Abstract
The spatial and temporal variation of SOL cluster bacteria was assessed in oligomesotrophic Lake Mondsee and adjacent lakes by fluorescence in situ hybridization over two annual cycles. The filamentous SOL bacteria were present in Lake Mondsee throughout the study period, and the seasonal dynamics of the SOL community were remarkably similar with respect to both abundance and composition in the two consecutive years. Only two of the three SOL subclusters were detected in Lake Mondsee and four connected lakes. These two populations significantly differed in size distribution and demonstrated pronounced but recurrent differences in seasonality and length of period of appearance in Lake Mondsee. Extensive sampling of the lakes in September 2003 revealed low horizontal variation in the composition of the SOL community within Lake Mondsee but marked variations with depth. Between connected habitats pronounced differences in the composition and abundance of the SOL community were detected. The interaction of SOL bacteria with bacterivorous protists, mesozooplankton, and phytoplankton was investigated in order to reveal variables controlling the structure and dynamics of SOL communities. No strong indication for a bottom-up influence of phytoplankton was found, while the estimated community grazing rates of mesozooplankton on SOL bacteria indicated a top-down control of SOL abundance during mesozooplankton peaks in spring and early autumn. Furthermore, species-specific differences in grazing of mesozooplankton on SOL bacteria were observed. In general, the overall composition of SOL communities was controlled by abiotic factors (water chemistry), while their dynamics seemed to be controlled by abiotic and biotic interactions.
Cultivation-independent methods roughly revealed the overall composition of bacterioplankton in freshwater ecosystems (4, 5, 12, 31, 34, 42). Several factors potentially influencing or controlling the bacterioplankton community composition (BCC) in freshwater habitats were discovered by interhabitat comparison of bacterioplankton composition, by seasonal studies, and by manipulation experiments (4, 13, 15, 17, 19-22, 32, 40, 41). Most of these studies investigated the environmental influences on the BCC at the community level by application of community fingerprinting methods, and only a few studies tried to reveal the environmental influences controlling the distribution and dynamics of species- or genus-like groups of freshwater bacteria (30, 38). The knowledge gained from these investigations allows understanding of why BCC in habitats differing in environmental conditions is different; however, it does not really allow understanding or predicting of the dynamics of particular populations of freshwater bacteria.
In the study presented here, we tried to reveal the crucial environmental factors controlling the dynamics of species-like populations of SOL cluster bacteria in an oligomesotrophic lake. We investigated the spatial and temporal distribution of the SOL community in Lake Mondsee and in four neighboring lakes connected by running waters to Lake Mondsee. The influence of predation (top-down factor) by protists and mesozooplankton was investigated by grazing experiments, and the grazing mortality of SOL bacteria caused by mesozooplankton was estimated for populations inhabiting Lake Mondsee. In order to reveal potential bottom-up (resource) factors, the dynamics of SOL cluster bacteria and the dynamics of phytoplankton in Lake Mondsee were compared statistically.
SOL cluster bacteria (29) are filamentous, heterotrophic bacteria affiliated with the monophyletic SOL cluster (Bacteroidetes phylum). This genus-like cluster was recently discovered and recognized as an undescribed taxon. Therefore, this phylogenetic cluster was designated with the preliminary name “SOL” (29). Bacteria affiliated with this cluster always possess a conspicuous filamentous morphology, characterized by rather stable diameters of 0.25 to 0.35 μm, and highly variable lengths of 5 to >100 μm (24, 29). The filaments are usually straight, and even the longest filaments do not show a segmentation in cell-like compartments. SOL bacteria are cosmopolitan inhabitants of the pelagic zones of inland waters and were also found in wastewater treatment plants. They were detected in many freshwater lakes, as well as in oligo- and polyhaline lakes (24, 29, 30, 42). SOL filaments comprise up to 11% of bacterioplankton cell numbers in inland waters (30), but due to their filamentous morphology they contribute overproportionally to total bacterioplankton biovolume (29). Within a range of 0 to 3% relative abundance of SOL bacteria, each percentage of cell number contribution equals a 10 to 20% contribution to the total bacterioplankton biovolume, and contributions of >40% to bacterioplankton biovolume have been observed repeatedly (24, 29). SOL bacteria contributed an average 17% of the total bacterioplankton biovolume in 84 inland waters inhabited by these bacteria (30).
The monophyletic SOL cluster can be divided into at least three subclusters (HAL, LD2, and GKS2-217) (29). Only the HAL subcluster contains a described species, namely, Haliscomenobacter hydrossis, known from wastewater treatment plants (35). The other subclusters consist exclusively of environmental sequences, and sequences obtained from mixed cultures (29). Sequence similarities within and between the three subclusters indicated that each subcluster represents a species-like group (27, 29). Previous studies provided first insights in the ecology of the SOL bacteria. Pernthaler et al. (24) characterized the LD2 bacteria as ephemeral bacteria, which bloomed during a phytoplankton spring peak and benefited from protection against grazing by protists. We recently demonstrated complete niche separation between GKS2-217 and the LD2 subclusters due to the adaptation to softwater and hardwater conditions, respectively. The members of the HAL subcluster possess intermediate adaptations, which partially overlap with those of members of the vicarious GKS2-217 and LD2 subclusters. LD2 and HAL bacteria are known to coinhabit Lake Mondsee and other lakes (29, 30); however, the temporal and spatial aspects of this coinhabiting, as well as the ecological factors influencing the potential interplay between the two populations, are not known. In the present study we further reveal the ecology of LD2 and HAL bacteria and especially investigate the co-occurrence of these two closely related populations in Lake Mondsee.
MATERIALS AND METHODS
Study sites and sampling.
Lake Mondsee is a deep (maximum depth, 68 m) oligomesotrophic lake situated in a pre-alpine region at an altitude of 481 m above sea level (Table 1). Samples were taken at the main sampling station from a depth of 1 m on a biweekly basis during the years 2002 and 2003. A more extensive sampling campaign was conducted in September 2003. Two depth profiles (0, 1, 3, 6, 9, 12, 15, 20, 30, and 40 and 0, 1, 3, 6, 9, 12, 15, 20, 30, 40, 50, and 60) were sampled at two sites (3 km apart) with maximum water depths of 48 and 60 m, respectively. In addition, Lake Mondsee was sampled (1-m depth) at sites close (30 to 50 m) to the apertures of three small rivers (Zeller Ache, Wangauer Ache, and Fuschler Ache) discharging into Lake Mondsee, as well as close (200 m) to the outlet forming the river Seeache. Furthermore, three lakes (Fuschlsee, Irrsee, and Attersee), which are directly connected by small rivers (3 to 22 km in length) to Lake Mondsee, were sampled. Lake Attersee was sampled at two sites separated by 20 km. Furthermore, a small montane lake (Lake Eibensee, 952 m), which is indirectly connected to Lake Mondsee via Lake Fuschlsee, as well as a creek flowing into Lake Eibensee, were also sampled. All of these lakes were sampled at 1 m depth on 4 September 2003. In addition, these habitats were sampled during the years 2002 to 2004. Sampling and processing of samples was performed as described previously (29). Temperature, conductivity, and pH were measured onsite with an automatic multiprobe set (Yellowsprings 6502 Profiler).
