Abstract
California poppy (Eschscholzia californica Cham.) cell cultures produce several benzophenanthridine alkaloids, such as sanguinarine, chelirubine, and macarpine, with potent pharmacological activity. Antisense constructs of genes encoding two enzymes involved in benzophenanthridine alkaloid biosynthesis, the berberine bridge enzyme (BBE) and N-methylcoclaurine 3′-hydroxylase (CYP80B1), were introduced separately into California poppy cell cultures. Transformed cell lines expressing antisense BBE or antisense CYP80B1 constructs and displaying low levels of BBE or CYP80B1 mRNAs, respectively, showed reduced accumulation of benzophenanthridine alkaloids compared with control cultures transformed with a β-glucuronidase gene. Pathway intermediates were not detected in any of the transformed cell lines. The suppression of benzophenanthridine alkaloid biosynthesis using BBE or CYP80B1 antisense RNA constructs also reduced the growth rate of the cultures. Two-dimensional 1H-nuclear magnetic resonance and in vivo 15N-nuclear magnetic resonance spectroscopy showed no difference in the abundance of carbohydrate metabolites in the various transgenic cell lines. However, transformed cells with reduced benzophenanthridine alkaloid levels contained larger cellular pools of several amino acids including alanine, leucine, phenylalanine, threonine, and valine compared with controls. The relative abundance of tyrosine, from which benzophenanthridine alkaloids are derived, was less than 2-fold higher in antisense-suppressed cells relative to controls. These results show that alterations in the metabolic flux through benzophenanthridine alkaloid biosynthesis can affect the regulation of amino acid pools. These data provide new insight into the metabolic engineering of benzophenanthridine alkaloid pathways.
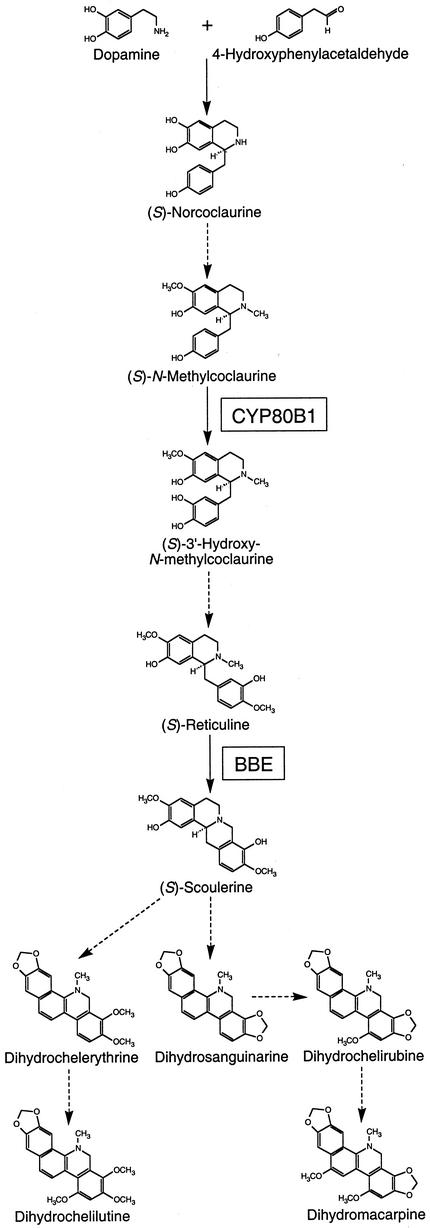
Alkaloids are a diverse group of low-Mr, nitrogenous compounds found in about 20% of plant species. Many of the approximately 12,000 alkaloids for which structures have been described function in the defense of plants against herbivores and pathogens. The potent biological activity of some alkaloids has also led to their widespread use as pharmaceuticals. The benzylisoquinoline alkaloid class, in particular, includes several important medicinal compounds such as morphine, codeine, papaverine, berberine, (+)-tubocurarine, and sanguinarine. All benzylisoquinoline alkaloids share a common biosynthetic origin beginning with a lattice of decarboxylations, ortho-hydroxylations, and deaminations that convert l-tyrosine into both dopamine and 4-hydroxyphenylacetaldehyde. Dopamine and 4-hydroxyphenylacetaldehyde condense to form the trihydroxylated alkaloid (S)-norcoclaurine (Fig. 1), which is the central precursor to all benzylisoquinoline alkaloids produced in plants (Stadler et al., 1987, 1989). (S)-Norcoclaurine is converted to (S)-reticuline by a 6-O-methyltransferase (Morishige et al., 2000), an N-methyltransferase (Choi et al., 2001), a P450 hydroxylase (Pauli and Kutchan, 1998), and a 4′-O-methyltransferase (Morishige et al., 2000). The aromatic-ring hydroxylation involved in the conversion of (S)-norcoclaurine to (S)-reticuline originally was thought to be catalyzed by a nonspecific phenol oxidase (Loeffler and Zenk, 1990). However, a P450-dependent monooxygenase (CYP80B1) isolated from California poppy (Pauli and Kutchan, 1998) exhibits a Km for (S)-N-methylcoclaurine considerably lower than that of the phenolase; thus, CYP80B1 is now known to convert (S)-N-methylcoclaurine to (S)-3′-hydroxy-N-methylcoclaurine (Fig. 1).
Figure 1.
Biosynthesis of the major benzophenanthridine alkaloids found in California poppy (Eschscholzia californica) cell cultures showing the sites of action of the berberine bridge enzyme (BBE) and (S)-N-methylcoclaurine 3′-hydroxylase (CYP80B1).
(S)-Reticuline is a branch-point intermediate involved in the biosynthesis of many structural types of benzylisoquinoline alkaloids. One such group, the benzophananthridine alkaloids, includes the antibiotics sanguinarine, which is used in various oral hygiene products, and marcarpine, the most highly oxidized alkaloid in its class. The first committed step in benzophenanthridine alkaloid biosynthesis involves conversion of the N-methyl group of (S)-reticuline into the methylene bridge moiety of (S)-scoulerine by the BBE (Fig. 1). (S)-Scoulerine is then converted by two P450-dependent oxidases, an N-methyltransferase, and two additional P450-dependent hydroxylases to dihydrosanguinarine, the first alkaloid with the benzophenanthridine nucleus (Facchini, 2001). Another P450-dependent monooxygenase and an O-methyltransferase are involved in the final steps leading to dihydromacarpine and related intermediates. Dihydrobenzophenanthridine oxidase converts these alkaloids to their corresponding oxidation products, such as sanguinarine and macarpine.
