Abstract
Utilizing a PCR-based subtractive cDNA approach, we demonstrated that the marine diatom Thalassiosira pseudonana exhibits a rapid response at the gene level to elevated concentrations of copper and that this response attenuates over 24 h of continuous exposure. A total of 16 copper-induced genes were identified, 11 of which were completely novel; however, many of the predicted amino acid sequences had characteristics suggestive of roles in ameliorating copper toxicity. Most of the novel genes were not equivalently induced by H2O2- or Cd-induced stress, indicating specificity in response. Two genes that could be assigned functions based on homology were also induced under conditions of general cellular stress. Half of the identified genes were located within two inverted repeats in the genome, and novel genes in one inverted repeat had mRNA levels induced by ∼500- to 2,000-fold by exposure to copper for 1 h. Additionally, some of the inverted repeat genes demonstrated a dose-dependent response to Cu, but not Cd, and appear to belong to a multigene family. This multigene family may be the diatom functional homolog of metallothioneins.
Copper (Cu) is an essential micronutrient required as a redox cofactor in a number of enzymes, including cytochrome oxidase, plastocyanin, and Cu/Zn superoxide dismutase. However, due to the redox chemistry of Cu, it is a potent toxin at elevated concentrations, and organisms utilize homeostatic mechanisms to tightly control both the intracellular concentration and activity of Cu (26).
One means of Cu detoxification includes the synthesis of metal-binding ligands. The primary types of described metal-binding ligands are metallothioneins and phytochelatins, which are cysteine-rich protein molecules found in the plant and animal kingdoms (9), with metallothioneins also occurring in the prokaryotic genus Synechococcus (36). The amino acid sequences of metallothioneins are gene encoded, while phytochelatins are enzymatically produced by phytochelatin synthase. Although both of these ligands have important roles in metal detoxification, additional functions have not been ruled out, including roles in essential metal ion homeostasis. (For a review of phytochelatins and metallothioneins, see reference 9.)
A number of transporter families contain members that specifically transport metals, such as the cation diffusion facilitator (CDF) family (40), a subgroup of P-type ATPase transporters referred to as P1B-type ATPases (initially named CPx-type ATPases) (4, 50), and certain ABC-type transporters (39, 53). Cu-transporting P-type ATPases have been identified in a wide variety of organisms and are important for Cu homeostasis. While most described Cu-transporting P-type ATPases are responsible for exporting Cu(I) (4), a possible role in metal uptake has also been suggested (35, 42). The action of Cu-specific transporters can ameliorate the effects of Cu stress by either excreting Cu from the cell or transporting it into compartments where it is sequestered away from sensitive cellular sites. Some prokaryotes (for a review of efflux-mediated metal resistance in prokaryotes, see reference 34) and the eukaryote Candida albicans (57) utilize dedicated extrusion transporters to remove excess Cu from the cytoplasm. Yeast (39), plants (56), and diatoms (33) sequester excess metals, such as cadmium, zinc, or copper, into vacuoles, and in Schizosaccharomyces pombe this mechanism relies on an ABC-type transporter localized to the vacuolar membrane (39). Escherichia coli utilizes a Cu-exporting P-type ATPase to transport excess Cu into the periplasmic space and away from the cytoplasm (45).
Eukaryotes can also down-regulate the synthesis of metal uptake transporters in response to excess Cu (27). In Saccharomyces cerevisiae, both the nutritional Cu sensor, Mac1p, and the toxic Cu sensor, Ace1p, are required for cell survival under toxic conditions (41). In this role, Mac1p is responsible for sensing excess Cu and down-regulating high-affinity Cu uptake transporters. Additionally, yeast proteolytically degrades high-affinity Cu uptake transporters at the plasma membrane when exposed to elevated levels of Cu (37). Down-regulation and proteolytic degradation of uptake transporters presumably prevent additional import of Cu into cells already experiencing toxic levels.
Cu stress has environmental ramifications. Cu pollution is increasing in coastal California waters (52) and is likely increasing in other coastal environments, which may affect the growth and species composition of phytoplankton at the base of aquatic food webs. Most abundant among these are diatoms, which are unicellular, eukaryotic microorganisms that are typically photosynthetic (47) and encased in a shell made of silica called a frustule. Diatoms are ecologically important because they are found throughout the world in both freshwater and marine systems, are major constituents of the base of aquatic food webs, are responsible for ∼20% of global carbon fixation (5, 58), and are the dominant contributors to biosilicification (55).
Until recently, little molecular information about diatoms was available. However, the genome sequence of the marine diatom Thalassiosira pseudonana (5) is now publicly available (www.jgi.doe.gov/). T. pseudonana was originally isolated from a polluted estuary, and it is highly resistant to Cu and cadmium (7). Molecular mechanisms for this resistance are undescribed, but the presence of a genome sequence facilitates the study of Cu homeostasis and detoxification mechanisms in this unexplored unicellular eukaryotic system. In order to understand the cellular response of these ecologically important organisms to Cu, we utilized in this study a PCR-based subtractive cDNA approach to identify Cu-responsive genes.
MATERIALS AND METHODS
Cell culture.
An axenic culture of Thalassiosira pseudonana (Hustedt) Hasle and Heimdal clone 3H (CCMP 1335) was obtained from the Provasoli-Guillard National Center for Culture of Marine Phytoplankton, Bigelow Laboratory for Ocean Sciences. Cultures were grown in f/2 medium (15) made with 0.2-μm-filtered, autoclaved, local seawater. f/2 vitamins and inorganic nutrients (15) were 0.2-μm-filter sterilized and added after autoclaving. Cultures were incubated at 18°C under continuous cool white fluorescent light at approximately 119 μmol quanta · m−2 · s−1. Sterility was monitored by occasional inoculation into tryptone-enriched media to check for bacterial growth (3). Cells intended for nucleic acid extraction were grown in batch cultures, and growth was monitored by cell counts with a Petroff Hausser counting chamber. Small-scale growth studies to test various stress conditions were performed by growing a 1-liter culture to early exponential phase and then aliquoting 20 ml into 50-ml glass tubes to which various concentrations of Cu, H2O2, or Cd were added. Growth was monitored by measuring chlorophyll fluorescence on a Turner Designs fluorometer (model 0-AU-000, with a red-sensitive photomultiplier) using filters with an excitation wavelength of ∼450 nm and an emission wavelength of ∼670 nm. Cell counts were also performed to confirm trends observed by measuring chlorophyll fluorescence.
Incubations to induce cell stress.
For cDNA subtraction, T. pseudonana was grown in three 8-liter batch cultures in polycarbonate bottles to a density of ∼3 × 105 cells · ml−1. Eight liters of exponentially growing culture were harvested as a control. CuSO4 was added to a final concentration of 10 μM to the remaining 16 liters of exponentially growing culture, and cells were incubated for 1 h.
Cells subjected to Cu stress that were intended for real-time reverse transcriptase PCR (RT-PCR) analysis were grown in three 8-liter batch cultures to a cell density of approximately 1.5 × 105 cells · ml−1. One culture was harvested as a control, and the other two cultures were incubated with 10 μM CuSO4 for 1 and 24 h. Cultures were treated with cycloheximide to arrest mRNA translation at a final concentration of 20 μg/ml at 10 min before cells were harvested by centrifugation for 15 min at 10,000 × g.
Cells subjected to H2O2 stress that were intended for RT-PCR analysis were grown in three 8-liter batch cultures to an approximate density of 4 × 105 cells · ml−1. One culture was harvested as a control, and the other two cultures were incubated with 0.1 mM H2O2 for 1 and 24 h. Cells were harvested by filtration onto 1-μm polycarbonate filters (GE Osmonics), resuspended in 30 ml of f/2, transferred to a 40-ml centrifuge tube, and pelleted at 10,000 × g for 10 min.
Metal titration incubations were performed as follows. Eight-liter batch cultures were grown to early exponential phase and divided into 1-liter aliquots, to which increasing concentrations of either Cu (0, 0.1, 0.3, 1, 3, and 10 μM) or Cd (0, 0.1, 0.3, 1, 3, 10, and 30 μM) were added. Cells were incubated with the metal for 1 h and harvested by filtration as described above.
RNA purification and reverse transcription.
