Abstract
The Cryptococcus neoformans NRG1 gene was identified using gene microarrays to define putative transcription factor genes regulated by the cyclic AMP (cAMP) signal transduction pathway. Disruption of NRG1 results in delayed capsule formation and mating, two phenotypes that are directly controlled by cAMP signaling. Putative targets of the Nrg1 transcription factor were identified using a second genome microarray to define differences in the transcriptomes of the wild-type and nrg1 mutant strains. These experiments implicate Nrg1 in the transcriptional control of multiple genes involved in carbohydrate metabolism and substrate oxidation, as well as the UGD1 gene encoding a UDP-glucose dehydrogenase required for polysaccharide capsule production and cell wall integrity. In addition to being under transcriptional control of the cAMP pathway, Nrg1 contains a putative protein kinase A phosphorylation site; mutation of this motif results in reduced Nrg1 activity. Consistent with prior studies in hypocapsular mutants, the nrg1 mutant strain is attenuated in an animal model of disseminated cryptococcal disease.
Microbial pathogens undergo dramatic cellular changes in order to adapt to the environment of the infected host. These adaptations are often initiated by host-specific signals, including nutrient deprivation, pH, oxidative stress, and nitrosative stress. In response to these environmental changes, the microorganism may need to coordinate the expression of multiple phenotypes to survive in this hostile environment.
Pathogenic bacteria have developed a number of mechanisms in order to coexpress genes required for survival after encountering the host. For example, the gastrointestinal pathogen Shigella flexneri contains multiple virulence genes on a single, coregulated plasmid. The importance of these genes for bacterial virulence is underscored by the observation that the introduction of this plasmid into a laboratory strain of Escherichia coli allows this nonpathogenic bacterium to invade human epithelial tissue (45). In contrast, virulence genes in the intracellular pathogen Salmonella enterica are clustered in regions called virulence islands. Not only are the physically clustered genes coregulated, but virulence-associated genes from different clusters may be regulated by the same signaling molecules, such as the PhoP/PhoQ two-component system in response to magnesium (5, 6).
Likewise, eukaryotic pathogens must be able to coordinate the expression of multiple genes in response to the host environment. However, these eukaryotic microorganisms do not share the regulatory mechanisms of pathogenic bacteria. Eukaryotic genes are not typically clustered in coregulated operons, nor are virulence plasmids described in these more complex pathogens. Therefore, eukaryotic microbial pathogens likely use different strategies to coordinate gene expression, allowing adaptation to the hostile environment of the infected host.
The human fungal pathogen Cryptococcus neoformans lives primarily in the external environment and is routinely isolated from decaying vegetation and bird excreta. When inhaled by a mammalian host, this fungus undergoes dramatic phenotypic changes in response to host conditions. These survival phenotypes include the induction of an antiphagocytic polysaccharide capsule as well as the production of dark, antioxidant pigments. Without the expression of these phenotypes, C. neoformans is crippled for survival within the host and for pathogenicity (32).
We have recently demonstrated that C. neoformans capsule and melanin genes are coregulated at the level of transcription by the cyclic AMP (cAMP) pathway (41). Therefore, strains with defects in the cAMP signaling cascade are unable to induce capsule formation or melanin, and these strains are highly attenuated for virulence (2, 3, 41). The coregulation of multiple virulence-associated phenotypes by a single signal transduction pathway allows the microorganism to coordinate an effective response to the host. However, the mechanism by which such transcriptional coordination occurs is as yet unknown. We have hypothesized that multiple transcription factors function downstream of the cAMP pathway in C. neoformans, controlling the expression of capsule and melanin in response to changing intracellular cAMP levels. By identifying C. neoformans transcription factors that are downstream targets of cAMP signaling, we will define precise mechanisms by which this eukaryotic pathogen coordinates its response to stress and adapts to the host environment. Here we identify one such transcription factor that is controlled by the C. neoformans cAMP pathway at the transcriptional and posttranslational levels to regulate multiple phenotypes, including pathogenesis.
MATERIALS AND METHODS
Strains and media.
The strains used are listed in Table 1. All strains in this study, except the serotype D strain JEC 20 used in mating experiments, were derivatives of the serotype A wild-type strain H99 (39). Standard yeast media were used as described previously (46). Niger seed extract agar and V8 mating medium (30) were also used to assess melanin production and mating, respectively. Capsule formation-inducing medium (Dulbecco's modified Eagle's medium containing 25 mM NaHCO3) was prepared as previously described (18).
TABLE 1.
Strain list
| Strain name | Genotype | Reference |
|---|---|---|
| H99 | Serotype A, MATα, wild type | 39 |
| KN99a | Serotype A, MATa, congenic with H99 | 37 |
| KLC27 | MATα nrg1::neo | This paper |
| QGC31 | MATα nrg1::neo NRG1-nat | This paper |
| QGC32 | MATα nrg1::neo NRG1-nat | This paper |
| QGC33 | MATα nrg1::neo nrg1-C529G-nat | This paper |
| QGC34 | MATα nrg1::neo nrg1-C529G-nat | This paper |
| QGC35 | MATα nrg1::neo nrg1-C529G-nat | This paper |
| AAC1 | MATα ade2 gpa1::ADE2 | 2 |
| JEC 20 | Serotype D, MATa, wild type | 30 |
Resistance to hydrogen peroxide was tested by a disc diffusion method. Strains were plated on yeast extract-peptone-dextrose (YPD) medium, and a sterile cotton diffusion disc was placed in the center of the plate. Ten microliters of 30% hydrogen peroxide was added to the disc, and the diameter of the zone of inhibition was measured at 48 h. Resistance to nitric oxide was quantified by determining the concentration of sodium nitrite that inhibited strain growth in minimal asparagine medium, pH 4.0 (1). Sodium nitrite concentrations of 60 μM to 500 μM were used, and all strains were completely growth inhibited at 125 to 250 μM.
RNA preparation.
Strains were incubated to mid-logarithmic phase at 30°C in synthetic complete (SC) medium with 2% glucose, pelleted, resuspended in SC medium with 0.1% glucose, and incubated for 30 min at 30°C. Cells were collected by centrifugation and flash frozen on dry ice. Total RNA was extracted from lyophilized cells using the TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA).
cDNA synthesis and labeling.
Fluorescently labeled cDNA was made by incorporating amino-allyl dUTP during reverse transcription of 10 μg of total RNA. Cy3 or Cy5 fluorescent dye (Amersham, Piscataway, NJ) was coupled to the amino-allyl group as previously described (15). We generated a reference sample by pooling an equal amount of RNA from the wild-type strain H99 incubated to mid-log phase in nine different inducing conditions (YPD medium, 30°C; YPD medium, 37°C; YPD medium, 39°C; synthetic low-ammonium dextrose medium [nitrogen limiting], 30°C; synthetic complete medium with 0.1% glucose [glucose limiting], 30°C; RPMI medium incubated in room air, 30°C; RPMI medium incubated in 5% CO2, 30°C; RPMI medium with 0.75 M NaCl, 30°C; YPD medium at 30°C for 48 h [saturated culture to stationary phase]). The reference pool RNA was converted to cDNA and labeled with Cy3. RNA from each time point was individually labeled with Cy5 and competitively hybridized against the reference sample.
Microarray hybridization.
Two distinct microarrays were used in these experiments. The first array was used to compare the transcriptome of the gpa1 mutant and wild-type strains, resulting in the identification of the NRG1 gene. This DNA microarray was described in detail previously (26, 41). Briefly, 6,144 PCR products were amplified from a 1.6- to 3.2-kb genomic library, made using strain H99 genomic DNA, and were printed on polylysine-coated glass slides. Slides were prehybridized at 42°C with 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate (SDS), and 1% bovine serum albumin, and hybridizations were performed at 42°C with 1× hybridization buffer (50% formamide, 5× SSC, and 0.1% SDS). Arrays were scanned with a GenePix 4000B scanner (Axon Instruments, Foster City, CA) and analyzed using GenePix Pro v4.0. Further data analysis was performed using CryptoArray, a Microsoft Excel macro for normalizing and formatting data.