Plant metabolic engineering is a relatively new field of research with the potential to create new opportunities for the improvement of plant metabolic, cellular, and physiological processes. In recent years, a number of impressive strategies for the genetic modification of several important plant metabolic pathways have been reported. These include the reduction of indole glucosinolate levels in Brassica napus seeds (Chavadej et al., 1994), the elevation of the α-tocopherol (vitamin E) content of Arabidopsis seeds (Shintani and DellaPenna, 1998), and the introduction of the entire β-carotene (provitamin A) biosynthetic pathway into rice endosperm cells (Ye et al., 2000). The first application of metabolic engineering to a plant alkaloid pathway involved the transformation of Atropa belladonna, which normally accumulates hyoscyamine, with the gene encoding hyoscyamine 6β-hydroxylase (H6H) from Hyoscyamus muticus (Yun et al., 1992). Plants expressing the H6H transgene accumulated high levels of scopolamine, the H6H reaction product, demonstrating that alkaloid metabolism can be altered in transgenic plants. However, the metabolic engineering of plant alkaloid pathways has generally been restricted by the limited availability of cloned biosynthetic genes and the inability to genetically transform many alkaloid-producing species. Recently, several new genes encoding enzymes involved in benzylisoquinoline alkaloid biosynthesis have been reported (Facchini, 2001) and protocols for the genetic transformation of the benzylisoquinoline alkaloid-producing species opium poppy (Papaver somniferum) and California poppy have been established (Belny et al., 1997; Park and Facchini, 2000a, 2000b, 2000c). These developments have created the opportunity to metabolically engineer benzylisoquinoline alkaloid pathways in plants.
In this paper, we report the suppression of benzophenanthridine alkaloid biosynthesis in cell cultures of California poppy transformed with antisense BBE and antisense CYP80B1 constructs. The consequences of restricting metabolic flux into benzophenanthridine alkaloid biosynthesis at these two key points in the pathway were evaluated by two-dimensional 1H-NMR and in vivo 15N-NMR spectroscopy. Our results show that changes in metabolic flux through benzylisoquinoline alkaloid biosynthesis can affect the regulation of amino acid pools. Our data also provide new insight into the development of metabolic engineering strategies that target benzylisoquinoline alkaloid pathways.
RESULTS
Suppression of BBE or CYP80B1 mRNAs Reduces Benzophenanthridine Alkaloid Accumulation in California Poppy Cell Cultures
Several independent, paromomycin-resistant callus lines were obtained from excised California poppy cotyledons exposed to Agrobacterium tumefaciens strain GV3101 harboring the 35S::GUS, 35S::antiBBE, or 35S::antiCYP80B1 constructs. The callus cultures were maintained on growth media containing paromomycin and timentin for several months to promote the selection of transgenic cells and the complete elimination of A. tumefaciens. PCR performed using genomic DNA isolated from each paromomycin-resistant cell line and primers specific to sequences in the NTPII selectable marker gene consistently produced a single amplicon with the expected size of 823 bp. No amplicons were obtained using genomic DNA isolated from wild-type California poppy cell cultures. Suspension cultures were subsequently initiated from several cell lines and eventually transferred to growth media lacking antibiotics. This last step was necessary because benzophenanthridine alkaloid accumulation in California poppy cultures is inducible by antibiotics (Schumacher et al., 1987). All experiments were performed several months after the establishment of antibiotic-free cell suspension cultures.
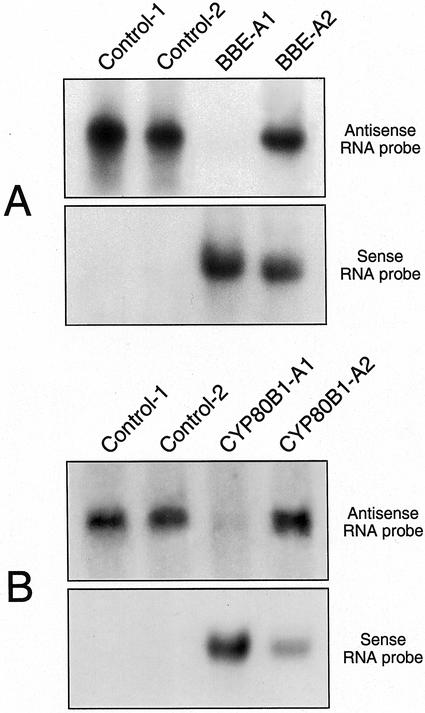
Northern-blot hybridization analysis using radiolabeled sense RNA probes demonstrated the presence of antisense BBE and antisense CYP80B1 transcripts in California poppy cell cultures transformed with 35S::antiBBE and 35S:antiCYP80B1, respectively (Fig. 2). Alternatively, the use of antisense RNA probes showed a dramatic reduction in BBE and CYP80B1 mRNA levels in some transgenic cell lines expressing 35S::antiBBE and 35S::antiCYP80B1, respectively, compared with control cultures expressing 35S::GUS (Fig. 2; cell lines BBE-A1 and CYP80B1-A1). BBE mRNAs were nearly undetectable in some transgenic cell lines, such as BBE-A1, that produced high levels of antisense BBE transcripts. Although CYP80B1 mRNA levels were extensively reduced in some transgenic cell lines that produced high levels of antisense CYP80B1 transcripts, such as CYP80B1-A1, the reduction was never as substantial as that found in BBE-A1 (Fig. 2) and other antisense BBE cell lines. However, BBE and CYP80B1 mRNA levels were not reduced in all cell lines with abundant antisense BBE or antisense CYP80B1 transcripts, respectively (Fig. 2; cell lines BBE-A2 and CYP80B1-A2). As expected, BBE activity was similar in the BBE-A2 (16 ± 8 pkat mg−1 protein) and control (19 ± 7 pkat mg−1 protein) cell lines, but was below the detection limit of the assay in the BBE-A1 cell line (data not shown).
Figure 2.
RNA gel-blot analysis showing the accumulation of sense and antisense BBE and CYP80B1 transcripts in California poppy cell suspension cultures transformed with 35S::GUS (control-1 and control-2), 35S::antiBBE (BBE-A1 and BBE-A2), and 35S::antiCYP80B1 (CYP80B1-A1 and CYP80B1-A2) constructs. Total RNA was extracted, and 15 μg was fractionated on a 1% (w/v) formaldehyde agarose gel, transferred to a nylon membrane, and hybridized at high stringency with 32P-labeled BBE or CYP80B1 sense or antisense RNA probes. Gels were stained with ethidium bromide before blotting to ensure equal loading.
GUS transcripts were abundant in control cultures transformed with 35S::GUS. Moreover, BBE and CYP80B1 mRNA levels were identical to those in wild-type cultures (data not shown). CYP80B1 transcript levels in the BBE-A1 and BBE-A2 cell lines were also identical to those in wild-type cultures. Similarly, BBE mRNA levels in the CYP80B1-A1 and CYP80B1-A2 cell lines were the same as those in wild-type cultures. Transcript levels of another alkaloid biosynthetic gene, Tyr/dopa decarboxylase, were identical in control, BBE-A1, and CYP80B1-A1 transgenic cell lines (data not shown).
