strain ARC2 were incubated in Czapek Dox broth for 24 h, when various concentrations of gliotoxin (0, 500 ng/ml, 5 μg/ml, and 20 μg/ml) were added and the cultures were incubated for an additional 24 h before total RNA extraction. Total RNA (500 ng) was reverse transcribed and used in a real-time RT-PCR. Data are presented as the mean fold change in expression (2−ΔΔCt) from wild-type AF293 (fold change = 1) ± the standard deviation. Expression of each gene under each condition is normalized to that of the gene for actin. Addition of 20 μg/ml gliotoxin restored expression of the gliotoxin biosynthesis gene cluster in ΔgliP mutant strain ARC2 but did not repress expression when added to the wild type.
This result suggested two possible competing mechanisms of gene regulation in the gliotoxin biosynthetic cluster. First, the gliP gene product itself could somehow regulate expression of the gene cluster. Alternatively, gliotoxin itself might control the induction of these genes. To distinguish between these two models and rule out damage to transcriptional regulators found in the gene cluster, we examined whether addition of gliotoxin to the ARC2 culture could restore expression of genes in the gliotoxin biosynthetic cluster. Addition of exogenous gliotoxin at concentrations of 500 ng/ml and 5 μg/ml had little effect on the expression of gliotoxin cluster genes in ARC2, and a direct relationship between gliotoxin concentration and gene transcription was not seen for all of the genes in the cluster. Interestingly, however, when gliotoxin was added to a final concentration of 20 μg/ml, expression of genes in the cluster was restored to wild-type or higher levels (Table 3). Addition of 20 μg/ml gliotoxin to a wild-type AF293 culture did not significantly alter cluster gene expression. These results suggest that disruption of gliP affects the expression of other genes in the gliotoxin biosynthetic cluster by eliminating production of gliotoxin, which subsequently positively regulates its own production by a dose-dependent mechanism.
Mast cell ionomycin chase β-hexosaminidase release assay.
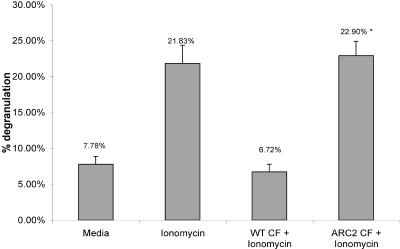
To begin to examine the potential effects of gliotoxin on A. fumigatus opportunistic parasitism, the effects of gliotoxin loss on effector cells of the innate immune system were examined. Fungal CFs from ARC2 and AF293 grown for 48 h in Czapek Dox broth were examined for their effects on RBL-2H3 mast cells. CFs from wild-type AF293 inhibited ionomycin-induced degranulation of the RBL-2H3 mast cells, while CFs from the gliotoxin mutant ARC2 did not (Fig. 4). Czapek Dox broth itself did not induce spontaneous degranulation. Light microscopy of the mast cells during treatment revealed that cells treated with wild-type CFs displayed morphological changes including rounding of the cells, which may indicate the beginning of cell death. However, these cells did not detach from the substrate, indicating that they were still viable. Trypan blue exclusion also revealed that the cells were still viable. Morphological changes in the cells were not observed with CFs from gliotoxin mutant strain ARC2. While CFs contain numerous fungal secondary metabolites, these results seem to confirm results previously reported by Niide et al. (24) that showed gliotoxin inhibition of mast cell functions. However, it should be noted that comparison of levels of all secondary metabolites in the CFs from the wild-type strain and strain ARC2 was not conducted, and thus changes in other secondary metabolite levels in the CFs may potentially account for this result. What is clear is that removal of gliotoxin production by disruption of the biosynthetic NRPS-encoding gene gliP significantly affects the interaction between A. fumigatus and RBL-2H3 mast cells.
FIG. 4.
Inhibition of ionomycin-dependent degranulation of RBL-2H3 mast cells by wild-type CF. CFs were added to RBL-2H3 mast cells, which were then incubated for 2 h. Mast cells were then stimulated for 1 h with 0.1 ml of 1 μM ionomycin. Significant inhibition of ionomycin-dependent degranulation is shown in wild-type CF versus the CF from ΔgliP mutant strain ARC2. *, P < 0.05 compared to wild-type (WT) CF plus ionomycin and a medium-alone control.
Inhalational neutropenic murine model of IA.