Total RNA was extracted with Tri Reagent (Sigma) as previously described (18). Contaminating genomic DNA was removed from RNA intended for RT-PCR by using the RNeasy Mini Kit (QIAGEN) in conjunction with the RNase-free DNase Set (QIAGEN). After DNase treatment, mRNA was purified from total RNA extracted from cells exposed to Cu for 1 and 24 h with the MicroPoly(A) Purist mRNA Purification Kit (Ambion) according to the manufacturer's recommendations. mRNA was not purified from cells incubated with H2O2 or increasing concentrations of Cu or Cd; instead, total RNA was used for reverse transcription and RT-PCR, yielding comparable results. RNA was quantified with a fluorometric assay using the RiboGreen RNA Quantitation Assay Kit (Molecular Probes, Inc.) in a 96-well microplate format according to the manufacturer's recommendations. Fluorescence as a result of excitation at 532 nm was measured with a Typhoon 9410 instrument (Amersham Pharmacia Biotech) using the 526SP emission filter. An equivalent amount of total RNA or mRNA from each time point or metal concentration in individual incubation experiments was reverse transcribed with Superscript II RNase H− reverse transcriptase (Invitrogen) according to the manufacturer's protocol. Reverse transcription was primed with oligo(dT)12-18 (Invitrogen) and carried out in the presence of RNasin (Promega) RNase inhibitor. We confirmed that the RNA was free of contaminating genomic DNA by PCR using the resulting cDNA as a template and primers that encompassed an intron.
cDNA subtraction.
cDNA subtraction was performed with the PCR-Select cDNA Subtraction Kit (Clontech) as described by the manufacturer. Forward and reverse subtraction libraries were designed to identify cDNAs whose expression was increased or decreased, respectively, by Cu exposure. Briefly, forward subtraction was performed by converting mRNA populations from control and Cu-exposed cells to double-stranded cDNA and digestion with RsaI to yield blunt ends. cDNA from Cu-exposed cells, termed the “tester” cDNA population, was divided into two portions that were ligated with different adapters. cDNA from control cells, or “driver” cDNA, was added in excess and allowed to hybridize with each of the adapter-ligated tester populations. A second hybridization was then performed between the two primary hybridization samples, allowing cDNAs unique to the tester population, and with different adapters, to hybridize. After hybridizations were complete, the overhanging adapters were filled in with Taq DNA polymerase (Roche). Only cDNAs unique to the tester population had different adapters at each end allowing exponential PCR amplification. cDNAs having only one adapter, or the same adapter at each end, would amplify linearly, or form a secondary structure and not amplify, respectively. The reverse subtraction was performed in the same way except that cDNA from control cells was used as the tester population and cDNA from Cu-exposed cells acted as the driver population.
cDNA screening by autoradiography.
Colony lifts of subtracted, cloned cDNAs onto Hybond N filters (Amersham) were performed as described previously (48) except that DNA was cross-linked to the filters by UV exposure with a UV Stratalinker 1800 instrument (Stratagene). Filters were stripped (0.4 N NaOH) for 30 min at 65°C to remove residual bacterial debris and then neutralized (0.2 M Tris [pH 7.5], 0.1% sodium dodecyl sulfate, 0.1× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate, pH 7]) for 30 min at 65°C. Filters were then rinsed with water and prehybridized in hybridization solution (6× SSC, 0.25% nonfat dried milk). All hybridizations were performed at 55°C.
Probes were created from both the forward and reverse subtracted cDNAs. Products of the secondary PCR performed with nested adapter primers and subtracted cDNAs as the template were digested with RsaI (New England Biolabs) to remove the adapter sequence. This material was then sequentially digested with SmaI and EagI (New England Biolabs) to ensure the removal of adapter sequence in the event that the RsaI site had been destroyed during ligation. Adapters were separated from cDNA with Sephacryl S-400 HR Microspin Columns (Amersham Biosciences). Probes were generated by random priming in the presence of [32P]dCTP with the Prime-It II kit from Stratagene according to the manufacturer's recommendations. Membranes containing forward and reverse subtracted cDNAs were first hybridized with the forward probe. Blots were then stripped as described above and hybridized with a probe made from the reverse cDNA library. Equivalent counts per minute of forward and reverse probes were added to the hybridization solution so that labeling intensity could be directly compared. Clone 76 was selected to generate a probe to identify how many of the forward subtracted clones corresponded to the same or similar genes. The insert from clone 76 was PCR amplified, and the adapters were removed by digestion with NotI and EagI. DNA was excised from a 1% agarose gel and purified with the Rapid Gel Extraction System (Marligen Bioscience, Inc.). The probe was labeled by random priming as described above.
After overnight hybridization with the probe, filters were washed twice at room temperature in 2× SSC and 0.1% sodium dodecyl sulfate for 5 min and then twice at 50°C for 40 min. Hybridization was imaged by exposing a general-purpose storage phosphor screen (Molecular Dynamics) to the radioactively labeled filters and subsequently scanning the screen with a Typhoon 9410 instrument (Amersham Pharmacia Biotech). The screen was exposed for equivalent amounts of time when forward and reverse subtracted cDNAs were used as probes so that labeling intensity could be compared. Densitometry of labeling intensity was performed with ImageQuant software (Molecular Dynamics).
RT-PCR.
mRNA levels corresponding to genes identified in the forward subtracted cDNA library were quantified after cells were exposed to Cu, Cd, or H2O2 by performing RT-PCR with a Lightcycler (Roche) and either the LightCycler DNA Master SYBR green I (Roche) or the Lithos qPCR MasterMix for SYBR green I (Eurogentec) DNA amplification kit. For genes on scaffold 73 that were highly induced, a 1:10 dilution was performed on the cDNA from 1 h of Cu exposure to prevent inhibition of target amplification due to excessive levels of transcript. Additionally, replicate RT-PCRs were performed on the highly induced genes on scaffold 73, and the results of the replicates were averaged. A 1:10 dilution of cDNA from cells exposed to H2O2 or increasing concentrations of Cu or Cd was also performed. Table 1 lists primers used for each gene and the size of the product amplified. A standard curve for each set of gene-specific primers was generated from dilutions of genomic DNA. The crossing point for each set of gene-specific primers was converted into nanogram equivalents with the standard curve. In all experiments, the mRNA levels were then normalized to mRNA levels in control cells. RT-PCR data are provided both graphically and in table format due to the logarithmic scale of the y axes in the figures.
TABLE 1.
Oligonucleotides used in this study
| Primer | Orientation | Sequence (5′ to 3′) | Fragment size (bp)a |
|---|---|---|---|
| 1B | Forward | TCTCCCGAACGTCTTCAAGCCA | |
| 1C | Reverse | ATCATTCCTCCACATAACAAAGG | 310 |
| 2C | Reverse | GCTTCCACACATGTTCACATTC | 318b |
| 3A | Forward | AGACGACGTGACGGTAATGGG | |
| 3B | Reverse | AACATCGCCACTCTCATCGGAT | 280 |
| 4A | Forward | ACTATGCTCTACAGAAGGAGCTGAA | |
| 4B | Reverse | ACTTCCAACCACCTTGTATGTCAC | 422 (618) |
| 6A | Forward | AGATGGGCTTTCTTTCCTCTTGG | |
| 6B | Reverse | CTATACAACCGCGTAAACAAATG | 148 |
| 7A | Forward | ACAAGGCACGCCATCCGCCG | |
| 7B | Reverse | TCGGTACTACTCAGTCGCGTC | 291 |
| 8A | Forward | TTGCTGTTATCTCTGCTACTGCG | |
| 8B | Reverse | TACCCTATTCCTCCTGGTTTAGG | 280 |
| 9A | Forward | TGGAGAGTTTCCTCCGGCGGT | |
| 9B | Reverse | AAGGCTGTATAAAGTCGCAAACAT | 278 |
| 10A | Forward | CAAGAGTTTGAGACATCATGGTGA | |
| 10B | Reverse | AACCATATGTCTCTACGGAAGGC | 280 |
| 11A | Reverse | ATCATAATCATCATCTTCTAAACTG | |
| 11B | Forward | ATGACAAAGAGCGAAGAGAAGG | 285 |
| 12A | ? | GTTGAAGGCGTGTGCCATAGG | |
| 12B | ? | ACAACCACACTTCGACGAGTAC | 119 |
| 13A | Reverse? | AGCGTTCTGTTGTAGGTCGAAG | |
| 13B | Forward? | GGGAATATTAACCCGTTGTCCAT | 207 |
| 14A | Forward | ACACACCAAGGCATCAACAAGC | |
| 14B | Reverse | GGCCTTATTGGAGAACGAGACA | 261 |
| 15A | Forward | CAACAATGTGCGTGCTCAGTATC | |
| 15D | Reverse | ACAGCACGGTTGAATCCGTAGT | 171 (283) |
| 16A | Reverse | TTGCTACCAGCACTCCTCCTCT | |
| 16B | Forward | ACGTCTCCGCATTGGATCGCA | 309 |
| 17A | Forward | GAGTGGAAAGGCCGTCACTCAA | |
| 17B | Reverse | ACTACCGCCCCAGGATGCGAT | 129 |
| 18B | Reverse | GCATGCATCTCCGTCGGGATC | |
| 18A | Forward | ACTGTCCAAATTGCGGAGGTGG | 322 |
| 19A | Forward | TCGCCTGATCAAGGCTCAAGGA | 277c |
| p150A | Forward | TTCAACGCATCTACTACAAG | |
| p150B | Reverse | GAGTGCAGCAGTGATGTAGAGC | 334 |
| p150-likeA | Forward | TCCTGTGTGCGACGACCTTTCA | |
| p150-likeB | Reverse | ACTTCGTGGTACTCCCTCAACAT | 306 |
| NR1 | Forward | CCTGGATACATTGGTGGAAGGAT | |
| NR2 | Reverse | GGAGGCAAGATACGATTATCATG | 106 |
| GPX1 F | Forward | CAAAGGCGACGTGCTATGCGTC | |
| GPX1 R | Reverse | GGCTCCTGAGCTCCAAACTGATT | 192 |
| GPX2 F | Forward | TTCGGAGCAACTTTCCAGAGGTG | |
| GPX2 R | Reverse | TGCCCATAACTCTTGACAGCCCT | 166 |
| GPX-like F | Forward | AAACTCCACCGCAAATACAAATCC | |
| GPX-like R | Reverse | CACGTAATCCTAGGAGGACCG | 229 |
| Catalase F | Forward | GTTGATGATTCGGTTGGCTTGGC | |
| Catalase R | Reverse | AGTTGAGAGGTGCAAGACGGATG | 101 (195) |
Genomic fragment sizes are in parentheses in cases in which the primers encompass an intron.
