Abstract
Ashbya gossypii is a natural riboflavin overproducer used in the industrial production of the vitamin. We have isolated an insertional mutant exhibiting higher levels of riboflavin production than the wild type. DNA analysis of the targeted locus in the mutant strain revealed that a syntenic homolog of the Saccharomyces cerevisiae BAS1 gene, a member of the Myb family of transcription factors, was inactivated. Directed gene disruption of AgBAS1 confirmed the phenotype observed for the insertional mutant, and the Δbas1 mutant also showed auxotrophy for adenine and several growth defects, such as a delay in the germination of the spores and an abnormally prolonged trophic phase. Additionally, we demonstrate that the DNA-binding domain of AgBas1p is able to bind to the Bas1-binding motifs in the AgADE4 promoter; we also show a clear nuclear localization of a green fluorescent protein-Bas1 fusion protein. Real-time quantitative PCR analyses comparing the wild type and the Δbas1 mutant revealed that AgBAS1 was responsible for the adenine-mediated regulation of the purine and glycine pathways, since the transcription of the ADE4 and SHM2 genes was virtually abolished in the Δbas1 mutant. Furthermore, the transcription of ADE4 and SHM2 in the Δbas1 mutant did not diminish during the transition from the trophic to the productive phase did not diminish, in contrast to what occurred in the wild-type strain. A C-terminal deletion in the AgBAS1 gene, comprising a hypothetical regulatory domain, caused constitutive activation of the purine and glycine pathways, enhanced riboflavin overproduction, and prolonged the trophic phase. Taking these results together, we propose that in A. gossypii, AgBAS1 is an important transcription factor that is involved in the regulation of different physiological processes, such as purine and glycine biosynthesis, riboflavin overproduction, and growth.
Ashbya gossypii is a filamentous hemiascomycete of considerable importance in biotechnology due to its natural ability to overproduce riboflavin (vitamin B2) (5), an essential factor for humans and animals that is frequently used as a food additive (38). A. gossypii overproduces riboflavin as a detoxifying and protective mechanism during the late growth phase, when the maximum mycelial mass has been reached (37). Thus, in terms of riboflavin production, two stages can be differentiated during A. gossypii culture: a trophic phase when riboflavin production is minimal and the growth rate increases, and a productive phase when the growth rate decreases and riboflavin is overproduced (37). Many physiological and morphological changes occur during the shift from the trophic to the productive phase (27, 37), but the mechanisms triggering the transition are not fully understood. The productive phase is associated with a characteristic intense yellow color of the mycelia, due to the accumulation of the vitamin in the vacuolar compartment (9). During the past few years, different studies have been carried out with a view to improving the excretion of the vitamin (8) and also to increasing the metabolic flux for riboflavin biosynthesis (16, 17, 25, 35).
Riboflavin is synthesized from GTP and ribulose 5-phosphate in a six-step pathway governed by the RIB genes (RIB1 to RIB5 and RIB7) (3). For A. gossypii, it has been shown that riboflavin production is correlated with the activity of the Rib3 protein (34). However, enhanced riboflavin overproduction has been reported only when GTP precursors are added to the medium or when the purine or glycine pathways are engineered to provide high concentrations of GTP precursors (16, 21, 25, 35, 38). This indicates that GTP availability is a limiting factor for riboflavin production in A. gossypii.
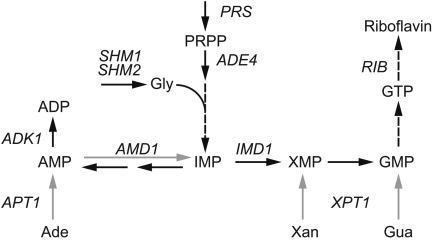
GTP is formed in the cell through the de novo purine pathway (Fig. 1), which starts with the conversion of 5′-phosphoribosyl-1-pyrophosphate (PRPP) to IMP, after which IMP can be transformed into AMP or GMP (30). Alternatively, purine salvage pathways (Fig. 1) allow the interconversion and recycling of purines with the consumption of PRPP (30). The purine salvage pathways contribute to a correct balance between adenyl and guanyl nucleotides. In fact, maintenance of this balance is essential for cell viability; hence, the biosynthesis of purines relies on several regulatory mechanisms at the transcriptional and metabolic levels (4, 10, 11, 13, 14, 22, 40).
FIG. 1.
Schematic representation of the purine biosynthetic pathway. Black arrows designate the de novo pathway, and gray arrows indicate the salvage pathways. Gly, glycine; GMP, guanosine 5′-monophosphate; Ade, adenine, Gua, guanine; Xan, xanthine. The gene names are italicized and correspond to the following enzymatic activities: PRS, PRPP synthetase; ADE4, PRPP amidotransferase; SHM1 and SHM2, serine hydroxymethyltransferase; IMD1, IMP dehydrogenase; AMD1, AMP deaminase; APT1, adenine phosphoribosyltransferase; XPT1, xanthine phosphoribosyltransferase; ADK1, adenylate kinase; RIB, riboflavin genes.
In Saccharomyces cerevisiae, it has been shown that a complex formed between the transcription factors Bas1 and Bas2 (Pho2) is required for the transcriptional activation of the de novo purine pathway under adenine deprivation (4). In addition, Bas1 and Bas2 also activate the biosynthesis of histidine, glutamine, glycine, and 10-formyl tetrahydrofolate (2, 7), which are purine-related pathways (see Fig. 5 in reference 31).
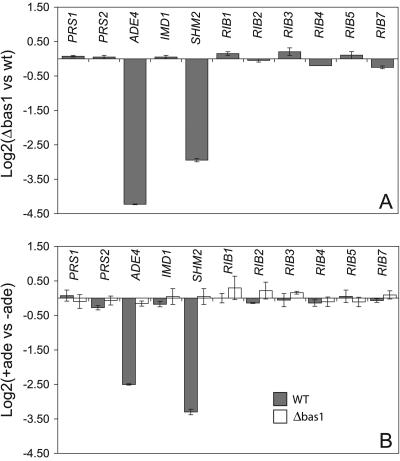
FIG. 5.
Comparison of the steady-state transcription levels of purine and riboflavin genes in wild-type and Δbas1 strains. (A) Differences in the transcription levels of the genes included in the assays between wild-type and Δbas1 strains grown in MA2 rich medium. (B) Relative transcription levels of the indicated genes measured in the wild-type (gray bars) and Δbas1 (white bars) strains grown in MA2 rich medium with (+ade) or without (−ade) an excess of adenine (0.1 g/liter). Transcription levels are normalized to the level of AgACT1 mRNA. The ratio of relative transcription of the target gene in panel A was calculated as 2−ΔΔCt, where ΔΔCt = ΔCtΔbas1 − ΔCtwild type. The ratio of the relative transcription of the target gene in panel B was calculated as 2−ΔΔCt, where ΔΔCt = ΔCtMA2 + ade − ΔCtMA2 − ade. An average of three separate cDNA dilutions from each target gene were obtained, and the relative transcription levels were expressed as log2 ± SD. vs, versus; WT, wild type.