TABLE 1.
Morphometric, hydrologic, and trophic characteristics of the investigated lakes
| Habitata | Maximum depth (m) | Lake area (ha) | Altitude (m) | Retention time (yr) | Trophic status |
|---|---|---|---|---|---|
| Lake Eibensee | NDb | 2.3 | 952 | ND | ND |
| Lake Fuschlsee | 67 | 265 | 662 | 2.6 | Oligotrophic |
| Lake Irrsee | 32 | 355 | 553 | 1.29 | Oligomesotrophic |
| Lake Mondsee | 68 | 1,378 | 481 | 1.82 | Oligomesotrophic |
| Lake Attersee | 169 | 4,620 | 469 | 7.13 | Oligotrophic |
All habitats are located in the Salzkammergut area of Austria.
ND, not determined.
Bacterial abundances.
Formaldehyde-fixed samples were stained with DAPI (4′,6′-diamidino-2-phenylindole; final concentration 6 μg ml−1) (26) and enumerated by epifluorescence microscopy as described previously (30). SOL filaments were identified by specific morphological characteristics (29, 30) and enumerated separately. The length of DAPI-stained SOL filaments was measured with an image analysis system as described previously (29). We sized 50 to >100 SOL filaments per sample.
FISH.
The fluorescence in situ hybridization (FISH) analyses were performed on polycarbonate filter sections according to the protocol of Alfreider et al. (1) as described previously (29, 30). The oligonucleotide probes used for FISH were SAP-309, SOL-852, LD2-1261, HAL-844, GKS-847, and HHY-441 (29, 30). The conspicuous morphology of SOL bacteria allowed for an easy and secure detection in both DAPI-stained and probe-hybridized samples. Following standard procedures (23, 26), it is possible to trace these cells down to very low abundances (<100 filaments ml−1). Probe HHY-441, specific for Haliscomenobacter hydrossis (29), was applied to selected samples to reveal the presence of this species.
Chlorophyll a and phytoplankton taxa.
Biweekly samples were taken with an integrating sampling device at a depth of 0 to 20 m in Lake Mondsee. Chlorophyll a was determined by using standard methods (14). Enumeration and biovolume measurements of lugol-fixed phytoplankton samples were done under an inverted microscope connected to an image analysis system (33). Fifty-five algal taxa (mainly species) were detected and quantified in the samples from 2002 and 2003.
Mesozooplankton abundance in Lake Mondsee and grazing impact on SOL filaments.
A total of 21 depth-integrated samples (30 liters) from Lake Mondsee were collected during 2002. Samples were filtered onto 50-μm-pore-size mesh filters, and mesozooplankton was preserved in a 4% sucrose-formalin solution (11). Mesozooplankton was enumerated in 10-ml chambers under a dissecting microscope (Reichert) and identified to the species level under a compound microscope (Neovar 2; Reichert-Jung). Species identification was done according to the methods of Flössner (8), Kiefer (16), Lilljeborg (18), and Einsle (6).
SOL filament specific grazing rates by Daphnia hyalina, Diaphanosoma brachyurum, Eudiaptomus gracilis, Cyclops abyssorum, and Mesocyclops leuckarti were determined in separate (except C. abyssorum and M. leuckarti) laboratory grazing experiments. Grazers were collected from Lake Mondsee and acclimatized to the experimental conditions prior to the experiments. During the acclimatization period, grazers were fed with the algae Cryptomonas sp. A SOL community from Lake Attersee was used for the grazing experiments. The size distribution of this community (range, 8 to 90 μm; average, 18 μm) was similar to the distribution of the SOL community in Lake Mondsee during September 2002. The predation experiments were performed in beakers receiving 55 ml of water from Lake Attersee and 9 to 15 acclimatized individuals of the respective mesozooplankton species or without the addition of the grazers (controls). The initial SOL abundance in the experiments was 1.5 × 104 filaments ml−1. Predation on SOL bacteria was investigated in the presence or absence of alternative food (Cryptomonas sp.) in triplicate experiments, respectively, over periods of 6 days. The growth rates of SOL bacteria (in the presence or absence of Cryptomonas sp.) were determined from grazer-free control treatments within the same grazing experiments. For the estimation of the in situ impact of the mesozooplankton community on SOL bacteria in Lake Mondsee, the SOL-specific mesozooplankton community grazing rate was calculated by using the experimentally determined grazing rates (corrected for temperature by assuming a Q10 of 2), the in situ SOL filament abundance, and the in situ mesozooplankton abundances. Grazing experiments could not be performed with Daphnia cuculata (contributing an average 2% of mesozooplankton biomass in Lake Mondsee) and Eubosmina coregoni (contributing an average 4% of mesozooplankton biomass). Therefore, grazing data measured in the experiments for Diaphanosoma brachyurum and Daphnia hyalina, which, respectively, share the filter mesh characteristics with the not investigated species, were used for the estimation of the SOL mortality caused by these two species.
Protistan grazing experiments.
The grazing of three cultivated bacterivorous protists on SOL bacteria from Lake Attersee was investigated in batch culture experiments. Axenic cultures of flagellated (Ochromonas sp. strain DS and Spumella sp. strain JBC07) and ciliated (Tetrahymena pyriformis strain ATTC205062) protists were precultured at 20°C with heat-killed bacteria (Listonella pelagia) (3). SOL bacteria were sampled from Lake Attersee. The size distribution of this community was similar to the distribution of the SOL community in Lake Mondsee during the spring and summer of 2002 (i.e., size range, 5 to 50 μm, with >90% of filaments being <20 μm [average, 14 μm]). The triplicated grazing experiments were performed in Erlenmeyer flasks, which received 40 ml of lake water (containing the SOL bacteria) and 10 ml of the respective axenic protist culture. As a control for grazing activity, three other treatments consisting of 10 ml of the respective protist culture and 50 ml of an inorganic medium (IBM medium) (10) were established and fed with nonfilamentous heat-killed bacteria (Listonella pelagia). Another control treatment (triplicates) consisted of lake water (containing the SOL bacteria) without the addition of protists and heat-killed bacteria. All treatments were incubated at 20°C without shaking in the dark. Subsamples were taken on days 0, 7, and 14; fixed with formaldehyde (2% final concentration); and analyzed by epifluorescence microscopy.