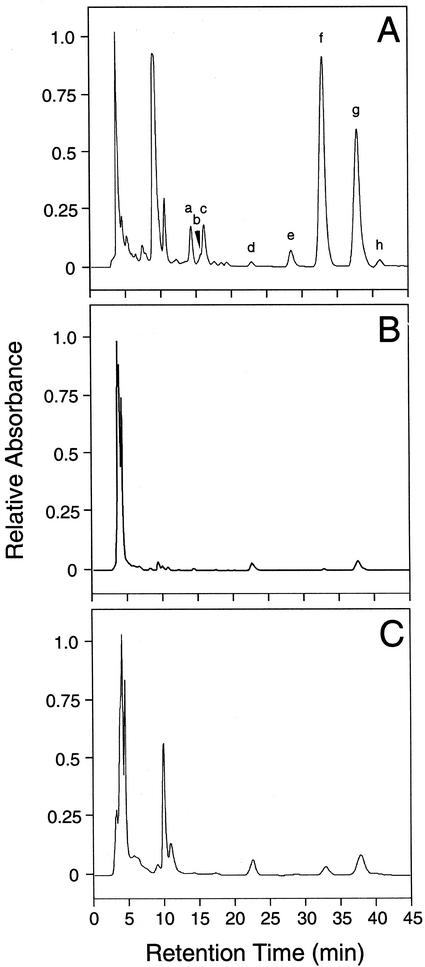
Cell lines transformed with 35S::GUS exhibited the same red color seen in wild-type California poppy cell cultures. However, transgenic cell lines that showed a substantial reduction in BBE and CYP80B1 mRNA levels (i.e. BBE-A1 and CYP80B1-A1) did not possess the red pigmentation found in wild-type and control cultures. HPLC analysis showed that benzophenanthridine alkaloid levels were substantially reduced in cell lines that displayed high levels of antisense BBE or antisense CYP80B1 transcripts and low levels of BBE and CYP80B1 mRNAs, respectively, compared with controls (Fig. 3). No new peaks were apparent on HPLC chromatograms of extracts from cell lines with low levels of BBE or CYP80B1 mRNAs and significantly reduced total alkaloid accumulation compared with controls (Fig. 3). In particular, a peak corresponding to (S)-reticuline was not detected on HPLC chromatograms of alkaloid extracts from the BBE-A1 cell line. The extraction of (S)-reticuline using the protocol described herein was confirmed by adding authentic (S)-reticuline to some samples as an internal standard. Moreover, (S)-reticuline was not detected in the original methanol extracts (data not shown).
Figure 3.
HPLC elution profiles of extracts from California poppy cell suspension cultures transformed with 35S::GUS (A, control-1), 35S::antiBBE (B, BBE-A1), and 35S::antiCYP80B1 (C, CYP80B1-A1) constructs. Peaks correspond to: a, chelilutine and chelirubine; b, chelerythrine and sanguinarine; c, marcarpine; d, dihydrochelilutine; e, dihydrochelerythrine; f, unknown alkaloid with m/z = 338; g, dihydromacarpine; and h, dihydrosanguinarine.
The total benzophenanthridine alkaloid content of the BBE-A1 cell line was 8- to 10-fold lower than that of control cells transformed with 35S::GUS (Table I). Similarly, the total alkaloid content of the CYP80B1-A1 cell line was 3- to 4-fold lower than that of controls (Table I). The level of most individual alkaloids was reduced in both the BBE-A1 and CYP80B1-A1 cell lines compared with controls, with the exception of dihydrochelilutine, which was present at marginally higher levels in cultures exhibiting a reduced abundance of BBE and CYP80B1 mRNAs (Table I). Cell lines that produced BBE and CYP80B1 antisense mRNAs but did not show a reduction in the corresponding sense mRNA levels (i.e. BBE-A2 and CYP80B1-A2) displayed total and individual levels of alkaloid accumulation that were not significantly different from those of controls (Table I).
Table I.
Accumulation of major benzophenanthridine alkaloids in cell suspension cultures of E. californica transformed with 35S∷GUS (control-1 and control-2), 35S∷antiBBE (BBE-A1 and BBE-A2), and 35S∷antiCYP80B1 (CYP80B1-A1 and CYP80B1-A2)
| Alkaloid | Cell Line
|
|||||
|---|---|---|---|---|---|---|
| Control-1 | Control-2 | BBE-A1 | BBE-A2 | CYP80B1-A1 | CYP80B1-A2 | |
| nmol g−1 (dry wt) | ||||||
| Macarpinea | 40.7 ± 47.1 | 14.5 ± 10.6 | Trace | 2.4 ± 2.7 | 7.2 ± 11.7 | 24.8 ± 16.5 |
| Dihydromacarpine | 278.9 ± 38.2 | 224.4 ± 128.5 | 55.3 ± 13.2 | 297.1 ± 175.7 | 76.3 ± 92.1 | 246.1 ± 141.4 |
| Chelerythrine | 3.7 ± 4.5 | 0.7 ± 0.7 | Trace | 2.4 ± 2.7 | 4.3 ± 4.0 | 0.7 ± 0.7 |
| Dihydrochelerythrine | 8.3 ± 0.7 | 8.0 ± 4.1 | Trace | Trace | 10.1 ± 5.5 | 10.0 ± 9.5 |
| Chelilutine/chelirubine | 28.4 ± 27.9 | 11.3 ± 8.3 | 2.8 ± 2.8 | 7.1 ± 7.8 | 3.7 ± 2.0 | 12.1 ± 9.8 |
| Dihydrochelilutine | 10.9 ± 4.1 | 7.0 ± 3.7 | 26.8 ± 6.8 | 109.7 ± 73.7 | 73.8 ± 75.8 | 193.7 ± 99.7 |
| Sanguinarine | 1.1 ± 1.5 | 0.4 ± 0.2 | Trace | 1.3 ± 0.7 | 2.1 ± 2.1 | 6.0 ± 9.9 |
| Dihydrosanguinarine | 10.9 ± 3.6 | 9.7 ± 7.4 | Trace | Trace | 4.4 ± 3.9 | 22.1 ± 38.3 |
| m/z = 338b | 484.4 ± 132.4 | 413.7 ± 245.8 | 0.6 ± 0.2 | 113.8 ± 161.4 | 57.1 ± 36.4 | 127.7 ± 38.0 |
| Totalc | 868.7 ± 248.6 | 690.0 ± 408.3 | 89.5 ± 21.1 | 544.5 ± 377.8 | 250.6 ± 207.6 | 627.2 ± 209.4 |
Values represent the mean ± sd of three independent experiments.
Unknown alkaloid with a Mr of 338.
Represents the sum of all major alkaloids.
Suppression of Benzophenanthridine Alkaloid Biosynthesis Reduces the Growth Rate of California Poppy Cell Cultures
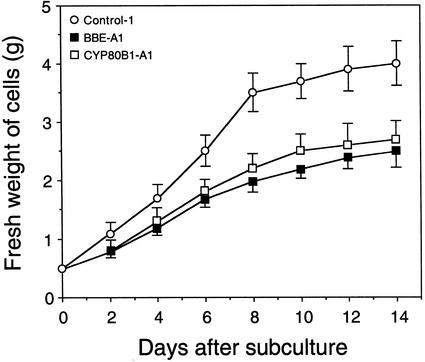
Transgenic cell lines with low levels of BBE or CYP80B1 mRNAs, and significantly reduced total alkaloid accumulation, displayed growth rates that were considerably slower than those of control cultures (Fig. 4). The growth rates of control cultures transformed with 35S::GUS were not significantly different from those of wild-type cultures (data not shown). Similarly, the growth rates of the BBE-A1 and CYP80B1-A1 cell lines were not significantly different from each other (Fig. 4).
