Abstract
Virulence-attenuating hypoviruses of the species Cryphonectria hypovirus 1 (CHV1) encode a papain-like protease, p29, that shares similarities with the potyvirus-encoded suppressor of RNA silencing HC-Pro. We now report that hypovirus CHV1-EP713-encoded p29 can suppress RNA silencing in the natural host, the chestnut blight fungus Cryphonectria parasitica. Hairpin RNA-triggered silencing was suppressed in C. parasitica strains expressing p29, and transformation of a transgenic green fluorescent protein (GFP)-silenced strain with p29 resulted in an increased number of transformants with elevated GFP expression levels. The CHV1-EP713 p29 protein was also shown to suppress both virus-induced and agroinfiltration-induced RNA silencing and systemic spread of silencing in GFP-expressing transgenic Nicotiana benthamiana line 16c plants. The demonstration that a mycovirus encodes a suppressor of RNA silencing provides circumstantial evidence that RNA silencing in fungi may serve as an antiviral defense mechanism. The observation that a phylogenetically conserved protein of related plant and fungal viruses functions as a suppressor of RNA silencing in both fungi and plants indicates a level of conservation of the mechanisms underlying RNA silencing in these two groups of organisms.
RNA-mediated, sequence-specific silencing of gene expression, termed RNA silencing, has been reported for plants (33, 48), fungi (39), and animals (16) and variously referred to as posttranscriptional gene silencing, quelling, and RNA interference, respectively. A common feature of RNA silencing is the processing of structured or double-stranded RNA into small interfering RNAs (siRNAs) of 21 to 25 nucleotides by members of the RNase III family of double-stranded RNA-specific endonucleases (Dicers). These siRNAs are then incorporated into an RNA-induced silencing complex that guides sequence-specific degradation of homologous RNA (reviewed in reference 56).
RNA silencing plays a key antiviral defense role in plants (reviewed in reference 52) and has been demonstrated to influence virus replication in animal cells (31). Viruses, in turn, produce proteins capable of suppressing host cell RNA silencing (reviewed in reference 46). RNA silencing is part of a larger set of regulatory pathways involving small RNAs that include microRNA (miRNA)-mediated developmental regulation in plants and animals (reviewed in reference 20). Recent findings (29) indicate that some viral suppressors of RNA silencing also contribute to virus-induced disease symptoms by interfering with miRNA-controlled developmental pathways.
Although RNA silencing in fungi is generally portrayed as having originated as an ancient antiviral defense mechanism, there is currently no experimental evidence to support this assumption. Our understanding of RNA silencing in fungi comes primarily from studies with the model filamentous fungus Neurospora crassa (4, 6, 11). However, no experimental virus system is available for this organism. In this regard, the chestnut blight fungus, Cryphonectria parasitica, is closely related phylogenetically to N. crassa (15) and has a well-established experimental system for the study of mycoviruses in the family Hypoviridae that reduce the virulence of this pathogenic fungus (35). Moreover, hypoviruses are phylogenetically related to plant potyviruses (30) and have been shown to encode a papain-like protease, p29, that shares sequence and functional similarities with the potyvirus-encoded suppressor of RNA silencing HC-Pro (47). We have taken advantage of the hypovirus/C. parasitica experimental system to test whether Cryphonectria hypovirus 1-EP713 p29 (CHV1-EP713 p29) can serve as a suppressor of RNA silencing in fungi and plants. The results are discussed as they relate to the role of RNA silencing in fungi and the conservation of RNA silencing mechanisms across phylogenetic kingdoms.
MATERIALS AND METHODS
Fungal strains, growth conditions, and transformation.
C. parasitica strains were maintained on potato dextrose agar (PDA; Difco) as described previously (21). Preparative cultures for RNA isolation were grown for 7 days at room temperature under ambient light, with cellophane covering the growth medium to facilitate mycelial harvest. Preparation and transformation of C. parasitica spheroplasts were carried out essentially as described by Churchill et al. (10). Hygromycin (40 μg/ml), benomyl (0.7 μg/ml), or blasticidin (300 μg/ml) was included in the growth medium to provide for selection of transformants.
Nucleic acid procedures.
General molecular biology procedures were carried out using standard techniques (41). Fungal genomic DNA was extracted as described by Choi et al. (8). Green fluorescent protein (GFP) transcript accumulation was measured using real-time reverse transcription-PCR (RT-PCR) with previously described primer/probe sets specific for 18S rRNA and GFP transcripts (36), and the values were normalized using the comparative ΔCt method relative to the amount of 18S rRNA in each sample, as described by Parsley et al. (36). RNA was prepared from colonies grown on cellophane-overlaid PDA as described previously (42).
DNA sequencing was performed by the Center for Biosystems Research sequencing facility using the ABI-Prism BigDye Terminator Ready-Reaction cycle sequencing kit (Applied Biosystems) to prepare the sequencing reaction mixtures, which were analyzed on an ABI 3100 genetic analyzer (Applied Biosystems).
Plasmid constructs.
Plasmid pGS was constructed using plasmid pCPX-NBn1, which is identical to pCPX-NBn2 (43) but has a polylinker with NotI, HpaI, HindIII, and SphI restriction sites in 5′-to-3′ order. The NsiI site of pCPX-NBn1 was removed by digestion with NsiI, filling in the ends with T4 DNA polymerase, and self-ligation. Subsequently, a SacI-NotI fragment containing the C. parasitica glyceraldehyde-3-phosphate dehydrogenase (GPD) promoter was replaced with a 900-bp SacI-NotI fragment containing the Aspergillus nidulans GPDA promoter (37). Finally, the second intron of the C. parasitica GPD gene was amplified from genomic DNA with primers intF (5′ TAT GCG GCC GCT CTA GAT TTC TGC AGT AAG TAA CAC ATA CCA AAC A 3′) and intR (5′ TAT AAG CTT GCT AGC GCT ATG CAT CTG TGG GGG GAA GAG TAA ATT G 3′). The primers were designed to have NotI, XbaI, and PstI restriction sites 5′ of the intron and NsiI, NheI, and HindIII 3′ of the intron. The NotI-HindIII-digested PCR fragment was cloned into the NotI-HindIII sites of the modified pCPX-NBn1 vector, resulting in plasmid pGS.
To construct pGS-2GFP, the enhanced GFP (EGFP) open reading frame was amplified from plasmid pPdEGFP (36) with primers GFP-NsF (5′ TAT ATG CAT AGC AAG GGC GAG GAG 3′) and GFP-NhR (5′ TAT GCT AGC TCG TCC ATG CCG AGA G 3′), digested with NsiI and NheI, and cloned into the NsiI/NheI-digested pGS vector, to create pGS-NNGFP. Subsequently, the digested GFP fragment was cloned into the XbaI-PstI sites of pGS-NNGFP, resulting in plasmid pGS-2GFP.