The second microarray, used to define potential targets of the Nrg1 transcription factor, is a publicly available whole-genome array of 70-mer oligonucleotides corresponding to each predicted gene in the C. neoformans genome (http://www.genome.wustl.edu/activity/ma/cneoformans/). All hybridizations and data acquisition were performed at the Duke Microarray Facility (http://microarray.genome.duke.edu/) according to their established protocols for custom spotted arrays.
Microarray data analysis.
Extracted microarray data from the second array were imported into a statistical software package (SAS v8; SAS Institute, Cary, NC), and initial background subtraction was performed. Mixed model analysis of microarray data (54) was used to evaluate differential expression data using approaches presented elsewhere (10, 40, 42). We calculated the statistical significance of each genetic element using least squares estimates of gene-specific treatment effects across pairs of treatments obtained for each gene under each treatment condition. Differences between treatment effects for pairs of inducing conditions were considered log2-transformed fold changes (54). Genes were selected for further evaluation if they possessed a P value of <0.02.
Northern analysis.
Fifteen micrograms of total RNA was analyzed for each sample. Gel electrophoresis, RNA transfer, hybridization, and autoradiography were performed as described previously (44). The probes used for Northern analysis included the 3′-most 190 bp of the coding sequences of the NRG1 and ACT1 (12) genes. The DNA for probes was labeled using a random-primed DNA labeling kit (Boehringer Mannheim) and [32P]dCTP (Amersham).
PCR.
All PCRs were performed using a Techne Genius thermocycler with 50 ng of template DNA, 100 ng of each oligonucleotide primer, and standard reagents from a TaKaRa kit (Takara Mirus Bio Co.). The PCR conditions were 95°C for 5 min followed by 35 cycles of 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 1 minute for each kilobase amplified in the reaction.
NRG1 gene disruption.
The NRG1 gene was disrupted by homologous recombination using biolistic transformation. A nrg1::neo mutant allele was constructed using PCR overlap extension as previously described (41) to completely replace the NRG1 coding region with the neomycin resistance gene (neo). The left and right fragments (corresponding to the regions of genomic DNA immediately upstream and downstream of the NRG1 open reading frame, respectively) were amplified from H99 genomic DNA using primers AA640 (CTTCACACACCCCTGTCCTT) and AA641 (GTCATAGCTGTTTCCTGGGTTTGGTGAGGTGATGCTT; M13 Rev sequence underlined) (left fragment) and primers AA642 (CTGGCCGTCGTTTTACTATCCTGCTGGGCCATACTC; M13 forward sequence underlined) and AA643 (GGACCCTGAAACCCAAGATA) (right fragment). The neo gene was PCR amplified from plasmid pJAF1 using M13 forward and reverse primers. The final disruption construct was PCR amplified using all three DNA fragments as template and primers AA640 and AA643. The nrg1 mutation was confirmed by PCR and Southern hybridization, as previously described (41). Three independent nrg1 mutants were isolated in this manner, all of which exhibited identical colony and growth phenotypes.
NRG1 reconstitution and creation of the nrg1-C529G mutant allele.
The entire NRG1 locus (including native promoter and terminator sequences) was PCR amplified from H99 genomic DNA using primers AA639 (ACCGATCCGTCATACTCTGC) and AA644 (TGCAGATAAAAGCCCGAGAT). The PCR fragment was TA cloned, digested with HindIII, and cloned adjacent to the nat selectable marker in the sole HindIII site of plasmid pCH233, to allow selection of transformants on nourseothricin-containing media.
The nrg1-C529G mutant allele was created by PCR overlap extension. Primer pairs AA639/AA697 (GCCGCTGAGCCCGAACCTCcACGGAGCGTAGGAG) and AA644/AA696 (CTCCTACGCTCCGTgGAGGTTCGGGCTCAGCGGC) (C529G mutation in lowercase) were used to amplify the left and right fragments, respectively, of the NRG1 gene from H99 genomic DNA. Primers AA639 and AA644 were used to generate the final overlap product that incorporated the C529G mutation. This mutant allele was isolated and cloned into plasmid pCH233 in an identical manner as the wild-type NRG1 allele, described above. Both alleles were sequenced to ensure that no unintended PCR-induced mutations were present.
Real-time PCR.
Total RNA was isolated from the relevant strains as described above. The RNA was treated with RNase-free DNase, and cDNA was synthesized using oligo(dT) primers from the SuperScript first-strand synthesis reverse transcription kit (Invitrogen). The resulting cDNA was used as template for quantitative real-time PCR using iQ SYBR Green Supermix (Bio-Rad) according to the manufacturer's specifications. The iCycler iQ multicolor real-time detection system was used as the fluorescence detector with the following PCR conditions: an initial denaturing cycle of 95°C for 3 min and 40 cycles of denaturation at 95°C for 10 s and annealing/extension at 53°C for 20 s. These cycles were followed by a standard melting curve from 53°C to 93°C with fluorescent monitoring each 0.5°C. These data confirmed the amplification of a single product for each primer pair and the lack of primer dimerization. Reactions were performed in triplicate, and the data were expressed as an average cycle threshold, ± one standard deviation. Standard PCRs were run with fivefold dilutions of the cDNA template to determine the optimal amount of template and optimal annealing temperature for the experimental and reference reactions, using 500 nM of each primer.
Gene amplification for each strain and condition was normalized against the constitutively expressed GPD gene (48). Induction (n-fold) was calculated relative to the wild-type strain H99 using the Bio-Rad iCycler software system, which utilizes the comparative cycle threshold statistical methods as previously described (47).
Virulence experiments.
In the murine inhalation model of systemic cryptococcosis, A/Jcr mice were intranasally inoculated with 5 × 105 C. neoformans cells as previously described (11). Groups of 5 to 10 mice were inoculated with one of three strains (NRG1 wild type, nrg1 mutant, or nrg1+NRG1 reconstituted strain) and observed twice daily for signs of infection. The statistical significance in the difference between the survival curves of the animals inoculated with each strain was evaluated using the log-rank test (JMP software; SAS Institute, Cary, NC).
In this model, mice develop meningoencephalitis after inhalation of C. neoformans, a course that mimics the natural history of human infection with this organism. Signs consistent with cryptococcosis in this experimental model include weight loss, lethargy, ruffled fur, and inability to maintain daily care. Mice were sacrificed prior to death from the infection at predetermined clinical end points that correlate with mortality, including weight loss of >15%, major neurological deficits, and inability to access food and water. All studies were performed in compliance with institutional guidelines for animal experimentation.
DNA sequences.
The NRG1 gene was identified by microarray analysis using the C. neoformans H99 sequencing project, Duke IGSP Center for Applied Genomics and Technology (http://cgt.duke.edu/). The gene was subsequently PCR amplified from genomic DNA, and the sequence was confirmed.
Nucleotide sequence accession number.
The gene sequence has been submitted to GenBank under accession number DQ641949.
RESULTS
The NRG1 gene is transcriptionally regulated by the cAMP pathway and by glucose availability.
We sought to identify transcription factors that acted downstream of the cAMP signal transduction pathway. Using a gene microarray approach, global gene expression was compared in the wild-type strain and the congenic gpa1 mutant, a strain lacking the G-alpha protein responsible for activating cAMP production (2). Among the genes that demonstrated altered expression in the gpa1 mutant compared to wild type, we identified one gene predicted to encode a homolog of the Candida albicans Nrg1 and Saccharomyces cerevisiae Nrg1/Nrg2 family of transcription factors. Reciprocal BLAST searches using the S. cerevisiae Nrg protein sequences to probe the C. neoformans genome similarly reveal that this gene encodes the closest C. neoformans homolog to these yeast transcription factors. Therefore, we designated this gene C. neoformans NRG1.