Cue2 was amplified by primers 1B and 2C.
Cue19 was amplified by primers 18B and 19A.
RACE.
To obtain full-length sequence, rapid amplification of cDNA ends (RACE) (13) was performed on a subset of copper-induced genes with the First Choice RLM-RACE kit from Ambion. Total RNA was ligated to an adapter at the 5′ end, and cDNA synthesis was initiated from an oligo(dT) primer containing a 3′ adapter sequence. Gene-specific primers were used in conjunction with provided adapter primers to amplify 5′ and 3′ cDNA ends. PCR products were excised from a 1% agarose gel and purified with the Rapid Gel Extraction System (Marligen Bioscience, Inc).
Cloning, sequence determination, and analysis.
PCR products resulting from RACE and cDNA subtraction were cloned into pCR 2.1-TOPO vector (Invitrogen) as recommended. RACE products were sequenced with BigDye Terminator Cycle Sequencing chemistry, version 3.1, on an ABI Prism 3100 Genetic Analyzer (Applied Biosystems). Cloned, subtracted cDNAs were sequenced either this way or with DYEnamic ET Dye Terminator chemistry on a MegaBACE DNA Analysis System (Amersham Biosciences) according to the manufacturer's instructions. The genome sequence of T. pseudonana is publicly available at www.jgi.doe.gov/. The browser window displays representations of genomic scaffolds and contiguous assembled DNA sequence, as well as gene models based on Grail and Genewise prediction software. Amino acid sequence alignments were performed with CLUSTAL W (54), and signal peptides were predicted with SignalP (6). PredictNLS (10) was used to identify potential nuclear localization signals (NLSs), and transmembrane domains were predicted with TMHMM (51). Sequence similarity searches were performed with BLAST (2), and conserved domains were identified with SMART (28).
Nucleotide sequence accession numbers.
cDNA sequences of characterized Cue genes have been deposited in the GenBank database (accession numbers DQ115006 to DQ115015). See Table 3 for a list of the Cue genes and corresponding accession numbers.
TABLE 3.
Features of Cue genes
| Gene | cDNA size (bp) | No. of amino acids | No. of introns | Inverted repeat | Putative gene function | E valuea | Accession no. |
|---|---|---|---|---|---|---|---|
| Genes for which complete cDNA was obtainedb | |||||||
| Cue1 | 560 | 134 | 0 | Yes | Unknown | NS | DQ115006 |
| Cue2 | 585 | 134 | 0 | Yes | Unknown | NS | DQ115007 |
| Cue4 | 766 | 198 | 2 | Yes | Unknown | NS | DQ115008 |
| Cue5 | 766 | 198 | 2 | Yes | Unknown | NS | DQ115009 |
| Cue6 | 1,035 | 280 | 0 | Yes | Unknown | NS | DQ115010 |
| Cue7 | 1,025 | 286 | 0 | Yes | Unknown | NS | DQ115011 |
| Cue10 | 1,336 | 365 | 0 | No | Unknown | NS | DQ115012 |
| Cue11 | 1,207 | 331 | 1 | No | Putative membrane protein (B. pseudomallei) | 8e-57 | DQ115013 |
| Cue15 | 853 | 243 | 1 | No | Unnamed protein product; a pathogenesis-related-1 homolog (N. tabacum) | 1e-4 | DQ115014 |
| Cue18* | 678 | 225 | 0 | Yes | Unknown | NS | |
| Cue19* | 831 | 276 | 0 | Yes | Unknown | NS | |
| Partially characterized genes | |||||||
| Cue3 | 1,326 | 441 | 2 | No | Trypsin protease (B. bacteriovorus) | 2e-37 | |
| Cue8 | 1,824 | 607 | NDc | No | Heat shock transcription factor 1 (D. rerio) | 3e-12 | |
| Cue9 | 2,013 | 670 | 1 | No | Ribonuclease II RNB family protein; dis3-like (S. pombe) | 3e-88 | |
| Cue12 | 115 | ND | ND | No | Unknown | NS | |
| Cue14 | 484 | 160 | ND | No | Unknown | NS | DQ115015 |
NS, not significant.
*, features of Cue18 and Cue19 are based on gene models.
ND, not determined.
RESULTS
Subtraction library results.
A forward subtracted cDNA library was constructed to identify Cu-induced genes, yielding a total of 143 clones. Fifty-six of the clones either could not be sequenced or yielded poor-quality sequence. Many of these clones had the same adapter ligated to both the 5′ and 3′ ends, which should have suppressed PCR amplification due to the formation of a secondary structure (49), thereby eliminating these cDNAs from the subtracted library. However, the subtraction process is imperfect. These cDNA fragments were present in the subtracted library due to undesirable amplification and were omitted from further analysis. Seven clones contained only vector sequence. In initial screening, we sequenced 39 correctly subtracted clones and determined that 25 were identical or nearly identical. One clone, 76, was selected to generate a probe to quantify the number of clones corresponding to the same or similar genes. In addition to the 25 sequenced clones, this probe strongly hybridized with an additional 41 clones (data not shown) which were not sequenced. Therefore, a total of 66 clones out of 80 correctly subtracted clones represented the same or similar genes. In total, 17 unique genes were identified in the forward subtracted cDNA library, and the number of sequenced clones representing each gene is shown in Table 2.
TABLE 2.
RT-PCR results
| Gene(s) | No. of clones sequenced from subtracted cDNA library | Cu-induced fold change in mRNA concn
|
H2O2-induced fold change in mRNA concn
|
||
|---|---|---|---|---|---|
| 1 h | 24 h | 1 h | 24 h | ||
| Potential Cu-expressed genesa | |||||
| 1 | 11 | 1,977 | 4 | 2.9 | 5.1 |
| 2 | 11 | 1,545 | 23 | 2 | 8.4 |
| 3 | 1 | 49 | 21 | 3.4 | 19.9 |
| 4, 5 | 1 | 1 | 2 | 0.4 | 5 |
| 6 | 14 | 1,826 | 11 | 2 | 0.9 |
| 7 | 14 | 1,249 | 7 | 1.1 | 0.7 |
| 8 | 1 | 46 | 5 | 1.5 | 1.1 |
| 9 | 1 | 4 | 1 | 2.7 | 2.1 |
| 10 | 1 | 7 | 2 | 3.7 | 3.2 |
| 11 | 2 | 1,101 | 203 | 2 | 2.3 |
| 12 | 1 | 8 | 3 | 1.4 | 2.2 |
| 13 | 1 | 0.4 | 0.4 | — | — |
| 14 | 1 | 2 | 2 | 1.8 | 2.6 |
| 15 | 2 | 13 | 2 | 1.5 | 5.1 |
| 16 | 1 | 0.9 | 0.1 | — | — |
| 17 | 1 | 0.6 | 0.8 | — | — |
| 18 | 0 | 514 | 16 | 0.8 | 4.6 |
| 19 | 0 | 772 | 25 | 0.8 | 11 |
| Cu-expressed genesb | |||||
| p150 | 0 | 2 | 26 | 1.3 | 4.7 |
| p150-like | 0 | 1 | 2 | 2 | 1.4 |
| Genes potentially involved in H2O2 degradation | |||||
| GPX1 | 0 | 0.8 | 0.3 | 1.4 | 0.7 |
| GPX2 | 0 | 1.5 | 1.1 | 1.3 | 1.6 |
| GPX-like | 0 | 2.7 | 2.3 | 2.1 | 2.2 |
| Catalase | 0 | 2 | 2.4 | 0.5 | 0.6 |
Details of 1-h forward subtracted cDNA library and RT-PCR results. —, not determined.
RT-PCR results for genes previously shown to be induced by 24 h of Cu exposure.
mRNA concentrations measured with RT-PCR.
RT-PCR was used to measure mRNA levels in Cu-exposed cells to confirm that genes identified in the forward subtracted library were up-regulated and to observe expression patterns over time. mRNA levels for each gene after 1 and 24 h of Cu exposure were normalized to levels measured in control cells and displayed in Table 2 and Fig. 1A. Potential Cu-induced genes numbered 1 to 12, 14, 15, 18, and 19 were confirmed by RT-PCR to be induced by 1 h of Cu exposure (Table 2, Fig. 1A; also see Fig. 3) and designated “Cu-expressed genes” (Cue genes).
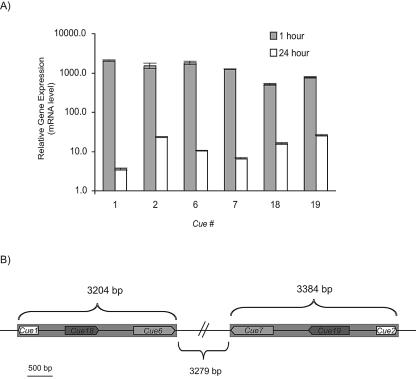
FIG. 1.