Current studies have reported that two intermediates of the purine pathway, namely, 5′-phosphoribosyl-4-succinocarboxamide-5-aminoimidazole (SAICAR) and 5′-phosphoribosyl-4-carboxamide-5-aminoimidazole (AICAR), are intracellular signals that promote the interaction between Bas1 and Bas2 and, hence, transcriptional activation (30, 31). Unlike SAICAR, AICAR is also synthesized through the histidine pathway, again confirming the cross talk between purine and histidine biosynthesis (4, 30).
Bas1 is a Myb-related transcription factor comprising an amino-terminal DNA-binding domain that binds to the sequence TGACTC (15, 39), an internal trans-activation domain, and a C-terminal domain called the Bas1 interaction and regulatory domain (BIRD) for interaction with Bas2 and regulation of the trans-activation domain (28). A recent report has suggested that Bas1 is permanently bound to DNA, with the trans-activation domain being inactive when adenine is present in the medium. Under conditions of adenine limitation, a regulatory signal, probably dependent on SAICAR and AICAR, increases the Bas1-Bas2 interaction; Bas2 is recruited to the promoters, and the trans-activation domains of Bas1 and Bas2 are unmasked for transcriptional activation (31, 36).
Here, we describe the identification and characterization of the transcription factor Bas1 in the filamentous fungus A. gossypii. AgBas1p participates in the regulated transcription of genes involved in the biosynthesis of purines and glycine. In addition, different bas1 mutants showed a significant increase in the production of riboflavin and other growth-related phenotypes. The involvement of AgBAS1 in different processes of the A. gossypii physiology and its implications for the biotechnological production of riboflavin are discussed.
MATERIALS AND METHODS
A. gossypii strains, media, and growth conditions.
The A. gossypii ATCC 10895 strain was used and considered a wild-type strain. A. gossypii strains were cultured in rich medium (MA2) (9) or synthetic minimal medium (SMM) (35) at 28°C. Amino acids adenine and guanine were purchased from Sigma (Steinheim, Germany) and were used at a final concentration of 0.1 g/liter. A concentration of 250 μg/ml for Geneticin (G418) (Sigma, Steinheim, Germany) or 200 μg/ml for hygromycin B (Phytotechnology Laboratories, Shawnee Mission, Kans.) was used when stated. Sporulation conditions and spore isolations were carried out as previously described (33). Liquid cultures were inoculated with 1 × 106 spores per liter of medium and were performed on a rotary shaker at 120 rpm. The determination of riboflavin production was performed by high-performance liquid chromatography as described previously (35).
Insertional mutagenesis and targeted locus identification.
A detailed description of the method has been previously reported elsewhere (33). Briefly, an in vitro transposition reaction using A. gossypii genomic DNA digested with PstI and a minitransposon R comprising the 5′ and 3′ inverted terminal repeats from the Himar1 transposon flanking the dominant G418 resistance marker and the bacterial replicon ColE1 was performed. After the transposition reaction, the genomic DNA carrying an integrated minitransposon R was self-ligated and used to transform the Escherichia coli DH10B strain.
To generate insertional mutants, the plasmid library was linearized by enzymatic digestion with PstI and used to transform the wild-type A. gossypii strain. The G418r A. gossypii colonies were sporulated and plated onto selective medium (MA2 plus G418) again. After 4 days, colonies exhibiting a bright yellow color were selected for further analyses. Genomic DNA from the insertional mutants was isolated, digested with XhoI, self-ligated, and transformed into E. coli DH10B. The transformants carrying the minitransposon R flanked by genomic DNA were selected on LB-kanamycin (50 μg/ml) plates. The identification of the targeted loci was achieved by sequence analysis using primers derived from the left and right ends of the minitransposon R, respectively. Determination of the full-length sequences was performed using primer-walking strategies.
Construction of the Δbas1 strain.
To construct the Δbas1 strain, a disruption cassette was engineered. An internal BamHI-SphI fragment of the AgBAS1 open reading frame (ORF) was replaced with the G418r marker obtained as a BamHI-SphI fragment (33). The disruption module was obtained as a XhoI-BglII fragment with a 356-bp 5′-flanking region and a 520-bp 3′-flanking region homologous to the AgBAS1 locus. This fragment was used to transform spores of the wild-type strain of A. gossypii. Primary G418r heterokaryotic transformants were sporulated and, after clonal selection, G418r Δbas1 homokaryotic colonies were isolated. The disruption of AgBAS1 was confirmed by analytical PCR and Southern blotting.
RNA extraction, reverse transcription-PCR, and real-time PCR.
A. gossypii mycelium (200 to 300 mg), previously frozen, was mechanically homogenized in liquid nitrogen using TRIzol reagent (Invitrogen, Carlsbad, Calif.), and total RNA was isolated as described by the manufacturer. RNA was incubated with 20 U of RNase-free DNase I (Roche, Basel, Switzerland), and 5 μg of total RNA was reverse transcribed using an oligo(dT) primer (Isogen, Ijsselstein, The Netherlands) and SuperScript II RT enzyme (Invitrogen, Carlsbad, Calif.).
For real-time quantitative PCR, three serial dilutions (1:10, 1:20, and 1:30) of the synthesized cDNA were amplified with target gene-specific primers (see Table S1 in the supplemental material) using an iCycler iQ system (Bio-Rad, Hercules, Calif.). The following conditions were used: heat activation of DNA polymerase for 15 min at 95°C; 42 cycles of PCR at 95°C for 30 s, 55°C for 30 s, and 72°C for 40 s; and a final incubation at 72°C for 10 min. For each PCR product, melting curves were determined according to the supplier's guidelines (Bio-Rad, Hercules, Calif.), ensuring specific amplification of the target gene. Quantitative values were obtained as the threshold PCR cycle number (Ct) when the increase in the fluorescent signal of the PCR product showed exponential amplification. The target gene mRNA level was normalized to that of AgACT1 (encoding β-actin) in the same sample. The relative transcription level of the target gene was calculated using the 2−ΔΔCt method, where ΔΔCt = (Cttarget gene − CtAgACT1)condition X − (Cttarget gene − CtAgACT1)condition Y (19). The mean of the results was obtained after the 2−ΔΔCt was calculated for three serial cDNA dilutions of each sample in triplicate, and the relative transcription levels were expressed as means ± standard deviations (SD).
N-terminal AgBAS1 tagging with GFP(S65T).
With PCR methods, an expression module containing a hygromycin resistance marker (Hygr) and the green fluorescent protein S65T [GFP(S65T)] coding region (20) in frame with the AgBAS1 ORF under the control of the constitutive promoter of the glyceraldehyde 3-phosphate dehydrogenase-encoding gene AgGPD was constructed. The wild-type AgBAS1 locus was replaced with the expression module described above, and the replacement was verified by Southern blot analysis.