Figure 4.
Growth rate of California poppy cell suspension cultures transformed with 35S::GUS (control-1), 35S::antiBBE (BBE-A1), and 35S::antiCYP80B1 (CYP80B1-A1) constructs. Each value represents the mean ± sd of the fresh weight of cells per flask from three independent experiments.
Antisense RNA-Mediated Reduction in BBE and CYP80B1 mRNA Levels Increases Free Amino Acid Pools in California Poppy Cells
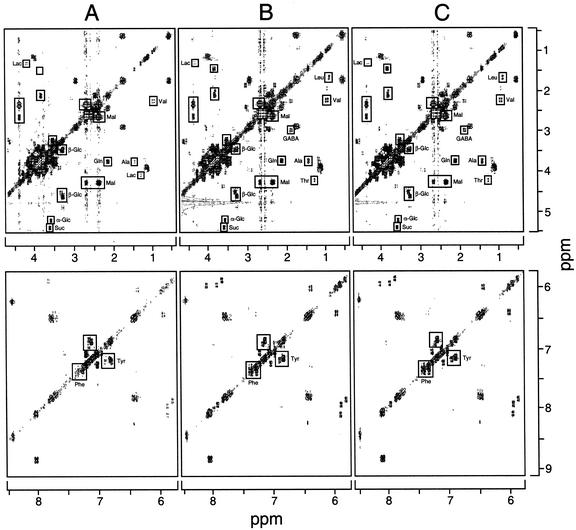
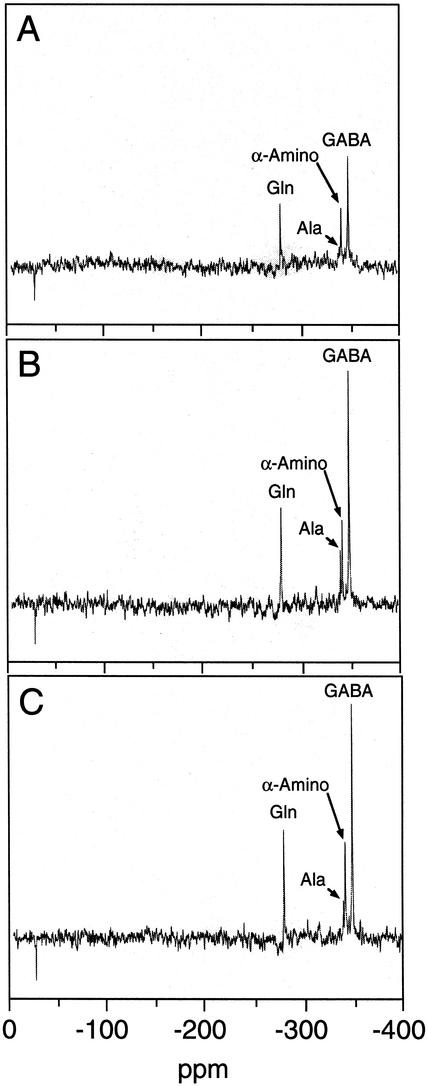
Two-dimensional 1H-NMR spectroscopy showed that the cellular pools of several amino acids were larger in cultures (i.e. BBE-A1 and CYP80B1-A1) exhibiting low levels of BBE or CYP80B1 mRNAs and significantly reduced total alkaloid accumulation (Fig. 5). Specifically, cellular pools of Ala, Leu, Phe, Thr, and Val were larger in cultures with reduced alkaloid levels compared with controls. In contrast, the relative abundance of Tyr appeared to be only marginally higher in the BBE-A1 and CYP80B1-A1 cell lines compared with controls (Fig. 5). Cellular levels of several carbohydrates including Suc, α-Glc, β-Glc, and malate were similar in the BBE-A1, CYP80B1-A1, and control lines. In vivo 15N-NMR spectroscopy also showed that the accumulation of detectable 15N-labeled amino acids, including α-amino acids, Ala, and Gln, was higher in cell lines exhibiting low levels of BBE or CYP80B1 mRNAs, and significantly reduced total alkaloid accumulation (Fig. 6). Two-dimensional 1H-NMR and noninvasive 15N-NMR also showed a substantial increase in the accumulation of γ-aminobutyrate (GABA) in transgenic cells lines with a reduced accumulation of benzophenenthridine alkaloids compared with controls (Figs. 5 and 6). An increase in the cellular pool size of several amino acids in the BBE-A1 and CYP80B1-A1 cell lines, compared with control cultures, was confirmed by direct HPLC analysis (data not shown). Using this method, Tyr levels were found to be less than 2-fold higher in antisense-suppressed cell cultures compared with controls.
Figure 5.
Two-dimensional 1H-NMR spectra of ethanol extracts from California poppy cell suspension cultures transformed with 35S::GUS (A, control-1), 35S::antiBBE (B, BBE-A1), and 35S::antiCYP80B1 (C, CYP80B1-A1) constructs. The spectra were normalized using sodium 2,2-dimethyl-2-silapentane-5-sulfonate as an internal standard. Lac, Lactate; Mal, malate.
Figure 6.
In vivo 15N-nuclear magnetic resonance spectra of California poppy cell suspension cultures transformed with 35S::GUS (A, control-1), 35S::antiBBE (B, BBE-A1), and 35S::antiCYP80B1 (C, CYP80B1-A1) constructs. Cultures were grown in standard B5 media containing K15NO3 rather than the corresponding 14N-containing compound.
DISCUSSION
Constitutive expression of antisense BBE or antisense CYP80B1 genes in transgenic California poppy cell cultures yielded several lines with high levels of antisense transcripts. In approximately 50% of these lines, corresponding sense mRNA levels were substantially reduced compared with controls transformed with 35S::GUS. In the remaining cell lines, little change in sense mRNA levels was detected relative to controls. Two transgenic lines, one displaying nearly undetectable and the other relatively normal sense mRNA levels, were selected from each set of cell lines producing antisense transcripts. Two transgenic control lines were also selected at random. The BBE-A1 and CYP80B1-A1 cell lines accumulated low to undetectable levels of BBE and CYP80B1 mRNAs, whereas the BBE-A2 and CYP80B1-A2 cell lines displayed mRNA levels similar to those found in controls (Fig. 2). The suppression of BBE mRNA accumulation in the BBE-A1 cell line was also accompanied by a substantial reduction in BBE activity. CYP80B1 activity was not measured because an enzymatic substrate was not available. However, it can be assumed that CYP80B1 activity was also substantially reduced in transgenic cell lines with low levels of CYP80B1 mRNA compared with controls.