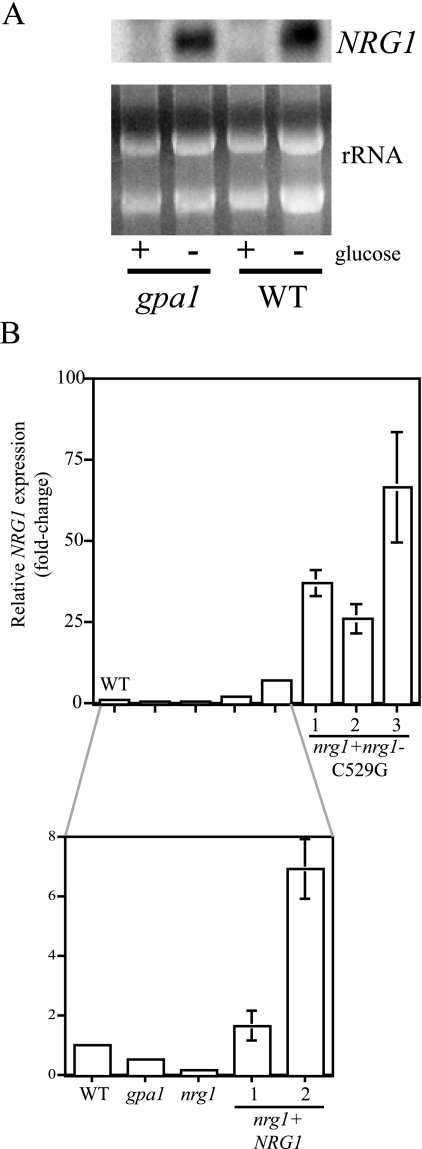
Northern hybridization and quantitative real-time PCR both confirmed the microarray observation that transcription of the C. neoformans NRG1 gene was twofold higher in the wild-type than in the isogenic gpa1 mutant strain (Fig. 1). In addition, we found that NRG1 expression is transcriptionally repressed by glucose. In the presence of 2% glucose, no NRG1 message was detected by Northern analysis. However, after 1 hour of incubation in glucose-poor conditions, the NRG1 message was detectable in the wild-type strain (Fig. 1A).
FIG. 1.
NRG1 is transcriptionally regulated by cAMP and glucose. (A) The wild-type (WT) and gpa1 mutant strains were incubated to mid-logarithmic phase in synthetic complete medium. The cells were then incubated in synthetic complete medium with either 2% glucose (+) or 0.1% glucose (−) for 30 min at 30°C. Total RNA was assessed by Northern analysis, using the NRG1 gene as a probe. Equal loading of RNA was assessed by ethidium bromide staining, with the rRNA bands demonstrated. (B) Total RNA was isolated from the following strains after 30 min of glucose deprivation of log-phase cells: wild type (WT; H99), gpa1 mutant (AAC1), nrg1 mutant (KLC27), two nrg1+NRG1 reconstituted strains (QGC31 and QGC32), and three nrg1+nrg1-C529G strains (QGC33, QGC34, and QGC35). Quantitative real-time PCR was performed on the corresponding cDNA samples to assess relative NRG1 transcript abundance compared to the wild-type strain ± one standard deviation.
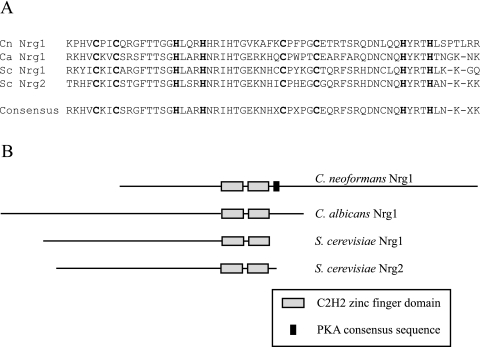
Like its homologs in S. cerevisiae, C. neoformans Nrg1 likely acts as a transcriptional regulator in response to carbohydrate availability. The predicted Nrg1 protein contains two Zn finger/DNA-binding domains common to the C2H2 family of transcription factors. Apart from the DNA-binding domains, the C. neoformans Nrg1 protein demonstrates little sequence similarity with the S. cerevisiae Nrg1/2 proteins or Candida albicans Nrg1p (Fig. 2). Also, C. neoformans Nrg1 contains a predicted consensus sequence for PKA activation not present in the other Nrg proteins (Fig. 2).
FIG. 2.
Alignment of C. neoformans Nrg1 and S. cerevisiae Nrg1 and Nrg2 proteins. (A) The C2H2 DNA-binding domains of the C. neoformans (Cn), C. albicans (Ca), and S. cerevisiae (Sc) Nrg proteins demonstrate notable sequence similarity within in this functional region. (B) A graphical representation of the Nrg proteins of C. neoformans, C. albicans, and S. cerevisiae, demonstrating the relative positions of the Zn finger domains and PKA consensus sequence for phosphorylation.
Disruption of NRG1.
To characterize the biological role of this putative transcriptional regulator, we disrupted the C. neoformans NRG1 gene by homologous recombination. Southern hybridization confirmed that the nrg1::neo mutant allele precisely replaced the native gene without additional ectopic integration events in the nrg1 mutant strain KLC27 (data not shown). Additionally, no NRG1 message was detectable in this mutant strain, even in carbohydrate-poor conditions in which the gene is induced in the wild-type strain (Fig. 1A).
To ensure that any observed phenotype of the nrg1 mutant strain was indeed due to the nrg1 mutation, an nrg1+NRG1 reconstituted strain (strain QGC31) was generated by integrating a wild-type copy of the NRG1 gene into the genome of the nrg1 mutant. Using quantitative real-time PCR, we documented that NRG1 expression in this reconstituted strain is similar to that in the wild-type strain (Fig. 1B).
The nrg1 mutant strain grows at an identical rate as the wild type on rich (YPD) and minimal (yeast nitrogen base) media at 25, 30, and 37°C. Also, the nrg1 mutant grows equally as well as the wild type on media containing either 2% or 0.1% glucose, the latter being a condition that typically induces transcription of NRG1. In addition, the nrg1 mutant strain produces amounts of melanin equivalent to those of the wild type on birdseed agar medium, a phenotype required for virulence and for protection from reactive oxygen/nitrogen species (31, 32, 53). There is also no significant difference between wild type and nrg1 mutant strains in their susceptibility to oxidative (hydrogen peroxide) or nitrosative (nitric oxide) stresses (data not shown).
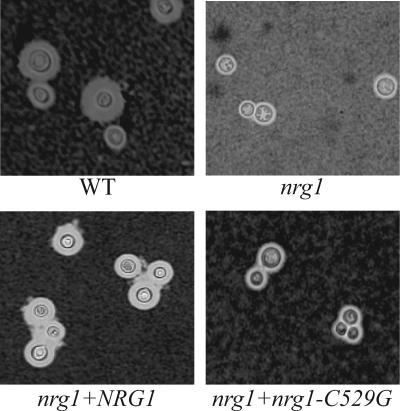
In contrast, the nrg1 mutant grows as a dry colony compared to the mucoid appearance of the wild-type strain. This suggested that the nrg1 mutant might have an alteration in capsule formation. The C. neoformans polysaccharide capsule can be induced in vitro by incubation in Dulbecco's modified Eagle's medium containing 25 mM NaHCO3 (18). Incubation in this medium typically results in the formation of large capsules, visible as a zone of exclusion on India ink preparations. The nrg1 mutant is able to make capsule when incubated in this medium, but it has a reduced capsule size compared to the wild type after 24 h of incubation (Fig. 3). Reintroduction of the wild-type NRG1 allele completely restores wild-type levels of capsule to the mutant strain (Fig. 3).
FIG. 3.