Nuclear DNA was stained with the DNA-specific dye Hoechst 33342 (HO) (Sigma, Steinheim, Germany). The fluorescence of the GFP-Bas1p fusion protein in living cells was monitored as previously described (26). Micrographs were acquired using a Photometrics Sensys charge-coupled-device camera coupled to a Leica DMXRA microscope equipped with Nomarski optics and epifluorescence.
Expression and purification of the N-terminal domain of the Bas1p from A. gossypii.
The DNA sequence corresponding to the N-terminal domain of the AgBas1 (amino acids 1 to 305) was PCR amplified and cloned into the NdeI and XhoI site of the pET-28b expression vector (Novagen, Darmstadt, Germany) and used to transform the E. coli BL21 (DE3) strain. A single positive colony was inoculated in LB medium plus kanamycin (50 μg/ml) and grown at 37°C until the A600 reached 1. Induction of the His-tagged recombinant protein was achieved by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside. After 1 h of induction, the cells were harvested and resuspended in lysis buffer (50 mM NaH2PO4, pH 8; 10 mM Tris-HCl, pH 8; 100 mM NaCl) containing protease inhibitor cocktail (Roche, Basel, Switzerland). Cells were disrupted by sonication for 30 min, and the homogenates were centrifuged at 10,000 × g for 10 min. The His-tagged recombinant protein was purified from the supernatant using a Talon (Clontech, Heidelberg, Germany) column, following the manufacturer's instructions. Pure fractions were stored at −20°C until use.
Electrophoretic mobility shift assay.
Electrophoretic mobility shift assays (EMSAs) were performed essentially as described previously (15). The binding reaction was performed over 30 min at room temperature. In each reaction, 1 × 105 to 5 × 105 cpm of a labeled DNA probe (0.3 to 0.5 pmol) and 0.1 to 1 μg of recombinant His-AgN305Bas1 peptide were used. The binding buffer was 20 mM Tris-HCl, 0.1 mM EDTA, 10% glycerol (vol/vol), 100 mM NaCl, 0.01% Triton X-100 (vol/vol), and protease inhibitor cocktail (Roche, Basel, Switzerland). For competition binding reactions, the unlabeled competitor at 100- and 200-fold molar excess was used. A DNA fragment corresponding to the AgGPD promoter (384 bp) was used as nonspecific competitor. The reaction mixtures were loaded onto a 5 to 8% native polyacrylamide gel in 1× TBE (89 mM Tris-HCl, 89 mM boric acid, 2 mM EDTA [pH 8.0]), electrophoresed, dried, and exposed to X-ray film at −80°C with an intensifying screen.
Construction of the ΔC631BAS1 strain.
The truncated AgBAS1 gene was generated by PCR-mediated modifications. A PCR-derived module containing the 50-bp fragment upstream from the codon encoding amino acid 631 of the AgBas1p followed by the ScADH1 terminator, the G418r marker, and the 50-bp fragment downstream from the AgBAS1 stop codon were used to transform wild-type A. gossypii spores. The truncated AgΔC631BAS1 gene was integrated in the BAS1 locus. As described above, homokaryotic clones were isolated, and the truncation of AgBAS1 gene was confirmed by Southern blotting (see Fig. 7A).
FIG. 7.
Characterization of the ΔC631BAS1 strain. (A) Growth pattern of ΔC631BAS1 strain grown in liquid MA2 rich medium in comparison with the wild-type and Δbas1 strains. (B) Relative transcription levels of purine and riboflavin genes in the wild-type (gray bars) and the ΔC631BAS1 (white bars) strains grown in MA2 rich medium with (+ade) or without (−ade) an excess of adenine (0.1 g/liter). Transcription levels are normalized to the level of AgACT1 mRNA. The ratio of the relative transcription of the target gene was calculated as 2−ΔΔCt, where ΔΔCt = ΔCtMA2 + ade − ΔCtMA2 − ade. An average of three separate cDNA dilutions from each target gene was obtained, and the relative transcription levels are expressed as log2 ± SD.
RESULTS
Identification and characterization of a riboflavin-overproducing mutant.
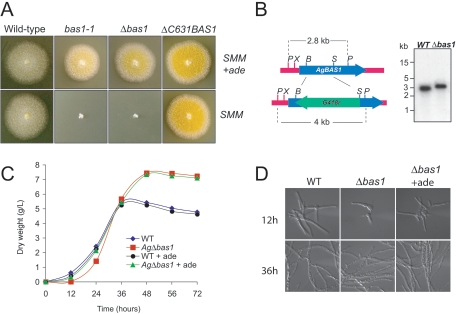
From a collection of A. gossypii mutants generated by an insertional mutagenesis method developed specifically for the purpose (33), a colony exhibiting a deep yellow color was selected (Fig. 2A); it proved to accumulate 5.5-fold (14.42 mg/g) more riboflavin than the wild-type strain. The presence in the Himar1-derived transposon module of a G418 resistance (G418r) selectable marker and the oriC replication origin allowed the recovery and identification of the genomic integration site (see Materials and Methods for details). Once the integration site had been cloned and sequenced, primers were made to amplify the targeted ORF. DNA analysis revealed that the Himar1-derived transposon had integrated into a TA dinucleotide located 697 bp downstream from the initiation codon. A BLAST search in the Ashbya Genome Database (http://agd.unibas.ch/) identified the target as a nonexperimentally characterized ORF (the database identified the gene as AFR297W, also designated AgBAS1), a syntenic homolog of the S. cerevisiae BAS1 gene, which encodes a transcription factor of the Myb family (39). The predicted amino acid sequence of AFR297W was aligned with ScBas1 and human c-Myb and showed low identity with ScBas1 (31.4%) and with human c-Myb (13.7%). However, the level of identity in the N-terminal domain was significantly higher (58.8% with ScBas1 and 21.5% with human c-Myb); using the ScanProsite tool (http://www.expasy.org/tools/scanprosite/), it was possible to identify the three signatures distinctive of the Myb family (15) (see Fig. S1 in the supplemental material). We also detected a hypothetical BIRD interaction domain within the Afr297w sequence (residues 630 to 664) compared with ScBas1 BIRD domain, although a low degree of identity (32.6%) was observed (see Fig. S1 in the supplemental material). We thus assumed that AFR297W ORF corresponded to the BAS1 homolog in A. gossypii and, consequently, the mutant strain described above was designated bas1-1.
FIG. 2.
Characterization of A. gossypii bas1 mutant strains. (A) Different A. gossypii strains were grown on solid SMM or SMM plus adenine (+ade; 0.1 g/liter). The bas1 mutants exhibit a yellow color due to riboflavin accumulation. (B) Right panel, schematic representation of the wild-type BAS1 and disrupted bas1::G418r loci. Left panel, Southern blot analysis to confirm correct BAS1 disruption. Genomic DNA of the wild-type and Δbas1 strains was digested with PstI. P, PstI; X, XhoI; B, BamHI; S, SphI. (C) Growth pattern of A. gossypii wild type and Δbas1 grown in liquid MA2 rich medium with (+ade) or without adenine supplementation (0.1 g/liter). (D) Microscopic phenotype of A. gossypii wild type and Δbas1 grown on liquid MA2 rich medium for 12 and 36 h. The germination delay of Δbas1 is restored by the addition of adenine (+ade; 0.1 g/liter). WT, wild type.