Cue genes located within an inverted repeat on genomic scaffold 73. (A) Gene induction after 1 and 24 h of copper exposure monitored by measuring mRNA levels with RT-PCR. mRNA concentrations are means normalized to control cells that were not treated with Cu. Error bars indicate standard deviations from two technical RT-PCR replicates. Note that the y axis is a logarithmic scale. (B) Schematic diagram of the inverted repeat region, indicated by gray bars. Cue gene designations and orientations are shown. The figure is shown to scale except for the intervening region between the inverted repeats, which is indicated by a double bar.
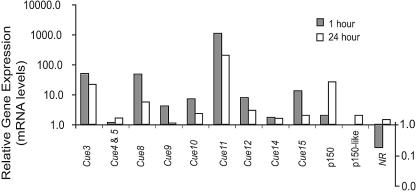
FIG. 3.
Changes in mRNA concentrations corresponding to identified genes, other than those presented in Fig. 1, measured by RT-PCR after 1 and 24 h of copper exposure. mRNA concentrations were normalized to control cells and are expressed in relative units. Note that the y axis is a logarithmic scale. RT-PCR results for p150, a gene whose induction by 24 h of copper exposure has already been described (11), as well as results for a similar gene within the genome, designated “p150-like,” are shown as examples of genes known to be induced by copper stress. Nitrate reductase (NR) was identified in a subtracted cDNA library enriched in genes down-regulated by 1 h of copper exposure and is included as an example of a down-regulated gene.
Cue genes located within an inverted repeat on scaffold 73 of the T. pseudonana genome.
RT-PCR revealed that after 1 h of Cu exposure, mRNA concentrations corresponding to the Cue genes located within an inverted repeat on genomic scaffold 73 were between 500- and 2,000-fold higher than in control conditions (Table 2, Fig. 1). Replicate RT-PCRs were performed for each gene, and the average result is shown in Fig. 1A. These genes, Cue1, Cue2, Cue6, and Cue7, were the ones that were numerically highly represented in the forward subtracted cDNA library (Table 2). An additional pair of genes located within this inverted repeat and designated Cue18 and Cue19 (Fig. 1B) were not represented in the subtracted library but were determined by RT-PCR to also be highly induced by Cu (Table 2, Fig. 1). After 24 h of Cu exposure, mRNA levels corresponding to the genes on scaffold 73 decreased to between 4- and 25-fold higher than control levels.
Scaffold 73 could not be unambiguously assigned to a position on the T. pseudonana chromosome map (5). The inverted repeat on scaffold 73 had a 3,279-bp single-copy spacer, and the individual repeat regions encompassed 3,204 bp and 3,384 bp (Fig. 1B). Gene models on the T. pseudonana website were available for Cue6 (grail.73.15.1), Cue7 (grail.73.16.1), Cue18 (grail.73.14.1), and Cue19 (grail.73.17.1) but not for Cue1 or Cue2.
5′ and 3′ RACE was performed to characterize full-length cDNA for Cue1, Cue2, Cue6, and Cue7. The full-length cDNAs of Cue1 and Cue2 were 560 bp and 585 bp, respectively; however, both cDNAs coded for 134 amino acids, and the difference in length occurred only in the 3′ untranslated region (UTR). Cue1 and Cue2 shared 99% similarity and 95% identity at the amino acid level. Full-length cDNAs of Cue6 and Cue7 were 1,035 bp and 1,025 bp, respectively, and Cue6 and Cue7 shared 96% similarity and 93% identity at the amino acid level. Although the coding sequences for these gene pairs were nearly identical, the 3′ UTRs for all four genes extended beyond the inverted repeat and therefore were distinct for each gene. Cue1, Cue2, Cue6, and Cue7 are related proteins, as determined by amino acid alignments (Fig. 2A), and collectively displayed 27% identity and 41% similarity. All of these proteins share a 73-amino-acid domain, and Cue6 and Cue7 are larger primarily due to a repeat of this domain. The domain contains a number of potential metal-binding residues, including six cysteines and four methionines as well as seven glutamic acid and seven aspartic acid residues.
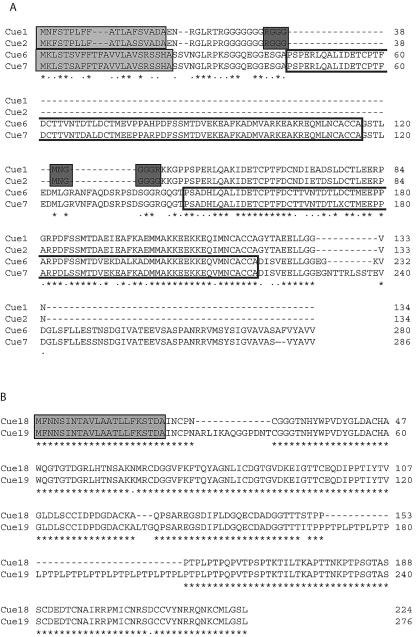
FIG. 2.
Amino acid sequence comparisons of predicted Cue proteins located within an inverted repeat on genomic scaffold 73. Alignments were performed with CLUSTAL W (1.81) (54). Sequences are identified on the left, and amino acid residue numbers are indicated on the right. Dashes in sequences were introduced to facilitate maximum alignment. Below the sequences, asterisks denote amino acid residues conserved in all sequences and dots indicate conservative replacements. Signal sequences were predicted with SignalP 3.0 (6) and are enclosed within light gray boxes. (A) Alignment of Cue1, Cue2, Cue6, Cue7. The repeated domain in Cue6 and Cue7 is encompassed within unshaded boxes. PredictNLS (10) identified a potential NLS in Cue1 and Cue2, denoted by dark gray boxes. (B) Alignment of Cue18 and Cue19.
Cue1 and Cue2 were predicted to contain an N-terminal signal sequence as well as a putative NLS (Fig. 2A). Cue6 and Cue7 were predicted to have a 23-amino-acid N-terminal signal sequence (Fig. 2A). Amino acid searches using BLAST failed to identify proteins with significant similarity (Table 3); therefore, all four of these proteins are novel. Cue1 and Cue2 contain weak homology to a domain (pfam07172, GRP) (E = 0.012) found in a protein family that includes glycine-rich proteins as well as proteins induced in response to various stresses. Statistically significant domains could not be identified in Cue6 or Cue7.
Cue18 and Cue19 were characterized based solely on their gene models and were fully contained within the inverted repeat. Amino acid sequence alignment revealed 81% similarity and 80% identity between them (Fig. 2B). Their predicted coding sequences differ primarily due to a stretch of amino acids consisting of multiple repeats of proline-threonine-proline-leucine (PTPL) within Cue19 that are absent from Cue18. Both proteins are predicted to have identical 22-amino-acid N-terminal signal sequences. No proteins with similar sequences were identified by BLAST searches (Table 3). Cue18 and Cue19 were both enriched in potential metal-binding and acidic amino acids. Statistically significant domains were not identified.
Cue genes located within an inverted repeat on scaffold 23 (chromosome 9) of the T. pseudonana genome.
Cue4 and Cue5 were represented by one clone in the forward subtracted cDNA library (Table 1). These genes are completely contained within an inverted repeat of 2,390 bp on genomic scaffold 23, located within chromosome 9, and were 100% identical at the amino acid level. Gene models for Cue4 and Cue5 were available: grail.23.58.1 and grail.23.68.1, respectively. However, full-length cDNA revealed that these models contained errors, including a miscalled start codon and an incorrectly designated intron which omitted part of the coding sequence from the gene model. The corrected amino acid sequence of Cue4 and Cue5 could not be assigned function based on BLAST analysis (Table 3). Due to the level of similarity of Cue4 and Cue5, it was not possible to develop unique primers and it could not be determined whether mRNA levels measured by RT-PCR represented one or both genes. Although also located within an inverted repeat, Cue4 and/or Cue5 had a substantially lower level of expression and a different expression pattern than genes located on scaffold 73. After a 1-h Cu treatment, mRNA levels of Cue4 and/or Cue5 remained close to those of control cells but displayed a longer-term response to Cu by increasing by twofold after 24 h (Table 2, Fig. 3).
Characterization of additional Cue genes for which full-length cDNA was acquired.
Cue10 was represented in the forward subtracted cDNA library by a single clone. To confirm the gene model prediction (grail.22.30.1), full-length cDNA for Cue10 was acquired. Cue10 is not predicted to have an N-terminal signal sequence but does have one transmembrane domain. Additionally, particular amino acids, such as glycine (13.2%), serine (12.9%), aspartic acid (10.1%), histidine (7.7%) and methionine (6.8%), are highly represented in Cue10, frequently occurring in uninterrupted stretches. This gene could not be assigned a function based on amino acid similarity searches (Table 3); however, the large proportion of amino acids that are well suited to bind metals suggests that Cue10 may have a role in metal binding. Statistically significant domains could not be identified in Cue10; however, it is predicted to contain a coiled-coil region at the C terminal end. mRNA levels for Cue10 were sevenfold higher than control cells after 1 h of Cu exposure, decreasing to twofold higher after 24 h (Table 2, Fig. 3).