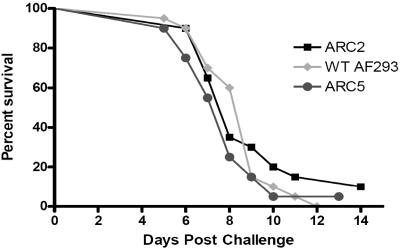
The potential inhibition of mast cell degranulation by gliotoxin suggested a possible role for gliotoxin in the development of IA. To test this hypothesis, the virulence of ΔgliP mutant strain ARC2 was compared to that of ectopic transformant ARC5 and wild-type strain AF293 in an inhalational neutropenic model of IA (Fig. 5). In this model, Aspergillus conidia are inhaled by immunosuppressed mice in a sterile chamber. This experimental model most closely reflects clinical Aspergillus infections in severely immunocompromised patients. Infection with wild-type strain AF293 in this model consistently results in mortality rates of 70 to 100%. In this study, no significant difference in the survival of mice infected with the wild-type strain or ΔgliP mutant strain ARC2 was seen (P > 0.05) (Fig. 5). The tissue burden in mouse lungs 1 h after inoculation revealed that all of the mice received approximately equal amounts of conidia from the wild-type strain and ΔgliP mutant strain ARC2 (P > 0.05) and the wild-type strain and ectopic transformant strain ARC5 (P > 0.05) (mean log10 CFU per gram ± the standard error of the mean: AF293, 4.34 ± 0.08; ARC2, 3.96 ± 0.06; ARC5, 4.50 ± 0.03). The tissue burden in mouse lungs was also measured 7 days after A. fumigatus infection, and no significant difference in CFU counts was seen among ARC2, ARC5, and wild-type AF293 (P > 0.05 for all pairwise comparisons) (mean log10 CFU per gram ± the standard error of the mean: AF293, 4.29 ± 0.11; ARC2, 4.56 ± 0.07; ARC5, 4.39 ± 0.11). Therefore, we conclude that gliotoxin production is not required for the development of IA in this murine model.
FIG. 5.
Inhalational neutropenic murine model of IA. Outbred ICR mice were used to assess the role of gliotoxin in fungal pathogenesis. Mice were immunosuppressed by i.p. injection of cyclophosphamide (250 mg/kg) and s.c. injection of cortisone acetate (250 mg/kg) 2 days prior to infection and injection of 200 mg/kg cyclophosphamide and 250 mg/kg cortisone acetate 3 days postinoculation. Mice were inoculated by exposure to a 109-conidium/ml suspension for 1 h in an inhalation chamber. A log rank test was used for pairwise comparisons of survival levels among the strain groups. P value for comparison between ARC2 and wild-type (WT) AF293, 0.642.
DISCUSSION
Gliotoxin has long been speculated to be a potential virulence factor of A. fumigatus (30). Gliotoxin is a strong immunosuppressant with effects on mature lymphocytes and macrophages and recently was detected in the serum of patients with IA (14, 21, 22, 29, 33). The disulfide bridge that characterizes ETP toxins like gliotoxin is responsible for the diverse array of biological activities of these toxins (31). The disulfide bridge can redox cycle, generating reactive oxygen species that can damage cells of the immune system, and additionally it can form mixed disulfide bonds with proteins that contain susceptible thiol residues (2, 34). Recently, it was shown that gliotoxin exerts much of its immunosuppressive biological activity through inhibition of the transcription factor NF-κΒ by inhibition of proteasome-mediated degradation of IκBα (12).
Given these biological effects on cells of the immune system, it has been hypothesized that gliotoxin is a significant factor in the pathogenesis of IA. However, direct genetic evidence in support of this hypothesis has been lacking, mainly because of the unknown identity of the gene or genes involved in the biosynthesis of gliotoxin. While many biologically active fungal secondary metabolites have been found, identifying the genes responsible for their complex biosynthesis can be extremely difficult. However, the completion of the A. fumigatus genome project has made it possible to identify candidate genes involved in secondary metabolism based on sequence similarity (23). Recently, identification of genes involved in the biosynthesis of the ETP toxin sirodesmin, from the plant pathogen Leptosphaeria maculans, made it possible to use sequence similarity to predict genes involved in gliotoxin production in A. fumigatus (9).