Plasmid pCPX-BSD1 was constructed by replacing the hygromycin phosphotransferase coding region of plasmid pCPX-Hy2 with the coding region for blasticidin-S-deaminase. pCPX-Hy2 is identical to pCPX-Hy3 (43) but has a polylinker as described above for pCPX-NBn1.
To construct plasmids pCPX-BSD/p29 and pCPX-BSD/mp29, the p29 coding region was amplified from pLDST (7) using primers NS26 and NS27 (47) for p29 and primer mp29F (5′ TAT AAG CTT AAT GGC TtA ATg AAG AAA ACC CAG 3′; nucleotides introducing mutations are indicated in lowercase) and NS27 for mp29. The p29 amplicon was digested with SacI, blunted with T4 DNA polymerase, digested with SphI, and cloned into the HpaI/SphI-digested pCPX-BSD1 vector. The mp29 amplicon was digested with HindIII and SphI and cloned into the HindIII/SphI-digested pCPX-BSD1 vector. The sequences of the inserts were confirmed by sequencing.
To test whether p29 would be biologically functional in planta, p29 was expressed as a fusion protein with GFP. The complete coding sequences of the CHV1-EP713 p29 gene and its mutated derivative were PCR amplified using primers pp215 (5′ GTC CGA Aat cgA TGG CTC AAT TAA GAA AAC CCA G 3′) and pp224 (5′ GAA CCT TTc gGc CGG GTA ACC TGT GGA G 3′) or pp224 and pp216 (5′ GTC CGA Aat cgA TGG CTt AAT gAA GAA AAC CCA G 3′), digested with ClaI and EagI, and cloned in frame to the GFP coding sequence in the ClaI/EagI sites of potato virus X (PVX)/GFP (5, 50) to produce PVX/p29-GFP and PVX/mp29-GFP, respectively. Introduced ClaI and EagI sites are underlined, modified nucleotides are in lowercase, and the start and mutated nonsense codons are in boldface. No p29-GFP fusion protein would be expressed from PVX/mp29-GFP due to the presence of two consecutive stop codons, TAA and TGA, that were introduced downstream of the p29 start codon. The integrity of the p29 and mp29 coding sequences was confirmed by nucleotide sequencing.
Constructs for transient expression of p29-GFP under a tandem repeat of the 35S promoter and polyadenylation sequences were also produced. Fragments encompassing p29-GFP and mp29-GFP coding sequences were excised from PVX/p29-GFP and PVX/mp29-GFP and cloned into a plant gene expression vector (18) to produce p35S/p29-GFP and p35S/mp29-GFP, respectively. The p29-GFP expression cassettes were then subcloned into pBINPLUS (49) to produce pBin35S/p29-GFP and pBin35S/mp29-GFP, which were electrotransformed into Agrobacterium tumefaciens LBA4404 (24).
GFP fluorescence measurements.
To measure GFP fluorescence of C. parasitica, colonies were grown for 4 days on PDA. Plugs from the growing edge of the colony were removed with a 5-mm cork borer and transferred to the wells of a black plastic 96-well plate, with the mycelium side facing up. Fluorescence at 507 nm was measured in a fluorescence plate reader (Perkin-Elmer, LS 55) with an excitation wavelength at 488 nm and cutoff set at 515 nm. As controls for background fluorescence, fluorescence of wild-type strain EP155 and EP155 transformed with pCPX/p29 was measured in triplicate. Relative fluorescence (RF) of transformants was calculated as follows: background fluorescence values of EP155, or EP155/pCPX/p29 in cases where pCPX/p29 was used, were deducted from the fluorescence values of the transformants. The resulting values were then expressed as percentages of the fluorescence of the recipient strain in the transformation.
Isolation and detection of siRNA.
Small-RNA purification and detection were performed essentially as described by Catalanotto et al. (3) using Trizol reagent (Invitrogen) following the manufacturer's instructions with minor modifications. Frozen mycelia were homogenized with liquid nitrogen in a mortar and pestle, and approximately 300 mg was transferred to a tube with 3 ml Trizol reagent. Before chloroform was added, the homogenate was centrifuged at 12,000 × g for 10 min. Following the first chloroform extraction, the top phase was reextracted with phenol-chloroform. The nucleic acids were precipitated by addition of 1/10 volume of 3 M NaAc, pH 5.2, and 3 volumes of 100% ethanol, incubation of the mixture at −20°C for 1 h, and centrifugation at 12,000 × g for 30 min. The pellet was resuspended in 1 ml water, and large RNA species were precipitated by addition of polyethylene glycol 8000 to 5% and NaCl to 0.5 M, incubation for 30 min on ice, and centrifugation at maximum speed in a microcentrifuge at 4°C for 10 min. The supernatant containing the siRNAs was extracted once with phenol-chloroform and once with chloroform, and the RNA was precipitated by addition of 3 volumes ethanol, incubation at −20°C for at least 2 h, and centrifugation for 30 min. The pellet was dissolved in 50 μl water.
The low-molecular-weight RNAs were separated on a 15% polyacrylamide gel in the presence of 7 M urea and 0.5× Tris-boric acid-EDTA (TBE) (1× TBE is 89 mM Tris, 89 mM boric acid, 2 mM EDTA). An RNA size ladder (Decade markers; Ambion) was run along with the samples to determine the size of the hybridizing band. The nucleic acids were electroblotted onto GeneScreen Plus membrane (Perkin-Elmer) in 1× TBE and cross-linked by UV irradiation.
Hybridizations were carried out overnight using Ultrahyb (Ambion) hybridization buffer at 40°C and a 32P-labeled GFP probe prepared using a random-primed labeling kit (Roche) according to the manufacturer's instructions. After hybridization, the blot was washed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate for 10 min at 40°C.
Virus infection assays.
PVX RNA transcripts were produced by in vitro transcription and mechanically inoculated onto Nicotiana benthamiana plants as described previously (5). Plants were maintained in an insect-free growth room at 25°C with continuous lighting to give a 12-h photoperiod. To investigate viral coat protein (CP) and p29-GFP fusion protein expression, total protein was extracted from leaf tissues as described by Hong et al. (25). Western blot analyses of protein aliquots (10 μg) were performed with a polyclonal antiserum raised against PVX CP or GFP and detected using a goat anti-rabbit immunoglobulin G conjugated with alkaline phosphatase (Sigma) and 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium substrates (Roche).
Seedlings of transgenic N. benthamiana line 16c constitutively expressing GFP (2) were mechanically inoculated with RNA transcripts produced by in vitro transcription from SpeI-linearized PVX/GFP, PVX/p29-GFP, and PVX/mp29-GFP. Alternatively, seedlings of transgenic N. benthamiana line 16c were infiltrated with A. tumefaciens strain C58C1 carrying a functional 35S-GFP expression cassette or A. tumefaciens strain LBA4404 carrying pBin35S/p29-GFP or pBin35S/mp29-GFP. Induction and suppression of RNA silencing of GFP expression were routinely examined under long-wavelength UV light and photographically recorded using a Nikon Digital Camera Coolpix 990 through a yellow filter.