Capsule formation induction defect of the nrg1 mutant. The following strains were incubated in capsule formation-inducing conditions (Dulbecco's modified Eagle's medium with 25 mM NaHCO3, 5% CO2) for 48 h: the NRG1 wild type (WT; strain H99), the nrg1 mutant (strain KLC27), the nrg1 mutant reconstituted with a wild-type copy of NRG1 (nrg1+NRG1 [strain QGC31]), and the nrg1 mutant reconstituted with an nrg1 allele with a mutation in the putative PKA phosphorylation sequence (nrg1+nrg1-C529G [strain QGC34]). Capsule size was assessed by counterstaining with India ink (×400).
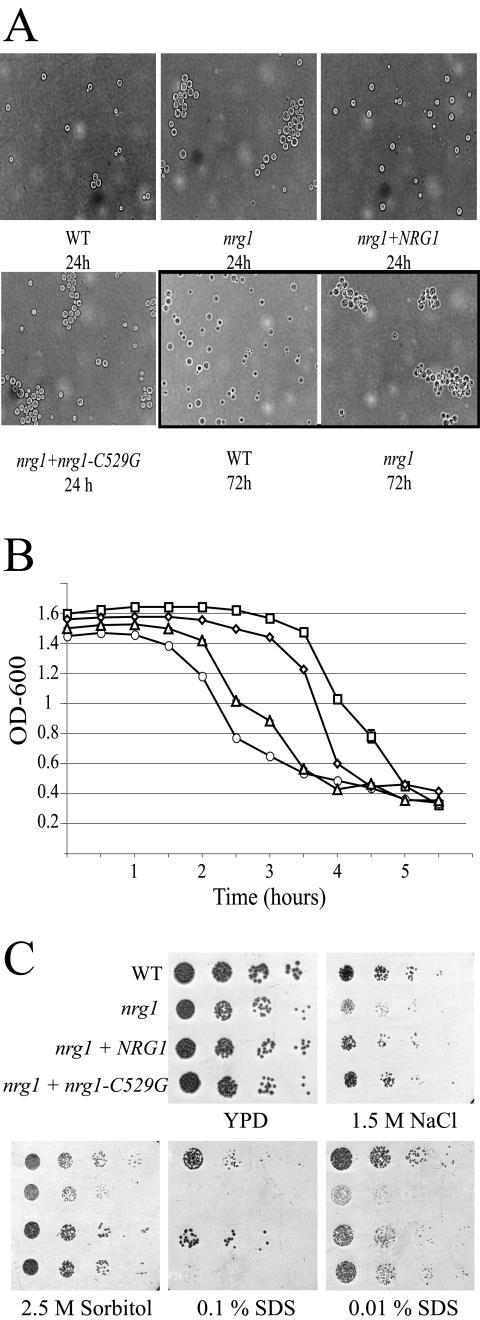
Microscopic analysis of the nrg1 mutant strain also revealed that the individual cells tended to aggregate (Fig. 4). Clusters of 10 to 20 cells were observed in cultures of the nrg1 mutant strain incubated in YPD medium for 12 to 24 h, and the number of cells in these aggregates increased with longer incubation times (Fig. 4). Although the clusters could not be disrupted by vortexing, the cells were easily separated by sonication. Therefore, the cell clustering was not a result of impaired cell separation. Similar degrees of cell clustering were observed when this mutant strain was incubated on either solid or liquid medium. Also, varying the glucose concentration in the medium from 0.01% to 2% did not alter the clustering observed in the nrg1 strain.
FIG. 4.
Altered cell surface phenotypes of the nrg1 mutant. (A) Cell-cell adherence. The wild-type (WT), nrg1, nrg1+NRG1, and nrg1+nrg1-C529G strains were incubated in YPD medium for 24 and 72 h. The cultures were vortexed and examined microscopically (×400). (B) Flocculation. The wild-type (squares), nrg1 (circles), nrg1+NRG1 (diamonds), and nrg1+nrg1-C529G (triangles) strains were incubated for 24 h in synthetic complete medium, and each culture was normalized to an OD600 of 1.5. The OD600 of the resulting cell suspension was serially measured to assess cell flocculation. Data points represent the means of three data points ± standard deviations. (C) Susceptibility to cell wall stresses. The wild-type (WT), nrg1, nrg1+NRG1, and nrg1+nrg1-C529G strains were serially diluted and incubated on YPD medium containing no additives (control), 0.01% SDS (48-h incubation), 0.1% SDS, 2.5 M sorbitol, or 1.5 M NaCl (7-day incubation).
This increased cell aggregation was not observed in cultures of the wild-type or nrg1+NRG1 reconstituted strains, directly implicating the Nrg1 protein in this process. Moreover, incubation of the wild-type strain in filtered, conditioned medium, in which the nrg1 mutant had grown for 72 h, did not induce cell aggregation in the wild-type strain. This suggests that the observed cell clustering is not the result of a transferable factor that is secreted into the medium.
Likely as a result of the increased cell clustering, nrg1 mutant cells incubated in liquid media displayed increased flocculation, settling out of suspension much more quickly than the wild type. To assess the rate of flocculation, cell suspensions of the various strains were normalized to an optical density at 600 nm (OD600) of 1.5 and allowed to stand without agitation. The absorbance at 600 nm of the suspension was measured every 30 minutes. The nrg1 mutant strain displayed enhanced flocculation, the majority of the cells settling out of suspension between 1 and 2 h (Fig. 4). In contrast, the wild-type cells did not display significant flocculation until 3 to 4 h.
Mating defect of the nrg1 mutant.
C. neoformans undergoes a complex series of cellular differentiation events during mating. Normally growing as a haploid budding yeast, cells of opposite mating type can fuse and transition to hyphal growth as a dikaryon. Fusion of the two nuclei, meiosis, and spore production occur in a terminal structure known as a basidium (29). Several environmental signals that influence C. neoformans mating include pheromone, light, and nutrient availability (21, 34).
In contrast to the MATα NRG1 wild-type strain that produces abundant mating hyphae when coincubated with a wild-type MATa mating partner, the MATα nrg1 mutant forms few hyphal structures during the first 7 days of a mating reaction (Fig. 5). After prolonged incubation, the mating reactions containing the MATα nrg1 mutant eventually display all of the structures associated with complete C. neoformans mating, including mature hyphae with fused clamp connections, basidia, and basidiospores. Reintroduction of the wild-type NRG1 allele completely suppresses the nrg1 mutant mating defect, supporting the observation that Nrg1 is required for efficient mating in C. neoformans (Fig. 5). However, the nrg1 mutant mating defect was not suppressed by the addition of 2% glucose or 10 mM cAMP to the mating medium.
FIG. 5.
Mating defect of the nrg1 mutant. The wild-type, nrg1 mutant, and nrg1+NRG1 reconstituted strains were incubated in mating reaction mixtures on V8 medium with the congenic MATa wild-type strain KN99a. The edges of the mating patches were microscopically assessed for mating filaments at 7 days and photographed (×40).
PKA control of Nrg1.
To test whether PKA phosphorylation affects the activity of the Nrg1 transcription factor, we created a mutant nrg1 allele encoding a protein with an altered PKA consensus sequence. The mutant nrg1 allele (nrg1-C529G) contains a single cytosine-to-guanine nucleotide substitution, a change predicted to abolish the PKA phosphorylation site in the encoded protein, resulting in amino acid changes from R-R-G-S to R-G-G-S. This allele was stably integrated into the genome of the nrg1 mutant strain by biolistic transformation. Three strains from this transformation were recovered and had identical phenotypes; one was chosen as a representative nrg1+nrg1-C529G strain (strain QGC34).