In this study, we definitively confirm the bioinformatic prediction that gliotoxin is synthesized by a two-module system consisting of NRPS and associated genes located in a gene cluster on chromosome 6 in the genome of A. fumigatus. Disruption of the NRPS gliP gene resulted in elimination of gliotoxin production, confirming the role of gliP in the biosynthesis of gliotoxin. Unexpectedly, we found that gliotoxin positively regulated the expression of genes in the biosynthetic cluster. Transcription of genes in the gliotoxin biosynthetic cluster was dramatically reduced in the gliotoxin mutant ARC2 (Table 3). Addition of gliotoxin to the culture medium restored expression of the genes in the biosynthetic cluster in a dose-dependent manner, with the threshold being approximately 20 μg/ml of gliotoxin (Table 3). A direct relationship between gliotoxin concentration and gene expression was not seen for all of the genes in the cluster (Table 3), although all of the genes were repressed without gliotoxin addition and expression was restored when the gliotoxin concentration reached 20 μg/ml. Thus, we cannot rule out the possibility that this apparent self-regulation is an indirect effect attributed to other not apparent effects on A. fumigatus physiology. Genome-wide expression profiling with various gliotoxin concentrations and time points may help clarify the underlying mechanism of this interesting regulation of secondary metabolite production. Examples of secondary metabolites regulating their own production, while rare, have been previously found in filamentous fungi. For example, the oxylipins produced by Aspergillus nidulans also seem to regulate their own production (32). However, to our knowledge, this is the first report of a fungal toxin involved in the regulation of its own production.
In an attempt to mechanistically examine the potential impact of gliotoxin on opportunistic fungal parasitism, we studied the impact of gliP loss on macrophage and mast cell interactions. Preliminary experiments with macrophage cell lines in our laboratory and CFs from wild-type AF293 and the gliotoxin mutant ARC2 suggested that loss of gliP does not affect the initial conidium-macrophage interaction but may impact macrophage cell viability later in the interaction when gliotoxin is present (data not shown). These results confirm previous reports of substantial effects of gliotoxin on macrophages and other phagocytes (8, 20). Next, we examined the potential effects of gliotoxin production on the A. fumigatus-mast cell interaction.
The role of mast cells in allergic disease is well established, but studies have also demonstrated a critical role for mast cells in the host defense against microorganisms (1, 17). Mast cells contain immunomodulating molecules that can accelerate the production of inflammation to clear invading microbes but can paradoxically lead to chronic inflammatory disease. Although the role of mast cells in invasive fungal infections has not been extensively explored, it was recently shown that gliotoxin can inhibit mast cell functions, including degranulation (24). Our results obtained with CFs from A. fumigatus confirm these results and suggest that gliotoxin is the main secondary metabolite responsible for inhibition of mast cell degranulation. Although it may be possible that changes in the presence or absence of other secondary metabolites in the CFs may account for the results seen with ARC2 CFs, it is clear from these results and other studies that gliotoxin has significant biological activity which may stimulate or depress immune system responses.
Since mast cells serve as sentinels of the immune system, we hypothesized that inhibition of degranulation by gliotoxin would contribute to the inhibition of neutrophil recruitment to sites of A. fumigatus infection and affect survival. However, we found no significant difference in mortality between mice infected with gliotoxin-deficient strain ARC2 and mice infected with gliotoxin-producing wild-type AF293 in an inhalational model of IA (Fig. 5). This result suggests that, despite its known immunosuppressant activities, including inhibition of mast cell degranulation, gliotoxin is not required for development of IA in immunosuppressed mice. This result is in agreement with a recent study by Lewis et al. (15) that found no correlation with gliotoxin production and the severity and dissemination of Aspergillus infections in patients at a tertiary cancer center. However, there are important additional issues to consider with regard to animal models and the pathophysiology of IA that may affect the ability to precisely define the role of gliotoxin in disease development.
The first issue for consideration is the immunosuppression regimen required for IA to develop in animal models. These regimens lead to profound immunosuppression in order to establish disease, which is an attempt to reproduce the immune status of patients with IA. Thus, it is questionable whether additional immunosuppression by gliotoxin would significantly alter the outcome of the fungus-host interaction. Furthermore, the timing of gliotoxin production in vivo may also be a critical factor in its possible role in IA. It is likely that Aspergillus infection is already established in immunocompromised hosts by the time gliotoxin production begins and gliotoxin accumulates to levels capable of affecting the already depleted host immune system. In vitro studies at 37°C generally first detect gliotoxin production 24 h after culture inoculation, with maximum production occurring between 48 and 72 h (10). Further studies are needed to address the possibility that gliotoxin production is altered somehow in vivo, which could affect its impact on disease development. However, our results with an inhalational murine model of IA strongly suggest that gliotoxin does not significantly affect the outcome of infection in an immunosuppressed murine model.