Cue11 and Cue15 were not predicted in the genome and were characterized only after cDNA was obtained. Cue11 did not contain a predicted N-terminal signal sequence, but it did contain seven predicted transmembrane segments. Although BLAST analysis identified proteins with similarity to Cue11 (highest E value at 8e-57), the sequences were bacterial, putative, or uncharacterized membrane proteins to which no function was assigned (Table 3). Cue11 contains a domain of unknown function (pfam03988, DUF347) (E = 8.1e-5) that occurs in a family of bacterial membrane proteins. After 1 h of Cu exposure, Cue11 was highly induced, at 1,101-fold, and then decreased to 203-fold above control levels after 24 h (Table 2, Fig. 3).
Cue15 was predicted to have a 19-amino-acid N-terminal signal sequence. BLAST analysis identified an unnamed protein product (E = 1e-4) that is a “pathogenesis-related-1” homolog from Nicotiana tabacum as being most significantly similar to Cue15. Cue15 and the pathogenesis-related-1 protein both contain a conserved domain (pfam00188, SCP), which belongs to a family of extracellular domains with no known function. Cue15 was induced by 13-fold after 1 h of Cu exposure and decreased to 2-fold after 24 h (Table 2, Fig. 3).
Characterization of additional Cue genes for which full-length cDNA was not acquired.
Cue3, Cue8, and Cue9 were represented in the genome by gene model numbers grail.35.48.1, grail.10.4.1, and grail.3.377.1, respectively. Although these gene models were incomplete, appearing to be truncated at their 5′ ends, they had reasonable similarity to genes of known function (Table 3); therefore, characterization of full-length cDNA was not pursued. These genes displayed a pattern of induction with Cu exposure similar to that of other Cue genes examined (Table 2, Fig. 3) in that they were more highly induced after 1 h than 24 h. Cue3 was most similar to a serine protease from Bdellovibrio bacteriovorus (E = 2e-37) and contained a domain found in serine protease, trypsin family proteins (IPR001254). The characterized portion of Cue8 contains the HSF-type DNA-binding domain (IPR000232) and a leucine zipper (IPR002158) and has significant similarity (E = 3e-12) to a HSTF from Danio rerio (zebrafish). Cue9 contains a RNase II domain (IPR001900) and was found to be significantly similar (E = 3e-88) to a hypothetical RNase II, RNB protein family member from Schizosaccharomyces pombe that is similar to the gene dis3.
Cue12 and Cue14 were not predicted in the genome. 5′ RACE was unsuccessful for Cue14; however, the 3′ end consisted of 730 bp, coding for 160 amino acids. BLAST analysis failed to identify amino acid sequences similar to this portion of the protein. Cue14 was induced by twofold at 1 h and remained at that level after 24 h of Cu exposure (Table 1, Fig. 3). Neither 5′ nor 3′ RACE was successful for Cue12. It was possible to translate the short DNA fragment obtained from the subtracted library in two frames; however, BLAST analysis failed to identify homologous amino acid sequences for either frame. Cue12 was induced by eightfold at 1 h and decreased to threefold after 24 h of Cu exposure (Table 1, Fig. 3).
Genes down-regulated by copper stress.
RT-PCR results revealed that potential Cu-expressed genes 13, 16 and 17 were false positives and demonstrated a decrease in mRNA concentration relative to controls after Cu exposure (Table 2). To serve as an example of a gene down-regulated by Cu stress, a clone was examined from the reverse subtracted cDNA library. Based on sequence similarity, this clone was found to represent nitrate reductase (NR) and was confirmed by RT-PCR to be down-regulated after Cu exposure for 1 h (Fig. 3). After 24 h, NR expression returned to levels slightly above those measured in control cells (Fig. 3).
Expression patterns: Cue genes and previously characterized, copper-induced proteins.
Most of the Cue genes were highly induced at 1 h and maintained an elevated but attenuating expression level after 24 h of exposure (Table 2, Fig. 1A, Fig. 3), which is consistent with a rapid and then attenuating metal stress response. We compared this response to that of p150, a protein known to accumulate on the cell surface of T. pseudonana after 24 h of Cu exposure, as well as with a p150-like gene identified in the genome (11). In contrast to most of the Cue genes characterized in this study, the expression patterns of the p150 and p150-like genes (Table 2, Fig. 3) suggest that they are part of a longer-term response that may include secondary effects of Cu toxicity, such as cell cycle arrest (11).
Behavior of Cue and other genes during stress induced by H2O2 exposure compared to Cu exposure.
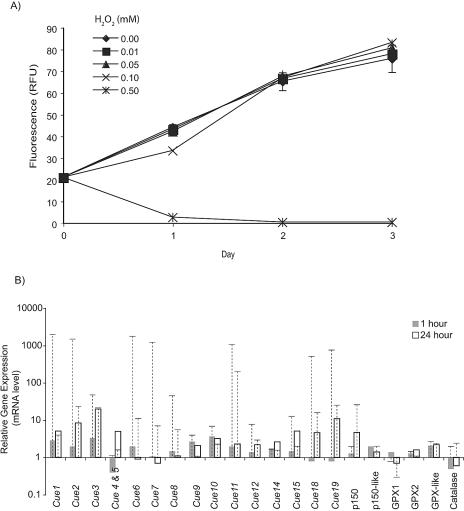
To determine whether Cue genes responded to other cellular stresses, their behavior was examined in cells experiencing H2O2-induced stress for 1 and 24 h. We first determined H2O2 levels that induced stress in T. pseudonana (Fig. 4A). H2O2 concentrations at or below 0.05 mM had no effect on growth relative to controls (Fig. 4A), and concentrations at 0.5 mM (Fig. 4A) or above (data not shown) resulted in cell death. Growth was inhibited by 0.1 mM H2O2 for at least 24 h after addition; however, cells were able to achieve control levels after 48 h probably due to the destruction of H2O2 in the culture tubes. Because 0.1 mM H2O2 induced cell stress but not cell death, similar to the Cu treatment, this concentration was used to monitor the response of the Cue genes to H2O2-induced stress for up to 24 h. In general, the timing and magnitude of the response of most of the Cue genes to H2O2 stress is different from that for Cu stress (Table 2, Fig. 4B). For example, Cue genes located in the inverted repeat on scaffold 73, Cue11, and Cue8 were much more highly induced by 1 h of Cu exposure than by H2O2 exposure. The response of these and most other Cue genes attenuates after 24 h of Cu exposure; however, attenuation does not generally occur with H2O2 exposure (Fig. 4B).
FIG. 4.
Effects of H2O2 exposure on T. pseudonana. (A) Growth of T. pseudonana exposed to various concentrations of H2O2. Cell growth was followed by measurement of chlorophyll fluorescence, and results are presented in relative fluorescence units (RFU). Results shown are of a representative growth experiment. Cells grown in batch culture were aliquoted into different treatment conditions on day 0. Error bars represent standard deviations for replicate cultures and are contained within the corresponding symbol when not visible. (B) Gene expression changes monitored by measuring mRNA levels with RT-PCR after 1 and 24 h of exposure to 0.1 mM H2O2. mRNA concentrations were normalized to control cells and are expressed in relative units. Note that the y axis is a logarithmic scale. mRNA levels from copper-exposed cells are also presented for comparison and are represented as dashed lines and bars.
The response of additional non-Cue genes to H2O2 stress was also examined. The p150 gene displayed a response over time (i.e., increasing mRNA level with length of exposure) similar to that with Cu stress but at a lower magnitude (Table 2, Fig. 4B). The p150-like gene was not highly induced by either stressor, but levels appeared to be slightly higher after a 1-h exposure to H2O2 than a 24-h exposure (Table 2, Fig. 4B). Genes potentially involved in H2O2 degradation, including glutathione peroxidase gene 1 (GPX1) and GPX2, as well as a glutathione peroxidase homolog (GPX-like), and a catalase gene were annotated in the T. pseudonana genome (newV2.0.genewise.27.59.1, newV2.0genewise.61.23.1, genewise.32.213.1, and grail.151.10.1, respectively) and were examined in an attempt to monitor oxidative stress at the transcription level (Table 2, Fig. 4B). The response of these genes to either stressor was minimal. GPX1 and GPX2 were either down-regulated or close to control levels upon exposure to either Cu or H2O2. The GPX-like gene appeared to be slightly induced (2- to 2.7-fold increase in mRNA) by both conditions of stress. The catalase gene was induced approximately twofold by Cu exposure but was down-regulated by H2O2 exposure. Multilevel (including posttranscriptional and posttranslational) regulation of antioxidant enzymes has been described for a number of organisms (8, 44), and the level of regulation of antioxidant genes in T. pseudonana is not known at this time, which may explain these results.
Responses of Cue genes to increasing concentrations of Cu and Cd.