Statistics.
Pearson correlations between SOL, LD2, and HAL abundances, physicochemical parameters, phytoplankton biovolumes (55 taxa), mesozooplankton abundances (9 taxa), and mesozooplankton-mediated SOL mortality estimated for three different grazer types were calculated. The best correlating parameters were further analyzed by multiple linear regressions. All statistics were determined with SIGMASTAT.
RESULTS
Detected SOL bacteria subclusters.
Bacteria of the SOL morphotype were detected in 86 of the 87 samples from the six different habitats (Table 2). A small creek feeding Lake Eibensee was the only system where no SOL bacteria could be detected. This creek was the only sampled habitat not fed by a lacustrin system. Members of all three SOL subclusters (HAL, LD2, and GKS2-217) have been detected in Austrian lakes (29, 30) but, despite the intensive sampling of Lake Mondsee over a period of 2 years (Fig. 1), only two of these three subclusters were detected in the lake. The same holds true for the other four sampled lakes, which all share with Lake Mondsee a similar water chemistry (hardwater lakes). Bacteria of the LD2 subcluster were found in all 63 hybridized samples. Apart from one sample from Lake Irrsee, members of the LD2 subcluster always represented the majority of the SOL cluster communities. Members of the HAL subcluster were present in 54% of the hybridized samples. The maximum share of members of this subcluster was 65% of the SOL bacteria (Lake Irrsee). Members of the GKS2-217 subcluster were not found in any of the 63 hybridized samples taken from the six freshwater systems. No hints on the presence of SOL bacteria not affiliated with subclusters LD2 or HAL were obtained, and all investigated filaments of the SOL morphotype gave positive hybridization signals with the probes SAP-309 and SOL-852 in all of the samples analyzed.
TABLE 2.
Variability of bacterial parameters in the six investigated habitats
| Habitat | No. of samples | Total bacteria (106 ml−1) | SOL bacteria (%)a | SOL bacteria (104 ml−1) | HAL (%)b | HAL (104 ml−1) | LD2 (%)b | LD2 (104 ml−1) |
|---|---|---|---|---|---|---|---|---|
| Inflow Lake Eibenseec | 1 | 0.34 | 0 | 0 | 0 | 0 | 0 | 0 |
| Lake Eibensee | 3 | 0.90-1.97 | 0.18-1.58 | 0.27-1.58 | 0 | 0 | 100 | 0.27-1.58 |
| Lake Fuschlsee | 3 | 1.27-1.94 | 0.15-1.93 | 0.30-2.45 | 0-23.9 | 0-0.27 | 76.1-100 | 0.30-2.45 |
| Lake Irrsee | 4 | 0.98-2.64 | 0.18-1.06 | 0.48-1.98 | 0-65.0 | 0-0.67 | 35.0-100 | 0.36-1.98 |
| Lake Mondsee | 68 | 1.05-6.04 | 0.01-1.42 | 0.02-2.29 | 0-47.8 | 0-0.39 | 52.2-100 | 0.02-2.29 |
| Lake Attersee | 8 | 0.52-1.74 | 0.27-6.91 | 0.32-5.07 | 0-10.0 | 0-0.03 | 90.0-100 | 0.29-5.07 |
That is, the percentage of total bacteria.
That is, the percentage of SOL bacteria.
No lacustrine habitat upstream.
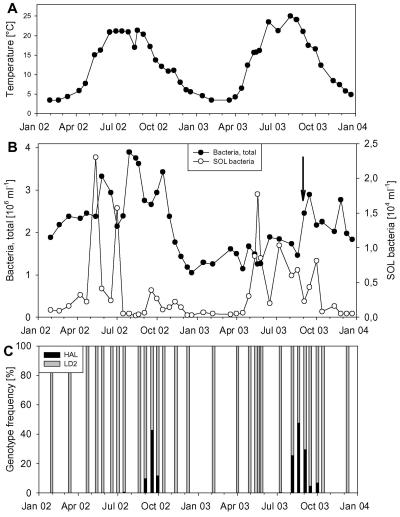
FIG. 1.
Biweekly sampling of Lake Mondsee in the years 2002 and 2003. All samples were taken from a depth of 1 m at the main sampling station. (A) Water temperature. (B) Total bacterial numbers and SOL bacteria numbers. The arrow marks the date of extensive sampling in September 2003 (see data presented in Fig. S1 in the supplemental material and Fig. 4). (C) Composition of SOL cluster community.
Temporal variations of SOL cluster bacteria in Lake Mondsee.
Lake Mondsee is characterized by two complete mixing events (holomixis) in spring and autumn and a stable stratification during summer. The data on water temperatures and total bacterial numbers during the two annual cycles are presented in Fig. 1. Lake Mondsee usually lacks marked phytoplankton blooms in spring followed by a clearwater phase as known from other temperate lakes, and its phytoplankton is dominated by the cyanobacterium Planktothrix rubescens (mean, 25.4% of the total phytoplankton biovolume). This species is forming a metalimnetic chlorophyll maximum during the stratification period. The heterotrophic SOL filaments contributed 0.01 to 1.4% of total bacterial numbers (0.02 × 104 to 2.3 × 104 filaments ml−1); thus, they were much more abundant than the filamentous P. rubescens.
The climatic conditions in the area of Lake Mondsee differed strongly between the two investigated years (Table 3). The summer of 2002 was characterized by major precipitation events causing extensive flooding of the region, while the summer of 2003 was dry and exceptionally hot. Despite these markedly different climatic situations, the dynamics of the SOL community in the years 2002 and 2003 was in respect to both abundance and composition remarkably similar (Fig. 1). In both years the highest observed abundances of SOL cluster bacteria occurred in late spring, and additional peaks of lower magnitude were detected during summer and autumn (Fig. 1B). Only low numbers of SOL cluster bacteria maintained in the lake during the winter period (0.02 × 104 to 0.1 × 104 filaments ml−1). In 66% of the hybridized samples the SOL community was exclusively composed of members of the LD2 subcluster (Fig. 1C). In the cold year 2002, members of the HAL subcluster only appeared for a period of 4 weeks, reaching 43.0% of the SOL cluster community. In this time span both populations increased simultaneously in numbers (see Fig. S1A in the supplemental material). In the following warmer year the HAL subcluster occurred during the same season but for a longer time (12 weeks, up to 47.8% of all SOL cluster bacteria). At the beginning of this period the HAL subcluster increased, while the LD2 subcluster decreased in numbers; however, in the second half the bloom of each population developed in the opposite way (see Fig. S1B in the supplemental material).
TABLE 3.