AgBAS1 inactivation causes riboflavin overproduction, adenine auxotrophy, and an extended trophic phase.
To confirm that the accumulation of riboflavin in the bas1-1 mutant was caused by the inactivation of the AgBAS1 ORF, we disrupted it in order to mimic the riboflavin overproduction phenotype found in the bas1-1 strain. An engineered disruption module containing the dominant G418r marker was used (see Materials and Methods). AgBAS1 disruption was confirmed by analytical PCR and Southern blotting (Fig. 2B). The new Δbas1 strain displayed the same yellow color and the same level of riboflavin production as the bas1-1 strain (Fig. 2A and Table 1), demonstrating that BAS1 inactivation was responsible for the increased production of vitamin B2 in A. gossypii.
TABLE 1.
Riboflavin levels in the different A. gossypii strains useda
| Strain | Concn of riboflavin (mg/g of biomass) at time indicated
|
|||
|---|---|---|---|---|
| 24 h | 48 h | 72 h | 96 h | |
| Wild type (ATCC 10895) | 0.54 ± 0.11 | 1.44 ± 0.13 | 2.09 ± 0.16 | 2.58 ± 0.13 |
| Agbas1-1 | 0.40 ± 0.08 | 3.15 ± 0.24 | 13.47 ± 0.08 | 14.42 ± 0.10 |
| AgΔbas1 | 0.49 ± 0.06 | 3.12 ± 0.11 | 14.78 ± 0.16 | 15.31 ± 0.23 |
| AgΔC631BAS1 | 2.23 ± 0.21 | 9.76 ± 0.33 | 23.66 ± 0.36 | 24.28 ± 0.37 |
Rich MA2 cultures were initiated from spores of the different A. gossypii strains. Ten-milliliter aliquots were removed at different times, and the riboflavin concentration was determined. Experiments were carried out in triplicate, and results are expressed as the average number of milligrams of riboflavin per gram of biomass ± SD.
To further characterize the bas1-1 and Δbas1 strains, we tested their ability to grow on minimal medium. As shown in Fig. 2A, the bas1-1 and Δbas1 strains were unable to grow unless adenine was present in the medium. However, guanine, pyrimidines, or amino acids failed to rescue the wild-type phenotype (not shown). The adenine requirement indicates that BAS1 is essential for the de novo biosynthesis of purines in A. gossypii and that purine salvage pathways are able to restore the synthesis of AMP in a bas1 mutant when adenine is supplied.
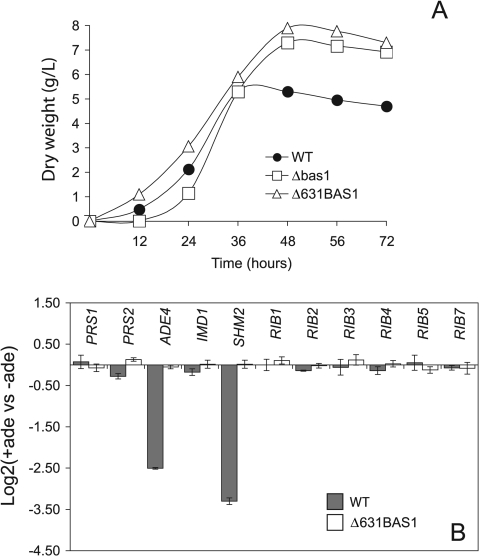
Another interesting phenotype was associated with BAS1 inactivation. When Δbas1 spores were grown on rich medium, a slight delay (4 to 6 h) in the time to germination, together with a significantly long trophic phase (approximately 12 h), was observed compared to the wild type (Fig. 2C-D). Supplementation of the medium with an excess of adenine corrected the delay in the time to germination for Δbas1 but failed to reduce the trophic-phase span (Fig. 2C-D).
AgBas1p is localized to the nucleus of A. gossypii and binds to the AgADE4 promoter.
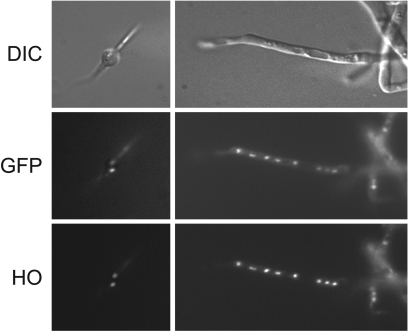
The subcellular localization of a transcription factor is expected to be nuclear. Accordingly, we examined the subcellular localization of AgBas1 using a GFP-BAS1 fusion under the control of the AgGPD promoter. The strain carrying the GFP-BAS1 fusion showed no differences from the wild-type strain in terms of riboflavin production, adenine requirement, and growth (not shown). The GFP-Bas1p fluorescence consistently colocalized with Hoechst 33342 staining of nuclear DNA, confirming that AgBas1p was localized to the nucleus (Fig. 3). This typical nuclear localization was invariably unaffected by the growth phase (Fig. 3).
FIG. 3.
AgBas1p is localized to the nuclei of A. gossypii. An A. gossyppi spore (left) and a hypha (right) expressing the GFP-Bas1p were stained with the DNA-selective dye Hoechst (HO) 33342 and viewed under epifluorescence optics. Upper images are the differential interference contrast (DIC) pictures; middle images represent the GFP channel, and lower images correspond to the HO channel of the same field.
Another essential feature of a transcription factor is its ability to bind to promoter sequences. We therefore wished to investigate whether AgBas1p was able to bind to the putative Bas1-binding sites located in the AgADE4 promoter (16). The AgBas1 DNA-binding domain (amino acids 1 to 305) was expressed in E. coli BL21(DE3) using the pET-28b expression vector, and specific DNA binding of the purified protein to different DNA probes from the AgADE4 promoter was monitored by EMSA analysis. In a first experiment, the complete AgADE4 promoter (363 bp upstream from the initiation codon) was analyzed using a distal 131-bp fragment (fragment A) and a proximal 232-bp fragment (fragment B) as radioactive probes (Fig. 4A). The analysis revealed two different DNA-protein complexes formed with the distal probe (Fig. 4B, lane 2), indicating that two molecules of AgBas1p could be recruited to the AgADE4 promoter. In Fig. 4, lane 2, the upper and lower bands correspond to fragment A, with two and one bound molecules of the AgBas1 DNA-binding domain, respectively. Addition of a 10-fold molar excess of unlabeled fragment completely abolished DNA-protein complex formation, confirming the specificity of the interaction (Fig. 4B, lane 3). In contrast, a similarly sized (384-bp) DNA fragment corresponding to the AgGPD promoter did not act as a competitor for AgBas1p (data not shown).