The suppression of BBE and CYP80B1 mRNAs in transgenic California poppy cell cultures resulted in a dramatic reduction in the accumulation of benzophenanthridine alkaloids (Fig. 3; Table I). BBE activity was below the detection limit of the assay. However, the accumulation of alkaloids at one-tenth the control level suggests that BBE activity was reduced by about 90% compared with controls. Alkaloid accumulation in transgenic lines that displayed control levels of BBE or CYP80B1 mRNAs was essentially the same as that in control cultures. Transgenic cell lines expressing 35S::GUS were used as controls to account for any metabolic or cellular alterations caused by the transformation process. Previously, we showed that the transformation of California poppy and opium poppy plants and root cultures did not alter the growth rate, anatomy, or benzylisoquinoline alkaloid content of these systems (Park and Facchini, 2000a, 2000b, 2000c). Moreover, transcript levels for other alkaloid biosynthetic genes were virtually identical in control and antisense-suppressed cell lines. These data indicate that the reduced accumulation of benzophenanthridine alkaloids in the BBE-A1 and CYP80B1 cell lines was specifically due to the silencing, or partial-silencing, of BBE or CYP80B1 mRNAs.
Several points about the effect of the antisense RNA-mediated suppression of benzophenanthridine alkaloid biosynthesis should be noted. First, a decrease in either BBE or CYP80B1 mRNA levels produced a similar reduction in alkaloid accumulation. Second, the metabolic intermediates (S)-reticuline and (S)-N-methylcoclaurine, which serve as substrates for BBE and CYP80B1, respectively, did not accumulate in antisense-suppressed cells (Fig. 3). The antisense suppression of BBE and CYP80B1 mRNAs caused an overall reduction in benzophenanthridine alkaloid accumulation, but the absence of additional peaks on HPLC chromatograms shows that pathway intermediates did not accumulate. Third, the silencing of BBE or CYP80B1 mRNAs resulted in a significant reduction in the growth rate of the cell cultures. The reduced growth rate occurred in all antisense-suppressed cell lines (Fig. 4), but not in transgenic cell lines exhibiting control levels of BBE and CYP80B1 mRNAs (data not shown). The reduced growth rate might have resulted from the low-level accumulation of cytotoxic pathway intermediates.
We suggest three hypotheses for the lack of a substantial accumulation of pathway intermediates in antisense-suppressed cell lines. The first involves feedback inhibition by one or more alkaloid intermediates on early biosynthetic enzymes. However, the inhibition of enzymes involved in benzylisoquinoline alkaloid biosynthesis by pathway intermediates or end products has not been reported. The second concerns the putative degradation of alkaloid intermediates by an as-yet-uncharacterized mechanism. A third hypothesis raises the possibility that benzylisoquinoline alkaloid biosynthetic enzymes operate as part of a metabolon, or metabolic channel. Although direct interactions between alkaloid biosynthetic enzymes have not yet been demonstrated, the existence of enzyme complexes has been detected in flavonoid biosynthesis (Burbulis and Winkel-Shirley, 1999; He and Dixon, 2000). In particular, He and Dixon (2000) have suggested that the association of an O-methyltransferase involved in isoflavonoid biosynthesis might result in an altered regiospecificity, compared with that exhibited by the isolated enzyme, by facilitating the direct presentation of substrates into the active site. Similarly, removal of components, such as BBE or CYP80B1, from a putative metabolic complex might preclude the accumulation of pathway intermediates because of the lack of coordination between integrated active sites of sequential biosynthetic enzymes. In addition, the apparent spatial segregation of benzophenanthridine alkaloid biosynthesis and end-product sequestration within a cell might also contribute to the lack of (S)-N-methylcoclaurine or (S)-reticuline accumulation. Specifically, benzylisoquinoline alkaloid end products, such as sanguinarine, accumulate in the vacuole (Kutchan et al., 1986), whereas BBE has been shown to contain a signal peptide (i.e. targeting the endoplasmic reticulum) and a vacuolar sorting determinant (Bird and Facchini, 2001). Thus, an engineered metabolic block in benzylisoquinoline alkaloid biosynthesis might sufficiently disrupt the normal metabolic and intracellular transport architecture to prevent the accumulation of pathway intermediates or end products. The inherent cytotoxicity of alkaloids is at least partially responsible for the extensive subcellular compartmentation of many biosynthetic pathways to sequester intermediates and end products away from the cytosol (De Luca and St-Pierre, 2000). The reduced growth rate of antisense-suppressed California poppy cells might be caused by the impaired metabolic flux of (S)-N-methylcoclaurine and (S)-reticuline, leading to the accumulation of these alkaloids to marginally cytotoxic levels in an improper subcellular environment. Clearly, a better understanding of the subcellular localization of biosynthetic enzymes, the trafficking of intermediates, and the mechanisms that control flux, is essential to design rational metabolic engineering strategies that target benzylisoquinoline alkaloid pathways.
The expanding collection of biosynthetic genes should contribute to an improved understanding of the metabolic mechanisms that regulate benzylisoquinoline alkaloid biosynthesis. For example, constitutive expression of the scoulerine 9-O-methyltransferase (SMT) gene in Coptis japonica cell cultures resulted in 20% higher SMT activity and a small increase in the accumulation of protoberberine alkaloids (Sato et al., 2001). SMT diverts (S)-scoulerine toward protoberberine, rather than benzophenanthridine, alkaloid biosynthesis. Expression of SMT in cultured California poppy cells led to a diversion of metabolic flux toward the protoberberine alkaloid columbamine and away from benzophenenthridine alkaloids. Columbamine is a novel product in this system since California poppy cell cultures do not possess native SMT activity. Hence, the modulation of a branch-point enzyme in benzylisoquinoline alkaloid biosynthesis can alter the flux of pathway intermediates toward native or novel products.
Relatively little attention has been focused on the relationship between benzylisoquinoline alkaloid and primary metabolic pathways. Certainly, an important consideration for any metabolic engineering strategy is the availability of primary metabolic precursors. The role of Tyr as the precursor to the benzylisoquinoline alkaloid pathway suggests that the regulation of the shikimate and aromatic amino acid pathways might contribute significantly to the ability of California poppy cell cultures to produce substantial quantities of products, such as sanguinarine and macarpine. NMR spectroscopy is a technique that can provide valuable insights into the integration and regulation of plant metabolism through both in vitro and in vivo measurements. We used two-dimensional 1H-NMR and noninvasive 15N-NMR to obtain a broad profile of metabolic alterations caused the antisense mRNA-mediated suppression of benzophenanthridine alkaloid biosynthesis. Recent applications of 1H-NMR include an analysis of the ligands of barley root exudates (Fan et al., 1997) and the metabolic composition of tomato fruits (Noteborn et al., 2000). Two-dimensional 1H-NMR of ethanol extracts from control and antisense-suppressed California poppy cell cultures showed the relative abundance of several amino acids, organic acids, and sugars. Similar internal concentrations of Suc, Glc, and malate suggest that many basic metabolic processes were not affected by the antisense suppression of BBE or CYP80B1 (Fig. 5). In contrast, cellular pools of several amino acids, especially Ala, Leu, Phe, Thr, Val, and to a lesser extent Gln were larger in antisense-suppressed cells compared with controls. An increase in the abundance of amino acids in antisense-suppressed cell lines might be expected because of the reduced flux of nitrogen into benzophenanthridine alkaloids. The increased abundance of Ala, Gln, and α-amino acids was confirmed by noninvasive 15N-NMR (Fig. 6). In vivo 15N-NMR has frequently been used to detect amino acids (Robinson et al., 1991) and some secondary metabolites, such as nicotine, agropine, and conjugated polyamines (Ford et al., 1994). Tyr levels also increased in antisense-suppressed cell lines compared with controls, but the increase was less than 2-fold. However, the elevated Tyr pool cannot account for the decrease in the accumulation of Tyr-derived benzylisoquinoline alkaloids in cell lines BBE-A1 and CYP80B1-A1. A decrease in the flux of Tyr into benzophenanthridine alkaloid biosynthesis clearly alters the steady-state levels of several amino acids. The increase in GABA levels in antisense-suppressed cultures should also be noted (Figs. 5 and 6). GABA is present in all plant tissues and accumulates under stress conditions, and has been shown to function as a major nitrogen sink in non-stressed carrot cell cultures (Robinson et al., 1991) and in the short-term maintenance of pH homeostasis (Carroll et al., 1994).