Climatic parameters of the study region and Lake Mondsee for the years 2002 and 2003
| Climatic parameter | Value for:
|
|
|---|---|---|
| 2002 | 2003 | |
| Mean air temp (°C)a | 19.00 | 21.43 |
| Mean water temp (°C)a,b | 20.02 | 22.56 |
| Precipitation (mm m−2)a | 514 | 312 |
| Mean annual precipitation (%)c | 119 | 82 |
For June, July, and August.
Lake Mondsee (1-m depth).
For the whole year.
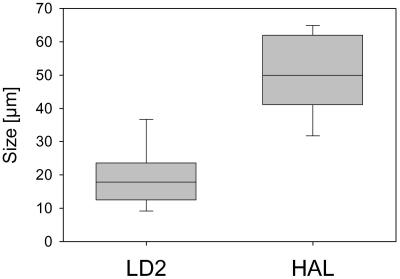
The size distribution of the SOL community was rather stable during the first nine months of 2002 but expanded toward larger filaments along with the appearance of HAL filaments (Fig. 2C). In part this change was caused by the significantly larger length of HAL bacteria (Fig. 3); however, the filament length of the LD2 bacteria was also larger during this period.
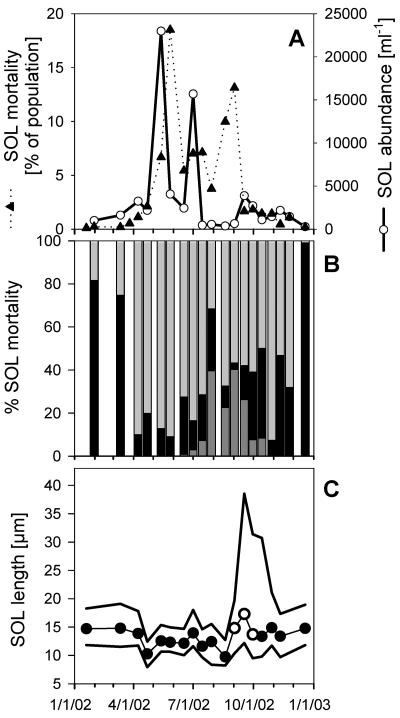
FIG. 2.
(A) Estimation of SOL bacteria mortality (as a percentage of the population per day) by mesozooplankton community grazing and SOL bacteria abundances in Lake Mondsee in 2002. (B) Estimated contribution of different metazooplankton groups to SOL mortality. Light gray bars (top) represent the mortality caused by cladocerons with coarse filter meshes (Daphnia hyalina and Eubosmina coregoni), the dark gray bars (bottom) represent the contribution of cladocerans with fine filter meshes (Diaphanosoma brachyurum and Daphnia cuculata), and the black bars depict the contribution of the calanoid copepod Eudiaptomus gracilis. (C) Length distribution of SOL filaments. Symbols: •, median length in samples exclusively containing LD2 filaments; ○, median length in samples containing LD2 and HAL filaments. The upper and lower lines depict the 75 and 25% percentiles, respectively, of the SOL length distributions.
FIG. 3.
Box plots depicting length distributions of LD2 and HAL populations in Lake Mondsee. Pooled data from 2002 and 2003 are presented. The 10th, 25th, 75th, and 90th percentiles and the medians are shown.
The abundance of HAL bacteria correlated with temperature (R2 = 0.20, P < 0.05), as well as with conductivity (R2 = 0.27; P < 0.01), whereas the abundance of LD2 bacteria correlated with pH (R2 = 0.22, P < 0.05).
Spatial variability of SOL community composition within Lake Mondsee.
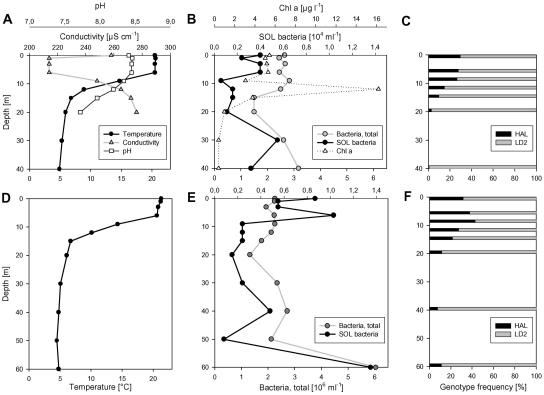
SOL bacteria numbers showed in the two investigated depths profiles a much higher vertical variability than total bacterial numbers (Fig. 4B and E). On average, SOL bacteria were more abundant in epilimnetic than in metalimnetic and hypolimnetic water layers. The exceptionally high values of both SOL bacteria numbers and total bacterial numbers in the deepest sample of the second depth profile (Fig. 4E) most likely resulted from the proximity of the sampled water layer to the sediment layer. The HAL subcluster constituted 29.9 and 32.0%, respectively, of the SOL community at the water surface, and both depth profiles showed a general trend of decreasing HAL frequencies with depth (Fig. 4C and F). The horizontal variation in Lake Mondsee (six samples, 1-m depth) of SOL abundance (0.2 × 104 to 1.1 × 104 filaments ml−1) (Fig. 5) was in a range similar to that of the vertical variation (0.1 × 104 to 1.4 × 104 filaments ml−1) (Fig. 4). By contrast, the horizontal variation of the SOL community composition in Lake Mondsee (26.4 to 32.0% HAL subcluster) (Fig. 5) was much more stable than the vertical variation (1.0 to 32.0% HAL subcluster).
FIG. 4.
Depth profiles of Lake Mondsee from September 2003. Samples were taken from 0 to 40 m (A, B, and C) and 0 to 60 m (D, E, and F), respectively. (A and D) Temperature, pH, and conductivity. (B and E) Total bacterial numbers, SOL bacteria numbers, and chlorophyll a. (C and F) Composition of SOL cluster community.
FIG. 5.
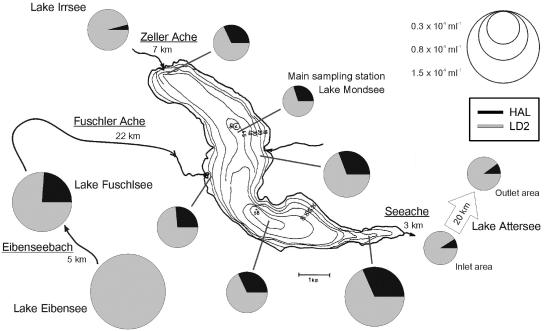
Abundance and composition of the SOL cluster community in Lake Mondsee and neighboring lakes on 3 and 4 September 2003. All samples were taken at a depth of 1 m. Sampling positions of Lake Mondsee are marked with gray lines. The names of sampled lakes and rivers connecting the lakes (underlined names with distances specified) are given. Arrowheads mark the directions of river flows. The sizes of the pie charts correspond to the total number of SOL bacteria. The scale bar (1 km) is only valid for Lake Mondsee. Sampling sites of Lake Attersee were 20 km apart.