Although the reintroduced NRG1 wild-type and nrg1-C529G mutant alleles are identical except for the single induced polymorphism, they are quite different in their expression patterns. Both alleles are controlled by the native NRG1 promoter and terminator, and both were subcloned into the same vector (pCH233) for transformation into C. neoformans. However, all three nrg1+nrg1-C529G strains express the reintroduced nrg1 allele at much higher levels (5- to 30-fold higher) than those strains expressing the reintroduced wild-type NRG1 allele (Fig. 1B).
As described above, reintroduction of the wild-type NRG1 allele complements the nrg1 mutant capsule, flocculation, and mating phenotypes. In contrast, the nrg1-C529G mutant allele is unable to fully suppress the nrg1 mutant defects in capsule production or cell-cell adherence (Fig. 3 and 4). Interestingly, reintroduction of this mutant allele into the nrg1 strain completely restores mating competence. Together these studies indicate that the nrg1-C529 allele is expressed in the nrg1 mutant strain. Furthermore, the mating restoration studies demonstrate that this allele encodes a functional protein. However, the failure of this allele to fully restore capsule induction and normal cell-cell interactions suggests that PKA phosphorylation of C. neoformans Nrg1 is required for full function of this protein. Alternatively, it is also possible that the nrg1-C529 mutation results in lower protein levels and that even low levels of Nrg1 protein are able to suppress the mating defect of the nrg1 mutant strain, but not its capsule or flocculation defects.
Gene microarrays to identify potential downstream targets of the Nrg1 transcription factor.
We used a whole-genome microarray to define those genes that are differentially expressed in the wild-type strain and the nrg1 mutant to begin to identify targets of the Nrg1 transcription factor. Both strains were incubated to mid-logarithmic phase in synthetic complete medium containing 2% glucose. The cells were transferred to synthetic complete medium containing 0.1% glucose for 60 min as a glucose-limiting inducing condition. Total RNA was harvested from these cells for whole-genome microarray analysis.
Seventy-one genes are either induced or repressed by at least 2.5-fold in the nrg1 mutant compared to wild type. Of these, 21 genes have no known functional homologs or identifiable protein domains. Many of the remaining genes that are differentially expressed in the wild-type and nrg1 mutant strains can be grouped into functional families based on the predicted function of the gene product (Table 2). Consistent with the role of the Nrg transcription factors in S. cerevisiae, the majority of the putative target genes of the C. neoformans Nrg1 pathway are involved in carbohydrate metabolism, sugar transport, and oxidation/oxidative stress response.
TABLE 2.
Gene microarray data indicating genes with transcript levels at least 2.5-fold higher or lower in the wild type than in the nrg1 mutant
| Annotated gene function | Locus ID/accession no.a | Fold change (WT/nrg1)b | Annotated gene function | Locus ID/accession no.a | Fold change (WT/nrg1)b | |
|---|---|---|---|---|---|---|
| Chitin biosynthesis | Catalase (Cat2) | CNL03740 | 0.22 | |||
| Chitin deacetylase | CND03580 | 15.1 | GMC oxidoreductase (Smg1) | CNL03730 | 0.22 | |
| Chitin synthase | CNN02100 | 4.3 | Glutathione transferase | CNC04660 | 0.15 | |
| Chitinase | CNI03860 | 4.2 | ||||
| Chitin synthase | CNE03240 | 3.6 | Cell cycle proteins | |||
| Chitin deacetylase-like mannoprotein MP98 | CND03490 | 3.2 | Cyclin hcs26 (G1-specific cyclin) | CNG01990 | 2.9 | |
| Chitin synthase | CNA05300 | 2.7 | Mih1p | CNC00870 | 2.7 | |
| Liz1p | CNE01880 | 0.12 | ||||
| Sugar transporters | ||||||
| Hexose transport-related protein | CNC03580 | 5.7 | Miscellaneous proteins | |||
| Sugar transporter | CNC01990 | 2.7 | Putative deacetylase | CNN00270 | 13.7 | |
| Hydrolase | CNN00140 | 6.7 | ||||
| Carbohydrate metabolism | Nucleus protein | CNK03430 | 3.3 | |||
| Tartrate transporter | CNI02800 | 4.4 | LSDR | CNA01160 | 3.1 | |
| Exo-beta-13-glucanase | CNL04840 | 3.7 | Endoplasmic reticulum-to-Golgi complex | CNG04370 | 2.8 | |
| Glucan 13-beta-glucosidase protein | CNN00660 | 3.5 | transport-related protein | |||
| Ribose/galactose isomerase | CNA08060 | 3 | WSC, yeast cell integrity and stress response | CNA04570 | 2.6 | |
| Sorbitol dehydrogenase | CNA02580 | 2.8 | Ugd1, UDP-glucose dehydrogenase | AF405548 | 0.34 | |
| Xylitol dehydrogenase | CNJ01090 | 2.7 | (capsule synthesis) | |||
| Galactose metabolism-related protein | CNM00600 | 0.29 | Tetrahydrofolylpolyglutamate synthase | CNC04840 | 0.32 | |
| Formate dehydrogenase | CNF03470 | 0.22 | Acid phosphatase | CNC05540 | 0.29 | |
| Signal transduction | Conserved hypothetical proteins | |||||
| Conserved protein of unknown function | CNC00480 | 3.4 | Protein of unknown function, ricin-type | CNG00700 | 15.2 | |
| with WD40 domain | beta-trefoil domain | |||||
| Serine/threonine protein kinase | CNK01630 | 3.4 | Protein of unknown function, ricin-type | CNA05710 | 4.8 | |
| Rho guanine nucleotide exchange factor | CNF03450 | 2.9 | beta-trefoil domain | |||
| Putative protein kinase | CNG03850 | 2.7 | Protein of unknown function, ricin-type | CNJ03160 | 4.3 | |
| beta-trefoil domain | ||||||
| Other transporters | Protein of unknown function, kinesin | CNA01600 | 3.2 | |||
| Myoinositol transporter | CND00070 | 3.3 | motor domain | |||
| Myoinositol transporter | CNH03060 | 2.9 | Protein of unknown function, DNA-binding/ | CND01440 | 3 | |
| ABC transporter, PMR5 | CNN00770 | 0.35 | transcriptional activation domain | |||
| Siderochrome-iron (ferrioxamine) uptake | CNC04510 | 0.31 | Protein of unknown function, ezrin/radixin/ | CNI03680 | 2.7 | |
| transporter activity | moesin domain | |||||
| Cytosine/purine permease | CNC01920 | 0.26 | Protein of unknown function, | CNF03630 | 2.7 | |
| rhomboid domain | ||||||
| Oxidative stress/substrate oxidation | Protein of unknown function, | CNH00360 | 2.7 | |||
| GMC oxidoreductase | CNM00900 | 4.3 | mannosyltransferase domain | |||
| Putative NADP-dependent oxidoreductases | CNK02860 | 0.36 |
GenBank locus identification (ID) or accession number.
Change between wild-type (WT) and nrg1 mutant strains.
We subsequently used real-time PCR to confirm that a subset of these genes is indeed differentially transcribed in the nrg1 mutant and wild-type strains, consistent with the microarray data. For example, the microarray results indicated that the SMG1 gene, encoding a putative GMC oxidoreductase, is induced 4.5-fold in the nrg1 mutant compared to wild type. Using primers directed against the coding region of the SMG1 gene, real-time PCR similarly indicated that SMG1 message is fourfold more abundant in the nrg1 mutant than in the wild-type strain (data not shown).
Both real-time PCR and the genome microarray studies also demonstrated that the UGD1 gene is induced 3.5-fold in the wild type compared to the nrg1 mutant (data not shown). UGD1 encodes a UDP-glucose dehydrogenase that is required for capsule formation and cell wall integrity (35). Altered expression of the UGD1 gene may therefore explain the mechanism of the delayed capsule formation in the nrg1 strain. Similarly to the nrg1 strain, the ugd1 mutant also demonstrates cell clustering (35). Additionally, the growth of the ugd1 mutant is inhibited by various cell wall stressors, including high concentrations of NaCl, sorbitol, and SDS (35). Likewise, the nrg1 mutant is more susceptible to 1.5 M NaCl, 2.5 M sorbitol, and SDS than is wild type (Fig. 4C).