RESULTS
RNA silencing in Cryphonectria parasitica.
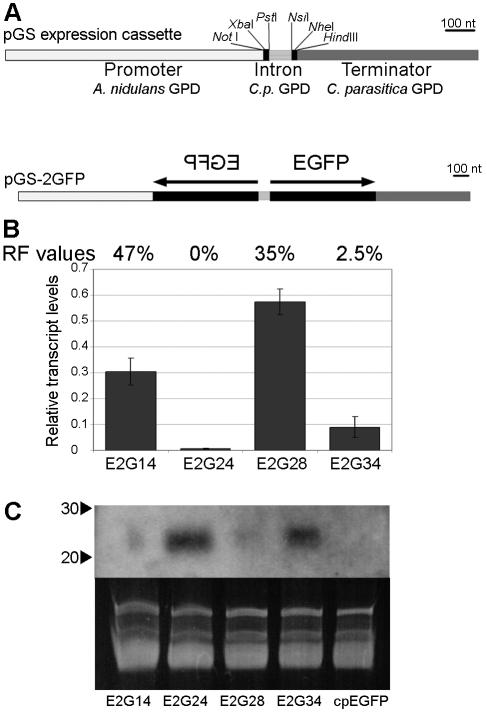
An experimental system was developed to test predictions that p29 may function as a suppressor of RNA silencing in C. parasitica by transforming a transgenic GFP-expressing C. parasitica strain (cpEGFP [36]) with a silencing vector, pGS-2GFP, designed to express an intron-containing hairpin RNA copy of the EGFP gene (Fig. 1A) and examining the resulting cpEGFP/pGS-2GFP (E2G) double transformants for GFP fluorescence. Reduced fluorescence was routinely observed in a high proportion of the double transformants, with a greater than 80% reduction in fluorescence observed for approximately 70% of the transformants (Fig. 2A). Southern analysis of several silenced transformants indicated intact transgenic EGFP promoter and coding regions (data not shown), indicating that the reductions in GFP fluorescence were not due to disruption of the GFP transgene. Real-time RT-PCR analysis of four selected transformants with various degrees of GFP silencing showed reduced accumulation of GFP transcript in each transformant with an apparent general correlation between the level of GFP silencing and GFP transcript accumulation (Fig. 1B).
FIG. 1.
RNA silencing in Cryphonectria parasitica. (A) Schematic representation of hairpin RNA silencing vector pGS and the pGS-2GFP construct used to silence an EGFP transgene in C. parasitica. Plasmid pGS has multiple cloning sites on either side of a small intron derived from the C. parasitica GPD gene to allow cloning of two copies of portions of the target gene in an inverted-repeat orientation similar to gene silencing vectors designed for RNA silencing in plants (55). Expression is driven by the Aspergillus nidulans GPD promoter, and transcription is terminated by the C. parasitica GPD terminator. The vector also contains the benomyl resistance cassette (not shown) as a selectable marker. The construct pGS-2GFP contains inverted repeats of the entire EGFP coding sequence in the pGS vector. nt, nucleotides. (B) Histogram of EGFP transcript accumulation in cpEGFP/pGS-2GFP (E2G)-silenced transformants, relative to EGFP transcript accumulation in the cpEGFP transgenic strain, as measured by real-time RT-PCR. The EGFP fluorescence values relative to that of the cpEGFP strain (RF values) are indicated above the bars. (C) Small-RNA Northern blot analysis of EGFP-specific siRNAs in the same silenced transformants as listed in panel B. Positions of 20- and 30-nucleotide RNA size markers are indicated by arrowheads.
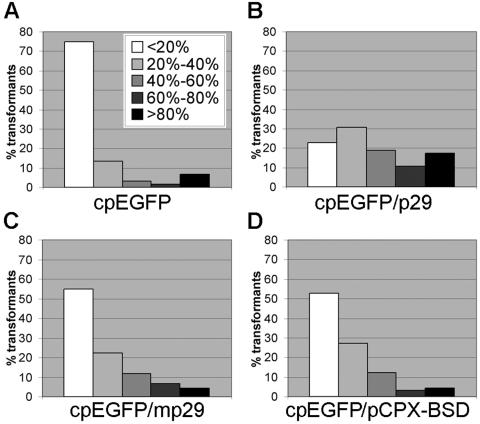
FIG. 2.
Effect of p29 expression on induction of pGS-2GFP hairpin-mediated RNA silencing. Hairpin RNA silencing plasmid pGS-2GFP was introduced into the cpEGFP strain and into double transformants expressing EGFP and either a functional p29 coding sequence, a mutated p29 sequence (mp29) that contained two stop codons immediately after the ATG start codon, or the empty expression vector pCPX-BDS1 used to introduce the p29 sequences. The transformants were divided into five groups based on their RF: <20% of the fluorescence of the corresponding reference strain, 20% to 40%, 40% to 60%, 60% to 80% and >80%. The bars in the histograms indicate the percentages of the total number of transformants that fall into each RF group. (A) Histogram representing 60 cpEGFP/pGS-2GFP transformants. (B) Histogram representing 180 transformants derived from three independent cpEGFP/p29 strains transformed with pGS-2GFP. (C) Histogram representing 180 transformants derived from three independent cpEGFP/mp29 strains transformed with pGS-2GFP. (D) Histogram representing 180 transformants derived from three independent cpEGFP/pCPX-BSD1 strains transformed with pGS-2GFP.
The generation of siRNAs is a hallmark of RNA silencing in plants and animals (56) and has been reported for RNA silencing in N. crassa (3), Magnaporthe oryzae (27), and Aspergillus nidulans (19). To confirm that the reduction in GFP fluorescence observed for the cpEGFP/pGS-2GFP transformants was a result of RNA silencing, low-molecular-weight RNAs were extracted and examined for the presence of GFP-specific siRNAs. As shown in Fig. 1C, GFP-specific siRNAs were observed for each of the RNA-silenced transformants, with a stronger signal observed for those strains showing the lowest fluorescence levels, i.e., the highest level of silencing.
Hypovirus papain-like protease p29 suppresses RNA silencing in C. parasitica.