Between-lake variability of SOL communities.
Sampling of freshwater lake ecosystems surrounding and connecting to Lake Mondsee in September 2003 resulted in larger between-lake differences in SOL community composition (see Table S1 in the supplemental material). In Lake Eibensee only members of the LD2 subcluster were found, while the samples from all other lakes contained members of both subclusters (Fig. 5). Despite the separation of the sampling sites of Lake Attersee by 20 km, the two samples from this lake displayed surprisingly similar values of total SOL numbers (0.3 × 104 SOL filaments ml−1 in both samples) and subcluster frequencies (9 and 10% HAL).
Besides the above-presented investigation (Fig. 5) the five lakes were also sampled at eight dates in the period of May to September during the years 2002, 2003 and 2004 (see Table S1 in the supplemental material). In all of the 18 samples members of the SOL cluster were present (see Fig. S2 in the supplemental material). Although bacteria of the LD2 subcluster were present in all samples, HAL subcluster bacteria were only found in six samples (33.3%) and were again absent from all of the samples from the headwater Lake Eibensee. All other lakes contained members of the HAL subcluster in various quantities in at least one of the samples taken. Of eight samples taken in the months August and September five (63%) contained bacteria of the HAL subcluster, while of ten samples taken from May to July only one sample (Lake Irrsee, 21 June 2004) was HAL positive (only 1% of the SOL cluster community). These observations indicate that the HAL bacteria showed in all four larger lakes a high seasonality with occurrence only in a short period during late summer and early autumn. In contrast, this group of bacteria seemed to be absent from the smaller, montane Lake Eibensee.
Grazing impact of protists.
A previous investigation demonstrated that not all filamentous bacteria are resistant to protistan predation (37). Therefore, we needed to verify the previously assumed protection of LD2 bacteria against protistan predation (24) by experimental investigations. Laboratory grazing experiments with three axenic protistan cultures demonstrated that SOL bacteria benefited from the activity of protistan predators (see Table S2 in the supplemental material). Grazing sensitive bacteria (i.e., small and medium sized single cells) were almost completely replaced by grazing-resistant types (floc-forming and filamentous bacteria) in all treatments with predators. The occurrence of these grazing-protected bacterial types resulted in a lack of decrease in total bacterial numbers. In contrast to the total bacterial numbers, the SOL bacteria strongly increased (by 283 to 1,818%) in all treatments with predators during the investigated 14-day period.
Mesozooplankton grazing on SOL filaments.
Over the annual cycle of 2002, the three mesozooplankton species Daphnia hyalina (41%), Cyclops abyssorum (34%), and Eudiaptomus gracilis (15%) contributed together 90% of total mesozooplankton biomass. In grazing experiments with five different mesozooplankton species, the cladoceron D. hyalina was identified as the most efficient predator on SOL bacteria (ingestion rate [I] = 5,381 ± 428 filaments individual−1 h−1). The cladoceron Diaphanosoma brachyurum (I = 1,854 ± 224 filaments individual−1 h−1) and the calonoid copepod E. gracilis (I = 1,381 ± 87 filaments individual−1 h−1) were found to feed less efficiently on SOL filaments, whereas the cyclopoid copepods Cyclops abyssorum and Mesocyclops leuckarti did not decrease SOL bacteria abundances at all. The herbivorous copepod E. gracilis was the only investigated species significantly selecting against SOL bacteria in the presence of alternative food. Furthermore, this species was the sole species significantly changing the size distribution of SOL bacteria in the course of the experiment. The copepod removed SOL filaments of >40 μm almost completely and also reduced filaments in the range of 30 to 40 μm overproportionally. The estimated mesozooplankton community-grazing rate on SOL filaments in Lake Mondsee peaked on 27 May 2002, when a calculated 18.5% of the SOL population were potentially removed by mesozooplankton per day (Fig. 2A). A second peak in grazing activity occurred on 2 September 2002, when 13% of the SOL population was potentially removed by zooplankton grazing per day.
The abundances of SOL, LD2, and HAL showed no significant correlations with the abundances of the investigated zooplankton taxa, as well as no significant correlations with the estimated SOL mortalities caused by the different zooplankton taxa. On the other hand, negative correlations of the average length of SOL filaments with the mortality caused by the copepod E. gracilis (R2 = 0.24; P < 0.05), as well as with the abundance of E. gracilis (R2 = 0.27; P < 0.05), were found.
Influence of phytoplankton on SOL bacteria.
No correlation was found between the SOL bacteria abundance and the total chlorophyll a content in Lake Mondsee. The SOL community dynamics significantly correlated with the dynamics of the phytoplankton group Chrysophytes (R2 = 0.17; P < 0.01). This group constituted on average 5% of the phytoplankton biovolume in Lake Mondsee during the years 2002 and 2003. Within the Chrysophytes the best correlations were found with the species Dinobryon divergens (R2 = 0.16; P = 0.009; mean, 2.8% of phytoplankton) and Dinobryon bavaricum (R2 = 0.23; P = <0.01; mean, 0.4% of phytoplankton). From a total of 55 taxa, 6 taxa were identified to significantly correlate with the marked seasonal dynamics of the HAL subcluster (“Microcystis aeruginosa” [R2 = 0.21; P < 0.05; mean, 0.4% of phytoplankton], Gomphosphaeria lacustris [R2 = 0.16; P < 0.05; mean, 0.003% of phytoplankton], Aphanitzomenon sp. [R2 = 0.4; P < 0.001; mean, 0.04% of phytoplankton]; Pseudanabaena catenata [R2 = 0.16; P < 0.05; mean, 0.35% of phytoplankton], Chrysochromulina parvus [R2 = 0.16; P < 0.05; mean, 0.001% of phytoplankton], and heterotrophic Katablepharis cf. ovalis [R2 = 0.15; P < 0.05; mean, 0.01% of phytoplankton]).
Combined influence of environmental factors on population dynamics.
Multiple linear regression analysis considering only the best correlating parameters indicated that pH and chrysophyte biovolume explained together 38% of the variability of the LD2 population during the two investigated years. In the case of the HAL population, the parameters temperature, conductivity, E. gracilis abundance (only data for 2002 were available), M. flos-aquae biovolume, and P. catenata biovolume explain together 82% of the observed variability in 2002. Without the parameter E. gracilis abundance, this set of parameters explained 79% of the variability of the HAL population in 2002 but only 32% of the variability in the investigated 2-year period.