We have shown that the transformation of California poppy cell cultures with antisense BBE or antisense CYP80B1 constructs can suppress the accumulation of benzophenanthridine alkaloids, which in turn affects cellular growth rate and amino acid metabolism. Our data provide insight into the complex regulation of benzylisoquinoline alkaloid biosynthesis and the integral relationship between amino acid and alkaloid metabolism. Much remains to be learned about the control of benzylisoquinoline alkaloid pathways before we can routinely design rational metabolic engineering strategies. A proposed model suggesting that a plant cell can be engineered to accumulate valuable benzylisoquinoline alkaloid intermediates using an antisense mRNA-mediated approach (Kutchan, 1995) does not appear to be feasible in some cases because of the complex and poorly understood overall metabolic regulation of the pathway.
MATERIALS AND METHODS
PCR
Plant genomic DNA was extracted as described by Edwards et al. (1991). Tissues (50 mg fresh weight) were homogenized in 200 μL of extraction buffer (0.5% [w/v] SDS, 250 mm NaCl, 100 mm Tris-HCl, pH 8.0, and 25 mm EDTA) and centrifuged. The supernatant was mixed with an equal volume of isopropanol, incubated on ice for 5 min, and then centrifuged. The pellet was dried at 60°C for 10 min, and resuspended in 100 μL of TE buffer (10 mm Tris-HCl, pH 7.4, and 1.0 mm EDTA). PCR was performed for 30 thermal cycles (95°C for 1 min, 55°C for 1 min, and 72°C for 1 min). BBE and CYP80B1 were amplified from California poppy (Eschscholzia californica Cham.) genomic DNA using primers corresponding to the beginning and end of each open reading frame (BBE, 5′-ATGGAAAACAAAACTCCC-3′ and 5′-CTATATTACAACTTCTCC-3′ [Dittrich and Kutchan, 1991]; CYP80B1, 5′-ATGGAGGTTGTCACAGTA-3′ and 5′-TCAAACCCTTGATTTAGG-3′ [Pauli and Kutchan, 1998]). PCR was also used to test the transformation of cell cultures using primers specific to sequences in NPTII (5′-CAAGATGGATTGCACGCA-3′ and 5′-TCACCCGAAGAACTCGTC-3′).
Construction of Transformation Vectors
BBE and CYP80B1 coding regions were re-amplified by PCR using primers designed to add KpnI and SalI restrictions sites to the 5′ and 3′ ends, respectively, of each clone. The PCR products were inserted into the pBI102 binary vector (Facchini et al., 1996) between KpnI and SalI sites to allow expression of antisense RNA driven by the cauliflower mosaic virus 35S promoter. Each construct was mobilized in Agrobacterium tumefaciens strain GV3101 carrying the helper plasmid pMP90 (Koncz and Schell, 1986). A. tumefaciens cultures were grown at 28°C on a gyratory shaker at 180 rpm in liquid Luria-Bertani medium (1% [w/v] tryptone, 0.5% [w/v] yeast extract, and 1% [w/v] NaCl, pH 7.0), containing 50 mg L−1 kanamycin, to mid-log phase (A600 = 0.5). The bacterial cells were collected by centrifugation for 10 min at 270g, and resuspended at a cell density of A600 = 1.0 in liquid inoculation medium (B5 salts and vitamins and 20 g L−1 Suc).
Production of Transgenic Cell Cultures
Seeds of California poppy (Richters Herbs, Goodwood, Canada) were surface-sterilized with 70% (v/v) ethanol for 30 s and 2% (v/v) sodium hypochlorite solution for 10 min, rinsed three times in sterilized water, and germinated on basal media consisting of B5 salts and vitamins (Gamborg et al., 1968), pH 5.8, solidified with 0.8% (w/v) Phytagar (Gibco, Burlington, Canada). All plant tissues were maintained in a growth chamber at 25°C under standard cool-white fluorescent tubes (Sylvania Gros-Lux Wide Spectrum, Mississauga, Canada) with a flux rate of 35 μmol s−1 m−2 and a 16-h photoperiod, unless otherwise noted. Excised cotyledons from 7-d-old seedlings were isolated by longitudinal bisection of the hypocotyl. Cotyledons were immersed for 15 min in the A. tumefaciens suspension cultures containing the various transformation constructs, blotted dry on sterile filter paper, and incubated in the dark at 25°C on callus induction media (B5 salts and vitamins, 30 g L−1 Suc, 1.0 mg L−1 2,4-dichlorophenoxyacetic acid, and 8 g L−1 Phytagar). After 2 d of cocultivation with A. tumefaciens, the cotyledons were transferred to callus induction media containing 50 mg L−1 paromomycin and 200 mg L−1 timentin. After 12 weeks, the callus was transferred to liquid B5 media containing 1.0 mg L−1 2,4-dichlorophenoxyacetic acid, 50 mg L−1 paromomycin, 200 mg L−1 timentin, and 1 g L−1 casein hydrolysate to produce suspension cultures. Cell suspensions were maintained in 30 mL of media in 125-mL Erlenmeyer flasks on a gyratory shaker at 120 rpm. The growth of suspension cultures was measured as the total fresh weight of tissue in one 125-mL Erlenmeyer flask.
RNA Gel-Blot Hybridization
Total RNA for gel-blot hybridization analysis was isolated using the method of Logemann et al. (1987). Fifteen micrograms was fractionated on a 1.0% (v/v) formaldehyde agarose gel before transfer to a nylon membrane. Duplicate blots were hybridized with sense or antisense RNA probes transcribed from linearized California poppy BBE and CYP80B1 open reading frames in pBluescript using T3 and T7 RNA polymerases, [α-32P]UTP, and an in vitro transcription kit (Stratagene, La Jolla, CA). Hybridization was performed at 70°C in 0.25 mm sodium phosphate buffer, pH 8.0, 7% (w/v) SDS, 1% (w/v) BSA, and 1 mm EDTA. The blot was washed at 70°C, twice with 2× SSC and 0.1% (w/v) SDS and twice with 0.2× SSC and 0.1% (w/v) SDS (1× SSC = 0.15 m NaCl and 0.015 m sodium citrate, pH 7.0), and autoradiographed with an intensifying screen at −80°C for 24 h.