To test for p29-mediated suppression of RNA silencing, we first asked whether the presence of p29 would reduce the frequency and level of GFP silencing conferred by the pGS-2GFP hairpin RNA silencing vector. This was accomplished by constructing three sets of transgenic cpEGFP C. parasitica strains that also contained either a functional p29 coding sequence, a mutated p29 sequence (mp29) that contained two stop codons immediately after the ATG start codon, or the empty expression vector pCPX-BSD1 used to introduce the p29 sequences. Only cpEGFP/p29 transformants that showed the reduced-pigmentation phenotype characteristic of p29 expression (14) were used for further analysis. The production of mp29 transcripts in cpEGFP/mp29 transformants, which does not result in phenotypic changes, was confirmed by RT-PCR (results not shown). Three independent transformants that exhibited a level of fluorescence similar to that of the original cpEGFP were selected for each construct, cpEGFP/p29, cpEGFP/mp29, and cpEGFP/pCPX-BDS1, and the nine double transformant strains were then transformed with the silencing vector pGS-2GFP. Sixty transformants were selected for each of the nine transformation reactions (180 transformants total for each of the p29, mp29, and pCPX-BSD1 constructs) and analyzed for GFP fluorescence with a fluorescence plate reader. The values were compared to the fluorescence level exhibited by the corresponding recipient strain and divided into five groups based on RF as shown in Fig. 2.
The pattern observed for transformants recovered from cpEGFP/m29 and cpEGFP/pCPX-BSD1 strains transformed with pGS-2GFP exhibited a slight but reproducible shift to the right, compared with the cpEGFP strain transformed with the same vector, with 52 to 56% of the transformants falling in the <20% RF group compared to 75% of the cpEGFP/pGS-2GFP transformants in this category (Fig. 2C and 2D). In contrast, only 20% of the transformants recovered from the pGS-2GFP-transformed cpEGFP/p29 strains fell within the <20% RF group with approximately 50% of the transformants in the 40% to 60% RF group and above (Fig. 2D). These results indicate that p29 is capable of suppressing hairpin RNA-triggered RNA silencing in C. parasitica.
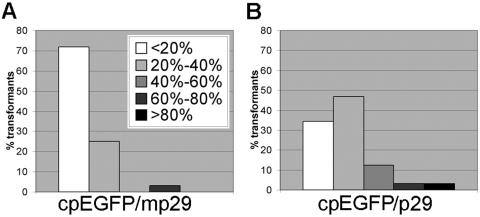
We next asked whether p29 could reverse RNA silencing. To this end, two pGS-2GFP-silenced cpEGFP strains (E2G24 and E2G34; Fig. 1B) were transformed with either pCPXp29 or pCPXmp29, programmed to express p29 protein and transcribe mp29 RNA, respectively. To determine the effect of the transformation procedure on silencing, spheroplasts of the E2G24 and E2G34 strains were taken through a mock transformation reaction and fluorescence was measured for portions of mass-regenerated colonies. The relative fluorescence values for the mock-transformed silenced strains and pCPXp29 and pCPXmp29 transformants were then normalized to the fluorescence level exhibited by the cpEGFP strain to determine relative fluorescence. The RF values of mock-transformed E2G24 and E2G34 showed a modest increase over the values observed for the original transformants (6% versus 0% for E2G24 and 16% versus 2.5% for E2G34) but were less than 20% of cpEGFP fluorescence levels. As indicated in Fig. 3A, transformation of GFP-silenced strains E2G24 and E2G24 with pCPXmp29 did not interfere with silencing, since the majority (approximately 70%) of the transformants still had RF values of less than 20% of the fluorescence level of cpEGFP (Fig. 3A). However, transformation with the nonmutated p29 coding region resulted in a shift in RF values, with fewer transformants (34%) showing the lowest level of GFP fluorescence, and an increasing number of transformants (47%) exhibiting RF values between 20% and 40% of cpEGFP fluorescence (Fig. 3B). All of the transformants containing the functional p29 coding region showed the reduced-pigmentation phenotype characteristic of p29 expression (14). These results suggest that p29 can also reverse established RNA silencing in C. parasitica.
FIG. 3.
Effect of p29 on established hairpin RNA silencing. Two pGS-2GFP-silenced cpEGFP strains, E2G24 and E2G34, were transformed with either pCPXp29 or pCPXmp29, programmed to express p29 or mp29, respectively. The fluorescence relative to that of untransformed strain cpEGFP was determined for 16 transformants selected from each transformation reaction. The percentage of transformants in each RF group was plotted as described for Fig. 2. (A) Histogram representing 32 transformants derived from E2G24 and E2G34 transformed with pCPXmp29. (B) Histogram representing 32 transformants derived from E2G24 and E2G34 transformed with pCPXp29.
Hypovirus CHV1-EP713 papain-like protease p29 suppresses RNA silencing in a heterologous plant system.
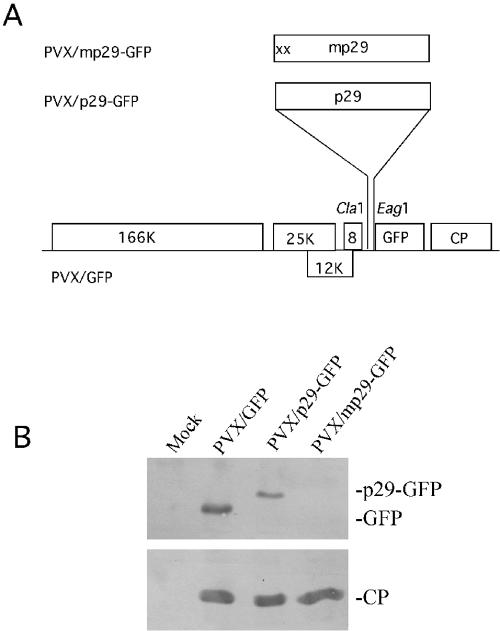
Since p29 shares significant amino acid sequence similarity with the well-documented potyvirus-encoded RNA silencing suppressor HC-Pro (9, 30, 47), we speculated that p29 could also function as an RNA silencing suppressor in plants. This was tested using the established PVX expression vector-based virus-induced gene silencing (VIGS) assay (40). Seedlings of gfp transgenic Nicotiana benthamiana line 16c plants were inoculated mechanically with in vitro-synthesized RNA transcripts of either the PVX expression vector containing the GFP gene, PVX/GFP (Fig. 4A); the GFP gene fused at the 5′ end with the p29 coding sequence, PVX/p29-GFP; or the GFP gene fused with the mutated p29 sequence that contained two stop codons immediately downstream of the start codon, PVX/mp29-GFP (Fig. 4A). As indicated in Fig. 4B, free GFP (26.9 kDa) and p29-GFP fusion protein (55.9 kDa) were detected by Western analysis in protein extracts from N. benthamiana plants infected with PVX/GFP or PVX/p29-GFP, respectively, but not in extracts of PVX/mp29-GFP-infected plants.
FIG. 4.