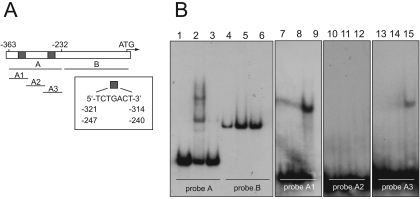
FIG. 4.
Identification of the DNA-binding site of AgBas1p by EMSA. (A) Structure of the AgADE4 promoter. Gray boxes indicate the Bas1-binding sites. The different probes used in the assays are depicted. (B) EMSA analyses using different probes: probe A, lanes 1 to 3; probe B, lanes 4 to 6; probe A1, lanes 7 to 9; probe A2, lanes 10 to 12, probe A3, lanes 13 to 15. An excess of the corresponding unlabeled probe was used in lanes 3, 6, 8, 11, and 14. No Bas1 DNA-binding domain peptide was included in lanes 1, 4, 7, 10, and 13.
When analyzing the sequence of the distal fragment A in detail, we were able to identify two putative heptanucleotide Bas1-binding motifs located at positions −247 and −321 from the start codon (Fig. 4A). Therefore, we explored the ability of the AgBas1 DNA-binding domain to bind to each heptanucleotide. We designed three overlapping oligonucleotides that expanded the fragment A to full-length. Oligonucleotide A1 contained the Bas1-binding motif located at position −321, oligonucleotide A2 did not contain any motif, and oligonucleotide A3 contained the heptanucleotide located at position −247 (Fig. 4A). EMSA analysis revealed that the AgBas1 DNA-binding domain caused gel retardation when combined with oligonucleotide A1 or A3 (Fig. 4B, lanes 9 and 15), but not with A2. Again, an excess of unlabeled probes completely abolished DNA-protein complex formation (Fig. 4B). Based on these results, it may be concluded that AgBas1p is a transcription factor that is able to bind to the two putative Bas1-binding motifs contained in the AgADE4 promoter.
AgBas1p is involved in the adenine-mediated regulation of AgADE4 and AgSHM2 in A. gossypii.
The above results suggested an important role for AgBAS1 in the biosynthesis of purine nucleotides and riboflavin. We therefore investigated the transcription profiles of ADE4 and SHM2, which have been reported to be regulated by adenine limitation in A. gossypii (16, 35); PRS1 and PRS2, which control the synthesis of the purine precursor PRPP (A. Jiménez, M. A. Santos, and J. L. Revuelta, unpublished results); the IMD1 gene, which directs the first step of GMP biosynthesis; and all six RIB genes in both wild-type and the Δbas1 strains.
Total RNA was isolated from cultures of the wild-type and Δbas1 strains in the exponential phase (24 h) grown in rich medium, and real-time quantitative PCR was performed to test the transcription levels of the ADE4, SHM2, PRS1, PRS2, IMD1, and RIB genes. As shown in Fig. 5A, ADE4 and SHM2 transcription was considerably lower in the Δbas1 strain than in the wild-type strain, whereas PRS1, PRS2, IMD1, and the six RIB genes were unaffected by the BAS1 inactivation. This result indicates that Bas1 is a transcription factor that activates the expression of ADE4 and SHM2 in A. gossypii.
We next analyzed whether the presence or absence of an excess of adenine in rich medium might affect the transcription level of the above genes in the wild-type and Δbas1 strains. Our results revealed that the addition of adenine clearly repressed the transcription of ADE4 and SHM2 in the wild-type strain (Fig. 5B). In contrast, adenine had no effect on ADE4 or SHM2 in the Δbas1 mutant, indicating that BAS1 was essential for the adenine-dependent regulation of both genes. The transcription levels of the other genes analyzed were unaltered by the concentration of adenine in both the wild-type and the Δbas1 strains, confirming that Bas1 did not modulate their transcription (Fig. 5B).
Purine and riboflavin genes are inversely regulated during the growth of A. gossypii.
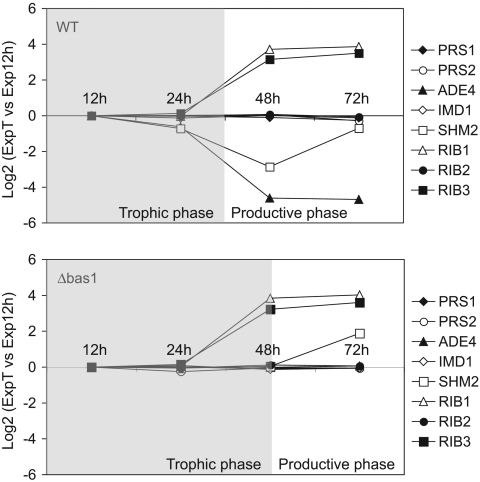
Riboflavin overproduction in A. gossypii occurs during the so-called production phase, when the cultures reach a stationary phase. In addition, the riboflavin-overproducing phenotype of the bas1 mutants suggests a direct link between the Bas1 function and riboflavin biosynthesis. We therefore checked the transcription pattern of purine and riboflavin genes in the A. gossypii wild-type and Δbas1 strains along the growth curve in rich medium. Real-time quantitative PCR showed that the mRNA levels of purine genes (ADE4 and SHM2) in the wild type were highest during the trophic phase (12 to 36 h of culture) (Fig. 6, upper panel), when riboflavin production was minimal and growth was active. Thereafter, the transcription of purine genes declined progressively, being lowest during the productive phase (48 to 72 h) (Fig. 6, upper panel). SHM2 transcription is increased at 72 h, but this effect seems to be independent of Bas1 regulation, since it also occurs in the Δbas1 strain (see below). The RIB1 and RIB3 genes were more actively transcribed than the purine genes during the productive phase (Fig. 6, upper panel). In fact, a previous report had already shown an increase in the promoter activity of RIB3 during the production phase (34). No differences in the transcription pattern of RIB genes between the wild type and Δbas1 strains were seen (Fig. 6). With regard to purine genes, a deregulated, low, but constitutive transcription was evident in the Δbas1 strain throughout the growth curve (Fig. 6, lower panel), demonstrating that Bas1 was essential for accurate regulation of the purine pathway. Remarkably, the transcription levels of purine genes were higher in the Δbas1 strain than in the wild type during the productive phase (Fig. 6, lower panel), indicating that a Bas1-independent basal transcription of the de novo purine genes was occurring in the Δbas1 strain. However, such a transcription level did not provide enough purines to enable the growth of the Δbas1 mutant in the absence of extracellular adenine.
FIG. 6.
Transcription profiles of purine and riboflavin genes in wild-type and Δbas1 strains during the trophic and productive phases. Transcription profiling of the wild-type (WT) and Δbas1 strains grown in MA2 rich medium was achieved at different time points. In the wild-type strain (upper panel), the transcription of ADE4 and SHM2 decreased along the growth curve, whereas the transcription of RIB1 and RIB3 increased during the productive phase. In the Δbas1 strain (lower panel), ADE4 and SHM2 show a constitutive deregulated transcription. Transcription levels are normalized with the transcription rate of AgACT1.Data are representative of four experiments with similar results and are presented as log2 of the 2−ΔΔCt (ΔΔCt = ΔCtT − ΔCt12 h). T, incubation time (h).