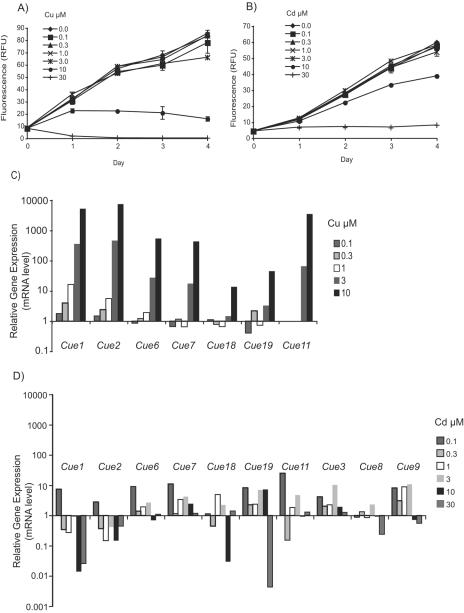
To further evaluate the specificity of the response of Cue genes to Cu, the dose-dependent behaviors of these genes to different concentrations of Cu and Cd were examined. Small-scale experiments were performed to determine the effects of various Cu or Cd concentrations on cell growth (Fig. 5A and B). Final Cu or Cd concentrations at or below 3 μM did not inhibit growth. However, 10 μM Cu halted cell growth and 30 μM Cu resulted in cell death (Fig. 5A). A concentration of 10 μM Cd inhibited growth compared to controls; however, 30 μM Cd was required to completely halt growth (Fig. 5B). The expression behaviors of select Cue genes were monitored in concentrations of Cu and Cd that did not result in cell death. For Cu, all six Cue genes within the inverted repeat on scaffold 73 were examined, as well as the highly Cu-induced Cue11 gene (Table 4, Fig. 5C). In addition to these genes, Cue3, Cue8, and Cue9 were monitored in Cd-exposed cells (Table 4, Fig. 5D).
FIG. 5.
Effects of Cu and Cd exposure on T. pseudonana. Growth of T. pseudonana exposed to various concentrations of Cu (A) and Cd (B) is shown. Cell growth was followed by measurement of chlorophyll fluorescence, and results are presented in relative fluorescence units (RFU). Results shown are of representative growth experiments. Cells grown in batch culture were aliquoted into various treatment conditions on day 0. Error bars represent standard deviations for replicate cultures and are contained within the corresponding symbol when not visible. Expression changes of a subset of Cue genes were monitored by measuring mRNA levels with RT-PCR after cells had been exposed for 1 h to either Cu (C) or Cd (D) at the concentrations indicated in the keys at right. Note that the y axes are logarithmic scales.
TABLE 4.
Changes in mRNA expression of selected Cue genes upon exposure to Cu or Cd for 1 h
| Gene | Change in mRNA expression at Cu or Cd concn (μM) ofa:
|
|||||
|---|---|---|---|---|---|---|
| 0.1 | 0.3 | 1 | 3 | 10 | 30 | |
| Cu exposure | ||||||
| Cue1 | 1.8 | 4 | 16.4 | 365.2 | 5376.1 | ND |
| Cue2 | 1.5 | 2.4 | 5.6 | 476.7 | 7585.3 | ND |
| Cue6 | 0.8 | 1.2 | 1.9 | 28.3 | 553.9 | ND |
| Cue7 | 0.7 | 1.2 | 0.6 | 18 | 442.6 | ND |
| Cue11 | 1 | 1 | 1 | 67.4 | 3578.6 | ND |
| Cue18 | 1.1 | 0.8 | 0.7 | 1.5 | 13.9 | ND |
| Cue19 | 0.4 | 2.2 | 0.7 | 3.3 | 46.3 | ND |
| Cd exposure | ||||||
| Cue1 | 7.5 | 0.3 | 0.3 | 0.9 | 0.01 | 0.03 |
| Cue2 | 2.8 | 0.4 | 0.1 | 0.4 | 0.1 | 0.4 |
| Cue3 | 4.2 | 2 | 2.2 | 10.4 | 2 | 1.3 |
| Cue6 | 9 | 1.4 | 1.9 | 2.7 | 0.7 | 1.1 |
| Cue7 | 11.1 | 1.1 | 3.4 | 4.3 | 2.5 | 1.2 |
| Cue8 | 0.9 | 1.3 | 0.8 | 2.4 | 0.9 | 0.2 |
| Cue9 | 8.2 | 3.1 | 8.9 | 10.9 | 0.7 | 0.6 |
| Cue11 | 25.1 | 0.2 | 1.8 | 4.8 | 0.9 | 1.3 |
| Cue18 | 1.1 | 0.4 | 4.9 | 2.3 | 0.03 | 1.4 |
| Cue19 | 8.4 | 2.3 | 2.4 | 7.1 | 7.4 | 0.004 |
ND, not determined.
Cue1 and Cue2 displayed a dose-dependent response to increasing Cu concentrations (Table 4, Fig. 5C); however, they were down-regulated by all but the lowest concentration of Cd, further suggesting their specificity for Cu. The pattern of induction of Cue6, Cue7, Cue11, Cue18, and Cue19 was slightly different in that mRNA levels were dependent on Cu concentration but only when cells were exposed to at least 3 μM Cu. All of the Cue genes examined were moderately induced in cells exposed to the lowest concentration of Cd (0.1 μM); however, mRNA levels did not consistently increase with increasing concentrations of Cd (Table 4, Fig. 5D), as was observed for Cu. In addition, the magnitude of the induction of these genes by Cd was substantially lower (Fig. 5D) than with Cu exposure (Fig. 5C). Although the two highest Cd concentrations tested (10 and 30 μM) inhibited cell growth, mRNA levels for most of the Cue genes at these concentrations were lower than in control cells. Cue3 was induced at all Cd concentrations without a consistent trend, with the highest induction being 10.4-fold at 3 μM Cd. Cue9 was induced at Cd concentrations at or below 3 μM, above which it was down-regulated.
DISCUSSION
Using subtractive hybridization, we identified 16 genes induced by 1 h of Cu stress. Two sets of them were located in inverted repeats. All of the genes located within the inverted repeat on scaffold 73 were functional and highly induced by Cu, did not contain introns, and encoded novel proteins (Table 3). Cue1, Cue2, Cue6, and Cue7 were related, based on amino acid alignments (Fig. 2A), and we refer to them as the Cue1 gene family. All of these proteins were predicted to contain N-terminal signal sequences (Fig. 2); however, Cue1 and Cue2 also contained a potential NLS (Fig. 2A), suggesting that members of this gene family may perform similar functions in different cellular compartments, such as the nucleus and chloroplast.
Many proteins that have the ability to sequester and transport heavy metals contain a CXXC metal-binding motif (where X is any amino acid) (12, 38), although this motif can also be found in some non-metal-binding domains. A repeated CXC sequence is a key structural motif present in all metallothionein classes (21, 32). Cue1, Cue2, Cue6, and Cue7 share a 73-amino-acid domain enriched in charged amino acids, such as aspartic and glutamic acid, and containing cysteine and methionine residues; the functional groups in the side chains of these amino acids are particularly well suited for metal coordination (22). Cue6 and Cue7 are longer than Cue1 and Cue2 primarily due to a repeat of this domain (Fig. 2A), which contains the sequence CXCC. Cue18 and Cue19 are also rich in cysteine and aspartic acid residues, and Cue18 contains a single CXXC motif. These characteristics, plus the high and apparently specific induction of the Cue1 gene family, suggest that they may bind Cu and possibly other metals with both characterized and novel metal-binding motifs.
Metallothioneins are characterized as low-molecular-weight, cysteine-rich, metal-binding proteins that contain CXC clusters. Metallothioneins are typically part of multigene families (9, 23, 24) and in plants occur in gene clusters, which have been described within inverted repeats (as seen with the Cue1 gene family [Fig. 1B]) (9) and tandem arrays (20). Amplification of metallothionein genes has been observed in a variety of organisms in cells selected for enhanced tolerance to certain trace metals (16, 46, 59). Munger et al. (31), working on the ascomycete Neurospora crassa, demonstrated that in response to Cu shock metallothionein mRNA increased, attaining maximum levels at 1 h after addition of Cu, and then rapidly decreased to initial concentrations (as also seen with the Cue1 gene family [Fig. 1A]) after metallothionein protein was synthesized. The transcription of metallothionein in Saccharomyces cerevisiae is similarly negatively autoregulated (17).
While metallothioneins from diatoms have not been characterized, a number of incomplete gene models in the T. pseudonana genome were identified as having some similarity to metallothioneins or metallothionein-like proteins. The most significantly matching metallothionein-like gene models in T. pseudonana were similar to tesmin, which is a metal-binding transcription factor (29). Similarity searches against the T. pseudonana genome using several described plant and fungal metallothioneins failed to identify strict homologs; however, similarity searches may not be effective due to the diverse nature of class II metallothioneins (9, 21). Metallothionein homologs were not represented in the subtracted cDNA library. If true homologs are absent in T. pseudonana, this suggests that the relatively small, cysteine-rich proteins from the Cue1 gene family could be functional homologs to the metallothioneins; however, direct proof of this function will require metal-binding studies.