(A) PVX-based p29-GFP expression vectors. The PVX/GFP expression vector was modified to express a p29-GFP fusion (PVX/p29-GFP) or to contain a nonfunctional mutant p29-GFP fusion sequence that contained two stop codons (x) immediately downstream of the start codon of the p29 gene (PVX/mp29-GFP). (B) Expression of the hypovirus p29 tagged with GFP in N. benthamiana plants. Proteins were extracted from young leaves of mock-inoculated plants and plants infected with PVX/GFP, PVX/p29-GFP, and PVX/mp29-GFP 10 dpi. An aliquot of 10 μg total proteins was separated on a sodium dodecyl sulfate-polyacrylamide gel and detected by Western blotting using antibodies specific to GFP (top) and PVX CP (bottom). The positions of CP, GFP, and GFP-tagged p29 are indicated.
Mock inoculation and infection with wild-type PVX had no effect on gfp transgene expression, and line 16c plants showed green fluorescence under long-wavelength UV light (Fig. 5A). However, plants inoculated with PVX/GFP first showed silencing of expression of both the gfp transgene and the gfp gene inserted into the PVX genome approximately 7 days postinoculation (dpi). Silencing was complete at 20 dpi and maintained. gfp-silenced plants displayed only red autofluorescence of chlorophyll under UV illumination (Fig. 5B). Similar silencing of gfp expression was seen in plants infected with PVX/mp29-GFP (Fig. 5D). These data indicate that insertion of a nontranslatable p29 RNA sequence and the silencing suppressor p25 encoded by PVX had no effect on PVX/GFP-based induction of RNA silencing. In contrast, expression of a p29-GFP fusion protein reproducibly produced a characteristic pattern of RNA silencing suppression in line 16c plants inoculated with PVX/p29-GFP. GFP expression (green fluorescence) was only sporadically visible in laminae or around the veins of older leaves but remained strong in stems and tissues of emerging and young developing leaves (Fig. 5C).
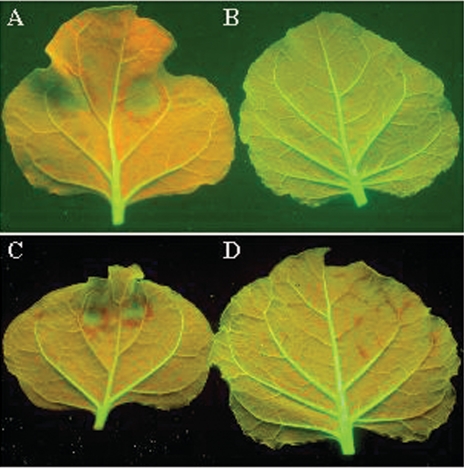
FIG. 5.
Hypovirus p29-mediated suppression of RNA silencing in plants. (A to D) Line 16c plants were mock inoculated (A) or inoculated with recombinant virus PVX/GFP (B), PVX/p29-GFP (C), or PVX/mp29-GFP (D) and observed under long-wavelength UV illumination through a yellow filter. Plant tissues without silencing or exhibiting silencing suppression showed green epifluorescence. gfp-silenced plant tissues showed red chlorophyll autofluorescence. (E to H) Systemic symptoms in line 16c plants mock inoculated (E) or infected with PVX/GFP (F), PVX/p29-GFP (G), or PVX/mp29-GFP (H). Plants were photographed 20 dpi at different angles to show the effects of induction and suppression of RNA silencing and symptom development. Silencing suppression and virus survival phenomena were observed in six to eight individual plants inoculated with the respective recombinant viruses in two independent experiments.
A common feature of VIGS in plants is the “symptom recovery” phenotype following initial viral infection due to degradation of viral genomes by an RNA silencing mechanism (13, 38). PVX/GFP, PVX/mp29-GFP, and PVX/p29-GFP all induced local lesions on inoculated leaves of line 16c plants 3 to 5 dpi and systemic mosaic and chlorotic symptoms on younger leaves 7 dpi. In addition to gfp transgene expression, local and systemic expression of GFP or p29-GFP from PVX/GFP or PVX/p29-GFP, respectively, was found in tissues showing symptoms of infection by either recombinant virus (data not shown). Consistent with the occurrence of RNA silencing, PVX/GFP-inoculated line 16c plants gradually recovered from the initial infection. They showed only sporadic mosaic symptoms on younger expanding leaves and were almost symptomless on newly emerging leaves, as in mock-inoculated plants, 20 dpi (Fig. 5E and F). Unexpectedly, no symptom recovery was observed in line 16c plants inoculated with PVX/mp29-GFP (Fig. 5H). Here, severe mosaic and chlorotic symptoms persisted, although gfp RNA silencing was induced efficiently and maintained throughout the whole plant (Fig. 5D and H). Significantly, PVX/p29-GFP-inoculated line 16c plants also showed no symptom recovery phenotype, consistent with the occurrence of reciprocal suppression of gfp transgene and viral RNA silencing by p29 (Fig. 5G). Interestingly, in these plants the laminae of young expanding and new emerging leaves that exhibited greater GFP fluorescence were less symptomatic. However, gfp RNA silencing subsequently occurred in those leaf sections that later became severely infected (Fig. 5C and G). To account for this observation, we propose that p29 probably blocked spread of silencing before the recombinant PVX delivered a sufficient amount of sequence-specific silencing inducer (i.e., gfp RNA) into nonsilencing plant cells and tissues.
Hypovirus CHV1-EP713 p29 does not alter cell-to-cell movement of RNA silencing but blocks long-distance spread.
The p25 movement protein of PVX has been shown to influence the spread of RNA silencing in N. benthamiana, potentially complicating the interpretation of suppression activity of an unknown silencing suppressor when using a PVX-based expression system (54). Thus, it is arguable that even if PVX p25 is unable to interfere with silencing induction in the aforementioned assay, this suppressor could still have synergistic effects with the CHV1-EP713 p29 protein. It was therefore important to demonstrate the silencing suppression activity of p29 in a background that excludes p25 and other silencing suppressors. To achieve this, we examined the effects of p29 on both local and systemic RNA silencing in plants by using an agroinfiltration assay. Agroinfiltration of an additional 35S-GFP transgene into line 16c plants resulted in local silencing of transient and resident GFP expression at the infiltrated sites and systemic gfp RNA silencing in young and noninfiltrated leaves. These tissues exhibited red fluorescence from chlorophyll rather than green fluorescence from GFP. Not surprisingly, a similar pattern of local and systemic gfp RNA silencing was triggered by the nontranslatable mp29-gfp RNA, in line 16c plants infiltrated with A. tumefaciens strain LBA4404 carrying pBin35S/mp29-GFP (Fig. 6C and D). Infiltration of A. tumefaciens strain LBA4404 carrying pBin35S/p29-GFP into line 16c plants also induced local gfp RNA silencing, showing the typical red fluorescent rings encircling the infiltration patches instead of green fluorescence (Fig. 6A), a hallmark associated with cell-to-cell movement of RNA silencing (22). However, consistently, no gfp RNA silencing was observed in noninfiltrated young leaves of these plants (Fig. 6B). Thus, expression of p29-GFP fusion protein from pBin35S/p29-GFP in line 16c plants did not interfere with induction or cell-to-cell spread of transient and resident gfp RNA silencing but blocked spread of gfp RNA silencing to distal plant tissues. These data reinforced the conclusion drawn from the VIGS assay.