AgBas1 contains a C-terminal regulatory BIRD domain.
As described above, we identified a hypothetical C-terminal BIRD domain within the AgBas1p sequence (residues 632 to 743) (see Fig. S1 in the supplemental material). In order to verify the functionality of this regulatory domain, we decided to remove it by PCR-mediated deletion using a G418r selectable marker (see Materials and Methods). Homokaryotic G418r clones carrying the deletion module were confirmed by analytical PCR and Southern blotting (not shown). The new strain lacking the AgBas1 C-terminal domain was designated ΔC631BAS1. Deletion of the BIRD domain enhanced the capacity to produce riboflavin—the ΔC631BAS1 mutant was able to produce almost twice (24.28 mg/g) more riboflavin than the Δbas1 mutant and 12-fold more riboflavin than the wild type, hence also affording an intense yellow color to the colonies (Fig. 2A and Table 1). Unlike Δbas1, ΔC631BAS1 was able to grow in minimal medium without adenine supplementation (Fig. 2A).
The ΔC631BAS1 strain also displayed a remarkable feature when the spores were cultured in rich medium. Like the Δbas1 mutant, ΔC631BAS1 displayed a longer trophic phase than the wild type; however, germination time was not affected, unlike results for Δbas1 (Fig. 7A).
Analysis of the transcription pattern of the purine and riboflavin genes in the ΔC631BAS1 strain revealed that ADE4 and SHM2 transcription was highly activated and that the activation was insensitive to extracellular adenine, since the mRNA levels of ADE4 and SHM2 were constitutively higher in the ΔC631BAS1 strain than in the wild type with adenine supplementation (Fig. 7B). However, the other genes analyzed did not show any change in transcription levels when the ΔC631BAS1 and the wild-type strains were compared (Fig. 7B). Our results suggest that the C-terminal BIRD domain of AgBas1p would be a regulatory domain and that the truncated ΔC631Bas1p form would induce a constitutive transcriptional activation of ADE4 and SHM2, an increase in riboflavin production, and a delayed entry into the productive stationary phase.
DISCUSSION
The overproduction of riboflavin in A. gossypii is a physiological process that occurs when active growth finishes and therefore must be somehow linked to the growth phase. It has been suggested that A. gossypii overproduces riboflavin as a detoxifying and protective mechanism (37), although the molecular mechanisms triggering the riboflavin overproduction are largely unknown.
In this report, we show that AgBAS1 is involved in the regulation of the glycine and purine biosynthesis, which have been shown to increase riboflavin production when added to the medium (21, 38). Furthermore, we constructed different bas1 mutants that exhibit remarkable differences in their growth patterns and higher levels of riboflavin production than the wild type. In light of this, we propose the AgBAS1 gene as a possible candidate for a link between the glycine and purine pathways, the growth profile, and riboflavin overproduction.
AgBAS1 was found as the target gene disrupted in a riboflavin-overproducing strain, which had been selected in a screening of random insertional mutants. Directed AgBAS1 gene disruption confirmed that BAS1 inactivation in A. gossypii afforded a riboflavin overproduction phenotype. In addition, the Δbas1 strain required adenine supplementation, suggesting an important role for Bas1p in the biosynthesis of purines, as described for other organisms (4). The asymmetry of the purine salvage pathways (Fig. 1) explains why only adenine supplementation, but not supplementation with either guanine or xanthine, enables the growth of the Δbas1 mutant, since adenyl nucleotides can be transformed into guanyl nucleotides, but not the opposite.
Real-time quantitative PCR experiments comparing mRNA profiles between the wild-type and Δbas1 strains showed that AgBas1p is essential for the transcriptional activation of ADE4 and SHM2 and also for the adenine-mediated repression of those genes. The role of AgBas1p as a transcription factor is consistent with the nuclear localization of a GFP-Bas1 fusion protein. Additionally, our EMSA experiments showed that the Myb-like DNA-binding domain present in AgBas1p is able to bind the heptanucleotide located in the ADE4 promoter, which has been reported to be a Bas1-binding motif (15, 16). Such a binding motif has also been found in the AgSHM2 promoter (M. A. Santos, L. Mateos, and J. L. Revuelta, unpublished results). Overall, this evidence prompts us to consider AgBas1p to be a transcription factor that regulates purine and glycine biosynthesis, as described for other organisms (4, 6). However, AgBas1p also seems to be either directly or indirectly involved, through the purine pathway, in other physiological processes, such as detoxifying mechanisms or growth and morphogenesis, as discussed below.
Homology analyses identified a C-terminal regulatory domain within the AgBas1p sequence, called the BIRD domain in the yeast ortholog (28), and the complete deletion of this hypothetical BIRD domain in the AgBas1p was achieved. The ΔC631BAS1 strain produces higher levels of riboflavin than the Δbas1 mutant, is able to grow in medium lacking adenine, and shows a constitutive activation of the ADE4 and SHM2 genes. In light of these observations, it may be concluded that the C-terminal domain of AgBas1 would be responsible for the adenine sensitivity, for masking the trans-activation domain when adenine is present and, presumably, for the Bas2 interaction, as described previously for the yeast BIRD domain (28, 36). In fact, a ScBAS2 ortholog in A. gossypii has recently been identified (L. Mateos, M. A. Santos, and J. L. Revuelta, unpublished results).
The Δbas1 and the ΔC631BAS1 strains also display a prolonged trophic growth phase in comparison with the wild type. In addition, the Δbas1 strain exhibits a slight delay in the time to germination, together with other morphological defects in the hyphal structure. These effects may be directly related to the deregulation of the purine pathway in the Δbas1 and ΔC631BAS1 strains. First, from the results described above, it may be assumed that the de novo purine pathway does not operate properly in the Δbas1 mutant and that purines must be formed mainly through the salvage pathways, delaying the germination during the early steps of growth. In fact, adenine supplementation corrected the germination defect in the Δbas1 mutant. Second, it has been reported that low levels of intracellular guanyl nucleotides are required for entry into the stationary phase in S. cerevisiae and Bacillus subtilis (29, 32). The riboflavin overproduction phenotype shown by the Δbas1 and the ΔC631BAS1 strains indicates that a high pool of GTP, the immediate precursor of riboflavin, must exist in both strains, producing a delay in entry into the stationary phase and extending the trophic phase. This hypothesis supports the notion that the guanyl nucleotide pool would be a signal that controls the transition from the trophic phase to the stationary production phase and suggests that an increased riboflavin production and a delayed entry into the stationary phase would be two aspects of the same phenomenon, i.e., a high GTP concentration. Indeed, the Δbas1 and ΔC631BAS1 strains were more resistant to mycophenolic acid than the wild type (see Fig. S2 in the supplemental material), a phenotype associated with high intracellular levels of GTP (1, 23).