HPLC Analysis
California poppy cell cultures were frozen in liquid N2, ground to a fine powder using a mortar and pestle, and extracted with methanol in a boiling water bath for 15 min. Extracts were reduced to dryness under vacuum, dissolved in 1.0 m sodium carbonate/bicarbonate (3:2, w/w), pH 10.0, and extracted three times with ethyl acetate. Pooled ethyl acetate fractions were reduced to dryness and the residue taken up in 1 mL of methanol. Extracts were analyzed using a System Gold 126 HPLC and 128 photodiode array detector (Beckman-Coulter, Mississauga, Canada). Alkaloids were separated at a flow rate of 0.75 mL min−1 on a C18 reverse phase column (4.6 × 250 mm, Ultrasphere, Beckman-Coulter) using methanol:water (3:1, v/v) containing 0.1% (v/v) triethylamine. The identity of peaks corresponding to various benzophenanthridine alkaloids was initially determined by liquid chromatography-mass spectroscopy. Subsequently, peaks were routinely analyzed by comparison of UV spectra and retention times with those of identified alkaloids. The expected retention time of (S)-reticuline was determined using an authentic standard.
Enzyme Assay
Two grams of cultured cells was ground to a fine powder under liquid N2 using a mortar and pestle and extracted in 50 mm Gly-NaOH, pH 8.9. Polyvinylpolypyrrolidone was added to remove phenolic compounds and the extract was desalted on a PD-10 column (Amersham-Pharmacia, Uppsala). Reaction mixtures consisting of 1 mm (S)-reticuline and 500 μL of the enzyme extract were incubated at 30°C for 1 to 2 h. Reactions were stopped by the addition of 10 μL of 1.0 n NaOH, and 100 μL was fractionated on a C18 reverse phase column (4.6 × 250 mm, Ultrasphere, Beckman-Coulter) using methanol:water (1:1, v/v) containing 0.1% (v/v) triethylamine as the mobile phase. The (S)-scoulerine peak was identified by comparison of its UV spectrum and retention time with those of an authentic standard.
15N- and Two-Dimensional 1H-NMR Spectroscopy
Whole-cell extracts for two-dimensional 1H-NMR analysis were prepared by grinding 100 mg of lyophilized California poppy cell cultures in three 10-mL aliquots of 80% (v/v) ethanol. After centrifugation of the pooled extracts, the supernatant was reduced to 5 mL under vacuum, de-ionized using 1 mL of Chelex-100, and lyophilized. The sample was dissolved in 100% (v/v) D2O to reduce the resonance for water. Nitrogen-15 NMR spectra were determined using cell cultures grown for 3 d in media containing 99.9% (w/w) K15NO3 rather than the equivalent 14N-containing compound. NMR(NMR) studies were performed with a Bruker AM400 widebore NMR spectrometer equipped with a 10-mm multinuclear broadband probe. NMR spectra were recorded at the following frequencies: 1H (400 MHz) and 15N (40 MHz). For in vivo measurements, cells were perfused in the NMR magnet by recirculating oxygen-saturated media.
Amino Acid Analysis
Cultured cells (1 g) were lyophylized and ground in 100% methanol (10:1 [v/w]). The homogenate was incubated at 60°C for 30 min and then centrifuged for 15 min at 12,000g. The supernatant was collected, and the pellet was extracted once more with 50% (v/v) methanol. The combined extracts were reduced to dryness and redissolved in 75 μL of dilution buffer containing 100 mm NaHCO3 and 100 mm H3BO3, pH 8.5. Twenty microliters of the resuspended solution was mixed with 20 μL of 9-fluorenylmethyl chloroformate (20.7 mg mL−1) and incubated at room temperature for 10 min to generate fluorescent amino acid derivatives. After extraction of the free fluorescent dye in 60 μL of pentane:ethyl acetate (8:2, v/v), 20 μL of the aqueous phase was subjected to HPLC. Amino acids were separated on an AminoTag column (Varian, Sugarland, TX) at a flow rate of 1.4 mL min−1 using the following solvent system: A, 50 mm sodium acetate buffer, pH 4.2, 20% (v/v) acetonitrile; B, 50 mm sodium acetate buffer, pH 4.2, 70% (v/v) acetonitrile (100% A ramped to 25% B over 22 min, 75% B over 20 min, 100% B over 1 min, and 100% A over 2 min). Amino acid derivatives were quantified using a Prostar 363 fluorescence detector (Varian) with excitation at 264 nm and emission at 340 nm.
ACKNOWLEDGMENTS
We thank Dean McIntyre for assistance with the NMR spectroscopy, Ikhlas Khan for the liquid chromatography-mass spectroscopy identification of benzophenanthridine alkaloids, and Fumihiko Sato for the gift of (S)-scoulerine. We also thank Tasmanian Alkaloids Pty Ltd for kindly providing a generous supply of (S)-reticuline.
Footnotes
This work was supported by the Natural Sciences and Engineering Research Council of Canada (grant to P.J.F.). S.U.P. was the recipient of Bettina-Bahlsen Memorial, Graduate Faculty Council, and J.B. Hyne Graduate Scholarships and a Dean's Special Doctoral Scholarship offered through the University of Calgary.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010741.