Cadmium is the most effective inducer of phytochelatin synthase in plants (14) and in the diatom T. weissflogii (1). Phytochelatin synthase genes were identified in the T. pseudonana genome (5) but were not represented in the subtracted cDNA library. In organisms equipped to synthesize both ligands, phytochelatins are induced by cadmium exposure while metallothioneins are produced in response to Cu (30, 60), which may explain the absence of phytochelatin synthase genes in the subtracted library. Alternatively, it is possible that in T. pseudonana phytochelatin synthase is constitutively expressed, as has been described for many organisms (9).
A gene similar to Cu-transporting P-type ATPases in other organisms was identified in the T. pseudonana genome (newV2.0.genewise.122.68.1); however, this gene was not represented in our subtracted cDNA library. While Cu-transporting P-type ATPases are present in most organisms and are involved in Cu homeostasis, the P-type ATPase homolog in T. pseudonana has not been characterized; therefore, the level of gene regulation and direction of Cu transport by the resulting protein have yet to be determined. However, Cue11 was identified in our cDNA library, was highly Cu inducible, and was predicted to contain seven transmembrane segments. Cue11 is significantly similar (E = 6e-51) to COG4705, which represents an uncharacterized membrane-anchored protein conserved in bacteria. Since transporters typically contain multiple transmembrane segments, it is tempting to speculate that Cue11 could be a novel type of Cu or Cu chelate transporter.
The Cue genes that had clear homologs were identified as a serine protease (Cue3), a heat shock-like transcription factor (HSTF) (Cue8), and an RNase (Cue9), all of which may facilitate a cell's response to changing conditions. Proteases are induced by cellular stress and are required to prevent damaged and abnormally formed proteins from accumulating within the cell (19), as well as to enable the cell to reduce the level of a protein if its presence becomes disadvantageous. Similarly, the induction of Cue8, an HSTF, may allow the cell to induce the transcription of genes required to respond to conditions of cellular stress. Cue9 was similar to dis3, which is essential for mitotic control (25) and is a component of the exosome, a complex consisting primarily of exoribonucleolytic proteins that is involved in maintaining correct RNA levels in eukaryotic cells (43). Induction of an exoribonucleolytic protein may facilitate changes in mRNA levels as a result of cellular stress.
Because Cue3 and Cue9 were also induced by H2O2 (Table 2, Fig. 4B) and most Cd concentrations examined (Table 4, Fig. 5D), they may represent general stress response genes. Although Cue4, Cue5, and Cue14 represent novel genes, they were similarly or more highly induced by H2O2 than by Cu at all times examined and may also represent genes that respond to general or oxidative stress. In contrast, the response of Cue8 to Cu was decidedly higher than to either H2O2 or Cd, suggesting that it may be a transcription factor involved in a Cu-specific response.
Evaluation of the responses of selected Cue genes to stress induced by H2O2 revealed that most of the Cue genes were much more highly induced by short-term Cu stress than by H2O2 stress. In general, the timing and magnitude of the response of the Cue genes to H2O2 was different from that for Cu exposure (Fig. 4B). Some of the Cue genes were similarly induced by both stressors after 24 h; however, the response to long-term exposure may include secondary effects. Overall, these results suggest that most of the Cue genes respond specifically to Cu.
To further evaluate specificity, we tested the responses of selected Cue genes to different concentrations of Cu and Cd. In the ascomycete N. crassa, Cu metallothionein mRNA levels were strongly dependent on the Cu ion concentration in the medium, and these metallothioneins did not respond to other metals, including Cd, Co, Ni, and Zn (31). Cue1 and Cue2 responded to Cu in a dose-dependent manner, and Cue6, Cue7, Cue18, Cue19, and Cue11 were strongly induced at a Cu concentration that did not significantly affect cell growth (Fig. 5A and C), a response consistent with the idea that these gene products ameliorate the effects of high Cu levels. The levels of induction of Cue genes examined in this titration experiment varied from previous results (Table 2, Fig. 1A, Fig. 3); however, these data were obtained from independent incubation experiments, and it is not unexpected that induction levels may vary due to slight differences between cultures, such as cell density upon metal exposure. In contrast to their strong Cu response, this subset of Cue genes did not demonstrate a strong response or pattern of induction upon Cd exposure. While it cannot be concluded that this subset of Cue genes is completely Cu specific, its involvement in a general stress response (with Cd and H2O2 used as examples) can be ruled out.
Acknowledgments
We thank Eric Allen, Jon Flowers, and Chris Willis for helpful advice and the University of California Toxic Substances Research and Training Program for providing a forum for discussing these research results at annual meetings and retreats. We thank anonymous reviewers for improving the manuscript.
Funding for this research was provided by an Environmental Protection Agency STAR Exploratory Research Grant (R827107-01-0) and a National Science Foundation graduate fellowship. Instrumentation used here was purchased with support from a National Science Foundation MRI Grant (0115801).
REFERENCES
- 1.Ahner, B., and F. M. M. Morel. 1995. Phytochelatin production in marine algae. 2. Induction by various metals. Limnol. Oceanogr. 40:658-665. [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Andersen, R. A., S. L. Morton, and J. P. Sexton. 1997. Provasoli-Guillard National Center for Culture of Marine Phytoplankton. J. Phycol. 33(Suppl.):1-75. [Google Scholar]
- 4.Argüello, J. M. 2003. Identification of ion-selectivity determinants in heavy-metal transport P1B-type ATPases. J. Membr. Biol. 195:93-108. [DOI] [PubMed] [Google Scholar]
- 5.Armbrust, E. V., J. A. Berges, C. Bowler, B. R. Green, D. Martinez, N. H. Putnam, S. G. Zhou, A. E. Allen, K. E. Apt, M. Bechner, M. A. Brzezinski, B. K. Chaal, A. Chiovitti, A. K. Davis, M. S. Demarest, J. C. Detter, T. Glavina, D. Goodstein, M. Z. Hadi, U. Hellsten, M. Hildebrand, B. D. Jenkins, J. Jurka, V. V. Kapitonov, N. Kroger, W. W. Y. Lau, T. W. Lane, F. W. Larimer, J. C. Lippmeier, S. Lucas, M. Medina, A. Montsant, M. Obornik, M. S. Parker, B. Palenik, G. J. Pazour, P. M. Richardson, T. A. Rynearson, M. A. Saito, D. C. Schwartz, K. Thamatrakoln, K. Valentin, A. Vardi, F. P. Wilkerson, and D. S. Rokhsar. 2004. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306:79-86. [DOI] [PubMed] [Google Scholar]
- 6.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 7.Brand, L. E., W. G. Sunda, and R. R. L. Guillard. 1986. Reduction of marine phytoplankton reproduction rates by copper and cadmium. J. Exp. Mar. Biol. Ecol. 96:225-250. [Google Scholar]
- 8.Clerch, L. B. 2000. Post-transcriptional regulation of lung antioxidant enzyme gene expression. Ann. N. Y. Acad. Sci. 899:103-111. [DOI] [PubMed] [Google Scholar]
- 9.Cobbett, C., and P. Goldsbrough. 2002. Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annu. Rev. Plant Biol. 53:159-182. [DOI] [PubMed] [Google Scholar]
- 10.Cokol, M., R. Nair, and B. Rost. 2000. Finding nuclear localization signals. EMBO Rep. 1:411-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis, A., M. Hildebrand, and B. Palenik. 2005. A stress-induced protein associated with the girdle band region of the diatom Thalassiosira pseudonana (Bacillariophyta). J. Phycol. 41:577-589. [Google Scholar]
- 12.DeSilva, T. M., G. Veglia, F. Porcelli, A. M. Prantner, and S. J. Opella. 2002. Selectivity in heavy metal-binding to peptides and proteins. Biopolymers 64:189-197. [DOI] [PubMed] [Google Scholar]
- 13.Frohman, M. A., M. K. Dush, and G. R. Martin. 1988. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85:8998-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grill, E., S. Loffler, E.-L. Winnacker, and M. H. Zenk. 1989. Phytochelatins, the heavy-metal-binding peptides of plants, are synthesized from glutathione by a specific gamma-glutamylcysteine dipeptidyl transpeptidase (phytochelatin synthase). Proc. Natl. Acad. Sci. USA 86:6838-6842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guillard, R. R. L. 1975. Culture of phytoplankton for feeding marine invertebrates, p. 29-60. In W. L. Smith and M. H. Chanley (ed.), Culture of marine invertebrate animals. Plenum Press, New York, N.Y.
- 16.Gupta, A., B. A. Whitton, A. P. Morby, J. W. Huckle, and N. J. Robinson. 1992. Amplification and rearrangement of a prokaryotic metallothionein locus smt in Synechococcus PCC6301 selected for tolerance to cadmium. Proc. R. Soc. Lond. B 248:273-281. [DOI] [PubMed] [Google Scholar]
- 17.Hamer, D. H., D. J. Thiele, and J. E. Lemontt. 1985. Function and autoregulation of yeast copperthionein. Science 228:685-690. [DOI] [PubMed] [Google Scholar]
- 18.Hildebrand, M., and K. Dahlin. 2000. Nitrate transporter genes from the diatom Cylindrotheca fusiformis (Bacillariophyceae): mRNA levels controlled by nitrogen source and by the cell cycle. J. Phycol. 36:702-713. [DOI] [PubMed] [Google Scholar]
- 19.Hilt, W., and D. H. Wolf. 1996. Proteasomes: destruction as a programme. Trends Biochem. Sci. 21:96-102. [PubMed] [Google Scholar]
- 20.Hudspeth, R. L., S. L. Hobbs, D. M. Anderson, K. Rajasekaran, and J. W. Grula. 1996. Characterization and expression of metallothionein-like genes in cotton. Plant Mol. Biol. 31:701-705. [DOI] [PubMed] [Google Scholar]
- 21.Kagi, J. H. R. 1993. Evolution, structure and chemical activity of class I metallothioneins: an overview, p. 1-55. In K. T. Suzuki, N. Imura, and M. Kimura (ed.), Metallothionein III: biological roles and medical implications. Birkhauser Verlag, Basel, Switzerland.