The microarray experiments suggest that none of the genes encoding elements of the C. neoformans pheromone response pathway are differentially expressed in the nrg1 mutant compared to wild type. The MFα1 pheromone gene (34), CPRa pheromone receptor gene (8), GPB1 G-β subunit gene (52), and STE11 gene (13) had equivalent transcript levels in the nrg1 mutant and wild-type strains (data not shown). Although this experiment assessed gene expression only under one condition and at one time point, it suggests that the nrg1 mutant mating defect is not a result of altered transcription of those genes specifically involved in the pheromone response pathway.
The Nrg1 transcription factor is required for C. neoformans pathogenesis.
We used the murine inhalation model of cryptococcosis to determine the role of Nrg1 in pathogenesis. Female A/Jcr mice were intranasally inoculated with 5 × 105 CFU of one of three strains (NRG1 wild-type, nrg1 mutant, or nrg1+NRG1 reconstituted strain). Mice were monitored for survival and sacrificed at predetermined clinical end points correlating with near-mortality (Fig. 6).
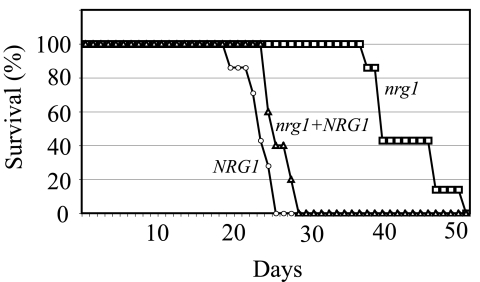
FIG. 6.
Virulence defect of the nrg1 mutant. The wild-type, nrg1 mutant, and nrg1+NRG1 reconstituted strains were intranasally inoculated into A/Jcr mice. Animal survival was measured over the course of the 52-day experiment.
Infection with either the wild-type or nrg1+NRG1 reconstituted strain resulted in complete lethality for the mice between 18 and 28 days; there was no statistically significant difference between the survival of these two groups of mice. In contrast, the nrg1 mutant strain is highly attenuated for virulence. The animals infected with the nrg1 mutant strain survived between 38 and 52 days (P = 0.0002, wild type compared to nrg1 mutant strain). Therefore, not only is Nrg1 important for growth and differentiation in vitro, but this transcription factor is also essential for pathogenicity in an animal model of C. neoformans disease.
DISCUSSION
The recent availability of microbial genome data provides new opportunities to explore the regulation of gene expression. In fact, given their relatively compact genome size, small gene number, and limited phenotypic responses, microorganisms are excellent systems in which to study how individual cells regulate their physiology by coordinated gene regulation.
cAMP regulation of Nrg1 transcription factor function in C. neoformans.
We recently demonstrated that multiple genes required for capsule and melanin production in C. neoformans are coordinately regulated at the level of transcription and that this regulation is influenced by cAMP signaling (41). We have chosen to identify transcriptional regulators that are targets of the C. neoformans cAMP pathway to elucidate the mechanisms by which such transcriptional coordination can occur. Here, we demonstrate that the Nrg1 transcription factor is likely regulated by the cAMP pathway in two ways. First, intact cAMP signaling is required for wild-type levels of transcription of the NRG1 gene, as evidenced by our microarray experiments with the gpa1 mutant strain. Second, the Nrg1 protein contains a consensus sequence for phosphorylation by the cAMP-dependent protein kinase, PKA. Targeted mutation of this putative phosphorylation site prevents this protein from being able to fully restore capsule and cell wall integrity to the nrg1 mutant strain. Also, three independent strains containing the nrg1-C529G mutant allele express NRG1 at much higher levels than similar strains reconstituted with the wild-type allele, suggesting that a phosphorylated form of Nrg1 may negatively regulate its own transcription. Together, these results support the idea that the putative PKA consensus sequence is a functional region of the Nrg1 protein.
Our data suggest that Nrg1 is not the only transcription factor by which the cAMP pathway controls capsule and melanin production. First, several of the capsule and melanin genes whose transcription is controlled by cAMP do not appear to be targets of Nrg1 (41). Additionally, the altered capsule phenotype of the nrg1 mutant strain is less severe than the acapsular phenotype of strains with defective cAMP signaling. Lastly, melanin is not significantly altered in the nrg1 mutant.
Nrg1 function in C. neoformans.
The closest homologs of C. neoformans NRG1 encode C2H2-type transcription factors in S. cerevisiae and C. albicans. These proteins contain two conserved cysteine residues (C2) and two conserved histidine residues (H2) in a 25- to 30-amino-acid domain that directs DNA binding (17). The Nrg1/Nrg2 proteins in S. cerevisiae are transcriptional repressors that control the expression of a large set of stress response genes (49). Originally recognized for their regulation of glucose-repressible genes, S. cerevisiae Nrg proteins also control the ability of budding yeast to undergo haploid invasive growth and biofilm formation (27, 38, 50). Their role in regulating gene expression in response to nutrients was further defined by documenting a physical interaction between the yeast Nrg proteins and the Snf1 kinase, a major effector of the glucose response pathway (51). In addition to their role in coordinating cellular adaptation to carbohydrate availability, the Nrg repressors are also involved in the pH response, interacting with the pH response regulator Rim101 (43).
The physiological relevance of the Nrg1/2-controlled stress response was recently confirmed in S. cerevisiae. In contrast to wild type, nrg1Δ nrg2Δ double mutant strains are more tolerant of oxidative and salt stress and more susceptible to cold stress. Both of these results were predicted by the pattern of gene repression/activation documented by the altered transcriptional profiles of the nrg1/2 mutant strains (49).
Similar to the studies in S. cerevisiae, an NRG1 ortholog also encodes a transcriptional regulator in the human-pathogenic fungus Candida albicans. The C. albicans Nrg1 protein represses hypha-specific genes. Accordingly, an nrg1/nrg1 homozygous mutant strain displayed precocious hyphal differentiation, even under conditions that typically promote yeast cell growth in wild-type strains (4, 36). Perhaps due to impairment of the normal switching between the yeast and hyphal states, this mutant strain was attenuated for virulence in a murine intravenous model of systemic candidiasis (36).
The NRG1 gene of C. neoformans shares many features with the NRG genes of S. cerevisiae and C. albicans. C. neoformans Nrg1 likely regulates gene expression in response to changing carbohydrate conditions. We demonstrated that NRG1 expression is transcriptionally repressed by glucose. Additionally, many of the genes with altered expression in the nrg1 mutant strain are involved in carbohydrate biosynthesis or transport. This observation does not necessarily indicate that the Nrg1 transcription factor directly interacts with the promoters of all of the genes with altered transcription in the nrg1 mutant; some of the altered gene transcription documented in the microarray experiments may also result from indirect, downstream effects of the nrg1 mutation. Future studies will establish which of these genes are directly regulated by this transcription factor by defining the cis-regulatory promoter elements that bind the Nrg1 protein. However, the current microarray studies strongly implicate Nrg1 in the regulation of carbohydrate metabolism.
In spite of potential functional similarities among the Nrg1 activities in these three divergent fungi, the predicted Nrg1 protein in C. neoformans is quite distinct from its closest homologs on S. cerevisiae and C. albicans. All of these proteins contain C2H2-type zinc finger binding domains. However, the S. cerevisiae and C. albicans proteins contain the DNA-binding regions at the C terminus; in contrast, C. neoformans Nrg1 contains these domains near the N terminus. Additionally, only the C. neoformans protein contains a classical consensus sequence for PKA phosphorylation.