LITERATURE CITED
- Belny M, Herouart D, Thomasset B, David H, Jacquin-Dubreuil A, David A. Transformation of Papaver somniferum cell suspension cultures with sam-1 from A. thaliana results in cell lines of different S-adenosyl-l-methionine synthase activity. Physiol Plant. 1997;99:233–240. [Google Scholar]
- Bird DA, Facchini PJ. Berberine bridge enzyme, a key branch-point enzyme in benzylisoquinoline alkaloid biosynthesis, contains a vacuolar sorting determinant. Planta. 2001;213:888–897. doi: 10.1007/s004250100582. [DOI] [PubMed] [Google Scholar]
- Burbulis IE, Winkel-Shirley B. Interactions among enzymes of the Arabidopsisflavonoid biosynthetic pathway. Proc Natl Acad Sci USA. 1999;96:12929–12934. doi: 10.1073/pnas.96.22.12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll AD, Fox GG, Laurie S, Phillips R, Ratcliffe RG, Stewart GR. Ammonium assimilation and the role of γ-aminobutyric acid in pH homeostasis in carrot cell suspensions. Plant Physiol. 1994;106:513–520. doi: 10.1104/pp.106.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavadej S, Brisson N, McNeil JN, De Luca V. Redirection of tryptophan leads to production of low indole glucosinolate canola. Proc Natl Acad Sci USA. 1994;91:2166–2170. doi: 10.1073/pnas.91.6.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K-B, Morishige T, Sato F. Purification and characterization of coclaurine N-methyltransferase from cultured Coptis japonicacells. Phytochemistry. 2001;56:649–655. doi: 10.1016/s0031-9422(00)00481-7. [DOI] [PubMed] [Google Scholar]
- De Luca V, St-Pierre B. The cell and developmental biology of alkaloid biosynthesis. Trends Plant Sci. 2000;4:168–173. doi: 10.1016/s1360-1385(00)01575-2. [DOI] [PubMed] [Google Scholar]
- Dittrich H, Kutchan TM. Molecular cloning, expression, and induction of berberine bridge enzyme, an enzyme essential to the formation of benzophenanthridine alkaloids in the response of plants to pathogen attack. Proc Natl Acad Sci USA. 1991;88:9969–9973. doi: 10.1073/pnas.88.22.9969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C. A simple and rapid method for preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991;19:1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchini PJ. Alkaloid biosynthesis in plants: biochemistry, cell biology, molecular regulation, and metabolic engineering applications. Annu Rev Plant Physiol Plant Mol Biol. 2001;51:29–61. doi: 10.1146/annurev.arplant.52.1.29. [DOI] [PubMed] [Google Scholar]
- Facchini PJ, Penzes C, Johnson AG, Bull D. Molecular characterization of berberine bridge enzyme genes from opium poppy. Plant Physiol. 1996;112:1669–1677. doi: 10.1104/pp.112.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan TW-M, Lane AN, Pedler J, Crowley D, Higashi RM. Comprehensive analysis of organic ligands in whole root exudates using nuclear magnetic resonance and gas chromatography-mass spectrometry. Anal Biochem. 1997;251:57–68. doi: 10.1006/abio.1997.2235. [DOI] [PubMed] [Google Scholar]
- Ford YY, Fox GG, Ratcliffe RG, Robins RJ. In vivo 15N NMR studies of secondary metabolism in transformed root cultures of Datura stramonium and Nicotiana tabacum. Phytochemistry. 1994;36:333–339. [Google Scholar]
- Gamborg OL, Miller RO, Ojima K. Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res. 1968;50:151–158. doi: 10.1016/0014-4827(68)90403-5. [DOI] [PubMed] [Google Scholar]
- He X-Z, Dixon RA. Genetic manipulation of isoflavone 7-O-methyltransferase enhances biosynthesis of 4′-O-methylated isoflavonoid phytoalexins and disease resistance in alfalfa. Plant Cell. 2000;12:1689–1702. doi: 10.1105/tpc.12.9.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J. The promoter TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacteriumvector. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- Kutchan TM. Alkaloid biosynthesis-the basis for metabolic engineering of medicinal plants. Plant Cell. 1995;7:1059–1070. doi: 10.1105/tpc.7.7.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutchan TM, Rush M, Coscia CJ. Subcellular localization of alkaloids and dopamine in different vacuolar compartments of Papaver bracteatum. Plant Physiol. 1986;81:161–166. doi: 10.1104/pp.81.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffler S, Zenk MH. The hydroxylation step in the biosynthetic pathway leading from norcoclaurine to reticuline. Phytochemistry. 1990;29:3499–3503. [Google Scholar]
- Logemann J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissues. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- Morishige T, Tsujita T, Yamada Y, Sato F. Molecular characterization of the S-adenosyl-l-methionine: 3′-hydroxy-N-methylcoclaurine-4′-O-methyltransferase of isoquinoline alkaloid biosynthesis in Coptis japonica. J Biol Chem. 2000;275:23398–23405. doi: 10.1074/jbc.M002439200. [DOI] [PubMed] [Google Scholar]
- Noteborn HPJM, Lommen A, van der Jagt RC, Weseman JM. Chemical fingerprinting for the evaluation of unintended secondary metabolic changes in transgenic food crops. J Biotechnol. 2000;77:103–114. doi: 10.1016/s0168-1656(99)00210-2. [DOI] [PubMed] [Google Scholar]
- Park S-U, Facchini PJ. Agrobacterium-mediated genetic transformation of California poppy, Eschscholzia californicaCham., via somatic embryogenesis. Plant Cell Rep. 2000a;19:421–426. doi: 10.1007/s002990000213. [DOI] [PubMed] [Google Scholar]
- Park S-U, Facchini PJ. Agrobacterium-mediated transformation of opium poppy, Papaver somniferumL., via shoot organogenesis. J Plant Physiol. 2000b;157:207–214. [Google Scholar]
- Park S-U, Facchini PJ. Agrobacterim rhizogenes-mediated transformation of opium poppy, Papaver somniferum L., and California poppy, Eschscholzia californicaCham., root cultures. J Exp Bot. 2000c;51:1005–1016. doi: 10.1093/jexbot/51.347.1005. [DOI] [PubMed] [Google Scholar]
- Pauli HH, Kutchan TM. Molecular cloning and functional heterologous expression of two alleles encoding (S)-N-methylcoclaurine 3′-hydroxylase (CYP80B1), a new methyl jasmonate-inducible cytochrome P-450-dependent mono-oxygenase of benzylisoquinoline alkaloid biosynthesis. Plant J. 1998;13:793–801. doi: 10.1046/j.1365-313x.1998.00085.x. [DOI] [PubMed] [Google Scholar]
- Robinson SA, Slade AP, Fox GG, Phillips R, Ratcliffe RG, Stewart GR. The role of glutamate dehydrogenase in plant nitrogen metabolism. Plant Physiol. 1991;95:509–516. doi: 10.1104/pp.95.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato F, Hashimoto T, Hachiya A, Tamura K, Choi K-B, Morishige T, Fujimoto H, Yamada Y. Metabolic engineering of plant alkaloid biosynthesis. Proc Natl Acad Sci USA. 2001;98:367–372. doi: 10.1073/pnas.011526398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher HM, Gundlach H, Fiedler F, Zenk MH. Elicitation of benzophenanthridine alkaloid synthesis in Eschscholtziacell cultures. Plant Cell Rep. 1987;6:410–413. doi: 10.1007/BF00272770. [DOI] [PubMed] [Google Scholar]
- Shintani D, DellaPenna D. Elevating the vitamin E content of plants through metabolic engineering. Science. 1998;282:2098–2100. doi: 10.1126/science.282.5396.2098. [DOI] [PubMed] [Google Scholar]
- Stadler R, Kutchan TM, Loeffler S, Nagakura N, Cassels B, Zenk MH. Revision of the early steps of reticuline biosynthesis. Tetrahedron Lett. 1987;28:1251–1254. [Google Scholar]
- Stadler R, Kutchan TM, Zenk MH. Norcoclaurine is the central intermediate in benzylisoquinoline alkaloid biosynthesis. Phytochemistry. 1989;28:1083–1086. [Google Scholar]
- Ye X, Al-Balili S, Klöti A, Zhang J, Lucca P, Beyer P, Potrykus I. Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science. 2000;287:303–305. doi: 10.1126/science.287.5451.303. [DOI] [PubMed] [Google Scholar]
- Yun D-J, Hashimoto T, Yamada Y. Metabolic engineering of medicinal plants: transgenic Atropa belladonnawith an improved alkaloid composition. Proc Natl Acad Sci USA. 1992;89:11799–11803. doi: 10.1073/pnas.89.24.11799. [DOI] [PMC free article] [PubMed] [Google Scholar]