- 22.Kaim, W., and B. Schwederski. 1991. Bioinorganic chemistry: inorganic elements in the chemistry of life. John Wiley & Sons, New York, N.Y.
- 23.Karin, M., and R. I. Richards. 1982. Human metallothionein genes—primary structure of the metallothionein-II gene and a related processed gene. Nature 299:797-802. [DOI] [PubMed] [Google Scholar]
- 24.Kawashima, I., T. D. Kennedy, M. Chino, and B. G. Lane. 1992. Wheat Ec metallothionein genes. Like mammalian Zn2+ metallothionein genes, wheat Zn2+ metallothionein genes are conspicuously expressed during embryogenesis. Eur. J. Biochem. 209:971-976. [DOI] [PubMed] [Google Scholar]
- 25.Kinoshita, N., M. Goebl, and M. Yanagida. 1991. The fission yeast dis3+ gene encodes a 110-kDa essential protein implicated in mitotic control. Mol. Cell. Biol. 11:5839-5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Labbe, S., and D. J. Thiele. 1999. Pipes and wiring: the regulation of copper uptake and distribution in yeast. Trends Microbiol. 7:500-505. [DOI] [PubMed] [Google Scholar]
- 27.Labbe, S., Z. Zhu, and D. J. Thiele. 1997. Copper-specific transcriptional repression of yeast genes encoding critical components in the copper transport pathway. J. Biol. Chem. 272:15951-15958. [DOI] [PubMed] [Google Scholar]
- 28.Letunic, I., R. R. Copley, S. Schmidt, F. D. Ciccarelli, T. Doerks, J. Schultz, C. P. Ponting, and P. Bork. 2004. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 32:D142-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuura, T., Y. Kawasaki, K. Miwa, S. Sutou, Y. Ohinata, F. Yoshida, and Y. Mitsui. 2002. Germ cell-specific nucleocytoplasmic shuttling protein, tesmin, responsive to heavy metal stress in mouse testes. J. Inorg. Biochem. 88:183-191. [DOI] [PubMed] [Google Scholar]
- 30.Mehra, R. K., J. R. Garey, T. R. Butt, W. R. Gray, and D. R. Winge. 1989. Candida glabrata metallothioneins. J. Biol. Chem. 264:19747-19753. [PubMed] [Google Scholar]
- 31.Münger, K., U. A. Germann, and K. Lerch. 1987. The Neurospora crassa metallothionein gene. J. Biol. Chem. 262:7363-7367. [PubMed] [Google Scholar]
- 32.Münger, K., and K. Lerch. 1985. Copper metallothionein from the fungus Agaricus bisporus: chemical and spectroscopic properties. Biochemistry 24:6751-6756. [Google Scholar]
- 33.Nassiri, Y., J. L. Mansot, J. Wery, T. Ginsburger-Vogel, and J. C. Amiard. 1997. Ultrastructural and electron energy loss spectroscopy studies of sequestration mechanisms of Cd and Cu in the marine diatom Skeletonema costatum. Arch. Environ. Contam. Toxicol. 33:147-155. [DOI] [PubMed] [Google Scholar]
- 34.Nies, D. H. 2003. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27:313-339. [DOI] [PubMed] [Google Scholar]
- 35.Odermatt, A., H. Suter, R. Krapf, and M. Solioz. 1993. Primary structure of two P-type ATPases involved in copper homeostasis in Enterococcus hirae. J. Biol. Chem. 268:12775-12779. [PubMed] [Google Scholar]
- 36.Olafson, R. W., W. D. McCubbin, and C. M. Kay. 1988. Primary- and secondary-structural analysis of a unique prokaryotic metallothionein from a Synechococcus sp. cyanobacterium. Biochem. J. 251:691-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ooi, C. E., E. Rabinovich, A. Dancis, J. S. Bonifacino, and R. D. Klausner. 1996. Copper-dependent degradation of the Saccharomyces cerevisiae plasma membrane copper transporter Ctr1p in the apparent absence of endocytosis. EMBO J. 15:3515-3523. [PMC free article] [PubMed] [Google Scholar]
- 38.Opella, S. J., T. M. DeSilva, and G. Veglia. 2002. Structural biology of metal-binding sequences. Curr. Opin. Chem. Biol. 6:217-223. [DOI] [PubMed] [Google Scholar]
- 39.Ortiz, D. F., L. Kreppel, D. M. Speiser, G. Scheel, G. McDonald, and D. W. Ow. 1992. Heavy metal tolerance in the fission yeast requires an ATP-binding cassette-type vacuolar membrane transporter. EMBO J. 11:3491-3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paulsen, I. T., and J. M. H. Saier. 1997. A novel family of ubiquitous heavy metal ion transport proteins. J. Membr. Biol. 156:99-103. [DOI] [PubMed] [Google Scholar]
- 41.Pena, M. M. O., K. A. Koch, and D. J. Thiele. 1998. Dynamic regulation of copper uptake and detoxification genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:2514-2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phung, L. T., H. Ajlani, and R. Haselkorn. 1994. P-type ATPase from the cyanobacterium Synechococcus 7942 related to the human Menkes and Wilson disease gene products. Proc. Natl. Acad. Sci. USA 91:9651-9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raijmakers, R., G. Schilders, and G. J. M. Pruijn. 2004. The exosome, a molecular machine for controlled RNA degradation in both nucleus and cytoplasm. Eur. J. Cell Biol. 83:175-183. [DOI] [PubMed] [Google Scholar]
- 44.Reimer, D. L., J. Bailley, and S. M. Singh. 1994. Complete cDNA and 5′ genomic sequences and multilevel regulation of the mouse catalase gene. Genomics 21:325-336. [DOI] [PubMed] [Google Scholar]
- 45.Rensing, C., and G. Grass. 2003. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 27:197-213. [DOI] [PubMed] [Google Scholar]
- 46.Robinson, N. J., A. M. Tommey, C. Kuske, and P. J. Jackson. 1993. Plant metallothioneins. Biochem. J. 295:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Round, F. E., R. M. Crawford, and D. G. Mann. 1990. The diatoms: biology and morphology of the genera. Cambridge University Press, Cambridge, England.
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 49.Siebert, P. D., A. Chenchik, D. E. Kellogg, K. A. Lukyanov, and S. A. Lukyanov. 1995. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 23:1087-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Solioz, M., and C. Vulpe. 1996. CPx-type ATPases: a class of P-type ATPases that pump heavy metals. Trends Biochem. Sci. 21:237-241. [PubMed] [Google Scholar]
- 51.Sonnhammer, E. L. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences, p. 175-182. In J. Glasgow, T. Littlejohn, F. Major, R. Lathrop, D. Sankoff, and C. Sensen (ed.), Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, Calif. [PubMed]
- 52.Stephenson, M. D., and G. H. Leonard. 1994. Evidence for the decline of silver and lead and the increase of copper from 1977 to 1990 in the coastal marine waters of California. Mar. Pollut. Bull. 28:148-153. [Google Scholar]
- 53.Tam, R., and J. Milton H. Saier. 1993. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol. Rev. 57:320-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tréguer, P., D. M. Nelson, A. J. V. Bennekom, D. J. DeMaster, A. Leynaert, and B. Queguiner. 1995. The silica balance in the world ocean: a reestimate. Science 268:375-379. [DOI] [PubMed] [Google Scholar]
- 56.Vogeli-Lang, R., and G. J. Wagner. 1990. Subcellular localization of cadmium and cadmium-binding peptides in tobacco leaves. Plant Physiol. 92:1086-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weissman, Z., I. Berdicevsky, B.-Z. Cavari, and D. Kornitzer. 2000. The high copper tolerance of Candida albicans is mediated by a P-type ATPase. Proc. Natl. Acad. Sci. USA 97:3520-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Werner, D. 1977. Introduction with a note on taxonomy, p. 1-498. In D. Werner (ed.), The biology of diatoms, vol. 13. University of California Press, Los Angeles, Calif. [Google Scholar]
- 59.Winge, D. R., K. B. Nielson, W. R. Gray, and D. H. Hamer. 1985. Yeast metallothionein. J. Biol. Chem. 260:14464-14470. [PubMed] [Google Scholar]
- 60.Zhou, J., and P. B. Goldsbrough. 1994. Functional homologs of fungal metallothionein genes from Arabidopsis. Plant Cell 6:875-884. [DOI] [PMC free article] [PubMed] [Google Scholar]