These transcription factors also clearly have distinctive roles in cellular differentiation. The C. albicans Nrg1 protein represses transcription of hypha-specific genes, and the corresponding nrg1 mutant strain is constitutively filamentous (4, 36). In contrast, the C. neoformans nrg1 mutant is moderately impaired in hyphal differentiation, as demonstrated in its delayed filamentation in mating reactions. Therefore, although these transcription factors may share some common functional features, they likely play quite distinct cellular roles.
One such distinctive feature of C. neoformans Nrg1 is its regulation of surface capsule. The nrg1 mutant strain displays delayed encapsulation compared to wild type. This effect does not appear to be due to an alteration of growth; the nrg1 mutant strain has no major growth defects in either liquid or solid media. However, the cell surface changes of the nrg1 mutant may help explain the altered capsule phenotype. This mutant strain is very flocculent, forming large cell clusters when incubated in liquid or solid medium. The nrg1 mutation is also associated with increased susceptibility to cell wall perturbations, such as detergent and osmotic stress. These mutant phenotypes may be due to impaired expression of the UGD1 gene, encoding UDP-glucose dehydrogenase. Mutation of this gene results in decreased capsule formation, poor growth at 37°C, and increased susceptibility to cell wall stress (35). In each case, the ugd1 mutant phenotypes are similar but more severe than those of the nrg1 mutant. Also, the nrg1 mutant does not share the temperature sensitivity of the ugd1 mutant strain. This variation in the phenotypic defects of the nrg1 and ugd1 mutants may be explained by our observation that UGD1 expression is only partially diminished in the nrg1 strain.
The altered capsule phenotype of the nrg1 mutant strain may alternatively be due to changes in carbohydrate metabolism and uptake. The surface capsule of C. neoformans is primarily composed of glucuronoxylomannan, a series of polymerized sugar residues on a mannose backbone (16). It is possible that the Nrg1 transcription factor regulates internal sugar stores that serve as the building blocks of the polysaccharide capsule, given the changes in carbohydrate homeostasis implied by the altered transcriptome of the nrg1 mutant strain.
C. neoformans capsule is required for this organism's virulence. Multiple studies have demonstrated that acapsular mutant strains are attenuated for virulence in animal models of cryptococcal disease (7, 9, 22, 32). Additionally, the polysaccharide capsule has numerous deleterious effects on the host and immune effector cells (23-25, 33). The altered capsule phenotype of the nrg1 mutant may explain the reduced virulence of this strain in the murine inhalational model of cryptococcosis. Additionally, nutrient acquisition is also an important aspect of survival for microbial pathogens in the host. The altered carbohydrate homeostasis resulting from the nrg1 mutation may also play a major role in the reduced virulence of this mutant strain.
Lastly, the nrg1 mutant cell surface abnormalities, as implied by the mutant's increased flocculation and susceptibility to various cell wall stresses, may be due to alterations in chitin or surface polysaccharides, especially given the transcriptional changes in chitin and carbohydrate metabolism suggested by the microarray data. Altered flocculation and cell adhesion are also often associated with changes in adhesins, surface proteins that have been extensively studied in S. cerevisiae and several Candida species. Adhesins in budding yeast, the Flo (flocculation) proteins, predominantly control flocculation and cell adhesion to substrates (19). In contrast, the C. albicans Als and Eap proteins and the Epa proteins of Candida glabrata mediate adhesion to epithelial tissue and are required for Candida pathogenesis (14, 20, 28). The cell surface changes in the C. neoformans nrg1 mutant may similarly alter this strain's ability to move from the lungs, through the systemic circulation, and to ultimately infect the central nervous system of the infected animal.
In conclusion, we have identified a new transcription factor gene in the human-pathogenic fungus C. neoformans. The Nrg1 protein appears to play a major role in several cellular processes, including carbohydrate acquisition and metabolism, capsule synthesis, and cell surface integrity. These studies also begin to define the complex ways in which conserved cAMP signaling pathways regulate the cellular response to external stress, allowing microbial pathogens to coordinate their defense against the hostile environment of the infected host.
Acknowledgments
This work was supported by PHS grant AI050128 (J.A.A.). J. Andrew Alspaugh is a Burroughs Wellcome Fund New Investigator in Molecular Pathogenic Mycology. Connie B. Nichols is supported by the Interdisciplinary Research Training Grant in AIDS (T32 AI007392), and Michael S. Price is supported by the Molecular Mycology and Pathogenesis Training Program (T32 AI52080).
We acknowledge the contributions of the C. neoformans H99 sequencing project, Duke IGSP Center for Applied Genomics and Technology (http://cgt.duke.edu/).
REFERENCES
- 1.Alspaugh, J. A., and D. L. Granger. 1991. Inhibition of Cryptococcus neoformans replication by nitrogen oxides supports the role of these molecules as effectors of macrophage-mediated cytostasis. Infect. Immun. 59:2291-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alspaugh, J. A., J. R. Perfect, and J. Heitman. 1997. Cryptococcus neoformans mating and virulence are regulated by the G-protein a subunit GPA1 and cAMP. Genes Dev. 11:3206-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alspaugh, J. A., R. Pukkila-Worley, T. Harashima, L. M. Cavallo, D. Funnell, G. M. Cox, J. R. Perfect, J. W. Kronstad, and J. Heitman. 2002. Adenylyl cyclase functions downstream of the G-alpha protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot. Cell 1:75-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun, B. R., D. Kadosh, and A. D. Johnson. 2001. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 20:4753-4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castelli, M. E., E. Garcia Vescovi, and F. C. Soncini. 2000. The phosphatase activity is the target for Mg2+ regulation of the sensor protein PhoQ in Salmonella. J. Biol. Chem. 275:22948-22954. [DOI] [PubMed] [Google Scholar]
- 6.Chamnongpol, S., M. Cromie, and E. A. Groisman. 2003. Mg2+ sensing by the Mg2+ sensor PhoQ of Salmonella enterica. J. Mol. Biol. 325:795-807. [DOI] [PubMed] [Google Scholar]
- 7.Chang, Y. C., and K. J. Kwon-Chung. 1994. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol. Cell. Biol. 14:4912-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, Y. C., G. F. Miller, and K. J. Kwon-Chung. 2003. Importance of a developmentally regulated pheromone receptor of Cryptococcus neoformans for virulence. Infect. Immun. 71:4953-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, Y. C., L. A. Penoyer, and K. J. Kwon-Chung. 1996. The second capsule gene of Cryptococcus neoformans, CAP64, is essential for virulence. Infect. Immun. 64:1977-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chhabra, S. R., K. R. Shockley, S. B. Conners, K. L. Scott, R. D. Wolfinger, and R. M. Kelly. 2003. Carbohydrate-induced differential gene expression patterns in the hyperthermophilic bacterium Thermotoga maritima. J. Biol. Chem. 278:7540-7552. [DOI] [PubMed] [Google Scholar]
- 11.Cox, G. M., J. Mukherjee, G. T. Cole, A. Casadevall, and J. R. Perfect. 2000. Urease as a virulence factor in experimental cryptococcosis. Infect. Immun. 68:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox, G. M., T. H. Rude, C. C. Dykstra, and J. R. Perfect. 1995. The actin gene from Cryptococcus neoformans: structure and phylogenetic analysis. J. Med. Vet. Mycol. 33:261-266. [DOI] [PubMed] [Google Scholar]
- 13.Davidson, R. C., C. B. Nichols, G. M. Cox, J. R. Perfect, and J. Heitman. 2003. A MAP kinase cascade composed of cell type specific and non-specific elements controls mating and differentiation of the fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 49:469-485. [DOI] [PubMed] [Google Scholar]