DISCUSSION
Pernthaler et al. (24) observed a short-term bloom of LD2 bacteria in a mesotrophic lake in Germany in spring and suggested that the ephemeral blooming was favored by selective predation of bacterivorous protists on bacteria competing with LD2 bacteria and that the blooming was terminated by the grazing of filter-feeding mesozooplankton. In temperate lakes, such a sequence of initially high protistan grazing pressure favoring predation-resistant filamentous bacteria and a later occurrence of high grazing pressure by mesozooplankton is typically occurring during phytoplankton spring blooms and subsequent clear water phases. In principle, this typical succession is also occurring in Lake Mondsee (28) (Fig. 1A and 2), but the mesozooplankton predation in the lake is usually not strong enough to cause a pronounced clear water phase. However, the scenario suggested by Pernthaler et al. (24) can also explain the spring peaks of LD2 in Lake Mondsee (Fig. 1 and 2), but this mechanism would not explain the LD2 and HAL peaks observed later in the annual succession. This seems to indicate that the mechanisms controlling the dynamics of LD2 and HAL populations are more complex as assumed previously. Therefore, we included potential bottom-up factors, i.e., phytoplankton as potential substrate sources, as well as more detailed investigations on potential top-down factors, i.e., specific impacts by different mesozooplankton species, in our study.
Possible factors triggering the dynamics of SOL bacteria.
Shaping of bacterioplankton communities is connected to both top-down (e.g., grazing by protists and mesozooplankton) and bottom-up factors (e.g., nutrient availability and phytoplankton bloom events) (9, 13, 17, 32). Höfle et al. (13) studied the dynamics of the bacterioplankton community structure in the eutrophic Lake Plussee and found strong changes in the overall diversity of the bacterioplankton connected to grazing pressure by zooplankton and associated with the dominance of particular phytoplankton species. Kent et al. (15) and Yannarell et al. (40) described similar influences on bacterioplankton composition. The potential influence of phytoplankton on bacterioplankton communities via the release of DOM is known (7, 13). Our statistical analysis revealed some correlations between phytoplankton taxa and LD2 or HAL bacteria; however, these findings should be interpreted cautiously in terms of potential bottom-up relationships. The observed correlations with the taxa chrysophytes, Dinobryon, Chrysochromulina, and Katablepharis may result from grazing-mediated reduction of bacteria competing with the grazing-resistant SOL bacteria by these mixotrophic or heterotrophic taxa (2, 25). We also observed that the dynamics of six phytoplankton species were correlated with the dynamics of HAL bacteria in Lake Mondsee. A closer look at the dynamics of these potential trigger species revealed that “Microcystis flos-aquae” shows several peaks in summer and autumn, which did not match the appearance of HAL filaments. The population dynamics of the other species correlated with the HAL appearance in the year 2003 but did not correlate in 2002. Thus, we did not observe any strong indication for a bottom-up effect mediated by phytoplankton on the SOL bacteria.
Subcluster-specific differences in the ecology of SOL bacteria.
Members of the GKS2-217 subcluster, i.e., the third SOL subcluster, were not detected in any of the samples hybridized in the present study. We conclude that members of the GKS2-217 subcluster are not present with detectable numbers in the investigated freshwater ecosystems throughout the year. The most likely complete absence of this subcluster in the investigated hardwater lakes well supports a recently proposed adaptation of the GKS2-217 subcluster to environments with circumneutral pH and low conductivity values (softwater) (30).
The populations of LD2 and HAL bacteria inhabiting Lake Mondsee demonstrated strong seasonal differences. LD2 bacteria were always detectable, whereas the appearance of HAL bacteria was restricted to a short period at the end of the summer. Furthermore, the set of samples from the other investigated lakes seems to indicate that the two populations behave similarly in these habitats to those in Lake Mondsee. Populations of these two groups significantly differed in filament lengths. Larger filament sizes, which were predominantly observed in the HAL population, were preferentially predated by the copepod Eudiaptomus gracilis. Although the grazing mortality mediated by this copepod, as well as its abundance was significantly correlated with the length of SOL filaments in Lake Mondsee, no correlation of these parameters with the HAL population was observed. Thus, size selective predation by zooplankton could potentially explain the differences in the population dynamics of LD2 and HAL bacteria; however, the analyzed data set does not strongly support this hypothesis. Furthermore, the two subcluster populations correlated differently with water temperature, conductivity, and pH. These correlations further supported previously proposed differences in ecophysiological adaptations of members of these two subclusters (30) and also indicated that physicochemical parameters may be factors responsible for the differences in the seasonal dynamics of the two closely related populations.
The bacterium Haliscomenobacter hydrossis, the only described species within the SOL cluster, is affiliated with the HAL subcluster (29). This species was thus far exclusively reported from wastewater treatment plants (36). Despite a low detection limit for SOL bacteria, Haliscomenobacter hydrossis was not detected in selected HAL subcluster-positive samples. This may indicate that this species is adapted to the hypertrophic and/or more turbulent conditions in wastewater treatment plants and also hints at ecological differentiations even within SOL subclusters.
Spatial variability of SOL bacteria in and between lakes.
Horizontal sampling of Lake Mondsee on one day in September 2003 revealed a quite constant genotypic composition of the SOL community (Fig. 5; see Fig. S1C in the supplemental material); however, the variability in SOL abundances was rather high. Thus, the genotype frequencies were independent from total SOL numbers, which could be an effect of unselective top-down factors (e.g., zooplankton grazing). The vertical profiles in Lake Mondsee displayed much more pronounced changes in the compositional structure of the SOL cluster community, pointing to strong changes in growth conditions with depth. Furthermore, the analysis of samples of Lake Mondsee and connected lakes indicate that the spatial variability of the epilimnetic SOL community composition within an ecosystem is much lower than between ecosystems (Fig. 5). Yannarell et al. (41) also found within-lake variability of the composition of bacterioplankton lower than variability between lakes. Lindström (21) studied the bacterioplankton community composition of five mesotrophic lakes and suggested the connectivity of the lakes and therewith the input of allochthonous bacteria by the inlet rivers to be a major structuring factor of bacterioplankton communities. Recently, she concluded that external factors (most likely bacterial import) dominate the control of bacterial community structures in lakes with hydraulic retention times of less than 200 days (22). The lakes investigated in our study, however, possess turnover times of between 470 and 2,600 days. The small inflow to the headwater Lake Eibensee was the only SOL-negative sample in the present study, while the lake contained SOL filaments. In addition, preliminary data suggest that the transport of SOL bacteria via the rivers connecting the studied lakes is limited (unpublished data). Therefore, we expect that the investigated systems harbor distinct bacterioplankton communities, which undergo only minor changes due to the allochthonous input of bacteria by the connecting rivers. The minor connectivity of these systems in regard to the SOL cluster community, together with apparent differences in the trophic status, may explain the differences in abundance and composition of the SOL cluster community in the investigated lakes. Even so, the river input of water and nutrients at different sites in Lake Mondsee may be responsible for a part of the within-lake variations in absolute numbers of SOL bacteria via potential direct (e.g., dilution) or potential indirect effects (e.g., nutrient input).