FIG. 6.
Effect of expression of the hypovirus p29-GFP on local and systemic RNA silencing. gfp-transgenic line 16c plants were observed under long-wavelength UV illumination through a yellow filter. Plants were agroinfiltrated with pBin35S/p29-GFP (A and B) or pBin35S/mp29-GFP (C and D), respectively. Photographs were taken 20 days postagroinfiltration for infiltrated leaves (A and C) and noninfiltrated fully developed young leaves (B and D). gfp RNA-silenced tissues show red autofluorescence.
DISCUSSION
Suppressors of RNA silencing have been identified for a large number of plant viruses and a growing number of animal viruses (recently reviewed in reference 53). Although RNA silencing has not been shown experimentally to serve as an antiviral defense mechanism in fungi, the results presented here show that a fungal virus, hypovirus CHV1-EP713, does encode a protein that is able to suppress RNA silencing in its fungal host. Moreover, the hypovirus-encoded p29 protein was able to suppress RNA silencing in a heterologous plant system, providing additional evidence for cross-kingdom conservation of RNA signaling mechanisms.
The RNA silencing assay developed for C. parasitica is characterized by a strong correlation between RNA silencing of a GFP transgene, reduced accumulation of GFP mRNA, and increased accumulation of GFP-specific siRNA (Fig. 1). The use of an intron-containing hairpin RNA-expressing vector (55) resulted in quite efficient RNA silencing with approximately 70% of transformants showing an 80% or greater reduction in fluorescence. This compares well with the silencing efficiency reported for two endogenous genes in N. crassa using a similarly designed silencing vector (17).
Since the cpEGFP/pGS-2GFP transformants exhibit a spectrum of silencing levels rather than an all-or-none response, we took a number of precautions in designing the experiments to test the ability of p29 to suppress hairpin RNA-induced RNA silencing. In the prevention assay, we carefully selected cpEGFP/p29, cpEGFP/mp29, and cpEGFP/pCPX-BDS1 transformants that exhibited a level of fluorescence similar to that of the original cpEGFP and three independent transformants of each construct were used for each pGS-2GFP-mediated silencing assay. Moreover, fluorescence levels for all of the subsequent pGS-2GFP transformants were normalized against the corresponding recipient strains. The presence of empty pCPX-BDS1 vector or vector containing the mutated p29 coding sequence in the cpEGFP recipient strains did result in a slight shift in the relative fluorescence distribution pattern for the pGS-2GFP transformants compared to the distribution of the cpEGFP/pGS-2GFP transformants (Fig. 2). We also observed minor changes in the distribution of silenced transformants when a silenced strain was transformed with the empty vector (not shown) or the vector containing the mutated p29 sequence (Fig. 2) in the reversal assay. Thus, there appears to be a level of noise associated with the transformation process. Nevertheless, very substantial changes in the relative fluorescence distribution pattern were observed in both the prevention (Fig. 2) and the reversal (Fig. 3) experiments for the vectors containing the functional p29 coding sequence.
The suppressor activity of p29 was predicted based on similarities to the well-characterized potyvirus-encoded suppressor of RNA silencing HC-Pro (9, 30) and previous reports that p29 could act in trans to increase accumulation and transmission of hypovirus RNA (47). Both p29 and HC-Pro exhibit papain-like protease activity and share striking similarities in the sequences surrounding the catalytic cysteine and histidine residues and the spacing of these essential residues relative to the respective cleavage sites (9). The N-terminal portions of the two proteins also contain four conserved cysteine residues, consistent with the prediction by Koonin et al. (30) of a common ancestry for hypoviruses and plant potyviruses. The in trans enhancement of CHV1-EP713 RNA accumulation (47) is reminiscent of the reported HC-Pro-mediated amplification of tobacco etch potyvirus RNA (28). Both proteins also appear to alter host developmental processes when expressed in the absence of virus infection. For example, C. parasitica transformants expressing p29 exhibit reduced levels of asexual sporulation and pigmentation (14), while expression of HC-Pro results in the formation of small tumors in N. benthamiana (1) and interference with development and miRNA function in Arabidopsis species (29). Although miRNAs have not been identified in fungi, phenotypic consequences of p29 expression in C. parasitica could be caused by disruption of an as-yet-unidentified endogenous RNA silencing regulatory pathways. In this regard, it will be of interest to see whether HC-Pro suppresses RNA silencing and causes phenotypic changes when expressed in fungi.
The similarities between p29 and HC-Pro extend to the ability of p29 to serve as a suppressor of RNA silencing in plants. This result does not necessarily mean that the two proteins use evolutionarily conserved mechanisms to suppress RNA silencing. Viruses have developed a surprisingly diverse array of strategies to counteract RNA silencing defense measures (reviewed recently in reference 53). The assays used for examining p29-mediated suppression of RNA silencing do not permit one to distinguish between suppression of initiation and maintenance of RNA silencing. It is clear, however, that p29 does suppress systemic spread of RNA silencing in plants. A comprehensive comparative study of p29 and HC-Pro in the reciprocal plant and fungal systems, now in progress, is required to elucidate the precise details of the suppression of RNA silencing by the related proteins. It is anticipated that such studies will provide new insight into the evolution of RNA silencing and virus-encoded suppressors of RNA silencing.
The observation that PVX/mp29-GFP-infected plants failed to establish the typical symptom recovery phenotype but continued to express severe symptoms in the presence of strong RNA silencing (Fig. 4) was unexpected and accompanied by the accumulation of high levels of PVX/mp29-GFP RNA (data not shown). In this regard, viroids, which consist of naked small circular single-stranded RNAs that do not encode any protein, as well as some plant viruses, can establish systemic infections in plants where RNA silencing is efficiently induced (23, 26, 32, 51). This suggests that an RNA-triggered escape or “getaway” mechanism (51) may operate to suppress RNA silencing in the absence of protein factor-mediated suppression of RNA silencing. Such RNA elements may facilitate the formation of structures that protect against or subvert targeting by activated RNA silencing surveillance systems. Alternatively, these RNA elements may contribute to increased replication and viral RNA accumulation, thus allowing some copies to escape RNA silencing. Experiments to test these possibilities and identify domains within the p29 RNA coding region that might contribute to the absence of a symptom recovery phase in PVX/mp29-GFP-infected plants are in progress.