- 14.De Las Penas, A., S. J. Pan, I. Castano, J. Alder, R. Cregg, and B. P. Cormack. 2003. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 17:2245-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 16.Doering, T. L. 2000. How does Cryptococcus get its coat? Trends Microbiol. 8:547-553. [DOI] [PubMed] [Google Scholar]
- 17.Evans, R. M., and S. M. Hollenberg. 1988. Zinc fingers: gilt by association. Cell 52:1-3. [DOI] [PubMed] [Google Scholar]
- 18.Granger, D. L., J. R. Perfect, and D. T. Durack. 1985. Virulence of Cryptococcus neoformans: regulation of capsule synthesis by carbon dioxide. J. Clin. Investig. 76:508-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo, B., C. A. Styles, Q. Feng, and G. R. Fink. 2000. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. USA 97:12158-12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoyer, L. L., T. L. Payne, M. Bell, A. M. Myers, and S. Scherer. 1998. Candida albicans ALS3 and insights into the nature of the ALS gene family. Curr. Genet. 33:451-459. [DOI] [PubMed] [Google Scholar]
- 21.Idnurm, A., and J. Heitman. 2005. Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol. 3:e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janbon, G., U. Himmelreich, F. Moyrand, L. Improvisi, and F. Dromer. 2001. Cas1p is a membrane protein necessary for the O-acetylation of the Cryptococcus neoformans capsular polysaccharide. Mol. Microbiol. 42:453-467. [DOI] [PubMed] [Google Scholar]
- 23.Kozel, T. R., and E. C. Gotschlich. 1982. The capsule of Cryptococcus neoformans passively inhibits phagocytosis of the yeast by macrophages. J. Immunol. 129:1675-1680. [PubMed] [Google Scholar]
- 24.Kozel, T. R., and R. P. Mastronianni. 1976. Inhibition of phagocytosis by cryptococcal polysaccharide: dissociation of the attachment and ingestion phases of phagocytosis. Infect. Immun. 14:62-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozel, T. R., G. S. Pfrommer, A. S. Guerlain, B. A. Highison, and G. J. Highison. 1988. Role of the capsule in phagocytosis of Cryptococcus neoformans. Rev. Infect. Dis. 10(Suppl. 2):S436-S439. [DOI] [PubMed] [Google Scholar]
- 26.Kraus, P. R., D. S. Fox, G. M. Cox, and J. Heitman. 2003. The Cryptococcus neoformans MAP kinase Mpk1 regulates cell integrity in response to antifungal drugs and loss of calcineurin function. Mol. Microbiol. 48:1377-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuchin, S., V. K. Vyas, and M. Carlson. 2002. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol. Cell. Biol. 22:3994-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumamoto, C. A., and M. D. Vinces. 2005. Alternative Candida albicans lifestyles: growth on surfaces. Annu. Rev. Microbiol. 59:113-133. [DOI] [PubMed] [Google Scholar]
- 29.Kwon-Chung, K. J. 1976. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia 68:821-833. [PubMed] [Google Scholar]
- 30.Kwon-Chung, K. J., and J. E. Bennett. 1992. Medical mycology, p. 397-446. Lea & Febiger, Malvern, Pa.
- 31.Kwon-Chung, K. J., I. Polacheck, and T. J. Popkin. 1982. Melanin-lacking mutants of Cryptococcus neoformans and their virulence for mice. J. Bacteriol. 150:1414-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwon-Chung, K. J., and J. C. Rhodes. 1986. Encapsulation and melanin formation as indicators of virulence in Cryptococcus neoformans. Infect. Immun. 51:218-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell, T. G., and L. Friedman. 1972. In vitro phagocytosis and intracellular fate of variously encapsulated strains of Cryptococcus neoformans. Infect. Immun. 5:491-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore, T. D., and J. C. Edman. 1993. The alpha-mating type locus of Cryptococcus neoformans contains a peptide pheromone gene. Mol. Cell. Biol. 13:1962-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moyrand, F., and G. Janbon. 2004. UGD1, encoding the Cryptococcus neoformans UDP-glucose dehydrogenase, is essential for growth at 37°C and for capsule biosynthesis. Eukaryot. Cell 3:1601-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murad, A. M., P. Leng, M. Straffon, J. Wishart, S. Macaskill, D. MacCallum, N. Schnell, D. Talibi, D. Marechal, F. Tekaia, C. d'Enfert, C. Gaillardin, F. C. Odds, and A. J. Brown. 2001. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 20:4742-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nielsen, K., G. M. Cox, P. Wang, D. L. Toffaletti, J. R. Perfect, and J. Heitman. 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect. Immun. 71:4831-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park, S. H., S. S. Koh, J. H. Chun, H. J. Hwang, and H. S. Kang. 1999. Nrg1 is a transcriptional repressor for glucose repression of STA1 gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:2044-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perfect, J. R., S. D. Lang, and D. T. Durack. 1980. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am. J. Pathol. 101:177-194. [PMC free article] [PubMed] [Google Scholar]
- 40.Price, M. S., S. B. Conners, S. Tachdjian, R. M. Kelly, and G. A. Payne. 2005. Aflatoxin conducive and non-conducive growth conditions reveal new gene associations with aflatoxin production. Fungal Genet. Biol. 42:506-518. [DOI] [PubMed] [Google Scholar]
- 41.Pukkila-Worley, R., Q. D. Gerrald, P. R. Kraus, M. J. Boily, M. J. Davis, S. S. Giles, G. M. Cox, J. Heitman, and J. A. Alspaugh. 2005. Transcriptional network of multiple capsule and melanin genes governed by the Cryptococcus neoformans cyclic AMP cascade. Eukaryot. Cell 4:190-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pysz, M. A., S. B. Conners, C. I. Montero, K. R. Shockley, M. R. Johnson, D. E. Ward, and R. M. Kelly. 2004. Transcriptional analysis of biofilm formation processes in the anaerobic, hyperthermophilic bacterium Thermotoga maritima. Appl. Environ. Microbiol. 70:6098-6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rothfels, K., J. C. Tanny, E. Molnar, H. Friesen, C. Commisso, and J. Segall. 2005. Components of the ESCRT pathway, DFG16, and YGR122w are required for Rim101 to act as a corepressor with Nrg1 at the negative regulatory element of the DIT1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 25:6772-6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Sansonetti, P. J., D. J. Kopecko, and S. B. Formal. 1982. Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect. Immun. 35:852-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 47.Vandesompele, J., K. De Preter, F. Pattyn, B. Poppe, N. Van Roy, A. De Paepe, and F. Speleman. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varma, A., and K. J. Kwon-Chung. 1999. Characterization of the glyceraldehyde-3-phosphate dehydrogenase gene and the use of its promoter for heterologous expression in Cryptococcus neoformans, a human pathogen. Gene 232:155-163. [DOI] [PubMed] [Google Scholar]
- 49.Vyas, V. K., C. D. Berkey, T. Miyao, and M. Carlson. 2005. Repressors Nrg1 and Nrg2 regulate a set of stress-responsive genes in Saccharomyces cerevisiae. Eukaryot. Cell 4:1882-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vyas, V. K., S. Kuchin, C. D. Berkey, and M. Carlson. 2003. Snf1 kinases with different beta-subunit isoforms play distinct roles in regulating haploid invasive growth. Mol. Cell. Biol. 23:1341-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vyas, V. K., S. Kuchin, and M. Carlson. 2001. Interaction of the repressors Nrg1 and Nrg2 with the Snf1 protein kinase in Saccharomyces cerevisiae. Genetics 158:563-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, P., J. R. Perfect, and J. Heitman. 2000. The G-protein beta subunit GPB1 is required for mating and haploid fruiting in Cryptococcus neoformans. Mol. Cell. Biol. 20:352-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, Y., P. Aisen, and A. Casadevall. 1995. Cryptococcus neoformans melanin and virulence: mechanism of action. Infect. Immun. 63:3131-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolfinger, R. D., G. Gibson, E. D. Wolfinger, L. Bennett, H. Hamadeh, P. Bushel, C. Afshari, and R. S. Paules. 2001. Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol. 8:625-637. [DOI] [PubMed] [Google Scholar]