However, since fungal toxins like gliotoxin have inhibitory and lethal effects on cells of the immune system, their role in noninvasive diseases such as allergic bronchopulmonary aspergillosis and invasive disease in other at-risk patient populations such as patients with chronic granulomatous disease needs further examination. Although we have shown that gliotoxin itself is not essential for fungal virulence in an invasive pulmonary infection, aspergillosis is a multifaceted disease characterized by a continuum of overstimulation of host responses in allergic bronchopulmonary aspergillosis and immune system repression in IA. Thus, we cannot rule out the possibility that gliotoxin contributes to the morbidity seen in patients with other forms of aspergillosis. Finally, we also cannot rule out the possibility that the immunosuppressive properties of gliotoxin contribute to the development of additional secondary infections in immunocompromised patients.
Acknowledgments
Robert A. Cramer, Jr., is funded by NIH/NIAID Molecular Mycology and Pathogenesis Training Program contract 5 T32 AI052080. This project was funded in part by NIH/NIAID K08 award to William J. Steinbach, contract 1 K08 A1061149-01. This project was also funded in part by federal funds from National Institute of Allergy and Infectious Diseases, National Institutes of Health, contract N01-AI-30041, Thomas F. Patterson, principal investigator. Carl J. Balibar is funded by a Department of Defense National Defense Science and Engineering Graduate Fellowship in the laboratory of Christopher T. Walsh.
Special thanks to Nancy Keller and Dawoon Chung, University of Wisconsin—Madison; Andrew Alspaugh, Yvonne Yamanaka, and Zack Perfect, Duke University Medical Center; and Leslie Eibest, Duke University. We also thank three anonymous reviewers for their time and suggestions that significantly improved the manuscript.
REFERENCES
- 1.Abraham, S. N., K. Thankavel, and R. Malaviya. 1997. Mast cells as modulators of host defense in the lung. Front. Biosci. 2:d78-87. [DOI] [PubMed] [Google Scholar]
- 2.Bernardo, P. H., N. Brasch, C. L. Chai, and P. Waring. 2003. A novel redox mechanism for the glutathione-dependent reversible uptake of a fungal toxin in cells. J. Biol. Chem. 278:46549-46555. [DOI] [PubMed] [Google Scholar]
- 3.Bhabhra, R., and D. S. Askew. 2005. Thermotolerance and virulence of Aspergillus fumigatus: role of the fungal nucleolus. Med. Mycol. 43(Suppl. 1):S87-S93. [DOI] [PubMed] [Google Scholar]
- 4.Bok, J. W., S. A. Balajee, K. A. Marr, D. Andes, K. F. Nielsen, J. C. Frisvad, and N. P. Keller. 2005. LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot. Cell 4:1574-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bok, J. W., and N. P. Keller. 2004. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 3:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, Y. C., H. F. Tsai, M. Karos, and K. J. Kwon-Chung. 2004. THTA, a thermotolerance gene of Aspergillus fumigatus. Fungal Genet. Biol. 41:888-896. [DOI] [PubMed] [Google Scholar]
- 7.Cramer, R. A., and C. B. Lawrence. 2003. Cloning of a gene encoding an Alt a 1 isoallergen differentially expressed by the necrotrophic fungus Alternaria brassicicola during Arabidopsis infection. Appl. Environ. Microbiol. 69:2361-2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eichner, R. D., M. Al Salami, P. R. Wood, and A. Mullbacher. 1986. The effect of gliotoxin upon macrophage function. Int. J. Immunopharmacol. 8:789-797. [DOI] [PubMed] [Google Scholar]
- 9.Gardiner, D. M., A. J. Cozijnsen, L. M. Wilson, M. S. Pedras, and B. J. Howlett. 2004. The sirodesmin biosynthetic gene cluster of the plant pathogenic fungus Leptosphaeria maculans. Mol. Microbiol. 53:1307-1318. [DOI] [PubMed] [Google Scholar]