The current view of RNA silencing in fungi is derived primarily from studies of N. crassa. Two RNA silencing pathways have been identified: quelling and meiotic silencing of unpaired DNA. The latter pathway silences the expression of genes within meiotically unpaired regions of DNA and occurs exclusively in sexual tissues (44, 45). Quelling operates during the vegetative phase of N. crassa growth and silences expression of tandemly integrated transgenes (12) and transposons (34). The demonstration in this report that a fungal virus encodes a suppressor of RNA silencing provides indirect evidence that RNA silencing also serves as an antiviral defense mechanism in fungi. The close phylogenetic relatedness to N. crassa, the availability of infectious cDNA clones for several hypoviruses (15), and a robust C. parasitica transformation protocol (10) make the C. parasitica hypovirus system uniquely suited to directly test this hypothesis.
Acknowledgments
We are grateful to D. C. Baulcombe for providing the original PVX vector and transgenic N. benthamiana line 16c seeds and to S. Santa Cruz for PVX coat protein and GFP antibodies. We thank T. M. A. Wilson for his encouragement throughout this work.
This study was supported by Warwick HRI core BBSRC funding to Y.H. and by Public Health Service grant GM55981 to D.L.N.
REFERENCES
- 1.Anandalakshmi, R., R. Marathe, X. Ge, J. M. Herr, C. Mau, A. Mallery, G. Pruss, L. Bowman, and V. B. Vance. 2000. A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science 290:142-144. [DOI] [PubMed] [Google Scholar]
- 2.Brigenti, G., O. Voinnet, W. X. Li, L. H. Ji, S. W. Ding, and D. C. Baulcombe. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739-6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Catalanotto, C., G. Azzalin, G. Macino, and C. Cogoni. 2002. Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes Dev. 16:790-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Catalanotto, C., M. Pallotta, P. ReFalo, S. Sachs, L. Vayssie, G. Macino, and C. Cogoni. 2004. Redundancy of the two Dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol. Cell. Biol. 24:2536-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapman, S., T. Kavanagh, and D. C. Baulcombe. 1992. Potato virus X as a vector for gene expression in plants. Plant J. 2:549-557. [DOI] [PubMed] [Google Scholar]
- 6.Chicas, A., and G. Macino. 2001. Characteristics of post-transcriptional gene silencing. EMBO Rep. 2:992-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi, G. H., and D. L. Nuss. 1992. Hypovirulence of chestnut blight fungus conferred by an infectious viral cDNA. Science 257:800-8003. [DOI] [PubMed] [Google Scholar]
- 8.Choi, G. H., T. G. Larson, and D. L. Nuss. 1992. Molecular analysis of the laccase gene from the chestnut blight fungus and selective suppression of its expression in an isogenic hypovirulent strain. Mol. Plant-Microbe Interact. 5:119-128. [DOI] [PubMed] [Google Scholar]
- 9.Choi, G. H., D. M. Pawlyk, and D. L. Nuss. 1991. The autocatalytic protease p29 encoded by a hypovirulence-associated virus of the chestnut blight fungus resembles the potyvirus-encoded protease HC-Pro. Virology 183:747-752. [DOI] [PubMed] [Google Scholar]
- 10.Churchill, A. C. L., L. M. Ciufetti, D. R. Hansen, H. D. Van Etten, and N. K. Van Alfen. 1990. Transformation of the fungal pathogen Cryphonectria parasitica with a variety of heterologous plasmids. Curr. Genet. 17:25-31. [Google Scholar]
- 11.Cogoni, C. 2001. Homology-dependent gene silencing mechanisms in fungi. Annu. Rev. Microbiol. 55:381-406. [DOI] [PubMed] [Google Scholar]
- 12.Cogoni, C., N. Romano, and G. Macino. 1994. Suppression of gene expression by homologous transgenes. Antonie Leeuwenhoek 65:205-209. [DOI] [PubMed] [Google Scholar]
- 13.Covey, S. N., N. S. Al-Kaff, A. Langara, and D. S. Turner. 1997. Plants combat infection by gene silencing. Nature 385:781-782. [Google Scholar]
- 14.Craven, M. G., D. M. Pawlyk, G. H. Choi, and D. L. Nuss. 1993. Papain-like protease p29 as a symptom determinant encoded by a hypovirulence-associated virus of the chestnut blight fungus. J. Virol. 67:6513-6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dawe, A. L., and D. L. Nuss. 2001. Hypoviruses and chestnut blight: exploiting viruses to understand and modulate fungal pathogenesis. Annu. Rev. Genet. 35:1-29. [DOI] [PubMed] [Google Scholar]
- 16.Fire, A. 1999. RNA-triggered gene silencing. Trends Genet. 15:358-363. [DOI] [PubMed] [Google Scholar]
- 17.Goldoni, M., G. Azzalin, G. Macino, and C. Cogoni. 2004. Efficient gene silencing by expression of double stranded RNA in Neurospora crassa. Fungal Genet. Biol. 41:1016-1024. [DOI] [PubMed] [Google Scholar]
- 18.Guerineau, F., L. Lucy, and P. Mullineaux. 1992. Effect of two consensus sequences proceeding the translation initiator codon on gene expression in plant protoplasts. Plant Mol. Biol. 18:815-818. [DOI] [PubMed] [Google Scholar]
- 19.Hammond, T. M., and N. P. Keller. 2005. RNA silencing in Aspergillus nidulans is independent of RNA-dependent RNA polymerase. Genetics 169:607-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hannon, G. J. 2002. RNA interference. Nature 418:244-251. [DOI] [PubMed] [Google Scholar]
- 21.Hillman, B. I., R. Shipira, and D. L. Nuss. 1990. Hypovirulence-associated suppression of host functions in Cryphonectria parasitica can be partially relieved by high light intensity. Phytopathology 80:950-956. [Google Scholar]
- 22.Himber, C., P. Dunoyer, G. Moissiard, C. Ritzenthaler, and O. Voinnet. 2003. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 22:4523-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiriart, J. B., E. M. Aro, and K. Lehto. 2003. Dynamics of the VIGS-mediated chimeric silencing of the Nicotiana benthamiana ChlH gene and of the tobacco mosaic virus vestor. Mol. Plant-Microbe Interact. 16:99-106. [DOI] [PubMed] [Google Scholar]
- 24.Hoekema, A., P. R. Hirsch, P. J. J. Hooykass, and R. A. Schilperoort. 1983. A binary plant vector strategy based on separation of the Vir- and T-region of the Agrobacterium tumefaciens Ti plasmid. Nature 303:179-180. [Google Scholar]
- 25.Hong, Y., K. Saunders, M. R. Hartley, and J. Stanley. 1996. Resistance to Gemini virus infection by virus-induced expression of dianthin in transgenic plants. Virology 220:119-1927. [DOI] [PubMed] [Google Scholar]