Pronounced and recurrent seasonality of SOL cluster bacteria in Lake Mondsee.
In spite of the fact that the years 2002 and 2003 were remarkably different in climatic conditions, a recurring seasonal development of the SOL cluster community with respect to both abundance and community composition was detected. A recurrent seasonality was also observed for a Polynucleobacter subcluster PnecB (Betaproteobacteria) population also inhabiting Lake Mondsee (39). For this species-like group the population dynamics was followed over a period of three consecutive years. The observation of recurrent seasonality of two different bacterial groups seems to be noteworthy in the light of studies by Kent et al. (15), Lindström (19), and Yannarell et al. (40), which did not observe pronounced recurrent seasonality in bacterioplankton communities of freshwater lakes. Although these studies revealed seasonal changes of bacterioplankton community composition, consecutive years were found to be rather different in terms of bacterioplankton community fingerprints. However, the fingerprinting methods applied in those studies (automated rRNA intergenic spacer analysis or denaturing gradient gel electrophoresis) presumably offer a higher taxonomic resolution than does FISH, with probes targeting species-like groups with minimal 16S rRNA gene sequence similarities of ca. 98% (39). This higher taxonomic resolution might result in the detection of different subsets of functionally similar phylotypes present in the lakes in different years, which would result in an overinterpretation of the actual ecological dynamics found in the bacterioplankton communities. Another possible explanation for the high variability in bacterioplankton community composition observed by the fingerprinting methods could be the size and hence the water retention times of the studied lakes, which are, except for Lake Mendota, far below that of Lake Mondsee. Therefore, the bacterioplankton in Lake Mondsee might be much less influenced by allochthonous factors (such as the import of bacteria and substrates) than lakes with shorter water retention times (22).
In conclusion, our study demonstrated that the basic composition of SOL communities is controlled by abiotic factors (related to conductivity and pH). Water chemistry (hardwater versus softwater) mainly determines which of the two vicarious subclusters, i.e., LD2 or GKS2-217, is present in a particular lacustrine habitat. Furthermore, the population dynamics of LD2 and HAL bacteria in Lake Mondsee seem to be differently influenced by water temperature and water chemistry. In addition, top-down influences by mesozooplankton and (indirectly) by protistan grazing are involved in the shaping of the dynamics of SOL bacteria. A strong indication for the involvement of phytoplankton in the bottom-up control of the dynamics of SOL, LD2, or HAL bacteria was not observed. Obviously, detailed knowledge of the substrate spectrum utilized by these bacteria, as well as detailed information of the impact by viruses, are required for a deeper understanding of the dynamics of these important freshwater bacteria.
Supplementary Material
Acknowledgments
We thank Katrin Teubner for kindly providing phytoplankton data for Lake Mondsee, Qinglong Wu for his support in sampling Lake Mondsee, Matthias Pöckl and Peter Stadler for excellent technical assistance, Ulrike Brandt for sizing of SOL bacteria, Rainer Kurmayer for an introduction to mesozooplankton species identification, and Jens Boenigk for kindly providing protist cultures.
This study was supported by the Austrian Science Fund (project P15655).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alfreider, A., J. Pernthaler, R. Amann, B. Sattler, F. O. Glöckner, A. Wille, and R. Psenner. 1996. Community analysis of the bacterial assemblages in the winter cover and pelagic layers of a high mountain lake using in situ hybridization. Appl. Environ. Microbiol. 62:2138-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bird, D. F., and J. Kalff. 1986. Bacterial grazing by planktonic algae. Science 231:493-495. [DOI] [PubMed] [Google Scholar]
- 3.Boenigk, J., P. Stadler, A. Wiedlroither, and M. W. Hahn. 2004. Strain-specific differences in the grazing sensitivity of closely related ultramicrobacteria affiliated with the Polynucleobacter cluster. Appl. Environ. Microbiol. 70:5787-5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crump, B. C., G. W. Kling, M. Bahr, and J. E. Hobbie. 2003. Bacterioplankton community shifts in an arctic lake correlate with seasonal changes in organic matter source. Appl. Environ. Microbiol. 69:2253-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eiler, A., and S. Bertilson. 2004. Composition of freshwater bacterial communities associated with cyanobacterial blooms in four Swedish lakes. Environ. Microbiol. 6:1228-1243. [DOI] [PubMed] [Google Scholar]
- 6.Einsle, U. 1993. Crustacea, Copepoda: Calanoida und Cyclopoida. Süsswasserfauna von Mitteleuropa, Band 8/4-1. Gustav Fischer Verlag, Stuttgart, Germany.
- 7.Fandino, L. B., L. Riemann, G. F. Steward, R. A. Long, and F. Azam. 2001. Variations in bacterial community structure during a dinoflagellate bloom analyzed by DGGE and 16S rDNA sequencing. Aquat. Microb. Ecol. 23:119-130. [Google Scholar]
- 8.Flössner, D. 1972. Krebstiere, Crustacea, Kiemen- und Blattfüβer, Branchiopoda, Fischläuse, Branchiura. Gustav Fischer Verlag, Stuttgart, Germany.
- 9.Hahn, M. W., and M. G. Höfle. 2001. Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiol. Ecol. 35:113-121. [DOI] [PubMed] [Google Scholar]
- 10.Hahn, M. W., P. Stadler, Q. L. Wu, and M. Pöckl. 2004. The filtration acclimatization method for isolation of an important fraction of the not readily cultivable bacteria. J. Microbiol. Methods 57:379-390. [DOI] [PubMed] [Google Scholar]
- 11.Haney, J. F., and D. J. Hall. 1973. Sugar-coated Daphnia: a preservation technique for Cladocera. Limnol. Oceanogr. 18:331-333. [Google Scholar]
- 12.Hiorns, W. D., E. A. Methé, S. A. Nierzwickibauer, and J. P. Zehr. 1997. Bacterial diversity in Adirondack mountain lakes as revealed by 16S rRNA gene sequences. Appl. Environ. Microbiol. 63:2957-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Höfle, M. G., H. Haas, and K. Dominik. 1999. Seasonal dynamics of bacterioplankton community structure in a eutrophic lake as determined by 5S rRNA analysis. Appl. Environ. Microbiol. 65:3164-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.International Organization for Standardization. 1992. Water quality—measurement of biochemical parameters—spectrometric determination of the chlorophyll-a concentration. ISO 10260. International Organization for Standardization, Geneva, Switzerland.