- 10.Gardiner, D. M., and B. J. Howlett. 2005. Bioinformatic and expression analysis of the putative gliotoxin biosynthetic gene cluster of Aspergillus fumigatus. FEMS Microbiol. Lett. 248:241-248. [DOI] [PubMed] [Google Scholar]
- 11.Kamei, K., and A. Watanabe. 2005. Aspergillus mycotoxins and their effect on the host. Med. Mycol. 43(Suppl. 1):S95-S99. [DOI] [PubMed] [Google Scholar]
- 12.Kroll, M., F. Arenzana-Seisdedos, F. Bachelerie, D. Thomas, B. Friguet, and M. Conconi. 1999. The secondary fungal metabolite gliotoxin targets proteolytic activities of the proteasome. Chem. Biol. 6:689-698. [DOI] [PubMed] [Google Scholar]
- 13.Latge, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis, R. E., N. P. Wiederhold, J. Chi, X. Y. Han, K. V. Komanduri, D. P. Kontoyiannis, and R. A. Prince. 2005. Detection of gliotoxin in experimental and human aspergillosis. Infect. Immun. 73:635-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis, R. E., N. P. Wiederhold, M. S. Lionakis, R. A. Prince, and D. P. Kontoyiannis. 2005. Frequency and species distribution of gliotoxin-producing Aspergillus isolates recovered from patients at a tertiary-care cancer center. J. Clin. Microbiol. 43:6120-6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liebmann, B., S. Gattung, B. Jahn, and A. A. Brakhage. 2003. cAMP signaling in Aspergillus fumigatus is involved in the regulation of the virulence gene pksP and in defense against killing by macrophages. Mol. Genet. Genomics 269:420-435. [DOI] [PubMed] [Google Scholar]
- 17.Malaviya, R., T. Ikeda, E. Ross, and S. N. Abraham. 1996. Mast cell modulation of neutrophil influx and bacterial clearance at sites of infection through TNF-α. Nature 381:77-80. [DOI] [PubMed] [Google Scholar]
- 18.Marr, K. A., R. A. Carter, F. Crippa, A. Wald, and L. Corey. 2002. Epidemiology and outcome of mould infections in hematopoietic stem cell transplant recipients. Clin. Infect. Dis. 34:909-917. [DOI] [PubMed] [Google Scholar]
- 19.McNeil, M. M., S. L. Nash, R. A. Hajjeh, M. A. Phelan, L. A. Conn, B. D. Plikaytis, and D. W. Warnock. 2001. Trends in mortality due to invasive mycotic diseases in the United States, 1980-1997. Clin. Infect. Dis. 33:641-647. [DOI] [PubMed] [Google Scholar]
- 20.Mullbacher, A., and R. D. Eichner. 1984. Immunosuppression in vitro by a metabolite of a human pathogenic fungus. Proc. Natl. Acad. Sci. USA 81:3835-3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mullbacher, A., A. F. Moreland, P. Waring, A. Sjaarda, and R. D. Eichner. 1988. Prevention of graft-versus-host disease by treatment of bone marrow with gliotoxin in fully allogeneic chimeras and their cytotoxic T cell repertoire. Transplantation 46:120-125. [DOI] [PubMed] [Google Scholar]
- 22.Mullbacher, A., P. Waring, and R. D. Eichner. 1985. Identification of an agent in cultures of Aspergillus fumigatus displaying anti-phagocytic and immunomodulating activity in vitro. J. Gen. Microbiol. 131:1251-1258. [DOI] [PubMed] [Google Scholar]
- 23.Nierman, W. C., A. Pain, M. J. Anderson, J. R. Wortman, H. S. Kim, J. Arroyo, M. Berriman, K. Abe, D. B. Archer, C. Bermejo, J. Bennett, P. Bowyer, D. Chen, M. Collins, R. Coulsen, R. Davies, P. S. Dyer, M. Farman, N. Fedorova, N. Fedorova, T. V. Feldblyum, R. Fischer, N. Fosker, A. Fraser, J. L. Garcia, M. J. Garcia, A. Goble, G. H. Goldman, K. Gomi, S. Griffith-Jones, R. Gwilliam, B. Haas, H. Haas, D. Harris, H. Horiuchi, J. Huang, S. Humphray, J. Jimenez, N. Keller, H. Khouri, K. Kitamoto, T. Kobayashi, S. Konzack, R. Kulkarni, T. Kumagai, A. Lafton, J. P. Latge, W. Li, A. Lord, C. Lu, W. H. Majoros, G. S. May, B. L. Miller, Y. Mohamoud, M. Molina, M. Monod, I. Mouyna, S. Mulligan, L. Murphy, S. O'Neil, I. Paulsen, M. A. Penalva, M. Pertea, C. Price, B. L. Pritchard, M. A. Quail, E. Rabbinowitsch, N. Rawlins, M. A. Rajandream, U. Reichard, H. Renauld, G. D. Robson, S. Rodriguez