- 26.Itaya, A., A. Folimonov, Y. Matsuda, R. S. Nelson, and B. Ding. 2001. Potato spindle tuber viroid as inducer of RNA silencing in infected tomato. Mol. Plant-Microbe Interact. 14:1332-1334. [DOI] [PubMed] [Google Scholar]
- 27.Kadotani, N., H. Nakayashiki, Y. Tosa, and S. Mayama. 2003. RNA silencing in the phytopathogenic fungus Magnaporthe oryzae. MPMI 16:769-776. [DOI] [PubMed] [Google Scholar]
- 28.Kasschau, K. D., S. Cronin, and J. C. Carrington. 1997. Genome amplification and long-distance movement functions associated with the central domain of tobacco etch potyvirus helper component-protease. Virology 228:251-262. [DOI] [PubMed] [Google Scholar]
- 29.Kasschau, K. D., Z. Xie, E. Allen, C. Llave, E. J. Chapman, K. A. Krizan, and J. C. Carrington. 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and mRNA function. Dev. Cell 4:205-217. [DOI] [PubMed] [Google Scholar]
- 30.Koonin, E. V., G. H. Choi, D. L. Nuss, R. Shapira, and J. C. Carrington. 1991. Evidence for common ancestry of a chestnut blight hypovirulence-associated double-stranded RNA and a group of positive-strand RNA plant viruses. Proc. Natl. Acad. Sci. USA 88:10647-10651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, H. W., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319-1321. [DOI] [PubMed] [Google Scholar]
- 32.Martinez de Alba, A. E., R. Flores, and C. Hernandez. 2002. Two chloroplastic viroids induce the accumulation of small RNAs associated with post-transcriptional gene silencing. J. Virol. 76:13094-13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Napoli, C., C. Lemieux, and R. Jorgensen. 1990. Introduction of a chalocone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2:279-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nolan, T., L. Braccini, G. Azzalin, A. De Toni, G. Macino, and C. Cogoni. 2005. The post-transcriptional gene silencing machinery functions independently of DNA methylation to repress a LINE1-like retrotransposon in Neurospora crassa. Nucleic Acids Res. 33:1564-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nuss, D. L. 2005. Hypovirulence: mycoviruses at the fungal-plant interface. Nat. Rev. Microbiol. 3:632-642. [DOI] [PubMed] [Google Scholar]
- 36.Parsley, T. B., B. Chen, L. M. Geletka, and D. L. Nuss. 2002. Differential modulation of cellular signaling pathways by mild and severe hypovirus strains. Eukaryot. Cell 1:401-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Punt, P. J., M. A. Dingemanse, A. Kuyvenhoven, R. D. Soede, P. H. Pouwels, and C. A. van den Hondel. 1990. Functional elements in the promoter region of the Aspergillus nidulans gpdA gene encoding glyceraldehyde-3-phosphate dehydrogenase. Gene 93:101-109. [DOI] [PubMed] [Google Scholar]
- 38.Ratcliff, F., B. D. Harrison, and D. C. Baulcombe. 1997. A similarity between viral defense and gene silencing in plants. Science 276:1558-1560. [DOI] [PubMed] [Google Scholar]
- 39.Romano, N., and G. Macino. 1992. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 6:3343-3345. [DOI] [PubMed] [Google Scholar]
- 40.Ruiz, M. T., O. Voinnet, and D. C. Baulcomb. 1998. Initiation and maintenance of virus-induced gene silencing. Plant Cell 10:937-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Segers, G. C., and D. L. Nuss. 2003. Constitutively activated Gα negatively regulates virulence, reproduction and hydrophobin gene expression in the chestnut blight fungus Cryphonectria parasitica. Fungal Genet. Biol. 38:198-208. [DOI] [PubMed] [Google Scholar]
- 43.Segers, G. C., J. C. Regier, and D. L. Nuss. 2004. Evidence for a role of the regulator of G-protein signaling protein CPRGS-1 in Gα subunit CPG-1-mediated regulation of fungal virulence, conidiation, and hydrophobin synthesis in the chestnut blight fungus Cryphonectria parasitica. Eukaryot. Cell 3:1454-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shiu, P. K., and R. L. Metzenberg. 2002. Meiotic silencing by unpaired DNA. Properties, regulation and suppression. Genetics 161:1483-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shiu, P. K., N. B. Raju, D. Zickler, and R. L. Metzenberg. 2001. Meiotic silencing by unpaired DNA. Cell 107:905-916. [DOI] [PubMed] [Google Scholar]
- 46.Silhavy, D., and J. Burgyan. 2004. Effects and side-effects of viral RNA silencing suppressors on short RNAs. Trends Plant Sci. 9:76-83. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki, N., K. Maruyama, M. Moriyama, and D. L. Nuss. 2003. Hypovirus papain-like protease functions in trans to enhance viral double-stranded RNA accumulation and vertical transmission. J. Virol. 77:11697-11707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Krol, A. R., L. A. Mur, M. Beld, J. N. Mol, and A. R. Stuitje. 1990. Flavonoid genes in petunia: addition of a limited number of gene copies may lead to suppression of gene expression. Plant Cell 2:291-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Engelen, F. A., J. W. Molthoff, A. J. Conner, J.-P. Nap, A. Pereira, and W. J. Stiekema. 1995. pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Res. 4:288-290. [DOI] [PubMed] [Google Scholar]
- 50.Van Wezel, R., H. Liu, P. Tien, J. Stanley, and Y. Hong. 2001. Gene C2 of the monopartite geminivirus Tomato yellow leaf curl virus-China encodes a pathogenicity determinant that is localized in the nucleus. Mol. Plant-Microbe Interact. 14:1125-1128. [DOI] [PubMed] [Google Scholar]
- 51.Van Wezel, R., and Y. Hong. 2004. Virus survival of RNA silencing without deploying protein-mediated suppression of Nicotiana benthamiana. FEBS Lett. 562:65-70. [DOI] [PubMed] [Google Scholar]
- 52.Voinnet, O. 2001. RNA silencing as a plant immune system against viruses. Trends Genet. 17:449-459. [DOI] [PubMed] [Google Scholar]
- 53.Voinnet, O. 2005. Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6:206-220. [DOI] [PubMed] [Google Scholar]
- 54.Voinnet, O., C. Lederer, and D. C. Baulcombe. 2000. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103:157-167. [DOI] [PubMed] [Google Scholar]
- 55.Wesley, S. V., C. A. Helliwell, N. A. Smith, M. Wang, D. T. Rouse, Q. Liu, P. S. Gooding, S. P. Singh, D. Abbott, P. A. Stoutjesdijk, S. P. Robinson, A. P. Gleave, A. G. Green, and P. M. Waterhouse. 2001. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27:581-590. [DOI] [PubMed] [Google Scholar]
- 56.Zamore, P. D. 2002. Ancient pathways programmed by small RNAs. Science 296:1265-1269. [DOI] [PubMed] [Google Scholar]