Nevertheless, the mechanisms inducing an increased intracellular concentration of GTP, and hence an enhanced riboflavin overproduction, in the Δbas1 and the ΔC631BAS1 strains must be different. ΔC631BAS1 shows a constitutive activation of the purine pathway that could be responsible for the high GTP levels and the increased riboflavin overproduction observed. Supporting this notion, we have previously demonstrated that the level of transcription of AgADE4 is correlated with the production of riboflavin (16). The explanation for the riboflavin overproduction phenotype in the Δbas1 strain is, however, less obvious. As evidenced by its adenine auxotrophy, purines must be formed mostly through the salvage pathways in the Δbas1 strain. Under these conditions, the AMP deaminase enzyme activity (Amd1) (24) establishes the asymmetry in the purine salvage pathways and redirects the metabolic flux toward guanyl nucleotides in competition with the adenylate kinase (Adk1) (18). In addition, the enzyme Xpt1 can metabolize xanthine and guanine toward GMP (12) (Fig. 1). The genes encoding these enzymes have been annotated in the Ashbya Genome Database (http://agd.unibas.ch/) as S. cerevisiae syntenic orthologs (AGD gene identification numbers AGR187W, ABR204C, and ABL070C for AgADK1, AgAMD1, and AgXPT1, respectively). However, an HPT1 homolog, which restores equilibrium by transforming hypoxanthine into IMP in other organisms (41), has not been identified; therefore, hypoxanthine cannot be transformed to IMP in A. gossypii. Accordingly, the purine salvage pathways in A. gossypii may lead to the production of higher amounts of guanyl than adenyl nucleotides; hence, the Δbas1 strain, which uses mostly the purine salvage pathways, may be able to synthesize higher amounts of the riboflavin precursor GTP than the wild type. Additionally, the AgSHM2 mRNA level during the first steps of growth was lower in the Δbas1 strain than in the wild type, and it has been reported that disruption of AgSHM2 produces glycine accumulation and enhanced riboflavin overproduction (35). Indeed, riboflavin overproduction in the Δbas1 strain was inversely correlated with the SHM2 transcription level.
In sum, the results presented here suggest that the mechanisms leading to a high concentration of GTP are responsible for riboflavin overproduction in A. gossypii. In addition, the BIRD domain found in AgBas1p could be considered an intracellular sensor of the GTP concentration. High levels of GTP could induce a repression signal of the de novo purine pathway through a mechanism in which the Bas1 BIRD domain may be involved, masking the trans-activation domain of Bas1. If the BIRD domain is indeed not functional, Bas1 must be constitutively active and the purine pathway must synthesize an excess of GTP, which must be detoxified through riboflavin production. Nevertheless, the bas1 mutants show several growth alterations affecting the transition from the trophic phase to the productive phase, and these effects indicate that Bas1 could be involved, either indirectly, through the purine pathway, or directly, in the regulation of other hitherto-undescribed growth-related pathways.
This work sheds further light on the complex role of BAS1 in the physiology of A. gossypii. A feasible link between purine and glycine biosynthesis, riboflavin overproduction, and the transition from the trophic to the stationary phase is established. Experiments are currently under way at our laboratory to explore the precise mechanisms underlying some of the effects induced by misregulation of the purine pathway.
Supplementary Material
Acknowledgments
This work was supported in part by BASF AG and grants GEN2001-4707-C08-01 and AGL2005-07245-C03-03 from the Ministerio de Educación y Ciencia, Spain. L.M. is supported by a predoctoral fellowship from the Universidad de Salamanca, Spain. A.J. is a recipient of a postdoctoral contract (Programa Juán de la Cierva) from the Spanish Ministerio de Educación y Ciencia.
We thank M. Martín and M. D. Sánchez for excellent technical help and N. Skinner for correcting the manuscript.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Allison, A. C., and E. M. Eugui. 2000. Mycophenolate mofetil and its mechanisms of action. Immunopharmacology 47:85-118. [DOI] [PubMed] [Google Scholar]
- 2.Arndt, K. T., C. Styles, and G. R. Fink. 1987. Multiple global regulators control HIS4 transcription in yeast. Science 237:874-880. [DOI] [PubMed] [Google Scholar]
- 3.Bacher, A., S. Eberhardt, M. Fischer, K. Kis, and G. Richter. 2000. Biosynthesis of vitamin B2 (riboflavin). Annu. Rev. Nutr. 20:153-167. [DOI] [PubMed] [Google Scholar]
- 4.Daignan-Fornier, B., and G. R. Fink. 1992. Coregulation of purine and histidine biosynthesis by the transcriptional activators BAS1 and BAS2. Proc. Natl. Acad. Sci. USA 89:6746-6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demain, A. L. 1972. Riboflavin oversynthesis. Annu. Rev. Microbiol. 26:369-388. [DOI] [PubMed] [Google Scholar]
- 6.Denis, V., H. Boucherie, C. Monribot, and B. Daignan-Fornier. 1998. Role of the myb-like protein bas1p in Saccharomyces cerevisiae: a proteome analysis. Mol. Microbiol. 30:557-566. [DOI] [PubMed] [Google Scholar]
- 7.Denis, V., and B. Daignan-Fornier. 1998. Synthesis of glutamine, glycine and 10-formyl tetrahydrofolate is coregulated with purine biosynthesis in Saccharomyces cerevisiae. Mol. Gen. Genet. 259:246-255. [DOI] [PubMed] [Google Scholar]
- 8.Forster, C., J. L. Revuelta, and R. Kramer. 2001. Carrier-mediated transport of riboflavin in Ashbya gossypii. Appl. Microbiol. Biotechnol. 55:85-89. [DOI] [PubMed] [Google Scholar]
- 9.Forster, C., M. A. Santos, S. Ruffert, R. Kramer, and J. L. Revuelta. 1999. Physiological consequence of disruption of the VMA1 gene in the riboflavin overproducer Ashbya gossypii. J. Biol. Chem. 274:9442-9448. [DOI] [PubMed] [Google Scholar]
- 10.Gedvilaite, A., and K. Sasnauskas. 1994. Control of the expression of the ADE2 gene of the yeast Saccharomyces cerevisiae. Curr. Genet. 25:475-479. [DOI] [PubMed] [Google Scholar]
- 11.Giani, S., M. Manoni, and D. Breviario. 1991. Cloning and transcriptional analysis of the ADE6 gene of Saccharomyces cerevisiae. Gene 107:149-154. [DOI] [PubMed] [Google Scholar]
- 12.Guetsova, M. L., T. R. Crother, M. W. Taylor, and B. Daignan-Fornier. 1999. Isolation and characterization of the Saccharomyces cerevisiae XPT1 gene encoding xanthine phosphoribosyl transferase. J. Bacteriol. 181:2984-2986.10217799 [Google Scholar]