de Cordoba, J. M. Rodriguez-Pena, C. M. Ronning, S. Rutter, S. L. Salzberg, M. Sanchez, J. C. Sanchez-Ferrero, D. Saunders, K. Seeger, R. Squares, S. Squares, M. Takeuchi, F. Tekaia, G. Turner, C. R. Vazquez de Aldana, J. Weidman, O. White, J. Woodward, J. H. Yu, C. Fraser, J. E. Galagan, K. Asai, M. Machida, N. Hall, B. Barrell, and D. W. Denning. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151-1156. [DOI] [PubMed] [Google Scholar]
- 24.Niide, O., Y. Suzuki, T. Yoshimaru, T. Inoue, T. Takayama, and C. Ra. 2006. Fungal metabolite gliotoxin blocks mast cell activation by a calcium- and superoxide-dependent mechanism: implications for immunosuppressive activities. Clin. Immunol. 118:108-116. [DOI] [PubMed] [Google Scholar]
- 25.Osbourn, A. E. 2001. Tox-boxes, fungal secondary metabolites, and plant disease. Proc. Natl. Acad. Sci. USA 98:14187-14188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perfect, J. R., G. M. Cox, J. Y. Lee, C. A. Kauffman, L. de Repentigny, S. W. Chapman, V. A. Morrison, P. Pappas, J. W. Hiemenz, and D. A. Stevens. 2001. The impact of culture isolation of Aspergillus species: a hospital-based survey of aspergillosis. Clin. Infect. Dis. 33:1824-1833. [DOI] [PubMed] [Google Scholar]
- 27.Reeves, E. P., C. G. Messina, S. Doyle, and K. Kavanagh. 2004. Correlation between gliotoxin production and virulence of Aspergillus fumigatus in Galleria mellonella. Mycopathologia 158:73-79. [DOI] [PubMed] [Google Scholar]
- 28.Shimizu, K., and N. P. Keller. 2001. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157:591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stanzani, M., E. Orciuolo, R. Lewis, D. P. Kontoyiannis, S. L. Martins, L. S. St John, and K. V. Komanduri. 2005. Aspergillus fumigatus suppresses the human cellular immune response via gliotoxin-mediated apoptosis of monocytes. Blood 105:2258-2265. [DOI] [PubMed] [Google Scholar]
- 30.Sutton, P., N. R. Newcombe, P. Waring, and A. Mullbacher. 1994. In vivo immunosuppressive activity of gliotoxin, a metabolite produced by human pathogenic fungi. Infect. Immun. 62:1192-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trown, P. W., and J. A. Bilello. 1972. Mechanism of action of gliotoxin: elimination of activity by sulfhydryl compounds. Antimicrob. Agents Chemother. 2:261-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsitsigiannis, D. I., T. M. Kowieski, R. Zarnowski, and N. P. Keller. 2005. Three putative oxylipin biosynthetic genes integrate sexual and asexual development in Aspergillus nidulans. Microbiology 151:1809-1821. [DOI] [PubMed] [Google Scholar]
- 33.Waring, P., R. D. Eichner, A. Mullbacher, and A. Sjaarda. 1988. Gliotoxin induces apoptosis in macrophages unrelated to its antiphagocytic properties. J. Biol. Chem. 263:18493-18499. [PubMed] [Google Scholar]
- 34.Waring, P., A. Sjaarda, and Q. H. Lin. 1995. Gliotoxin inactivates alcohol dehydrogenase by either covalent modification or free radical damage mediated by redox cycling. Biochem. Pharmacol. 49:1195-1201. [DOI] [PubMed] [Google Scholar]
- 35.Wilson, D. M., W. Mubatanhema, and Z. Jurjevic. 2002. Biology and ecology of mycotoxigenic Aspergillus species as related to economic and health concerns. Adv. Exp. Med. Biol. 504:3-17. [DOI] [PubMed] [Google Scholar]
- 36.Xue, T., C. K. Nguyen, A. Romans, D. P. Kontoyiannis, and G. S. May. 2004. Isogenic auxotrophic mutant strains in the Aspergillus fumigatus genome reference strain AF293. Arch. Microbiol. 182:346-353. [DOI] [PubMed] [Google Scholar]