- 15.Kent, A. D., S. E. Jones, A. C. Yannarell, J. M. Graham, G. H. Lauster, T. K. Kratz, and E. W. Triplett. 2004. Annual patterns in bacterioplankton community variability in a humic lake. Microb. Ecol. 48:550-560. [DOI] [PubMed] [Google Scholar]
- 16.Kiefer, F. 1978. Das Zooplankton der Binnengewässer. Teil 2, E. Schweizerbart'sche Verlagsbuchhandlung, Stuttgart, Germany.
- 17.Langenheder, S., and K. Jürgens. 2001. Regulation of bacterial biomass and community structure by metazoan and protozoan predation. Limnol. Oceanogr. 46:121-134. [Google Scholar]
- 18.Lilljeborg, W. 1982. Cladocera sueciae I u. II. Almqvist & Wiksell, International, Stockholm, Sweden.
- 19.Lindström, E. S. 1998. Bacterioplankton community composition in a boreal forest lake. FEMS Microbiol. Ecol. 27:163-174. [Google Scholar]
- 20.Lindström, E. S. 2000. Bacterioplankton community composition in five lakes differing in trophic status and humic content. Microb. Ecol. 40:104-113. [DOI] [PubMed] [Google Scholar]
- 21.Lindström, E. S. 2001. Investigating influential factors on bacterioplankton community composition: results from a field study of five mesotrophic lakes. Microb. Ecol. 42:598-605. [DOI] [PubMed] [Google Scholar]
- 22.Lindström, E. S., M. Forslund, G. Algesten, and A.-K. Bergström. 2006. External control of bacterial community structure in lakes. Limnol. Oceanogr. 51:339-342.
- 23.Pernthaler, J., F. O. Glöckner, W. Schönhuber, and R. Amann. 2001. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. Methods Microbiol. 30:207-226. [Google Scholar]
- 24.Pernthaler, J., E. Zöllner, F. Warnecke, and K. Jürgens. 2004. Bloom of filamentous bacteria in a mesotrophic lake: identity and potential controlling mechanism. Appl. Environ. Microbiol. 70:6272-6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Porter, K. G. 1988. Phagotrophic phytoflagellates in microbial food webs. Hydrobiologia 159:89-97. [Google Scholar]
- 26.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 27.Rosello-Mora, R., and R. Amann. 2001. The species concept for prokaryotes. FEMS Microbiol. Rev. 25:39-67. [DOI] [PubMed] [Google Scholar]
- 28.Salbrechter, M., and H. Arndt. 1994. The annual cycle of protozooplankton in the alpine, mesotrophic Lake Mondsee (Austria). Mar. Microb. Food Webs 8:217-234. [Google Scholar]
- 29.Schauer, M., and M. W. Hahn. 2005. Diversity and phylogenetic affiliations of morphologically conspicuous large filamentous bacteria occurring in the pelagic zones of a broad spectrum of freshwater habitats. Appl. Environ. Microbiol. 71:1931-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schauer, M., C. Kamenik, and M. W. Hahn. 2005. Ecological differentiation within a cosmopolitan group of planktonic freshwater bacteria (SOL cluster, Saprospiraceae, Bacteroidetes). Appl. Environ. Microbiol. 71:5900-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sekiguchi, H., M. Watanabe, T. Nakahara, B. Xu, and H. Uchiyama. 2002. Succession of bacterial community structure along the Changjiang River determined by denaturing gradient gel electrophoresis and clone library analysis. Appl. Environ. Microbiol. 68:5142-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Šimek, K., J. Pernthaler, M. G. Weinbauer, K. Hornák, J. R. Dolan, J. Nedoma, M. Mašín, and R. Amann. 2001. Changes in bacterial community composition and dynamics and viral mortality rates associated with enhanced flagellate grazing in a mesoeutrophic reservoir. Appl. Environ. Microbiol. 67:2723-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teubner, K., M. Tolotti, S. Greisberger, H. Morscheid, M. T. Dokulil, and H. Morscheid. 2003. Steady state phytoplankton in a deep pre-alpine lake: species and pigments of epilimnetic versus metalimnetic assemblages. Hydrobiologia 502:49-64. [Google Scholar]
- 34.Urbach, E., K. L. Vergin, L. Young, A. Morse, G. L. Larson, and S. J. Giovannoni. 2001. Unusual bacterioplankton community structure in ultraoligotrophic crater lake. Limnol. Oceanogr. 46:557-572. [Google Scholar]
- 35.Van Veen, W. L., D. van der Kooij, E. C. W. A. Geuze, and A. W. van der Vlies. 1973. Investigations of the sheathed bacterium Haliscomenobacter hydrossis gen.n., sp.n., isolated from activated sludge. Antonie Leeuwenhoek 39:207-216. [DOI] [PubMed] [Google Scholar]
- 36.Wagner, M., R. Amann, P. Kämpfer, B. Assmus, A. Hartmann, P. Hutzler, N. Springer, and K. H. Schleifer. 1994. Identification and in situ detection of gram-negative filamentous bacteria in activated sludge. Syst. Appl. Microbiol. 17:405-417. [Google Scholar]
- 37.Wu, Q. L., Boenigk, J., and M. W. Hahn. 2004. Successful predation of filamentous bacteria by a nanoflagellate challenges current models of flagellate bacterivory. Appl. Environ. Microbiol. 70:332-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu, Q. L., and M. W. Hahn. 7 March 2006, posting date. Differences in structure and dynamics of Polynucleobacter communities in a temperate and a subtropical lake, revealed at three phylogenetic levels. FEMS Microbiol. Ecol. [Online.] doi: 10.1111/j.1574-6941.2006.00105.x. [DOI] [PubMed]
- 39.Wu, Q. L., and M. W. Hahn. High predictability of the seasonal dynamics of a species-like Polynucleobacter population in a freshwater lake. Environ. Microbiol., in press. [DOI] [PubMed]
- 40.Yannarell, A. C., A. D. Kent, G. L. Lauster, T. K. Kratz, and E. W. Triplett. 2003. Temporal patterns in bacterial communities in three temperate lakes of different trophic status. Microb. Ecol. 46:391-405. [DOI] [PubMed] [Google Scholar]
- 41.Yannarell, A. C., and E. W. Triplett. 2004. Within- and between-lake variability in the composition of bacterioplankton communities: investigations using multiple spatial scales. Appl. Environ. Microbiol. 70:214-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zwart, G. J. M., B. C. Crump, M. Agterveld, F. Hagen, and S. K. Han. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28:141-155. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.