- 13.Holmes, E. W., J. A. McDonald, J. M. McCord, J. B. Wyngaarden, and W. N. Kelley. 1973. Human glutamine phosphoribosylpyrophosphate amidotransferase. Kinetic and regulatory properties. J. Biol. Chem. 248:144-150. [PubMed] [Google Scholar]
- 14.Holmes, E. W., D. M. Pehlke, and W. N. Kelley. 1974. Human IMP dehydrogenase. Kinetics and regulatory properties. Biochim. Biophys. Acta 364:209-217. [DOI] [PubMed] [Google Scholar]
- 15.Hovring, I., A. Bostad, E. Ording, A. H. Myrset, and O. S. Gabrielsen. 1994. DNA-binding domain and recognition sequence of the yeast Bas1 protein, a divergent member of the Myb family of transcription factors. J. Biol. Chem. 269:17663-17669. [PubMed] [Google Scholar]
- 16.Jiménez, A., M. A. Santos, M. Pompejus, and J. L. Revuelta. 2005. Metabolic engineering of the purine pathway for riboflavin production in Ashbya gossypii. Appl. Environ. Microbiol. 71:5743-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato, T., and E. Y. Park. 2005. Expression of alanine:glyoxylate aminotransferase gene from Saccharomyces cerevisiae in Ashbya gossypii. Appl. Microbiol. Biotechnol. [Epub ahead of print.] doi: 10.1007/s00253-005-0124-5. [DOI] [PubMed]
- 18.Konrad, M. 1988. Analysis and in vivo disruption of the gene coding for adenylate kinase (ADK1) in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 263:19468-19474. [PubMed] [Google Scholar]
- 19.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 20.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 21.Mac, L. J. 1952. The effects of certain purines and pyrimidines upon the production of riboflavin by Eremothecium ashbyii. J. Bacteriol. 63:233-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mantsala, P., and H. Zalkin. 1984. Glutamine nucleotide sequence of Saccharomyces cerevisiae ADE4 encoding phosphoribosylpyrophosphate amidotransferase. J. Biol. Chem. 259:8478-8484. [PubMed] [Google Scholar]
- 23.McPhillips, C. C., J. W. Hyle, and D. Reines. 2004. Detection of the mycophenolate-inhibited form of IMP dehydrogenase in vivo. Proc. Natl. Acad. Sci. USA 101:12171-12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merkler, D. J., A. S. Wali, J. Taylor, and V. L. Schramm. 1989. AMP deaminase from yeast. Role in AMP degradation, large scale purification, and properties of the native and proteolyzed enzyme. J. Biol. Chem. 264:21422-21430. [PubMed] [Google Scholar]
- 25.Monschau, N., H. Sahm, and K. Stahmann. 1998. Threonine aldolase overexpression plus threonine supplementation enhanced riboflavin production in Ashbya gossypii. Appl. Environ. Microbiol. 64:4283-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niedenthal, R. K., L. Riles, M. Johnston, and J. H. Hegemann. 1996. Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast 12:773-786. [DOI] [PubMed] [Google Scholar]
- 27.Philippsen, P., A. Kaufmann, and H. P. Schmitz. 2005. Homologues of yeast polarity genes control the development of multinucleated hyphae in Ashbya gossypii. Curr. Opin. Microbiol. 8:370-377. [DOI] [PubMed] [Google Scholar]
- 28.Pinson, B., T. L. Kongsrud, E. Ording, L. Johansen, B. Daignan-Fornier, and O. S. Gabrielsen. 2000. Signaling through regulated transcription factor interaction: mapping of a regulatory interaction domain in the Myb-related Bas1p. Nucleic Acids Res. 28:4665-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ratnayake-Lecamwasam, M., P. Serror, K. W. Wong, and A. L. Sonenshein. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 15:1093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rebora, K., C. Desmoucelles, F. Borne, B. Pinson, and B. Daignan-Fornier. 2001. Yeast AMP pathway genes respond to adenine through regulated synthesis of a metabolic intermediate. Mol. Cell. Biol. 21:7901-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rebora, K., B. Laloo, and B. Daignan-Fornier. 2005. Revisiting purine-histidine cross-pathway regulation in Saccharomyces cerevisiae: a central role for a small molecule. Genetics 170:61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sagot, I., J. Schaeffer, and B. Daignan-Fornier. 2005. Guanylic nucleotide starvation affects Saccharomyces cerevisiae mother-daughter separation and may be a signal for entry into quiescence. BMC Cell Biol. 6:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santos, M. A., L. Mateos, K. P. Stahmann, and J. L. Revuelta. 2005. Insertional mutagenesis in the vitamin B2 producer fungus Ashbya gossypii, p. 283-300. In J. L. Barredo (ed.), Methods in biotechnology: microbial process and products, vol. 18. Humana Press, Inc., Totowa, N.J. [Google Scholar]
- 34.Schlosser, T., G. Schmidt, and K. P. Stahmann. 2001. Transcriptional regulation of 3,4-dihydroxy-2-butanone 4-phosphate synthase. Microbiology 147:3377-3386. [DOI] [PubMed] [Google Scholar]
- 35.Schlupen, C., M. A. Santos, U. Weber, A. de Graaf, J. L. Revuelta, and K. P. Stahmann. 2003. Disruption of the SHM2 gene, encoding one of two serine hydroxymethyltransferase isoenzymes, reduces the flux from glycine to serine in Ashbya gossypii. Biochem. J. 369:263-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Som, I., R. N. Mitsch, J. L. Urbanowski, and R. J. Rolfes. 2005. DNA-bound Bas1 recruits Pho2 to activate ADE genes in Saccharomyces cerevisiae. Eukaryot. Cell 4:1725-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stahmann, K. P., H. N. Arst, Jr., H. Althofer, J. L. Revuelta, N. Monschau, C. Schlupen, C. Gatgens, A. Wiesenburg, and T. Schlosser. 2001. Riboflavin, overproduced during sporulation of Ashbya gossypii, protects its hyaline spores against ultraviolet light. Environ. Microbiol. 3:545-550. [DOI] [PubMed] [Google Scholar]
- 38.Stahmann, K. P., J. L. Revuelta, and H. Seulberger. 2000. Three biotechnical processes using Ashbya gossypii, Candida famata, or Bacillus subtilis compete with chemical riboflavin production. Appl. Microbiol. Biotechnol. 53:509-516. [DOI] [PubMed] [Google Scholar]
- 39.Tice-Baldwin, K., G. R. Fink, and K. T. Arndt. 1989. Bas1 has a Myb motif and activates HIS4 transcription only in combination with Bas2. Science 246:931-935. [DOI] [PubMed] [Google Scholar]
- 40.Van der Weyden, M. B., and W. N. Kelly. 1974. Human adenylosuccinate synthetase. Partial purification, kinetic and regulatory properties of the enzyme from placenta. J. Biol. Chem. 249:7282-7289. [PubMed] [Google Scholar]
- 41.Woods, R. A., D. G. Roberts, T. Friedman, D. Jolly, and D. Filpula. 1983. Hypoxanthine: guanine phosphoribosyltransferase mutants in Saccharomyces cerevisiae. Mol. Gen. Genet. 191:407-412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.